Abstract
The Drosophila seven in absentia (sina) gene was initially discovered because its inactivation leads to R7 photoreceptor defects. Recent data indicate that Sina binds to the Sevenless pathway protein Phyllopod, and together they mediate degradation of Tramtrack, a transcriptional repressor of R7 cell fate. Independent studies have shown that Sina and its highly related mammalian homologues Siah-1 and Siah-2 bind to the DCC (deleted in colorectal cancer) protein and promote its proteolysis via the ubiquitin-proteasome pathway. To determine the roles of mammalian Siahs in proteolysis and their interactions with target proteins, we sought to define Siah-1 domains critical for regulation of DCC. Mutant Siah-1 proteins, harboring missense mutations in the carboxy (C)-terminal domain analogous to those present in Drosophila sina loss-of-function alleles, failed to promote DCC degradation. Point mutations and deletion of the amino (N)-terminal RING finger domain of Siah-1 abrogated its ability to promote DCC proteolysis. In the course of defining Siah-1 sequences required for DCC degradation, we found that Siah-1 is itself rapidly degraded via the proteasome pathway, and RING domain mutations stabilized the Siah-1 protein. Siah-1 was found to oligomerize with itself and other Sina and Siah proteins via C-terminal sequences. Finally, evidence that endogenous Siah-1 regulates DCC proteolysis in cells was obtained through studies of an apparent dominant negative mutant of Siah-1, as well as via an antisense approach. The data indicate that the Siah-1 N-terminal RING domain is required for its proteolysis function, while the C-terminal sequences regulate oligomerization and binding to target proteins, such as DCC.
The development of the R7 photoreceptor cell in the Drosophila compound eye has been highly amenable to study, and many genes that specify its fate have been identified and characterized. The neuronal specification of the R7 cell requires a receptor tyrosine kinase encoded by the sevenless gene, the interaction of the Sevenless protein with the Boss ligand on the neighboring R8 cell, and downstream signaling molecules, including the Ras, Raf, and MAPK (mitogen-activated protein kinase) proteins (23, 27). In R7 cells, activation of the sevenless pathway results in gene expression changes, including the induction of the phyllopod (phyl) gene (5, 6). In addition to genes in the sevenless pathway, others, such as seven in absentia (sina) and tramtrack (ttk), have critical roles in R7 cell fate determination (4, 12, 26).
Until recently, however, the relationship of the Sina and Ttk proteins to the sevenless pathway was poorly understood. Recent studies have demonstrated that the Sina and Phyl proteins form a ternary complex with Ttk and promote ubiquitination and rapid degradation of Ttk through the proteasome pathway (14, 22). This is a critical event in R7 determination, because Ttk is a potent repressor of neuronal cell fate. The results of the Drosophila studies are consistent with the following hypotheses: (i) Phyl functions as bridging factor between Ttk and Sina; and (ii) Sina has the critical role in promoting ubiquitination and proteasome degradation of Ttk. Further support for this model and for a more general role for Sina in ubiquitin-proteasome proteolysis has been obtained through independent studies of Sina and its highly related mammalian homologues Siah-1 and Siah-2. Specifically, Sina and Siah proteins were found to bind to the cytoplasmic domain of the DCC (deleted in colorectal cancer) protein and to promote its degradation via the proteasome pathway (9). In addition, evidence was obtained that the Sina and Siah proteins may interact directly with ubiquitin-conjugating proteins (9, 22).
The sequences of the Sina and Siah proteins do not offer clues with respect to their specific biochemical function in proteolysis. Drosophila Sina is 314 amino acids long, and the only sequence motif of Sina with obvious similarity to other well-characterized proteins is an N-terminal cysteine-rich domain of the C3HC4 or RING zinc finger type (4). The human Siah-1 protein is 282 amino acids long, and human Siah-2 is 324 amino acids long (10). The two human Siah proteins differ from one another and from Drosophila Sina essentially only in the length and sequence content of their most amino (N)-terminal sequences (10). Over their carboxy (C)-terminal 250 amino acids, the three proteins share more than 85% amino acid identity.
While recent studies have implicated the Sina and Siah proteins in the degradation of specific target proteins, few definitive insights have been obtained into the specific means by which the Sina and Siah proteins carry out this function, particularly in mammalian cells. To further explore this issue, we sought to define domains in the human Siah-1 protein that are critical for promoting DCC protein degradation. We first generated three different mutated Siah-1 proteins, each with a missense substitution in the C-terminal domain analogous to those present in three previously described Drosophila sina mutant alleles (4). The basis for this approach was that these particular mutated alleles are the only known sina alleles with localized inactivating mutations. Two of the three Siah-1 proteins with missense mutations failed to promote DCC degradation. Missense mutations and deletion of the N-terminal RING domain of Siah-1 abrogated its ability to promote DCC proteolysis. Through our studies, we found that Siah-1 is itself rapidly degraded, and RING domain mutations greatly stabilized its expression. Siah-1 was found to oligomerize with itself, as well as Sina and Siah-2, via its C-terminal sequences. Further evidence that Siah-1 regulates DCC expression in cells was obtained by employing an antisense approach as well as a mutant Siah-1 protein with dominant negative activity. Using immunofluorescence microscopy, we found that the RING domain of Siah-1 regulates its localization in the cell. Our results indicate that the N-terminal RING domain of Siah-1 is required for its proteolysis function, while the C-terminal sequences of Siah-1 may regulate its oligomerization and binding to target proteins.
MATERIALS AND METHODS
Siah-1 mutant expression constructs.
