Abstract
Cryptococcus neoformans is a fungus that is pathogenic in humans and that can produce melanin in vitro. Melanization is associated with virulence, but there is no evidence that melanin is made during infection. Melanins are difficult to study because they are amorphous and insoluble. Melanin-binding peptides from a phage display library were used to demonstrate that C. neoformans makes melanin-like compounds in tissue. Melanin-binding peptides were characterized by a high proportion of positively charged and aromatic residues. Two other methods, demonstration of an antibody response to melanin in mice infected with C. neoformans and analysis of yeast cell walls in infected tissue by light microscopy, were used to support these findings. The demonstration that C. neoformans melanizes in tissue has important implications for pathogenesis and drug discovery.
Melanins are pigments of biological origin that conform to a unique electron spin resonance pattern (4, 19). In contrast to the other great natural pigments, such as the hemoglobins, chlorophylls, flavonoids, and carotenoids, little is known about the structure of naturally occurring melanin (2, 4, 7, 10, 11, 15, 27). It has been difficult to use existing biochemical and biophysical techniques to study melanins because they are insoluble and amorphous. Although the pigments are usually black or brown, blue, green, and red melanins also exist (27). This indicates that melanins are structurally heterogeneous. Some chemical and biological properties of melanin include electron exchange, free radical production and absorbtion, protection from UV light, and drug binding (7). In addition, some biological species utilize melanin for camouflage or sexual display (7). Melanin is also believed to play a significant role in the pathogenesis of malignant melanoma (6) and may interfere with the efficacy of melanoma therapy by modifying the effects of ionizing radiation (17) or by binding and chelating antineoplastic drugs.
Cryptococcus neoformans is an encapsulated fungus that causes life-threatening meningoencephalitis in 6 to 8% of patients with AIDS (3). C. neoformans has a laccase that catalyzes the synthesis of melanin in the presence of phenolic compounds, such as l-dopa (14, 28). The ability of C. neoformans to melanize in vitro has been associated with virulence (16, 18), but melanin synthesis in vivo has not been demonstrated. Melanin has been shown to protect C. neoformans against oxidants (25), amphotericin B (23), and macrophages in vitro (22). Some drugs that bind melanin are toxic to eukaryotic cells containing melanin. For example, the melanin-binding compound trifluoperazine has greater fungicidal activity against melanized than nonmelanized cryptococcal cells (26). Establishing whether melanization occurs in vivo is important for understanding the relationship of phenoloxidase activity, pigment production, and virulence. To date, this has not been possible, because stains for melanin are not specific for this compound (9). Melanin forms a shell in the cell wall of C. neoformans that is resistant to acid hydrolysis (24). Melanin “ghosts” in the shape of cells can be isolated from melanized cells by treatment with detergent and acid, and this permits biochemical studies of this pigment (24).
Phage display libraries are powerful tools with which to identify peptide sequences with affinity for a specific ligand because they contain a vast array of protein sequences (20). Some applications of phage display include epitope mapping, identification of ligands, cDNA expression screening, generation of immunogens, and drug development (8). We screened a random decapeptide phage display library (21) with purified C. neoformans melanin in search of melanin-binding peptides that could serve as novel tools with which to investigate melanin and melanization. We also studied surface characteristics of melanin by analyzing the structure of the peptides expressed by melanin-selected phage and examining their binding to C. neoformans melanin by scanning electron microscopy.
MATERIALS AND METHODS
Peptide library and host bacteria.
The decapeptide library (L100) has been described previously and contains approximately 400 million different peptides (21). The library contains random peptide inserts at the N terminus of the pIII coat protein of the phage fd-tet (13). Phage were grown in the kanamycin-resistant Escherichia coli strain K91kan.
Selection of melanin-binding phage.
