Abstract
5-Fluorouracil (5-FU)-based chemotherapy is the conventional treatment approach for patients with colorectal cancer (CRC). However, de novo and acquired resistance to 5-FU are frequently observed during treatment, which eventually lead to patients succumbing to the disease. Accumulating data have revealed an association of CRC resistance to 5-FU with aberrant expression of microRNAs (miRs). In the present study, Cell Counting Kit-8 was performed to measure cell viability, flow cytometry was performed to detect cell apoptosis, reverse transcription-quantitative PCR was conducted to measure proviral integration site for Moloney murine leukemia virus 1 (PIM1) and miR-3135b expression, western blotting was conducted to measure PIM1 expression. Microarray data analysis indicated that the level of miR-3135b expression was decreased in patients with recurrent CRC that were treated with 5-FU when compared with non-recurrent cases. Overexpression of miR-3135b increased the sensitivity of CRC cells to 5-FU treatment. Moreover, PIM1 was identified as a target gene of miR-3135b using bioinformatics analysis, reverse transcription-quantitative PCR and western blotting. The direct interaction between these two targets was confirmed by luciferase reporter assays. Notably, PIM1 overexpression compensated the effect of miR-3135b in CRC cells. Furthermore, an inverse correlation between PIM1 mRNA expression levels and miR-3135b expression was observed in clinical samples. Therefore, the present study identified miR-3135b as a novel regulator of 5-FU sensitivity in CRC.
Keywords: colorectal cancer, 5-fluorouracil sensitivity, proviral integration site for Moloney murine leukemia virus 1
Introduction
Colorectal cancer (CRC) contributes to 1.2 million new cases and 700,000 mortalities every year (1). This disease remains the fourth leading cause of cancer-related mortality worldwide after lung, stomach and liver cancer (1,2). CRC is considered a primary public health issue in the majority of industrialized countries (3). Unfortunately, ≥80% of patients with CRC are diagnosed at advanced stages, leading to low response to treatment and poor survival (4). This suggests the necessity for identifying novel molecular targets to specifically inhibit oncogenic processes.
Based on the recent advances of medical science and technology, as well as the improved understanding of the progression and molecular pathogenesis of CRC, additional targeted therapies have been applied for the treatment of patients with this disease (5). Despite these achievements, the clinical prognosis of patients with CRC remains unsatisfactory (6). 5-Fluorouracil (5-FU) is an important constituent of the systemic chemotherapy in the palliative and adjuvant treatments used for patients with CRC (7). However, its clinical efficacy in patients with CRC is low due to increased chemoresistance (8).
MicroRNAs (miRs) are ~22 nucleotides in length and inhibit protein expression by targeting coding genes (9). Generally, miRs are complementary to the site of the 3'untranslated region (UTR) in their target mRNAs (10). miR-based target therapies that exhibit substantial efficacy in cancer treatment have been identified in a previous study, including breast cancer, glioma and lung cancer (11). Numerous miRNAs have been reported to regulate the chemoresistance in CRC. For instance, miR-27a promotes CRC resistance to chemotherapy by promoting aerobic glycolytic metabolism, which results in excessive proliferation (12). In addition, exosome-transmitted miR-128-3p has been shown to promote chemosensitivity of oxaliplatin-resistant CRC by inhibiting epithelial-mesenchymal transition and inducing intracellular oxaliplatin accumulation (13). However, the interaction of miR-3135b with the sensitivity of CRC tumors to 5-FU remains unknown. The present study aimed to investigate the potential association between these two parameters.
Materials and methods
Clinical samples
In total, 40 tumor tissue samples and their corresponding adjacent tissue samples, which were ≥5 cm distal to the tumor margins, were extracted from 40 patients (28 males and 12 females) with CRC who underwent surgery in the ChinaJapan Union Hospital of Jilin University (Changchun, China) between February 2018 and May 2019. These patients had stage III CRC tumors and were aged between 45 and 85 years (63.5±11.7 years). Among these patients, 34 received 5-FU treatment. A total of 17 patients who received 5-FU treatment experienced recurrence during follow-up. The present study was approved by the Ethics Committee of China-Japan Union Hospital of Jilin University. Written informed consent was obtained from all patients with CRC with the following inclusion criteria: i) Aged >18 years; ii) Confirmed diagnosis of CRC by surgical pathological diagnosis (14); iii) Are aware they will receive adjuvant chemotherapy; and iv) Can communicate with the medical professional team. In the present study, patients with CRC who received neoadjuvant therapy were excluded from the study.