Three Siah-1 proteins with missense mutations in their C termini were generated by site-directed mutagenesis on a SIAH-1 cDNA, using two rounds of PCR (8). In brief, to generate each mutant, two overlapping PCR fragments of SIAH1 cDNA were amplified in the first round of PCR, using outer primers and specific internal primers derived from the sequence region to be mutated. In the second round of PCR, the mutagenized full-length SIAH1 coding region was generated with only outer primers and the first-round PCR products as templates. A Flag epitope tag sequence (DYKDDDDK) was present in the outer forward primer. The primer sequences were as follows: SH1-1S, 5′-AGGAATTCACAGAAATGAGCGACTACAAGGACGACGATGACAAGCGTCAGACTGCTACAGCATTAC-3′ (outer forward primer); SH1-2A, 5′-GCACTAGTTGATTGCCATTTCAACACATGG-3′ (outer reverse primer); SH1-4S, 5′-CTGATGCATCAGTATAAGTCCATTAC-3′ (inner forward primer for SIAH1-Y152); SH1-4A, 5′-GTAATGGACTTATACTGATGCATCAG-3′ (inner reverse primer for SIAH1-Y152); SH1-6S, 5′-AAATACGATGGTTACCAGCAGTTCT-3′ (inner forward primer for SIAH1-Y202); SH1-6A, 5′-AGAACTGCTGGTAACCATCGTATTT-3′ (inner reverse primer for SIAH1-Y202); SH1-7S, 5′-CAATCGTACAGCGGATAGGAACAC-3′ (inner forward primer for SIAH1-R211); and SH1-7A, 5′-GTGTTCCTATCCGCTGTACGATTG-3′ (inner reverse primer for SIAH1-Y211). The SIAH1 cDNAs with these specific C-terminal missense mutations were then cloned downstream of the cytomegalovirus promoter-enhancer in the mammalian expression vector pcDNA3 (Invitrogen, San Diego, Calif.). The Siah-1 mutant with the RING finger deleted (SIAH1-dR) was also constructed by two rounds of PCR. During the first round of PCR, two SIAH1 fragments were amplified using the outer forward and reverse primers described above and the following two internal primers: SH1dRP2S (5′-TTTATCGATCCTCTCGAGCCTTTGGGATCCATTCGCAACTTGGCTA-3′) and SH1dRP1A (5′-TCCCTCGAGAGGATCGATAAAACTCGCCAAGTCATTGTTGG-3′). The full-length cDNA fragment of SIAH1-dR, generated by a second round of PCR with outer primers and the first-round PCR fragments as templates, was inserted into pcDNA3. Constructs encoding Siah-1 mutant proteins with four single and three double missense mutations in the RING domain were generated by site-specific mutagenesis with the QuikChange kit (Stratagene, La Jolla, Calif.) and specific oligonucleotides for each of the single and double missense mutations. Further details of the generation of the missense mutations in the RING domain are available from the authors. A hemagglutinin (HA) epitope-tagged SIAH1-dR expression construct was generated by switching the Flag-tagged SIAH1-dR fragment in pcDNA3 with a PCR-amplified SIAH1-dR cDNA fragment containing the HA epitope tag (YPYDVPDYA). Amino-terminal Flag-tagged SIAH1-dC1 (amino acids 2 to 262) and SIAH1-dC2 (amino acids 2 to 230) cDNA fragments were obtained by PCR by using similar methods and cloned into the pcDNA3 vector. Pfu DNA polymerase (Stratagene) was used for all PCR, and the authenticity of the sequences of all PCR products was confirmed by sequencing. The construction of pcDNA3-Flag-SIAH1-dN (previously termed FLAG-Siah-1Δ, aa 77-282) and pcDNA-Myc-Sina-dC (previously termed SinaTNMyc, aa 2-199) has been described elsewhere (9).
Cell culture and DNA transfection.
COS-1 and 293 cells were obtained from the American Type Culture Collection (Rockville, Md.). Cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics. DNA transfections were carried out by using Lipofectamine reagent (GIBCO BRL/Life Technologies, Gaithersburg, Md.) and Opti-MEM reduced serum medium, per the manufacture’s protocol.
RNA isolation and Northern blot analysis.
Total RNA was isolated with Trizol reagent (GIBCO BRL). Fifteen micrograms of total RNA from each sample was electrophoresed on a 1.5% agarose-formaldehyde gel. After electrophoresis, RNA was transferred by capillary action onto a Zeta-Probe GT membrane (Bio-Rad Laboratories, Hercules, Calif.) in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate; pH 7.0) buffer for 12 h. The membrane was rinsed in 2× SSC, air dried briefly, and baked at 80°C for 0.5 h, and Northern hybridization was performed as described previously (10).
In vitro binding assay.
Radiolabeled Siah-1 wild-type and mutant proteins were generated by in vitro transcription and translation, using TNT T7 Quick Coupled Transcription/Translation system (Promega, Madison, Wis.) and [35S]methionine (Amersham Life Science, Arlington Heights, Ill.). The GST-Sina fusion construct was generated by cloning the full-length Drosophila Sina sequence in frame with glutathione transferase (GST) sequences in the pGX-2TK vector (Pharmacia Biotech, Uppsala, Sweden). The construct was verified by sequencing. A GST-DCC construct containing 300 amino acids of the DCC cytoplasmic domain has been previously described (18). The GST-Sina and GST-DCC fusion proteins were purified, using protocols provided by the manufacturer. The in vitro binding assays with the GST-DCC or GST-Sina fusion proteins were carried out as previously described (9).
Western blotting and immunoprecipitation.
Forty-eight hours after transfection of COS-1 or 293 cells with the Siah-1, DCC, and/or control pcDNA3 expression constructs, the cells were lysed in TBS-Triton lysis buffer (Tris-buffered saline [TBS] [pH 8.0], 1% Triton X-100, 10 μg of phenylmethylsulfonyl fluoride per ml, 50 μg of antipain per ml, 5 μg of aprotinin per ml, 2 μg of leupeptin per ml). Cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to Immobilon membranes (Millipore, Marlborough, Mass.), followed by Western blot analysis with DCC monoclonal antibody (G97-13; PharMingen, San Diego, Calif.), anti-Flag M2 antibody (Eastman Kodak, New Haven, Conn.), or anti-HA antibody (12CA5; Boehringer Mannheim, Indianapolis, Ind.). Immunoprecipitation of Siah-1 proteins with the anti-Flag M2 antibody was carried out in TBS-Triton lysis buffer supplemented with 1% bovine serum albumin (BSA) by standard protocols. For the MG132 studies, the transfected cells were treated for 6 h at 37°C with various concentrations of MG132 (Calbiochem-Novabiochem, La Jolla, Calif.) in dimethyl sulfoxide or with dimethyl sulfoxide alone, as a control.
Immunofluorescence studies.
Transfected COS-1 cells in chamber slides (Nalge Nunc International, Naperville, Ill.) were fixed in freshly prepared 2% paraformaldehyde–phosphate-buffered saline (PBS) at room temperature. Fixed cells were washed twice with PBS, preincubated in the staining solution of PBS with 1% goat serum and 0.1% saponin (Sigma, St. Louis, Mo.) for 10 min, and then incubated with the anti-Flag M2 monoclonal antibody (1:1,000 dilution) for 1.5 h. Subsequently, the cells were rinsed twice with PBS, incubated in the staining solution for 5 min, and stained with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse antibody (double-labeling grade; Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.) for 1 h in the dark. Cells were twice rinsed with PBS and incubated with a 1-μg/ml solution of 4′,6′-diamidine-2′-phenylindole dihydrochloride (DAPI; Boehringer Mannheim) in PBS for 15 min at 37°C. Cells were then rinsed with PBS once and air dried briefly, and coverslips were mounted with Vectashield H-1000 (Vector Laboratories, Inc., Burlingame, Calif.) medium. Slides were viewed with an immunofluorescence AX-70 microscope (Olympus, Lake Success, N.Y.), and the images were obtained with a PM-30 photomicrographic system (Olympus).
RESULTS
Siah-1 mutant proteins fail to promote DCC degradation.
As noted above, the human Siah-1 and Siah-2 proteins are highly conserved with one another and Drosophila Sina, with the three proteins sharing >85% identity over their C-terminal 250 amino acids (10). Because of the high degree of conservation, we generated three SIAH-1 alleles encoding single missense substitutions in the C-terminal half of Siah-1 (Fig. 1), each analogous to a mutated sina allele previously seen in Drosophila mutants with R7 defects (4). When homozygous, each of the sina missense alleles displays a weak eye morphology phenotype, in contrast to the strong phenotypes resulting from premature truncation of the Sina protein product. These three Siah-1 missense mutations were present at positions with identical sequence in all known vertebrate homologues of Sina, and the mutations generated were as follows: histidine-to-tyrosine substitution in codon 152, histidine-to-tyrosine substitution in codon 202, and leucine-to-arginine substitution in codon 211. In addition to the missense mutations, we also generated a mutated Siah-1 protein with an in-frame deletion of the roughly 40-amino-acid RING domain (i.e., SIAH1-dR [Fig. 1]). Despite repeated attempts, we have failed to produce polyclonal antibodies against the Siahs. Thus, the wild-type and mutated Siah-1 proteins were tagged with a Flag epitope at their N termini to allow their detection.
