Abstract
Analysis of autoinflammatory and immunodeficiency disorders elucidates human immunity and fosters the development of targeted therapies. Oligoadenylate synthetase 1 is a type I interferon-induced, intracellular dsRNA sensor that generates 2’−5’-oligoadenylate to activate RNase L as a means of antiviral defense. We identified four de novo heterozygous OAS1 gain-of-function variants in six patients with a polymorphic autoinflammatory immunodeficiency characterized by recurrent fever, dermatitis, inflammatory bowel disease, pulmonary alveolar proteinosis, and hypogammaglobulinemia. To establish causality, we applied genetic, molecular dynamics simulation, biochemical, and cellular functional analyses in heterologous, autologous and inducible pluripotent stem cell derived macrophages. We found that upon interferon-induced expression, OAS1 variant proteins displayed dsRNA-independent activity, which resulted in RNase L-mediated RNA-cleavage, transcriptomic alteration, translational arrest, and dysfunction and apoptosis of monocytes, macrophages, and B-cells upon interferon stimulation. RNase L-inhibition with curcumin modulated and allogeneic hematopoietic cell transplantation cured the disorder. Together, these data suggest that human OAS1 is a regulator of interferon-induced hyperinflammatory monocyte and B-cell pathophysiology.
One Sentence Summary
OAS1-GOF/RNase L-mediated RNA-cleavage caused an interferon-induced hyperinflammatory monocyte and B-cell immunodeficiency.
Introduction
Inborn errors of immunity (IEI) cause autoinflammation, infection susceptibility, autoimmunity, malignancy, and extra-hematopoietic syndromic features (1). The IEI subgroup of predominantly antibody deficiencies is a paradigm of how genetic and molecular dissection contribute to our understanding of human immunity and foster precision medicine (2). The IEI subgroup of autoinflammatory disorders, as exemplified by the activated stimulator of interferon genes (STING) syndrome (3), similarly provides fundamental insight into inflammation that has impacted the anti-inflammatory treatment of high prevalence disorders such as coronavirus disease 2019 (COVID-19)(4, 5).
The type I interferon-inducible oligoadenylate synthetase 1 (OAS1) is a member of the OAS protein family sharing structural and functional homology to cyclic GMP-AMP (cGAMP) synthetase (cGAS) (6). Both OAS1 and cGAS initiate an antiviral immune response upon recognition of cytoplasmic foreign nucleic acids (6), i.e. viral double-stranded RNA (dsRNA) (7) and viral double-stranded DNA (dsDNA) (8), respectively. Both proteins are template-independent nucleotidyltransferases and produce the second messengers 2’−5’-oligoadenylate (2–5A) (9) and cGAMP (10), respectively. 2–5A activates RNase L which degrades viral and cellular RNA thereby interfering with viral propagation (11). cGAS activates STING and induces the expression of multiple antiviral genes (12). Activating variants in STING cause an autoinflammatory IEI (3). OAS1 is linked to pulmonary alveolar proteinosis (PAP) with hypogammaglobulinemia (13), however, specific mechanistic and pathophysiological insights necessary to demonstrate causality of this genotype-phenotype association are lacking.
Here, we demonstrated that four different heterozygous OAS1 gain-of-function (GOF) variants cause a polymorphic autoinflammatory immunodeficiency that can be cured by allogeneic hematopoietic cell transplantation (HCT). We identified RNase L-mediated RNA cleavage as the underlying disease mechanism and human OAS1 as critical regulator of interferon-induced hyperinflammatory monocyte and B-cell pathophysiology.
Results
A polymorphic autoinflammatory immunodeficiency with de novo heterozygous OAS1 variants
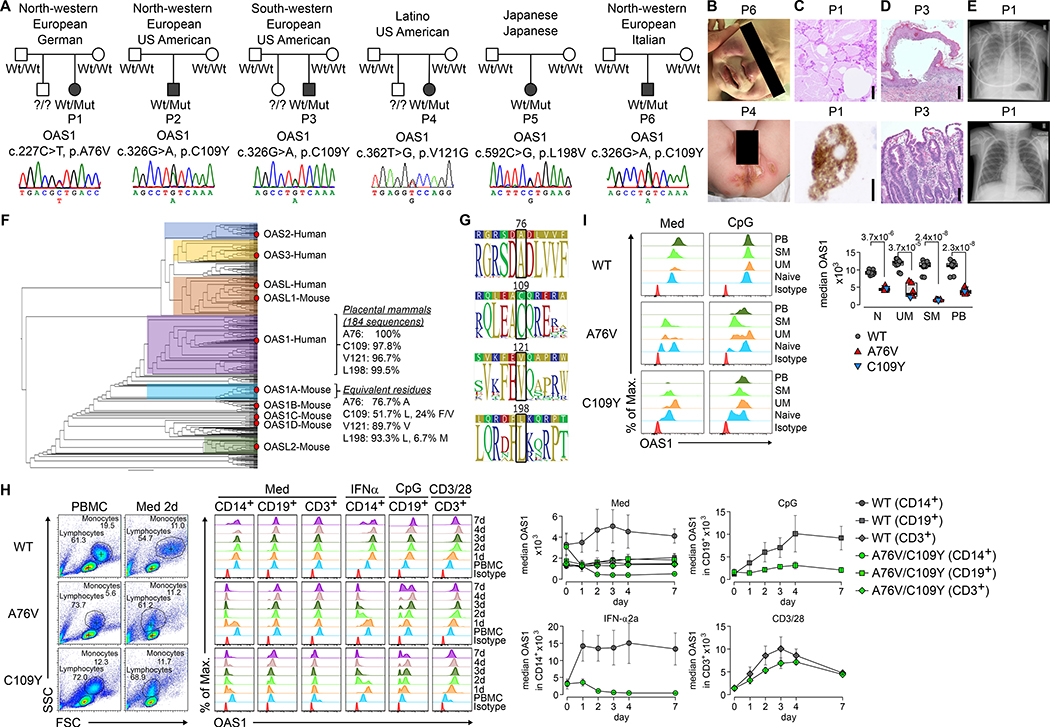
We evaluated six unrelated patients (patient 1–6, abbreviated in the figures as P1-P6; patient 5 has been previously reported (13)) born to non-consanguineous healthy parents of variable ethnicities (Fig. 1A and Tab. S1). All six patients presented with an early-onset disorder characterized by recurrent fever, ulcerative skin rash (Fig. 1B), viral-like airway infection, and diarrhea (Tab. S1). Radiological and histological analyses showed mild interstitial lung disease and PAP, dermatitis with mononuclear inflammation, and non-specific gastrointestinal inflammation (Fig. 1C–E, Tab. S1 and Fig. S1). Peripheral blood T-cell count was normal, but monocytes and B-cells were low (during flares) to normal (in remission), and all patients had hypogammaglobulinemia (Tab. S1). All patients had respiratory insufficiency and poor weight gain or failure to thrive despite multiple lines of conservative treatment (Tab. S1). Four patients were treated with allogeneic HCT leading to correction of the clinical phenotype in two (Fig. 1E, Fig. S1B), while one succumbed to complications, and another to chronic graft-versus-host disease (Tab. S1). Patient 5 (13) deceased prior to and patient 6 was awaiting allogeneic HCT at the time of this study (Tab. S1).
Fig. 1. A polymorphic autoinflammatory immunodeficiency with de novo heterozygous OAS1 variants.
(A) Ancestry of patient 1–6 (abbreviated P1–6 throughout the figures) and electropherograms [fibroblasts (patient 1–4), PBMCs (patient 5), and buccal swabs (patient 6)] of de novo germline heterozygous OAS1 variants. (B) Clinical photographs of facial rash (patient 6) and skin erosions (patient 4). (C) Lung histology (patient 1) of PAP with granular material, cholesterol cleft-like cracks (upper image, bar = 250 μm), and foamy alveolar macrophages (lower image, bar = 50 μm). (D) Skin histology (patient 3) with intra- and subepidermal blister formation, epidermal lymphocytes, and dermal chronic inflammation (upper image, bar = 500 μm); duodenal histology (patient 3) with marked villous atrophy, crypt hyperplasia and epithelial regeneration (lower image, bar = 100 μm). (E) Chest radiography (patient 1) with PAP 14 days before (upper image) and resolution 1 year after HCT (lower image). (F) OAS superfamily phylogenetic analysis indicating conservation of A76, C109, V121, and L198 for the full OAS1 and the human OAS1 subclade. (G) Consensus and sequence logos within the human OAS1 subclade surrounding A76, C109, V121, and L198. (H) Pseudo-color plots, overlay histograms, and summary graphs of PBMCs, monocytes (CD14+), B-cells (CD19+), and T-cells (CD3+) either unstimulated (Med) or after IFNα-, CpG- and CD3/CD28-stimulation showing time-dependent changes in median intracellular OAS1 expression. (I) Overlay histograms and summary graph of intracellular OAS1 expression in naïve (IgD+CD27-, Naive), un-switched memory (IgD+CD27+, UM), class-switched memory (IgD-CD27+, SM) B-cells, and plasmablasts (CD27+CD38+, PB) in unstimulated (Med) and CpG-stimulated PBMCs. For H and I, summary graphs of 6 different OAS1-WT donors, patient 1 (A76V) and patient 6 (C109Y) from 4 independent experiments (error bars, mean ± s.d.) are shown.
Exome sequencing from DNA extracted from PBMCs (patient 1–6) and Sanger validation from DNA extracted from fibroblasts (patient 1–4), buccal swabs (patient 6) and PBMCs (patient1,5–6) identified four de novo heterozygous OAS1 missense variants (OAS1-A76V, OAS1-C109Y, OAS1-V121G, OAS1-L198V) and excluded hematopoietic mosaicism (Fig. 1A, Fig. S1A). Minor allele frequencies of 0.00000 (Tab. S2), the Combined Annotation-Dependent Depletion (CADD) (14), and the Mutation Significance Cutoff (MSC) (15) algorithms predicted all but OAS1-L198V to be disease-causing (Tab. S2). Phylogenetic analysis showed close relation of the human OAS1 gene with those of non-human placental mammals (Fig. 1F) and logo plot analysis revealed high conservation of all four amino acids and their adjacent sequences (Fig. 1G).
OAS1 wild-type (WT) protein expression in unstimulated monocytes, B-cells, and T-cells from healthy donors (HD) peaked around day four of culturing and was further increased upon interferon (IFN)-α-, CpG-, or CD3/CD28-stimulation (Fig. 1H, Fig. S2A). Primary cells were available from patient 1 (having OAS1 A76V) and patient 6 (having OAS1 C109Y) and we detected A76Vlow and C109Ylow monocytes and B-cells, but not T-cells (Fig. 1H, Fig. S2A–B). When comparing unstimulated and CpG-stimulated B-cell subsets, we identified higher OAS1 expression in non-class-switched memory B-cells (IgD+CD27+), class-switched memory B-cells (IgD-CD27+), and plasmablasts (CD27+CD38+), as compared to naïve B-cells (IgD+CD27-) (Fig. 1I, Fig. S2C–D), but all B-cell subsets contained apparently pre-apoptotic-looking A76Vlow and C109Ylow cells (Fig. 1I) based on FSC and SSC (16).
Thus, the severe autoinflammatory immunodeficiency correlates with de novo heterozygous OAS1 missense variants and OAS1low monocyte and B-cell subsets and can be cured by allogeneic HCT.