Cell culture
Normal human colon epithelial NCM460 cells, human colon adenocarcinoma SW480 cell line and human colon carcinoma HCT116 cells were purchased from the American Type Culture Collection. The cells were grown in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.) and 50 U/ml penicillin-streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) in a 5% CO2 humidified atmosphere at 37˚C.
Bioinformatics analysis
Clinical and miR microarray expression data from 119 non-recurrent and 73 recurrent patients receiving 5-FU treatment were retrieved from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/; accession. no. GSE81653). The differential expression analysis was conducted using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/). mRNAs that could be potential targets of miR-3135b were predicted using TargetScan software 7.2 (http://www.targetscan.org/vert_72/).
Transfection
The cells were transfected with miR-3135b mimic (30 nM, 5'-GGCUGGAGCGAGUGCAGUGGUG-3'), pcDNA3.1- PIM1 (20 µg), miR-negative control (NC; 60 nM, 5'-UUCUCCGAACGUGUCACGUTT-3') and pcDNA3.1, which were obtained from Shanghai GenePharma Co., Ltd., using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The cells were seeded in six-well plates (2x105 cells/well) and incubated in DMEM for 48 h at 37˚C until they reached 60-70% confluence. Subsequently, the transfection efficiency was assessed by reverse transcription-quantitative PCR (RT-qPCR) to evaluate the expression levels of miR-3135b or PIM1 at 48 h post-transfection.
Treatment with 5-FU
To determine 5-FU (Selleck Chemicals) sensitivity, the cells were treated for 48 h at 37˚C with various concentrations (5, 10, 20, 50 and 100 µM) of 5-FU.
Cell Counting Kit-8 (CCK8) assay
The cells were resuspended and seeded in 96-well plates (5x103 cells/well). CCK-8 (Dojindo Molecular Technologies, Inc.) was performed according to the manufacturer's instructions to detect cell viability. Briefly, 10 µl CCK-8 reagent was added to each well at 0, 24, 48, 72 and 96 h. Following incubation for 2 h at 37˚C, the absorbance was recorded at 450 nm in each well using a microplate reader.
Flow cytometric analysis
The induction of cellular apoptosis was quantified by FITClabeled Annexin V and PI reagents from an Annexin VFITC Apoptosis Detection Kit (EMD Millipore). The cells were seeded into 12-well plates (3x105 cells/well) and cultured for 48 h at 37˚C. Subsequently, they were resuspended in 100 µl HEPES buffer, followed with incubation of 5 µl Annexin V and 5 µl PI at room temperature for 10 min in the dark according to the manufacturer's instructions. The samples were analyzed using a BD FACSCalibur™ flow cytometer (BD Biosciences) within 1 h. The results were analyzed using FlowJo 10.2 software (FlowJo LLC).
RTqPCR
Total RNA from CRC tumor tissue samples and cell lines was extracted using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Subsequently, the RNA was reverse transcribed into cDNA using a PrimeScript™ RT Reagent Kit (Takara Bio, Inc.) according to the following thermal cycling protocols: A total three cycles of 37˚C at 15 min, termination at 85˚C for 5 sec, and maintenance at 4˚C. qPCR was performed with a SYBR® Premix Ex Taq™ kit (Takara Bio, Inc.) on a CFX-96 Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.) according to the manufacturer's instructions. The thermal cycling protocols were as follows: Denaturation at 95˚C for 30 sec; annealing at 95˚C for 5 sec; elongation at 60˚C for 30 sec for a total of 40 amplification cycles. The primer sequences used were as follows: Forward, 5'-CCCGACAGTTTCGTCCTGAT-3' and reverse, 5'-ACCCGAAGTCGATGAGCTTG-3' for PIM1; forward, 5'-ACAGAGCCTCGCCTTTGCCGAT-3' and reverse, 5'-CTTGCACATGCCGGAGCCGTT-3' for β-actin; forward, 5'-GGCTGGAGCGAGTGCAGTGGTG-3' and reverse, 5'-CACCACTGCACTCGCTCCAGCC-3' for miR-3135b; forward, 5'-CTCGCTTCGGCAGCACA-3' and reverse, 5'-AACGCTTCACGAATTTGCGT-3' for U6. β-actin and U6 served as internal controls to normalize PIM1 mRNA and miR-3135b expression levels, respectively. The relative PIM1 mRNA and miR-3135b expression levels were calculated using the comparative 2-ΔΔCq method (15).