Phage expressing melanin-binding peptides were selected by consecutive cycles of selection and amplification (20). Briefly, a 1-mg sample of purified melanin was washed three times with biopanning buffer (BPB) (10 mM Tris-HCl [pH 7.5], 150 mM NaCl [Tris-buffered saline {TBS}], 0.1% [wt/vol] bovine serum albumin [BSA], 0.1% [vol/vol] Tween 20, and 0.02% NaN3) at room temperature (RT). Between washes, the melanin particles were collected by centrifugation at 6,000 rpm in a DAMON centrifuge (IEC Division) for 5 min. For the first incubation, melanin was incubated with 1.2 × 1011 transducing units (TU) of the library. For subsequent rounds of selection, ∼1 × 109 TU from the preceding biopan were used. The first three biopans were incubated overnight. To minimize nonspecific binding, the fourth-round biopan was of 20-min duration. Following incubation, the melanin was washed seven times with BPB, eluted in 100 μl of 0.1 M glycine-HCl (pH 2.2), and neutralized with 15 μl of 2 M Tris-HCl (pH 8.0). Titration, amplification, and purification of phage were done according to the method of Smith and Scott (20).
Sequencing.
Purification of phage DNA was done as described previously (20). The primer PIIIP (TGAATTTTCTGTATGAGG) was used for annealing. An automated sequencer (model ABI 377; Perkin-Elmer Corp., Foster City, Calif.) was used for sequencing. Peptide sequences were deduced from the DNA sequence.
Peptide synthesis.
The decapeptide coded for by phage 4B4 was synthesized in the Laboratory for Macromolecular Analysis (Albert Einstein College of Medicine, Bronx, N.Y.), and the peptide structure was verified by mass spectrometry and amino acid sequencing. Peptide 4B4 was also synthesized with biotin at the C terminus.
ELISA.
Melanin enzyme-linked immunosorbent assay (ELISA) plates were produced as described previously (12). All incubations were done at 37°C, and the plates were washed three times with TBS, 1% (wt/vol) BSA, 0.05% (vol/vol) Tween 20, and 0.02% NaN3 between steps. A phage lacking the decapeptide insert (φ33) and a phage containing a nonspecific insert (φA1 [LQYTPSWMLV]) were used as negative controls (21). Phage (5 × 1010 virions) were incubated for 2 h. Sheep anti-M13 antibody (5 Prime, 3 Prime, Inc., Boulder, Colo.) was diluted 1:4,000 in TBS and applied for 1 h. An alkaline-phosphatase- conjugated donkey anti-sheep immunoglobulin G (IgG) (Sigma Chemical Co., St. Louis, Mo.) was added at a 1:4,000 dilution to TBS and left for 1 h. The reaction was developed with phosphatase substrate, and absorbance at 405 nm was measured with a Ceres 900HDi (Bio-Tek Instruments Inc., Winooski, Vt.). The titer was defined as the highest dilution that gave an absorbance measurement two times greater than the background.
A competition ELISA using 5 × 1010 virions of φ4B4 and serial dilutions of either the synthetic peptide 4B4 (YERKFWHGRH) or an irrelevant control peptide, P601G (DGASYSWMYGA), was performed. The samples were then tested in accordance with the method described above.
Serum was obtained at various times from A/JCr and C57BL/6 mice (6 to 14 weeks old) infected as described previously (5). Analysis of the serum by ELISA for antimelanin antibodies was tested as described previously (12).
Scanning electron microscopy.
C. neoformans melanin was incubated with or without φ4B4 or φ4A3 for 20 min at RT. The sample was washed three times with TBS and then incubated in 2.5% glutaraldehyde for 1 h at RT. The sample was then applied to a polylysine-coated coverslip and serially dehydrated in alcohol. The sample was dried (Samdri-790; Tousimis, Rockville, Md.), coated with gold palladium (Desk-1; Denton Vacuum, Inc., Cherry Hill, N.J.), and viewed using a JEOL (Tokyo, Japan) JAM-6400 electron microscope. Negative controls were prepared with phage without an insert (φ33).
Immunocytochemistry.