Functional and transcriptomic alteration in OAS1-A76V and C109Y monocytes, B-cells, and T-cells
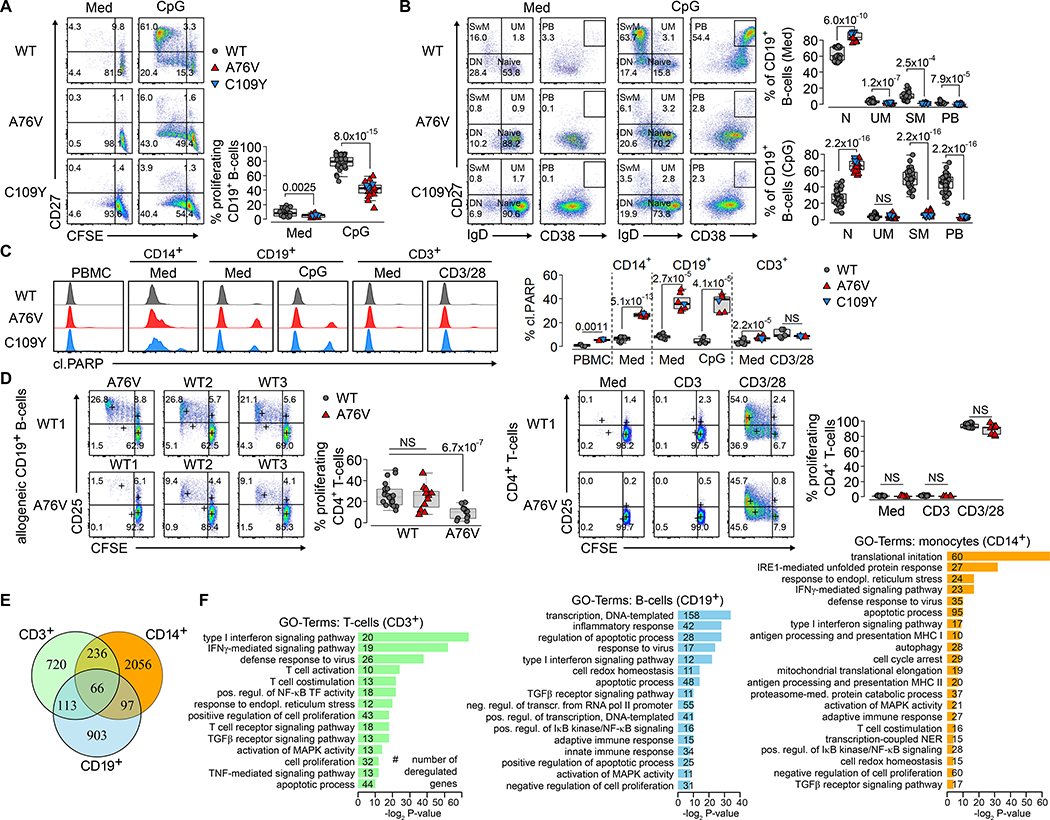
We performed cellular and transcriptomic studies on primary monocytes, B-cells, and T-cells to analyze the link between the autoinflammatory immunodeficiency and the OAS1 variant genotype. First, we assessed the capacity of OAS1-WT, A76V and C109Y CD19+ B-cells to proliferate and differentiate. While unstimulated and CpG-stimulated WT B-cells showed normal responses, A76V and C109Y B-cell proliferation and differentiation into non-class-switched memory B-cell, class-switched memory B-cells, and plasmablasts were impaired as indicated by CFSE-dilution and up-regulation of CD27 and CD38 (Fig. 2A–B). Next, we used unstimulated, CD3-, CD3/CD28-, and PMA/ionomycin-stimulated PBMCs to analyze CD4+ and CD8+ T-cell proliferation. As opposed to OAS-WT, A76V and C109Y T-cells had impaired activation and proliferation and activation as indicated by CD25 up-regulation and CFSE-dilution, especially after CD3- and, to a lesser extent, after CD3/CD28-stimulation (Fig. S3). To distinguish a defect in proliferation and differentiation from increased apoptosis, we probed the same conditions for apoptosis using cleaved PARP staining. While apoptosis in OAS1-WT, A76V, and C109Y total PBMCs and T-cells was comparable, there was increased apoptosis in A76V and C109Y monocytes and especially in differentiated B-cell subsets (Fig. 2C, Fig. S4–S5).
Fig. 2. Functional and transcriptomic alteration in OAS1-A76V and OAS1-C109Y monocyte, B- cells, and T-cells.
(A) Pseudo-color plots showing B-cell proliferation of unstimulated (Med) and CpG-stimulated PBMCs from OAS1-WT, A76V, and C109Y after 5 days by CFSE-dilution and up-regulation of CD27. Graphs show summary of B-cell proliferation percentages for 6 different OAS1-WT, A76V and C109Y from 5 independent experiments. (B) Pseudo-color plots showing B-cell differentiation of naïve B-cells (IgD+CD27-, Naive) into un-switched memory (IgD+CD27+, UM), class-switched memory (IgD-CD27+, SM) B-cells, and plasmablasts (CD27+CD38+, PB)òf unstimulated (Med) and CpG-stimulated PBMCs from OAS1-WT, A76V, and C109Y after 5 days. Graphs show summary of B-cell differentiation for 6 different OAS1-WT, A76V and C109Y from 5 independent experiments. (C) Histogram plots of cleaved PARP-1 (cl.PARP) in unstimulated and medium- (Med), CpG-, and CD3/CD28–48h-stimulated PBMCs, monocytes (CD14+), B-cells (CD19+), and T-cells (CD3+), and summary of percentages of cleaved PARP for 4 different OAS1-WT, A76V, and C109Y from 4 independent experiments. (D) Pseudo-color plots of CD25 expression and CFSE-dilution on CD4+ T-cells in a mixed lymphocyte reaction after allogeneic B-cell stimulation of 3 different allogeneic OAS1-WT donors and A76V without (Med) and after stimulation with anti-CD3 and anti-CD3/CD28 for 5 days. Summary graph of CD4+ T-cell proliferation percentages and median CD25 expression for 4 different OAS1-WT (gray) and A76V (red) from 3 independent experiments. (E) Venn diagram intersection of DEGs (adjusted p-value <0.05 and log2FC>0.5) in monocytes, B-cells, and T-cells from 3 OAS1-WT and A76V. (F) GO-term enrichment analysis of DEGs specific for monocytes, B-cells, and T-cells (non-redundant terms related to immune function with Fisheŕs exact P-value <0.05 containing >10 genes; a complete GO-term list is presented in Tab. S5). (A-D) Significance levels are calculated with two-sided Welchś t-test and indicated in the summary graphs (NS = non-significant).
Reduced A76V and C109Y T-cell proliferation (Fig. S3) was not attributable to increased apoptosis (Fig. S5) in the assays using total PBMCs that, in addition to T-cells, contained monocytes and B-cells. Therefore, we analyzed FACS-sorted CD4+ and CD8+ T-cell proliferation in a mixed lymphocyte reaction with allogeneic B-cell-stimulation (Fig. S6). In the absence of monocytes and B-cells, CD3-stimulation alone did not induce activation and proliferation of OAS1-WT or A76V T-cells as illustrated by CD25 up-regulation and CFSE-dilution (Fig. 2D). However, A76V T-cell activation and proliferation were intact as indicated by CD3/CD28- and allogeneic OAS1-WT B-cell-stimulation, but A76V B-cells failed to induce activation and proliferation of allogeneic wild-type T-cells that properly responded to CD3/CD28-stimulation (Fig. 2D, Fig. S7), pointing toward defective co-stimulation of OAS1 A76V and C109Y B-cells and monocytes.
Next, we performed poly-A-selection based RNA-sequencing on FACS-sorted OAS1-WT and A76V monocytes, B-cells, and T-cells (Fig. S8). We found A76V allele expression in 50% of T-cells, 43% of B-cells and 17% of monocytes (Fig. S9) thereby excluding nonsense mediated decay. The reduced A76V allele expression in monocytes might reflect a selective survival disadvantage inferred by the variant allele as these cells showed the strongest propensity towards apoptosis (Fig. 1H and 2C). Compared to OAS1-WT, A76V monocytes, B-cells, and T-cells differentially expressed 2,455, 1,179, and 1,135 genes, respectively (Fig. 2E, Tab. S5). Gene ontology (GO) term enrichment analysis revealed 21 functional groups for monocytes, 16 for B-cells, and 14 for T-cells when focusing on clusters related to immune function with >10 differentially expressed genes (DEG) with a P-value <0.05 (Fig. 2F, Tab. S5). Heat maps for particular GO clusters further illustrated a global defense response to virus and a type I IFN signature and detailed the apoptotic process in monocytes and B-cells (Fig. S10). Translation was globally suppressed in monocytes, and to a lesser extent in B-cells, while T-cell co-stimulation and antigen processing and presentation signatures were altered in monocytes and T-cells (Fig. S10).
Therefore, and as illustrated by primary A76V and C109Y cells, the autoinflammatory immunodeficiency was linked to the OAS1 genotype by increased spontaneous and stimulation-dependent apoptosis of monocytes and B-cells. This resulted in impaired B-cell proliferation, differentiation, and co-stimulation; impaired monocyte translation initiation, co-stimulation, and antigen presentation; and a type I IFN- and defense to virus-signature in monocytes, T-cells, and B-cells.
Interferon-α induced RNase L-dependent dysfunction of monocytes and iPSC-derived macrophages
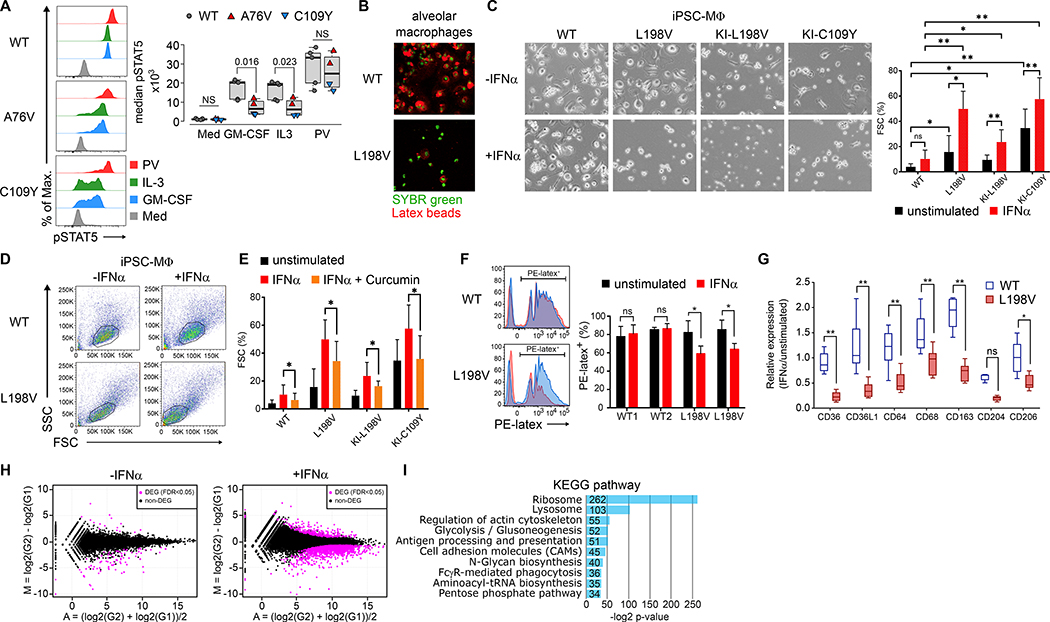
To understand the pathophysiology of PAP, we analyzed primary CD14+ monocyte function by measuring STAT5 phosphorylation in unstimulated, GM-CSF-, IL-3-, and pervanadate (PV)-stimulated OAS1-WT, A76V, and C109Y cells. GM-CSF-receptor-expression (Fig. S11) and STAT5 phosphorylation after GM-CSF- and IL-3-stimulation were reduced in A76V and C109Y monocytes, but normal in OAS1-WT and unstimulated and PV-stimulated cells (Fig. 3A). Additionally, primary alveolar macrophages from patient 5 (having OAS1 L198V) that were derived from CD14+ monocytes and were uniquely available after bronchoalveolar lavage, showed reduced phagocytosis as compared to OAS1-WT cells (Fig. 3B).
