Abstract
Genes encoding the Phe-Gly (FG) repeat-containing nucleoporins NUP98 and CAN/NUP214 are at the breakpoints of several chromosomal translocations associated with human acute myeloid leukemia (AML), but their role in oncogenesis is unclear. Here we demonstrate that the NUP98-HOXA9 fusion gene encodes two nuclear oncoproteins with either 19 or 37 NUP98 FG repeats fused to the DNA binding and PBX heterodimerization domains of the transcription factor HOXA9. Both NUP98-HOXA9 chimeras transformed NIH 3T3 fibroblasts, and this transformation required the HOXA9 domains for DNA binding and PBX interaction. Surprisingly, the FG repeats acted as very potent transactivators of gene transcription. This NUP98-derived activity is essential for transformation and can be replaced by the bona fide transactivation domain of VP16. Interestingly, FG repeat-containing segments derived from the nucleoporins NUP153 and CAN/NUP214 functioned similarly to those from NUP98. We further demonstrate that transactivation by FG repeat-rich segments of NUP98 correlates with their ability to interact functionally and physically with the transcriptional coactivators CREB binding protein (CBP) and p300. This finding shows, for the first time, that a translocation-generated fusion protein appears to recruit CBP/p300 as an important step of its oncogenic mechanism. Together, our results suggest that NUP98-HOXA9 chimeras are aberrant transcription factors that deregulate HOX-responsive genes through the transcriptional activation properties of nucleoporin-specific FG repeats that recruit CBP/p300. Indeed, FG repeat-mediated transactivation may be a shared pathogenic function of nucleoporins implicated human AML.
An expanding subgroup of chromosomal translocation-generated oncoproteins in human acute myeloid leukemias (AML) involve the FG repeat-containing nuclear pore complex (NPC) proteins NUP98 (39) and CAN/NUP214 (13, 22). The NUP98 gene is found at the breakpoints of two distinct chromosomal rearrangements: t(7;11)(p15;p15) (7, 18, 33), and inv(11)(p15;q22) (2), which link NUP98 to the class I homeotic transcription factor HOXA9 and the putative RNA helicase DDX10, respectively. In each rearrangement, the chromosomal breakpoints are located within two flanking introns of the NUP98 gene that separate the FG repeat-rich N terminus of NUP98 from its C terminus, which contains a ribonucleoprotein (RNP)-binding motif (39). Although each translocation generates two reciprocal chimeric products, only those driven by the NUP98 promoter and containing the FG repeat region are predicted to mediate leukemogenesis (2, 7, 33). Another nucleoporin gene, CAN/NUP214, is found at the breakpoint of two independent chromosomal rearrangements: t(6;9)(p23;q34), which fuses CAN/NUP214 to DEK (49), and inv(9;9)(q34;q34), which links it to SET (50). The leukemia-specific transcripts, DEK-CAN/NUP214 and SET-CAN/NUP214, both encode nuclear fusion proteins. The proteins contain identical C-terminal portions of CAN/NUP214, including its FG repeat-rich region, and a coiled-coil domain (13, 22). DEK and SET are both nuclear proteins that have no sequence similarity other than the presence of acidic motifs that may participate in DNA binding (13, 14, 31).
The involvement of two FG repeat-containing nucleoporins in multiple translocations associated with human leukemia raises intriguing questions about their role in leukemogenesis. In particular, the consistent presence of FG repeat regions suggests that such domains could serve a common function in the transformation of hematopoietic cells. Many of the known components of the NPC have regions rich in FXFG, GLFG and/or FG repeats (amino acids are given in single-letter code, with X indicating any amino acid). Such repeats (called FG for simplicity) are presumed contact sites for soluble nucleocytoplasmic transport factors carrying different kinds of cargo; however, their precise functions in vivo remain to be determined (35, 36).
HOXA9, expressed in both the primitive pluripotent precursors and the myeloid progenitors of human bone marrow (42), is the only nucleoporin fusion partner with an established physiological role in hematopoietic development. HOXA9 knockout mice have multiple hematopoietic defects, including reduced numbers of peripheral blood granulocytes and lymphocytes, as well as myeloid and pre-B-cell progenitors, and their spleens and thymuses are smaller than normal (25). Besides its involvement in t(7;11)-mediated myeloid leukemogenesis, HOXA9 has been implicated in the formation of myeloid leukemias in the BXH-2 strain of mice (29). BXH-2 mice carry an endogenous murine leukemia virus that acts as a viral mutagen predisposing the animals to myeloid malignancies (4, 5). In this experimental tumor model, about 3% of all leukemias in BXH-2 mice display proviral activation of HOXA9 (34). Constitutive expression of HOXA9 alone is not sufficient for efficient transformation of murine hematopoietic cells; it requires coexpression of MEIS1 (23, 34), a PBX1-related divergent homeodomain-containing protein that cooperatively binds DNA with HOXA9 in vitro (44).
In this study, we show that the t(7;11)-derived fusion gene generates two chimeric proteins via alternative splicing within NUP98. Investigation of the structural and functional regions of the chimeric NUP98-HOXA9 proteins demonstrated that HOXA9-mediated DNA binding and interaction with PBX are essential for transformation of NIH 3T3 fibroblasts. In both chimeras, the NUP98 portions contained very potent transcription activation domains, which replace a strong transcriptional repressor domain within the amino-terminal half HOXA9. Interestingly, the transcriptional coactivators CREB binding protein (CBP) and potentially p300 interacted and functionally cooperated with the NUP98 FG-repeat-rich portions. Abbrogation of NUP98-HOXA9-mediated transformation corresponded to the loss of NUP98-mediated transcriptional activity and CBP binding. Thus, NUP98-HOXA9 seems to recruit CBP/p300 as part of its oncogenic mechanism. Because CBP and p300 are coactivators for a number of gene-specific transcription factors, they could also be critical accessory factors for other fusion proteins that deregulate transcription.
MATERIALS AND METHODS
RT-PCR.
First-strand cDNA was synthesized from 1 μg of total RNA with avian myeloblastosis virus reverse transcriptase (Promega) and primer p1 or p4 in a total volume of 20 μl. The mixtures were incubated 1.5 h at 42°C. Reverse transcription RT mixture was used at 2 μl per PCR (total volume, 50 μl). The PCR primer combinations were as indicated in Fig. 1A (p1, 5′-GGCTGTCGTAGTATTAAATCCAGGGG-3′; p2, 5′-GATTATCAGGCTAACAGGAAGGGC-3′; p3, 5′-CCACAACTGGCTTGTTTGGGTC-3′; p4, 5′-CGTGAAGCCAGTTGGCTG-3′). The PCRs were performed with AmpliTaq Gold (Perkin-Elmer Cetus) in buffer supplied by the manufacturer and supplemented with 2.5 mM MgCl2. The PCR cycles were 30 s at 94°C, 1 min at 50°C, and 3 min at 72°C.
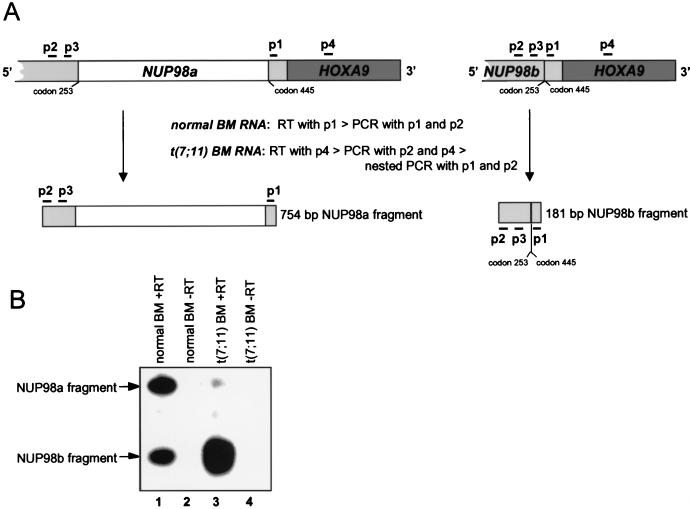
FIG. 1.