Western blot analysis
Total protein from the CRC cell lines was isolated using Mammalian Protein Extraction solution (Thermo Fisher Scientific, Inc.) supplemented with a protease inhibitor cocktail (Merck KGaA). The protein concentration was analyzed using the Bio-Rad DC Protein Assay (Bio-Rad Laboratories, Inc.) according to the manufacturer's instructions. A total of 20 µg protein was separated by 8% polyacrylamide gels and transferred to 0.2-µm nitrocellulose membranes (Bio-Rad Laboratories, Inc.). Subsequently, the membranes were blocked with 5% non-fat milk for 60 min at 37˚C and incubated overnight at 4˚C with primary antibodies against PIM1 (cat. no. ab245417; dilution, 1:2,000) and β-actin (cat. no. ab8227; dilution, 1:2,000), which were obtained from Abcam. The following day, the membranes were incubated for 60 min at 37˚C with HRP goat anti-rabbit secondary antibody IgG H&L preadsorbed (cat. no. ab7090; dilution, 1:5,000) obtained from Abcam. The bound antibodies were visualized by enhanced chemiluminescence (Thermo Fisher Scientific, Inc.). Densitometry was used for protein semi-quantification using ImageJ Software version 1.8.0 (National Institutes of Health) and β-actin served as the internal control.
Luciferase reporter gene assay
The sequence of PIM1 3'-UTR with the miR-3135b seed region was amplified from the HCT116 cell cDNA and cloned into the pGL2-control vector (Promega Corporation) and termed the PIM1-wild-type (WT). The sequence of PIM13'-UTR with the mutated miR-3135b seed region was obtained by introducing two-point mutations into the pGL2-PIM1 plasmid using a Quick Site-Directed Mutation Kit (Agilent Technologies Deutchland GmbH), which was defined as PIM1-mutant (MUT). Subsequently, HCT116 cells were seeded into a 24-well plate (3x104 cells/well) and co-transfected with the corresponding vector (100 ng pGL2-PIM1-WT or pGL2-PIM1-MUT) and miR-NC (100 ng) or miR-3135b mimic. (100 ng) Following 48 h of incubation at 37˚C, luciferase activity was determined using the Dual-Luciferase® Reporter Assay (Promega Corporation) on an FB12 Luminometer (Titertek-Berthold). Renilla luciferase activity served as the internal control for normalizing the luciferase activity levels.
Statistical analysis
Statistical analysis was performed using Student's t-test or one-way ANOVA followed by Tukey's multiple comparison post hoc test. Tumor and adjacent non-tumor samples were analyzed using paired Student's t-test. Analysis was conducted using GraphPad Prism 6.0 (GraphPad Software, Inc.). The association between miR-3135b and PIM1 mRNA levels was evaluated using the Pearson correlation coefficient. P<0.05 was considered to indicated a statistically significant difference and all values are presented as means ± SD. For each assay, there were three independent experimental repeats.
Results
Increased miR-3135b expression levels enhance the sensitivity of CRC cells to 5-FU treatment
To investigate the key miRs involved in the mechanism of CRC resistance to 5-FU, the microarray data of miR expression levels were processed (accession. no. GSE81653). The analysis involved 192 patients with CRC who received 5-FU treatment and comprised two main groups defined as 5-FU non-recurrence (n=119) and 5-FU recurrence (n=73). miR-3135b was identified as one of the differentially expressed miRs between these two groups. The level of miR-3135b expression was lower in recurrent patients with 5-FU (n=73) when compared with that of those who were non-recurrent (n=119) as determined by the microarray data (Fig. 1A). In addition, miR-3135b expression levels were lower in tumor tissue samples (n=40) when compared with non-tumor tissue samples (n=40; Fig. 1B and C). The level of miR-3135b expression was not associated with age, sex, tumor site and lymph node metastasis of patients with CRC (Table I). Similar findings were noted in the tumor recurrent (n=17) and non-recurrent (n=17) groups (Fig. 1B and C). Subsequently, the miR-3135b expression levels were lower in CRC cell lines, including HCT116 and SW480 when compared with those observed in the NCM460 cells (Fig. 1D). The results indicated the tumor suppressive function of miR-3135b levels in CRC and the recurrence of CRC in patients that were administered with 5-FU.