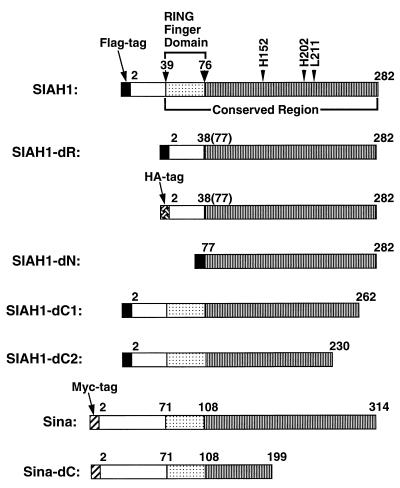
FIG. 1.
Schematic representation of Siah-1 and Sina proteins encoded by the expression vector constructs. For each protein, the locations of the N-terminal Flag (black box), HA (cross-hatched box), and c-Myc (hatched box) epitope tags are shown, as are the specific Siah or Sina sequences. Note that the full-length Siah-1 protein has 282 amino acids, and the full-length Drosophila Sina protein has 314 amino acids. The positions of the C-terminal missense substitutions are indicated, as are the sequences deleted in the various deletion mutants.
We assessed the ability of these four Siah-1 mutant proteins to regulate DCC protein expression. While the Flag-tagged wild-type Siah-1 protein was readily able to reduce DCC expression in transfected COS-1 cells (Fig. 2A, lane 2), two of the Siah-1 C-terminal missense mutants (Y152 and R211 [lanes 3 and 5]) and the RING-deleted form (lane 6) failed to decrease DCC expression. In fact, not only did the RING-deleted form of Siah-1 fail to reduce DCC expression, it appeared to increase expression of DCC over that of the empty vector control (Fig. 2A, compare lanes 1 with 6). This apparent dominant negative effect of the RING-deleted Siah-1 protein will be further explored below. No effect on the ability to regulate DCC was seen when the Flag-tagged wild-type Siah-1 protein was compared to a Siah-1 protein lacking an epitope tag (data not shown).
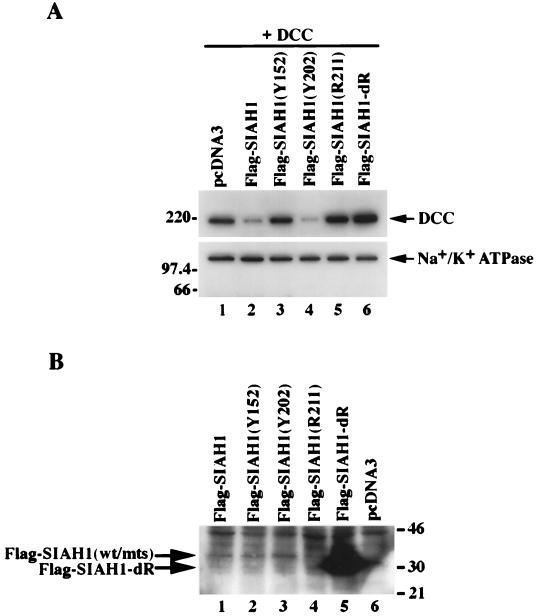
FIG. 2.
Regulation of DCC protein expression by wild-type and mutant Siah-1 proteins. (A) Wild-type Siah-1, but not certain mutated forms, promote DCC degradation. Western blot analysis of DCC expression in COS-1 cells cotransfected with pcDNA3 expression vectors containing the following cDNAs was done: DCC (lanes 1 to 6) and Flag epitope-tagged wild-type SIAH1 (lanes 2), Flag epitope-tagged and mutated SIAH1 cDNAs (lanes 3 to 6), or the empty pcDNA3 expression vector (lanes 1). Forty-eight hours after transfection, enhanced chemiluminescence (ECL)-Western blot analysis was carried out on the cell lysates, using the DCC extracellular domain monoclonal antibody G92-13. The membrane was then stripped, and Western blotting with a polyclonal antibody against Na+/K+ ATPase was performed to confirm equal loading of the lanes. The migration positions (in kilodaltons) of selected molecular mass markers are indicated to the left of the blots. (B) Western blot analysis of expression of wild-type (wt) and mutant (mts) forms of Siah-1. The cell lysates described in panel A were analyzed by ECL-Western blotting using the anti-Flag M2 monoclonal antibody. Extremely low levels of expression of the Flag-Siah-1 protein (lane 1) and three missense mutants (lanes 2 to 4) were detected only after long exposure to film. The strong signal in lane 5 represents the faster-migrating, RING-deleted form of Siah-1 (Flag-Siah-1-dR). The migration positions (in kilodaltons) of selected protein molecular mass markers are indicated to the right of the blot.
To establish that the mutated Siah-1 proteins were expressed following transfection, we carried out Western blotting with an antibody directed against the Flag epitope. The Flag-tagged wild-type Siah-1 protein was expressed at low levels (Fig. 2B, lane 1). Attempts to immunoprecipitate detectable amounts of a 35S-labeled, Flag-tagged wild-type Siah-1 protein were unsuccessful (data not shown), suggesting that Siah-1 is rapidly degraded in mammalian cells. Similar to wild-type Siah-1, low-level expression of Siah-1 proteins with the C-terminal missense substitutions was seen (Fig. 2B, lanes 2 to 4). In contrast, the RING-deleted Siah-1 protein was highly expressed (lane 5). To establish that the elevated expression of the RING-deleted protein was not attributable to differences in gene expression following transfection, we performed Northern blot studies of the transfected cells, using a SIAH-1 cDNA probe. No differences in expression of the transfected SIAH-1 cDNAs were observed (Fig. 3). Hence, the increased expression of the RING-deleted Siah-1 protein results form posttranscriptional events and likely reflects an increase in its stability relative to the wild-type and missense forms of Siah-1.
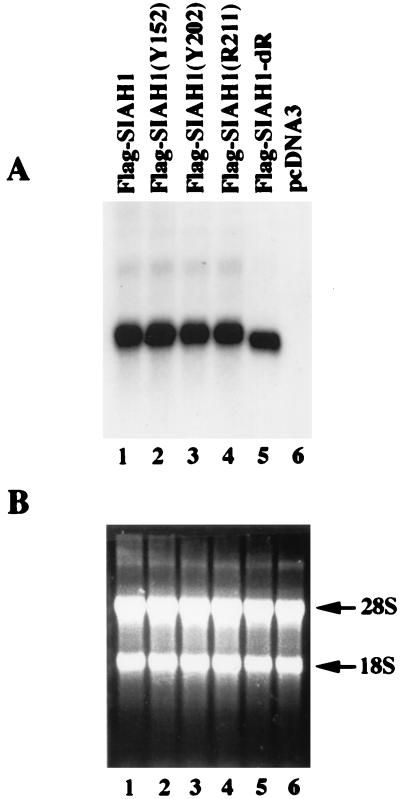
FIG. 3.