Fig. 3. Interferon-α induced RNase L-dependent dysfunction of monocyte and iPSC-derived macrophages.
(A) Overlay histograms of phosphorylated STAT5 (pSTAT5) in monocytes of unstimulated (Med), GM-CSF-, IL-3-, and pervanadate- (PV) stimulated PBMCs and summary of median pSTAT5 from 5 different OAS1-WT (gray), A76V (red), and C109Y (blue) from 3 independent experiments. (B) Fluorescence microscopy of OAS1-WT and L198V alveolar macrophages (green nuclear staining) and phagocytosed PE-latex beads (red). (C) Bright-field microscopy of OAS1-WT, L198V, KI-L198V, and KI-C109Y iPSC-MΦ either unstimulated or IFNα-stimulated (−/+IFNα) for 48 hours, and summary graph of proportions of floating single cells (FSC) of total live iPSC-MΦ unstimulated (black) or IFNα-stimulated (red) from 3 independent experiments. (D) Pseudo-color plots of OAS1-WT and L198V iPSC-MΦ unstimulated or IFNα-stimulated for 7 days. (E) Proportions of floating single cells (FSC) of total live OAS1-WT, L198V, KI-L198V iPSC-MΦ, and KI-C109Y iPSC-MΦ unstimulated (black), IFNα- (red), and curcumin (orange) stimulated/inhibited for 48 hours from 3 independent experiments. (F) Histogram overlays of PE-latex beads phagocytosed by OAS1-WT and L198V iPSC-MΦ unstimulated (black) or IFNα- (red) stimulated for 48 hours and summary graph of 2 OAS1-WT and 2 L198V clones analyzed in 3 independent experiments. (G) Relative expression levels of surface scavenger receptors of OAS1-WT (white) and L198V (red) iPSC-MΦ unstimulated and IFNα-stimulated for 48 hours from 3 independent experiments. (H) MA-plot visualization of DEGs (magenta) with a FDR <0.05 between unstimulated and 48 hours IFNα-stimulated OAS1-WT and L198V iPSC-MΦ. (I) KEGG-based GSEA of down-regulated DEG in IFNα-stimulated L198V iPSC-MΦ as compared to OAS1-WT. Significance levels are calculated with two-sided paired t-test and indicated in the summary graphs (ns = non-significant, * p<0.05, ** p<0.005, error bars, mean ± s.d.).
To overcome the scarcity of primary cells and to better understand the monocyte/macrophage disorder, we generated primary OAS1-WT and L198V or gene-edited knock-in (KI) L198V and C109Y CD34+ hematopoietic stem cell-derived inducible pluripotent stem cells and differentiated them into macrophages (iPSC-MΦ) (Fig. S12). As viral-like bronchitis preceded the development of PAP (Tab. S1) and OAS1 expression is induced by type I IFNs, we compared unstimulated and IFNα-stimulated iPSC-MΦ. Unstimulated primary L198V iPSC, and L198V and C109Y KI iPSC-MΦ showed decreased adhesion, cell clustering, and increased floating single cells (FSC) as compared to OAS1-WT iPSC-MΦ (Fig. 3C). IFNα-stimulation further augmented the frequency of FSC (Fig. 3C) and lead to a pre-apoptotic phenotype based on FSC and SSC (16) and as exemplified by L198V iPSC-MΦ (Fig. 3D). To test whether the increase in FSC was mediated by RNase L, we treated iPSC-MΦ with curcumin, a noncompetitive inhibitor of RNase L (17). Curcumin inhibited the IFNα-induced increase of FSC in OAS1-WT, L198V and C109Y iPSC-MΦ (Fig. 3E). As observed in primary L198V alveolar macrophages, L198V iPSC-MΦ showed reduced phagocytosis of PE-latex beads (Fig. 3F), and reduced expression of multiple surface scavenger receptors (Fig. 3G) after IFNα-stimulation.
To understand the transcriptional consequences of variant OAS1 mediated monocyte/macrophage dysfunction, we performed poly-A-selection based RNA-sequencing from unstimulated and IFNα-stimulated OAS1-WT and L198V iPSC-MΦ. Mean average (MA) plot visualization of DEG with a false discovery rate (FDR) <0.05 showed a few DEG between unstimulated (124 down- and 15 up-regulated), but numerous DEG between IFNα-stimulated (1960 down- and 299 up-regulated) OAS1-WT and L198V iPSC-MΦ (Fig. 3H). Kyoto Encyclopedia of Genes and Genomes (KEGG)-based gene set enrichment analysis (GSEA) identified no up-regulated, but 10 down-regulated gene sets in IFNα-stimulated L198V iPSC-MΦ (Fig. 3I). In particular, expression of ribosomal and transfer RNAs, phagocytosis-, lysosomal-, antigen-presentation-, cytoskeletal, and glucose metabolism-related gene sets were decreased (Fig. 3I, Fig. S13).
Thus, primary monocytes/macrophages and IFNα-stimulated variant OAS1 iPSC-MΦ displayed impaired cell adhesion and clustering, scavenger receptor expression, and phagocytosis in a RNase L-dependent manner that correlated to down-regulated concomitant gene sets.
OAS1-GOF 2–5A synthetase activity- and RNase L-dependent cellular RNA degradation, protein translational arrest, and apoptosis
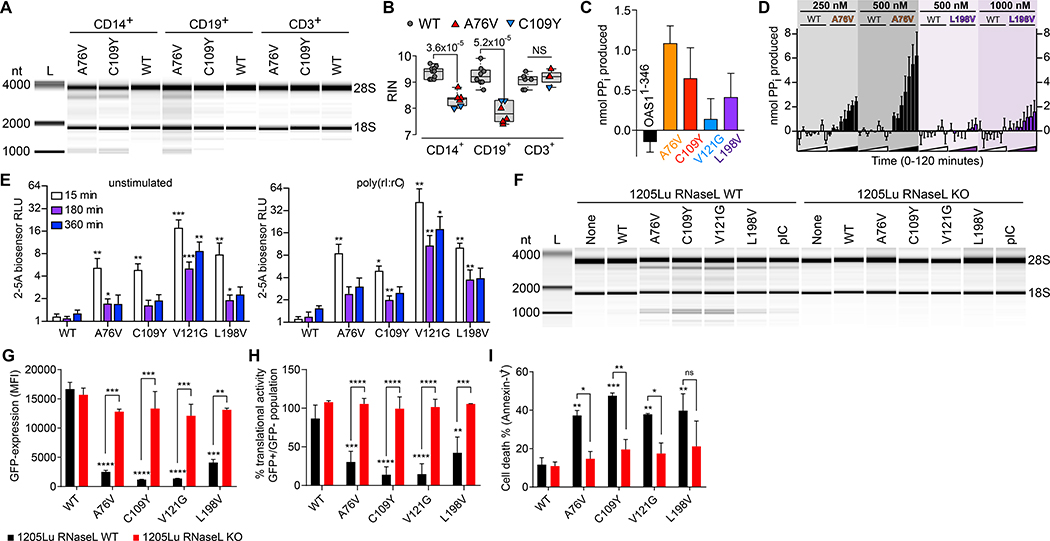
To understand the molecular mechanisms of monocyte/macrophage and B-cell dysfunction and apoptosis, we FACS-sorted unstimulated primary OAS1-WT, A76V and C109Y cells (Fig. S8), isolated cellular RNA from monocytes, B-cells, and T-cells, and analyzed RNA degradation to calculate the RNA integrity number (RIN) (18). While OAS1-WT cells uniformly had intact cellular RNA, A76V and C109Y monocytes and B-cells, but not T-cells, displayed RNA degradation (Fig. 4A–B) that resembled RNase L-mediated degradation (19).
Fig. 4. OAS1-GOF 2–5A synthetase activity- and RNase L-dependent cellular RNA degradation, protein translational arrest, and apoptosis.
(A) RNA chip analysis from OAS1-WT, A76V, and C109Y monocytes (CD14+), B-cells (CD19+), and T-cells (CD3+) of total RNA degradation and (B) summary graph of RIN from 5 independent experiments (NS = non-significant). (C) In vitro activity for OAS1-WT and OAS1-MUT (A76V, C109Y, V121G, L198V) proteins in the absence of poly(rI:rC) dsRNA after 60 minutes (error bars, mean ± s.e.m). (D) Chromogenic analysis of reaction time courses for purified OAS1-WT (white bars), A76V (black bars), and L198V (purple bars) protein in the absence of poly(rI:rC) dsRNA for a range of protein concentrations (error bars, mean ± s.e.m). (E) 2–5A biosensor analysis of unstimulated and poly(rI:rC) dsRNA-stimulated HEK293T cells transiently transfected with OAS1-WT and OAS1-MUT (error bars, mean ± s.d.). (F) RNA chip analysis of total RNA isolated from 1205Lu melanoma RNase L wild-type (WT) and RNase L knockout (KO) cell lines untransfected (None) and transiently transfected with OAS1-WT, OAS1-MUT, or poly(rI:rC) (pIC). (G) Expression levels of OAS1-WT or OAS1-MUT in transfected 1205Lu melanoma wild-type (WT) and RNase L knockout (KO) cell lines determined by eGFP co-expression 48h after transfection (error bars, mean ± s.d.). (H and I) Translational activity (GFP-positive/GFP-negative) and frequency of dead (Annexin-V+) cells 48h after transfection (error bars, mean ± s.d., * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001).
To test the hypothesis of OAS1-GOF-based activation of RNase L, we expressed recombinant OAS1-WT and OAS1-MUT (A76V, C109Y, V121G, L198V) proteins in E. coli BL21(DE3) as N-terminal 6xHis- SUMO fusions, purified them as described previously(20–22) nd tested their ability to synthesize 2–5A in the presence and absence of poly(rI:rC)-dsRNA. In the presence of dsRNA, each OAS1-MUT protein except L198V, showed reduced 2–5A synthesis compared to OAS1-WT indicating a potential protein instability-related reduced ligand-dependent activation (Fig. S14A). As the effects of OAS1-MUT observed in cell-based assays, however, did not appear to have their origin in dsRNA-induced reduced enzyme activity, we inspected the basal activity of each OAS1-MUT enzyme in the absence of dsRNA. As compared to OAS1-WT, OAS1-MUT enzyme activity consistently appeared above background (Fig. 4C). We compared OAS1-WT, A76V, and L198V activity in the absence of dsRNA at different protein concentrations (C109Y and V121G could not be analyzed because of instability of the purified proteins). We observed 2–5A synthesis in a protein concentration- and time-dependent manner for A76V and L198V, but not OAS1-WT (Fig. 4D) further confirming their GOF 2–5A synthetase activity.
As the ligand-independent OAS1-MUT activity produced very small amounts of 2–5A in vitro, we asked if this was sufficient to explain the observed RNase L-activity of OAS1-MUT cells. We transiently co-transfected HEK293T cells with the 2–5A biosensor V6 or the non-functional variant V6-Y312A (23) together with OAS1-WT or OAS1-MUT, and analyzed these cells in the presence and absence of exogenous poly(rI:rC) dsRNA (Fig. 4E). As compared to OAS1-WT, OAS1-MUT cells had increased 2–5A production, that further increased after poly(rI:rC)-dsRNA treatment (Fig. 4E). To check for RNase L-dependency, we transiently co-transfected 1205Lu melanoma wild-type (WT) and RNase L knockout (KO) cells that have no constitutive OAS1, 2, and 3 expression, with OAS1-WT or OAS1-MUT in combination with an eGFP reporter gene (Fig. S14B). While OAS1-WT did not induce RNA degradation, OAS1-MUT proteins induced the same RNA degradation pattern as observed in primary monocytes and B-cells in a RNase L-dependent manner (Fig. 4F).
