Abstract
The orphan receptor germ cell nuclear factor (GCNF) is a member of the superfamily of nuclear receptors. During development, GCNF exhibits a restricted brain-specific expression pattern, whereas GCNF expression in the adult is germ cell specific. Therefore, the receptor may participate in the regulation of neurogenesis and reproductive functions. No natural GCNF target gene has yet been identified, but recent data demonstrate specific and high-affinity binding of GCNF either to the direct repeat DNA element AGGTCAAGGTCA (DR0) or to extended half-sites, such as TCAAGGTCA. In this study, we show that murine GCNF (mGCNF) can bind as a homodimer to extended half-sites, thus describing a novel property within the nuclear receptor superfamily. Homodimeric binding to extended half-sites requires the presence of a dimerization function within the mGCNF DNA-binding domain (DBD) and a novel dimerization surface encompassing the putative helix 3 and the helix 12 region of the mGCNF ligand-binding domain (LBD). In addition, the mGCNF LBD has the potential to adopt different conformations with distinct dimerization properties. The helix 12 region of the mGCNF LBD not only regulates the switch between these dimerization conformations but also dictates the DNA-binding behavior and transcriptional properties of the different dimerization conformations. In summary, our findings describe unique DNA-binding and dimerization properties of a nuclear receptor and suggest a novel mechanism that allows mGCNF to modulate target gene activity.
Nuclear receptors form a superfamily of ligand-activated transcription factors that play important roles in development, differentiation, and homeostasis (7, 12, 16, 28, 32). Nuclear receptors share a common modular structure and are composed of several domains that mediate DNA-binding, dimerization, ligand-binding, and transcriptional activities (12). Cooperative and high-affinity dimeric DNA binding of nuclear receptors requires two independent dimerization functions, one located within the DNA-binding domain (DBD) (17, 24, 30, 35, 52, 53) and the second located in the ligand-binding domain (LBD) (31, 34, 55). Depending on the particular DNA-binding and dimerization properties, members of the nuclear receptor superfamily bind either as homodimers or heterodimers to palindromic or direct repeat response elements, or even as monomers to extended half-site sequences (27).
In addition to contributing to receptor dimerization, the LBD performs a number of functions that include ligand binding, transcriptional activation, and repression (3, 7, 16, 28, 32). Elucidation of the crystal structures of the LBDs of unliganded (apo form) hRXRα (human retinoid X receptor α) and liganded (holo form) hRARγ (human retinoic acid receptor γ), hTRα (human thyroid receptor α), and hERα (human estrogen receptor α) demonstrated that the LBDs fold as antiparallel α-helical sandwiches composed of 12 α helices (H1 to H12) (5, 6, 36, 45). Crystallographic data and structure-function analysis indicate that the homodimeric interactions of RXR LBDs as well as heterodimeric interactions between the LBDs of RXR and RAR are mediated mainly by H10 and to a lesser extent by H9 and the loop between H7 and H8 (5, 31). Recently, a novel dimerization motif comprising the putative H5 to H7 of the LBD of the nuclear orphan receptor SHP (short heterodimer partner) has been found to mediate interactions between SHP and RXR, RAR, or TR (40).
Upon ligand binding, several major structural changes are induced within the LBD. One obvious difference between the unliganded (apo form) and liganded (holo form) LBD structures is a positional reorientation of H12. H12 is indispensable for the transcriptional activation function of the LBD and contains the so-called activation function 2 core motif (AF2 AD core) (2, 10, 11, 44). The ligand-induced conformational changes most likely result in the formation of novel surfaces in the holo-LBD which, in turn, allow direct protein-protein interactions with cofactors and transcriptional intermediary factors (reviewed in references 7 and 14).
In addition to receptors for steroid hormones, retinoids, vitamin D, and thyroid hormone, various reports have described numerous related gene products for which no ligands have been identified and that therefore are referred to as nuclear orphan receptors. The nuclear orphan receptor germ cell nuclear factor (GCNF) was first cloned from murine tissue (8, 18). Recently, cloning of Xenopus laevis GCNF (20) and human GCNF (hGCNF) (26, 41) has been reported. Both mouse GCNF (mGCNF) and hGCNF are highly expressed in the germ cells of the ovary and testis (8, 18, 22, 41) as well as in embryonic stem cells and embryonic carcinoma cells (26). Furthermore, GCNF expression has been observed in the developing nervous system during neurulation of X. laevis (20). The highly restricted expression pattern suggests a role for GCNF in the control of gene expression during early embryogenesis (neurogenesis) and gametogenesis.
Sequence analysis of GCNF revealed that the DBD is most closely related to that of RXR, whereas the LBD of GCNF shows considerable homology to those of the nuclear orphan receptor COUP-TFI and RXR (8). GCNF binds to DNA sequences containing an AGGTCA core motif oriented as a direct repeat with 0-bp spacing (DR0) or to a single extended core motif (XRE), like TCAAGGTCA (8, 51). However, the exact binding mode of GCNF to XREs is unclear. While homodimeric binding of GCNF to a DR0 has been established (4, 8, 51), Chen et al. (8) also suggested homodimeric binding to the XRE sequence TCAAGGTCA. In contrast, monomeric binding to this sequence was favored by Yan et al. (51). A dimerization motif was hypothesized to be located in the putative H9 and H10 of the GCNF LBD (4). However, since deletion of this region did not influence homodimeric binding of GCNF to a DR0 (4), the location of potential dimerization motifs in GCNF is unclear. Furthermore, in contrast to most nuclear receptors, critical residues in the AF2 AD core that are essential for transcriptional activation are not conserved in GCNF. Therefore, the function of the H12 region located at the very C terminus of the GCNF LBD remains to be elucidated.
In this study, we analyzed in detail the DNA-binding, dimerization, and transcriptional properties of mGCNF. We provide evidence that the LBD of mGCNF has the potential to adopt different conformations. One conformation appears to be similar to that of the holo-LBDs of hRARγ, hTRα, or hERα (6, 36, 45), whereas the other may resemble the conformation of the apo-LBD of hRXRα (5). The two different GCNF conformations exhibit distinct dimerization properties. The transition between these two dimerization conformations may be controlled by a mechanism that involves the H12 region of the mGCNF LBD. Structural alterations such as deletion, replacement, or spatial dislocation of the H12 region are accompanied by a transition of the mGCNF LBD into the apo-like conformation and affect the dimerization properties of the receptor. As a consequence, these alterations also drastically reduce both homodimeric and monomeric binding of mGCNF to an XRE and thus affect the transcriptional properties of the receptor. Other selective point mutations within the H12 region affect only homodimeric DNA binding of mGCNF; monomeric DNA binding is unaltered. Accordingly, these point mutations do not alter the holo-like conformation of the LBD. Furthermore, we show that mGCNF has the potential to bind as a homodimer to extended half-site sequences. Homodimeric binding of mGCNF to an XRE depends on two dimerization functions, one located in the DBD and the other located in the LBD. While in the apo-like conformation homodimeric LBD interactions are mainly mediated by the putative H3, the holo-like conformation of mGCNF most likely utilizes additional contacts mediated by the H12 region. Our results suggest that both dimerization and DNA binding of the nuclear orphan receptor mGCNF can be regulated by novel mechanisms, thus revealing additional strategies by which the activities of nuclear receptors might be governed.
MATERIALS AND METHODS
Recombinant plasmids.
The expression plasmid CMX.ATG was derived from CMX.PL2 (43) by replacing the EcoRV-EcoRI fragment of the multiple cloning site with a double-stranded oligonucleotide (5′-GATATCCACCATGGAATCC-3′) containing an optimized Kozak sequence followed by an ATG start codon. The expression plasmid CMX.ATG-mGCNF was generated by PCR amplification of a cDNA encoding full-length mGCNF from a mouse testis cDNA library (Stratagene). The PCR product was cloned as an EcoRI-BamHI fragment into CMX.ATG. Truncated receptors were produced by PCR amplification or by the excision of mGCNF fragments from CMX.ATG-mGCNF, using internal restriction sites. The mGCNF mutants were cloned as EcoRI-BamHI fragments into CMX.ATG. The following expression plasmids were produced: CMX.ATG-mGCNF1–483, CMX.ATG-mGCNF1–468, CMX.ATG-mGCNF1–406, CMX.ATG-mGCNF1–324, CMX.ATG-mGCNF1–284, CMX.ATG-mGCNF1–268, CMX.ATG-mGCNF1–160, CMX.ATG-mGCNF69–495, CMX.ATG-mGCNF69–483, CMX. ATG-mGCNF69–324, CMX.ATG-mGCNF69–284, CMX.ATG-mGCNF69–160, CMX.ATG-mGCNF69–149, and CMX.ATG-mGCNF69–140. (Numbers indicate the first and last amino acids of mGCNF encoded in these constructs.)
The mGCNF-hRXRα swap mutants and mGCNF–mouse steroidogenic factor 1 (mSF-1) swap mutants were generated by PCR amplification and ligation into CMX.ATG. Exact details will be provided upon request. The following expression plasmids were produced: CMX.ATG-mGCNF1–258-RXR222–462 (SWAP1), CMX.ATG-mGCNF1–326-RXR293–462 (SWAP2), CMX.ATG-mGCNF1–258-RXR222–261-mGCNF296–326-RXR293–462 (SWAP3), CMX.ATG-mGCNF1–258-RXR222–291 (SWAP4), mGCNF69–139-mSF179–97 [mGCNF-DBD(SF1-TA)], and mGCNF1–139-mSF179–97-mGCNF161–495 [mGCNF(SF1-TA)]. The mGCNF single- or double-point mutants mGCNF(S487A), mGCNF(C488A/K489A), mGCNF(T490A), mGCNF(S491A), mGCNF(T492A/V493A), mGCNF(K494A/E495A), and mGCNF(R113E/D114L) were generated by PCR amplification. PCR fragments were inserted at the EcoRI-BamHI restriction site of CMX.ATG. The vector CMX.ATG-mGCNF1–483-RXR446–462 [mGCNF-H11(RXR-H12)] was generated by PCR amplification of amino acids 446 to 462 of hRXRα and the simultaneous introduction of BamHI restriction sites at both ends of the PCR fragment. The PCR fragment was then cloned at the BamHI restriction site of CMX.ATG-mGCNF1–483. The double-point mutant CMX.ATG-mGCNF(V484D/L485P) was generated by PCR amplification of amino acids 486 to 495 and simultaneous introduction of BamHI restriction sites at the 5′ and 3′ ends of the PCR product. The PCR product was cloned at the BamHI restriction site of CMX.ATG-mGCNF1–483. Expression plasmids CMX.PL2-VP16-mGCNF, CMX.PL2-VP16-mGCNF1–483, and CMX.PL2-VP16-mGCNF(V484D/L485P) were generated by inserting the VP16 transactivation domain at the HindIII restriction site of CMX.PL2 (43) and subsequently cloning the cDNAs for mGCNF, mGCNF1–483, or mGCNF(V484D/L485P) at the EcoRI-BamHI restriction site of CMX.PL2-VP16. Expression plasmids pRSETB-mGCNF257–318, pRSETB-mGCNF296–318, and pRSETB-mGCNF415–468 were generated by PCR amplification of the indicated mGCNF cDNA segments, which were cloned at the EcoRI-HindIII restriction site of pRSETB (Invitrogen). The reporter plasmid TK-LUC has been described previously (41). Reporter plasmids XRE11x-TK-LUC, XRE13x-TK-LUC, XRE18x-TK-LUC, and DR02x-TK-LUC were obtained by cloning the indicated number of double-stranded XRE1 or DR0 oligonucleotides at the BamHI restriction site (for XRE1) or the HindIII restriction site (for DR0) of TK-LUC. The reporter plasmid XRE13x-TATA-LUC contains three copies of the XRE1 in front of the β-globin minimal promoter (33). All constructs were verified by sequencing using Sequenase (U.S. Biochemical).