Reduced expression of wild-type Siah-1 protein and certain mutants is not due to reduced gene expression. (A) Northern blot analysis of the expression of SIAH1 transcripts following transfection of the SIAH1 cDNA constructs into COS-1 cells. The indicated SIAH1 expression constructs (lanes 1 to 5) or the empty vector control (lane 6) were transfected into COS-1 cells. Forth-eight hours after transfection, total RNA was extracted with Trizol reagent and Northern blotting was carried out. The blot was probed with a 32P-labeled SIAH1 cDNA fragment. Similar levels of wild-type and mutated SIAH1 transcripts were observed, in all lanes except the negative control (lane 6). (B) Ethidium bromide staining of the RNAs verifies equivalent loading. The 28S and 18S ribosomal bands are indicated.
Proteasome degradation of Siah-1 requires the RING domain.
Because our previous studies demonstrated that Sina and Siah proteins regulate DCC expression through the proteasome pathway (9), we inferred that deletion of the RING domain inactivated the function of the mutated Siah-1 protein in proteasome-mediated degradation of DCC. The markedly increased expression of the RING-deleted Siah-1 protein implied that the RING domain might also be critical in regulating the stability of Siah-1 itself via the proteasome pathway. Hence, we sought to determine whether Siah-1 expression was increased following treatment of cells with the peptide aldehyde MG132, a potent inhibitor of proteasome function. A 6-h treatment of transfected cells with various doses of MG132 resulted in a clear increase in wild-type Siah-1 expression, but MG132 was not able to further elevate expression of the RING-deleted form of Siah-1 (Fig. 4A). Like wild-type Siah-1, increased expression of the three Siah-1 proteins with C-terminal missense mutations was also seen following treatment of the cells with MG132 (Fig. 4B). The 20 to 50 μM concentrations of MG132 are well within the range in which MG132 specifically inhibits proteasome activity in cells (17, 19), supporting the view that the proteasome pathway has a major role in regulating turnover of Siah-1.
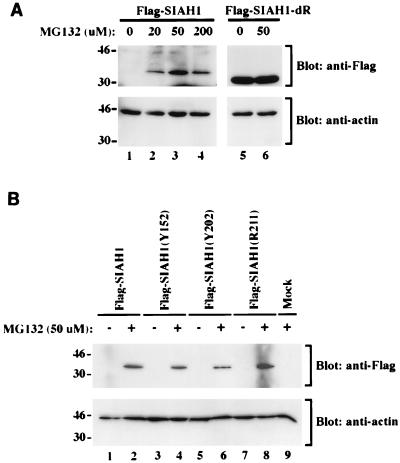
FIG. 4.
Regulation of Siah-1 protein expression by the proteasome pathway. (A) Increased expression of wild-type Siah-1 expression, but not the RING-deleted form, following treatment of cells with the proteasome inhibitor MG132. cDNAs encoding Flag-tagged wild-type Siah-1 and the RING-deleted form of Siah-1 (SIAH1-dR) were transfected into COS-1 cells, as indicated. Forty-eight hours after transfection, the cells were treated for 6 h with various concentrations of MG132, as indicated. Cell lysates were prepared, and ECL-Western blot analysis with the anti-Flag M2 antibody was performed. The blot was then stripped and reprobed with anti-actin polyclonal antibody C11 to verify the loading. The migration positions (in kilodaltons) of selected markers are shown to the left of the blots. (B) Expression of Siah-1 proteins with missense mutations is also increased following treatment of cells with MG132. Studies essentially identical to those in panel A were carried out. A no DNA “mock” negative-control transfection is shown in lane 9. The migration positions (in kilodaltons) of selected protein markers are shown to the left of the blots.
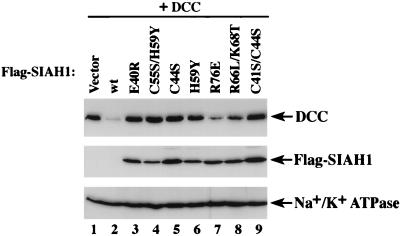
Because deletions may affect the folding and function of proteins in unexpected ways, we also carried out studies to demonstrate that localized missense mutations of highly conserved residues in the RING domain, including those at the cysteine or histidine residues that define the motif, abrogated the ability of Siah-1 to promote DCC degradation as well as the rapid turnover of the Siah-1 protein itself. As shown in Fig. 5, single missense and double missense mutations in the RING domain abrogated the ability of Siah-1 to promote DCC degradation. In addition, like the RING-deleted form, stable expression of Siah-1 proteins with missense mutations in the RING domain was seen. Therefore, we conclude that the RING domain is required for the proteolysis function of Siah-1.
FIG. 5.
Localized mutations in the RING domain abrogate the ability of Siah-1 to promote DCC degradation and result in stable expression of the mutant Siah-1 protein. ECL-Western blot studies of lysates from COS-1 cells harvested 48 h after cotransfection with an expression construct for DCC and a pcDNA3 expression construct containing no insert (vector) (lane 1), wild-type (wt) Siah-1 (lane 2), or a cDNA for a specific mutant Siah-1 protein with a single or double missense substitution in the RING domain (lanes 3 to 9) are shown. The various missense mutants of Siah-1 studied are indicated above the respective lane. DCC protein expression was detected with the G92-13 monoclonal antibody and expression of Siah-1 proteins was detected by Western blotting with an anti-Flag M2 monoclonal antibody reactive with the epitope tag at their N termini. The DCC and Flag Western blots were generated in parallel, and the Flag immunoblot was stripped and reprobed with an antibody against Na+/K+ ATPase to confirm equal loading of the lanes.
Siah-1 binding to DCC and oligomerization occurs via C-terminal sequences.