Since OAS1/RNase L pathway-activation leads to translational arrest and subsequently to apoptosis, we studied the impact of OAS1-MUT on cellular translational activity and viability. First, we transfected 1205Lu melanoma WT and RNase L KO cells with OAS1-WT and OAS1-MUT. Next we analyzed eGFP reporter gene expression, translational activity through surface sensing of translation (SUnSET) (24), that relies on the amount of puromycin incorporated into newly translated proteins, and apoptosis by flow cytometry. While there was no difference in eGFP expression and translational activity between RNase L WT and KO cells upon expression of OAS1-WT, transfection of OAS1-MUT proteins led to significantly reduced expression of eGFP and translational activity in WT, but not KO cells (Fig. 4G–H). Concurrently, and in line with our finding in primary monocytes and B-cells (Fig. 2B), we observed apoptosis upon OAS1-MUT transfection in RNase L WT but not in KO cells (Fig. 4I, Fig. S14C–D).
Thus, OAS1-MUT proteins displayed dsRNA-independent GOF 2–5A synthetase activity that led to RNase L-dependent RNA degradation, translational arrest, and cellular apoptosis. While L198V displayed the weakest GOF activity followed by A76V, C109Y and V121G had stronger activity which was hampered by protein instability.
dsRNA-independent OAS1-GOF 2–5A synthetase activity favored by allosteric loss of structural constraints
Residues A76, C109, and V121 distributed across the OAS1 N-lobe β-sheet-core and adjacent α-helices, while residue L198 located in the OAS1 C-lobe. All four residues located in regions of the protein that under dsRNA-induced conformational changes or local reorganization (25–27). To gain initial insight into the potential disruption of local protein structure for each OAS1-MUT substitution, we first generated in silico models of each amino acid substitution (Fig. S15). Although there is a structure available of human OAS1 bound to dsRNA, we used porcine OAS1 (pOAS1) in its free and dsRNA-bound forms for direct comparisons of both protein conformational states since the overall structure, the OAS1-MUT residues, and their interaction partners were conserved (Fig. S15) (25–27).
Upon dsRNA-binding, A76 shifted into a hydrophobic pocket formed by the adjacent β-strands and α2-helix, forming new interactions that might have been more readily promoted by the bulkier V76 side chain in the absence of dsRNA (Fig. S15A–F). Thus, the A76V substitution could promote formation of a more dsRNA bound-like structure in this region of the protein. C109 was located on the α2: α3 interface in a hydrophobic pocket and substitution with the bulkier Y109 would have disrupted these interactions and force the two α-helices apart (Fig. S15G–H). Similarly, V121 was sequestered in a predominantly hydrophobic pocket formed by residues of the α3-helix and the β-sheet-core but, in this case, its substitution with the smaller G121 would have eliminated these interactions destabilizing the protein in this critical region (Fig. S15I–J). Thus, both Y109 and G121 may have disrupted the protein structure in a manner that relieves constraints on the dsRNA-free OAS1 structure that ordinarily requires dsRNA binding. Finally, L198 was located in a predominantly hydrophobic pocket of the OAS1 C-lobe that upon dsRNA-binding underwent complex conformational changes mediated by E233 and resulting in the displacement of K65 and R194 (Fig. S15K–M). The dsRNA-free and catalytically inactive OAS1 conformation was stabilized by an inhibitory clash between L198 and K65 the pro-catalytic repositioning of which might have been facilitated by the smaller V198 (Fig. S15N).
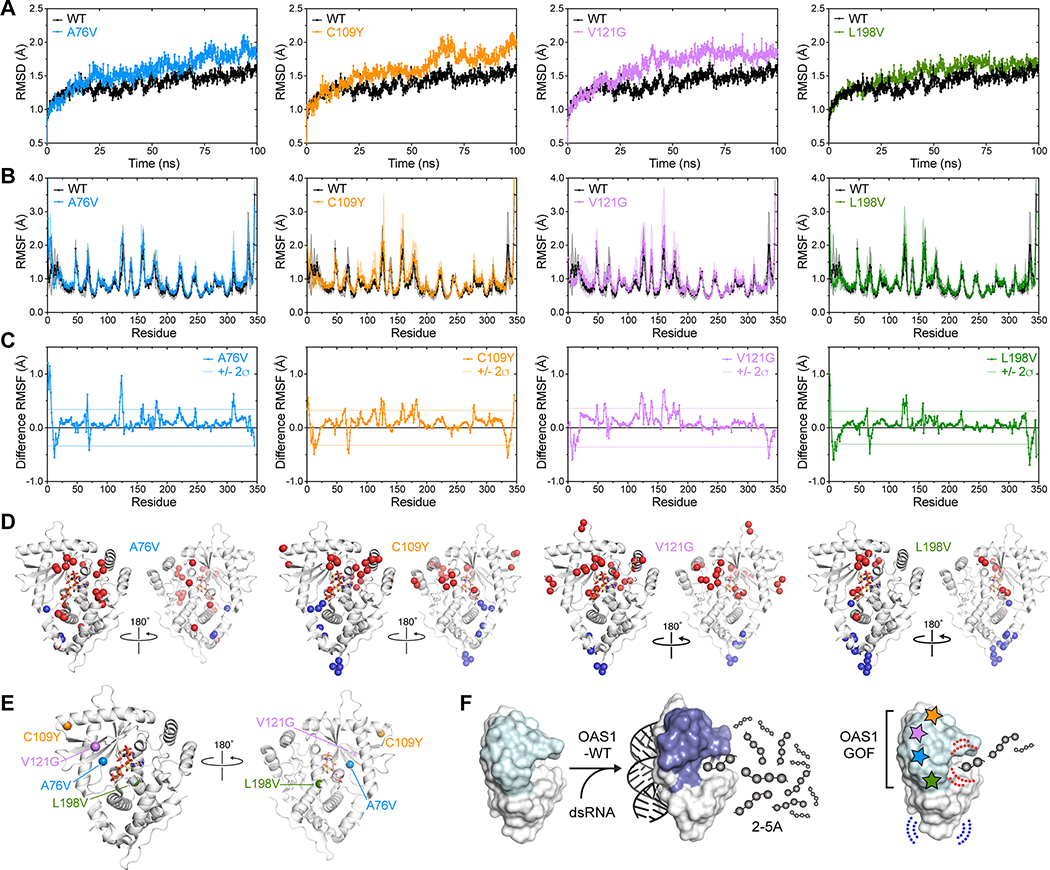
To test how the different OAS-MUT substitutions might have manifested as a common GOF outcome, we used molecular dynamics simulations to assess and compare protein dynamics of the wild-type and the four variant proteins. First, we compared overall root-mean-square deviation (RMSD) for all protein backbone atoms over the 100 ns production run of the MD simulation (Fig. 5A). These analyses revealed that each of the OAS-MUT proteins exhibited higher RMSD over the simulation reflecting a globally more dynamic nature than OAS-WT. The differences were most apparent for A76V, C109Y and V121G, and less in L198V, consistent with its lower measure activity in the absence of dsRNA. To identify specific regions of OAS-MUT that exhibited altered dynamics, we examined the residue root-mean-square fluctuation (RMSF) for backbone atoms and calculated difference RMSF for each variant by subtraction of the OAS-WT values (Fig. 5B–C). For each protein’s difference RMSF, we calculated the standard deviation for all residues and used a 2σ cut-off for the most significant changes in each OAS-MUT and mapped these residues onto the pOAS1 structure in its dsRNA-bound conformation (Fig. 5D–E). This analysis revealed that despite their distinct locations in OAS1, each OAS-MUT substitution caused changes in protein dynamics shared by each OAS-MUT. While some substitutions induced local perturbations as expected (e.g., C109Y in α3 and V121G in the β-sheet core of the OAS1 N-lobe), most increases in protein dynamics were induced allosterically in more distant protein regions, in particular surrounding the enzyme active site (Fig. 5D). Further supporting the idea that such induced changes were one pathway for weak GOF activity, we also observed allosterically induced decreases in protein flexibility in the core of the protein (centered around the loop containing R70) and more distantly at the base of the C-lobe. Again, many of these distant changes were common to all OAS-MUT substitutions and some were located in regions that form more extensive interactions in the dsRNA-bound conformation compared to free OAS1 (such as the R70 loop).
Fig. 5. dsRNA-independent OAS1-GOF 2–5A synthetase activity favored by allosteric loss of structural constraints.
MD simulation analysis of OAS-WT and OAS-MUT: (A) overall protein root-mean-square deviation (RMSD) over the 100 ns MD production run, (B) residue backbone atom root-mean-square fluctuation (RMSF), and (C) difference RMSF (OAS-WT subtracted from each OAS-MUT). Individual proteins are as noted in the plot legends (left to right): A76V (blue), C109Y (orange), V121G (purple) and L198V (green); OAS-WT (black) is shown in panels A and B for comparison to each OAS-MUT. Shaded regions on plots in panel B are the error in each value (SD). In panel C, dotted lines denote two standard deviations (+/− 2σ) from the average difference RMSF for each protein (i.e., the most significant changes in residue dynamics). (D) Residues with most significant changes in dynamics mapped onto OAS1 structure (shown in the dsRNA-bound conformation; PDB code 4RWN) as Cα spheres; color coding denotes increased dynamics (> 2σ; red) and decreased dynamics (< −2σ; blue) compared to OAS-WT. (E) Locations of the OAS-MUT substituted residues shown in the same views as panel D. (F) Model for common action of OAS1-GOF variants. OAS1-WT (left) strictly regulated by dsRNA binding-induced conformational changes, whereas OAS1-MUT amino acid substitutions (right) relieve this strict control by allosteric induction of changes around the enzyme catalytic center (red dotted lines) and mirroring changes that occur upon dsRNA binding (blue dotted lines). These changes promote low-level GOF activity in the absence of dsRNA.
In summary, while each of the local structural perturbations caused by the OAS-MUT substitutions were likely distinct, a common feature of all OAS1-MUT proteins appeared to be the loss of constraints that favor adoption of the activated conformation by OAS1, independent of dsRNA. As such, these variant OAS1 proteins might have been partially able to adopt a dsRNA-bound-like conformation which resulted in the observed constitutive OAS1-GOF 2–5A synthetase activity (Fig. 5F).
Discussion
We report a polymorphic autoinflammatory immunodeficiency with recurrent fever, dermatitis, inflammatory bowel disease, pulmonary alveolar proteinosis, and hypogammaglobulinemia that was caused by de novo heterozygous OAS1 GOF variants. Upon type I IFN-induced expression in response to common infectious agents, OAS1-GOF variants inappropriately synthetized 2–5A independent of dsRNA-binding and induced RNase L-mediated cleavage of cellular RNA leading to transcriptomic alteration, translational arrest, dysfunction and apoptosis of monocytes, iPSC-derived macrophages, and B-cells. The ensuing hypogammaglobulinemia may cause susceptibility to infection while monocyte and tissue macrophage dysfunction may cause accumulation of pro-inflammatory damage-associated molecular patterns. Recent findings indicate that dsRNA-mediated activation of the OAS1/RNase L pathway induces rapid degradation of global mRNAs, but spares innate immunity mRNAs such as IFNs and IFN-stimulated genes (ISGs) (23, 28, 29). The resulting selective transcription and translation of innate immunity mRNAs and proteins combined with a translational arrest of metabolic and housekeeping genes may create a positive feedback loop that builds up an inflammatory immune response (28, 29). In line with this, our findings on OAS1-GOF point towards an initial IFN-induced expression of ISGs including OAS1 that was followed by a self-sustaining autoinflammatory and immunodeficiency vicious circle. Based on the mechanistic findings of OAS1/RNase L-mediated systemic inflammation that have been detailed before (23, 28, 30), we thus described a disease entity of IFN-induced OAS1/RNase L pathway-mediated hyperinflammatory monocyte and B-cell dysfunction that in vitro could be modulated by RNase L-inhibition with curcumin and that could cured by allogeneic HCT in some patients. In parallel with the previously identified STING-associated vasculopathy with onset in infancy (SAVI) (3), we name this IEI OAS1-associated polymorphic autoinflammatory immunodeficiency disorder (OPAID).