C. neoformans serotype D strain 24067 obtained from the American Type Culture Collection (Rockville, Md.) and a well-characterized melanin-deficient mutant, 24067, mel− (22) were grown at 30°C with shaking in a defined chemical medium (15 mM glucose, 10 mM MgSO4, 29.4 mM KH2PO4, 13 mM glycine, 3 μM thiamine). Cultures were grown for 8 days with and without the addition of 1 mM l-dopa (Sigma) as a substrate for melanin production. The cells were washed twice in TBS, collected by centrifugation, and then embedded in Tissue Freezing Media (Triangle Biomedical Sciences, Durham, N.C.) and sectioned. The samples were washed in TBS and then blocked with 2% BSA (ICN Biomedicals, Inc., Aurora, Ohio)–5% fetal calf serum (Harlan, Indianapolis, Ind.) in TBS (pH 7.2) (BSA/FCS) for 1 h at RT. Phage were biotinylated by incubating sulfoNHS-biotin (Sigma) (1:10 ratio of biotin to phage [wt/wt]) with 0.1 M sodium borate (pH 8.8) for 4 h. The reaction was terminated by 0.5 μl of diethanolamine. Biotinylated phage (1010 virions) were incubated on the samples for 1 h at RT. The specimens were washed three times. Streptavidin conjugated with fluorescein isothiocyanate (FITC) (Southern Biotech, Birmingham, Ala.) diluted 1:4,000 with TBS was added and left for 1 h at RT. The samples were again washed three times, coverslips were applied using Gel/mount (Biomeda Corp., Foster City, Calif.), and the slides were viewed. Negative controls included cells grown without l-dopa and biotinylated nonspecific phage (φA1).
A/JCr and C57BL/6 mice (6 to 14 weeks old) were infected as described previously (5). Mice were sacrificed at 2 h, 24 h, 48 h, 7 days, 14 days, and 28 days, and the lungs were embedded in paraffin. Sections 4 μm thick were stained with hematoxylin and eosin and viewed by light microscopy. The tissue sections were also used to test for biotinylated melanin-binding phage 4B4 or biotinylated synthetic melanin-binding peptide 4B4 binding to cryptococcal cells. The synthetic peptides were biotinylated during synthesis (see above). Tissue sections were removed from paraffin and incubated with proteinase K (Sigma) (20 μg/ml) for 2 h at RT. The samples were then microwaved in 10 mM citric acid for 5 min. After cooling, the samples were blocked with BSA/FCS for 1 h at RT. A solution of monoclonal antibodies specific for cryptococcal polysaccharide [2H1(1)] (5 μg) with either phage (1010 virions), synthetic peptide (50 μg), or TBS was applied overnight at RT. The specimens were washed three times. Streptavidin-FITC (Southern Biotech) diluted 1:4,000 with TBS and goat anti-mouse IgG tetramethylrhodamine isothiocyanate (Southern Biotech) diluted 1:4,000 with TBS were added and left for 1 h RT. The samples were washed three times at coverslips, were applied, and the samples were then viewed. Negative controls included the use of irrelevant phage (φA1), TBS, and tissue from 2 h after infection with C. neoformans.
RESULTS AND DISCUSSION
Selection of melanin-binding peptides.
Phage containing melanin-binding peptides were obtained using a random decapeptide library expressed in the N-terminal part of the pIII coat protein of the phage fd-tet (21). The protocol involved biopanning on C. neoformans melanin performed in four duplicate rounds. The yield of phage selected increased with each biopan (increasing from 1.5 × 10−5 to 30 to 32% in round 4 in the duplicate biopans). Phage yield was calculated by dividing the number of eluted TU by the number of TU introduced into the round multiplied by 100. Phage were amplified in E. coli K91kan between rounds. Four third-round and 15 fourth-round clones were randomly chosen for amplification, purification, and sequence analysis. Two of the fourth-round phage clones from the same biopan expressed the same peptide sequence (φ4A7), and the remaining phage clones expressed unique peptide sequences (Table 1).
TABLE 1.
Peptide sequences of melanin-binding phage and reactivity by ELISA
| Phagea | Sequenceb | Reactivity (OD405) |
|---|---|---|
| Motif | ++**Hc | |
| φ4A4 | L H K L V R H G R W | 1.94 |
| φ4B4 | Y E R K F W H G R H | 1.67 |
| φ4A3 | Y L R R H T H V F W | 1.65 |
| φ4B1 | K K H S H Y W V R Y | 1.63 |
| φ4A1 | E F G T R H M R H R | 1.47 |
| φ4B3 | Y R H H A H G G R G | 1.24 |
| φ4A5 | R K K W H G W T R W | 1.18 |
| φ4A6 | P K W R H G Y T R F | 1.12 |
| φ4A2 | R H G T V K H A R H | 0.94 |
| φ4B5 | R R H W H P P V Q I | 0.90 |
| φ4B7 | E A Y K R R W H W P | 0.86 |
| φ4A7 (2 clones) | R W P K R H L S G H | 0.73 |
| φ4B6 | S R V P F R H Y H H | 0.62 |
| φ4B2 | R R P E H T K A R W | 0.44 |
| φ3B2 | W R A F L P R W H A | 0.22 |
| φ3B3 | W N R G W R W W M G | 0.18 |
| φ3B1 | G F F W K W R I G R | 0.16 |
| φ3A1 | H I R W K G H I S W | 0.09 |
The first number in the phage nomenclature represents the round of biopanning, the letter indicates the biopan repetition number, and the last number indicates a unique clone within the biopan.