Figure 1.
Decreased expression of miR-3135b in CRC tissues and cell lines. (A) miR-3135b expression was lower in patients in the 5-FU recurrence group when compared with those in the 5-FU non-recurrence group. (B) miR-3135b expression was lower in tumor tissue samples when compared with that in non-tumor tissue samples. (C) miR-3135b expression was lower in patients in the tumor recurrent group when compared with those in the tumor non-recurrent group. (D) miR-3135b expression was lower in HCT116 and SW480 CRC cell lines when compared with that in NCM460. **P<0.01 vs. NCM460; ***P<0.001 vs. NCM460, 5-FU non-recurrence, non-tumor tissues and tumor of non-recurrence. miR, microRNA; CRC, colorectal cancer; 5-FU, 5-fluorouracil.
Table I.
Association between miR-3135b expression levels and clinical factors of patients with colorectal cancer.
| Relative miR-3135b expression | ||||
|---|---|---|---|---|
| Variable | Total | High | Low | P-value |
| Age (years) | 0.527 | |||
| <60 | 19 | 8 | 11 | |
| ≥60 | 21 | 12 | 9 | |
| Sex | 0.082 | |||
| Male | 28 | 11 | 17 | |
| Female | 12 | 9 | 3 | |
| Tumor site | 0.999 | |||
| Colon | 23 | 11 | 12 | |
| Rectum | 17 | 9 | 8 | |
| Lymph node metastasis | 0.515 | |||
| Yes | 15 | 6 | 9 | |
| No | 25 | 14 | 11 | |
miR, microRNA.
The function of miR-3135b in CRC cell proliferation and the sensitivity of CRC cells to 5-FU were also investigated. Initially, HCT116 and SW480 cells were transiently transfected with miR-3135b mimic and miR-NC. The increase in miR-3135b expression indicated successful transfection (Fig. 2A). The miR-3135b mimic significantly reduced HCT116 and SW480 cell viability (Fig. 2B and C). In addition, the miR-3135b mimic caused a significant increase in the sensitivity of HCT116 and SW480 cells to 5-FU (Fig. 2D and E).
Figure 2.
Overexpression of miR-3135b increased sensitivity of CRC cells to 5-FU. (A) Higher miR-3135b expression was observed in the miR-3135b mimic group when compared with the miR-NC group. Significantly lower cell viability of (B) HCT116 and (C) SW480 cells was observed in the miR-3135b mimic group when compared with the miR-NC group. The miR-3135b mimic significantly enhanced sensitivity of (D) HCT116 and (E) SW480 cells to 5-FU. *P<0.05, **P<0.01 and ***P<0.001 vs. miR-NC. miR, microRNA; CRC, colorectal cancer; 5-FU, 5-fluorouracil; NC, negative control.
PIM1 is a target gene of miR-3135b
Bioinformatics analysis identified two different complementary sites of PIM1 that could bind to miR-3135b (Fig. 3A). miR-3135b mimic significantly inhibited the relative mRNA expression levels of PIM1 when compared with those of the miR-NC group (Fig. 3B). In addition, similar effects were induced by miR-3135b mimic on the expression levels of PIM1 (Fig. 3C).
Figure 3.
miR-3135b inhibited PIM1 expression. (A) PIM1 was complementary to miR-3135b. The miR-3135b mimic significantly inhibited the (B) relative mRNA and (C) protein expression of PIM1 when compared with miR-NC. ***P<0.001 vs. miR-NC. miR, microRNA; PIM1, proviral integration site for Moloney murine leukemia virus 1; NC, negative control; UTR, untranslated region.
To verify the direct interaction between miR-3135b and PIM1, luciferase reporter assays were performed in HCT116 cells. Three sequences were constructed including one full-length (FL) PIM1 3'-UTR and two truncated forms, namely the truncate 1 (Trun1) form of PIM1 3'-UTR and Trun2 (Fig. 4A). HCT116 cells were co-transfected with FL, Trun1 or Trun2 of PIM1 3'-UTR and the miR-NC or miR-3135b mimic. The luciferase activity was assessed and the data indicated that the lowest activity levels were observed in FL, followed by Trun1 (Fig. 4B). The activity levels were not decreased in the Trun2 group (Fig. 4B), suggesting that the first binding site between PIM1 3'-UTR and miR-3135b was more important. The combining sites between miR-3135b and PIM1, as well as the Mut sequences of PIM1, are presented in Fig. 4C. The miR-3135b mimic significantly inhibited the relative luciferase activity of HCT116 cells transfected with PIM1 3'-UTR-WT when compared with miR-NC, whereas it did not have a significant effect on HCT116 cells transfected with PIM1 3'UTR-Mut (Fig. 4D).