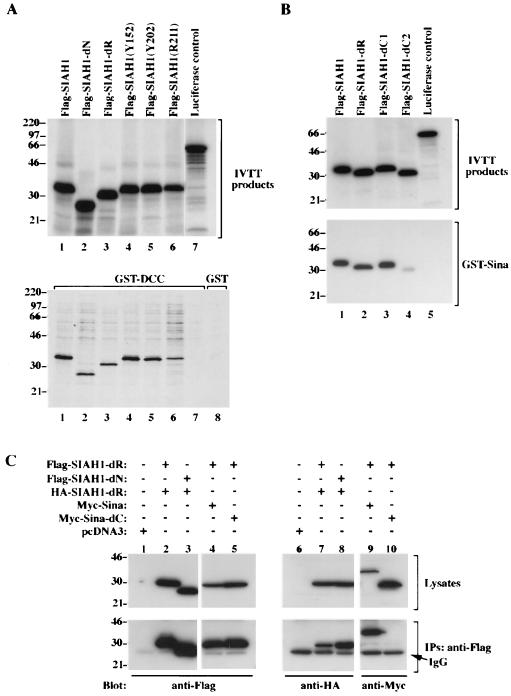
In our prior Saccharomyces cerevisiae two-hybrid studies of Siah-2 binding to DCC, we found that sequences between Siah-2 amino acid 184 and the C terminus were sufficient for binding to DCC (9). These Siah-2 sequences correspond to amino acid 144 of the C terminus of Siah-1 (i.e., the C-terminal 138 amino acids of Siah-1) (10). Given the high conservation between the C-terminal Siah-1 and Siah-2 sequences, we expected that the Siah-1 N-terminal region, including the RING domain, was dispensable for binding to DCC. Nevertheless, we sought to confirm this directly. In addition, we sought to determine whether the three Siah-1 proteins with missense mutations in the C-terminal region (i.e., codons 152, 202, and 211) retained binding to DCC. The wild-type and mutated Siah-1 proteins were synthesized in vitro and tested for their binding to a recombinant GST-DCC protein immobilized on glutathione-agarose beads. Wild-type Siah-1 bound strongly to DCC (Fig. 6A, lane 1). As predicted, Siah-1 proteins with N-terminal deletions retained strong binding to DCC (Fig.1 and Fig. 6A, lanes 2 and 3). In addition, while sequences in the C-terminal 138 amino acids of Siah-1 appeared to be required for DCC binding and two of the three C-terminal missense mutants failed to promote DCC degradation in transfected cells, all three missense mutants retained binding to DCC in vitro (Fig. 6A).
FIG. 6.
In vitro and in vivo interactions of Sina and Siah proteins. (A) Wild-type and mutant Siah-1 proteins bind to DCC in vitro. In the upper panel, the products of coupled in vitro transcription and translation (IVTT) of wild-type and mutant SIAH1 cDNAs are shown. Essentially equivalent amounts of [35S]methinonine-labeled wild-type and mutated Siah-1 proteins were generated, and the proteins are Flag-Siah-1 (lane 1), Flag-Siah-1-dN (lane 2), Flag-Siah-1-dR (lane 3), Flag-Siah-1(Y152) (lane 4), Flag-Siah-1(Y202) (lane 5), Flag-Siah-1(R211) (lane 6), and a luciferase control (lane 7). In the lower panel, the in vitro binding of these proteins to a recombinant GST-DCC protein (lanes 1 to 8) is shown. In lane 8, the absence of binding of the Flag-Siah-1 protein to a control GST protein is shown. (B) In vitro binding of Siah-1 to the Sina protein. Wild-type and mutated Siah-1 proteins as well as a control luciferase protein were synthesized in vitro and are shown in the upper panel. The in vitro binding assay was carried out with a recombinant GST protein containing full Sina sequences (lower panel). (C) Siah-1 and Sina proteins form homo- and heterooligomers in cells. 293 cells were contransfected with expression constructs containing cDNAs for Flag-Siah-1-dR (Flag, amino acids [aa] 2 to 38 and 77 to 282), Flag-Siah-1-dN (Flag, aa 77 to 282), HA-Siah-1-dR (HA, aa 2 to 38 and 77 to 282), Myc-Sina (c-Myc-tagged full-length Sina), Myc-Sina-dC (Myc aa 2 to 199), or control expression vector (pcDNA3), as indicated. Forty-eight hours after transfection, cell lysates were prepared and a portion of lysates was used for immunoprecipitation with anti-Flag M2 monoclonal antibody. The cell lysates (Lysates) and immunoprecipitates (IPs) were electrophoresed, and Western blotting studies were carried out with anti-Flag M2, anti-HA 12CA5, and anti-c-Myc 9E10.2 monoclonal antibodies, respectively. Flag-tagged Siah-1 proteins coprecipitated with HA-tagged Siah-1-dR (lanes 7 and 8) as well as c-Myc-tagged Sina protein (lane 9). However, the Flag-tagged Siah-1 protein did not coprecipitate with a C-terminal truncated form of Sina (lane 10). The migration positions (in kilodaltons) of selected molecular mass markers are indicated to the left of the blots. IgG, immunoglobulin heavy chain.
Interestingly, our results from pilot yeast two-hybrid studies suggested that Siah-1 and Siah-2 proteins, as well as Sina, form homo- and heterooligomers (data not shown). To substantiate this observation and to localize the sequences involved, we tested the abilities of the wild-type Siah-1 protein and various deletion mutants to complex with the Drosophila Sina protein in an in vitro binding assay. As demonstrated in Fig. 6B, deletion of the RING domain of Siah-1 did not affect its binding to Sina (lane 2 of the lower blot). Similarly, deletion of the C-terminal 20 amino acids of Siah-1 did not alter binding to Sina (lane 3). However, a mutant Siah-1 protein lacking the C-terminal 52 amino acids had reduced binding to Sina (lane 5). The results of these studies were confirmed in mammalian cells. The upper panels in Fig. 6C indicate the expression of the various epitope-tagged proteins in lysates from transfected COS-1 cells. As shown in Fig. 6C, expression of each Flag and HA epitope-tagged Siah-1 proteins was readily detected. Likewise, Myc epitope-tagged wild-type and mutant Sina proteins were strongly expressed. Following cotransfection of Flag- and HA-tagged Siah-1 proteins and the Myc-tagged Sina protein, immunoprecipitation was carried out with the anti-Flag antibodies. The immunoprecipitates were found to contain the HA-tagged Siah-1 mutant protein (lanes 7 and 8). Similarly, the Flag immunoprecipitates contained the wild-type Sina protein (lane 9). Consistent with the results of the in vitro binding studies of Siah-1 and Sina, deletion of the C-terminal sequences of Sina abrogated its interaction with Siah-1 in cells (lane 10). These findings implicate the C-terminal sequences of Siah and Sina proteins in oligomerization.
Test of the model of Siah-1 structure and function.
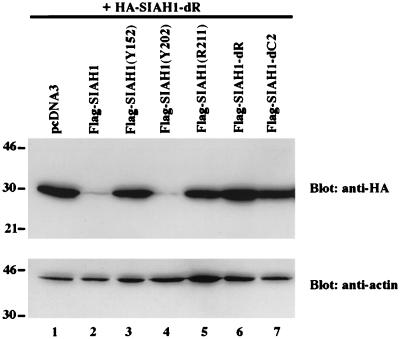
We sought to develop additional support for our model that the RING domain of Siah-1 is critical for its function in mediating proteolysis of target proteins. Our findings also implicated the C-terminal sequences of the Siah and Sina proteins in oligomerization. Hence, we predicted that wild-type Siah-1, but not certain Siah-1 mutants, would promote degradation of a heterologous Siah-1 target protein. To test this hypothesis, the heterologous Siah-1 target protein that we used was an HA-tagged Siah-1 mutant lacking its RING domain. This particular Siah-1 mutant was chosen, because deletion of the RING domain results in stable Siah-1 expression (Fig. 2B), and the HA tag allowed us to distinguish the Siah-1 target protein from the Flag-tagged Siah-1 proteins whose function we were testing. As predicted, expression of the Flag-tagged wild-type Siah-1 protein promoted degradation of the HA-tagged, RING-deleted Siah-1 protein (Fig. 7, lane 2). Consistent with the results of the studies of DCC degradation by Siah-1 mutants, the RING-deleted form of Siah-1 (lane 6) and the Y152 and R211 missense mutants (lanes 3 and 5) failed to promote degradation of the HA-tagged Siah-1 target protein. Finally, consistent with the model, deletion of the C-terminal oligomerization domain of Siah-1 also abrogated its ability to promote degradation of the HA-tagged Siah-1 target protein (lane 7).
