Abstract
The high-risk human papillomaviruses (HPVs) are associated with carcinomas of the cervix and other genital tumors. Previous studies have identified two viral oncoproteins, E6 and E7, which are expressed in the majority of HPV-associated carcinomas. The ability of high-risk HPV E6 protein to immortalize human mammary epithelial cells (MECs) has provided a single-gene model to study the mechanisms of E6-induced oncogenic transformation. In this system, the E6 protein targets the p53 tumor suppressor protein for degradation, and mutational analyses have shown that E6-induced degradation of p53 protein is required for MEC immortalization. However, the inability of most dominant-negative p53 mutants to induce efficient immortalization of MECs suggests the existence of additional targets of the HPV E6 oncoprotein. Using the yeast two-hybrid system, we have isolated a novel E6-binding protein. This polypeptide, designated E6TP1 (E6-targeted protein 1), exhibits high homology to GTPase-activating proteins for Rap, including SPA-1, tuberin, and Rap1GAP. The mRNA for E6TP1 is widely expressed in tissues and in vitro-cultured cell lines. The gene for E6TP1 localizes to chromosome 14q23.2-14q24.3 within a locus that has been shown to undergo loss of heterozygosity in malignant meningiomas. Importantly, E6TP1 is targeted for degradation by the high-risk but not the low-risk HPV E6 proteins both in vitro and in vivo. Furthermore, the immortalization-competent but not the immortalization-incompetent HPV16 E6 mutants target the E6TP1 protein for degradation. Our results identify a novel target for the E6 oncoprotein and provide a potential link between HPV E6 oncogenesis and alteration of a small G protein signaling pathway.
The human papillomaviruses (HPVs) are associated with epithelial tumors or benign lesions especially those of anogenital origin (64, 65). These viruses are grouped into low-risk HPVs, such as HPV6 and HPV11, which are usually associated with benign warts, and high-risk HPVs, such as HPV16 and HPV18, which are associated with carcinomas (64, 65). Similar to other DNA tumor viruses (14, 17, 58), two early genes of the high-risk HPVs, encoding E6 and E7, associate with and functionally inactivate cellular tumor suppressor proteins p53 and retinoblastoma protein (Rb), respectively (18, 25–27, 40, 46, 47, 57). This is thought to provide a basis for why the high-risk but not the low-risk HPVs promote oncogenesis.
In recent years, a distinct mechanism of viral oncoprotein-induced inactivation of tumor suppressor protein function has emerged, involving the targeting of tumor suppressor protein(s) to ubiquitin-proteasome-mediated degradation machinery (6, 12, 25–27, 37, 47, 50). The ubiquitin-proteasome pathway participates in physiological regulation of the levels of cell cycle-related proteins such as cyclins and cdks as well as tumor suppressor proteins such as p53 and Rb (6, 12, 37). Apparently, the viral oncoproteins have attained an ability to target cellular tumor suppressor proteins for enhanced degradation by the same pathway (6, 25, 47, 50). The first such example was provided by the high-risk HPV E6 oncoprotein, which associates with E6AP, a ubiquitin ligase, which in turn interacts with p53 and targets it for degradation (25, 47). Recently, we found that the high-risk HPV16 E7 enhances degradation of the bound Rb protein, again via the ubiquitin-proteasome pathway (6). More recently, the simian virus 40 large T antigen was shown to interact, through an N-terminal J domain, with the Rb-related protein p130 and to induce its degradation (50).
Characterization of oncogenesis-related cellular targets for the HPV oncoproteins has been facilitated by their ability to dominantly immortalize primary human cells in vitro (3, 23, 29, 39, 41, 60). Both the E7 and E6 proteins of high-risk HPVs are required for efficient immortalization of cervical keratinocytes (23, 39), whereas E6 alone is insufficient. Surprisingly, we observed that HPV E6 alone is sufficient to immortalize a subtype of normal human mammary epithelial cells (MECs), providing an experimental system to analyze the targets of E6-mediated cellular transformation (2). Mutational analysis of HPV16 E6 revealed a direct correlation between MEC immortalization and the ability of E6 proteins to induce in vivo p53 degradation in MECs, supporting a crucial role for p53 degradation in E6-mediated immortalization of MECs (13). However, compared to a highly efficient immortalization by the HPV16 E6, dominant-inhibitory p53 mutants were either unable to immortalize MECs (9 of 12 tested) or were relatively inefficient (7, 20). These observations suggested that additional cellular targets exist for the HPV E6 proteins. Consistent with this possibility, HPV16 E6 has been recently found to interact with three proteins: ERC55, a putative calcium binding protein; paxillin, a protein involved in transducing signals from the plasma membrane to the actin cytoskeleton; and hDlg, the human homologue of the Drosophila discs large tumor suppressor protein (9, 31, 35, 52). Binding of these proteins to HPV E6 mutants correlated with the immortalizing ability of E6 proteins, suggesting the potential role of these non-p53 E6-binding proteins in cellular transformation. However, unlike p53, none of these newly identified E6-binding proteins has been reported to be targeted for degradation by HPV E6. In this study, we have used the yeast two-hybrid system to identify a novel putative GTPase-activating protein (GAP), E6TP1 (for E6-targeted protein 1), that interacts with the high-risk HPV E6 proteins and is targeted for in vitro and in vivo degradation. The ability of E6TP1 to selectively interact with immortalizing HPV16 E6 mutants implicates this protein in E6-mediated oncogenesis.
MATERIALS AND METHODS
Yeast two-hybrid constructs and screening.
A two-hybrid library, representing mRNAs expressed in normal MECs, was constructed in pGAD10 vector (custom made through Clontech, Palo Alto, Calif.). Briefly, mRNA purified from normal MEC strain 76N was used to synthesize cDNA in the presence of a mixture of oligo(dT) and random hexanucleotide primers. The cDNA was cloned into the EcoRI site of pGAD10, and a library of 1.5 × 106 primary recombinants with an average insert size of 1.5 kb was obtained. The bait plasmid, pGBT9-E6, was constructed by cloning HPV16 E6 residues 2 to 158, derived by PCR, as a SalI-SmaI fragment into pGBT9 (11). The two-hybrid library screen was performed according to the Matchmaker two-hybrid system protocol (Clontech) to identify E6-interacting proteins. The Saccharomyces cerevisiae yeast strain CG-1945 (five transformations) or HF7c (one transformation) was simultaneously transformed with pGBT9-E6 and the pGAD10 library DNA. HPV16 E6-interacting proteins were identified by growth on Trp−, Leu−, and His− selection medium and expression of β-galactosidase (β-Gal) activity. Clones that remained positive in both assays were retested for E6-specific interaction by assessing their interaction with pGB9-E6 versus two control baits, pLam 5′ (which encodes a human lamin-GAL4 DNA binding domain hybrid in pGBT9) and pVA3 (which encodes a murine p53-GAL4 DNA binding domain hybrid in pGBT9) (Clontech).
Molecular cloning and sequencing of full-length E6TP1 and plasmid constructs.
The 1,403-bp E6TP1 DNA fragment isolated through two-hybrid screening was 32P labeled and used as a probe for colony hybridization of the pGAD10 library. Two longer clones (2,419 and 2,262 bp) obtained through this approach were sequenced. Based on this sequence, two rounds of Marathon PCR (Clontech) were performed with normal mammary gland cDNA as templates to clone the additional 5′ sequences (first round Marathon PCR primers: 5′-ccggcggccgcGAAAGCTGGCAGTACCTTTGATACTGC-3′ and 5′-ccggcggccgcAGGTCCTCTATAACTGTAAGCCATCTG-3′ [for re-PCR]; second round Marathon PCR primers: 5′-ccggcggccgcTCACTCTATCTAGGTGCAACACCAAGTT C-3′ and 5′-ccggcggccgcAATGGGAACTAAGGGTAGACTCAAAGGAG-3′ [for re-PCR]) (10). PCR products were cloned into pBluescript, and the complete sequence was determined based on multiple independent clones. The full-length E6TP1α cDNA used for expression studies was obtained by PCR from mammary gland cDNA with primers that included a NotI restriction site (sense primer, 5′-ccggcggccgcGGTGTGGACG TTGTCTAAATTTCGGTAGCC-3′; antisense primer, 5′-ccggcggccgcAGGTGCTCTGAGGATGCTTTCTATGG-3′). This PCR product was cloned into pBluescript and sequenced. The PCR-generated mutations were corrected by restriction fragment swap with a separate clone. The corrected full-length sequence was cloned through blunt-end ligation into BamHI sites of pSG5 to yield PSG5-E6TP1, which was used for in vitro transcription-translation and in vivo expression in mammalian cells. PSG5-E6TP1α lacking the C-terminal 540 amino acids (aa) (PSG5-E6TP1-ΔC-540) and 129 aa (PSG5-E6TP1-ΔC-129) were constructed by deleting the BamHI and PstI fragments, respectively, from the full-length E6TP1α. PSG5-E6TP1-C-378 and pSG5E6TP1-C-194 were derived from two independent pGAD10 clones obtained from the two-hybrid screen. An N-terminal myc tag, together with an initiation codon and a consensus Kozak sequence for optimized translation, was appended to the 5′ end of E6TP1-C-378 and C-194. GST-E6TP1-C-378 and GST-E6TP1-C-194 were constructed by subcloning the respective cDNA inserts from pGAD10 clones into pGEX2TK. GST-E6TP1-393-1058 was constructed by cloning nucleotides 1524 to 3517 of E6TP1 as an EcoRI fragment into pGEX4T-3. The HPV16 E6 open reading frame was cloned into pGEX2TK to produce GST-E6. A glutathione S-transferase (GST)-fusion protein of E6AP (aa 37 to 865), GSTE6AP, and its mutant lacking aa 391 to 408, GSTE6APmut, were provided by Peter Howley (27). HPV16, -18, -11, and -6 E6 constructs for in vitro translation have been previously described (9). PSG5-HPV16 E6 was provided by Elliot Androphy. The HPV18 and -11 E6 open reading frames were cloned into pSG5 as EcoRI-HindIII fragments, and the HPV6 E6 open reading frame was cloned into pSG5 as an EcoRI-PstI fragment for in vivo expression.