Underlining indicates positive and aromatic residues or positive H pairs in either the 3′→5′ or 5′→3′ direction. Boldface indicates amino acids in the motif.
The motif ++**H is composed of two positive amino acids (+) followed by either positive or aromatic residues (*) and an H.
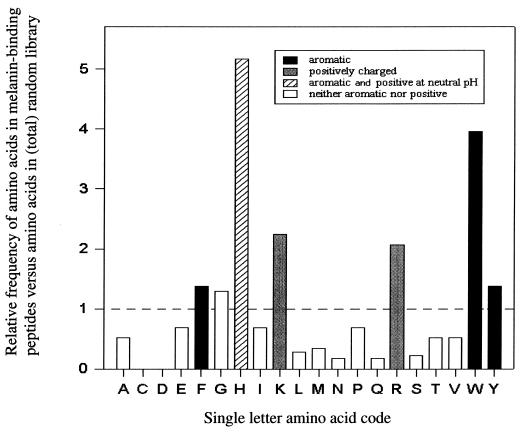
Amino acid sequence analysis of the peptides expressed by the melanin-binding phage revealed an excess of positive and aromatic amino acid residues. Figure 1 shows the frequency of amino acid residues in the melanin-binding peptide relative to the frequency of the amino acid in the peptide inserts in the total library. The sequences of the fourth-round melanin-binding peptide contained an average of 4.8 positive charges per peptide. Histidine (H), which is positively charged and aromatic at neutral pH, was the second most prevalent residue and increased with the greatest frequency compared to the original library (Fig. 1). Two positively charged amino acids, arginine (R) and lysine (K), were found at a frequency more than twofold higher than expected, and R was the most common residue in the melanin-binding peptide. All aromatic amino acids were present in higher frequencies than expected, particularly tryptophan (W). Negatively charged amino acids were rare and most melanin-binding peptides had none.
FIG. 1.
Ratio of the frequency of an amino acid in the melanin-binding peptide to the frequency of the amino acid in the library (21). The graph was constructed from the analysis of 15 melanin-binding peptides, which comprise 150 amino acids.
Analysis of individual sequences revealed that a motif of a positively charged residue followed by either an H or an aromatic residue was common in the fourth-round phage (Table 1). Each sequence had a minimum of one such pair, and several phage contained three such pairs (e.g., φ4A2, which contained the motif RHXXXKHXRH). The combination of a positive amino acid and H was not seen in the third-round phage. Instead, these peptides had a positive amino acid followed by a W. Thus, a positive amino acid followed by H may represent a more specific combination.
Phage from the fourth-round biopans contained an amino acid motif of ++∗∗H, where + represents a positive residue and ∗ is either a positive or aromatic residue (Table 1). The finding of a common motif, ++∗∗H, and the overall aromaticity and charge of the peptide suggest that the surface of the C. neoformans melanin contains structural determinants that are both aromatic and negatively charged.
