Abstract
What is known about this topic?
Few major outbreaks of coronavirus disease 2019 (COVID-19) have occurred in China after major non-pharmaceutical interventions and vaccines have been deployed and implemented. However, sporadic outbreaks that had high possibility to be linked to cold chain products were reported in several cities of China..
What is added by this report?
In July 2020, a COVID-19 outbreak occurred in Dalian, China. The investigations of this outbreak strongly suggested that the infection source was from COVID-19 virus-contaminated packaging of frozen seafood during inbound unloading personnel contact.
What are the implications for public health practice?
Virus contaminated paper surfaces could maintain infectivity for at least 17–24 days at -25 ℃. Exposure to COVID-19 virus-contaminated surfaces is a potential route for introducing the virus to a susceptible population. Countries with no domestic transmission of COVID-19 should consider introducing prevention strategies for both inbound travellers and imported goods. Several measures to prevent the introduction of the virus via cold-chain goods can be implemented.
Keywords: COVID-19, outbreak, imported cold-chain products, SARS-CoV-2-contaminated packaging
On July 22, 2020, Dalian City, a major city in Liaoning Province, reported one confirmed coronavirus disease 2019 (COVID-19) case in a cold-chain products processing and storage company (Company K) who seek healthcare in hospital because of having a fever. Active case finding and management was conducted according to prevention and control protocols. On July 23, another 12 cases or asymptomatic infections with nucleic acid test positive results from Company K and 2 community cases who had no apparent relation to Company K were reported. On July 25, the affected streets in Ganjingzi District (approximately 1.83 million residents) were elevated to the “high-risk level” and locked down. Population quantitative real-time reverse transcription PCR (RT-PCR) screening was initiated for the high-risk region and was then conducted for other regions of Dalian.
This was the first local outbreak reported after having no new local infections in Dalian for 111 consecutive days. The Dalian CDC, Liaoning CDC, and China CDC together conducted a detailed epidemiological investigation to sort out the sources of the outbreak.
INVESTIGATION AND RESULTS
COVID-19 cases were diagnosed by a local hospital in Dalian according to the Protocol for Prevention and Control of COVID-19 (Edition 6) issued by China CDC (1). In this study, a confirmed case was defined as having a throat swab that tested positive for COVID-19 virus RNA by RT-qPCR; an undiscovered infected case was defined as having a sera sample that tested positive for COVID-19 antibodies but negative for COVID-19 virus RNA. Detailed epidemiological investigation for early cases were conducted through in-person interviews for their travel history, activity, work history, and contact history starting 14 days (incubation period) before the onset of illness. Environmental samples and cold-chain product samples collected from Company K were further tested with RT-qPCR. Individuals who had contact with the positive environment samples and cold-chain products samples (mostly the dockworkers who had handled the cold-chain products) were quarantined and tested for COVID-19 virus nucleic acid and anti-COVID-19 virus antibodies.
Environmental smear samples were collected from the cold-chain seafood processing work area of Company K, which was the workplace of the first confirmed case of the Dalian outbreak, including samples from the operating table, floor, tools, sinks, sewers, and other environmental locations. According to records from the electronic cold storage management system of Company K, cold-chain products imported before the COVID-19 outbreak from the importing country were excluded. Surface swab samples of the inner and outer packaging of the cold-chain seafood imported by Company K from June 22 to July 8 (incubation period before the onset date of the first case) that were suspected to be contaminated by COVID-19 virus were carefully smeared and collected. In addition to the imported cold-chain seafood by Company K, all other imported cold-chain seafood products from the two cargo ships that were temporarily stored at Company K starting on June 1 were sampled and tested.
Viral RNA was extracted directly from 200-μL swab samples with a QIAamp Viral RNA Mini Kit (QIAGEN, Germany). RT-qPCR was conducted using the commercially available Novel Coronavirus 2019 Nucleic Acid Test Kit (Bojie, Shanghai, China) with an Applied Biosystems QuantStudio 5 instrument (Applied Biosystems, Hong Kong, China) following the manufacturers’ instructions. A TaqMan probe-based kit was designed to detect the ORF1ab and N genes of COVID-19 virus in 1 reaction. The viral copy number was determined according to the Certified Reference Material of the COVID-19 virus Ribonucleic Acid Genome (No. GBW(E)091099) obtained from the National Institute of Metrology using RT-qPCR.