PSG5-HPV16 E6-FLAG was constructed by amplifying HPV16 E6 by PCR with the following primers: sense primer, 5′gcggaattcATGGACTACAAGG ACGACGATGACAAGTTTCAGGACCCACAGGAGCGACCCAG; anti-sense primer, 5′-gccggatccTTACAGCTGGGTTTCTCTACGTGTTCTTGATGA. The PCR sense primer introduced a FLAG tag between the first and second amino acids of the HPV16 E6. The PCR product was digested with the EcoRI and BamHI enzymes and ligated to the EcoRI-BamHI-digested pSG5.
Northern hybridization.
Nylon membranes with 2 μg of poly(A)+ RNA derived from various tissues (tissue blot from Clontech) or 20 μg of total RNA from various cell lines per lane were probed with the 32P-labeled full-length E6TP1α probe and visualized by autoradiography as previously described (3). Hybridization with the 36B4 probe was used as a loading control (34).
In vitro binding assay.
The indicated proteins were translated in vitro in the presence of [35S]cysteine (HPV E6 proteins, E6TP1α, E6TP1α-ΔC-540, and E6TP1-ΔC-129) or [35S]methionine (E6TP1-C-378 and C-194) (NEN, Boston, Mass.) by using a wheat germ lysate-based coupled transcription-translation system (TNT wheat germ lysate system; Promega, Wis.) according to the supplier’s recommendations. The 35S-labeled in vitro-translated proteins were incubated with 1 μg of appropriate GST fusion proteins noncovalently bound to glutathione beads in 300 μl of lysis buffer (100 mM Tris [pH 8.0], 100 mM NaCl, 0.5% Nonidet P-40 [NP-40]) for 2 h at 4°C, and bound 35S-labeled proteins were resolved on a sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gel and visualized by fluorography (9).
In vivo binding assay.
Cos-7 cells were transfected with pSG5E6TP1-C-194, encoding the myc-tagged E6TP1-C194 fragment, and pSG5-16E6, encoding the FLAG-tagged 16E6, by using the Lipofectamine reagent according to the manufacturer’s recommended method (Life Technologies, Bethesda, Md.). Forty-eight hours after transfection, cells were harvested in lysis buffer (100 mM Tris [pH 8.0], 100 mM NaCl, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride), precleared twice with protein G-agarose, and incubated with anti-myc antibody (9E10, obtained from Hamid Band, Harvard Medical School) for 4 h at 4°C. The samples were washed six times with lysis buffer, resuspended in sample buffer, resolved on an SDS–17% PAGE gel, transferred to a polyvinylidene difluoride (PVDF) membrane, blotted with anti-FLAG monoclonal antibody M2 (Sigma, St. Louis, Mo.), and detected by using the enhanced chemiluminescence (ECL) detection system (Amersham, Piscataway, N.J.).
E6-induced in vitro degradation assay.
For the in vitro degradation assay, the various proteins were translated in vitro in the presence of [35S]cysteine (HPV E6 proteins or their mutants and E6TP1α, E6TP1α ΔC-540, E6TP1-ΔC-129, or p53) or [35S]methionine (E6TP1-C-378 and E6TP1-C-194) by using a rabbit reticulocyte lysate-based coupled transcription-translation system (TNT rabbit reticulocyte lysate system; Promega). Five microliters of each in vitro-translated 35S-labeled protein was incubated together with 5 μl of HPV E6 or water-primed (control) lysate. After 5 h at 30°C, the degradation reaction was stopped by adding 100 μl of sample buffer, and the proteins were resolved by SDS-PAGE and visualized by fluorography.
E6-induced in vivo degradation of E6TP1.
A total of 5 × 105 293T cells in a 100-mm-diameter dish were transfected with 10 μg of DNA of the pSG5 vector or pSG5 constructs encoding the indicated proteins (as shown in Fig. 5B and 6B) by using the polyamine reagent (Panvera, Madison, Wis.), according to the manufacturer’s protocol. The total amount of DNA was held constant at 20 μg per dish by adding vector DNA. Cells were harvested after 48 h, and 400 μg of total protein was resolved on an SDS–6% PAGE gel and transferred to a PVDF membrane. Membranes were blotted with a rabbit anti-E6TP1 antiserum (raised against GST-E6TP1-C-378) and detected by ECL.
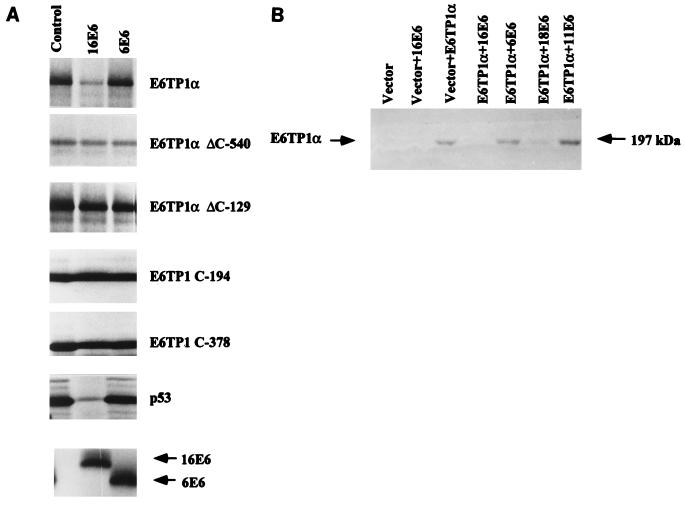
FIG. 5.
E6-induced degradation of E6TP1α. (A) E6-induced degradation of E6TP1α in vitro. HPV16 or -6 E6 proteins (indicated on top) and E6TP1α were translated in vitro in rabbit reticulocyte lysate in the presence of [35S]methionine or -cysteine (see Materials and Methods). 35S-labeled E6TP1α (5 μl) was incubated with water-primed lysate (control) or various E6 proteins (5 μl each) for 5 h. The E6TP1 remaining at the end of the degradation assay was analyzed by SDS-PAGE and visualized by fluorography. Upper panels show E6TP1 and various fragments of E6TP1; p53 was used as a control. The bottom panel shows the HPV16 or 6 E6 proteins used in the degradation assay. (B) E6-induced degradation of E6TP1α in vivo. A total of 5 × 105 293T cells in 100-mm-diameter dishes were transfected with 10 μg each of indicated HPV E6 constructs together with E6TP1α in pSG5 vector with the polyamine reagent. The total amount of DNA per dish was kept constant at 20 μg. Cells were harvested after 48 h, and 400-μg aliquots of lysates were resolved by SDS–6% PAGE and immunoblotted with rabbit anti-E6TP1 antibody followed by ECL detection.
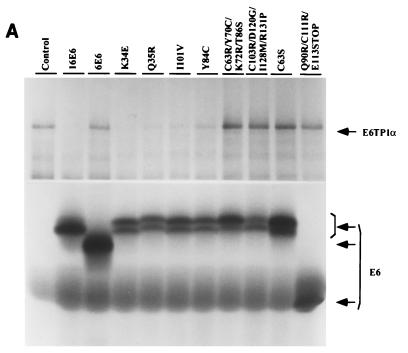
FIG. 6.