Figure 4.
miR-3135b targeted the PIM1 3'UTR. (A) FL, Trun1 and Trun2 of PIM1 3'UTR are presented. (B) Compared with miR-NC, the relative luciferase activity of HCT116 cells co-transfected with FL was significantly decreased by the miR-3135b mimic, whereas in the HCT116 cells co-transfected with miR-3135b mimic and Trun1 the decrease was significant, but the difference was less. The HCT116 cells co-transfected with Trun2 were not affected by the miR-3135b mimic. (C) The combining sites between miR-3135b and PIM1, as well as the Mut sequences of PIM1 are presented. (D) The miR-3135b mimic significantly inhibited the relative luciferase activity of HCT116 cells transfected with PIM1 3'UTR-WT, but not PIM1 3'UTR-Mut when compared with miR-NC. *P<0.05 and ***P<0.001 vs. miR-NC. miR, microRNA; PIM1, proviral integration site for Moloney murine leukemia virus 1; UTR, untranslated region; FL, full-length; Trun, truncated; Mut, mutant; WT, wild-type; NC, negative control.
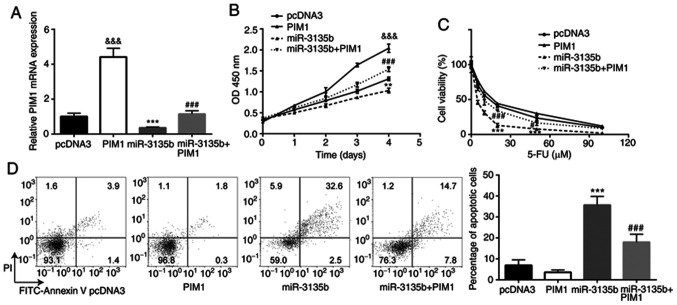
PIM1 overexpression compensates the effects of miR-3135b in CRC cells
The interaction between miR-3135b and PIM1 was assessed with regard to the sensitivity of HCT116 cells to 5-FU. HCT116 cells were transiently transfected with pcDNA3-PIM1 and pcDNA3. The higher PIM1 mRNA expression suggested that successful transfection was achieved (Fig. 5A). pcDNA3-PIM1 also reversed the downregulation of PIM1 expression in cells treated with miR-3135b mimic (Fig. 5A). PIM1 overexpression promoted cell proliferation in HCT116 cells, and miR-3135b overexpression significantly reduced the cell viability of HCT116 cells when compared with the pcDNA3 group, which was significantly compensated by PIM1 overexpression (Fig. 5B). In addition, while PIM1 overexpression showed little effect on 5-FU sensitivity, miR-3135 overexpression significantly increased the sensitivity of HCT116 cells to 5-FU, which was compensated by PIM1 overexpression (Fig. 5C). PIM1 overexpression showed little effect on cell apoptosis, whereas miR-3135b overexpression caused a significant increase in cell apoptosis of HCT116 cells when compared to the pcDNA3 group, which was compensated by PIM1 overexpression (Fig. 5D).
Figure 5.
PIM1 overexpression compensated the effect of miR-3135b in CRC cells. (A) In HCT116 cells, transfection with pcDNA3-PIM1 increased PIM1 expression when compared with pcDNA3. pcDNA3-PIM1 also reversed the miR-3135b mimic-induced downregulation of PIM1. In HCT116 cells, PIM1 overexpression (B) promoted cell proliferation and compensated the miR-3135b overexpression-induced cell viability reduction; (C) upregulated 5-FU sensitivity; and (D) downregulated cell apoptosis. &&&P<0.001 vs. pcDNA3. **P<0.01, ***P<0.001 vs. pcDNA3. #P<0.05 and ###P<0.001 vs. miR-3135b mimic. PIM1, proviral integration site for Moloney murine leukemia virus 1; miR, microRNA; CRC, colorectal cancer; 5-FU, 5-fluorouracil.