FIG. 7.
Oligomerization of Siah-1 proteins requires C-terminal sequences, and degradation function requires the RING finger domain. Regulation of HA-SIAH1-dR protein expression by SIAH1. COS-1 cells were cotransfected with constructs encoding an HA-tagged RING-deleted Siah-1 mutant (i.e., Siah-1-dR) (lanes 1 to 7) and Flag-tagged wild-type and mutant Siah-1 proteins (lanes 2 to 7, and indicated), or empty vector (lane 1). The expression of HA-Siah-1-dR was analyzed by Western blotting with anti-HA monoclonal antibody 12CA5. The same blot was then stripped and blotted with anti-actin polyclonal antibody C11 to verify equivalent loading in the lanes. The migration positions (in kilodaltons) of selected molecular mass markers are indicated to the left of the blots.
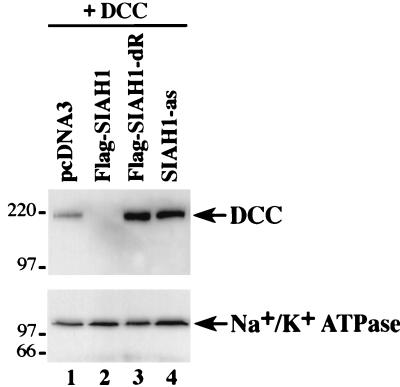
We also sought further evidence that our studies were reflecting the function of the endogenous Siah-1 protein in cells. As noted above, we obtained preliminary evidence that the RING-deleted form of Siah-1 not only failed to degrade DCC, but in fact, the RING-deleted Siah-1 mutant appeared to result in increased expression of DCC in COS-1 cells over that of a control transfection (Fig. 2A, compare lanes 1 and 6). We were able to confirm the apparent dominant negative effect of the RING-deleted Siah-1 protein in further studies (Fig. 8, lanes 1 and 3). We believe that the basis for the dominant negative activity of the RING-deleted Siah-1 protein is the following. (i) Like wild-type Siah-1, the RING-deleted form can bind to target proteins via its C-terminal sequences. (ii) In contrast to wild-type Siah-1, the RING-deleted form accumulates to high levels in cells, particularly when overexpressed, because it cannot be degraded by the proteasome pathway. More importantly, because the RING-deleted form is in greater excess than wild-type Siah-1 and it binds to but does not promote the degradation of target proteins, it strongly inhibits the activity of the limiting amounts of wild-type Siah-1 in the cell. Similar to the results obtained with the Ring-deleted Siah-1 mutant, transfection of COS-1 cells with a SIAH-1 antisense expression construct resulted in increased DCC expression (Fig. 8, lane 4). Therefore, the data argue that the endogenous Siah-1 protein regulates DCC degradation in cells.
FIG. 8.
Endogenous Siah-1 functions to regulate DCC proteolysis in cells. Western blot studies were carried out on lysates of COS-1 cells cotransfected with a DCC expression construct and a pcDNA3 construct containing no cDNA insert (lane 1), a Flag-tagged wild-type SIAH-1 cDNA (lane 2), a Flag-tagged RING-deleted SIAH-1 cDNA (lane 3), or SIAH-1 cDNA in the antisense orientation (lane 4). Increased expression of DCC relative to the control lane was seen with the RING-deleted form of Siah-1 as well in the antisense studies. The blot was stripped and reprobed with an antibody against Na+/K+ ATPase to confirm equal loading. The migration positions (in kilodaltons) of selected molecular mass markers is indicated to the left of the blots.
RING domain and cellular localization of Siah-1.
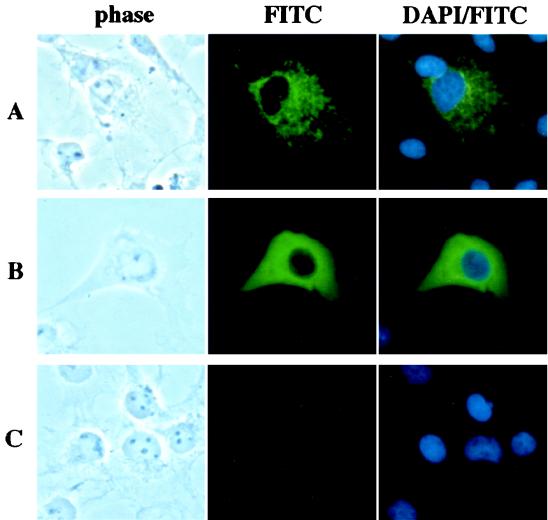
Though Sina has been suggested to localize predominantly in the nucleus in some studies (4, 15, 22), our prior studies have indicated that the Sina and Siah proteins localize predominantly in the cytoplasm with a punctate or speckled pattern (9, 10). To investigate the role of the RING finger domain in this localization, we expressed the RING-deleted form of Siah-1 and compared its localization to that of the full-length protein. The wild-type and RING-deleted proteins were each tagged at their amino terminus with a Flag epitope, and their expression and localization in transfected COS cells were studied by immunofluorescence microscopy, using an anti-Flag monoclonal antibody. Because only low levels of wild-type Siah-1 protein were detected in cells, prior to staining, the transfected cells were treated with the proteasome inhibitor MG132 to stabilize Siah-1 expression. As in our previous studies (9, 10), the wild-type Siah-1 protein showed a punctate cytoplasmic staining pattern (Fig. 9A). Deletion of the RING domain resulted in a much more uniform distribution of the Siah-1 protein throughout the cytoplasm (Fig. 9B), indicating that the RING finger motif is important for the punctate pattern. MG132 had no effect on the expression or localization of the RING-deleted form of Siah-1 in our immunofluorescence studies (data not shown), consistent with the resistance of the RING-deleted form of Siah-1 to degradation by the proteasome pathway.
FIG. 9.
The RING domain regulates the localization of Siah-1 in cells. COS-1 cells were transiently transfected with expression plasmids encoding Flag-Siah-1 (A), Flag-Siah-1-dR (B), or an empty pcDNA3 vector as a control (C). Forty-eight hours after transfection, cells were incubated with Dulbecco’s modified Eagle medium containing 10% fetal bovine serum and 50 μM MG132 for 4 h at 37°C and then fixed. Cellular localization of the expressed proteins was detected by indirect immunofluorescence staining of the cells with anti-Flag M2 monoclonal antibody and a FITC-conjugated goat anti-mouse immunoglobulin secondary antibody. Cells were also stained with DAPI to visualize the nuclei.