Corresponding serum samples were tested for anti-COVID-19 virus antibodies using a chemiluminescence immunoassay (CLIA, Bioscience, Chongqing, China). All these tests were conducted by the National Institute for Viral Disease Control and Prevention, China CDC.
Libraries were prepared using a Nextera XT Library Prep Kit (Illumina, San Diego, CA, USA), and the resulting DNA libraries were sequenced on either a MiSeq or an iSeq platform (Illumina) using a 300-cycle reagent kit. Mapped assemblies were generated using the COVID-19 virus genome (accession number NC_045512) as a reference. Variant calling, genome alignment, and sequence illustrations were generated with CLCBio software (Qiagen, Hilden, Germany).
Each of the selected RT-qPCR-positive samples was seeded separately in Vero cells. The cells were monitored daily with light microscopy for cytopathic effects. COVID-19 virus genome sequences were submitted to GISAID database, accession numbers: EPI_ISL_2170885-2170894.
RESULTS
A total of 135 COVID-19 cases were identified in the present outbreak, 64 cases (47.4%) were employees of Company K and 67 (49.6%) were their contacts or residents living around Company K. The date of onset of the primary case (Case A) was July 9, and the infection was confirmed on July 25 by RT-qPCR testing. The index case was confirmed on July 23. A total of 4 early cases had onset by July 14, and the peak occurred from July 22 to 23 (Figure 1). Serum samples collected from Case A on August 5 tested positive for higher IgM level and lower IgG level (geometric mean titer=1:53.427 and 1:15.139, respectively).
Figure 1.
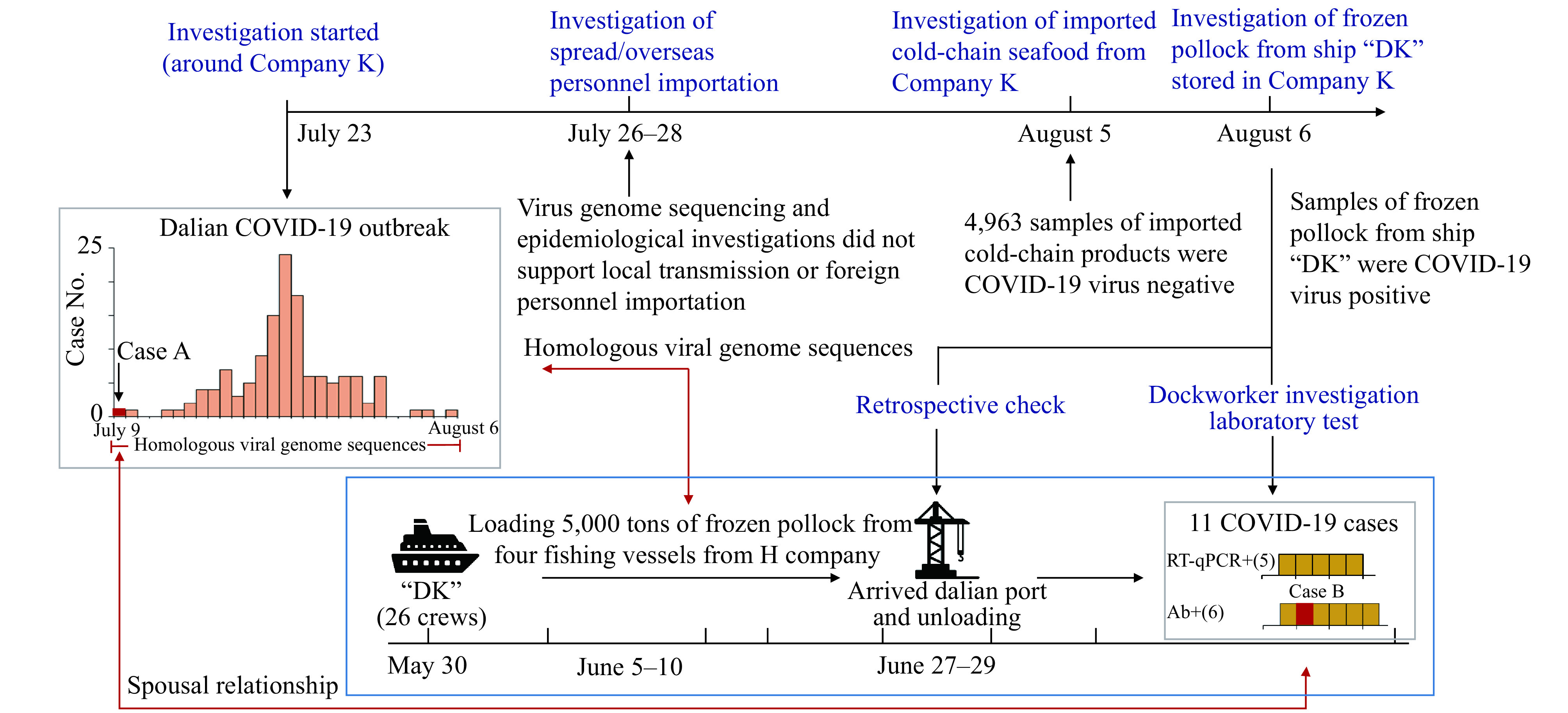