Expression of recombinant mGCNF proteins.
Expression plasmids pRSETB-mGCNF257–318 (for the peptide His/H1-3), pRSETB-mGCNF296–318 (for the peptide His/H3), and pRSETB-mGCNF415–468 (for the peptide His/H9-10) were grown in Escherichia coli BL21pLys(DE3). Expression and purification of the various mGCNF proteins were performed according to the standard protocol (Clontech). Purity of the peptides was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
DNA-binding studies.
For the electrophoretic mobility shift assays (EMSAs), the double-stranded oligonucleotides XRE1 (5′-GATCCCCCTCAAGGTCAATGAGATC-3′) and DR0 (5′-AGCTTCAGGTCAAGGTCAGAGAGCT-3′) were used. mGCNF proteins were in vitro translated by using CMX.ATG-mGCNF or various mutants thereof and TNT coupled reticulocyte lysate (Promega). Primed lysate was incubated in buffer containing 10 mM HEPES (pH 7.9), 1 mM MgCl2, 50 mM KCl, 5% glycerol, 1 μg of poly(dI-dC), and 50 ng of retinoid Z receptor response element (15) as an unrelated competitor. Approximately 1 ng of the 32P-labeled oligonucleotide probe was added to the reaction mixture and incubated at room temperature (RT) for 20 min. For the competition experiments, in vitro-translated mGCNF proteins were preincubated for 10 min at RT with the purified His-tagged peptides (70 pmol) or 2 μg of bovine serum albumin (BSA). After addition of the 32P-labeled oligonucleotide probe, the reaction mixture was incubated for 20 min at RT. Subsequently, the reactions were loaded on a 5% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA running buffer at 4°C. EMSAs of small mGCNF mutants (e.g., mGCNF69–160) were resolved on a 7% polyacrylamide gel.
Limited proteolysis.
mGCNF and mGCNF mutants were in vitro translated in the presence of [35S]methionine. Then 5-μl aliquots of primed lysates were mixed with 1 μl of trypsin (100 μg/ml) or chymotrypsin (400 μg/ml) and incubated for the indicated time at RT. Proteolytic digests were stopped by mixing 1.8 μl of the reaction mixture with SDS gel-loading buffer (50 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, 10% glycerol) and boiling for 5 min. Subsequently, the samples were subjected to SDS-PAGE on a 15% polyacrylamide gel; 1.5 μl of untreated primed lysate served as a loading control. After electrophoresis, gels were first incubated for 20 min in fixation solution (25% isopropanol, 65% H2O, 10% acetic acid) and for an additional 20 min in Amplify solution (Amersham).
Modeling of the mGCNF homodimer.
The mGCNF LBD model was constructed by using the academic version of Modeller, version 4.0 (38), by taking the hRARγ crystal structure as a template and following the sequence alignment shown in Fig. 1. The mGCNF homodimer interface centered around H3, as suggested by our experiments, was generated with H3 of each LBD monomer providing the key contacts and respecting a twofold symmetry axis between the LBD monomers. The Cα traces of H3 of each LBD monomer were positioned at distances similar to those observed in the homodimer interface of the hRXRα crystal structure, in which H10 mediates key dimer contacts.
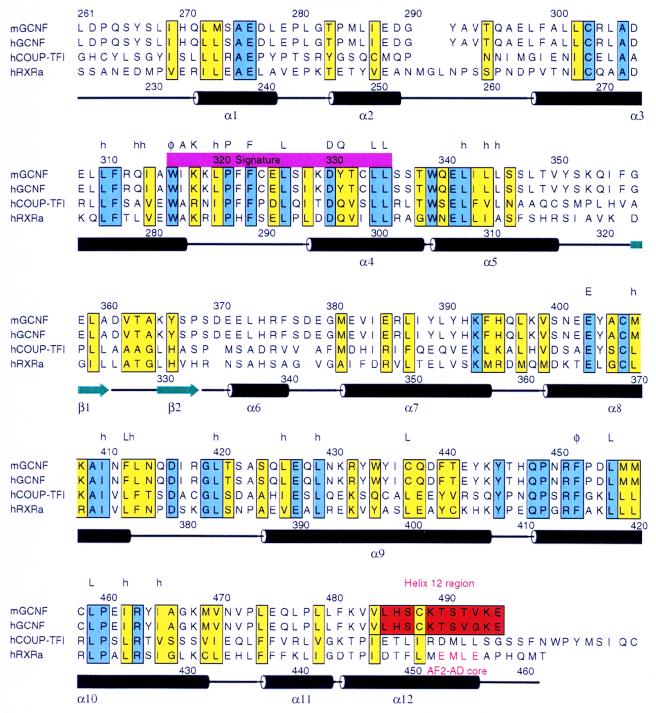
FIG. 1.
Alignment of the LBDs of mGCNF, hGCNF, hCOUP-TFI, and hRXRα. Conserved and similar residues are boxed in blue and yellow, respectively. The H12 region located at the very C terminus of the GCNF LBDs is colored in red. Secondary structure elements found in the crystal structure of the apo-hRXRα LBD (5) are indicated; α helices are depicted as black cylinders, and β strands are shown as green arrows. Regions of highest homology between mGCNF and hRXRα encompass H3 to H5 and H8 to H10. The position of AF2 AD core is indicated. Letters above the amino acid sequence of hRXRα mark residues that are highly conserved in the canonical fold of nuclear receptor LBDs (50). Abbreviations: h, hydrophobic; φ, aromatic; A, alanine; K, lysine; P, proline; F, phenylalanine; L, leucine; D, aspartic acid; Q, glutamine; E, glutamic acid.
Cell culture and transient transfection assays.
293 and BHK cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with either 10% (293 cells) or 5% (BHK cells) fetal calf serum. Transient transfection assays were carried out by the standard calcium phosphate coprecipitation method as described by Greiner et al. (15). Luciferase activity was assayed as recommended by the manufacturer (Promega) in a Luminometer ML3000 (Dynatech). Relative light units were normalized to β-galactosidase activity (43) and protein concentration, using the Bradford dye assay (Bio-Rad). All experiments were repeated at least three times. Standard deviations were <10%.
RESULTS
Sequence alignment of the LBDs of GCNF, hCOUP-TFI, and hRXRα.
The LBDs of mGCNF, hGCNF, hCOUP-TFI, and hRXRα were aligned according to the reported crystal structure of the hRXRα LBD (5). As shown in Fig. 1, regions of highest homology encompass H3 to H5 and H8 to H10 of the hRXRα LBD. In addition, H1, which defines the N terminus of the LBD, reveals similar patterns in mGCNF and hRXRα. Thus, the salt bridge formed in the hRXRα LBD between Glu239 located in H1 and Arg371 located in H8 (5) may also be present in mGCNF (Glu276 and Lys408). Since almost all residues that stabilize the LBD core, including the signature residues defined by Wurtz et al. (50), are conserved, the alignment suggests that the overall folding of the LBD is conserved between mGCNF and hRXRα. Additional secondary structure predictions (using the programs SOPMA and PHD [13, 37]) also support a folding similar to the α-helical sandwich structure observed for the LBDs of hRXRα, hRARγ, hTRα, and hERα (references 5, 6, 36, and 45 and data not shown). However, in the homology model of the GCNF LBD, the exact folding of the C terminus remains unclear. Secondary structure predictions suggest a folding into coil and β-sheet structures rather than an α helix found in the liganded LBDs of hRARγ, hTRα, and hERα (6, 36, 45). However, for reasons of simplicity we will refer to the C terminus of the GCNF LBD as the H12 region. Interestingly, the AF2 AD core located at the C termini of the LBDs of hRXRα and most other nuclear receptors is conserved neither in mGCNF (Fig. 1) nor in human or X. laevis GCNF (reference 21 and data not shown). The AF2 AD core plays a central role in transcriptional activation by nuclear receptors (2, 10, 11, 44). The lack of conservation of the AF2 AD core in the GCNF LBD and the difficulties to accurately predict the folding and the orientation of the H12 region suggest that the H12 region may serve functional roles distinct from those in other nuclear receptors.
mGCNF binds specifically as a homodimer to a DR0 and to an extended half-site.
To initiate a structure-function analysis of mGCNF, we first examined the binding properties of in vitro-translated mGCNF to the extended half-site XRE1 (CCCTCAAGGTCA) and the direct repeat element DR0 (AGGTCAAGGTCA) in EMSAs (Fig. 2A). In agreement with previous reports (4, 8, 51), mGCNF binds as an apparent homodimer to the DR0 (Fig. 2A, lane 8), whereas on the XRE1, mGCNF forms two specific protein-DNA complexes (Fig. 2A, lane 2). The slower-moving mGCNF-XRE1 complex comigrates with the apparent homodimeric mGCNF-DR0 complex. This suggests that mGCNF can bind either as a homodimer or as a monomer to the XRE1. Binding of mGCNF to both the DR0 and the XRE1 is specific, since it is competed with a 50-fold molar excess of unlabeled DNA (Fig. 2A, lanes 3 and 9) but not with an unrelated control oligonucleotide containing a retinoid Z receptor (RZR)/Rev-Erbα binding site (Fig. 2A, lanes 4 and 10). In addition, the mGCNF-DNA complexes are recognized by an anti-mGCNF antibody (Fig. 2A, lanes 5 and 11).