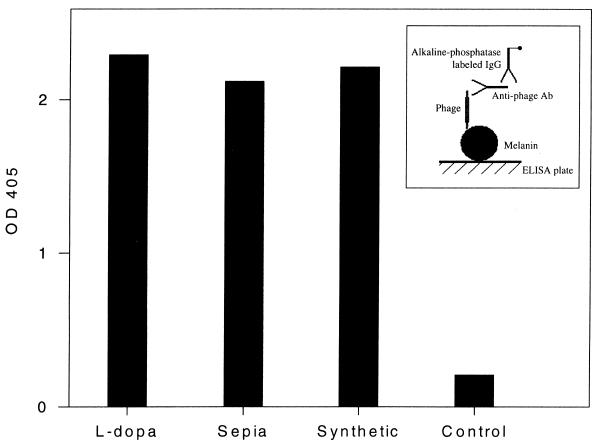
An ELISA to measure binding of the phage expressing melanin-binding peptide to cryptococcal melanin was developed. Phage from the fourth round of biopanning demonstrated significantly better binding than phage from the previous round (Table 1) or nonspecific phage (data not shown). An inhibition ELISA using phage φ4B4 with either the decapeptide coded for by this phage or an irrelevant peptide (P601G) used as a negative control was performed. The results demonstrate that the binding by the melanin-binding phage is specifically inhibited by melanin-binding peptides (data not shown). An ELISA to examine the reactivity of phage expressing melanin-binding peptide to C. neoformans melanin and melanin from other sources was also used. The two other melanins tested were synthetic melanin derived from the oxidation of tyrosine (Sigma) and melanin from the invertebrate Sepia (Sigma). The phage φ4B4 bound to each of the three melanins (Fig. 2), suggesting that each melanin had similar, if not identical, peptide-binding surface motifs.
FIG. 2.
Binding to l-dopa C. neoformans melanin, Sepia melanin, and synthetic melanin was tested by ELISA using φ4B4. Binding of φ33 to l-dopa was used as a control. The experiment was done twice, with similar results. Inset shows the configuration of the ELISA used to detect melanin binding. OD 405, optical density at 405 nm. Ab, antibody.
Scanning electron microscopy.
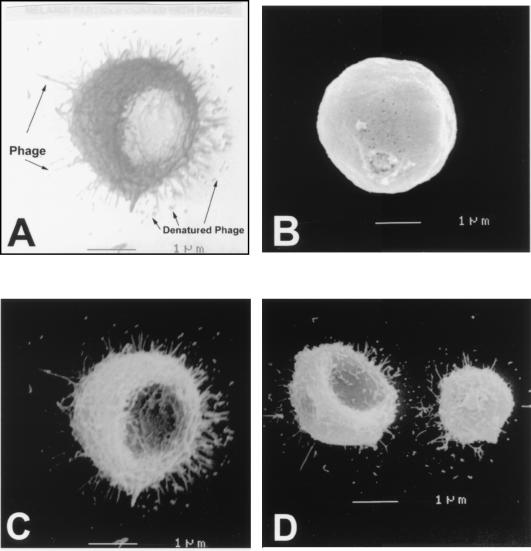
Scanning electron microscopy was used to study the interaction of phage expressing melanin-binding peptide with melanin particles. The scanning electron images revealed that melanin particles incubated with phage expressing melanin-binding peptide contained numerous filaments with dimensions corresponding closely to those of the filamentous phage and that most of the filaments were binding by their ends (Fig. 3). End binding is consistent with an interaction of the peptide displayed by the phage protein pIII with a binding site on the surface of melanin. The finding of multiple phage bound to each melanin particle indicates the presence of multiple peptide binding sites on the surface of melanin and suggests that the motif that the phage bind is a repeating structural constituent. No binding of phage to melanin was observed when melanin particles were incubated with control phage (Fig. 3B).
FIG. 3.
Melanin particle bound by phage (A). Scanning electron photographs of C. neoformans melanin particles with nonspecific phage (φ33) (B) or with melanin-binding phage (φ4B4 [C] and φ4A3 [D]). Magnification, ×15,000. A bud scar in the melanin particle is seen in panel B.
Immunohistochemistry.