All reported pathogenic OAS1 variants are de novo, absent from genomic databases and cause highly lethal early-onset OPAID, thereby indicating that OAS1 activity requires tight limitation. A human splice site polymorphism reducing OAS1 activity has been reported (30) and several primates have evolved OAS1 loss-of-function orthologs (31). However, reduced OAS1 activity is associated with increased risk of viral infections (32) and immune dysregulation (33). Taking into account the cost-benefit ratio of immune defense versus longevity and fecundity, there is no obvious evolutionary reason for human OAS1-low-activity alleles (31). The severe systemic and cellular consequences of OAS1-GOF showed here that monocytes and B-cells were particularly susceptible towards OAS1/RNase L pathway activation. Both cell types are professional antigen presenting cells, but also exert immune cell effector functions that depend upon proliferation, differentiation and migration into lymphoid and non-lymphoid tissues (34, 35). While further analyses are needed to understand the molecular mechanisms and functional properties of the OAS1/RNase L system in monocytes and B-cells as opposed to T-cells and extra-hematopoietic cells, we speculate that it might constitute a cell intrinsic immune checkpoint to balance the need for antigen-presentation and the risk of antigen-spreading by deleting cells that exceed a certain signaling threshold induced by foreign dsRNA.
Besides its role in anti-viral immunity, OAS1 has been found to mediate apoptosis of thymic pre-T-cells that have not succeeded in productive TCR-beta recombination (36). We show that differentiating B-cells undergoing class-switch-recombination and somatic hypermutation have increased OAS1 expression and are prone to OAS1/RNase L-mediated apoptosis. Although the mechanism of interferon-receptor expression and signaling, OAS1 up-regulation, putative endogenous ligands and negative-regulators in pre-T-cells, differentiating B-cells, and likely also pre-B-cells, are not clear, we envision a critical role for OAS1 in shaping the adaptive immunity TCR- and BCR-repertoire. In line with that is the current identification of STING as a critical regulator of TCR-dependent calcium homeostasis and the finding that STING-GOF variants lead to increased T-cell apoptosis and inflammatory lung disease (37). Additional analyses are necessary to study the impact of OAS1 GOF variants in thymocyte, B-cell precursor and plasma cell differentiation models.
In acute COVID-19 germinal centers are lost in lymph nodes and spleens upon increased tumor necrosis factor (TNF) α production and this leads to an inefficient humoral immune response (38), a mechanism that is reminiscent of B-cell apoptosis and hypogammaglobulinemia in OAS1 GOF. A systematic review describes OAS1 as a gene influencing coronavirus-associated phenotypes (39). A single cell atlas and a systems biology assessment identify enhanced ISG-expression including OAS1 and downregulation of genes related to antigen presentation in monocytes in severe COVID-19 (40, 41). Consequently, we envision that OAS1/RNase L-targeted anti-inflammatory treatment with curcumin (5, 42) in IFN-induced hyperinflammatory monocyte and B-cell pathophysiology might allow for a more successful clinical management of these high prevalence disorders.
Materials and methods
Study design
The objective of this study was to elucidate the pathophysiology of an unexplained polymorphic autoinflammatory immunodeficiency associated with de novo heterozygous OAS1 variants. We evaluated in silico genetic and molecular dynamics simulation analyses. We cloned, expressed and purified variant OAS1 proteins and determined their enzymatic activity in vitro. We analyzed their impact on RNA metabolism and cell survival in a heterologous cell system. To further study the consequences of mutant OAS1 on B-cells, monocytes, and macrophages, we used primary human cells and inducible pluripotent stem cell derived macrophages by RNA-sequencing and particular biochemical and cell biological techniques. Based on these findings we established the rationale for curative patient treatment with allogeneic hematopoietic cell transplantation and delivered this service to eligible patients.
Patients
Legal guardians of the patients gave written informed consent according to current ethical and legal guidelines and the Declaration of Helsinki. The Institutional Review Boards at the Medical Faculty of the Ludwig-Maximilians-Universität München (project number 381–11), the Children’s Hospital of Philadelphia (CHOP 2001–4-2405), the Children’s Hospital Los Angeles (CCI 2006–00093), and the Tokyo Medical and Dental University (protocol number 103 and 676) approved the study. Ethnic groups are defined as described in (43).
Genetics and sequence conservation analysis
Genomic DNA from peripheral blood mononucleated cells (PBMC) was used for whole exome sequencing (Agilent enrichment with Illumina sequencing). Rare variant detection and filtering were described previously (44). OAS1 sequence variants were validated by Sanger sequencing using specific primers (Tab. S3).
For phylogenetic analysis, OAS sequences were retrieved by BLAST search using human OAS1, 2, 3, and L as queries in UniProt. A set of 669 unique amino acid sequences was then selected by applying a 0.99% sequence identity cut-off in CD-HIT (45). This unique sequence set, corresponding to both known and hypothetical OAS1, 2, 3, and L proteins, were aligned with CLUSTAL omega and the alignment was subjected to phylogenetic inference using the unweighted pair group method with arithmetic mean (UPGMA) method in MEGA 6.0 to generate an OAS superfamily evolutionary tree (46). Calculations of A76, C109, V121, and L198 conservation were made using Geneious Prime for all OAS1 sequences resulting from the process (336 sequences) and for the subclade containing human OAS1 (184 sequences) (47). Evolutionary distances (amino acid substitutions per site) were computed using the JTT matrix-based method and the rate variation among sites was modeled with a gamma distribution (shape parameter = 1). All positions with less than 95% site coverage were eliminated from the calculations.
Cell culture and stimulation
PBMC were isolated by density gradient centrifugation using Ficoll-Hypaque (Biochrom, Berlin, Germany) and maintained in RPMI1640 supplemented with 2mM Glutamax, 100U/ml penicillin, 100μg/ml streptomycin and 10% FCS (Thermo Fisher Scientific, Waltham, USA) at 37°C and 5% CO2. For T- and B-cell proliferation PBMC were labeled with 2.5μM carboxyfluorescein succinimidyl ester (CFSE, Thermo Fisher Scientific). For analysis of T-cell proliferation and apoptosis PBMC were stimulated with anti-CD3-coupled beads (anti-Biotin MACSiBeads, Miltenyi Biotec coupled with Biotin-anti-CD3, OKT3) at a ratio of 5:1 with and without 1μg/ml CD28 (CD28.2, both Thermo Fisher Scientific) or with 0.5ng/ml phorbol 12-myristate 13-acetate (PMA) and 1μM ionomycin (Sigma Aldrich, St. Louis, USA). For analysis of B-cell proliferation and apoptosis PBMC were stimulated with 50nM CpG (ODN 2006, InvivoGen, San Diego, USA). OAS1-expression was induced by stimulation with 1×104U/ml IFN-α2a (Miltenyi Biotec, Bergisch Gladbach, Germany). For detection of phosphorylated STAT5 PBMC were stimulated with GM-CSF (10ng/ml), IL-3 (200ng/ml), and pervanadate (500μM) for 20min at 37°C. Titration curves of stimulation reagents (Fig. S16) and gating strategy (Fig. S17) are shown in the Supplementary Appendix.
Flow cytometry and fluorescence activated cell sorting
B-cell proliferation and differentiation were measured by labeling PBMC with PC7-anti-CD19 (J3–119, 1:50, Beckman Coulter, Brea, USA), PE-anti-CD27 (L128, 1:25), APC-anti-CD38 (HIT2, 1:12.5) and BV421-anti-IgD (IA6–2, 1:100, Becton Dickinson (BD), San Jose, USA). Intracellular staining of cleaved-PARP was performed using Cytofix/Cytoperm buffer set and A647-anti-cleaved-PARP (Asp-214, 1:20) after surface staining with APC-H7-anti-CD3 (SK7, 1:50), PE-anti-CD14 (MφP9, 1:6.25) all BD and PC7-anti-CD19 (J3–119, 1:50, Beckman Coulter). T-cell proliferation was measured by labeling PBMC with APC-H7-anti-CD3 (SK7, 1:50), APC-anti-CD4 (SK3, 1:50), PacB-anti-CD8 (RPA-T8, 1:50) and PE-anti-CD25 (M-A251, 1:25) all from BD. For analysis of T- and B-cell apoptosis PBMC were additionally stained with FITC-anti-Annexin-V (1:25) and 7-amino-actinomycin-D (7-AAD, 2.5μg/ml) all BD. For OAS1-expression analysis in PBMC Cytofix/Cytoperm (BD) buffer set and anti-OAS1 (F-3, 10μg/ml, Santa Cruz, Dallas, USA), mouse-IgG (MG1–45, 10μg/ml, BioLegend, San Diego, USA) as isotype control and mouse FITC-conjugated secondary antibody (RMG1–1, 1:200, BioLegend) were used followed by blocking with normal mouse IgG (Thermo Fisher Scientific) and surface staining with APC-H7-anti-CD3 (SK7, 1:25), PE-anti-CD14 (MφP9, 1:6.25) and PC7-anti-CD19 (J3–119, 1:50, Beckman Coulter). For phospho-flow analysis PBMC were fixed and permeabilized with fixation buffer (Thermo Fisher Scientific) in 90% methanol followed by staining with A647-anti-phospho-STAT5 (47/Stat5, 1:10), PerCP-anti-CD3 (SK7, 1:20) and FITC-anti-CD14 (MφP9, 1:20) all BD. Gates were set on single CD14+ monocytes. Data was acquired on a BD FACSCanto II flow cytometer and data analysis was performed with FlowJo software (TreeStar, Ashland, USA). Gating strategies are shown in Fig. S17. Monocytes, T- and B-cells were stained with APC-anti-CD14 (M5E2, 1:10), FITC-anti-CD3 (SK7, 1:10) and PC7-anti-CD19 (J3–119, 1:10, Beckman Coulter) and sorted using a BD FACSAria cell sorter II (Fig. S6 and S8).
Protein expression, in vitro activity, and structural analyses
Human OAS1 proteins were expressed in E. coli BL21(DE3) as N-terminal 6xHis-SUMO fusions and purified as described previously (20–22). More information is available in the supplement.
OAS1 RNA expression analysis
Briefly, cell lineages and B-cell subpopulations were sorted on a BD FACSAria cell sorter II and total RNA was purified by RNeasy Micro Kit (Qiagen). The RNA-seq library was constructed by NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs) and sequenced by Hiseq 1500 with single read 50 cycle kit (Illumina). The obtained sequence data was mapped to human genome (hg38) by TopHat2 and transcripts by RefSeq data. Fragments per kilobase of transcript per million mapped reads (FPKM) calculation was performed with cufflinks 2.2.1. More information is available in the supplement.
Human iPSC generation and gene editing
Human CD34+ hematopoietic stem cell-derived inducible pluripotent stem cells (iPSC) were generated as previously described (48). To verify iPSC integrity, karyotype (G-banding), pluripotency (qPCR for OCT4 and NANOG), and tri-lineage differentiation capacity [qPCR for PAX6, SOX1, SOX17, FOXA2, TBXT, and NCAM (STEMdiff™ Trilineage Differentiation Kit, STEM CELL Technologies, Vancouver, Canada)] were analyzed. T- and B-cell origin of the iPSCs was excluded by TCRB or IGH Gene Clonality Assay (Invivoscribe Technologies, San Diego, USA).
Gene editing of iPSCs was done with the single nicks in the target gene and donor plasmid (SNGD) method as previously described (49). More information is available in the supplement.
Human iPSC-derived macrophage differentiation
Differentiation of iPSCs into hematopoietic stem cells (HSC) was performed as previously described (50). More information is available in the supplement.