Investigation workflow and the Dalian COVID-19 outbreak. Imported cold-chain seafood products with COVID-19 virus infected wharf workers and then were introduced into the seafood product workshop of Company K before spreading further. The epidemiological investigation strategy was shown bolded in blue. COVID-19 case data were represented by a grey dotted box, and virus introduction from the DK ship was represented by a blue dotted box.
Among 64 Company K employees, 63 were exposed to the environment of the cold-chain seafood processing workspace of Company K. On July 23, we collected 39 environmental smear samples from this location, including samples from the operating table, floor, tools, sinks, sewers, and other environmental areas. COVID-19 virus nucleic acid was tested in these samples, with a positive rate of 35.9% (14/39). In addition, the workers in the same and adjacent working areas to case A were identified as the first infected cases through work contact.
A total of 24 throat swab samples from confirmed cases and 2 environmentally positive samples from Company K were further sequenced, and full-length genomic sequences were acquired. Whole-genome analysis revealed that the sequences of 24 cases and two environmental samples were highly homologous. Compared with the reference sequence of COVID-19 virus, strain Wuhan-1 (GenBank No. NC_045512), all the sequences shared a total of 14 nucleotide mutations (C241T, C2091T, C3037T, A5128G, A8360G, C13860T, C14408T, T19839C, G19999T, A23403G, G28881A, G28882A, G28883C, and C28905T), belonging to PANGO lineage B.1.1.317 (2) (Figure 2).
Figure 2.