NUP98 and NUP98-HOXA9 genes generate an alternatively spliced transcript in human BM cells. (A) Overview of the RT-PCR procedure with representations of relevant NUP98-HOXA9 cDNA portions and the expected PCR products. The positions of the primers used in the various RT reactions and PCRs are indicated by horizontal bars. To detect NUP98a and NUP98b transcripts in normal BM cells, we used primer 1 (p1) to generate cDNA and primer set p1 and p2 for PCR amplification (40 cycles). Seminested PCR was used to detect NUP98-HOXA9-specific transcripts in BM cells from a patient with t(7;11)-positive leukemia; p4 was used for the RT step, p2 and p4 were used in the first round of amplification (40 cycles), and p1 and p2 were used in the seminested round of PCR (30 additional cycles). The oligonucleotide probe for detection of NUP98a- and NUP98b-derived PCR fragments is depicted as p3. (B) Autoradiogram of RT-PCR products detected by probe p3. Lanes: 1, 754- and 181-bp fragments from NUP98a and NUP98b transcripts in normal human BM; 2, negative control for lane 1 (same sample as in lane 1 but without reverse transcriptase); 3, 754- and 181-bp fragments from NUP98a-HOXA9 and NUP98b-HOXA9 transcripts in BM from a patient with t(7;11)-positive leukemia; 4, negative control for lane 3. Since our RT-PCR technique was not quantitative, the intensities of the 181- and 754-bp PCR bands may not be correlated with the actual transcript levels. Moreover, Northern blot analysis revealed that the BM RNA from the t(7;11) patient was partially degraded.
Expression constructs and mutagenesis.
All cDNA fragments were initially cloned in pTZ19 containing the synthetic sequence GAATTCGCCGCCACCATGTATGACGTCCCAGATTACGCAAGTTTGCCCGGGTATGATTTCCTGATTATGCTAGC derived from pHA1triple-tag#10 (13). This sequence contained a 5′ Kozak consensus motif, sequences encoding two consecutive influenza virus HA1 epitopes (51) (indicated in italics), and a 3′ NheI site. NUP98b cDNA was isolated from a λgt10 human bone marrow (BM) cDNA library (no. HL1168a; Clontech, Palo Alto, Calif.). Full-length NUP98a cDNA was reconstructed from NUP98b cDNA and sequences encoding amino acids 254 to 444 obtained from a partial NUP98a cDNA clone that was isolated from a human placental library (Hu2002B#29203). A cDNA encoding the HOXA9 portion of the t(7;11)-generated fusion protein was produced by PCR amplification of DNA extracted from our λgt10 human BM cDNA library. cDNA clones encoding NUP98a-HOXA9 and NUP98b-HOXA9 were created from HOXA9, NUP98a, and NUP98b cDNA constructs by overlap extension PCR. A cDNA encoding the NUP98b portion of NUP98b-HOXA9 was generated by insertion of an oligonucleotide in the NUP98b cDNA so that codon 469 is followed by a stop codon. NUP98b-HOXA9(FIKI) cDNA was generated from NUP98b-HOXA9 by inserting the oligonucleotide TTTATAAAGATC between codons 47 and 48 of the homeodomain. NUP98a-HOXA9(W−6>A) cDNA was obtained by standard PCR-based site-directed mutagenesis. NUP98a(Δ51–223)-HOXA9 and NUP98b(Δ51–223)-HOXA9 mutants were constructed by deleting a 819-bp StuI-KpnI fragment from NUP98a-HOXA9 and NUP98b-NUP98b cDNA, respectively. To generate NUP98b(Δ51–469)-HOXA9, NUP98b(Δ1–223)-HOXA9, CAN/NUP214(1864–2090)-HOXA9, NUP153(1121–1479)-HOXA9, and VP16(413–490)-HOXA9 cDNAs, we cloned PCR-amplified NheI fragments containing the desired nucleoporin and VP16 portions into the NheI site of our clone encoding the HOXA9 portion of NUP98-HOXA9. For NUP98b(Δ51–469)-HOXA9, we generated a 156-bp NheI fragment encoding the first 50 amino acids of NUP98b, which was PCR amplified from NUP98b cDNA. For NUP98b(Δ1–223)-HOXA9, we PCR amplified a 171-bp NheI fragment from NUP98b cDNA encoding amino acids 224 to 253 and 445 to 469. For CAN/NUP214(1864–2090)-HOXA9, a 684-bp NheI fragment encoding the last 226 amino acids of human CAN/NUP214 was generated by RT-PCR from BM RNA. For NUP153(1121–1479)-HOXA9, a 1,695-bp NheI fragment encoding the last 563 amino acids of human NUP153 was obtained by PCR amplification from vector pHA1-NUP153 (3). For VP16(413–490)-HOXA9 a 220-bp NheI fragment encoding amino acids 413 to 490 of the herpes simplex virus type 1 VP16 protein was amplified from plasmid pCMX-VP16 (12). All DNA fragments generated by PCR amplification were sequenced. pTZ19 cDNA fragments were cloned into pSRαMSVtkCD8 (for transformation studies), pSP64 or pSP73 (for in vitro transcription-translation purposes), and the pUH10-3 derivative (16) termed pUHD10S (13) (for overexpression in HtTA cells).
Retroviral stocks and transformation analysis.
cDNA inserts were cloned into the ClaI site of the ecotropic retroviral expression vector pSRαMSVtkCD8 (17). 293T cells were cotransfected with various pSRαMSVtkCD8 constructs and the ecotropic packaging pSV-Ψ−E-MLV (30). Culture supernatants containing viral particles were harvested at 6- to 10-h intervals from days 2 to 4 posttransfection, filtered (Acrodisc 13 syringe filter [pore size, 0.45 μm]; Gelman Sciences, Ann Arbor, Mich.), and supplemented with 6 μg of Polybrene per ml. Low-passage-number NIH 3T3 cells were seeded at low cell density and infected the next day with virus at 8- to 12-h intervals for 48 h. Infected NIH 3T3 cells were harvested 24 to 72 h later and immunostained with anti-CD8 antibodies. CD8-positive cells were then isolated by fluorescence-activated cell sorting (17). To test for anchorage-independent growth, we suspended 2 × 104 CD8+ NIH 3T3 cells in Iscove’s medium supplemented with 15% fetal bovine serum and 0.3% soft agar and plated this mixture in a 3-cm-diameter dish. The cells were then cultured at 37°C under 8% CO2, and colonies were counted after 3 weeks. The cells were plated in triplicate for each experiment, with total number of colonies reported as the average of the counts from the three dishes. BM transformation assays were performed as described previously (24).
Indirect immunofluorescence.
NIH 3T3 or HtTA cells were seeded on 12-well microscope slides for at least 12 h, fixed in 3% paraformaldehyde for 15 min on ice, washed three times with phosphate-buffered saline (PBS), permeabilized in PBS–0.2% Triton X-100 for 10 min, washed five times in PBS–2% nonfat milk (Kroger Co., Cincinnati, Ohio), and incubated overnight with the first antibodies. Monoclonal antibody 12CA5 (Boehringer Mannheim) was incubated at 4 μg per ml in PBS–2% nonfat milk. Affinity-purified anti-hNUP98 polyclonal antibodies (raised in rabbits against NUP98 amino acids 51 to 223) were diluted 1:20, and anti-hCRM1 antiserum (raised in rabbits against the peptide sequence EREIALRQADEEK) was diluted 1:1000. Primary antibodies were visualized with fluorescein isothiocyanate (Sigma)- or Texas red-conjugated goat anti-mouse or goat anti-rabbit antibodies at 4 μg per ml in PBS–2% nonfat milk. The cells were examined by confocal laser-scanning microscopy.
Coimmunoprecipitations.