Phenotypic and functional analyses of human iPSC-derived macrophages
Surface marker expression of iPSC-derived macrophages was analyzed by flow cytometry on a LSRFortessa flow cytometer (BD, Franklin Lakes, USA) with CD11b-PE/Cy5 (#301307), CD14-APC (#301807), CD36-APC/Cy7 (#336213), CD36L1-PE (#363203), CD64-Pacific Blue (#305017), CD68-PE/Cy7 (#333815), CD115-APC (#347305), CD116-PE (#305908), CD163-FITC (#333617), CD204-APC (#371905), CD206-Alexa Fluor 700 (#321131) (all Bio Legend, San Diego, USA), and CD16-FITC (#130–106-761) (Miltenyi Biotec, Bergisch Gladbach, Germany) antibodies and evaluated with FlowJo (BD, Franklin Lakes, USA). iPSC-derived macrophages were stimulated with 1,000 IU/mL (f/c) of human recombinant IFNα (R&D Systems, Minneapolis, USA) for indicated time points. Singlet live cells were gated by FSC/SSC profile, and then CD14 positive macrophages were FACS-sorted. Mean fluorescence Intensity (MFI) of these surface markers were compared within the samples.
For analysis of phagocytosis activity, iPSC-derived macrophages were incubated with PE-labeled latex beads (Sigma #L3030, Sigma-Aldrich, St. Louis, USA) for three hours with intermittent stirring. Then, all cells were harvested, washed once, and stained with CD14-APC (#301807, BioLegend). Intracellular PE-labeled latex beads were analyzed by flow cytometry on a LSR Fortessa flow cytometer. Singlet live cells were gated by FSC/SSC profile, and then CD14 positive macrophages were FACS-sorted. Percentages of PE-positive cells were compared within the samples.
Phagocytosis activity of alveolar macrophages was analyzed as previously described (51). In brief, isolated alveolar macrophages were incubated with 0.5% PE-labelled latex beads (Sigma-Aldrich) for 30 minutes and fixed in 4% in paraformaldehyde. Then, cells were stained with Syber green (Dojindo Laboratories, Kumamoto, Japan) and analyzed by a confocal laser microscope (Carl Zeiss Inc., Oberkochen, Germany).
Cell culture analysis of OAS1 variants
The OAS1 gene was amplified from cDNA of IFN-stimulated 1205Lu human melanoma cells (provided by R. Besch, University Hospital, LMU Munich, Germany) and subcloned into pCR4. OAS1 mutants were produced by Q5 mutagenesis (New England Biolabs, Ipswich, USA), verified by Sanger sequencing, and a T2A-eGFP cassette was introduced by Gibson Assembly cloning (New England Biolabs). OAS1-WT and mutant OAS1-T2A-eGFP were cloned into pcDNA3.1 and transiently transfected into 1205Lu cells using TransIT-X2 Transfection Reagent (Mirus, Wisconsin, USA).
1205Lu RNaseL knockout cells were generated by transient expression of rationally engineered enhanced Cas9(1.1) nucleases with improved specificity together with a single-guide (sg) RNA (5’-GCCGAGTTGCTGTGCAAACG-3’) targeting exon 2 of RNASEL from a T2A-Puro-modified eSpCas9(1.1) plasmid [a gift from Feng Zhang (Addgene plasmid # 71814)] (52). Transfected cells were enriched by puromycin selection and single-cell clones were established by limiting-dilution cultivation. Knockout cells were validated by next-generation sequencing of the target locus applying the OutKnocker web tool and via western blotting (anti-RNaseL antibody from CST, 27281S) (53). More information is available in the supplement. 1205Lu cells were cultivated in DMEM supplemented with 10% bovine fetal calve serum (Thermo Fisher Scientific), L-glutamine (Sigma-Aldrich) and penicillin/streptomycin (Sigma-Aldrich) at 37°C and 5% CO2. 1205Lu cells were seeded at 12×104 cells per well in 96 well plates a day before transfection. Transfection was performed at a ratio of 250ng of plasmid DNA to 0.75μl TransIT-X2 Transfection Reagent (Mirus) per ml culture volume according to the manufacturer’s protocol. For dsRNA stimulation, 1μg of poly(rI:rC) [poly(I:C) (HMW), Invivogen] was complexed using 2.5μl of Lipofectamine RNAiMAX (Thermo Fisher Scientific) per ml of culture medium according to manufacturer’s recommendations. Cells were STR-typed and regularly tested for mycoplasma contamination using MycoAlert™ Mycoplasma Detection Kit (Lonza, Basel, Switzerland).
To quantify apoptosis induction and GFP-expression, cells were stained 48h after plasmid transfection with PE- or APC- conjugated Annexin-V (1:50; Immunotools, Friesoythe, Germany) or Violet Live Cell Caspase Probe (1:1250; BD) together with Fixable Viability Dye eFluor 780 (1:5000, Thermo Fisher Scientific) and analyzed by flow cytometry (BD LSR Fortessa). To measure RNA integrity, total RNA was isolated 20h after plasmid transfection using Total RNA Kit, peqGOLD (VWR International, Radnor, USA). 5ng of RNA were analyzed on an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, USA) using Agilent RNA 6000 Pico Kit (Agilent Technologies).
Global translational activity was measured by SUnSET (24). Cells were seeded at 2×105 per well in 6 well plates one day prior to treatment. Transfection of plasmid DNA was performed in 1mL of culture medium at 250ng/ml. 24h after plasmid transfection, all experimental conditions were pulsed with 1μg/ml puromycin in complete medium for 30min, followed by a chase in puromycin-free medium for 1h to facilitate detection of labeled proteins at the cell surface. For FACS analysis cells were detached by scraping, washed once and stained with anti-puromycin-AF647 antibody (12D10, 1:200; Merck).
RNA degradation and sequencing
Total RNA was isolated from sorted cells using Qiazol lysis reagent and RNeasy Mini Kit (Qiagen, Hilden, Germany). RNA degradation was analyzed on RNA Pico chips using Agilent Bioanalyzer 2100 (Agilent) and RNA integrity number (RIN) was calculated (18). For primary PBMCs, Illumina RNA-Seq and data analysis was done as previously described (54). For iPSC-derived macrophages, poly-A tailed mRNA isolation, library preparation and quantification were performed with NEBNext kits (New England Biolabs, Ipswich, USA) following manufacturer’s instructions. Libraries were sequenced on a Lumin (Illumina, San Diego, USA) and transcriptome analysis was performed as previously described (55). Differentially expressed genes were assessed by edgeR and pathway analysis was performed by gene set enrichment analysis (GSEA) as previously described (56, 57).
Immunoblotting
Whole cell lysates were prepared from T-lymphoblasts, total protein was resolved by SDS-PAGE, transferred to nitrocellulose membrane, and probed with anti-OAS1 (sc-374656, 1:1,000), anti-Actin (sc-8432, 1:1,000) and goat anti-mouse IgG-HRP (sc-2005, 1:10,000, Santa Cruz, USA) as previously described (54).
In vivo 2–5A biosensor assay
Measurement of in vivo 2–5A synthesis was performed as previously described by Chitrakar et al (23). Briefly, we designed the 2–5A biosensor V6 (wild type) and V6-Y312A variant (lacking 2–5A binding capacity) and cloned them into pcDNA3.1 vector (GenScript, Piscataway, NJ, USA). HEK 293T cells were seeded into flat bottom white 96-well plates and grown to semiconfluency, and each V6 or V6–312A plasmid with OAS1 expressing plasmid (WT, C109Y, A76V, V121G, or L198V) were transfected using Lipofectamine 3000 (Thermo Fischer Scientific). 24 hours after transfection, the cells were treated with D-luciferin ethyl ester (Marker Gene Technologies, Eugene, OR, USA) and with 2 μg/ml poly(rI:rC) dsRNA and luminescence was measured in 15-minute intervals over 6 hours on a TriStar LB941 plate reader (Berthold Technologies, Baden-Württemberg, DE).
Curcumin RNase-L inhibition assays
0.2–0.3 × 106 iPSCs (OAS1-WT and OAS1-MUT) were subjected to terminal differentiation to macrophages in 24 well plates. Curcumin (FUJIFILM Wako Pure Chemical Corporation, Tokyo, Japan) was added to the culture at 25mM at 24 hours after initiation of differentiation and incubation was continued for another 24 hours. The cells were then cultured in the presence of IFN-α (1,000 IU/mL, R&D Systems, Minneapolis, USA) for 48 hours, at the end of which floating cells in the culture medium and viable adherent cells were counted under by microscopy. Cells were cultured in duplicate and the ratio of floating single cells (FSCs) and viable adherent cells were calculated for three independent experiments.
Molecular Dynamics (MD) Simulations
pOAS1 (PDB code 4RWQ) was used to generate in silico models for A75V, C108Y, V120G, and L197V variants, corresponding to the human OAS-MUT proteins A76V, C109Y, V121G, and L198V, respectively. The resulting structures were prepared using the Protein Preparation Wizard and molecular dynamics (MD) simulation were performed in Desmond using OPLS3e force field of the Schrödinger software (Schrödinger, New York, NY, USA). MD simulations were run in triplicate for all proteins with each set of runs initiated using a different initial velocity. Each system was first neutralized by adding sodium ions around the protein using the System Builder module. The neutralized protein was placed in TIP3P water, and random water molecules were substituted in order to obtain an ionic strength of 150 mM. Each solvated system was relaxed using a series of restrained minimization stages each of 1 ns duration: 1) all heavy atoms with force constant 1000 kcal/molÅ2, all protein backbone atoms with 2) 100 kcal/molÅ2 and 3) 5 kcal mol kcal/molÅ2, and finally 4) with no constrains. Unrestrained MD simulations were then performed for 120 ns, comprising 20 ns for equilibration followed by a 100 ns production run in the isothermal-isobaric (NPT) ensemble using Langevin thermostat and barostat with relaxation times of 1 and 2 ps, respectively. The equations of motion were integrated using multiple time steps for short-range (2 fs) and long-range (6 fs) interactions with a 10 Å cutoff applied for non-bonded interactions. RMSD and RMSF values were output directly from the production runs and plotted as the average of triplicates for each protein. Difference RMSF was calculated by subtracting the average RMSF for each residue in OAS-WT from the equivalent residue in each OAS-MUT. The standard deviation in each set of difference RMSF values was calculated after excluding five residues at each protein terminus and a cut-off of two standard deviations (2σ) used to identify the most significant changes within each difference RMSF calculation.
In silico substitutions of amino acids and structural images were made using PyMOL (Schrödinger, New York, NY, USA). These analyses used the structures of dsRNA-bound human OAS1 (PDB code 4IG8), dsRNA-bound porcine OAS1 (PDB code 4RWN), and free porcine OAS1 (PDB code 4RWQ).
Statistics
Statistical analysis was done with the software environment R (version 4.0.2) or GraphPad Prism 8 (GraphPad Software, San Diego, USA). Two-sided Welch’s t-test was performed after testing for normal Gaussian distribution using Shapiro Wilk normality test. To compare two independent variables a Two-way ANOVA was performed followed by Tukey’s multiple comparisons or by Sidak’s post-hoc test. P values < 0.05 were considered as significant.
Supplementary Material
Table S5. Differentially expressed genes in T-cells, B-cells and monocytes.
Table S6. Raw data table
Fig. S1. Genetic, radiological and histopathological findings.
Fig. S2. OAS1-expression.
Fig. S3. T-cell proliferation.
Fig. S4. B-cell apoptosis.
Fig. S5. T-cell apoptosis.
Fig. S6. Purity and viability of FACS sorted T-cells, B-cells, and monocytes.
Fig. S7. Proliferation of allogeneic B-cell- and CD3/CD28-stimulated CD8+ T-cells.
Fig. S8. Purity of FACS sorted T-cells, B-cells, and monocytes for RNAseq and RNA degradation.
Fig. S9. Allele frequency of the heterozygous OAS1 A76V variant.