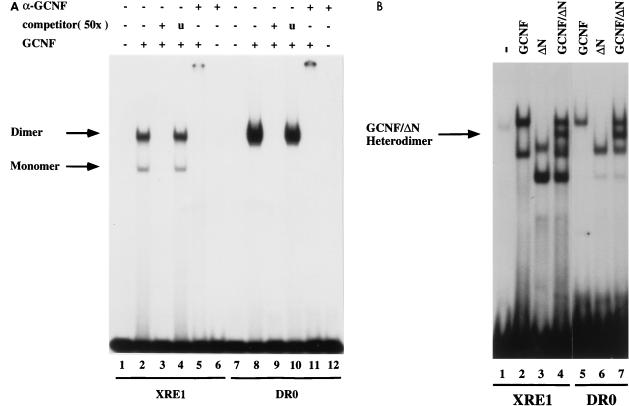
FIG. 2.
Binding of mGCNF to the XRE1 and the DR0. (A) Equal amounts of in vitro-translated mGCNF were assayed for binding to the XRE1 (lanes 2 to 5) and the DR0 (lanes 8 to 11) in EMSA. Unprimed reticulocyte lysate served as a control (lanes 1 and 7). Apparent monomeric or homodimeric mGCNF-DNA complexes are marked. DNA binding of mGCNF was competed with the XRE1 (lane 3), the DR0 (lane 9), or a control oligonucleotide containing an unrelated RZRβ binding site (lanes 4 and 10). DNA-bound mGCNF can be upshifted with a mGCNF-specific antibody (lanes 5 and 11). Lanes 6 and 12 contain only the antibody. (B) Heterodimer formation between mGCNF and ΔN-mGCNF on the XRE1 (lanes 2 to 4) and the DR0 (lanes 5 to 7). The position of the DNA-bound mGCNF–ΔN-mGCNF complex (lanes 4 and 7) is indicated by an arrow.
To demonstrate homodimeric binding of mGCNF to the XRE1 and the DR0, we constructed a mutant in which the entire N-terminal domain (NTD) of mGCNF was deleted (ΔN-mGCNF). We then performed mixing experiments with this mutant and mGCNF. ΔN-mGCNF binds to both binding sites with affinities similar to those of mGCNF and forms protein-DNA complexes with a mobility higher than that of the wild-type receptor (Fig. 2B; compare lane 2 with lane 3 and lane 5 with lane 6). As expected, the mixing of mGCNF with ΔN-mGCNF results in the formation of DNA-bound mGCNF/ΔN-mGCNF heterodimers which migrate with a mobility intermediate between those of the homodimeric mGCNF and ΔN-mGCNF complexes (Fig. 2B, lanes 4 and 7). In contrast, the faster-migrating complexes of mGCNF and ΔN-mGCNF do not give rise to a complex with an intermediate mobility, providing further evidence that both correspond to DNA-bound monomers. Taken together, these results demonstrate that mGCNF can form homodimers on both binding sides. Since coimmunoprecipitation studies revealed that mGCNF homodimer formation is dependent on DNA (data not shown), EMSAs were used to study further the DNA-binding and dimerization properties of the receptor.
C-terminal deletion mutants of mGCNF display altered DNA-binding affinities.
To identify motifs within the LBD that contribute to dimeric binding of mGCNF to the XRE1 and the DR0, we constructed a series of C-terminal deletion mutants (Fig. 3A) and analyzed them in EMSAs. The nomenclature of the mutants follows the assumption that the mGCNF LBD adopts an α-helical sandwich structure composed of 11 helices and the H12 region (for which the exact folding is unclear). All mutants were in vitro translated in equal amounts (Fig. 3A), and equal amounts of wild-type or mutant receptors were used in the EMSA shown in Fig. 3B. Deletion of the C-terminal 12 amino acids (H12 region) significantly reduces DNA binding of the mutant receptor mGCNF-H11 to the XRE1 and the DR0 (Fig. 3B; compare lane 2 with lane 3 and lane 11 with lane 12). The affinity of mGCNF-H10 to DNA (Fig. 3B, lanes 4 and 13) is similar to that of mGCNF-H11, whereas DNA binding of mGCNF-H8 and mGCNF-H3 gradually increases to about wild-type levels (Fig. 3B, lanes 5, 6, 14, and 15). Interestingly, all mutants described so far have the potential to bind as apparent homodimers to the XRE1 and the DR0. These results suggest that H9 to H10, the dimerization motif in RAR, RXR, or TR, does not contribute to homodimeric binding of mGCNF on either binding site.
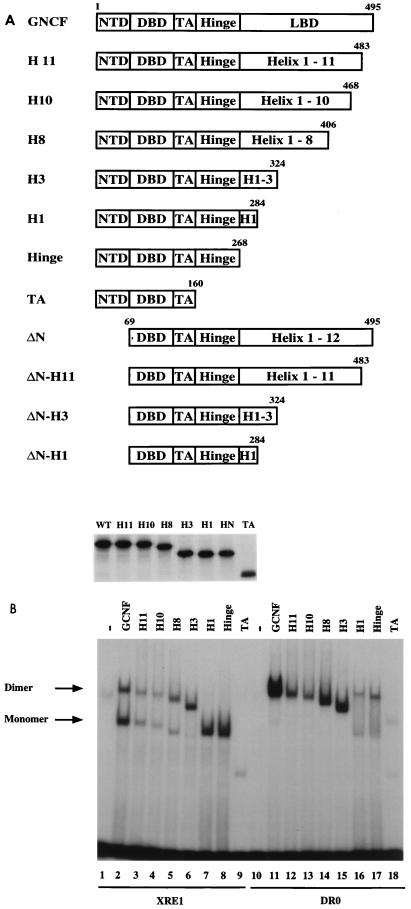
FIG. 3.
mGCNF deletion mutants display reduced DNA-binding affinities. (A) Schematic representation of mGCNF deletion mutants. Numbers represent the first and last amino acids of mGCNF. The domain organization of mGCNF is indicated. The full-length mGCNF protein is composed of the NTD, DBD, TA box, hinge region, and putative LBD. The nomenclature for the deletion mutants refers to the assumed folding of the mGCNF LBD into an α-helical sandwich structure based on the homology with other nuclear receptors. In vitro translation carried out in reticulocyte lysate in the presence of [35S]methionine confirmed equal expression levels of mGCNF and the deletion mutants. (B) Binding of mGCNF and mGCNF deletion mutants to the XRE1 (lanes 2 to 9) and the DR0 (lanes 11 to 18) in EMSA. Equal amounts of primed reticulocyte lysate were used in each lane. Unprimed lysate served as a control (lanes 1 and 10). Apparent homodimeric and monomeric protein-DNA complexes are indicated.
Further C-terminal deletions result in mutants that apparently bind as monomers to the XRE1, with either high (mGCNF-H1 and mGCNF-HINGE) or low (mGCNF-TA) affinities (Fig. 3B, lanes 7 to 9). Interestingly, mGCNF-H1 and mGCNF-HINGE migrate with a reduced mobility compared to mGCNF-H3. This may be due to a conformational change or differences in the surface charge distribution within the truncated receptors mGCNF-M1 and mGCNF-HINGE. On the DR0, mGCNF-H1, mGCNF-HINGE, and mGCNF-TA bind as monomers and also as apparent homodimers since additional slower-migrating complexes can be detected (Fig. 3B, lanes 16 to 18). Mixing experiments using the corresponding N-terminal deletion mutants of mGCNF-H3 and mGCNF-H1 reveal that the slower-migrating complexes on the DR0 indeed correspond to DNA-bound homodimers, while the faster-migrating complexes of mGCNF-H1 on the XRE1 and the DR0 represent DNA-bound monomers (data not shown). These results suggest that the deletion of the putative H2 and H3 abrogates homodimeric binding to the XRE1. In contrast, dimeric binding of mGCNF-H1, mGCNF-HINGE, and mGCNF-TA to the DR0 probably results from additional protein-protein interactions mediated by a putative dimerization motif located in the mGCNF DBD. Such a dimerization motif is commonly found in the DBDs of other nuclear receptors (17, 19, 24, 25, 30, 35, 39, 52, 53).
The TA box contributes to homodimeric DNA binding of mGCNF.
The previous experiments indicate that mGCNF contains two dimerization functions, one located in the DBD (including the TA box) and the other located in the N-terminal part of the LBD. To characterize the DNA-binding and dimerization properties of the mGCNF DBD and the TA box independently from other receptor domains, we generated the mutants mGCNF-DBD-TA, mGCNF-DBD-T, and mGCNF-DBD (Fig. 4A). In the latter two mutants, either the A box or the TA box is deleted. In addition, we replaced the mGCNF TA box with the highly homologous TA box of mSF-1 (8) (Fig. 4A). If the TA box of mGCNF is involved in protein-protein interactions of a DR0-bound DBD-TA homodimer, the swap mutant mGCNF-DBD(SF1-TA) is expected to display severely impaired dimerization properties. Involvement of the TA box in homodimer contacts has been previously described for the DBD heterodimer of RXR and TR (17, 35) and has been postulated to be important for homodimer formation of the RXR DBD on a DR1 (19, 25). All mGCNF mutants were in vitro translated in similar amounts (Fig. 4A) and tested in EMSAs (Fig. 4B).
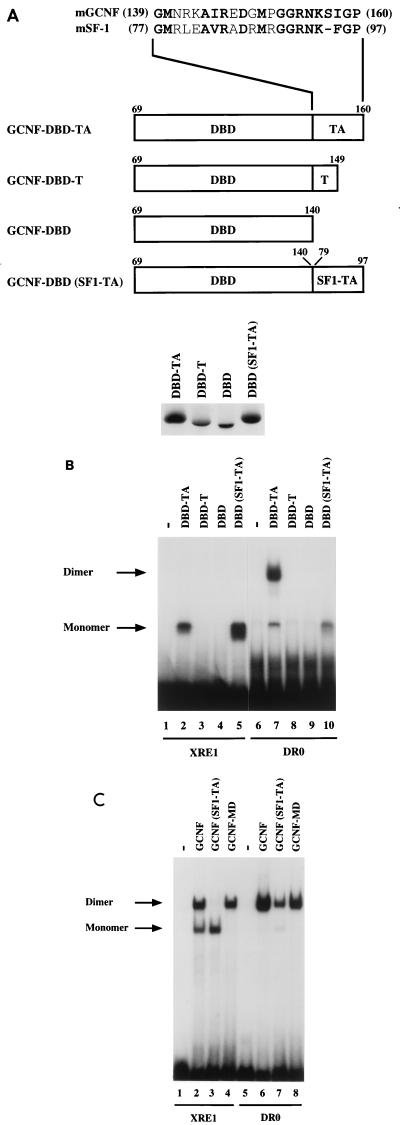
FIG. 4.