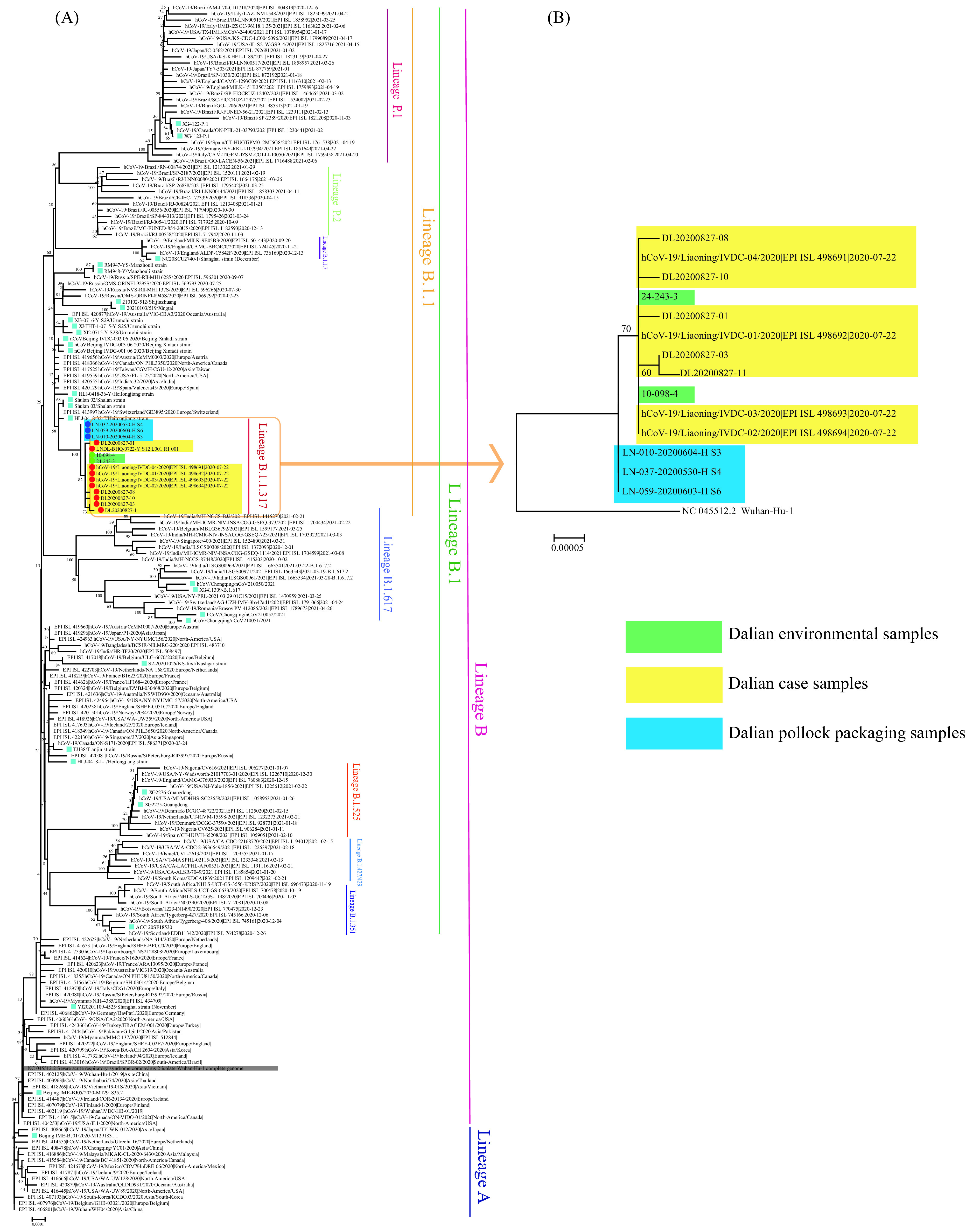
Phylogenetic tree based on Dalian outbreak-related samples. (A) Phylogenetic tree based on the representative full-length genome sequences of the COVID-19 virus. The genomes of the COVID-19 virus from Dalian case samples were highlighted in yellow, environmental samples from cold-chain seafood processing work area of Company K were highlighted in green and pollock packaging samples were highlighted in blue. The genome of the reference COVID-19 virus from Wuhan in December 2019 was shaded in grey. Strains associated with specific outbreaks in China were indicated by blue squares. The PANGO lineages were marked and colored on the right. The tree was rooted using strain WH04 (EPI_ISL_406801) in accord with the root of PANGO tree. (B) Phylogenetic tree based on the full-length genome sequences of Dalian COVID-19 virus samples, rooted using the Wuhan reference strain.
From June 24 to July 8, which was the 14 days of the COVID-19 incubation period before the onset date of Case A, a total of 8 imported cases were reported in Dalian. Based on the measures to prevent the risk of importation from overseas implemented in China, all the overseas arrivals were swabbed and tested for COVID-19 upon their entry at customs, were then transferred to the centralized quarantine location (usually a designated hotel) for 14 days for medical observation, and were tested again before their release. These 8 cases had no historical contact with early cases of the Dalian outbreak, and the whole-genome sequences from the samples of these cases were distinct from those of the Dalian outbreak (Figure 2). In addition, during this period, smeared swab samples were taken from the inner and outer packaging of a total of 2,119 suspicious packages of cold-chain seafood imported by Company K, and a total of 4,963 samples were collected for COVID-19 virus nucleic acid testing. The results revealed that all samples were negative.