Degradation of E6TP1 by immortalizing and nonimmortalizing HPV16 E6 mutants. (A) In vitro degradation. Degradation of E6TP1 in the presence of mutant E6 proteins is shown (see Table 2 for details). E6TP1α and HPV16 E6, and its mutants were translated in vitro in rabbit reticulocyte lysate in the presence of [35S]cysteine. Degradation assays were performed as described in the legend for Fig. 5A. (B) In vivo degradation. HPV16 E6 and its mutants were cotransfected with vector or E6TP1 into 293T cells followed by assessment of E6TP1 protein levels by anti-E6TP1 immunoblotting as described in the legend for Fig. 5B. (C) Expression of mutant E6 proteins in transfected 293T cells. Paired dishes of 293T transfectants shown in panel B were labeled with [35S]cysteine and lysates were immunoprecipitated with an anti-E6 antibody. Bound proteins were resolved by SDS–12% PAGE and visualized by fluorography.
Immunoprecipitation of transfected HPVE6.
293T cells transfected as described above were labeled with 300 μCi of [35S]cysteine (ICN)/ml for 4 h, and lysates were prepared in radioimmunoprecipitation assay buffer (0.15 M NaCl, 50 mM Tris [pH 7.4], 1 mM EDTA, 1% Triton X-100, 1% deoxycholate, 0.1% SDS) (1a). HPV16 E6 protein or its mutants were immunoprecipitated with a rabbit anti-HPV16 E6 antiserum (55). The immunoprecipitated E6 proteins were resolved on an SDS–12% PAGE gel and visualized by fluorography.
Nucleotide sequence accession number. The nucleotide sequences of human E6TP1α and -β have been deposited in GenBank under accession no. AF090989 and AF090990, respectively.
RESULTS
Identification of HPV16 E6-interacting protein by using the yeast two-hybrid system.
To identify novel HPV E6 targets, the HPV16 E6 open reading frame was cloned in the bait vector pGBT9 (11) and used to screen for interacting proteins encoded by normal MEC cDNAs (strain 76N) cloned in the yeast two-hybrid vector pGAD10 (11). Out of a total of 3.96 × 106 transformant clones screened in six transformations, 221 colonies grew on selection medium, and 91 of these were positive in a subsequent β-Gal assay. To identify E6-interacting proteins among these 91 clones, an interaction assay was performed with pGB9-E6 versus two control baits that included human lamin- or murine p53-Gal4 hybrids in pGBT9. Twenty-eight clones were found to interact with HPV16 E6 specifically and were sequenced. Of these 28 clones, 1 weakly positive clone encoded the 476 carboxyl-terminal aa of E6AP, a known E6-binding protein, including its 18-aa E6-binding motif (27). This result indicated that the two-hybrid library and the method of screening were suitable for isolating E6-binding proteins. Interestingly, a set of 11 identical and 1 distinct strongly positive clone identified overlapping regions of a single cDNA. These represented 194 and 378 carboxyl-terminal residues, respectively, of a novel polypeptide referred to as E6TP1, which is described here. The remaining 15 clones encode four novel and one known protein.
Expression pattern of E6TP1.
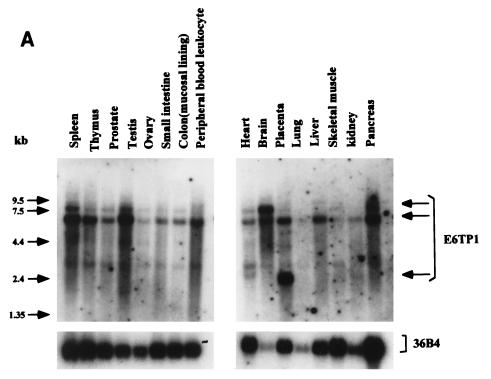
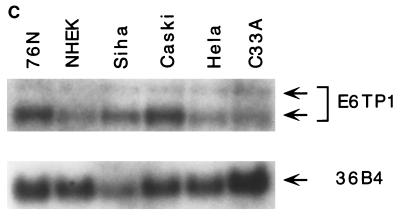
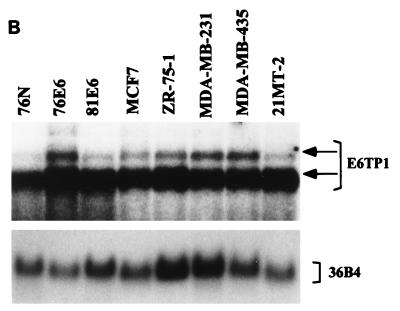
To verify that E6TP1 corresponded to an expressed human gene and to assess its expression, we performed Northern blot analysis of various tissues and cell lines. As shown in Fig. 1, E6TP1 mRNA was expressed in all the tissues and in vitro-established cell lines tested, although the levels varied. The cell lines included the 76N MEC strain from which the two-hybrid library was constructed, human primary foreskin keratinocytes, immortal and tumor MEC lines, and HPV-positive and HPV-negative cervical cancer cell lines (Fig. 1B and C). Two major transcript of 7.5 and 6.0 kb are evident in essentially all tissues and cell lines, with the 6.0-kb transcript predominating in most cases. The larger 7.5-kb transcript is more abundant in brain tissue. A 2.2-kb transcript is abundant in placenta (Fig. 1A). These analyses show that the E6TP1 cDNA fragments isolated by yeast two-hybrid screening represent an authentic mRNA that is widely expressed in human tissues and in vitro-grown cell lines.
FIG. 1.
E6TP1 mRNA expression in human tissues (A) and cultured cell lines (B) and (C). A tissue blot with 2 μg of poly(A+) mRNA per lane (A) (Clontech) and blots with 20 μg of total mRNA from MEC strain 76N, HPV16 E6-immortalized MECs 76E6 and 81E6, and the indicated breast cancer cell lines (B) and from normal human epithelial keratinocytes (HKEC), HPV-positive cervical carcinoma cell lines (HeLa, Siha, and Caski), and a HPV-negative cervical carcinoma cell line (C33A) (C) were probed with a 32P-labeled full-length E6TP1 probe followed by autoradiography. Hybridization with the 36B4 probe was used as a loading control. The major E6TP1 mRNA species are indicated by arrows. Size markers in kilobases are shown at the left of panel A.
Cloning of full-length E6TP1 cDNA.
In order to obtain a full-length cDNA for E6TP1, we utilized a combination of DNA hybridization screening of the 76N pGAD10 library and Marathon PCR cloning from normal mammary gland cDNA (see Materials and Methods). A 5,965-bp cDNA predicting a 1,783-aa polypeptide (E6TP1α) was obtained by using these strategies. The size of this cDNA and the presence of several in-frame stop codons 5′ to the initiation methionine indicate that this cDNA represents the major 6.0-kb transcript expressed in the 76N MEC cell strain (Fig. 2A). One cDNA library-derived clone revealed a 63-bp in-frame insertion after nucleotide 3993, predicting a 1,804-aa polypeptide (E6TP1β) (Fig. 2B). A recent rat cDNA entry in the National Center for Biotechnology Information (NCBI) database (accession no. 2555183) showed a 95.4% amino acid identity with E6TP1α except for a 39-aa insertion in the rat sequence after aa 630. This cDNA sequence is likely to be the rat homologue of E6TP1. The rat homologue lacks the extra 21 aa present in E6TP1β. A human cDNA fragment entry that encodes the C-terminal 1,138 aa of E6TP1β was also noticed (accession no. 2662161).
FIG. 2.