The TA box contributes to homodimeric DNA binding of mGCNF. (A) Schematic representation of mGCNF deletion and swap mutants. Numbers indicate the first and the last amino acids of mGCNF or mSF-1. The alignment shows the amino acid sequence of the TA box of mGCNF and mSF-1. All mutants were in vitro translated in similar amounts in reticulocyte lysate in the presence of [35S]methionine. Note that the mutants DBD-TA and DBD(SF1-TA) contain five methionine residues, whereas DBD-T and DBD contain only four and three methionine residues, respectively. (B) Binding of in vitro-translated mGCNF mutants to the XRE1 (lanes 2 to 5) and the DR0 (lanes 7 to 10) in EMSA. Unprimed reticulocyte lysate served as a control (lanes 1 and 6). (C) Binding of in vitro-translated mGCNF, mGCNF(SF1-TA), and mGCNF-MD to the XRE1 (lanes 2 to 4) and the DR0 (lanes 6 to 8) in EMSA. In mGCNF(SF1-TA), the TA box of mGCNF (amino acids 139 to 160) was replaced with the TA box of mSF-1 (amino acids 77 to 97). mGCNF-MD contains two point mutations (R113E and D114L) in the D box. Unprimed reticulocyte lysate served as a control in lanes 1 and 5.
As expected, mGCNF-DBD-TA binds as a monomer to the XRE1 and as a homodimer to the DR0 (Fig. 4B, lanes 2 and 7). In agreement with results of Borgmeyer (4), deletion of the A box or the TA box abolishes DNA binding of the mutant receptors mGCNF-DBD-T and mGCNF-DBD (Fig. 4B, lanes 3, 4, 8, and 9), suggesting that the TA box is critically involved in protein-DNA interactions. In contrast, replacement of the mGCNF TA-box with that of mSF-1 enhances monomeric binding of mGCNF-DBD(SF1-TA) to the XRE1 (Fig. 4B, lane 5) but completely abolishes homodimeric binding to the DR0 (Fig. 4B, lane 10). Importantly, mGCNF-DBD(SF1-TA) binds as a monomer to the DR0, indicating that the TA box of mGCNF is indeed involved in homodimeric interactions.
Since replacement of the mGCNF TA box with that of SF-1 blocks the dimerization function of DBD-TA, we addressed the question of whether this TA swap would also affect DNA binding or dimerization in the context of full-length mGCNF. The mutant mGCNF(SF1-TA), in which the mGCNF TA box is replaced with that of SF-1, no longer binds as a homodimer to the XRE1 (Fig. 4C; compare lanes 2 and 3). Together with results in Fig. 3B, these findings suggest that the dimerization functions of both DBD-TA and the LBD are required for homodimeric binding of mGCNF to XRE1. In contrast, mGCNF(SF1-TA) still binds as a homodimer to the DR0, although with a significantly reduced affinity (Fig. 4C, lane 7). Since the DBD(SF1-TA) mutant binds only as a monomer to the DR0, homodimeric binding of mGCNF(SF1-TA) to this site probably results from protein-protein interactions within the LBD-dimer interface. Interestingly, DNA binding of the mutant mGCNF-MD, which contains two point mutations (R113E and D114L) in the D box, is not significantly affected (Fig. 4C, lanes 4 and 8). This finding indicates that in contrast to other nuclear receptors such as ER (39), the D box is not involved in homodimeric DNA binding of mGCNF.
Taken together, our data demonstrate the presence of two dimerization functions in mGCNF, one located in the DBD-TA and the other found in the LBD. The loss of either of the two dimerization functions abolishes homodimeric binding of mGCNF to the XRE1.
The LBDs of mGCNF and mGCNF-H11 adopt different conformations.
Next we asked how the mGCNF LBD controls DNA binding and dimerization of the receptor. The results presented in Fig. 3B demonstrate that deletion of the C-terminal 12 amino acids of mGCNF significantly reduces the ability of the receptor to bind DNA. This effect is specific, since deletion of homologous regions in other nuclear orphan receptors, such as COUP-TFII or RZRβ does not alter the DNA-binding capabilities of these receptors (references 1 and 15 and data not shown). Our results indicate that the H12 region of the mGCNF LBD might have a structural and/or a regulatory role in the control of DNA binding. Furthermore, the dimerization properties of truncated receptors suggest that the region spanning the putative H2 and H3 is involved in homodimeric binding of mGCNF to the XRE1 and the DR0.
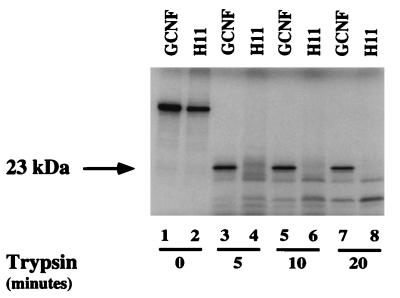
The reduced DNA binding upon deletion of the H12 region prompted us to investigate whether possible conformational changes between mGCNF and mGCNF-H11 were responsible for the observed differences in DNA binding. To address this issue, we used a limited proteolytic digestion assay (23, 42). Comparisons of the crystal structures of the holo-LBDs of hRARγ, hTRα, and hERα (6, 36, 45) with that of the apo-LBD of hRXRα (5) suggest that the reduced susceptibility of holo-LBDs to proteolytic degradation correlates with their increase in compactness resulting from ligand binding and the structural reorientation of H12, which folds back onto the LBD surface. Limited proteolysis of mGCNF with trypsin generates a protected fragment of about 23 kDa (Fig. 5, lanes 3, 5, and 7). In contrast, mGCNF-H11 is rapidly degraded by the protease (Fig. 5, lanes 4, 6, and 8), suggesting that mGCNF and mGCNF-H11 adopt different conformations. Identical results were obtained by limited proteolysis of mGCNF and mGCNF-H11 with chymotrypsin (data not shown). The conformational differences between mGCNF and mGCNF-H11 are most likely related to the structure of the LBD, which in the case of mGCNF may adopt a compact conformation similar to that of the holo-LBD of hRARγ or hTRα. In contrast, the mGCNF-H11 LBD seems to be in a more relaxed conformation similar to that of the apo-LBD of hRXRα. In addition, the spatial orientation of the H12 region of the mGCNF LBD probably contributes to the compactness of the holo-like conformation. Thus, in the holo-like conformation, the H12 region of mGCNF might fold back onto the LBD surface and be near H2 and H3, similar to the folding of H12 observed in the holo-hRARγ, -hTRα, or -hERα LBD structures (6, 36, 45).
FIG. 5.
mGCNF and mGCNF-H11 adopt different conformations. mGCNF and mGCNF-H11 were incubated with trypsin for the indicated time. Lanes 1 and 2 represent the input for mGCNF and mGCNF-H11, respectively. The position of the protected 23-kDa fragment is marked by an arrow.
mGCNF and mGCNF-H11 display distinct dimerization properties.
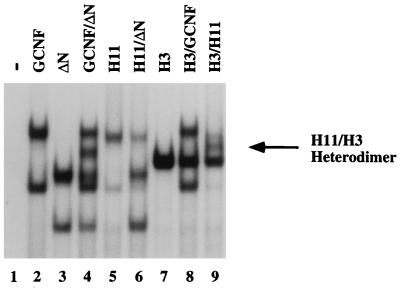
Next, we questioned whether the different conformations represented by mGCNF and mGCNF-H11 not only influence DNA binding but also regulate the dimerization properties of the receptor. The mixing of mGCNF with ΔN-mGCNF in EMSAs results in the formation of the previously observed, intermediate-mobility heterodimeric complex on the XRE1 (Fig. 6, lane 4). It is important to note that deletion of the NTD does not influence the dimerization properties of the truncated receptor. Interestingly, mGCNF-H11 does not form heterodimeric complexes with ΔN-mGCNF (or mGCNF) (Fig. 6, lane 6). Furthermore, mGCNF-H3 forms heterodimers with mGCNF-H11 (Fig. 6, lane 9) but not with mGCNF (Fig. 6, lane 8). The formation of mGCNF-H11–mGCNF-H3 heterodimers further strengthens the idea that the putative H2 and H3 are involved in dimeric interactions. In addition, the different LBD conformations represented by mGCNF and mGCNF-H11 are most likely responsible for the inability of these two proteins to form heterodimers with each other. Taken together, the data suggest that mGCNF and mGCNF-H11 adopt two distinct dimerization conformations.
FIG. 6.
mGCNF and mGCNF-H11 adopt distinct dimerization conformations. mGCNF, ΔN-mGCNF, mGCNF-H11, and mGCNF-H3 were assayed for heterodimer formation on the XRE1 in EMSA. Various amounts of protein were used in each lane to compensate for the different DNA-binding affinities of the deletion mutants. Only mGCNF and ΔN-mGCNF (lane 4) and mGCNF-H11 and mGCNF-H3 (lane 9) form heterodimers; ΔN-mGCNF–mGCNF-H11 (lane 6) and mGCNF–mGCNF-H3 (lane 8) heterodimers are not observed. The position of the mGCNF-H11–mGCNF-H3 heterodimer (H11/H3) is marked by an arrow. Deletion of the amino-terminal domain in ΔN-mGCNF does not influence the dimerization properties of the truncated receptor.
Mutations that either replace or dislocate the H12 region result in a conformational change of the mGCNF LBD.
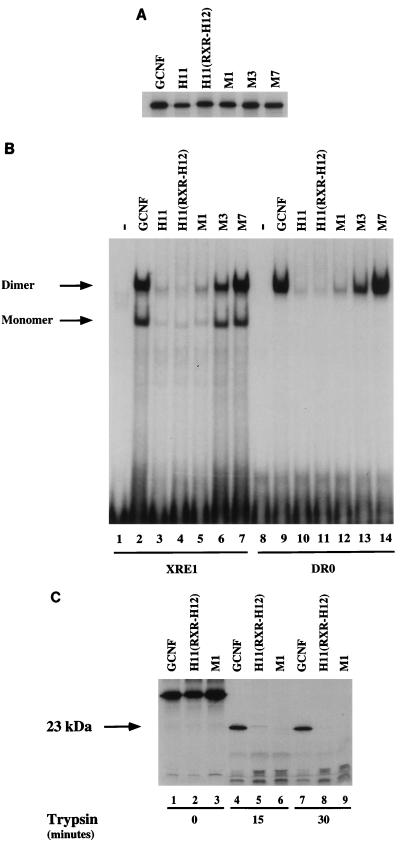
Since mGCNF and mGCNF-H11 can adopt distinct dimerization conformations, we assumed that the H12 region of mGCNF, in addition to controlling the DNA-binding affinity, might also critically influence and/or regulate the ability of the receptor to dimerize on DNA. In this case, either replacement of the H12 region or specific point mutations influencing the spatial orientation of the H12 region should alter the dimerization properties of the receptor. Accordingly, we replaced the H12 region of mGCNF with the corresponding segment of hRXRα [mGCNF-H11(RXR-H12)]. In addition, we generated the double-point mutant mGCNF-M1 (V484D/L485P). The introduction of a proline residue is expected to dislocate the position of the H12 region of mGCNF. As a control, the mutants mGCNF-M3 (C488A/K489A) and mGCNF-M7 (K494A/E495A) were generated (for details, see Fig. 8A). All mutants were in vitro translated in equal amounts (Fig. 7A) and subjected to EMSAs. As expected, the binding of mGCNF-H11(RXR-H12) and mGCNF-M1 to the XRE1 (Fig. 7B, lanes 4 and 5) and the DR0 (Fig. 7B, lanes 11 and 12) is significantly less than that of mGCNF (Fig. 7B, lanes 2 and 9) and comparable to the weak binding of mGCNF-H11 (Fig. 7B, lanes 3 and 10). In contrast, the control mutants mGCNF-M3 and mGCNF-M7 exhibit DNA binding similar to that of wild-type mGCNF (Fig. 7B, lanes 6, 7, 13, and 14).