In addition to importing and processing imported cold-chain seafood, Company K temporarily stored bonded cold-chain products for other companies. Imported bonded cold-chain products loaded from two overseas cargo ships were temporarily stored by Company K during that time. We traced the location of these cold-chain seafood products and conducted nucleic acid testing of their inner and outer packaging for COVID-19 virus and detected positive samples from the outer packaging of some cold-chain pollock loaded on one of the cargo ships (cargo ship DK). No positive samples were found from the other cargo ship. We further conducted RT-qPCR testing of the cold-chain pollock packaging that had been stored in four different freezers of DK, and the positive rates were 19.0% (19/100), 36.3% (37/102), 43.9% (29/66), and 49.0% (49/100).
DK was a large ocean-going refrigerated transport ship. From June 5 to 10, the ship DK carried 5,000 tons of cold-chain pollock from four fishing vessels that belonged to the same overseas company H. During the marine operation, the crews of the four overseas fishing vessels participated in processing and loading the cold chain pollock into the freezers of ship DK (temperature minus 25 degrees Celsius), while the crews of ship DK neither participated in cargo handling nor contacted the overseas crew. When the cargo ship arrived at the Dalian terminal on June 27–29, a total of 216 dockworkers from Company K were divided into three shifts; each shift spent 10 hours handling the cold-chain products, while the crews from ship DK did not participate in handling. According to custom inspection records, the ship DK crew’s temperatures were normal, and they presented no symptoms of infection, but they were not tested with RT-qPCR while in China. All of them were quarantined on the ship until the ship left the port, which was confirmed by video from the dock around the ship. We also interviewed the crew of the DK ship, who maintained a distance of at least 10 metres from overseas crews; they had neither talked with the overseas crews nor used the toilet on their vessels. All the dockworkers of Company K wore labour gloves and disposable masks but carried the seafood products close to their faces and drank water and wiped sweat with their face mask off (Figure 1).
We further investigated the COVID-19 virus infection status of the workers who handled the cold-chain pollock. From July 23 to August 6 (14 days since the index case was confirmed), a total of 245 dockworkers and 13 cold storage stevedores of Company K who were exposed to DK cargo were quarantined and screened with RT-qPCR several times until August 5. Among these 258 potential contacts, five dock workers tested positive for COVID-19 virus nucleic acid, and all the others were negative. In addition, on August 11, sera samples of 195 dock workers and 13 cold storage stevedores of Company K were collected for antibody detection, and 50 dock workers were not sampled because they had been released from quarantine and were lost to follow up. Among the 208 potential contacts, 6 dockworkers who tested negative for nucleic acid were positive for COVID-19 IgG and IgM antibodies. Among the 5 confirmed cases and 6 undiscovered cases who had been infected with COVID-19, 2 of them reported physical soreness or cold symptoms at the end of June and early July, and the other nine were asymptomatic. These 11 people were all among the 216 dockworkers who participated in the actual handling of cold-chain pollock carried by the overseas DK cargo ship, and the infection rate was 5.1% (11/216). It is worth noting that 1 of the 6 dockworkers who were positive for COVID-19 IgG and IgM antibodies was the husband of Case A (designated Case B). The geometric mean titer of anti COVID-19 virus IgM and IgG of Case B were 1:3.899 and 1:13.677,respectively (Figure 1).
Most importantly, the COVID-19 whole-genome sequences from 7 nucleic acid-positive samples of the packaging of cold-chain pollock from the ship DK were obtained, all of which had very high homology with the sequences of the Dalian cases, also belonging to PANGO lineage B.1.1.317, only lacking a mutation site (A5128G), which was strongly suggestive of the parental virus of the Dalian cases (Figure 2).