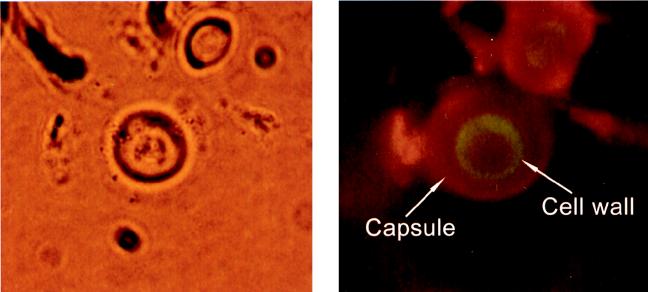
C. neoformans 24067 cells grown in defined chemical media with l-dopa were bound by the melanin-binding phage 4B4 but not by control phage (data not shown). Melanin-binding phage did not bind cells from cultures of strain 24067 grown without l-dopa or the 24067 mel− grown with or without l-dopa. Both phage expressing melanin-binding peptide (Fig. 4) and synthesized melanin-binding peptides bound cryptococcal cells in tissue from chronically infected mice. Analysis of phage binding to tissue from mice at various times of infection revealed reactivity after 48 h. Tissue collected at 2 or 24 h postinfection demonstrated no reactivity. Presumably this reflects the fact that melanization of cells takes time. Cultures of C. neoformans in defined chemical media with l-dopa take 3 to 5 days for melanization to occur (25). No binding occurred when control phage (φA1) was used with tissue from day 14 or 28. These experiments were also performed using brain tissue from chronically infected mice with similar results (data not shown).
FIG. 4.
Demonstration of the binding of phage φ4B4 to melanized C. neoformans cells by immunohistochemistry. The left panel shows a cryptococcal cell as viewed by bright-field microscopy, and the right panel shows the same cell visualized using an FITC and rhodamine filter (magnification, ×400). The green and red fluorescence represents phage binding to melanin-like compounds in the cell wall and monoclonal antibody to polysaccharide capsule, respectively.
Specificity of peptides for melanin.
In this study, the specificity of the melanin-binding peptides for melanin was an important concern. The inhibition ELISA demonstrated that the binding of the melanin-binding phage 4B4 was specifically inhibited by the synthetic peptide 4B4. The scanning electron microscopy studies confirm that the phage actually bind to the purified melanin particles. The immunohistochemical studies used nonmelanized cells, melanin-deficient mutants, and nonspecific reagents as controls. No binding of melanin-binding phage to cells without melanin was demonstrated. Control phage did not bind either melanized or nonmelanized C. neoformans. Furthermore, the experiments were all reproducible. Hence, the melanin-binding peptides bind melanin or melanin-like compounds in the cell wall of C. neoformans.
Antibody response and light microscopy.
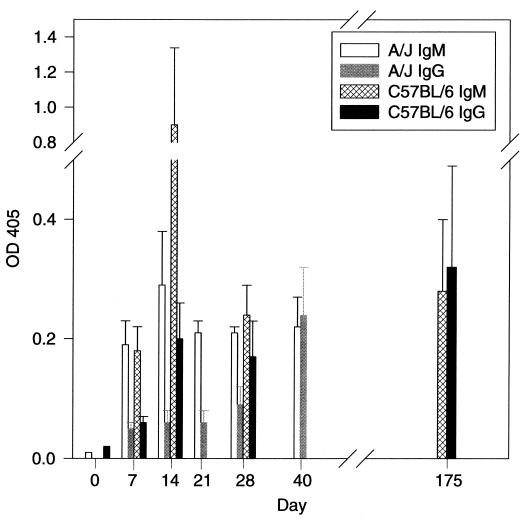
Although immunohistochemistry with phage containing melanin-binding peptides provided direct evidence for production of melanin-like compounds in tissue, we sought to confirm this result by other independent techniques. We assayed sera for the presence of melanin-binding antibodies and analyzed the cell wall by light microscopy. Fungal melanin is immunogenic and can elicit an antibody response when injected into mice (12). Analysis of sera from two strains of mice infected with C. neoformans revealed the production of IgM and IgG antibodies to melanin (Fig. 5). Melanin-binding antibodies appeared by day 7 of C. neoformans infection and were predominantly of the IgM isotype. We interpret this result as an indication that C. neoformans produces a melanin-like compound during infection that elicits an antibody response in mice. Examination by light microscopy of C. neoformans cells in murine tissue stained with hematoxylin and eosin at various times after infection revealed a progressive increase in thickness and darkening of the cell wall. These changes in the cell wall are similar to those seen over time in cells cultured in vitro with l-dopa (data not shown). The combination of positive immunohistochemical staining with melanin-binding peptides, the development of an antibody response to melanin with infection, and progressive cell wall changes during infection provides strong evidence that melanin-like compounds are synthesized by C. neoformans during infection.