FIG. 8.
A double-point mutation reduces homodimeric DNA binding of mGCNF. (A) Schematic representation of mutations (M1 to M7) introduced into the H12 region of the mGCNF LBD. All mutants were in vitro translated in equal amounts in reticulocyte lysate in the presence of [35S]methionine. (B) Binding of mGCNF, mGCNF-H11, and the mGCNF point mutants to the XRE1 in EMSA. Lane 1 contains unprimed reticulocyte lysate. The double-point mutant mGCNF-M6 shows significantly reduced homodimeric DNA binding (lane 8), whereas monomer binding is not affected.
FIG. 7.
Replacement or dislocation of the H12 region of the LBD reduces DNA binding of mGCNF. (A) In vitro translation of mGCNF mutants carried out in reticulocyte lysate in the presence of [35S]methionine. All mutants were translated in equal amounts. A detailed representation of the mGCNF mutants is shown in the Fig. 3A and 8A. (B) Binding of mGCNF and mGCNF mutants to the XRE1 (lanes 2 to 7) and the DR0 (lanes 9 to 14) in EMSA. Lanes 1 and 8 contain unprimed reticulocyte lysate. The mutants mGCNF-M3 (C488A/K489A) and mGCNF-M7 (K494A/E495A) served as controls. mGCNF-H11, mGCNF-H11(RXR-H12), and mGCNF-M1 display reduced monomeric and homodimeric binding to the XRE1 (lanes 3 to 5) and the DR0 (lanes 10 to 12). (C) Limited proteolytic digestion assay of mGCNF, mGCNF-H11(RXR-H12), and mGCNF-M1 with trypsin. The position of the protected 23-kDa fragment is marked by an arrow.
Next, we compared the limited proteolysis pattern of mGCNF with that of the various mutants. mGCNF-H11(RXR-H12) and mGCNF-M1 are drastically more susceptible to degradation by trypsin (Fig. 7C; compare lanes 1 to 3 with lanes 7 to 9) and chymotrypsin (data not shown) than mGCNF. These results strongly support the idea that like mGCNF-H11, both mutants adopt an apo-like conformation that is distinct from the holo-like conformation of wild-type mGCNF. Since mGCNF-H11 and the double-point mutant mGCNF-M1 behave identically in limited proteolysis assays and EMSAs, we finally tested if the conformational change in mGCNF-M1 also influenced the dimerization properties of this mutant. As expected, mGCNF-M1 homodimerizes on an XRE1 and forms heterodimers with ΔN-mGCNF-H11 (or mGCNF-H11) (data not shown). Importantly, however, mGCNF-M1 does not heterodimerize with ΔN-mGCNF (or mGCNF) (data not shown). Again, the presence or absence of the NTD does not influence the dimerization properties of the receptor mutants. In summary, these studies demonstrate that the very C terminus of mGCNF (H12 region) controls the abilities of mGCNF to both dimerize and bind DNA.
Double-point mutations in the H12 region of mGCNF show reduced dimerization properties.
The previous experiments demonstrate the influence of the relative position of the H12 region on both the DNA-binding capabilities and the dimerization conformation of mGCNF. The apo-receptor-like conformation represented by mGCNF-H11 or mGCNF-M1 displays reduced monomeric and dimeric binding to the XRE1. However, the contribution of the H12 region to homodimeric DNA binding of wild-type mGCNF remained unclear. To identify specific amino acids within the H12 region which could potentially affect homodimeric DNA binding of mGCNF without inducing conformational changes of the LBD, we introduced a series of alanine mutations in the H12 region (Fig. 8A) and assayed these mutants in EMSAs (Fig. 8B). As demonstrated before, mGCNF-H11 shows a significantly reduced affinity to the XRE1 in comparison with mGCNF (Fig. 8B, lanes 2 and 3). In contrast, DNA binding of the mGCNF mutants M2, M4, M5, and M7 is not altered (Fig. 8B, lanes 4, 6, 7, and 9). Dimeric binding of the mutant mGCNF-M3 is weakly affected (Fig. 8B, lane 5). Importantly, dimeric binding of the mutant mGCNF-M6 to the XRE1 is drastically reduced, whereas monomer binding is not affected (Fig. 8B, lane 8). These results argue for an involvement of the H12 region in homodimeric interactions of wild-type mGCNF. Limited proteolysis experiments reveal that mGCNF and all alanine scan mutants adopt the same holo-like conformation (data not shown), indicating that these particular mutations within the H12 region are not sufficient to induce the switch into the apo-like dimerization conformation. Accordingly, the mutation in mGCNF-M6 blocks only dimeric DNA binding; monomeric binding is unaffected. In contrast, the mutant mGCNF-M1 (V484D/L485P), which most likely dislocates the position of the H12 region, adopts an apo-like conformation similar to that of mGCNF-H11. Consequently, this mutant does not bind efficiently to DNA either as a dimer or as a monomer (Fig. 7B, lanes 5 and 12).
Dimeric binding of mGCNF-H11 and mGCNF-H3 to the XRE1 can be specifically competed with a peptide encompassing H3.
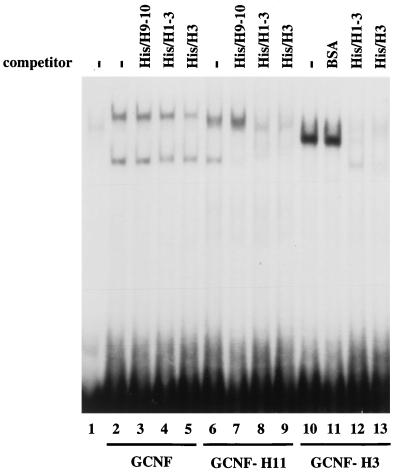
Analysis of the mGCNF deletion mutants presented in Fig. 3B revealed that deletion of the putative H2 and H3 of the mGCNF LBD generated truncated receptors which had lost the ability to homodimerize on the XRE1. These results argued for an involvement of H2 and H3 in homodimeric binding of mGCNF to extended half-sites. Consequently, we attempted to block dimeric binding to the XRE1 with His-tagged peptides comprising H1 to H3 (His/H1-3) or H3 (His/H3). As a control, we used either BSA or a peptide that encompasses the putative H9 and H10 of the mGCNF LBD (His/H9-10). Homodimeric binding of wild-type mGCNF to the XRE1 is not competed by the peptide His/H9-10 and is only weakly competed by the peptides His/H1-3 and His/H3 (Fig. 9, lanes 2 to 5). In contrast, homodimeric binding of mGCNF-H11 and mGCNF-H3 is efficiently competed by the His/H1-3 and His/H3 peptides but not by the control peptide His/H9-10 or BSA (Fig. 9, lanes 6 to 13). These results demonstrate that H3 is critically involved in the dimerization surface of the apo-like LBD conformation that is represented by mGCNF-H11. In contrast, mGCNF adopts a distinct holo-like dimerization conformation that is only weakly blocked under these conditions. This result further supports the idea that the H12 region as well as H3 may stabilize the dimerization interface of the DNA-bound wild-type receptor. A peptide comprising the H12 region of the LBD failed to compete dimeric binding of mGCNF and did not enhance the weak competition of His/H1-3 and His/H3 (data not shown). This result indicates that a distinct spatial orientation of H3 and the H12 region could determine the dimerization surface of the mGCNF LBD.
FIG. 9.
Dimeric binding of GCNF-H11 and GCNF-H3 to the XRE1 can be blocked with an H3 peptide. His-tagged mGCNF polypeptides (70 pmol of His/H9-10, His/H1-3, or His/H3) were used in EMSAs to interfere with homodimeric binding of mGCNF, mGCNF-H11, and mGCNF-H3 to the XRE1. Peptides His/H1-3 and His/H3 contain the putative mGCNF dimerization motif. His/H9-10 and BSA (2 μg) served as controls.
The dimerization motif of mGCNF is transferable to the hRXRα LBD.
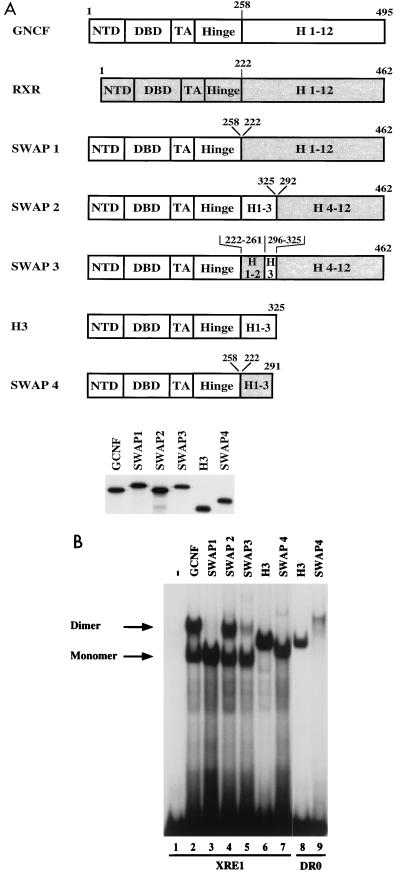
In the next set of experiments, we addressed the question of whether the region spanning H3, the novel mGCNF dimerization motif, is transferable to a different nuclear receptor. Due to the conserved pattern in H1 and the high similarity in H3, hRXRα is a good candidate for such swap experiments. Therefore, mGCNF-hRXRα swap mutants were constructed, expressed in vitro in equal amounts (Fig. 10A), and tested in EMSAs (Fig. 10B). A swap mutant in which the entire LBD of mGCNF is replaced by that of hRXRα (SWAP1) is no longer able to form homodimers on the XRE1 (Fig. 10B, lane 3). Even the addition of specific RXR agonists that were reported to enhance RXR homodimer formation (54) fails to induce the formation of SWAP1 dimers (data not shown). This result demonstrates that neither the dimerization motif located in H9 to H10 nor H3 of the hRXRα LBD is able to promote dimeric interactions under these conditions. Importantly, when we replace H1 to H3 of hRXRα with the corresponding segment of mGCNF, the resulting mutant (SWAP2) homodimerizes efficiently (Fig. 10B, lane 4). Finally, a mutant (SWAP3) in which only H3 of hRXRα is exchanged with H3 of mGCNF regains, at least partially, the ability to homodimerize (Fig. 10B, lane 5). These results clearly demonstrate that the mGCNF dimerization surface can be transferred to a heterologous LBD. In addition, H1 to H3 of hRXRα do not promote dimerization on their own since the transfer of this region onto mGCNF-HINGE results in a mutant (SWAP4) that binds exclusively as a monomer to the XRE1 (Fig. 10B, lane 7), whereas the corresponding mutant mGCNF-H3 binds as a homodimer (Fig. 10B, lane 6). As expected, the additional dimerization contacts mediated by the mGCNF DBD allow homodimeric binding of mutant SWAP4 to the DR0 (Fig. 10B, lane 9).