Retrovirally transduced NIH 3T3 cells (7 × 106 to 8 × 106) expressing HA1-NUP98a-HOXA9, HA1-NUP98b-HOXA9, or the HOXA9 portion of these fusion proteins (designated HA1-HOXA9[portion]) were washed three times with PBS and then incubated for 20 min in 10 ml of short-term labeling medium (methionine-free Dulbecco’s modified Eagle’s medium, 5% dialyzed fetal bovine serum) at 37°C to deplete the intracellular pools of methionine. The cells were then incubated in 6.5 ml of short-term labeling medium containing 0.18 mCi of [35S]methionine per ml and harvested 4 h later. Four to five million HtTA cells (16) were transfected with 40 μg of pUHD10S vectors encoding HA1-NUP98a-HOXA9 or HA1-HOXA9[portion] (by using Superfect transfection reagent [Qiagen]). At 40 h posttransfection, the cells were metabolically labeled with [35S]methionine for 4 h as described above. Labeled NIH 3T3 or HtTA cells were washed three times with ice-cold PBS and scraped from the dishes and total-cell lysates were prepared in 1 ml of NP-40 lysis buffer (1% Nonidet P-40 [NP-40], 50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EGTA, 5 mM EDTA, 15 mM MgCl2, 60 mM β-glycerolphosphate, 1 mM dithiothreitol, 0.1 mM Na3VO4, 0.1 mM NaF, 15 mM p-nitrophenylphosphate, 1.8 μg of aprotinin per ml, 1 μg of leupeptin per ml, 1 μg of pepstatin per ml, 10 μg of soybean trypsin inhibitor per ml, 0.1 mM benzamidine, and 1 mM phenylmethylsulfonyl fluoride). Lysates were kept on ice for 10 min, vortexed gently at 2-min intervals, and centrifuged for 5 min at 14,000 × g. The supernatants were filtered through a 0.45-μm-pore-size low-protein-binding syringe filter (SuperAcrodisc; Gelman Sciences), and precleared in 100 μl of protein A-sepharose (50% slurry in NP-40 lysis buffer) for 1.5 h at 4°C. Primary immunoprecipitations were performed by adding 50 μl of Sepharose-coupled 12CA5 antibody (50% slurry) or 50 μl of protein A-Sepharose (50% slurry) preincubated with either 40 μl of affinity-purified NUP98 antiserum or 50 μl of a CBP/p300 cocktail consisting of equal parts of CBP (A-22), CBP (C-20), CBP (415), p300 (N-15), and p300 (C-20) antisera (purchased from Santa Cruz Biotechnology). Following a 3-h rotation at 4°C, the pellets were washed five times in NP40 lysis buffer and antigens were released by boiling for 2 min in 100 μl of boiling buffer (20 mM Tris-HCl [pH 8.0], 0.5% SDS, 1 mM dithiothreitol). After centrifugation, the supernatants were collected and split into 20- and 80-μl aliquots. The 20-μl aliquots were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted to a polyvinylidene difluoride membrane (MSI, Westboro, Mass.), and the amount of HA1-tagged protein was assessed by Western blot analysis. The blots were blocked for 1 h in PBS–5% nonfat milk and subsequently incubated with 1 μg of 12CA5 antibody per ml in PBS for 1 h. Bound antibodies were visualized with an ECL chemiluminescence kit (Amersham Life Science Inc., Cleveland, Ohio). Washings between antibody incubations were performed in PBS–0.05% Tween 20 (five washings for 10 min each). The 80-μl aliquots were diluted with 4 volumes of RIPA buffer (52) and incubated overnight at 4°C with 50 μl of protein A-Sepharose (50% slurry) preincubated with 50 μl of our CBP/p300 antiserum cocktail (see above). After five washes with RIPA buffer, the beads were boiled in SDS-PAGE sample buffer and the precipitated proteins were analyzed by SDS-PAGE and fluorography.
Transactivation assays.
GAL4 fusion proteins were tested for transcriptional activation properties on the GAL4-responsive reporter construct G5B/pGL2. This reporter was constructed by inserting a XhoI-SmaI fragment from G5BCAT (27), containing five GAL4 DNA binding sites and the adenovirus E1b TATA box, into pGL-2 (Promega Corp., Madison, Wis.) cut with HindIII (repaired with Klenow polymerase) and XhoI. For transcriptional repression studies, we used a GAL4-responsive reporter construct that contained five tandem copies of the GAL4 DNA binding site upstream of the simian virus 40 (SV40) early promoter (a kind gift from C. Abate-Shen). Blunt-ended cDNA fragments encoding relevant portions of FG repeat-rich nucleoporins or HOXA9 were cloned into the SmaI sites of either pM1 or pM2, such that in-frame fusions were created between sequences encoding the GAL4 DNA binding domain and the FG repeat-rich nucleoporin motifs or HOXA9 sequences. NIH 3T3 cells (2 × 104) were seeded in 24-well dishes. The next day, cells in each 24-well dish were cotransfected (with Superfect) with 0.66 μg of GAL4 fusion protein expression plasmid, 0.33 μg of G5B/pGL2 reporter plasmid, 25 or 2.5 ng of CMV-E1A vectors (where applicable), and 10 ng of pRL-CMV internal control plasmid (Promega Corp.). The cells were harvested after about 16 h, and enzyme assays were performed to assess reporter gene expression. Reporter gene-derived luciferase activity was normalized to Renilla luciferase derived from pRL-CMV. Transfections and analyses with the GAL4-NUP and GAL4-HOXA9 fusion constructs were performed in triplicate. To test for a potential effect of E1A on GAL4-NUP98 protein levels, 0.5 × 106 NIH 3T3 cells were seeded in 10-cm dishes. A day later, the cells were cotransfected with 15.8 μg of GAL4 fusion protein expression plasmid and 60 ng of CMV-E1A vectors [these DNA concentrations correspond to the 0.66 μg of GAL4-NUP98(1–469) plasmid and 2.5 ng of CMV-E1A vector per 2 × 104 NIH 3T3 cells as used in our transactivation assay]. At 24 h posttransfection, the cells were harvested and immunoprecipitations and Western blot analysis with 12CA5 antibodies were performed as described above.
For CBP potentiation experiments NIH 3T3 cells were cotransfected with 0.1 μg of GAL4-NUP98b(1–469) or 0.5 μg of GAL4-CREB(160–284), 0.25 μg of G5B/pGL2 reporter plasmid, 0, 0.2, or 0.6 μg of RSV-CBP (or equimolar amounts of empty Rous sarcoma virus [RSV] vector as a negative control), and 10 ng of pRL-TK internal control plasmid (Promega Corp.). Transactivation assays in Ba/F3 cells were performed as previously described in detail (28). All transfection experiments were done either in duplicate or in triplicate and repeated several times.
In vitro DNA binding assays.
Proteins for electrophoretic mobility shift assays (EMSAs) were produced in vitro from SP6 expression plasmids with a coupled reticulocyte lysate system as previously described (11). To ensure that approximately equal amounts of each lysate were added to the DNA binding reaction mixtures, proteins were synthesized in parallel in the presence of [35S]methionine, subjected to SDS-PAGE, quantitated on a PhosphorImager (Molecular Dynamics), and normalized for the number of methionine residues. DNA binding reactions were performed as previously described (11). Single-stranded oligonucleotides were labeled with [γ-32P]ATP, annealed, purified, and used with core sequences matching the consensus TGATTTAT.
RESULTS
The NUP98-HOXA9 fusion gene encodes two proteins.
To construct a NUP98-HOXA9 fusion cDNA encoding the predicted chimeric oncoprotein (7, 33), we first isolated NUP98 cDNA clones from a human BM library. We identified two independent cDNA clones containing the 5′ and 3′ ends of the human NUP98 coding sequence (7); however, both cDNAs lacked a segment that encodes amino acids 254 to 444 of the NUP98 protein, including 18 FG repeats. We named this alternatively spliced product NUP98b. The full-length NUP98 cDNA, referred to as NUP98a, was reconstructed from partial BM and placental cDNAs (see Fig. 2A). To confirm expression of both mRNA transcripts from the NUP98 locus, we analyzed human BM RNA by RT-PCR (Fig. 1A). In this assay, NUP98a and NUP98b RNAs should generate diagnostic PCR fragments of 754 and 181 bp, respectively. Both amplification products were obtained (Fig. 1B, lane 1). Sequencing of the fragments indicated that they indeed differed in the 573-bp segment encoding amino acids 254 to 444 of the full-length NUP98 protein (not shown).
FIG. 2.