We tried to isolate the virus from COVID-19 virus nucleic acid-positive samples detected in the cold-chain pollock from the DK cargo ship, but the virus was not successfully isolated. However, the result of quantitative fluorescence RT-qPCR detection was 1,090 copies, which was equivalent to 13,080 copies of the gene in the 200-µL environmental swab. Even though the virus was not isolated, such high viral copy numbers indicated that the viral load was large.
DISCUSSION
COVID-19 virus is transmitted from person to person through respiratory droplets and close contact during coughing, sneezing and speaking. In China, there have been some reports of COVID-19 virus nucleic acid detection on imported cold-chain chicken and cold-chain shrimp packaging material (3). Based on epidemiological investigations, laboratory testing (nucleic acid and antibody detection) and full-length genome sequencing, our study strongly suggested that the Dalian COVID-19 outbreak was caused by COVID-19 virus-contaminated imported cold-chain seafood products, sugggesting the possibility that COVID-19 virus can be transmitted through cold-chain products.
We investigated the 14-day incubation period of COVID-19 before the onset date of the first case (June 24 to July 8), first ruling out the possibility that the COVID-19 virus that caused the Dalian COVID-19 outbreak was from overseas personnel on the ground based on epidemiological investigations and whole-genome sequence differences. Second, 4,963 suspicious imported and processed frozen white shrimp samples from Company K tested negative for COVID-19 virus nucleic acid, which excluded the possibility that COVID-19 virus came from cold-chain goods imported and processed by Company K.
After detecting COVID-19 virus nucleic acid-positive cold-chain pollock samples from ship DK, an overseas cargo ship that had temporarily stored sealed imported cold-chain seafood for Company K, the investigation was clearer. The relatively high COVID-19 virus nucleic acid positivity rate in the four different freezers of the ship DK indicated a high proportion of virus contamination, and the 216 dockworkers who participated in the actual handling of cold-chain pollock carried by the overseas ship DK had a COVID-19 virus infection rate of 5.1% (11/216). This finding suggested that the source of COVID-19 virus infection in the Dalian outbreak was very likely the outer packaging of cold-chain pollock loaded on the DK cargo ship. Furthermore, the serological results of Cases B (high IgG level and low IgM level) and A (high IgM level) strongly suggested that Case A was infected after Case B, which further indicated that Case A was infected through family transmission from contact with Case B.
The timeline of the Dalian outbreak could be well summarized based on the detailed retrospective investigation. Case B was asymptomatically infected with COVID-19 virus while handling the COVID-19 virus contaminated packaging of imported cold-chain seafood from the ship DK from June 27 to 29 and then transmitted COVID-19 virus to his wife, Case A. Case A presented symptoms of fatigue on July 9, which was 10 to 12 days since her husband was infected. Case B tested negative for nucleic acid from July 23 to August 5, indicating a transient infection. The serological evidence further demonstrated that Case B had been infected for some time (higher IgG level and low IgM level) but that Case A was newly infected (positive for nucleic acid and very high IgM level). After becoming infectious, Case A transmitted COVID-19 to her co-workers who had been working with Case A from July 6 to 23 in the cold-chain seafood processing workspace of Company K and contaminated the work environment. The specialized environment, with poor ventilation, humidity and low temperature in the work space, made it easier to contaminate with the virus (4-6). Therefore, the rapid spread through human-to-human transmission and environmental exposure triggered the outward spread of COVID-19 virus in the Company K, which led to the Dalian outbreak. The whole-genome sequences from positive pollock packaging samples lacked a unique mutation site compared with the sequences from the Dalian case samples, indicating that COVID-19 virus from pollock packaging was the originator of COVID-19 virs for the Dalian cases.
The results of this study have important public health significance for international organizations and countries for taking measures to prevent and control the importation of COVID-19 virus. The seafood product industry and trading market could act as a “virus amplifier” that causes COVID-19 outbreaks. COVID-19 virus was more stable on plastic and stainless steel than on copper and cardboard, and live virus was detected within 72 hours after application to these surfaces (7). Therefore, frequent exposure to contaminated surfaces in public places is a potential route for the spread of COVID-19 virus. One study found that when COVID-19 virus was added to chicken, salmon, and pork slices, the virus could survive (8). After 21 days of storage at 4 °C (standard refrigeration) and –20 °C (standard freezing), the titre of infectious virus did not decrease ( 9). According to a study published by the British Food Standards Agency on April 29, 2020, although consumers are less likely to be infected with the virus by eating food or handling packaging, COVID-19 virus in cold-chain or refrigerated transport and storage products may still be infectious.