FIG. 10.
The mGCNF dimerization motif can be transferred to the heterologous hRXRα LBD. (A) Schematic representation of mGCNF-hRXRα swap mutants. Numbers represent the first and last amino acids of the fragments of mGCNF or hRXRα present in the swap mutants. All mutants were in vitro translated in similar amounts in reticulocyte lysate in the presence of [35S]methionine. (B) Binding of mGCNF-hRXRα swap mutants to the XRE1 (lanes 2 to 7) and the DR0 (lanes 8 and 9) in EMSA. Lane 1 represents the reticulocyte lysate control. Replacement of the mGCNF LBD by the hRXRα LBD (SWAP1) abolishes dimeric binding to the XRE1 (lane 3). Homodimeric binding can be restored in SWAP2 (lane 4) and SWAP3 (lane 5). SWAP4 binds as a monomer to the XRE1 (lane 7) and as a homodimer to the DR0 (lane 9), whereas mGCNF-H3 binds as a homodimer to both binding sites (lanes 6 and 8).
Three-dimensional illustration of the novel mGCNF dimerization interface.
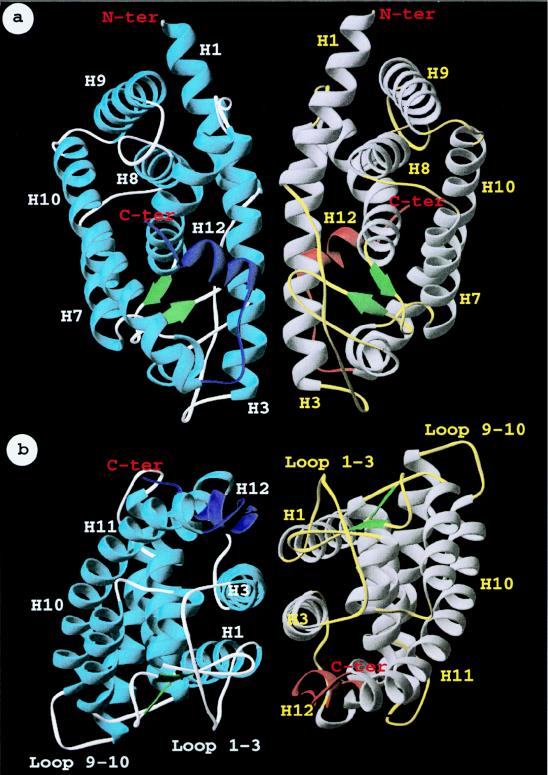
Our results demonstrate that mGCNF uses a novel dimerization surface. In the apo-like conformation, the dimerization function is generated by H3. In the holo-like conformation, additional interactions, such as with H12, contribute to the formation of the dimerization surface. The elucidation of these novel dimerization properties of mGCNF prompted us to model the putative interface used by a mGCNF dimer. We first considered if structural features might exclude dimerization of mGCNF via H10. Three-dimensional modeling suggests that dimer formation via H10 would be hindered by bulky side chains like that of Tyr464 in mGCNF (Ser427 in hRXRα), the loss of hydrophobic contacts, and unfavorable contacts due to clusters of similarly charged residues (data not shown). In contrast, the construction of a symmetrical mGCNF homodimer complex in which H3 is the key contact region revealed a complementarity of shape and charge which could accommodate the homodimer interface (data not shown). In support of our experimental data, the 3D illustration predicts that in addition to H3, other regions of the LBD come in close contact with each other and contribute to the dimer interface. Indeed, as depicted in Fig. 11, the H12 region of one monomer is near the loop connecting H1 to H3 and the β-sheet region of the other monomer. A calculation of the dimer interface (3.5-Å distance cutoff between the two molecular surfaces) with Grasp (29) reveals a contact area of 1,450 Å2 between the two LBD monomers similar to that observed in the hRXRα LBD dimer.
FIG. 11.
Three-dimensional illustration of the GCNF LBD homodimer, using H3 as the dimer interface. Views with H3 parallel to the drawing plane (a) and rotated by 90° around the x axis (b) are shown. Putative secondary structure elements of the LBD are shown as ribbon drawings and numbered according to the crystal structures of hRXRα and hRARγ (5, 36). The spatial position of the H12 region is shown for illustrative purposes as an α helix similar to the crystal structure of the ligand-bound hRARγ LBD (36). This region is colored in dark blue or orange in the two monomers.
Deletion or dislocation of the H12 region alters the transcriptional properties of mGCNF.
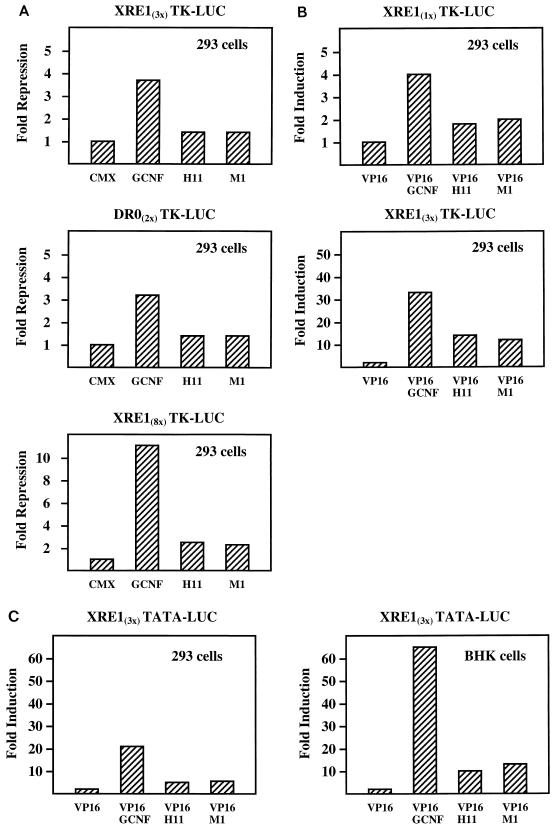
In the final set of experiments, we examined the potential effects of the deletion or dislocation of the H12 region on the transcriptional properties of mGCNF in transient transfection assays. In 293 cells, expression of mGCNF results in 3- to 4-fold repression of XRE13x-TK-LUC and DR02x-TK-LUC activities, while the high number of mGCNF binding sites in the XRE8x-TK-LUC reporter results in an approximate 10-fold repression (Fig. 12A). No repression is observed with a TK-LUC reporter that does not contain mGCNF binding sites (data not shown). mGCNF-mediated transcriptional repression has also recently been reported by Cooney et al. (9). However, these authors observe repression only on reporter plasmids containing DR0 binding sites. Therefore, our results establish that mGCNF can also repress transcription from extended half-sites. Importantly, the mutants mGCNF-H11 and mGCNF-M1 have a significantly reduced ability to repress reporter gene activity (Fig. 12A). Since all mGCNF proteins are expressed at equal levels (data not shown), the altered receptor conformation that results from the deletion or dislocation of the H12 region impairs the repression potential of both mGCNF mutants.
FIG. 12.
Transcriptional properties of mGCNF and mutant receptors in transient transfection assays. (A) Transcriptional repression by mGCNF, mGCNF-H11, and mGCNF-M1 in 293 cells. Cells were transfected with 250 ng of reporter plasmid (XRE13x-TK-LUC, DR02x-TK-LUC, or XRE18x-TK-LUC) and 25 ng of CMX.ATG expression plasmid coding for mGCNF, mGCNF-H11, or mGCNF-M1; 50 ng of the different CMX.ATG expression plasmids were used for cotransfection with the XRE18x-TK-LUC reporter. The empty expression vector served as a control. Results are expressed as fold repression relative to the CMX.ATG vector control. (B and C) Transcriptional activation by VP16-mGCNF, VP16-mGCNF-H11, and VP16-mGCNF-M1 in 293 or BHK cells. 293 cells were transfected with 250 ng of reporter plasmid (XRE11x-TK-LUC or XRE13x-TK-LUC) and 25 ng of CMX.VP16 expression plasmid coding for mGCNF, mGCNF-H11, or mGCNF-M1 as described for panel A. Alternatively, 293 or BHK cells were cotransfected with 500 ng of the XRE13x-TATA-LUC reporter plasmid and 50 ng of the different CMX.VP16 expression plasmids. The empty CMX.VP16 vector served as a control. Results are expressed as fold induction relative to the CMX.VP16 vector control.
The impaired capability of mGCNF-H11 and -M1 to repress transcription may be a consequence of reduced DNA binding or be due to the loss of functional interactions with putative corepressors. To distinguish between these potential mechanisms, we generated constitutively active wild-type and mutant mGCNF proteins by fusing a VP16 transactivation domain to their N termini. In EMSAs, the resulting fusion proteins VP16-mGCNF-H11 and VP16-mGCNF-M1 bound to the XRE1 or the DR0 with the expected reduced affinity compared to VP16-mGCNF (data not shown). All VP16 fusion proteins were then characterized in transient transfection assays. VP16-mGCNF strongly activates transcription from XRE1- as well as DR0-containing reporter plasmids (Fig. 12B and C and data not shown). Compared to VP16-mGCNF, the mutants VP16-mGCNF-H11 and VP16-mGCNF-M1 activate transcription to a significantly lower degree on all tested reporters (Fig. 12B and C). Since all three VP16-mGCNF fusion proteins are expressed to equal levels (data not shown), these results indicate that reduced DNA binding is the major cause for the reduced transcriptional activation of the mutants VP16-mGCNF-H11 and -M1. Taken together, the results of the transient transfection assays are in agreement with the in vitro properties of mGCNF proteins and suggest that the H12 region may also control the DNA-binding and transcriptional properties of the receptor in vivo.