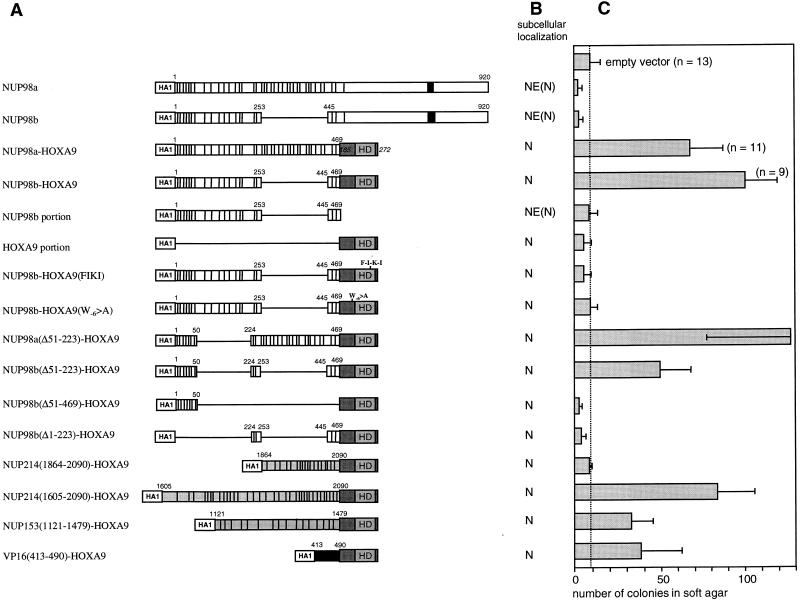
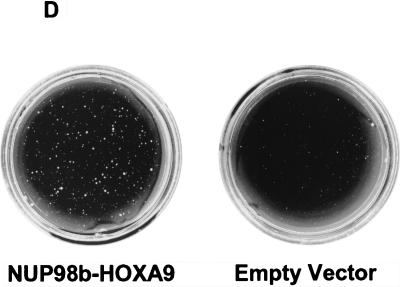
Structural and functional properties of NUP98-HOXA9 isoforms and mutants. (A) Schematic representations of the NUP98 and NUP98-HOXA9 isoforms and mutants used in this study. Indicated NUP98 motifs include the nucleoporin-specific FG repeats (each repeat is denoted by a vertical bar), the NUP98 RNP-binding motif (solid box), and NUP153 and CAN/NUP214 sequences (indicated in grey). Numbers correspond to the amino acids of the full-length NUP98 (7). Indicated in the HOXA9 portion of the fusion protein is the homeodomain (HD). Numbers in italics correspond to the amino acids of the full-length HOXA9 (44). (HA1) represents two consecutive HA1 epitopes recognized by monoclonal antibody 12CA5. (B) Summary of the subcellular localization studies (N, nuclear localization; NE, nuclear envelope localization) of the NUP98 and NUP98-HOXA9 isoforms and mutants. (C) Anchorage-independent growth of NIH 3T3 cells expressing NUP98-HOXA9 isoforms and mutants. The number of colonies (per 2 × 104 CD8+ cells with a diameter of at least 90 to 100 μm) was determined after 3 weeks of growth in soft agar. The values are the means and standard deviations of 4 to 13 experiments (if more than 4 experiments were performed, the actual number of experiments is indicated in the figure). (D) Soft agar dishes demonstrating efficient colony formation by NIH 3T3 cells infected with NUP98b-HOXA9. In contrast, NIH 3T3 cells infected with pSRαMSVtkCD8 (empty vector) displayed only minimal anchorage-independent growth.
We then adapted the above RT-PCR strategy to screen for alternative NUP98-HOXA9 fusion transcripts. A nested RT-PCR assay based on RNA from a patient with t(7;11)(p15;p15)-positive acute myeloid leukemia (AML) yielded the expected amplification products of 754 and 181 bp (Fig. 1B, lane 3). Subcloning and DNA sequence analysis of the fragments revealed that they had originated from NUP98a-HOXA9 and NUP98b-HOXA9 fusion transcripts (data not shown), indicating that the leukemic cells express both chimeric isoforms.
NUP98-HOXA9 chimeras are nuclear proteins that transform NIH 3T3 fibroblasts.
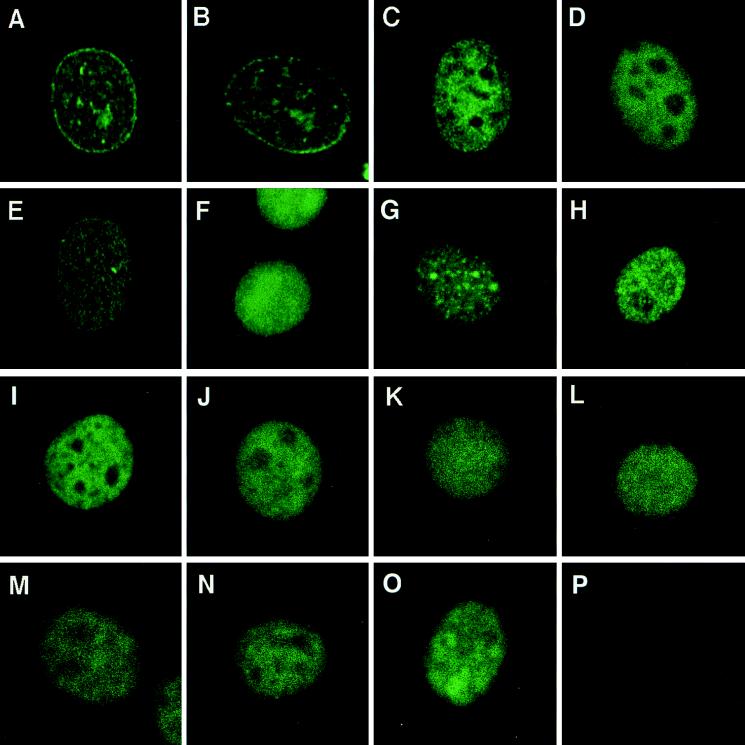
Hemagglutinin (HA1) epitope-tagged versions of cDNAs containing NUP98a, NUP98b, NUP98a-HOXA9, NUP98b-HOXA9, and the separate NUP98b and HOXA9 portions of NUP98b-HOXA9 (Fig. 2A) were cloned into the pSRαMSVtkCD8 retroviral vector (17) and expressed in NIH 3T3 cells by retroviral gene transfer. To determine the subcellular localization of the HA1-tagged proteins, NIH 3T3 fibroblasts were immunostained with 12CA5 monoclonal antibody against the HA1 epitope. HA1-NUP98a and HA1-NUP98b both showed punctate staining of the nuclear rim (Fig. 3A and B), typical of NPC-associated proteins (39). By contrast, the HA1-NUP98a-HOXA9 and HA1-NUP98b-HOXA9 chimeras were found located in the nuclei of NIH 3T3 cells (Fig. 3C and D). Typically, such nuclei displayed a very fine granular staining pattern whereas the nucleoli did not stain at all. To determine whether the HA1-NUP98-HOXA9 chimeras also associate with NPCs, cells were double stained for HA1 and CRM1, a transport factor localized mainly at the nuclear envelope (NE). Our HA1-specific signals did not co-localize with CRM1 signals at the NE, indicating that there is no detectable localization of chimeric protein at the NE (data not shown). Like the fusion proteins, the HOXA9 portion without NUP98 showed nuclear localization but the staining pattern was less granular (Fig. 3F). The NUP98b fragment derived from the chimera was concentrated at the NE, although a substantial proportion of this truncated protein was found in the nucleus, even at low to moderate levels of expression (Fig. 3E). In all cases, protein expression was confirmed by performing immunoprecipitations with anti-HA1 monoclonal antibodies on lysates of metabolically labeled cells (data not shown).
FIG. 3.
NUP98-HOXA9 isoforms and engineered mutants are localized in the nucleus. NIH 3T3 cells were transduced with retroviral expression vectors and immunostained with 12CA5 monoclonal antibodies that recognize the HA1 epitope encoded by the 5′ end of the various cDNA constructs. (A) NUP98a; (B) NUP98b; (C) NUP98a-HOXA9; (D) NUP98b-HOXA9; (E) NUP98b portion; (F) HOXA9 portion; (G) NUP98b-HOXA9(FIKI); (H) NUP98b-HOXA9(W−6>A); (I) NUP98a(Δ51–223)-HOXA9; (J) NUP98b(Δ51–223)-HOXA9; (K) NUP98b(Δ51–4693)-HOXA9; (L) NUP98a(Δ1–223)-HOXA9; (M) CAN/NUP214(1864–2090)-HOXA9; (N) NUP153(1121–1479)-HOXA9; (O) VP16(413–490)-HOXA9; (P) empty pSRαMSVtkCD8 vector (negative control).
To determine the oncogenic potential of the NUP98-HOXA9 proteins, we used an NIH 3T3 fibroblast transformation assay. This assay is commonly used in the field for structure-function studies on leukemia oncoproteins, and the results with fibroblasts are generally in agreement with those of other transformation assays. NIH 3T3 fibroblasts expressing NUP98a-HOXA9 or NUP98b-HOXA9 formed colonies efficiently in soft agar (Fig. 2C and D), indicative of cellular transformation. By contrast, NIH 3T3 fibroblasts overexpressing NUP98a, NUP98b, or the NUP98b or HOXA9 portions of the chimeric constructs, did not form colonies above background levels observed with NIH 3T3 cells infected with retroviral stocks prepared from the empty vector pSRαMSVtkCD8 (Fig. 2C and D). These results indicate that the in vitro cellular oncogenicity of the NUP98-HOXA9 proteins requires both the C-terminal HOXA9 sequences and the N-terminal region of NUP98 containing the nucleoporin-specific FG repeats.
NUP98-HOXA9-mediated transformation depends on its ability to bind DNA and heterodimerize with PBX.