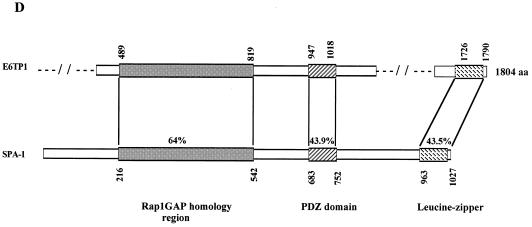
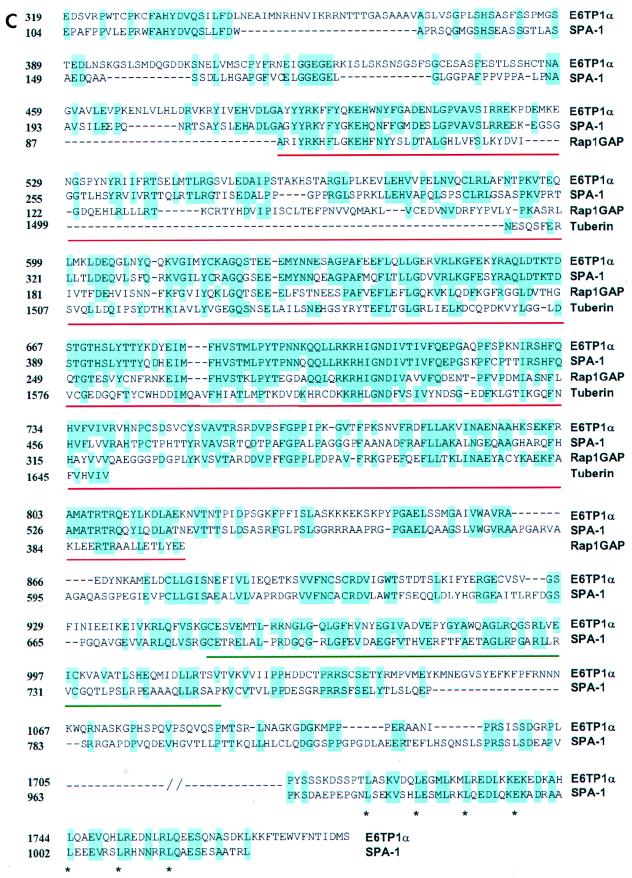
(A) The predicted amino acid sequence of E6TP1α. (B) The additional 21-aa acid sequence present in the E6TP1β sequence immediately after amino acid 1215. Abbreviations for amino acids are A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr. (C) Amino acid sequence alignment of E6TP1β to known GAP proteins. Homology comparisons were made by using the Clustral algorithm with a gap penalty of 3 and were refined by manual adjustment. Identical amino acids are shown in blue. Red underlining, GAP domain; green underlining, PDZ domain; ∗, leucine zipper motif. The GAP proteins shown are human tuberin, human SPA-1, and human Rap1GAP. (D) Schematic alignment of E6TP1 with human SPA-1. Numbers indicate beginning and ending amino acid positions of homologous regions. The percent homology is indicated for each region.
Homology analysis of the E6TP1 polypeptide sequence.
Although the E6TP1 sequence represents a novel cDNA sequence, a Gapped BLASTP search of the NCBI database showed that E6TP1 residues 489 to 819 share high sequence identity with GAP domains of known and putative Rap GTPase-activating proteins. The proteins with the highest degrees of homology to E6TP1 include the mammalian GAPs Rap1GAP (42, 44, 45), tuberin (the tuberous sclerosis complex 2 product, TSC2) (51, 28, 32, 59, 62), and SPA-1 (22, 33) (Fig. 2C and D), as well as Drosophila Rapgap1 (8) and two putative RapGAPs in Caenorhabditis elegans, predicted by the open reading frames identified in the genomic sequences (Table 1). Homology with SPA-1 extends beyond the GAP domain and includes the putative leucine zipper region (47% identity between E6TP1α aa 1705 to 1779 or E6TP1β aa 1726 to 1790 and SPA-1 residues 963 to 1027), as well as other regions, with an overall 42% amino acid identity between E6TP1 residues 319 to 1205 and SPA-1 residues 104 to 930.
TABLE 1.
Comparison of E6TP1 with Rap1GAP homologous proteins
| GAP protein | Species | Accession no. | aa | % Identity with E6TP1 aa 489–819 |
|---|---|---|---|---|
| SPA-1 | Homo sapiens | 2389009 | 216–542 | 64 |
| T27F2.2 | C. elegans | 1403291 | 185–506 | 56 |
| Drosophila Rapgap1 | D. melanogaster | 2655096 | 230–540 | 40 |
| CELF47F6 | C. elegans | 1707164 | 33–343 | 41 |
| Rap1GAP | H. sapiens | 106198 | 13–326 | 38 |
| Tuberin | H. sapiens | 450352 | 1499–1650 | 31 |
Profilescan analysis using the ISREC profilescan server (http://www.ch.embnet.org/software/PFSCAN_form.html) detected a PDZ domain at aa 947 to 1018 in E6TP1. PDZ domains, named after proteins in which they were originally identified, i.e., the synaptic protein PSD-95, the Drosophila discs large gene product (Dlg), and the tight junction protein ZO1, have been found in over 50 different proteins (43). One known function of these modular domains is to promote submembranous protein complexes by binding to C termini of target proteins (43). The E6TP1 PDZ domain showed 43% sequence identity with residues 683 to 752 of SPA-1, indicating that SPA-1 also contains a PDZ domain and that this domain is conserved between E6TP1 and SPA-1. The E6TP1 PDZ domain also showed a 36% identity with the second PDZ domain of the neuronal protein X11 (aa 613 to 683) (5, 16). Overall, the comparison of the amino acid sequences of E6TP1 with known proteins suggests that E6TP1 may also function as a GAP towards Rap and/or other small GTPases.
A search of the NCBI sequence tagged site (STS) database (http://www.ncbi.nlm.nih.gov/cgi-bin/SCIENCE96/ssrch), containing 20,043 gene-based STSs and 5,264 polymorphic STSs in the Genethon genetic map (15, 48), revealed DNA sequence matches between E6TP1 and STS A006F03 (accession no. G20753), SHGC-2959 (accession no. T17112), and WI-9040 (accession no. G06003). These STSs have been localized to human chromosome 14 region 14q23.2-14q24.3. Interestingly, this region is known to undergo loss of heterozygosity in malignant meningiomas (38, 49, 53).
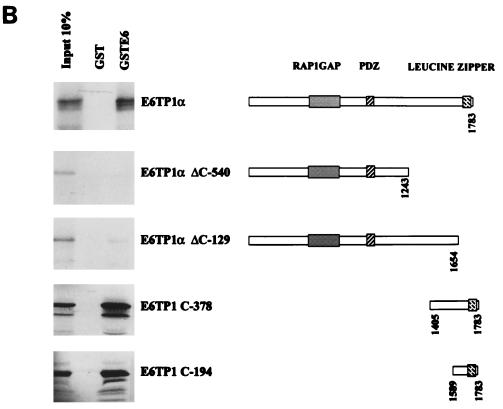
Binding of E6TP1 and HPV E6 proteins in vitro.
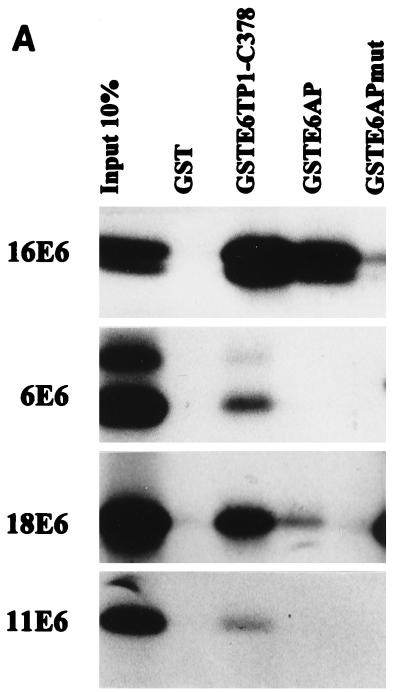
To further confirm the interaction between HPV E6 proteins and E6TP1, we prepared a GST fusion protein encoding the C-terminal 378 residues of E6TP1, representing the sequences present in the longer cDNA clone isolated in the yeast two-hybrid screening. GST-E6TP1-C-378 was used for in vitro-binding experiments with the in vitro wheat germ lysate-translated 35S-labeled HPV16, -18, -6, or -11 E6 polypeptides. GST-E6AP (aa 37 to 865), used as a positive control (27), showed substantial binding to HPV16 and -18 E6 proteins but little binding to HPV6 or -11 E6 proteins, as expected. Neither GST nor a GST-E6AP mutant (Δ391-408, which lacks the 18-aa E6 binding motif [27]) bound to the HPV E6 proteins above background level, demonstrating the specificity of binding. Importantly, the GST-E6TP1 protein showed a substantially higher level of binding to the high-risk HPV E6 proteins (16E6 and 18E6), with relatively low binding to the low-risk HPV E6 proteins (6E6 and 11E6) (Fig. 3A). Thus, similar to E6AP, E6TP1 appears to selectively bind to E6 proteins of high-risk HPVs.
FIG. 3.