DISCUSSION
As reviewed above, a failure to degrade the Ttk transcriptional repressor appears to underlie the defects in R7 photoreceptor development seen in flies harboring loss-of-function mutations in sina (14, 22). Moreover, Sina and its mammalian Siah-1 and Siah-2 homologs have been found to have a more general role in proteolysis (9). The data imply that the Sina and Siah proteins mediate degradation of target proteins via the ubiquitin-proteasome pathway, perhaps through the interaction of Sina and Siah amino-terminal sequences with ubiquitin-conjugating enzymes (9, 22). The studies in this manuscript were undertaken to characterize further the functions of the Sina and Siah proteins in promoting degradation of target proteins. The design of our studies was guided by prior analyses of Drosophila sina mutants. Specifically, two classes of sina mutant alleles, with either strong or weak effects on eye morphology, have been previously defined (4). The two strong alleles encode prematurely truncated Sina proteins, lacking the C-terminal 103 or 105 amino acids. In addition to their effects of R7 formation, the strong alleles result in other phenotypes, including a marked reduction of adult life span, lethargic and uncoordinated behavior, and infertility. Less information is available about the functions of the three known weak alleles of sina, other than that they encode Sina proteins with missense mutations in the C-terminal region. Nevertheless, we hypothesized that the localized missense mutations might be useful for defining specific functions. Indeed, by generating mutated Siah-1 proteins analogous to Sina proteins encoded by the weak alleles and using the DCC protein as the target, we were able to demonstrate specific effects on Siah-1 function in mammalian cells.
Two of three Siah-1 proteins with missense mutations in their C-termini (Y152 and R211, equivalent to the Y184 and R243 substitutions in Sina, respectively) failed to degrade DCC, and they also failed to degrade the heterologous RING-deleted Siah-1 target protein. These findings further support the proposal that the critical function of the Sina and Siah proteins is degradation of specific protein targets. The missense mutations likely cause a partial loss of function, since the mutant proteins retained their ability to associate with DCC and to form homo- and heterooligomers. One of the Siah-1 missense mutations (Y202 [equivalent to Y234 in Sina]) appeared to retain the ability to regulate both DCC and Siah-1 protein expression in cells. There are at least three possible explanations for the effects we observed with the Siah-1 missense mutants, including the Y202 mutant. First, our in vitro assays may not be sufficiently robust for identifying subtle effects on Siah-1 proteolysis function and/or binding to target proteins. Second, some of the proteins with missense mutations, such as the Siah-1 Y202 mutated form (and the corresponding Sina Y234 mutant), may fail to promote the degradation of certain critical targets, like Ttk, but may retain full activity on other protein targets (e.g., DCC and Siah-1 itself). Third, while the interaction of Sina and Siah proteins with DCC appears to be direct, Sina and Siah proteins may require bridging factors to promote the degradation of particular target proteins. Indeed, this appears to be the case with Ttk, where Phyl is required for formation of the complex between Sina and Ttk (14, 22). Hence, some mutations in the C-terminal regions of the Sina and Siah proteins, such as the Y202 substitution, may alter the ability of the mutated proteins to bind to critical cellular bridging factors.
Missense mutations and deletion of the RING domain inactivated the ability of Siah-1 to regulate DCC degradation, as well as the degradation of Siah-1 itself through the proteasome pathway. RING domains are present in a large and functionally diverse group of proteins (for a review, see reference 20). In a number of cases, the RING motif appears to have a critical role in protein-protein interactions (2). For example, the RING finger domain of BRCA1 is critical for its function and interaction with BARD1 (1, 24, 25). In addition, activation of NF-κB effector function by the TRAF (tumor necrosis factor receptor-associated factor) family of proteins depends on their RING finger domains (13, 20). Similiarly, deletion or point mutations of the RING finger domain abolish the transactivation potential of the VZ61 viral protein from varicella-zoster virus (16). While our previous studies have suggested that the N-terminal regions of the Sina and Siah proteins are required for their interaction in yeast with ubiquitin-conjugating proteins (9), further studies will be needed to define the critical in vivo interactions of the Siah-1 RING domain that underlie its function in promoting proteolysis.
In our prior studies, as well as this report, extremely low levels of wild-type Siah-1 protein expression were uniformly seen following transfection of COS or 293 cells, and metabolic labeling studies of epitope-tagged Siah-1 also indicated that the protein is very unstable (data not shown). A plausible explanation for these observations has been obtained. Sina and Siah proteins oligomerize in vitro and in cells via their C-terminal sequences. Following transfection and overexpression, the RING-deleted form of Siah-1 probably forms predominantly homooligomers with itself, and as a result, it is stably expressed in cells. However, because the RING-deleted form of Siah-1 can still oligomerize with wild-type Siah-1, the RING-deleted form can be degraded when wild-type Siah-1 is overexpressed. These findings, together with the results of the MG132 proteasome inhibitor studies, establish that the degradation of Siah-1 oligomers is regulated via the proteasome pathway. While it is likely that Siah-1 is ubiquitinated prior to its degradation by the proteasome pathway, we have not yet been able to detect specific ubiquitination of the Siah-1 protein in cotransfection studies with a cDNA encoding a Myc epitope-tagged form of ubiquitin (reference 9 and data not shown).
A number of immunofluorescence studies of RING finger-containing proteins have suggested that they are components of multiprotein complexes (reviewed in reference 20) and that these complexes can be visualized as distinct nuclear or cytosolic bodies. The size of these macromolecular assemblages varies from large bodies, such as the nuclear bodies formed by acute promyelocytic leukemia proto-oncoprotein (PML) (2, 3, 7), to small speckles, such as those formed by the RING1 and BRCA1 proteins (11, 20, 21). When transiently expressed in cells, Sina as well as Siah-1 and Siah-2 form distinct cytosolic structures reminiscent of other RING finger proteins (9, 10). Mutations in the RING finger domain of PML dramatically affect the size and shape of PML nuclear bodies (2, 3), suggesting that its RING motif is required for the formation of the structural bodies. Similarly, we found that the RING domain of Siah-1 is required for the formation of punctuate structures in the cytoplasm. Moreover, oligomerization of the Sina and Siah proteins may also be required for their localization to the cytoplasmic structures, because C-terminal truncations of the Sina and Siah proteins that abolish their oligomerization also abolish their localization in the cytoplasmic structures (reference 9, and data not shown). While the cytosolic particles may not entirely reflect the physiological distribution of Siah proteins, the fact that Siah proteins with mutations in critical N-terminal and C-terminal domains results in a diffuse cytoplasmic localization is a reasonable initial test of their significance. However, the precise nature of the subcellar structures and their specific role in protein degradation await further investigation.
In summary, the findings presented here provide convincing evidence that the Siah-1 N-terminal RING domain is required for its proteolysis function, while the C-terminal sequences regulate oligomerization and binding to target proteins. Evidence has also been provided that endogenous Siah-1 regulates DCC expression and that Siah-1 RING domain mutants with apparent dominant negative activity have been defined. Further studies of Sina and Siah proteins will provide more-detailed insights into the specific mechanisms through which the Sina and Siah RING domain regulates proteolysis, as well as additional proteins that are regulated by Sina and Siah proteins. Such studies will greatly improve our understanding of cell fate specification in the nervous system and other tissues. Moreover, given the important role of the Ras-Raf-MAPK pathway in growth regulation and neoplastic transformation, further studies of the function of mammalian Siah proteins may also shed considerable light on these important cellular processes as well.