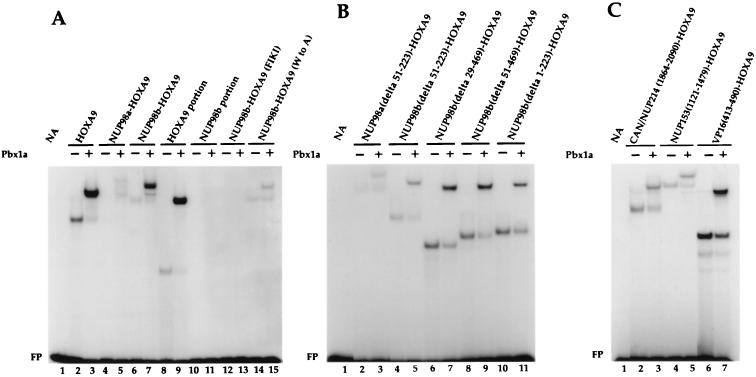
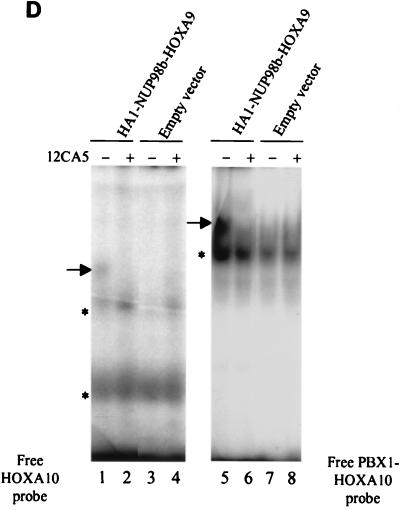
The nuclear localization of NUP98-HOXA9 proteins suggested that DNA binding activity might be important for their oncogenic potential. The HOXA9 portion contained within the NUP98-HOXA9 oncoprotein has two distinct regions implicated in DNA binding: (i) the homeodomain and (ii) a short tryptophan-containing motif required for binding to the PBX transcriptional cofactors. This motif is located immediately N-terminal of the homeodomain. HOX-PBX heterodimers have increased DNA binding affinity and specificity, allowing selective recognition and activation of genes containing HOX response elements (9–11, 26, 37, 38, 40, 47, 48). We determined the in vitro DNA binding activity of the NUP98-HOXA9 oncoproteins by EMSAs. Because the consensus DNA binding site for HOXA9 is unknown, we selected the PBX1-HOXA10 consensus DNA binding sequence as a target for interaction with the HOXA9 homeodomain. This consensus sequence has a TTAT core motif, preferentially recognized by homeobox proteins of the Abd-B-like gene family, including HOXA9 (6, 45). DNA binding of in vitro-synthesized HOXA9, NUP98-HOXA9 isoforms, or their truncated derivatives to the PBX1-HOXA10 bipartite probe was determined in the presence or absence of in vitro-translated PBX1a protein. Both full-length HOXA9 and the HOXA9 portion of NUP98-HOXA9 bound the DNA target alone, while addition of PBX1a to the binding-reaction mixtures increased their activities (Fig. 4A, compare lane 2 with lane 3 and lane 8 with lane 9). Both NUP98-HOXA9 chimeras displayed cooperative DNA binding with PBX1a; however, the DNA binding of PBX1a/NUP98b-HOXA9 heterodimers to the consensus site was considerably greater than that of PBX1a/NUP98a-HOXA9 dimeric complexes (compare lanes 4 and 5 with lanes 6 and 7). As expected, the NUP98b portion failed to demonstrate any activity by itself (lanes 10 and 11). To show formation of PBX/NUP98-HOXA9 heterodimers in vivo, we prepared lysates from NIH 3T3 cells expressing HA1-NUP98b-HOXA9 and performed EMSAs with HOXA10 and PBX1-HOXA10 probes. Typically, a slower-migrating complex was formed with the PBX1-HOXA10 probe than with the HOXA10 consensus probe (Fig. 4D, compare lanes 1 and 5), indicative for the formation of PBX/NUP98b-HOXA9 heterodimers in vivo (Fig. 4D).
FIG. 4.
NUP98-HOXA9 fusion proteins bind cooperatively with PBX1a to a PBX1-HOXA10 bipartite DNA sequence. (A to C) EMSAs of in vitro-translated proteins whose identities are indicated above the gel lanes. EMSAs were performed with a radiolabeled probe containing a PBX1-HOXA10 bipartite binding site in the absence (−) or presence (+) of in vitro-translated PBX1a. Cooperative DNA binding was observed for fusion proteins with an intact HOXA9 homeodomain. Typically, the binding affinity for the bipartite probe increased when the NUP98 portion fused to HOXA9 became smaller. The differences in intensities of the protein-DNA complexes within each panel (A or B) represent true variations in DNA binding activity. FP, free probe. (D) NUP98b-HOXA9 and PBX form heterodimers in NIH 3T3 cells. Radiolabeled probe containing the PBX1-HOXA10 or HOXA10 binding site was added to lysates of cells expressing the HA1-tagged NUP98-HOXA9 fusion proteins, and the formation of protein-DNA complexes in the absence (−) or presence (+) of 12CA5 monoclonal antibody was studied. For each lysate, the shifted complexes formed with the PBX1-HOXA10 probe (arrow to the left of lane 5) are larger than those formed with the HOXA10 probe (arrow to the left of lane 1), indicating that NUP98-HOXA9 and PBX form heterodimers in vivo. The shifted complex ablates when incubated with 12CA5 antibody, confirming the presence of HA1-NUP98b-HOXA9 in such complexes.
To confirm that NUP98-HOXA9 requires an intact homeodomain as well as a functional N-terminal PBX-interaction motif for binding to the bipartite DNA probe, we constructed two NUP98b-HOXA9 mutants. In one [NUP98b-HOXA9(FIKI) (Fig. 2A)], the oligopeptide FIKI was inserted in the third helix of the HOXA9 homeodomain, while in the other [NUP98b-HOXA9(W−6>A) (Fig. 2A)], the conserved tryptophan in the PBX-interaction motif was replaced with an alanine. The NUP98b-HOXA9(FIKI) mutant completely failed to interact with the DNA probe (Fig. 4A, lanes 12 and 13). The DNA-binding capacity of the NUP98b-HOXA9(W−6>A) mutant was unaffected in the absence of PBX, but its cooperative binding to the probe was severely impaired (compare lane 6 with lane 14 and lane 7 with lane 15).
We then asked whether NUP98-HOXA9 proteins mediate their oncogenic effects by a specific DNA-binding mechanism. When the NUP98b-HOXA9(FIKI) and NUP98b-HOXA9(W−6>A) mutants were assayed for their ability to induce NIH 3T3 colonies in soft agar, both mutants showed a complete loss of transforming potential (Fig. 2C), despite their effective expression (data not shown) and localization in the nucleus (Fig. 3G and H). Like NUP98b-HOXA9, both mutants displayed the fine granular nuclear staining pattern. Thus, HOXA9-mediated DNA binding and interaction with PBX are essential for transformation, suggesting that NUP98-HOXA9 chimeras are altered transcription factors rather than altered transport factors.
NUP98 FG repeats are critical for transactivation and transformation.
To evaluate the role of the NUP98 portions in NUP98-HOXA9-mediated transformation, we prepared a series of N-terminal deletion constructs and tested their ability to transform NIH 3T3 cells. Mutant NUP98a(Δ51–223)-HOXA9, which lacks 8 of 37 FG repeats in the NUP98a segment (Fig. 2A), retained its ability to transform NIH 3T3 fibroblasts (Fig. 2C). Moreover, mutant NUP98b(Δ51–223)-HOXA9, which lacked the same 8 FG repeats and had only 11 repeats left, remained transforming, although the number of colonies formed was only half of that induced by NUP98b-HOXA9 (Fig. 2A and C). However, when the seven N-terminal and four C-terminal FG repeats of mutant NUP98b(Δ51–223)-HOXA9 were separately fused to HOXA9 [designated NUP98b(Δ51–469)-HOXA9 and NUP98b(Δ1–223)-HOXA9, respectively], all transforming ability was abolished (Fig. 2C), despite their proper expression in NIH 3T3 cells (data not shown). Each of the above mutants was localized in the nucleus (Fig. 3I to L) and bound with PBX1a to the PBX1-HOXA10 bipartite DNA probe (Fig. 4B, lanes 2 to 11). Hence, the lack of oncogenicity of NUP98b(Δ51–469)-HOXA9 and NUP98b(Δ1–223)-HOXA9 did not arise from aberrant localization within cells or an inability to bind specific cellular DNA targets.