Correlation analysis between PIM1 mRNA and miR-3135b expression levels in CRC tumor samples
PIM1 expression levels were higher in tumor tissue samples (n=40) when compared with those in non-tumor tissue samples (n=40; Fig. 6A). Similar findings were observed in PIM1 expression when comparing tumor recurrent cases (n=17) with non-recurrent cases (n=17; Fig. 6B). In addition, an inverse correlation (r=-0.583) was identified between PIM1 mRNA levels and miR-3135b expression in CRC tumor tissue samples (Fig. 6C).
Figure 6.
PIM1 mRNA levels were inversely correlated with miR-3135b expression in CRC tumor samples. (A) PIM1 expression was higher in tumor tissue samples when compared with non-tumor tissue samples. (B) PIM1 expression was higher in the tumor recurrent group when compared with that of the tumor non-recurrent group. (C) An inverse correlation was observed between PIM1 mRNA levels and miR-3135b expression in CRC tumor tissue samples. ***P<0.001. PIM1, proviral integration site for Moloney murine leukemia virus 1; miR, microRNA; CRC, colorectal cancer.
Discussion
In the past decade, significant advances have been made with regard to investigating the molecular mechanisms of tumor resistance to 5-FU. Substantial evidence has been derived from pre-clinical models and clinical trials (16). Considerable attention has been paid to miR dysregulation and the molecular events associated with this process (17). For example, miR-552 downregulation promotes 5-FU resistance by targeting SMAD2 in CRC (18). miR-543 deficiency enhances 5-FU chemosensitivity of CRC cells by targeting PTEN (19). miR-200c can be used as a predictive biomarker for 5-FU sensitivity in CRC (20). In the present study, microarray data analysis indicated that the level of miR-3135b expression was decreased in patients with CRC who experienced disease recurrence following 5-FU treatment compared with those who were non-recurrent. However, to date, to the best of our knowledge, the role of miR-3135b in the sensitivity of CRC to 5-FU has not been investigated.
In the present study, a decrease was observed in the expression levels of miR-3135b in patients with recurrent CRC who were treated with 5-FU when compared with those who were non-recurrent to the disease. Moreover, overexpression of miR-3135b was shown to increase the sensitivity of CRC cells to 5-FU. Taken together, the aforementioned findings demonstrated for the first time, to the best of our knowledge, the tumor suppressive role of miR-3135b in CRC and the effects of its increased expression on the sensitivity of CRC to 5-FU.
miRs are complementary to their target mRNA 3'-UTR (10). The present study identified PIM1 as a novel target gene of miR-3135b in CRC cells. As a member of the constitutively activated serine/threonine kinases, PIM1 was initially identified as a proviral integration site for the Moloney murine leukemia virus in 1989(21). Overexpression of PIM1 is observed in multiple cancer cell types, including glioblastoma (22), lung cancer (23) and CRC (24), and is associated with their development and progression. In addition, PIM1 is associated with cell apoptosis (25), cell proliferation (26), tumor growth (26) and chemotherapy response (27) in various cancer types. Furthermore, PIM1 inhibits cancer cell sensitivity to 5-FU. Human antigen R induces hypoxia-mediated chemoresistance by upregulation of PIM1 protein in pancreatic cancer (28). Based on this evidence, the development of combinatorial therapies for CRC treatment can be mediated by increasing 5-FU sensitization following PIM1 knockdown, which is targeted by miR-15b (29). However, to the best of our knowledge, the interaction between PIM1 and miR-3135b in CRC and its effects on the sensitivity of CRC cells to 5-FU have not previously been reported.
The present study demonstrated that PIM1 overexpression compensated the effect of miR-3135b in CRC cells. An inverse correlation between PIM1 mRNA expression and miR-3135b expression was also observed. The downregulation of miR-3135b in CRC cell lines and clinical tumor samples has recently been reported; ectopic expression of miR-3135b sensitized cells to 5-FU by targeting Golgi phosphoprotein-3(30). These observations are consistent with the present findings on the effects of miR-3135b on the sensitivity of CRC cells to 5-FU.
Therefore, the present study identified miR-3135b as a novel regulator of 5-FU sensitivity in CRC by targeting PIM1, implicated a potential new therapeutic target for the treatment of patients with CRC. However, there were certain limitations of the present study. Further investigation into whether miR-3135b regulates 5-FU resistance in established 5-FU resistance cell lines is required. Furthermore, the function and mechanism of miR-3135b in animals, such as nude mice, will be investigated in future work.