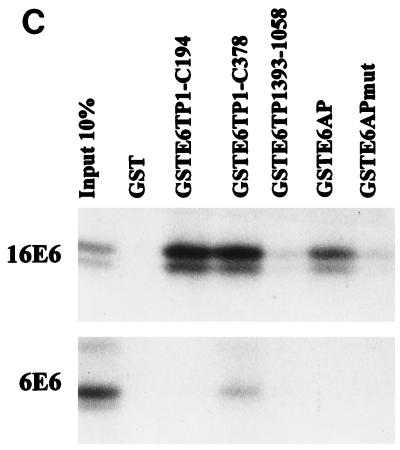
In vitro binding of HPV16 E6 proteins to E6TP1. (A) Various E6 proteins were translated in vitro in wheat germ lysate, and 35S-labeled E6 proteins were allowed to bind to GST, GSTE6TP1 (C-terminal 378 aa of E6TP1 fused to GST), GSTE6AP, or GSTE6APmut (E6AP lacking aa 391 to 408). Input 10%, an aliquot of 10% labeled E6 proteins used in the binding reactions. Bound HPV E6 proteins were analyzed by SDS-PAGE and visualized by fluorography. (B) Full-length E6TP1α or its truncated versions (see amino acid designations in the schematics at right) were translated in vitro in wheat germ lysate, and 35S-labeled proteins were allowed to bind to GST or GST-HPV16 E6. Binding reactions were analyzed as described for panel A. (C) Binding between GST fusion proteins incorporating different regions of E6TP1 (as indicated) and 35S-labeled HPV16 or 6 E6 generated by in vitro translation in wheat germ lysate. Binding reactions were analyzed as described for panel A.
A recent study showed that the human homologue of the Dlg tumor suppressor protein binds to the E6 oncoprotein of the high-risk HPVs through a PDZ domain (31) and mutation of this domain abolished the hDlg interaction with the E6 protein (31). Therefore, it became important to determine if E6TP1 also uses its PDZ domain for interaction with the HPV E6 oncoprotein. Although the binding experiments described above demonstrated that the C-terminal 378 residues of E6TP1, which lack the PDZ domain, were sufficient for E6 binding, it remained possible that the PDZ domain of E6TP1 could also bind to E6, either independently or cooperatively with the C-terminal region. To further define the binding domain(s) of E6TP1, we utilized two series of reciprocal binding experiments. In one approach, a set of E6TP1 mutants were in vitro translated in wheat germ lysate and incubated with either GST or GST-HPV16 E6 fusion proteins. As shown in Fig. 3B, the full-length E6TP1, as well as E6TP1-C-194 or E6TP1-C-378, prominently bound to E6, whereas relatively poor binding was observed with the E6TP1-ΔC-540 and E6TP1-ΔC-129 proteins. In a reciprocal experiment, different regions of E6TP1 were expressed as C-terminal fusions with GST and used for binding assays with in vitro-translated HPV16 E6 (Fig. 3C). These analyses confirmed the results shown in Fig. 3B. Together, these analyses localize the E6 binding site in E6TP1 within the C-terminal 194 residues and show that the PDZ domain contributes little if any toward E6 binding. Thus, E6TP1 utilizes a mechanism distinct from that of the hDlg protein to interact with E6.
Binding of E6TP1 and HPV16 E6 protein in vivo.
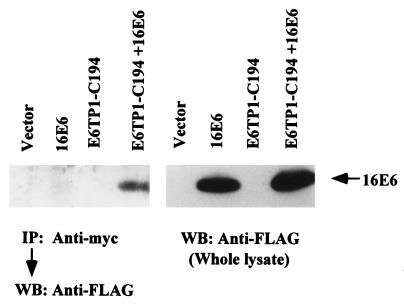
In order to demonstrate that E6 binds to E6TP1 in vivo, Cos-7 cells were transfected with plasmids expressing the myc-tagged E6TP1-C194 fragment and FLAG-tagged E6 protein either alone or together. Forty-eight hours after transfection, the E6TP1-E6 complex was immunoprecipitated by using anti-myc monoclonal antibody (9E10), followed by Western blotting for E6 with the anti-FLAG antibody. As expected, no E6 protein was detected in immunoprecipitates of cells transfected with either vector alone or with the two constructs transfected individually (Fig. 4, left). In contrast, E6 protein was clearly detected in anti-myc immunoprecipitates of cells cotransfected with two constructs (Fig. 4, left). Western blotting of whole lysates (Fig. 4, right) indicated that HPV16 E6 was expressed when transfected with and without E6TP1. These results clearly demonstrate that E6 is able to bind E6TP1 in vivo. Similar results were observed upon transfection of these constructs in 293T cells (data not shown).
FIG. 4.
In vivo binding of HPV16 E6 protein to E6TP1. Cos-7 cells were transfected with 10 μg each of pSG5, myc-tagged E6TP1-C-194, FLAG-tagged-HPV16 E6, or myc-tagged E6TP1-C-194 plus FLAG-tagged HPV16 E6, as indicated. Forty-eight hours after transfection cells were lysed in lysis buffer, and anti-myc (9E10 antibody) immunoprecipitation was carried out, followed by Western blotting with anti-FLAG antibody (left). Direct anti-FLAG Western blotting of whole-cell lysate (right) indicates the expression of HPV16 E6 in the transfected cells.
In vitro and in vivo degradation of E6TP1 induced by high-risk HPV E6 proteins.
The high-risk HPV E6 proteins are known to target p53 for degradation via the E6AP-mediated ubiquitination pathway (25–27, 46, 47). Since E6TP1 showed a selective interaction with high-risk HPV E6 proteins, we wished to examine if E6TP1 was also a target of E6-induced degradation. For this purpose, HPV16 E6, HPV6 E6, p53, full-length E6TP1, and various fragments of E6TP1 were translated in vitro in a rabbit reticulocyte lysate system and subjected to degradation assays as described in Materials and Methods. The high-risk HPV16 E6 induced the degradation of E6TP1 as well as that of p53 (Fig. 5A), but the low-risk HPV6 E6 did not induce degradation of either E6TP1 or p53. The water-primed control lysate (Fig. 5, lanes 1) did not show any effect, as expected. Figure 5A (bottom) shows that similar amounts of HPV16 and 6 E6 proteins were present in the degradation assay. Additional experiments showed that the E6 protein of a second high-risk HPV (HPV 18) was also able to degrade E6TP1, whereas that of the low-risk virus HPV11 was unable to do so (data not shown). Interestingly, all four partial fragments of E6TP1 (ΔC-540, ΔC-129, C-194, and C-378) were deficient in degradation even though E6TP1-C-194 and E6TP1-C-378 efficiently bind to E6 (as shown in Fig. 3B). These results indicate that binding of E6 to E6TP1 by itself is not sufficient for degradation.
In order to assess the in vivo degradation of E6TP1 protein, we coexpressed the E6TP1 and E6 proteins of HPV16, -18, -11, and -6 in 293T cells. The lysates of these cells were subjected to immunoblotting with a rabbit polyclonal antibody specific to E6TP1. This antibody did not detect endogenous E6TP1 in 293T cells, allowing an assessment of the effect of E6 coexpression on the levels of introduced E6TP1. Coexpression of E6 proteins of high-risk HPVs (HPV16 and -18) drastically reduced the level of E6TP1, whereas the levels of E6TP1 were similar to those of controls upon coexpression of E6TP1 with low-risk HPV6 and -11 E6 proteins (Fig. 5B). Immunoprecipitation with antibodies to HPV16 E6, -6 E6, and -18 E6 showed that these proteins were expressed at comparable levels (data not shown); the levels of HPV11 E6 could not be assessed due to lack of an antibody. We conclude from these experiments that E6 proteins from high-risk but not low-risk HPVs can induce in vitro as well as in vivo degradation of E6TP1.
Immortalizing but not the nonimmortalizing E6 mutants target E6TP1 for degradation.
Previous analyses of HPV16 E6 have identified immortalizing and nonimmortalizing mutants of HPV16 E6; immortalizing mutants selectively induce degradation of p53 protein (13). Therefore, we examined if the ability of E6 protein to induce E6TP1 degradation correlates with their immortalizing abilities by using in vitro and in vivo degradation assays. As shown in Fig. 6A and B and Table 2, both in vitro and in vivo experiments demonstrated that, similar to wild-type E6, E6 mutants capable of immortalizing MECs were also efficient in inducing E6TP1 degradation. In contrast, E6 mutants that are deficient in MEC immortalization were incapable of inducing E6TP1 degradation, even though all E6 proteins were expressed at easily detectable levels (Fig. 6C). Thus, the ability of HPV16 E6 mutants to target E6TP1 for degradation directly correlates with their immortalizing ability.
TABLE 2.