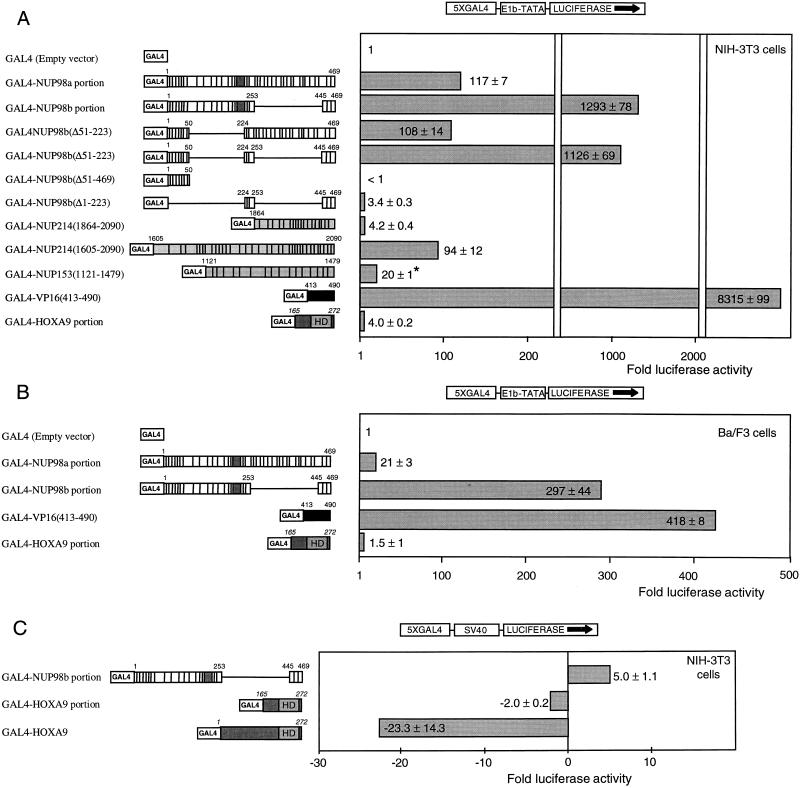
We reasoned that if NUP98-HOXA9 proteins operate as deregulated transcription factors in t(7;11)-associated leukemia, the NUP98 portion may provide a transcription-regulatory domain. To overcome interference from endogenous homeodomain proteins, we created fusion proteins containing the GAL4 DNA binding domain and various FG repeat-containing NUP98 segments to test for potential transcriptional activity. Each GAL4 fusion construct was cotransfected into NIH 3T3 cells with a reporter plasmid containing five GAL4 DNA binding sites (Fig. 5A). The HOXA9 portion of the chimeras fused to the GAL4 DNA binding domain displayed very little ability to activate transcription of this reporter. By contrast, both the NUP98a and the NUP98b portions, when fused to GAL4, were very strong activators of gene transcription. GAL4-NUP98a(Δ51–223) and GAL4-NUP98b(Δ51–223), which both transformed NIH 3T3 cells when linked to the HOXA9 C terminus, were also efficient transcriptional activators (Fig. 5A). However, NUP98b(Δ51–469) and NUP98b(Δ1–223), the two short segments derived from NUP98b that failed to transform fibroblasts as fusion partners of HOXA9, lacked transcriptional activation potential as GAL4 fusions (Fig. 2C).
FIG. 5.
Transcriptional regulatory properties of HOXA9 and FG repeat-containing segments of nucleoporins. (A) Schematic representation of the fusion proteins containing the GAL4 DNA binding domain and HOXA9, NUP98, NUP153, CAN/NUP214, and VP16 protein regions corresponding to the amino acids indicated (the various protein domains are indicated as described in the legend to Fig. 2). Transfection assays were performed in NIH 3T3 cells with reporter plasmid p5GB/GL2. Synthesis of the GAL4 fusion proteins was verified by Western blot analysis (data not shown). The asterisk indicates that there were no major differences in the levels of the various effector proteins, with the exception of NUP153(1121–1479), which was consistently expressed at approximately 10-fold-lower levels (data not shown). (B) Schematic representation of GAL4 fusion proteins tested for transcriptional properties in Ba/F3 cells (IL-3-dependent myeloid progenitor cells). The reporter used was p5GB/GL2. (C) HOXA9 is a strong repressor of gene transcription in NIH 3T3 cells. The reporter was the pGL2-promoter vector with five GAL4 DNA-binding sites ligated upstream of the SV40 promoter (43). Data in all panels are expressed as the fold difference in luciferase activity obtained with the GAL4 fusion proteins compared to that obtained with the GAL4 DNA binding domain alone. Values are the means and standard deviations of three independent experiments.
Furthermore, a selection of the above GAL4 fusion constructs stimulated the GAL4 reporter in Ba/F3 cells (myeloid progenitors that are dependent on interleukin-3 for growth [Fig. 5B]), demonstrating that FG repeats of NUP98 can act as transactivators in myeloid cells, where NUP98-HOXA9 chimeras are implicated in myeloid leukemia. Taken together, these studies suggest that the FG repeat-rich portions of NUP98a and NUP98b function as novel transactivation domains critical for NUP98-HOXA9-mediated transformation.
FG repeats of CAN/NUP214 and NUP98 are functionally exchangeable.
FG repeats from the other nucleoporin implicated in AML, CAN/NUP214 (49, 50), might function similarly to those from NUP98 in our transformation and transactivation assays. To investigate this possibility, we prepared two CAN/NUP214 cDNA fragments, one encoding the last 227 amino acids of CAN/NUP214 [CAN/NUP214(1864–2090)] and including 19 FG repeats (2 FXFG and 17 FG) and another encoding the last 469 amino acids [CAN/NUP214(1605–2090)] and including all 34 FG repeats from DEK-CAN/NUP214 and SET-CAN/NUP214 (3 FXFG and 32 FG). In addition, to test the generality of FG repeats in these assays, we generated a cDNA fragment encoding the last 359 amino acids of NUP153 [NUP153(1121–1479)], which contains 16 FG repeats (8 FXFG and 8 FG). The NUP153 and NUP214 segments prepared showed no significant sequence similarity to NUP98 other than FG repeats and a prevalence for Asn, Gln, Ser, and Thr residues in the spacer sequences between the FG repeats (the nucleoporin portion of NUP98a-HOXA9 consists of 17 GLFG, 2 FXFG, and 18 FG repeats; that of NUP98b-HOXA9 has 10 GLFG, 1 FXFG, and 8 FG repeats). CAN/NUP214(1605–2090)-HOXA9, CAN/NUP214(1864–2090)-HOXA9, and NUP153(1121–1479)-HOXA9 were all nuclear proteins (Fig. 3M and N), displayed the predicted molecular weight (data not shown), and efficiently bound DNA (Fig. 4C, lanes 2 to 5). When they were assayed for their colony-forming properties, CAN/NUP214(1605–2090)-HOXA9 and NUP153(1121–1479)-HOXA9, but not CAN/NUP214(1864–2090)-HOXA9, induced colony formation in soft agar [Fig. 2C; the average number of colonies generated by CAN/NUP214(1605–2090)- and NUP153(1121–1479)-HOXA9 expression was 78 and 35%, respectively, of that seen with NUP98b-HOXA9]. We then generated fusion proteins of GAL4 and the CAN/NUP214 and NUP153 portions to determine their transactivation potential. GAL4-CAN/NUP214(1605–2090) strongly activated reporter gene transcription (Fig. 5A), whereas GAL4-CAN/NUP214(1864–2090) had very little transactivation potential (comparable to the HOXA9 portion of the NUP98-HOXA9 fusion). GAL4-NUP153(1121–1479) displayed moderate transactivation of the reporter. We noticed that the levels of GAL4-NUP153(1121–1479) protein were about 5- to 10-fold lower than those of the other GAL4-NUP fusion proteins (data not shown), possibly accounting for its lower effect. Thus, our observations suggest that FG repeat-rich regions from heterologous nucleoporins can replace NUP98 in the NUP98-HOXA9 chimeras if they contain appreciable transcriptional activation potential.
We also swapped the NUP98 portion of NUP98b-HOXA9 for the transactivation domain of VP16 (amino acids 413 to 490 [Fig. 2A]) (12). The resulting VP16(413–490)-HOXA9 protein showed nuclear localization (Fig. 3O), bound to the PBX1-HOXA10 DNA probe with PBX1a (Fig. 4C, lanes 6 and 7), and transformed NIH 3T3 cells (about 40% of the efficiency of NUP98b-HOXA9 [Fig. 2C]). Thus, a heterologous transactivation domain fused to HOXA9 DNA binding and PBX heterodimerization domains is sufficient for transformation of the NIH 3T3 fibroblasts. This result supports the idea that NUP98-HOXA9 proteins are oncogenic transcription factors rather than oncogenic transport factors.