FIG. 5.
The graph illustrates the development of antimelanin antibodies during infection of two mouse strains (A/JCr and C57BL/6) with C. neoformans. Each bar represents 3 mice. The experiment was done twice with similar results. OD 405, optical density at 405 nm.
Conclusions.
A decapeptide phage display library was used to demonstrate that melanin-binding peptides exist and that melanin-like compounds are produced during infection by C. neoformans. Melanin-binding peptides contain a high proportion of positively charged and aromatic amino acid residues, suggesting that they bind epitopes on the surface of melanin that are negatively charged and aromatic. The results also suggest that although melanins are heterogeneous compounds, some melanins contain similar structures that permit the binding of C. neoformans melanin-selected peptides. It was demonstrated that C. neoformans cells in mouse tissue make melanin-like compounds. The demonstration of melanin-like compounds in fungal cells in tissue suggests that the protective mechanisms ascribed to melanin in vitro may also apply in vivo. Furthermore, the synthesis of melanin-like compounds during infection suggests that melanogenesis may be a target for researchers interested in drug discovery, since drugs that bind melanin and/or inhibit melanization could be therapeutically beneficial.
ACKNOWLEDGMENTS
This work was supported by grants from the NIH (RO1-AI33774, AI13342, and HL59842 to A.C.; K08AI01341 to M.F.; and K08AI01489 to J.D.N.), the Burroughs Wellcome Fund (to A.C.), the Philippe Foundation (to P.V.), and the Infectious Disease Society of America (to J.D.N.).
We thank L. Pirofski and M. D. Scharff for their critical reviews.
REFERENCES
- 1.Casadevall A, Scharff M D. The mouse antibody response to infection with Cryptococcus neoformans: VH and VL usage in polysaccharide binding antibodies. J Exp Med. 1991;174:151–160. doi: 10.1084/jem.174.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chedekel M R, Ahene A B, Zeiss L. Melanin standard method: empirical formula 2. Pigm Cell Res. 1992;5:240–246. doi: 10.1111/j.1600-0749.1992.tb00543.x. [DOI] [PubMed] [Google Scholar]
- 3.Currie B P, Casadevall A. Estimation of the prevalence of cryptococcal infection among patients infected with the human immunodeficiency virus in New York City. Clin Infect Dis. 1994;19:1029–1033. doi: 10.1093/clinids/19.6.1029. [DOI] [PubMed] [Google Scholar]
- 4.Enochs W S, Nilges M J, Swartz H M. A standardized test for the identification and characterization of melanins using electron paramagnetic (EPR) spectroscopy. Pigm Cell Res. 1993;6:91–99. doi: 10.1111/j.1600-0749.1993.tb00587.x. [DOI] [PubMed] [Google Scholar]
- 5.Feldmesser M, Casadevall A. Effect of serum IgG1 to Cryptococcus neoformans glucuronoxylomannan on murine pulmonary infection. J Immunol. 1997;158:790–799. [PubMed] [Google Scholar]
- 6.Hill H Z. Melanins in the photobiology of skin cancer and the radiobiology of melanomas. In: Wilson S H, editor. Cancer biology and biosynthesis. Caldwell, N.J: Telford Press; 1991. pp. 31–53. [Google Scholar]
- 7.Hill H Z. The function of melanin or six blind people examine an elephant. Bioessays. 1992;14:49–56. doi: 10.1002/bies.950140111. [DOI] [PubMed] [Google Scholar]
- 8.Kay B K, Hoess R H. Principles and applications of phage display. In: Kay B K, Winter J, McCafferty J, editors. Phage display of peptides and proteins. A laboratory manual. San Diego, Calif: Academic Press, Inc.; 1996. pp. 21–34. [Google Scholar]
- 9.Kwon-Chung K J, Hill W B, Bennett J E. New, special stain for histopathological diagnosis of cryptococcosis. J Clin Microbiol. 1981;13:383–387. doi: 10.1128/jcm.13.2.383-387.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mason H S, Ingram D J E, Allen B. The free radical property of melanins. Arch Biochem Biophys. 1960;86:225–230. doi: 10.1016/0003-9861(60)90409-4. [DOI] [PubMed] [Google Scholar]