Mutational analysis of HPV16 E6 to induce in vitro and in vivo degradation of E6TP1α
| Construct | Degradation
|
Immorta-lizationa | |
|---|---|---|---|
| In vitro | In vivo | ||
| HPV16 E6 | + | + | 6 (6) |
| Lys-34 Glu | + | + | 3 (3) |
| Gln-35 Arg | + | NDb | 3 (3) |
| Ile-101 Val | + | ND | 3 (3) |
| Tyr-84 Cys | + | + | 3 (3) |
| Cys-63 Ser | − | − | 1 (3) |
| Cys-63 Arg/Tyr-70 Cys/Lys-72 Arg/Thr-86 Ser | − | − | 0 (4) |
| Cys-103 Arg/Asp-120 Gly/Ile-128 Met/Arg-131 Pro | − | − | 0 (4) |
| Gln-90 Arg/Cys-111 Arg/Glu-113 Stop | − | − | 0 (4) |
All HPV16 E6 mutants and immortalization experiments are described in detail in reference 13. Immortalization is shown as the number of experiments that yielded immortal cells. The numbers of experiments performed are in parentheses.
ND, not done.
DISCUSSION
It is now amply clear that viral oncogenes target specific cellular proteins that are involved in normal cell growth control. In many cases, such viral target proteins are also altered in human malignancies, thus enhancing the rationale to search for new targets of viral oncoproteins. Notable examples of viral oncogene targets that play eminent roles in cell growth control and are also involved in human cancer are the tumor suppressor proteins Rb and p53 (36, 56). The human papillomavirus E6 oncoprotein targets the p53 tumor suppressor protein, and this trait is crucial for immortalization of human MECs by E6 (13). Surprisingly, dominant-inhibitory p53 mutants are either unable or very inefficient at immortalizing MECs (7, 20), suggesting the existence of additional E6 targets that are involved in cellular transformation. Using the yeast two-hybrid screening method, we have identified a novel putative GAP that selectively binds to high-risk HPV E6 proteins and is a target of E6-induced degradation.
E6TP1 represents a novel gene whose mRNA is expressed widely. The presence of multiple transcripts with tissue-specific differences in relative abundance (e.g., the high abundance of 7.5-kb transcript in brain and 2.2-kb transcript in placenta), together with the isolation of two isoforms that differ in the presence of an insert of 21 aa (present in E6TP1β and absent in E6TP1α) suggests that several forms of E6TP1 protein may be expressed and the relative expression of these forms may be regulated. The isoform cloned and analyzed in this study corresponds to the 6-kb mRNA that represents the major RNA species in most tissues.
Comparison of E6TP1 polypeptide with other proteins showed that E6TP1 possesses a region with extensive homology with GAP domains of several previously characterized GAPs. Those with highest homology are either documented or likely members of the RapGAP family with selectivity towards Raps, the ras-related small G proteins. In particular, high homologies were observed between E6TP1 and SPA-1 (22, 33), rap1GAP (42, 44, 45), tuberin (50a, 28, 32, 59, 62), and a Drosophila Rapgap1 (8), as well as two putative C. elegans rapGAPs (Table 1). Rap1GAP1 exhibits strong GAP activity towards Rap1 but relatively weak activity towards Rap2 (45). SPA-1, on the other hand, exhibits GAP activity towards both Rap1 and Rap2 (33). While it has been suggested that tuberin acts as a Rab5GAP in vivo (61), it also exhibits in vitro GAP activity towards Rap1 (59). These comparisons strongly suggest that E6TP1 may also function as a GAP toward Rap and/or other small GTPases. Further analyses will be required to confirm the putative GAP activity of E6TP1 and to determine its preferred targets.
Homology analysis revealed that E6TP1 is most closely related to SPA-1. In addition to a high level of sequence identity between their GAP domains, strong sequence conservation was noted in other regions (Fig. 2). A C-terminal leucine zipper previously identified in SPA-1 is conserved in E6TP1 (E6TP1α, aa 1705 to 1779; E6TP1β, aa 1726 to 1790). Another region for which there is a high level of sequence identity between these proteins is predicted to form a PDZ domain in each protein. PDZ domains are modular structures that have been identified in a number of distinct proteins (43). One notable function of such domains is to promote submembranous protein assemblies by binding to C termini of transmembrane proteins (43). At present, the subcellular localization of E6TP1 remains to be determined. Nonetheless, these extensive sequence comparisons strongly suggest that SPA-1 and E6TP1 represent a subfamily of RapGAPs.
Interestingly, the expression of SPA-1 mRNA is upregulated quickly following mitogenic stimulation of lymphocytes, suggesting a potential role of this subfamily of proteins in cell growth regulation and/or cell cycle events. In fact, introduction of SPA-1 into NIH 3T3 cells followed by serum starvation led to cell death resembling the mitotic catastrophes of the S phase (22). Notably, the gene that encodes tuberin (TSC2) is frequently mutated in the autosomal dominant syndrome of tuberous sclerosis and therefore represents a clear example of a tumor suppressor protein. Furthermore, when TSC2 was introduced into a renal carcinoma cell line derived from the Eker rat, a model of hereditary renal carcinoma, tumorigenicity was suppressed (28). Similarly, expression of TSC2 as a transgene in the Eker rat rescued these animals from embryonic lethality and renal carcinogenesis (32). Thus, it is tempting to speculate that the E6 oncoprotein-binding partner E6TP1 may also be involved in cell growth regulation and carcinogenesis. This would suggest a potential link between HPV E6 oncogenesis and alteration of a small G protein signaling pathway. Notably, E6TP1 localizes to chromosome 14q23.2-14q24.3. This region has been previously shown to undergo loss of heterozygosity in malignant meningiomas (38, 49, 53), suggesting that this locus may harbor a tumor suppressor gene. It will be of obvious significance to determine if E6TP1 represents the relevant tumor suppressor gene. Interestingly, another protein that has been recently identified as an HPV E6-binding protein, hDlg, is a homologue of a protein identified in Drosophila as a tumor suppressor protein (31, 35).
How E6TP1 may contribute to cell growth regulation and toward HPV E6-induced cellular transformation are matters of speculation at present. Rap1 was originally identified by its ability to revert the phenotype of viral K-ras-transformed NIH 3T3 cells (30); however, the potential physiological function of Rap1 in signaling pathways remains poorly defined. Rap1 has been shown to bind in vitro to many of the same effector proteins as Ras, such as Raf-1, the catalytic subunit of p110 of phosphatidylinositol 3-kinase, and the ral guanine nucleotide exchange factors (19, 21, 24). Thus, it is hypothesized that Rap1 antagonizes Ras function by sequestering Ras effectors in inactive complexes. However, Vossler et al. showed that Rap1, activated either by mutation or by the cyclic AMP-dependent protein kinase PKA, is a selective activator of B-Raf, which activates the mitogen-activated protein kinase cascade and then activates transcription factor Elk-1 in PC12 cells (54). Also, Yoshida et al. showed that the microinjection of Rap1 into Swiss 3T3 cells enhanced the mitogenic signaling pathways in response to insulin (63). Recently, transfection of Rap1 into Swiss 3T3 cells was shown to increase cell proliferation and oncogenically transform the cells (1). Sequence homology analysis strongly suggested E6TP1 to be a RapGAP. As a putative negative regulator of Rap1 GTPase, E6TP1 could negatively regulate the mitogenic signaling pathways mediated by Rap1 or related proteins. Degradation of E6TP1 by HPV16 E6 or inactivation of E6TP1 by another mechanism such as mutation would then be expected to promote mitogenic signaling and thus may contribute to oncogenic transformation.
The ability of HPV E6 protein to bind and target E6TP1 for degradation is reminiscent of the similar degradation of p53 tumor suppressor protein upon its indirect E6AP-mediated interaction with E6. Previous analyses of E6-p53 interaction have suggested that E6 binding is sufficient to target bound proteins for degradation. On the other hand, recently identified E6-binding proteins ERC55, paxillin, and hDlg have not been shown to undergo E6-induced degradation. This result would suggest that degradation is not an obligatory consequence of E6 binding. Notably, the truncated mutants of E6TP1 that efficiently bound to E6 were not degraded. Perhaps, other regions of E6TP1 are required to target it for degradation. Thus, distinct regions of cellular target proteins may be crucial for E6 binding and E6-dependent targeting to cellular degradation machinery. The precise degradation pathway for E6TP1 remains to be determined, although parallels with E6-induced p53 interaction suggest a potential role for proteasome-dependent degradation. It will also be of interest to determine if E6TP1 degradation induced by E6 utilizes E6AP, a ubiquitin ligase, similar to the E6-induced p53 degradation. In this context, we have observed that E6 can simultaneously bind E6TP1 and E6AP (20a). Further analyses are required to address the role of E6AP in E6-induced degradation of E6TP1 protein.
Using the available antisera against E6TP1, we have not yet been able to detect the endogenous E6TP1 protein. Therefore, it is not clear at present whether the endogenous E6TP1 is also regulated by degradation. Generation of more suitable reagents, such as monoclonal antibodies, should allow detection of the endogenous E6TP1 protein as well as assessment of its degradation in naturally E6-expressing and nonexpressing cervical carcinoma cell lines.