Acknowledgements
Not applicable.
Funding Statement
Funding: The present study was supported by the International Cooperation of Jilin Provincial Science & Technology Department (grant. nos. 20150101175JC and 20190201221JC) and the National Natural Science Foundation of China (grant. nos. 81172000 and 30772488).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YW, XC and SM performed the experiments, analyzed the data, and confirm the authenticity of all the raw data. HZ designed the experiments. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of ChinaJapan Union Hospital of Jilin University (Changchun, China). Written informed consent was provided by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Brody H. Colorectal cancer. Nature. 2015;521(S1) doi: 10.1038/521S1a. [DOI] [PubMed] [Google Scholar]
- 3.Pourhoseingholi MA. Increased burden of colorectal cancer in Asia. World J Gastrointest Oncol. 2012;4:68–70. doi: 10.4251/wjgo.v4.i4.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.García-Osogobio S, Téllez-Ávila FI, Méndez N, Uribe-Esquivel M. Results of the first program of colorectal cancer screening in Mexico. Endoscopia. 2015;27:59–63. [Google Scholar]
- 5.Arnold D, Prager GW, Quintela A, Stein A, Vera SM, Mounedji N, Taieb J. Beyond second-line therapy in patients with metastatic colorectal cancer: A systematic review. Ann Oncol. 2018;29:835–856. doi: 10.1093/annonc/mdy038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stintzing S. Management of colorectal cancer. F1000Prime Rep. 2014;6(108) doi: 10.12703/P6-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vodenkova S, Buchler T, Cervena K, Veskrnova V, Vodicka P, Vymetalkova V. 5-fluorouracil and other fluoropyrimidines in colorectal cancer: Past, present and future. Pharmacol Ther. 2020;206(107447) doi: 10.1016/j.pharmthera.2019.107447. [DOI] [PubMed] [Google Scholar]
- 8.Liu C, Zhao Y, Wang J, Yang Y, Zhang Y, Qu X, Peng S, Yao Z, Zhao S, He B, et al. FoxO3 reverses 5-fluorouracil resistance in human colorectal cancer cells by inhibiting the Nrf2/TR1 signaling pathway. Cancer Lett. 2020;470:29–42. doi: 10.1016/j.canlet.2019.11.042. [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Wang XJ, Reyes JL, Chua NH, Gaasterland T. Prediction and identification of arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5(R65) doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trang P, Weidhaas JB, Slack FJ. MicroRNAs as potential cancer therapeutics. Oncogene. 2008;27 (Suppl 2):S52–S57. doi: 10.1038/onc.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barisciano G, Colangelo T, Rosato V, Muccillo L, Taddei ML, Ippolito L, Chiarugi P, Galgani M, Bruzzaniti S, Matarese G, et al. miR-27a is a master regulator of metabolic reprogramming and chemoresistance in colorectal cancer. Br J Cancer. 2020;122:1354–1366. doi: 10.1038/s41416-020-0773-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu T, Zhang X, Du L, Wang Y, Liu X, Tian H, Wang L, Li P, Zhao Y, Duan W, et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer. 2019;18(43) doi: 10.1186/s12943-019-0981-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Choi Y, Choi HS, Jeon WK, Kim BI, Park DI, Cho YK, Kim HJ, Park JH, Sohn CI. Optimal number of endoscopic biopsies in diagnosis of advanced gastric and colorectal cancer. J Korean Med Sci. 2012;27:36–39. doi: 10.3346/jkms.2012.27.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Xie P, Mo JL, Liu JH, Li X, Tan LM, Zhang W, Zhou HH, Liu ZQ. Pharmacogenomics of 5-fluorouracil in colorectal cancer: Review and update. Cell Oncol (Dordr) 2020;43:989–1001. doi: 10.1007/s13402-020-00529-1. [DOI] [PubMed] [Google Scholar]