- 11.Nosanchuk J D, Casadevall A. Cellular charge of Cryptococcus neoformans: contributions from the capsular polysaccharide, melanin, and monoclonal antibody binding. Infect Immun. 1997;65:1836–1841. doi: 10.1128/iai.65.5.1836-1841.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nosanchuk J D, Rosas A L, Casadevall A. The antibody response to fungal melanin in mice. J Immunol. 1998;160:6026–6031. [PubMed] [Google Scholar]
- 13.Parmley S R, Smith G P. Antibody-selectable filamentous fd phage vectors: affinity purification of target genes. Gene. 1988;73:305–318. doi: 10.1016/0378-1119(88)90495-7. [DOI] [PubMed] [Google Scholar]
- 14.Polacheck I, Kwon-Chung K J. Melanogenesis in Cryptococcus neoformans. J Gen Microbiol. 1988;134:1037–1041. doi: 10.1099/00221287-134-4-1037. [DOI] [PubMed] [Google Scholar]
- 15.Porebska-Budny M, Sakina N L, Stepien K B, Dontsov A E, Wilczok T. Antioxidative activity of synthetic melanins. Cardiolipin liposome model. Biochim Biophys Acta. 1992;1116:11–16. doi: 10.1016/0304-4165(92)90121-a. [DOI] [PubMed] [Google Scholar]
- 16.Rhodes J C, Polacheck I, Kwon-Chung K J. Phenoloxidase activity and virulence in isogenic strains of Cryptococcus neoformans. Infect Immun. 1982;36:1175–1184. doi: 10.1128/iai.36.3.1175-1184.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rofstad E K. Radiation biology of malignant melanoma. Acta Radiol Oncol. 1986;25:1–10. doi: 10.3109/02841868609136368. [DOI] [PubMed] [Google Scholar]
- 18.Salas S D, Bennett J E, Kwon-Chung K J, Perfect J R, Williamson P R. Effect of the laccase gene, CNLAC1, on virulence of Cryptococcus neoformans. J Exp Med. 1996;184:377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sealy R C. Free radicals in melanin formation, structure and reactions. In: Armstrong D, et al., editors. Free radicals in molecular biology, aging and disease. New York, N.Y: Raven Press; 1984. pp. 67–76. [Google Scholar]
- 20.Smith G P, Scott J K. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 1993;217:228–257. doi: 10.1016/0076-6879(93)17065-d. [DOI] [PubMed] [Google Scholar]
- 21.Valadon P, Nussbaum G, Boyd L F, Margulies D H, Scharff M D. Peptide libraries define the fine specificity of anti-polysaccharide antibodies to Cryptococcus neoformans. J Mol Biol. 1996;261:11–22. doi: 10.1006/jmbi.1996.0438. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Aisen P, Casadevall A. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect Immun. 1995;63:3131–3136. doi: 10.1128/iai.63.8.3131-3136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Casadevall A. Growth of Cryptococcus neoformans in presence of l-dopa decreases its susceptibility to amphotericin B. Antimicrob Agents Chemother. 1994;38:2648–2650. doi: 10.1128/aac.38.11.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Aisen P, Casadevall A. Melanin, melanin “ghosts,” and melanin composition in Cryptococcus neoformans. Infect Immun. 1996;64:2420–2424. doi: 10.1128/iai.64.7.2420-2424.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Casadevall A. Susceptibility of melanized and nonmelanized Cryptococcus neoformans to nitrogen- and oxygen-derived oxidants. Infect Immun. 1994;62:3004–3007. doi: 10.1128/iai.62.7.3004-3007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Casadevall A. Susceptibility of melanized and nonmelanized Cryptococcus neoformans to the melanin-binding compounds trifluoperazine and chloroquine. Antimicrob Agents Chemother. 1996;40:541–545. doi: 10.1128/aac.40.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wheeler M H, Bell A A. Melanins and their importance in pathogenic fungi. Curr Top Med Mycol. 1988;2:338–387. doi: 10.1007/978-1-4612-3730-3_10. [DOI] [PubMed] [Google Scholar]
- 28.Williamson P R. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J Bacteriol. 1994;176:656–664. doi: 10.1128/jb.176.3.656-664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]