ACKNOWLEDGMENTS
We thank Kathleen R. Cho and David Ginsburg for their comments on the manuscript.
This work was supported by Public Health Service grant CA-70097 from the National Cancer Institute.
REFERENCES
- 1.Blackwood M A, Weber B L. BRCA1 and BRCA2: from molecular genetics to clinical medicine. J Clin Oncol. 1998;16:1969–1977. doi: 10.1200/JCO.1998.16.5.1969. [DOI] [PubMed] [Google Scholar]
- 2.Borden K L, Boddy M N, Lally J, O’Reilly N H, Martin S, Howe K, Solomon E, Freemont P S. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J. 1995;14:1532–71541. doi: 10.1002/j.1460-2075.1995.tb07139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borden K L B, Freemont P S. The RING finger domain: a recent example of a sequence-structure family. Curr Opin Struct Biol. 1996;6:395–401. doi: 10.1016/s0959-440x(96)80060-1. [DOI] [PubMed] [Google Scholar]
- 4.Carthew R W, Rubin G M. seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell. 1990;63:561–577. doi: 10.1016/0092-8674(90)90452-k. [DOI] [PubMed] [Google Scholar]
- 5.Chang H C, Solomon N M, Wassarman D A, Karim F D, Therrien M, Rubin G M, Wolff T. phyllopod functions in the fate determination of a subset of photoreceptors in Drosophila. Cell. 1995;80:463–472. doi: 10.1016/0092-8674(95)90497-2. [DOI] [PubMed] [Google Scholar]
- 6.Dickson B. Nuclear factors in Sevenless signaling. Trends Genet. 1995;11:106–111. doi: 10.1016/S0168-9525(00)89011-3. [DOI] [PubMed] [Google Scholar]
- 7.Grimwade D, Solomon E. Characterization of the PML/RAR alpha rearrangement associated with t(15;17) acute promyelocytic leukemia. Curr Top Microbiol Immunol. 1997;220:81–112. doi: 10.1007/978-3-642-60479-9_6. [DOI] [PubMed] [Google Scholar]
- 8.Higuchi R. Recombinant PCR. In: Ennis M A, editor. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 177–183. [Google Scholar]
- 9.Hu G, Zhang S, Vidal M, La Baer J, Xu T, Fearon E R. Mammalian homologs of seven in absentia regulate DCC via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:2701–2714. doi: 10.1101/gad.11.20.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu G, Chung Y-L, Glover T, Valentine V, Look T A, Fearon E R. Characterization of human homologs of the Drosophila seven in absentia (sina) gene. Genomics. 1997;46:103–111. doi: 10.1006/geno.1997.4997. [DOI] [PubMed] [Google Scholar]
- 11.Jin Y, Xu X L, Yang M C, Wei F, Ayi T C, Bowcock A M, Baer R. Cell cycle-dependent colocalization of BARD1 and BRCA1 proteins in discrete nuclear domains. Proc Natl Acad Sci USA. 1997;94:12075–12080. doi: 10.1073/pnas.94.22.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai Z-C, Harrison S d, Karim F, Li Y, Rubin G M. Loss of tramtrack gene activity results in ectopic R7 cell formation, even in a sina mutant background. Proc Natl Acad Sci USA. 1996;93:5025–5030. doi: 10.1073/pnas.93.10.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S Y, Lee S Y, Choi Y. TRAF-interacting protein (TRIP): a novel component of the tumor necrosis factor receptor (TNFR)- and CD30-TRAF signaling complexes that inhibits TRAF2-mediated NF-kB activation. J Exp Med. 1997;185:1275–1285. doi: 10.1084/jem.185.7.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Li Y, Carthew R W, Lai Z-C. Photoreceptor cell differentiation requires regulated proteolysis of the transcriptional repressor tramtrack. Cell. 1997;90:469–478. doi: 10.1016/s0092-8674(00)80507-3. [DOI] [PubMed] [Google Scholar]
- 15.Matsuzawa S, Takayama S, Froesch B A, Zapata J M, Reed J C. P53-inducible human homologue of Drosophila seven in absentia (Siah) inhibits cell growth: suppression by BAG-1. EMBO J. 1998;17:2736–2747. doi: 10.1093/emboj/17.10.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moriuchi H, Moriuchi M, Cohen I J. The RING finger domain of the varicella-zoster virus open reading frame 61 protein is required for its transregulatory functions. Virology. 1994;205:238–246. doi: 10.1006/viro.1994.1639. [DOI] [PubMed] [Google Scholar]
- 17.Palombella V J, Rando O J, Goldberg A L, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NG-kB1 precursor protein and the activation of NG-kB. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 18.Reale M A, Hu G, Zafar A I, Getzenberg R H, Levine S M, Fearon E R. Expression and alternative splicing of the deleted in colorectal cancer (DCC) gene in normal and malignant tissues. Cancer Res. 1994;54:4493–4501. [PubMed] [Google Scholar]
- 19.Rock K L, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 20.Saurin A J, Borden K L B, Boddy M N, Freemont P S. Does this have a familiar ring? Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- 21.Scully R, Ganesan S, Brown M, De Caprio J A, Cannistra S A, Feunteun J, Schnitt S, Livingston D M. Location of BRCA1 in human breast and ovarian cancer cells. Science. 1996;272:123–126. doi: 10.1126/science.272.5258.123. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 22.Tang A H, Neufeld T P, Kwan E, Rubin G M. PHYL acts to down-regulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell. 1997;90:459–467. doi: 10.1016/s0092-8674(00)80506-1. [DOI] [PubMed] [Google Scholar]
- 23.Wassarman D A, Therrien M, Rubin G M. The Ras signaling pathway in Drosophila. Curr Opin Genet Dev. 1995;5:44–50. doi: 10.1016/s0959-437x(95)90052-7. [DOI] [PubMed] [Google Scholar]
- 24.Wu L C, Wang W, Tsan J T, Spillman M A, Phung A, Xu X L, Yang M C, Hwang L Y, Bowcock A M, Baer R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 25.Xu C F, Solomon E. Mutations of the BRCA1 gene in human cancer. Semin Cancer Biol. 1996;7:33–40. doi: 10.1006/scbi.1996.0005. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto D, Nihonmatsu I, Matsuo T, Miyamoto H, Kondo S, Hirata K, Ikegami Y. Genetic interactions of pokkuri with seven in absentia, tramtrack and downstream components of the sevenless pathway in R7 photoreceptor induction in Drosophila melanogaster. Roux’s Arch Dev Biol. 1996;205:215–224. doi: 10.1007/BF00365799. [DOI] [PubMed] [Google Scholar]
- 27.Zipursky S L, Rubin G M. Determination of neuronal cell fate: lessons from the R7 neuron of Drosophila. Annu Rev Neurosci. 1994;17:373–397. doi: 10.1146/annurev.ne.17.030194.002105. [DOI] [PubMed] [Google Scholar]