Virus isolation is usually difficult, and it is not easy to isolate live virus even from respiratory tract samples of COVID-19 patients (10). Although the virus was not successfully isolated in this study, when the viral load of the contaminated goods was high and the virus was stored and transported in the cold chain, the survival rate of the virus was very high.
The key to strengthening the prevention and control of cold-chain seafood product import of the virus is to regularly carry out nucleic acid screening for COVID-19 virus in dockworkers engaged in the import of cold-chain goods and special personnel engaged in seafood product production and sales; carry out health education for dockworkers and seafood product production and sales personnel; and take protective and hygiene measures to reduce the potential risk of the recurrence of the epidemic.
Conflicts of interest:No conflicts of interest were reported.
Acknowledgments
We thank Dalian, Liaoning and China CDC for the epidemiological investigation.
Funding Statement
The National Key Research and Development Project, Ministry of Science and Technology of the People’s Republic of China (2021YFC0863000, 2018ZX10713002, and 2020YFC0846900), National Natural Science Foundation of China (No.72042012, and No. 82041032) and National Key Technology R&D Program of China (2017ZX10104001)
Contributor Information
Wei Yao, Email: dlyaowei@126.com.
Wenqing Yao, Email: wenqingyao@sina.com.
Zijian Feng, Email: fengzj@chinacdc.cn.
Wenbo Xu, Email: xuwb@ivdc.chinacdc.cn.
References
- 1.National Health Commission of the People’s Republic of China Protocol for prevention and control of COVID-19 (Edition 6) China CDC Wkly. 2020;2(19):321–6. doi: 10.46234/ccdcw2020.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rambaut A, Holmes EC, O'Toole Á, Hill V, McCrone JT, Ruis C, et al A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5(11):1403–7. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li YH, Fan YZ, Jiang L, Wang HB Aerosol and environmental surface monitoring for SARS-CoV-2 RNA in a designated hospital for severe COVID-19 patients. Epidemiol Infect. 2020;148:e154. doi: 10.1017/S0950268820001570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arslan M, Xu B, El-Din MG Transmission of SARS-CoV-2 via fecal-oral and aerosols-borne routes: environmental dynamics and implications for wastewater management in underprivileged societies. Sci Total Environ. 2020;743:140709. doi: 10.1016/j.scitotenv.2020.140709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye GM, Lin HL, Chen S, Wang SC, Zeng ZK, Wang W, et al Environmental contamination of SARS-CoV-2 in healthcare premises. J Infect. 2020;81(2):e1–e5. doi: 10.1016/j.jinf.2020.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matson MJ, Yinda CK, Seifert SN, Bushmaker T, Fischer RJ, van Doremalen N, et al Effect of environmental conditions on SARS-CoV-2 stability in human nasal mucus and sputum. Emerg Infect Dis. 2020;26(9):2276–8. doi: 10.3201/eid2609.202267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, et al Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382(16):1564–7. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlottau K, Rissmann M, Graaf A, Schön J, Sehl J, Wylezich C, et al SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. Lancet Microbe. 2020;1(5):e218–e25. doi: 10.1016/S2666-5247(20)30089-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhonghua Liu Xing Bing Xue Za Zhi Novel Coronavirus Pneumonia Emergency Response Epidemiology T. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. 2020;41(2):145–51. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [Google Scholar]
- 10.Kucharski AJ, Klepac P, Conlan AJK, Kissler SM, Tang ML, Fry H, et al Effectiveness of isolation, testing, contact tracing, and physical distancing on reducing transmission of SARS-CoV-2 in different settings: a mathematical modelling study. Lancet Infect Dis. 2020;20(10):1151–60. doi: 10.1016/S1473-3099(20)30457-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