DISCUSSION
In this study, we analyzed in detail the DNA-binding, dimerization, and transcriptional properties of mGCNF. In agreement with previous reports (4, 8, 51), we observe homodimeric binding of mGCNF to a DR0. In addition, our data clearly demonstrate that mGCNF can bind as a homodimer to an extended half-site like the XRE1 (CCCTCAAGGTCA). Homodimeric binding is not dependent on particular nucleotides in positions −4 to −6 of the extended half-site, since mGCNF binds as a homodimer to the XRE sequence AAATCAAGGTCA as well (data not shown). Nevertheless, dimeric binding of mGCNF to an extended half-site is unexpected and in marked contrast to the DNA-binding properties of other orphan receptors such as NGFI-B, SF-1, or ROR/RZR, which bind exclusively as monomers to extended half-sites (15, 48, 49). To unravel further the dimerization properties of mGCNF, we generated a series of mGCNF deletion mutants and analyzed their DNA-binding and dimerization properties on both the XRE1 and the DR0. Surprisingly, deletion of the C-terminal 12 amino acids drastically reduces the ability of the receptor to bind to the XRE1 and the DR0. Since deletions of corresponding segments in other nuclear receptors such as COUP-TFII and RZRβ do not alter their DNA-binding properties (1, 15), this effect appears to be specific to mGCNF.
Structure-based nuclear receptor sequence alignments suggest that the mGCNF LBD adopts an α-helical sandwich fold, commonly found for nuclear receptor LBDs (references 36 and 50 and data not shown). However, the folding of the H12 region at the C terminus of the mGCNF LBD cannot be accurately predicted. Interestingly, in contrast to most members of the nuclear receptor superfamily, the transcriptional activation motif (AF2 AD core) is not conserved in the H12 region of X. laevis, human, or mouse GCNF. Upon ligand binding, the AF2 AD core, located in H12 of the LBDs of other nuclear receptors such as hRARγ, hTRα, and hERα, is involved in significant conformational changes (6, 36, 45). It is currently believed that the ligand-induced conformational changes of the LBD result in the disruption of apo-receptor–corepressor interfaces while simultaneously creating novel interaction surfaces for coactivators (44). The equilibrium between the apo- and holo-LBD conformation is thought to be influenced either by the binding of ligand, by mutations that mimic the effect of ligand binding, or by secondary modifications such as phosphorylation (44, 46, 47). The absence of a conserved AF2 AD core in mGCNF already suggested the possibility that the H12 region of GCNF has functional roles distinct from those in other nuclear receptors. We show that the limited proteolysis pattern of mGCNF is similar to that of liganded (holo-type) receptors. In contrast, mGCNF-H11 behaves like an unliganded (apo-type) receptor and thus adopts a conformation different from that of mGCNF. Importantly, either replacement of the H12 region of mGCNF by the corresponding segment of hRXRα or dislocation of the H12 region by the introduction of a proline residue (in mGCNF-M1) induces a mGCNF-H11-like conformation. Although the mechanism(s) by which the conformation of mGCNF is regulated in vivo is not known, our data suggest a dynamic model of the mGCNF LBD structure. Thus, upon potential secondary modifications and/or binding or dissociation of potential ligands, the relative position of the H12 region might be efficiently redirected similar to the position of the H12 region in mGCNF-M1. Consequently, the LBD would adopt a holo- or apo-like conformation that in turn regulates the ability of the receptor to either bind to or dissociate from specific response elements like the XRE1 or the DR0. Since both conformations are still able to bind DNA (although with markedly different affinities), such a strategy might contribute to the fine-tuning of target gene expression.
As a consequence of the observed in vitro properties, the transcriptional properties of mGCNF differ significantly from the behavior of the mutants mGCNF-H11 and mGCNF-M1. Both mutants display a reduced ability to repress reporter gene activity in comparison with the wild-type receptor. In addition, transcriptional activation by VP16-mGCNF-H11 or -M1 is greatly diminished relative to VP16-mGCNF. Together, these results indicate that reduced DNA binding of the mutant receptors mainly accounts for the differences in transcriptional properties. Recently, Cooney et al. (9) suggested that mGCNF-mediated transcriptional control was specific for reporter systems that contain DR0 binding sites. In contrast, we also observe mGCNF-mediated repression as well as VP16-mGCNF-mediated activation from reporters containing the XRE1. Thus, transcriptional control by mGCNF appears not to be limited to the presence of DR0 binding sites but may also occur in the presence of extended half-sites.
The conformational change of the mGCNF LBD that results from the deletion or dislocation of the H12 region is also accompanied by a change of the dimerization properties of the receptor on DNA: mGCNF does not form heterodimers with mGCNF-H11 or mGCNF-M1 on the XRE1. Thus, the H12 region of mGCNF not only influences the DNA-binding affinity but also is involved in the transition from one dimerization conformation to the other, thereby controlling the ability of the different conformations to dimerize on DNA. To date no potential transcriptional cofactors or heterodimerization partners for mGCNF are known. In preliminary studies, we observed neither heterodimerization between mGCNF and other members of the nuclear receptor superfamily nor interactions of mGCNF with published coactivators or corepressors for nuclear hormone receptors (data not shown). Nevertheless, it is tempting to suggest that the dimerization conformation of mGCNF-H11 or mGCNF-M1 could provide altered surfaces for protein-protein interactions with novel sets of transcriptional cofactors or putative heterodimerization partners. This scenario may be similar to that reported for the holo-hERα LBD (6), where alternative positioning of H12 by different ligands determines the transcriptional properties of the hormone-bound receptor. Taken together, our results suggest two novel functions for the H12 region of mGCNF: regulation of dimerization and DNA binding.
The dimerization properties of a number of nuclear receptors including RAR, RXR, and ER are well established (5, 6, 19, 25, 31, 39). Apart from dimerization functions found in the DBDs of these receptors, a second dimerization motif is found in the C-terminal part of their LBDs. The crystal structures of the LBDs of hRXRα, hRARγ, hTRα and hERα (5, 6, 36, 45) show that this dimerization motif is mainly located in H9 and H10. In fact, crystal structure data demonstrate that homodimeric interactions of hRXRα LBDs are mediated mainly by H10 and to a lesser extent by H9 and the loop between H7 and H8 (5, 6). Recently, a different dimerization motif has been described for the nuclear orphan receptor SHP; this motif is located within the putative H5 to H7 of the LBD (40). Our data indicate that mGCNF, like other nuclear receptors, contains two dimerization functions, one located in the DBD (including the TA box) and the other located in the LBD. Both dimerization functions appear to be necessary for homodimeric binding of the receptor to the XRE1. While the existence of two dimerization functions has been anticipated, the dimerization surface of the mGCNF LBD is unexpected. Our analyses of mGCNF mutants suggest that the putative H3 in the mGCNF LBD is critically involved in homodimeric interactions. In agreement with the results of Borgmeyer (4), we observe that deletion of H9 and H10 does not influence the dimerization properties of mGCNF. In providing further evidence that a surface generated by H3 is part of the dimerization interface, we are able to block homodimeric DNA binding of mGCNF-H11 and mGCNF-H3 with a peptide spanning H1 to H3 or H3. In marked contrast, a peptide spanning H9 to H10 fails to do so. We also transferred H1 to H3 or H3, the dimerization interface of mGCNF, into the context of the hRXRα LBD, changing a protein which is unable to dimerize (SWAP1) into a chimera which is competent to homodimerize (SWAP2 and SWAP3). The novel mGCNF dimerization motif is also clearly distinct from that of SHP, where H5 to H7 have been reported to be involved in heterodimeric contacts between SHP and RXR, RAR, or TR (40). Together, these results indicate that receptor-receptor interaction surfaces may be more diverse than initially expected from the studies of the LBD dimerization motifs of RXR, RAR, TR, and ER.
The results of the competition experiments suggest that in the wild-type receptor, H3 requires additional regions of the LBD to form the dimerization surface. Consequently, we could demonstrate that the H12 region is involved in the stabilization of the wild-type mGCNF dimer interface. Our predicted model of the mGCNF LBD indicates that the H12 region may be in contact with H1 to H3, thus influencing the exact conformation of the dimerization surface centered around H3. In addition, the illustration of the dimerization interface suggests that the H12 region of one molecule may contact loop regions between H1 and H3 or between the β sheet and H6 of the other molecule and therefore contribute directly to the dimerization surface. Consequently, mutations such as in mGCNF-M6 would affect the dimerization properties without influencing the holo-like conformation of the LBD and monomeric DNA binding of the receptor. In addition to the contribution of the H12 region, a potential bending of H3 might also account for the distinct dimerization properties of mGCNF compared to mGCNF-H11. Such a bending of H3 has been observed in the hormone-bound LBD of hRARγ (36) but not in the unliganded LBD of hRXRα (5). Interestingly, H3, the center of the dimerization interface in mGCNF and H9 to H10, the dimerization surface in RXR, are located on almost opposite sides of the LBDs. Together, these data reveal unexpected properties of mGCNF and suggest novel mechanisms by which the receptor may control transcriptional processes.
ACKNOWLEDGMENTS
We thank Erich F. Greiner and Oliver Werner for plasmids, Darren Daniels, Laszlo Tora, Jutta Kirfel, Judith Müller, Erich F. Greiner, and the members of the Schüle laboratory for discussions and critical reading of the manuscript, and Corina Schüle for providing the artwork and administrative assistance.
This work was supported in part by grants from the Schering AG and the Deutsche Forschungsgemeinschaft to R.S.