Fig. S10. Altered transcriptome of OAS1-GOF monocytes, B-cells, and T-cells.
Fig. S11. GM-CSF-receptor expression in monocytes.
Fig. S12. Generation and functional validation of human iPSC-derived macrophages.
Fig. S13. Interferon-α induced transcriptomic changes of OAS1-GOF iPSC-derived macrophages.
Fig. S14. Analysis of OAS1-WT and OAS1-MUT in vitro activity, transfection efficacy and mechanism of cell death.
Fig. S15. Impact of OAS1-GOF 2-5A amino substitutions V76A, C109Y, V121G and L198V on local protein structure.
Fig. S16. Titration curves of stimulation reagents.
Fig. S17. Flow cytometry gating strategies.
Table S1. Genetic, clinical, and immunological patient characteristics
Table S2. OAS1 variant in silico analysis
Table S3. OAS1 variant sequencing primers
Table S4a. gRNA sequences.
Table S4b. Knock-in variant sequencing primers.
Acknowledgments:
We value the personal contribution of the patients and their families as well as the excellent clinical care by the nursing and medical teams. We thank Eva Eisl, Mayumi Hofmann, and Irmgard Eckerlein for technical assistance. We acknowledge the expert services of the sequencing facility at the Dr. von Hauner Children’s Hospital and the associated computational biology unit. We thank Matt Pankratz, Biorepository Supervisor at the Phoenix Children’s Hospital, for providing a paraffine embedded patient bone marrow sample.
Funding:
F.H. received funding from the Care-for-Rare Foundation (C4R, 160073), the German Centre for Infection Research (DZIF, TTU 07.909), the Else Kröner-Fresenius Stiftung (EKFS, 2017_A110), and the German Federal Ministry of Education and Research (BMBF, 01GM1910C). T.Mo. received funding from the JSPS Grant-in-Aid for Scientific Research (26293244, 18H02778), the AMED Rare/Intractable Disease Project (1010139), and the Health and Labor Science Research Grant (201911006B, 201911013B). T.O. received funding from the JSPS Grant-in-Aid for Scientific Research (19K23861 and 20K16847). C.K. received funding from the German Research Foundation (DFG, Gottfried Wilhelm Leibniz program). L.M.K received funding from the Else Kröner-Fresenius Stiftung (EKFS, 2017_A50), the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, Project 442265435) and the Friedrich-Baur-Stiftung (48/17). G.L.C. received funding from the National Institutes of Health (NIH, R01-AI144067), the Emory University Research Council, and the Emory University School of Medicine Bridge Funding Program. S.L.S. received funding from the National Institutes of Health (NIH, T32-GM008367 and F31-AI133950). S.L.S. and B.M.C. also acknowledge support from the Atlanta Chapter of the ARCS Foundation. D.D. received funding from the Cystic Fibrosis Foundation (#DEY18F0). M.G. received support from the BMBF for the chILD-EU register and biobank project (HCQ4Surfdefect) under the ERA-Net for Research on Rare Diseases, the European Cooperation in Science and Technology (COST A16125) and the Deutsche Forschungsgemeinschaft (DFG, Gr970–9/1). M.H.A., C.K. and F.H. are members of the European Reference Network for Rare Immunodeficiency, Autoinflammatory and Autoimmune Diseases -Project ID No 739543.
Footnotes
Competing interests:
We declare that none of the authors has competing financial or non-financial interests.
Data and materials availability:
Single-cell RNA-sequencing data are available at the GEO accession number GSE173667. All data is available in the main text or the supplementary materials.
References and Notes:
- 1.Tangye SG, Al-Herz W, Bousfiha A, Chatila T, Cunningham-Rundles C, Etzioni A, Franco JL, Holland SM, Klein C, Morio T, Ochs HD, Oksenhendler E, Picard C, Puck J, Torgerson TR, Casanova JL, Sullivan KE, Human Inborn Errors of Immunity: 2019 Update on the Classification from the International Union of Immunological Societies Expert Committee. J Clin Immunol 40, 24–64 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Durandy A, Kracker S, Fischer A, Primary antibody deficiencies. Nat Rev Immunol 13, 519–533 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Liu Y, Jesus AA, Marrero B, Yang D, Ramsey SE, Sanchez GAM, Tenbrock K, Wittkowski H, Jones OY, Kuehn HS, Lee CR, DiMattia MA, Cowen EW, Gonzalez B, Palmer I, DiGiovanna JJ, Biancotto A, Kim H, Tsai WL, Trier AM, Huang Y, Stone DL, Hill S, Kim HJ, St Hilaire C, Gurprasad S, Plass N, Chapelle D, Horkayne-Szakaly I, Foell D, Barysenka A, Candotti F, Holland SM, Hughes JD, Mehmet H, Issekutz AC, Raffeld M, McElwee J, Fontana JR, Minniti CP, Moir S, Kastner DL, Gadina M, Steven AC, Wingfield PT, Brooks SR, Rosenzweig SD, Fleisher TA, Deng Z, Boehm M, Paller AS, Goldbach-Mansky R, Activated STING in a vascular and pulmonary syndrome. N Engl J Med 371, 507–518 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.R. C. Group, Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, Staplin N, Brightling C, Ustianowski A, Elmahi E, Prudon B, Green C, Felton T, Chadwick D, Rege K, Fegan C, Chappell LC, Faust SN, Jaki T, Jeffery K, Montgomery A, Rowan K, Juszczak E, Baillie JK, Haynes R, Landray MJ, Dexamethasone in Hospitalized Patients with Covid-19 - Preliminary Report. N Engl J Med, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bronte V, Ugel S, Tinazzi E, Vella A, De Sanctis F, Cane S, Batani V, Trovato R, Fiore A, Petrova V, Hofer F, Barouni RM, Musiu C, Caligola S, Pinton L, Torroni L, Polati E, Donadello K, Friso S, Pizzolo F, Iezzi M, Facciotti F, Pelicci PG, Righetti D, Bazzoni P, Rampudda M, Comel AC, Mosaner W, Lunardi C, Olivieri O, Baricitinib restrains the immune dysregulation in severe COVID-19 patients. J Clin Invest, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hornung V, Hartmann R, Ablasser A, Hopfner KP, OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat Rev Immunol 14, 521–528 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chebath J, Benech P, Revel M, Vigneron M, Constitutive expression of (2’−5’) oligo A synthetase confers resistance to picornavirus infection. Nature 330, 587–588 (1987). [DOI] [PubMed] [Google Scholar]
- 8.Sun L, Wu J, Du F, Chen X, Chen ZJ, Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clemens MJ, Williams BR, Inhibition of cell-free protein synthesis by pppA2’p5’A2’p5’A: a novel oligonucleotide synthesized by interferon-treated L cell extracts. Cell 13, 565–572 (1978). [DOI] [PubMed] [Google Scholar]
- 10.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ, Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han Y, Donovan J, Rath S, Whitney G, Chitrakar A, Korennykh A, Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response. Science 343, 1244–1248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ, Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341, 1390–1394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho K, Yamada M, Agematsu K, Kanegane H, Miyake N, Ueki M, Akimoto T, Kobayashi N, Ikemoto S, Tanino M, Fujita A, Hayasaka I, Miyamoto S, Tanaka-Kubota M, Nakata K, Shiina M, Ogata K, Minakami H, Matsumoto N, Ariga T, Heterozygous Mutations in OAS1 Cause Infantile-Onset Pulmonary Alveolar Proteinosis with Hypogammaglobulinemia. Am J Hum Genet 102, 480–486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J, A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46, 310–315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itan Y, Shang L, Boisson B, Ciancanelli MJ, Markle JG, Martinez-Barricarte R, Scott E, Shah I, Stenson PD, Gleeson J, Cooper DN, Quintana-Murci L, Zhang SY, Abel L, Casanova JL, The mutation significance cutoff: gene-level thresholds for variant predictions. Nat Methods 13, 109–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lecoeur H, Ledru E, Prevost MC, Gougeon ML, Strategies for phenotyping apoptotic peripheral human lymphocytes comparing ISNT, annexin-V and 7-AAD cytofluorometric staining methods. J Immunol Methods 209, 111–123 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Gupta A, Rath PC, Curcumin, a natural antioxidant, acts as a noncompetitive inhibitor of human RNase L in presence of its cofactor 2–5A in vitro. Biomed Res Int 2014, 817024 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T, The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 7, 3 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li G, Xiang Y, Sabapathy K, Silverman RH, An apoptotic signaling pathway in the interferon antiviral response mediated by RNase L and c-Jun NH2-terminal kinase. J Biol Chem 279, 1123–1131 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Calderon BM, Conn GL, A human cellular noncoding RNA activates the antiviral protein 2’−5’-oligoadenylate synthetase 1. J Biol Chem 293, 16115–16124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyazaki K, MEGAWHOP cloning: a method of creating random mutagenesis libraries via megaprimer PCR of whole plasmids. Methods Enzymol 498, 399–406 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Vachon VK, Calderon BM, Conn GL, A novel RNA molecular signature for activation of 2’−5’ oligoadenylate synthetase-1. Nucleic Acids Res 43, 544–552 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chitrakar A, Rath S, Donovan J, Demarest K, Li Y, Sridhar RR, Weiss SR, Kotenko SV, Wingreen NS, Korennykh A, Real-time 2–5A kinetics suggest that interferons beta and lambda evade global arrest of translation by RNase L. Proc Natl Acad Sci U S A 116, 2103–2111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt EK, Clavarino G, Ceppi M, Pierre P, SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6, 275–277 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Donovan J, Dufner M, Korennykh A, Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc Natl Acad Sci U S A 110, 1652–1657 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC, Crystal structure of the 2’-specific and double-stranded RNA-activated interferon-induced antiviral protein 2’−5’-oligoadenylate synthetase. Mol Cell 12, 1173–1185 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Lohofener J, Steinke N, Kay-Fedorov P, Baruch P, Nikulin A, Tishchenko S, Manstein DJ, Fedorov R, The Activation Mechanism of 2’−5’-Oligoadenylate Synthetase Gives New Insights Into OAS/cGAS Triggers of Innate Immunity. Structure 23, 851–862 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Burke JM, Moon SL, Matheny T, Parker R, RNase L Reprograms Translation by Widespread mRNA Turnover Escaped by Antiviral mRNAs. Mol Cell 75, 1203–1217 e1205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rath S, Prangley E, Donovan J, Demarest K, Wingreen NS, Meir Y, Korennykh A, Concerted 2–5A-Mediated mRNA Decay and Transcription Reprogram Protein Synthesis in the dsRNA Response. Mol Cell 75, 1218–1228 e1216 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonnevie-Nielsen V, Field LL, Lu S, Zheng DJ, Li M, Martensen PM, Nielsen TB, Beck-Nielsen H, Lau YL, Pociot F, Variation in antiviral 2’,5’-oligoadenylate synthetase (2’5’AS) enzyme activity is controlled by a single-nucleotide polymorphism at a splice-acceptor site in the OAS1 gene. Am J Hum Genet 76, 623–633 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carey