The aberrant function of NUP98-HOXA9 could also be due to loss of a transcriptional repression function present in full-length HOXA9. Indeed, the similar HOXA7 is a repressor of gene transcription (43). To test for HOXA9-dependent transcriptional repression activity, we cotransfected into NIH 3T3 cells GAL4-HOXA9 and a reporter plasmid containing five GAL4 DNA binding sites upstream of a luciferase gene driven by an SV40 promoter. As shown in Fig. 5C, GAL4-HOXA9 functioned as a very strong transcriptional repressor. For comparison, the HOXA9 portion from NUP98-HOXA9 fused to GAL4 did not display appreciable transcriptional repression whereas GAL4-NUP98b activated the SV40 promoter severalfold. These data suggest that the N terminus of HOXA9 contains a motif that functions as an inhibitor of transcription and is replaced by a nucleoporin-mediated transactivation function in t(7;11) leukemic cells.
CBP/p300 is necessary for NUP98-mediated transactivation.
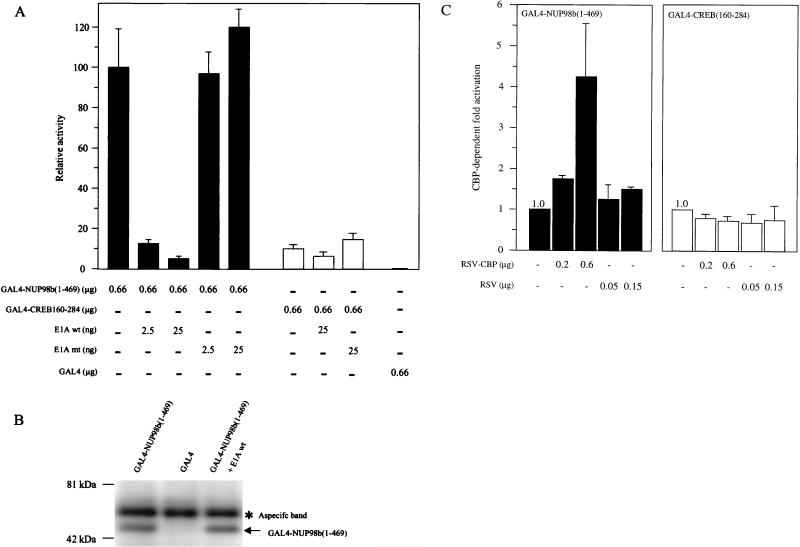
To further elucidate the mechanism whereby NUP98 FG repeats act as transactivators, we first examined the role of the CBP and p300 in this process. CBP and p300 are direct targets of chromosomal rearrangements in human leukemias, and both function as coactivators of a variety of gene-specific activators (20). To this end, we tested if the adenovirus 12S E1A protein inhibits GAL4-NUP98 function (12S E1A binds to the CH3 domain of CBP/p300 and inhibits associated transactivator function). Surprisingly, expression of 12S E1A protein dramatically repressed NUP98-dependent transcription activity in a dose-dependent manner (Fig. 6A). As shown in Fig. 6B, expression of E1A did not reduce the overall level of GAL4-NUP98(1–469) protein in NIH 3T3 cells. When we expressed a 12S E1A(Δ2–36) mutant that cannot bind to CBP/p300, we observed no inhibition of FG repeat-mediated transcriptional activation. As expected, 12S E1A expression had no effect on the activity of CBP/p300-independent transactivation mediated by CREB residues 160 to 284 [GAL4-CREB(160–284)] (Fig. 6A) (52), confirming that E1A specifically targeted CBP/p300-dependent transactivation in our assay.
FIG. 6.
NUP98 transactivation function requires CBP/p300. (A) GAL4-NUP98b(1–469) transcriptional activity is repressed by E1A, an inhibitor of CBP/p300. NIH 3T3 cells were transfected with p5GB/GL2 firefly luciferase reporter plasmid, and expression vectors for GAL4(1–147), GAL4-NUP98b(1–469), or GAL4-CREB(160–284) (a CBP/p300-independent-glutamine-rich activator from CREB; amino acids 160 to 284), and wild-type or mutant E1A [mutant E1A(Δ2–36)]. Luciferase activity derived from the p5GB/GL2 luciferase reporter was normalized to Renilla luciferase derived from the internal control reporter pRL-CMV. The mean activity and standard deviation is presented relative to the activity of GAL4-NUP98b(1–469) in the absence of E1A. (B) GAL4-NUP98 levels do not change in the presence of E1A. Western blot analysis of proteins precipitated with 12CA5 antibody from lysates of UCLA cells transiently transfected with GAL4-NUP98(1–469) (predicted molecular mass, ∼50 kDa), GAL4, or NUP98(1–469) plus E1A. Precipitated proteins were visualized with 12CA5. The asterisk indicates a protein precipitated nonspecifically with 12CA5. (C) Full-length CBP potentiates GAL4-NUP98b(1–469) transcriptional activity in NIH 3T3 cells. The p5GB/GL2 luciferase reporter was cotransfected with expression vectors for GAL4-NUP98b(1–469) or GAL4-CREB(160–284), and equimolar amounts of CBP (RSV-CBP) or empty vector (RSV). Luciferase activity derived from the p5GB/GL2 luciferase reporter was normalized to Renilla luciferase derived from the internal control reporter pRL-TK. Mean activation and standard deviation is expressed as the ratio of luciferase activity in the presence of RSV-CBP plasmid DNA divided by the level of luciferase activity in its absence. The results represent at least two independent experiments.
These results suggested that CBP would potentiate GAL4-NUP activity. We determined this by cotransfecting GAL4-NUP98b(1–469) expression vector with increasing amounts of RSV-CBP plasmid. CBP was found to stimulate GAL4-NUP98b(1–469)-induced reporter activity in a dose-dependent fashion (Fig. 6C). By contrast, the CBP/p300-independent transactivator GAL4-CREB(160–284) was not potentiated by RSV-CBP. Together, the above data implicate CBP/p300 in NUP98-dependent transcriptional activity.
FG repeat-rich portions of NUP98 bind CBP/p300 in vitro and in vivo.
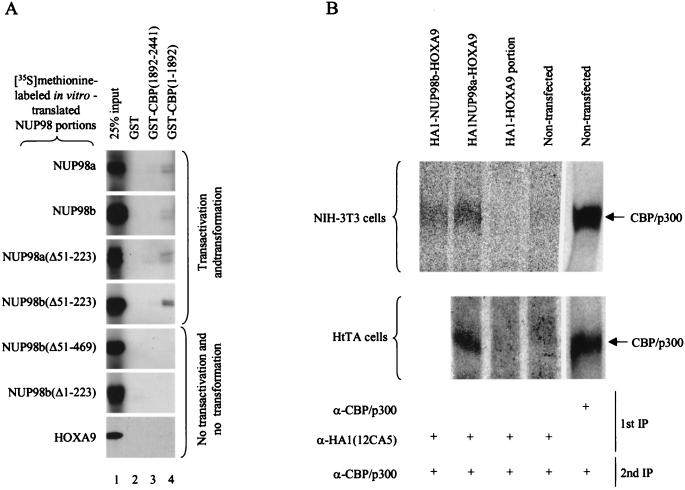
GAL4-NUP98 activity was inhibited by E1A and synergized with CBP, suggesting that FG repeat-rich portions of NUP98 may bind to CBP/p300. To test this possibility, we performed pull-down assays with glutathione S-transferase (GST)–CBP fusion proteins purified from Escherichia coli and in vitro-translated NUP98 fragments. Both NUP98 portions of the NUP98-HOXA9 chimeras bound effectively to GST-CBP(1–1892), a CBP portion that contains three major CBP domains for transcription factor binding (Fig. 7A, lanes 4). By contrast, no substantial binding was detected with GST-CBP(1892–2441), a CBP portion containing a glutamine-rich domain that binds p53 and SRC-1 (lane 3). Given that the NUP98 portions of both NUP98-HOXA9 chimeras bound to CBP(1–1892), we investigated whether NUP98-dependent transactivation correlates with physical binding to CBP. In vitro-translated NUP98 fragments corresponding to GAL4-NUP98 mutants with known transactivation activities (Fig. 5A) were tested for binding to GST-CBP(1–1892) and GST-CBP(1892–2441). As shown in Fig. 7A, NUP98 mutants that displayed transcriptional activity as GAL4 fusion proteins bound GST-CBP(1–1892) whereas those that lacked transactivation potential as GAL4 fusions failed to interact significantly with GST-CBP(1–1892). None of the NUP98 mutants showed significant interaction with GST-CBP(1892–2441). The HOXA9 portion of the fusion, which is not transcriptionally active when fused to GAL4, also failed to demonstrate noticeable association with either GST-CBP(1–1892) or GST-CBP(1892–2441). Thus, it appears that efficient binding of NUP98 fragments to CBP in vitro correlates with their ability to activate transcription.