The recently identified E6-binding protein, hDlg, was shown to utilize its PDZ domain to bind to E6. In contrast, E6TP1 requires its C-terminal 194 residues for its interaction with E6. Furthermore, when the C-terminal 194 residues were deleted, E6TP1 could not bind to E6. Thus, the E6TP1 PDZ domain does not contribute to E6 binding and would be available for other interactions. Given the ability of PDZ domains to mediate protein-protein interactions, we speculate that the E6TP1-E6 complex may also include other proteins bound to the E6TP1 PDZ domain. Thus, the E6TP1 PDZ domain may help in localizing E6 to other cellular targets.
Similar to E6AP, ERC55, paxillin, and hDlg (9, 25, 26, 31, 52), E6TP1 selectively bound to high-risk HPV E6 proteins in vitro. Furthermore, only high-risk HPV E6 proteins promoted E6TP1 degradation in vitro and in vivo. In addition, E6TP1 binding and degradation were observed with E6 mutants that are competent at cellular immortalization but not with those mutants that fail to immortalize cells. Altogether, these results strongly argue that E6TP1, similar to other known E6-binding proteins, is a transformation-relevant target of HPV E6. It appears likely that an oncoprotein, such as E6, concurrently targets a number of cellular metabolic pathways en route to efficient transformation. Given multiple checkpoints that cells utilize to ensure proper cellular proliferation, the strategy of oncogenic viruses to target multiple cellular pathways is likely to reflect the natural tumorigenesis which is well documented to be a multistep process.
In conclusion, we have identified a novel putative RapGAP as a cellular target of the high-risk HPV E6 oncoproteins. Given that previously identified targets of HPV E6 and other DNA tumor virus oncogenes are tumor suppressor proteins and several GAP proteins (such as tuberin and the Neurofibromatosis type 1 gene product) are known to function as tumor suppressors (4, 51), E6TP1 may represent a novel tumor suppressor protein whose inactivation contributes to oncogenesis. Localization of E6TP1 to chromosome 14q23.2-14q24.3, a site for loss of heterozygosity in malignant meningiomas (38, 49, 53), further points to this possibility.
ACKNOWLEDGMENTS
We thank E. J. Androphy and P. Howley for plasmids, D. Galloway for HPV 6E6 antiserum, and H. Band for critical reading of the manuscript and suggestions throughout these studies.
This work was supported by NIH grants CA56803 and CA64823 to V.B.
REFERENCES
- 1.Altschuler D L, Ribeiro-Neto F. Mitogenic and oncogenic properties of the small G protein Rap1b. Proc Natl Acad Sci USA. 1998;95:7475–7479. doi: 10.1073/pnas.95.13.7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Androphy E J, Hubbert N L, Schiller J T, Lowy D R. Identification of the HPV-16 E6 protein from transformed mouse cells and human cervical carcinoma cell lines. EMBO J. 1987;6:989–992. doi: 10.1002/j.1460-2075.1987.tb04849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Band V, De C J A, Delmolino L, Kulesa V, Sager R. Loss of p53 protein in human papillomavirus type 16 E6-immortalized human mammary epithelial cells. J Virol. 1991;65:6671–6676. doi: 10.1128/jvi.65.12.6671-6676.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Band V, Zajchowski D, Kulesa V, Sager R. Human papilloma virus DNAs immortalize normal human mammary epithelial cells and reduce their growth factor requirements. Proc Natl Acad Sci USA. 1990;87:463–467. doi: 10.1073/pnas.87.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basu T N, Gutmann D H, Fletcher J A, Glover T W, Collins F S, Downward J. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature. 1992;356:713–715. doi: 10.1038/356713a0. [DOI] [PubMed] [Google Scholar]
- 5.Borg J P, Ooi J, Levy E, Margolis B. The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol Cell Biol. 1996;16:6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer S N, Wazer D E, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 7.Cao Y, Gao Q, Wazer D E, Band V. Abrogation of wild-type p53-mediated transactivation is insufficient for mutant p53-induced immortalization of normal human mammary epithelial cells. Cancer Res. 1997;57:5584–5589. [PubMed] [Google Scholar]
- 8.Chen F, Barkett M, Ram K T, Quintanilla A, Hariharan I K. Biological characterization of Drosophila Rapgap1, a GTPase activating protein for Rap1. Proc Natl Acad Sci USA. 1997;94:12485–12490. doi: 10.1073/pnas.94.23.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J J, Reid C E, Band V, Androphy E J. Interaction of papillomavirus E6 oncoproteins with a putative calcium-binding protein. Science. 1995;269:529–531. doi: 10.1126/science.7624774. [DOI] [PubMed] [Google Scholar]
- 10.Chenchik A, Moqadam F, Siebert P. A new method for full-length cDNA cloning by PCR. In: Krieg P A, editor. A laboratory guide to RNA: isolation, analysis, and synthesis. New York, N.Y: John Wiley & Sons, Inc.; 1996. pp. 273–321. [Google Scholar]
- 11.Chien C T, Bartel P L, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 13.Dalal S, Gao Q, Androphy E J, Band V. Mutational analysis of human papillomavirus type 16 E6 demonstrates that p53 degradation is necessary for immortalization of mammary epithelial cells. J Virol. 1996;70:683–688. doi: 10.1128/jvi.70.2.683-688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeCaprio J A, Ludlow J W, Figge J, Shew J Y, Huang C M, Lee W H, Marsilio E, Paucha E, Livingston D M. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 15.Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 16.Duclos F, Boschert U, Sirugo G, Mandel J L, Hen R, Koenig M. Gene in the region of the Friedreich ataxia locus encodes a putative transmembrane protein expressed in the nervous system. Proc Natl Acad Sci USA. 1993;90:109–113. doi: 10.1073/pnas.90.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dyson N, Bernards R, Friend S H, Gooding L R, Hassell J A, Major E O, Pipas J M, Vandyke T, Harlow E. Large T antigens of many polyomaviruses are able to form complexes with the retinoblastoma protein. J Virol. 1990;64:1353–1356. doi: 10.1128/jvi.64.3.1353-1356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyson N, Howley P M, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 19.Frech M, John J, Pizon V, Chardin P, Tavitian A, Clark R, McCormick F, Wittinghofer A. Inhibition of GTPase activating protein stimulation of Ras-p21 GTPase by the Krev-1 gene product. Science. 1990;249:169–171. doi: 10.1126/science.2164710. [DOI] [PubMed] [Google Scholar]
- 20.Gao Q, Hauser S H, Liu X L, Wazer D E, Madoc-Jones H, Band V. Mutant p53-induced immortalization of primary human mammary epithelial cells. Cancer Res. 1996;56:3129–3133. [PubMed] [Google Scholar]
- 20a.Gao, Q., and V. Band. Unpublished data.