CM, Govande AA, Cooper JM, Hartley MK, Kranzusch PJ, Elde NC, Recurrent Loss-of-Function Mutations Reveal Costs to OAS1 Antiviral Activity in Primates. Cell Host Microbe 25, 336–343 e334 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim JK, Lisco A, McDermott DH, Huynh L, Ward JM, Johnson B, Johnson H, Pape J, Foster GA, Krysztof D, Follmann D, Stramer SL, Margolis LB, Murphy PM, Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS Pathog 5, e1000321 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Reksten TR, Ice JA, Kelly JA, Adrianto I, Rasmussen A, Wang S, He B, Grundahl KM, Glenn SB, Miceli-Richard C, Bowman S, Lester S, Eriksson P, Eloranta ML, Brun JG, Goransson LG, Harboe E, Guthridge JM, Kaufman KM, Kvarnstrom M, Cunninghame Graham DS, Patel K, Adler AJ, Farris AD, Brennan MT, Chodosh J, Gopalakrishnan R, Weisman MH, Venuturupalli S, Wallace DJ, Hefner KS, Houston GD, Huang AJW, Hughes PJ, Lewis DM, Radfar L, Vista ES, Edgar CE, Rohrer MD, Stone DU, Vyse TJ, Harley JB, Gaffney PM, James JA, Turner S, Alevizos I, Anaya JM, Rhodus NL, Segal BM, Montgomery CG, Scofield RH, Kovats S, Mariette X, Ronnblom L, Witte T, Rischmueller M, Wahren-Herlenius M, Omdal R, Jonsson R, Ng WF, U. K. P. S. s. S. R. for, Nordmark G, Lessard CJ, Sivils KL, Identification of a Sjogren’s syndrome susceptibility locus at OAS1 that influences isoform switching, protein expression, and responsiveness to type I interferons. PLoS Genet 13, e1006820 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wacleche VS, Tremblay CL, Routy JP, Ancuta P, The Biology of Monocytes and Dendritic Cells: Contribution to HIV Pathogenesis. Viruses 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Getahun A, Cambier JC, Non-Antibody-Secreting Functions of B Cells and Their Contribution to Autoimmune Disease. Annu Rev Cell Dev Biol 35, 337–356 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein F, Mitrovic M, Roux J, Engdahl C, von Muenchow L, Alberti-Servera L, Fehling HJ, Pelczar P, Rolink A, Tsapogas P, The transcription factor Duxbl mediates elimination of pre-T cells that fail beta-selection. J Exp Med 216, 638–655 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu J, Chen YJ, Dobbs N, Sakai T, Liou J, Miner JJ, Yan N, STING-mediated disruption of calcium homeostasis chronically activates ER stress and primes T cell death. J Exp Med, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaneko N, Kuo HH, Boucau J, Farmer JR, Allard-Chamard H, Mahajan VS, Piechocka-Trocha A, Lefteri K, Osborn M, Bals J, Bartsch YC, Bonheur N, Caradonna TM, Chevalier J, Chowdhury F, Diefenbach TJ, Einkauf K, Fallon J, Feldman J, Finn KK, Garcia-Broncano P, Hartana CA, Hauser BM, Jiang C, Kaplonek P, Karpell M, Koscher EC, Lian X, Liu H, Liu J, Ly NL, Michell AR, Rassadkina Y, Seiger K, Sessa L, Shin S, Singh N, Sun W, Sun X, Ticheli HJ, Waring MT, Zhu AL, Alter G, Li JZ, Lingwood D, Schmidt AG, Lichterfeld M, Walker BD, Yu XG, Padera RF Jr., Pillai S, G. Massachusetts Consortium on Pathogen Readiness Specimen Working, Loss of Bcl-6-Expressing T Follicular Helper Cells and Germinal Centers in COVID-19. Cell 183, 143–157 e113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Maria E, Latini A, Borgiani P, Novelli G, Genetic variants of the human host influencing the coronavirus-associated phenotypes (SARS, MERS and COVID-19): rapid systematic review and field synopsis. Hum Genomics 14, 30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arunachalam PS, Wimmers F, Mok CKP, Perera R, Scott M, Hagan T, Sigal N, Feng Y, Bristow L, Tak-Yin Tsang O, Wagh D, Coller J, Pellegrini KL, Kazmin D, Alaaeddine G, Leung WS, Chan JMC, Chik TSH, Choi CYC, Huerta C, Paine McCullough M, Lv H, Anderson E, Edupuganti S, Upadhyay AA, Bosinger SE, Maecker HT, Khatri P, Rouphael N, Peiris M, Pulendran B, Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 369, 1210–1220 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martinez-Colon GJ, McKechnie JL, Ivison GT, Ranganath T, Vergara R, Hollis T, Simpson LJ, Grant P, Subramanian A, Rogers AJ, Blish CA, A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med 26, 1070–1076 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoang TN, Pino M, Boddapati AK, Viox EG, Starke CE, Upadhyay AA, Gumber S, Busman-Sahay K, Strongin Z, Harper JL, Tharp GK, Pellegrini KL, Kirejczyk S, Zandi K, Tao S, Horton TR, Beagle EN, Mahar EA, Lee MY, Cohen J, Jean SM, Wood JS, Connor-Stroud F, Stammen RL, Delmas OM, Wang S, Cooney KA, Sayegh MN, Wang L, Weiskopf D, Filev PD, Waggoner J, Piantadosi A, Kasturi SP, Shakhshir HA, Ribeiro SP, Sekaly RP, Levit RD, Estes JD, Vanderford TH, Schinazi RF, Bosinger SE, Paiardini M, Baricitinib treatment resolves lower airway inflammation and neutrophil recruitment in SARS-CoV-2-infected rhesus macaques. bioRxiv, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, Walters RK, Tashman K, Farjoun Y, Banks E, Poterba T, Wang A, Seed C, Whiffin N, Chong JX, Samocha KE, Pierce-Hoffman E, Zappala Z, O’Donnell-Luria AH, Minikel EV, Weisburd B, Lek M, Ware JS, Vittal C, Armean IM, Bergelson L, Cibulskis K, Connolly KM, Covarrubias M, Donnelly S, Ferriera S, Gabriel S, Gentry J, Gupta N, Jeandet T, Kaplan D, Llanwarne C, Munshi R, Novod S, Petrillo N, Roazen D, Ruano-Rubio V, Saltzman A, Schleicher M, Soto J, Tibbetts K, Tolonen C, Wade G, Talkowski ME, C. Genome Aggregation Database, Neale BM, Daly MJ, MacArthur DG, The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schober T, Magg T, Laschinger M, Rohlfs M, Linhares ND, Puchalka J, Weisser T, Fehlner K, Mautner J, Walz C, Hussein K, Jaeger G, Kammer B, Schmid I, Bahia M, Pena SD, Behrends U, Belohradsky BH, Klein C, Hauck F, A human immunodeficiency syndrome caused by mutations in CARMIL2. Nat Commun 8, 14209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu L, Niu B, Zhu Z, Wu S, Li W, CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S, MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A, Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okumura T, Horie Y, Lai CY, Lin HT, Shoda H, Natsumoto B, Fujio K, Kumaki E, Okano T, Ono S, Tanita K, Morio T, Kanegane H, Hasegawa H, Mizoguchi F, Kawahata K, Kohsaka H, Moritake H, Nunoi H, Waki H, Tamaru SI, Sasako T, Yamauchi T, Kadowaki T, Tanaka H, Kitanaka S, Nishimura K, Ohtaka M, Nakanishi M, Otsu M, Robust and highly efficient hiPSC generation from patient non-mobilized peripheral blood-derived CD34(+) cells using the auto-erasable Sendai virus vector. Stem Cell Res Ther 10, 185 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakajima K, Zhou Y, Tomita A, Hirade Y, Gurumurthy CB, Nakada S, Precise and efficient nucleotide substitution near genomic nick via noncanonical homology-directed repair. Genome Res 28, 223–230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takayama N, Nishimura S, Nakamura S, Shimizu T, Ohnishi R, Endo H, Yamaguchi T, Otsu M, Nishimura K, Nakanishi M, Sawaguchi A, Nagai R, Takahashi K, Yamanaka S, Nakauchi H, Eto K, Transient activation of c-MYC expression is critical for efficient platelet generation from human induced pluripotent stem cells. J Exp Med 207, 2817–2830 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tazawa R, Hamano E, Arai T, Ohta H, Ishimoto O, Uchida K, Watanabe M, Saito J, Takeshita M, Hirabayashi Y, Ishige I, Eishi Y, Hagiwara K, Ebina M, Inoue Y, Nakata K, Nukiwa T, Granulocyte-macrophage colony-stimulating factor and lung immunity in pulmonary alveolar proteinosis. Am J Respir Crit Care Med 171, 1142–1149 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F, Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmid-Burgk JL, Schmidt T, Gaidt MM, Pelka K, Latz E, Ebert TS, Hornung V, OutKnocker: a web tool for rapid and simple genotyping of designer nuclease edited cell lines. Genome Res 24, 1719–1723 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Magg T, Wiebking V, Conca R, Krebs S, Arens S, Schmid I, Klein C, Albert MH, Hauck F, IPEX due to an exon 7 skipping FOXP3 mutation with autoimmune diabetes mellitus cured by selective TReg cell engraftment. Clin Immunol 191, 52–58 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Kubara K, Yamazaki K, Ishihara Y, Naruto T, Lin HT, Nishimura K, Ohtaka M, Nakanishi M, Ito M, Tsukahara K, Morio T, Takagi M, Otsu M, Status of KRAS in iPSCs Impacts upon Self-Renewal and Differentiation Propensity. Stem Cell Reports 11, 380–394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Justesen J, Kjeldgaard NO, Spectrophotometric pyrophosphate assay of 2’,5’-oligoadenylate synthetase. Anal Biochem 207, 90–93 (1992). [DOI] [PubMed] [Google Scholar]
- 59.Shearer WT, Rosenblatt HM, Gelman RS, Oyomopito R, Plaeger S, Stiehm ER, Wara DW, Douglas SD, Luzuriaga K, McFarland EJ, Yogev R, Rathore MH, Levy W, Graham BL, Spector SA, Pediatric ACTG, Lymphocyte subsets in healthy children from birth through 18 years of age: the Pediatric AIDS Clinical Trials Group P1009 study. J Allergy Clin Immunol 112, 973–980 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Blanco E, Perez-Andres M, Arriba-Mendez S, Contreras-Sanfeliciano T, Criado I, Pelak O, Serra-Caetano A, Romero A, Puig N, Remesal A, Torres Canizales J, Lopez-Granados E, Kalina T, Sousa AE, van Zelm M, van der Burg M, van Dongen JJM, Orfao A, P. I. D. g. EuroFlow, Age-associated distribution of normal B-cell and plasma cell subsets in peripheral blood. J Allergy Clin Immunol 141, 2208–2219 e2216 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S5. Differentially expressed genes in T-cells, B-cells and monocytes.
Table S6. Raw data table
Fig. S1. Genetic, radiological and histopathological findings.
Fig. S2. OAS1-expression.
Fig. S3. T-cell proliferation.
Fig. S4. B-cell apoptosis.
Fig. S5. T-cell apoptosis.
Fig. S6. Purity and viability of FACS sorted T-cells, B-cells, and monocytes.
Fig. S7. Proliferation of allogeneic B-cell- and CD3/CD28-stimulated CD8+ T-cells.
Fig. S8. Purity of FACS sorted T-cells, B-cells, and monocytes for RNAseq and RNA degradation.
Fig. S9. Allele frequency of the heterozygous OAS1 A76V variant.
Fig. S10. Altered transcriptome of OAS1-GOF monocytes, B-cells, and T-cells.
Fig. S11. GM-CSF-receptor expression in monocytes.
Fig. S12. Generation and functional validation of human iPSC-derived macrophages.
Fig. S13. Interferon-α induced transcriptomic changes of OAS1-GOF iPSC-derived macrophages.
Fig. S14. Analysis of OAS1-WT and OAS1-MUT in vitro activity, transfection efficacy and mechanism of cell death.
Fig. S15. Impact of OAS1-GOF 2-5A amino substitutions V76A, C109Y, V121G and L198V on local protein structure.
Fig. S16. Titration curves of stimulation reagents.
Fig. S17. Flow cytometry gating strategies.
Table S1. Genetic, clinical, and immunological patient characteristics
Table S2. OAS1 variant in silico analysis
Table S3. OAS1 variant sequencing primers
Table S4a. gRNA sequences.
Table S4b. Knock-in variant sequencing primers.
Data Availability Statement
Single-cell RNA-sequencing data are available at the GEO accession number GSE173667. All data is available in the main text or the supplementary materials.