REFERENCES
- 1.Achatz G, Holzl B, Speckmayer R, Hauser C, Sandhofer F, Paulweber B. Functional domains of the human orphan receptor ARP-1/COUP-TFII involved in active repression and transrepression. Mol Cell Biol. 1997;17:4914–4932. doi: 10.1128/mcb.17.9.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barettino D, Vivanco Ruiz M M, Stunnenberg H G. Characterization of the ligand-dependent transactivation domain of thyroid hormone receptor. EMBO J. 1994;13:3039–3049. doi: 10.1002/j.1460-2075.1994.tb06603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beato M, Herrlich P, Schütz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 4.Borgmeyer U. Dimeric binding of the mouse germ cell nuclear factor. Eur J Biochem. 1997;244:120–127. doi: 10.1111/j.1432-1033.1997.00120.x. [DOI] [PubMed] [Google Scholar]
- 5.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXRα. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 6.Brzozowski A M, Pike A C W, Dauter Z, Hubbard R E, Bonn T, Engström O, Öhman L, Greene G L, Gustafsson J-A, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 7.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 8.Chen F, Cooney A J, Wang Y, Law S W, O’Malley B W. Cloning of a novel orphan receptor (GCNF) expressed during germ cell development. Mol Endocrinol. 1994;8:1434–1444. doi: 10.1210/mend.8.10.7854358. [DOI] [PubMed] [Google Scholar]
- 9.Cooney A J, Hummelke G C, Herman T, Chen F, Jackson K J. Germ cell nuclear factor is a response element-specific repressor of transcription. Biochem Biophys Res Commun. 1998;245:94–100. doi: 10.1006/bbrc.1998.8391. [DOI] [PubMed] [Google Scholar]
- 10.Danielian P S, White R, Lees J A, Parker M G. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durand B, Saunders M, Gaudon C, Roy B, Losson R, Chambon P. Activation function 2 (AF-2) of retinoic acid receptor and 9-cis retinoic acid receptor: presence of a conserved autonomous constitutive activating domain and influence of the nature of the response element on AF-2 activity. EMBO J. 1994;13:5370–5382. doi: 10.1002/j.1460-2075.1994.tb06872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans R M. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geourjon C, Deleage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 14.Glass C K, Rose D W, Rosenfeld M G. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 15.Greiner E F, Kirfel J, Greschik H, Dörflinger U, Becker P, Mercep A, Schüle R. Functional analysis of retinoid Z receptor β, a brain-specific nuclear orphan receptor. Proc Natl Acad Sci USA. 1996;93:10105–10110. doi: 10.1073/pnas.93.19.10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gronemeyer H, Laudet V. Transcription factors 3: nuclear receptors. Protein Profile. 1995;2:1173–1308. [PubMed] [Google Scholar]
- 17.Gronemeyer H, Moras D. How to finger DNA. Nature. 1995;375:190–191. doi: 10.1038/375190a0. [DOI] [PubMed] [Google Scholar]
- 18.Hirose T, O’Brien D A, Jetten A M. RTR: a new member of the nuclear receptor superfamily that is highly expressed in murine testis. Gene. 1995;152:247–251. doi: 10.1016/0378-1119(94)00656-d. [DOI] [PubMed] [Google Scholar]
- 19.Holmbeck S M A, Foster M P, Casimiro D R, Sem D S, Dyson H J, Wright P E. High-resolution structure of the retinoid X receptor DNA-binding domain. J Mol Biol. 1998;281:271–284. doi: 10.1006/jmbi.1998.1908. [DOI] [PubMed] [Google Scholar]
- 20.Joos T O, David R, Dreyer C. xGCNF, a nuclear orphan receptor is expressed during neurulation in Xenopus laevis. Mech Dev. 1996;60:45–57. doi: 10.1016/s0925-4773(96)00599-0. [DOI] [PubMed] [Google Scholar]
- 21.Kapelle M, Kratzschmar J, Husemann M, Schleuning W D. cDNA cloning of two closely related forms of human germ cell nuclear factor (GCNF) Biochim Biophys Acta. 1997;1352:13–17. doi: 10.1016/s0167-4781(97)00043-2. [DOI] [PubMed] [Google Scholar]
- 22.Katz D, Niederberger C, Slaughter G R, Cooney A J. Characterization of germ cell-specific expression of the orphan nuclear receptor, germ cell nuclear factor. Endocrinology. 1997;138:4364–4372. doi: 10.1210/endo.138.10.5444. [DOI] [PubMed] [Google Scholar]
- 23.Keidel S, LeMotte P, Apfel C. Different agonist- and antagonist-induced conformational changes in retinoic acid receptors analyzed by protease mapping. Mol Cell Biol. 1994;14:287–298. doi: 10.1128/mcb.14.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurokawa R, Yu V C, Naar A, Kyakumoto S, Han Z, Silverman S, Rosenfeld M G, Glass C K. Differential orientations of the DNA-binding domain and carboxy-terminal dimerization interface regulate binding site selection by nuclear receptor heterodimers. Genes Dev. 1993;7:1423–1435. doi: 10.1101/gad.7.7b.1423. [DOI] [PubMed] [Google Scholar]
- 25.Lee M S, Kliewer S A, Provencal J, Wright P E, Evans R M. Structure of the retinoid X receptor α DNA binding domain: a helix required for homodimeric DNA binding. Science. 1993;260:1117–1121. doi: 10.1126/science.8388124. [DOI] [PubMed] [Google Scholar]
- 26.Lei W, Hirose T, Zhang L-X, Adachi H, Spinella M J, Dmitrovsky E, Jetten A M. Cloning of the human orphan receptor germ cell nuclear factor/retinoid receptor-related testis-associated receptor and its differential regulation during embryonal carcinoma cell differentiation. J Mol Endocrinol. 1997;18:167–176. doi: 10.1677/jme.0.0180167. [DOI] [PubMed] [Google Scholar]
- 27.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 28.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicholls A, Sharp K A, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 30.Perlmann T, Rangarajan P N, Umesono K, Evans R M. Determinants for selective RAR and TR recognition of direct repeat HREs. Genes Dev. 1993;7:1411–1422. doi: 10.1101/gad.7.7b.1411. [DOI] [PubMed] [Google Scholar]
- 31.Perlmann T, Umesono K, Rangarajan P N, Forman B M, Evans R M. Two distinct dimerization interfaces differentially modulate target gene specificity of nuclear hormone receptors. Mol Endocrinol. 1996;10:958–966. doi: 10.1210/mend.10.8.8843412. [DOI] [PubMed] [Google Scholar]
- 32.Perlmann T, Evans R M. Nuclear receptors in Sicily: all in the famiglia. Cell. 1997;90:391–397. doi: 10.1016/s0092-8674(00)80498-5. [DOI] [PubMed] [Google Scholar]
- 33.Pfitzner E, Becker P, Rolke A, Schüle R. Functional antagonism between the retinoic acid receptor and the viral transactivator BZLF1 is mediated by protein-protein interactions. Proc Natl Acad Sci USA. 1995;92:12265–12269. doi: 10.1073/pnas.92.26.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi J S, Desai-Yajnik V, Greene M E, Raaka B M, Samuels H H. The ligand-binding domains of the thyroid hormone/retinoid receptor gene subfamily function in vivo to mediate heterodimerization, gene silencing, and transactivation. Mol Cell Biol. 1995;15:1817–1825. doi: 10.1128/mcb.15.3.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rastinejad F, Perlmann T, Evans R M, Sigler P B. Structural determinants for nuclear receptor assembly on DNA direct repeats. Nature. 1995;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 36.Renaud J-P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 37.Rost B. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 38.Sali A, Blundell T L. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 39.Schwabe J W R, Chapman L, Finch J T, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- 40.Seol W, Chung M, Moore D D. Novel receptor interaction and repression domains in the orphan receptor SHP. Mol Cell Biol. 1997;17:7126–7131. doi: 10.1128/mcb.17.12.7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Süsens U, Borgmeyer U. Characterization of the human germ cell nuclear factor gene. Biochim Biophys Acta. 1996;1309:179–182. doi: 10.1016/s0167-4781(96)00157-1. [DOI] [PubMed] [Google Scholar]
- 42.Trapp T, Holsboer F. Ligand-induced conformational changes of the mineralcorticoid receptor analyzed by protease mapping. Biochem Biophys Res Commun. 1995;215:286–291. doi: 10.1006/bbrc.1995.2464. [DOI] [PubMed] [Google Scholar]
- 43.Umesono K, Murakami K K, Thompson C C, Evans R M. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vivat V, Zechel C, Wurtz J-M, Bourguet W, Kagechika H, Umemiya H, Shudo K, Moras D, Gronemeyer H, Chambon P. A mutation mimicking ligand-induced conformational change yields a constitutive RXR that senses allosteric effects in heterodimers. EMBO J. 1997;16:5697–5709. doi: 10.1093/emboj/16.18.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 46.Weis K E, Ekena K, Thomas J A, Lazennec G, Katzenellenbogen B S. Constitutively active human estrogen receptors containing amino acid substitutions for tyrosine 537 in the receptor protein. Mol Endocrinol. 1996;10:1388–1398. doi: 10.1210/mend.10.11.8923465. [DOI] [PubMed] [Google Scholar]
- 47.White R, Sjöberg M, Kalkhooven E, Parker M G. Ligand independent activation of the oestrogen receptor by mutation of a conserved tyrosine. EMBO J. 1997;16:1427–1435. doi: 10.1093/emboj/16.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson T E, Paulsen R E, Padgett K A, Milbrandt J. Participation of non zinc-finger residues in DNA binding by two nuclear orphan receptors. Science. 1992;256:107–110. doi: 10.1126/science.1314418. [DOI] [PubMed] [Google Scholar]
- 49.Wilson T E, Fahrner T J, Milbrandt J. The orphan receptors NGFI-B and steroidogenic factor 1 establish monomer binding as the third paradigm of nuclear receptor-DNA interaction. Mol Cell Biol. 1993;13:5794–5804. doi: 10.1128/mcb.13.9.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wurtz J-M, Bourguet W, Renaud J-P, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 51.Yan Z H, Medvedev A, Hirose T, Gotoh H, Jetten A M. Characterization of the response element and DNA binding properties of the nuclear orphan receptor germ cell nuclear factor/retinoid receptor-related testis-associated receptor. J Biol Chem. 1997;272:10565–10572. doi: 10.1074/jbc.272.16.10565. [DOI] [PubMed] [Google Scholar]
- 52.Zechel C, Shen X Q, Chambon P, Gronemeyer H. Dimerization interfaces formed between the DNA binding domains determine the cooperative binding of RXR/RAR and RXR/TR heterodimers to DR5 and DR4 elements. EMBO J. 1994;13:1414–1424. doi: 10.1002/j.1460-2075.1994.tb06395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zechel C, Shen X Q, Chen J Y, Chen Z P, Chambon P, Gronemeyer H. The dimerization interfaces formed between the DNA binding domains of RXR, RAR and TR determine the binding specificity and polarity of the full-length receptors to direct repeats. EMBO J. 1994;13:1425–1433. doi: 10.1002/j.1460-2075.1994.tb06396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X K, Lehmann J, Hoffmann B, Dawson M, Cameron J, Graupner G, Hermann T, Tran P, Pfahl M. Homodimer formation of retinoid X receptor induced by 9-cis retinoic acid. Nature. 1992;358:587–591. doi: 10.1038/358587a0. [DOI] [PubMed] [Google Scholar]
- 55.Zhang X K, Salbert G, Lee M O, Pfahl M. Mutations that alter ligand-induced switches and dimerization activities in the retinoid X receptor. Mol Cell Biol. 1994;14:4311–4323. doi: 10.1128/mcb.14.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]