- 21.Hata Y, Kikuchi A, Sasaki T, Schaber M D, Gibbs J B, Takai Y. Inhibition of the ras p21 GTPase-activating protein-stimulated GTPase activity of c-Ha-ras p21 by smg p21 having the same putative effector domain as ras p21s. J Biol Chem. 1990;265:7104–7107. [PubMed] [Google Scholar]
- 22.Hattori M, Tsukamoto N, Nur-e-Kamal M S, Rubinfeld B, Iwai K, Kubota H, Maruta H, Minato N. Molecular cloning of a novel mitogen-inducible nuclear protein with a Ran GTPase-activating domain that affects cell cycle progression. Mol Cell Biol. 1995;15:552–560. doi: 10.1128/mcb.15.1.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawley-Nelson P, Vousden K H, Hubbert N L, Lowy D R, Schiller J T. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989;8:3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrmann C, Horn G, Spaargaren M, Wittinghofer A. Differential interaction of the ras family GTP-binding proteins H-Ras, Rap1A, and R-Ras with the putative effector molecules Raf kinase and Ral-guanine nucleotide exchange factor. J Biol Chem. 1996;271:6794–6800. doi: 10.1074/jbc.271.12.6794. [DOI] [PubMed] [Google Scholar]
- 25.Huibregtse J M, Scheffner M, Howley P M. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus type 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huibregtse J M, Scheffner M, Howley P M. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol Cell Biol. 1993;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huibregtse J M, Scheffner M, Howley P M. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol Cell Biol. 1993;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin F, Wienecke R, Xiao G H, Maize J C, Jr, DeClue J E, Yeung R S. Suppression of tumorigenicity by the wild-type tuberous sclerosis 2 (Tsc2) gene and its C-terminal region. Proc Natl Acad Sci USA. 1996;93:9154–9159. doi: 10.1073/pnas.93.17.9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaur P, McDougall J K. Characterization of primary human keratinocytes transformed by human papillomavirus type 18. J Virol. 1988;62:1917–1924. doi: 10.1128/jvi.62.6.1917-1924.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitayama H, Sugimoto Y, Matsuzaki T, Ikawa Y, Noda M. A ras-related gene with transformation suppressor activity. Cell. 1989;56:77–84. doi: 10.1016/0092-8674(89)90985-9. [DOI] [PubMed] [Google Scholar]
- 31.Kiyono T, Hiraiwa A, Fujita M, Hayashi Y, Akiyama T, Ishibashi M. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:11612–11616. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi T, Mitani H, Takahashi R, Hirabayashi M, Ueda M, Tamura H, Hino O. Transgenic rescue from embryonic lethality and renal carcinogenesis in the Eker rat model by introduction of a wild-type Tsc2 gene. Proc Natl Acad Sci USA. 1997;94:3990–3993. doi: 10.1073/pnas.94.8.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurachi H, Wada Y, Tsukamoto N, Maeda M, Kubota H, Hattori M, Iwai K, Minato N. Human SPA-1 gene product selectively expressed in lymphoid tissues is a specific GTPase-activating protein for Rap1 and Rap2. Segregate expression profiles from a rap1GAP gene product. J Biol Chem. 1997;272:28081–28088. doi: 10.1074/jbc.272.44.28081. [DOI] [PubMed] [Google Scholar]
- 34.Laborda J. 36B4 cDNA used as an estradiol-independent mRNA control is the cDNA for human acidic ribosomal phosphoprotein PO. Nucleic Acids Res. 1991;19:3998. doi: 10.1093/nar/19.14.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee S S, Weiss R S, Javier R T. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:6670–6675. doi: 10.1073/pnas.94.13.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 37.Maki C G, Huibregtse J M, Howley P M. In vivo ubiquitination and proteasome-mediated degradation of p53. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 38.Menon A G, Rutter J L, von Sattel J P, Synder H, Murdoch C, Blumenfeld A, Martuza R L, von Deimling A, Gusella J F, Houseal T W. Frequent loss of chromosome 14 in atypical and malignant meningioma: identification of a putative ‘tumor progression’ locus. Oncogene. 1997;14:611–616. doi: 10.1038/sj.onc.1200853. [DOI] [PubMed] [Google Scholar]
- 39.Munger K, Phelps W C, Bubb V, Howley P M, Schlegel R. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munger K, Werness B A, Dyson N, Phelps W C, Harlow E, Howley P M. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pirisi L, Yasumoto S, Feller M, Doniger J, DiPaolo J A. Transformation of human fibroblasts and keratinocytes with human papillomavirus type 16 DNA. J Virol. 1987;61:1061–1066. doi: 10.1128/jvi.61.4.1061-1066.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polakis P, Rubinfeld B, McCormick F. Phosphorylation of rap1GAP in vivo and by cAMP-dependent kinase and the cell cycle p34cdc2 kinase in vitro. J Biol Chem. 1992;267:10780–10785. [PubMed] [Google Scholar]
- 43.Ponting C P, Phillips C, Davies K E, Blake D J. PDZ domains: targeting signalling molecules to sub-membranous sites. Bioessays. 1997;19:469–479. doi: 10.1002/bies.950190606. [DOI] [PubMed] [Google Scholar]
- 44.Rubinfeld B, Crosier W J, Albert I, Conroy L, Clark R, McCormick F, Polakis P. Localization of the rap1GAP catalytic domain and sites of phosphorylation by mutational analysis. Mol Cell Biol. 1992;12:4634–4642. doi: 10.1128/mcb.12.10.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rubinfeld B, Munemitsu S, Clark R, Conroy L, Watt K, Crosier W J, McCormick F, Polakis P. Molecular cloning of a GTPase activating protein specific for the Krev-1 protein p21rap1. Cell. 1991;65:1033–1042. doi: 10.1016/0092-8674(91)90555-d. [DOI] [PubMed] [Google Scholar]
- 46.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 47.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 48.Schuler G D, Boguski M S, Stewart E A, Stein L D, Gyapay G, Rice K, White R E, Rodriguez-Tome P, Aggarwal A, Bajorek E, Bentolila S, Birren B B, Butler A, Castle A B, Chiannilkulchai N, Chu A, Clee C, Cowles S, Day P J, Dibling T, Drouot N, Dunham I, Duprat S, East C, Hudson T J, et al. A gene map of the human genome. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- 49.Simon M, von Deimling A, Larson J J, Wellenreuther R, Kaskel P, Waha A, Warnick R E, Tew J M, Jr, Menon A G. Allelic losses on chromosomes 14, 10, and 1 in atypical and malignant meningiomas: a genetic model of meningioma progression. Cancer Res. 1995;55:4696–4701. [PubMed] [Google Scholar]
- 50.Stubdal H, Zalvide J, Campbell K S, Schweitzer C, Roberts T M, DeCaprio J A. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol Cell Biol. 1997;17:4979–4990. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.The European Chromosome 16 Tuberous Sclerosis Consortium. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell. 1993;75:1305–1315. doi: 10.1016/0092-8674(93)90618-z. [DOI] [PubMed] [Google Scholar]
- 52.Tong X, Howley P M. The bovine papillomavirus E6 oncoprotein interacts with paxillin and disrupts the actin cytoskeleton. Proc Natl Acad Sci USA. 1997;94:4412–4417. doi: 10.1073/pnas.94.9.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tse J Y, Ng H K, Lau K M, Lo K W, Poon W S, Huang D P. Loss of heterozygosity of chromosome 14q in low- and high-grade meningiomas. Hum Pathol. 1997;28:779–785. doi: 10.1016/s0046-8177(97)90149-0. [DOI] [PubMed] [Google Scholar]
- 54.Vossler M R, Yao H, York R D, Pan M G, Rim C S, Stork P J. cAMP activates MAP kinase and Elk-1 through a B-Raf- and Rap1-dependent pathway. Cell. 1997;89:73–82. doi: 10.1016/s0092-8674(00)80184-1. [DOI] [PubMed] [Google Scholar]
- 55.Wazer D E, Liu X L, Chu Q, Gao Q, Band V. Immortalization of distinct human mammary epithelial cell types by human papilloma virus 16 E6 or E7. Proc Natl Acad Sci USA. 1995;92:3687–3691. doi: 10.1073/pnas.92.9.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 57.Werness B A, Levine A J, Howley P M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 58.Whyte P, Buchkovich K J, Horowitz J M, Friend S H, Raybuck M, Weinberg R A, Harlow E. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 59.Wienecke R, Konig A, DeClue J E. Identification of tuberin, the tuberous sclerosis-2 product. Tuberin possesses specific Rap1GAP activity. J Biol Chem. 1995;270:16409–16414. doi: 10.1074/jbc.270.27.16409. [DOI] [PubMed] [Google Scholar]
- 60.Woodworth C D, Doniger J, DiPaolo J A. Immortalization of human foreskin keratinocytes by various human papillomavirus DNAs corresponds to their association with cervical carcinoma. J Virol. 1989;63:159–164. doi: 10.1128/jvi.63.1.159-164.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao G H, Shoarinejad F, Jin F, Golemis E A, Yeung R S. The tuberous sclerosis 2 gene product, tuberin, functions as a Rab5 GTPase activating protein (GAP) in modulating endocytosis. J Biol Chem. 1997;272:6097–6100. doi: 10.1074/jbc.272.10.6097. [DOI] [PubMed] [Google Scholar]
- 62.Yeung R S, Xiao G H, Jin F, Lee W C, Testa J R, Knudson A G. Predisposition to renal carcinoma in the Eker rat is determined by germ-line mutation of the tuberous sclerosis 2 (TSC2) gene. Proc Natl Acad Sci USA. 1994;91:11413–11416. doi: 10.1073/pnas.91.24.11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshida Y, Kawata M, Miura Y, Musha T, Sasaki T, Kikuchi A, Takai Y. Microinjection of smg/rap1/Krev-1 p21 into Swiss 3T3 cells induces DNA synthesis and morphological changes. Mol Cell Biol. 1992;12:3407–3414. doi: 10.1128/mcb.12.8.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zur Hausen H. Papillomaviruses in human cancer. Appl Pathol. 1987;5:19–24. [PubMed] [Google Scholar]
- 65.Zur Hausen H, Schneider A. The Papillomaviruses. In: Howley P M, Salzman N P, editors. The papovaviridae. Vol. 2. New York, N.Y: Plenum; 1987. pp. 245–263. [Google Scholar]