- 17.Blondy S, David V, Verdier M, Mathonnet M, Perraud A, Christou N. 5-Fluorouracil resistance mechanisms in colorectal cancer: From classical pathways to promising processes. Cancer Sci. 2020;111:3142–3154. doi: 10.1111/cas.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao P, Ma YG, Zhao Y, Liu D, Dai ZJ, Yan CY, Guan HT. MicroRNA-552 deficiency mediates 5-fluorouracil resistance by targeting SMAD2 signaling in DNA-mismatch-repair-deficient colorectal cancer. Cancer Chemother Pharmacol. 2019;84:427–439. doi: 10.1007/s00280-019-03866-7. [DOI] [PubMed] [Google Scholar]
- 19.Liu G, Zhou J, Dong M. Down-regulation of miR-543 expression increases the sensitivity of colorectal cancer cells to 5-Fluorouracil through the PTEN/PI3K/AKT pathway. Biosci Rep. 2019;39(BSR20190249) doi: 10.1042/BSR20190249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dermani FK, Najafi R. miR-200c as a predictive biomarker for 5-Fluorouracil chemosensitivity in colorectal cancer. J Gastrointest Cancer. 2018;49:102–103. doi: 10.1007/s12029-017-0038-3. [DOI] [PubMed] [Google Scholar]
- 21.van Lohuizen M, Verbeek S, Krimpenfort P, Domen J, Saris C, Radaszkiewicz T, Berns A. Predisposition to lymphomagenesis in pim-1 transgenic mice: Cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell. 1989;56:673–682. doi: 10.1016/0092-8674(89)90589-8. [DOI] [PubMed] [Google Scholar]
- 22.Herzog S, Fink MA, Weitmann K, Friedel C, Hadlich S, Langner S, Kindermann K, Holm T, Böhm A, Eskilsson E, et al. Pim1 kinase is upregulated in glioblastoma multiforme and mediates tumor cell survival. Neuro Oncol. 2015;17:223–242. doi: 10.1093/neuonc/nou216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang R, Wang X, Jin Z, Li K. Association of nuclear PIM1 expression with lymph node metastasis and poor prognosis in patients with lung adenocarcinoma and squamous cell carcinoma. J Cancer. 2016;7:324–334. doi: 10.7150/jca.13422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang M, Liu T, Sun H, Weng W, Zhang Q, Liu C, Han Y, Sheng W. Pim1 supports human colorectal cancer growth during glucose deprivation by enhancing the warburg effect. Cancer Sci. 2018;109:1468–1479. doi: 10.1111/cas.13562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu JJ, Wang Z, Reeves R, Magnuson NS. PIM1 phosphorylates and negatively regulates ASK1-mediated apoptosis. Oncogene. 2009;28:4261–4271. doi: 10.1038/onc.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Chen L, Luo C, ChenYan Ge J, Zhu Z, Wang K, Yu X, Lei J, Liu T, et al. TAB3 upregulates PIM1 expression by directly activating the TAK1-STAT3 complex to promote colorectal cancer growth. Exp Cell Res. 2020;391(111975) doi: 10.1016/j.yexcr.2020.111975. [DOI] [PubMed] [Google Scholar]
- 27.Braso-Maristany F, Filosto S, Catchpole S, Marlow R, Quist J, Francesch-Domenech E, Plumb DA, Zakka L, Gazinska P, Liccardi G, et al. PIM1 kinase regulates cell death, tumor growth and chemotherapy response in triple-negative breast cancer. Nat Med. 2016;22:1303–1313. doi: 10.1038/nm.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanco FF, Jimbo M, Wulfkuhle J, Gallagher I, Deng J, Enyenihi L, Meisner-Kober N, Londin E, Rigoutsos I, Sawicki JA, et al. The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene PIM1 in pancreatic cancer cells. Oncogene. 2016;35:2529–2541. doi: 10.1038/onc.2015.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weirauch U, Beckmann N, Thomas M, Grünweller A, Huber K, Bracher F, Hartmann RK, Aigner A. Functional role and therapeutic potential of the pim-1 kinase in colon carcinoma. Neoplasia. 2013;15:783–794. doi: 10.1593/neo.13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Núñez-Olvera SI, Chávez-Munguía B, Del Rocío Terrones-Gurrola MC, Marchat LA, Puente-Rivera J, Ruíz-García E, Campos-Parra AD, Vázquez-Calzada C, Lizárraga-Verdugo ER, Ramos-Payán R, et al. A novel protective role for microRNA-3135b in golgi apparatus fragmentation induced by chemotherapy via GOLPH3/AKT1/mTOR axis in colorectal cancer cells. Sci Rep. 2020;10(10555) doi: 10.1038/s41598-020-67550-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.