FIG. 7.
Transcriptionally active FG repeat-rich portions of NUP98 bind CBP/p300. (A) GAL4-NUP98 transcriptional activity correlates with binding to CBP in vitro. GST pull-down assays were performed with the indicated [35S]methionine-labeled NUP98 or HOXA9 portions produced in vitro and GST-CBP fusion proteins purified from E. coli. The 25% input (lane) shows 25% of the NUP98 or HOXA9 segments used in each pull-down assay. Typically, in vitro-translated NUP98 portions appear as doublets, representing fragments with and without a HA1 tag (the NUP98 portions were cloned in pSP73 as HA1 fusion genes that retained the endogenous NUP98 translation initiation codon). GST acts as a negative control for binding. Comparable amounts of GST and GST-CBP fusion proteins were used in each pull-down assay. The experiment shown is representative of three independent experiments. (B) In vivo interaction between NUP98-HOXA9 oncoproteins and CBP/p300. Total-cell lysates were prepared from [35S]methionine-labeled HtTA or NIH 3T3 cells expressing the indicated HA1-tagged proteins. Two sequential immunoprecipitations were performed as detailed in Materials and Methods. The first and second immunoprecipitations (IP) were performed with the indicated antisera. A CBP/p300 antiserum cocktail was used for most efficient detection of these transcriptional coactivators. The positions of CBP/p300 and a molecular mass marker are indicated.
To confirm that the NUP98-HOXA9 proteins do interact with CBP/p300 in vivo, we prepared lysates of [35S]methionine-labeled NIH 3T3 cells expressing either HA1-NUP98a-HOXA9, HA1-NUP98b-HOXA9, or the HA1-tagged HOXA9 portion and performed two sequential immunoprecipitations, first with 12CA5 monoclonal antibody then with an CBP/p300-specific antibodies (52). An in vivo interaction between each of the NUP98-HOXA9 chimeras and CBP/p300 (seen as a broad band) was detected (Fig. 7B; the results shown are representative of three independent experiments). CBP/p300 was not detectable in an immunocomplex with the HOXA9 portion of the fusion protein alone, demonstrating that the FG repeat-rich portions of NUP98 mediate binding of CBP/p300 to NUP98a-HOXA9 and NUP98b-HOXA9. Confirming our findings in NIH 3T3 cells, a specific interaction between NUP98 sequences and CBP/p300 was also detected in lysates of [35S]methionine-labeled HtTA cells transiently transfected with an HA1-NUP98a-HOXA9 expression vector (Fig. 7B; the results are representative of two experiments). Western blot analysis with 12CA5 antibody revealed that the various HA1-tagged proteins studied for coimmunoprecipitation of CBP/p300 were expressed at comparable levels. This excludes the possibility that gross variations in HA1-tagged protein levels underlie the observed differences in binding to CBP/p300.
DISCUSSION
Although several distinct chromosomal translocations in human leukemias disrupt FG repeat-containing nucleoporins, a role for these components in the transformation of blood cells has not been established. Remarkably, we demonstrate that FG repeat-rich segments, which normally function in transport of macromolecules across the NE, perform a critical role in NUP98-HOXA9-mediated transformation, where they function as potent activators of transcription that physically and functionally interact with the transcriptional coactivators CBP and p300. Additional evidence to support a model in which NUP98-HOXA9 proteins act as oncogenic transcription factors includes (i) their nuclear localization, (ii) the specific DNA binding activity of the chimeras, and (iii) the requirement for intact HOXA9 DNA binding and PBX cofactor interaction. Interestingly, FG repeats from NUP98-HOXA9 could be replaced by CAN/NUP214 FG repeats, indicating that FG repeat-mediated transactivation may be a shared pathogenic function of nucleoporins in AML.
Deregulation of HOX activity by NUP98-HOXA9.
Vertebrate HOX genes are developmental regulators that also mediate key steps in the proliferation and differentiation of fetal and adult hematopoiesis in a lineage- and stage-specific manner (25). Although the repertoire of HOX gene expression during hematopoiesis is beginning to emerge, the genetic targets regulated by HOX proteins remain elusive, as is the case for the PBX and MEIS family members, which may cooperate with the implicated HOX proteins. NUP98-HOXA9-mediated transformation depends on its ability to bind DNA with members of the PBX family of HOX cofactors, implying that NUP98-HOXA9 affects the control of PBX/HOXA9-regulated target genes.
Like HOXA7, HOXA9 can function as a very efficient inhibitor of gene transcription. The HOXA9 portion of the NUP98-HOXA9 chimeras displayed no such repressor function, indicating that residues in the N terminus of full-length HOXA9 mediate gene repression. It is unlikely that the mere loss of HOXA9 repressor status is sufficient for the development of AML, because overexpression of a carboxy-terminal HOXA9 fragment does not transform NIH 3T3 cells. However, to gain oncogenic properties, the carboxy-terminal HOXA9 portion requires a transcriptional activator, which favors a model of leukemogenesis in which the FG repeats of NUP98-HOXA9 strongly activate HOXA9-responsive target genes whose deregulated expression interferes with proper execution of myelopoiesis. Transformation driven by cooverexpression of HOXA9 and MEIS1 does not seem to require a gain of transactivation function of HOXA9, suggesting that the mechanism of target gene deregulation involved may be different from that of NUP98-HOXA9-induced leukemias.
FG repeat-rich nucleoporins as potent oncogenic transactivators.
Most striking is the finding that FG repeat-containing portions from the N terminus of NUP98a and NUP98b can function as transactivation domains. Indeed, the oncogenic potential of NUP98-HOXA9 correlates with NUP98-mediated transcriptional competence. A certain threshold level of transactivation function is apparently required for oncogenicity, after which the correlation between transactivation and transformation is qualitative. However, we cannot exclude the possibility that NUP98 contributes another function to the NUP98-HOXA9 chimeras beside transcriptional activation. Nevertheless, our finding that the completely unrelated VP16 transactivation domain can functionally replace NUP98 suggests that other potential NUP98 functions are not necessary for oncogenesis in NIH 3T3 cells. Moreover, specific FG repeat-rich domains of two heterologous nucleoporins, CAN/NUP214 and NUP153, were oncogenic and contained transcriptional activation properties, further strengthening the correlation between transformation and transactivation for these domains. The transactivating FG repeat-rich segment of CAN/NUP214 is retained in the DEK-CAN/NUP214 and SET-CAN/NUP214 leukemic fusion proteins (49, 50). This suggests that the shared pathogenic mechanism of FG repeat-containing nucleoporins is to activate gene transcription. Although the precise function of DEK remains to be established, there is evidence to suggest that DEK is a sequence-specific DNA binding protein (15). Interestingly, the nuclear protein SET interacts directly with MLL (also called HRX) (1), which binds to DNA and positively regulates HOX gene expression (53). The MLL region that binds SET is consistently retained in all MLL leukemic fusion proteins (1). Thus, SET-CAN/NUP214 may contribute to leukemogenesis by interfering with the proper expression of MLL-controlled HOX proteins.
CBP/p300: the link between NUP98 FG repeats and the transcription machinery.
How does a protein that mediates macromolecule transport through the nuclear pores act as a regulator of transcription? Thus far, no evidence supporting a direct connection between NPC proteins and the basal transcription machinery has been reported, suggesting that the FG repeat segments of NUP98 may be fortuitous rather than genuine activators of gene transcription. Our results show that transactivation of FG repeat-rich segments of NUP98 correlates with their ability to interact and functionally collaborate with the transcriptional coactivators CBP/p300. Thus far, NUP98-HOXA9 is the first translocation-generated fusion protein known to recruit CBP/p300 as part of its oncogenic mechanism. However, there is additional evidence to suggest a more central role for CBP/p300 in both hematopoiesis and leukemia. First, CBP and p300 are direct targets of at least three independent chromosomal rearrangements in human leukemias. Both CBP and p300 are fused to MLL through, respectively, the t(11;16) (41, 46) and t(11;22) (19) translocations. In addition, CBP is linked to MOZ through the t(8;16) translocation associated with AML (8). Second, CBP/p300 interacts and functionally cooperates with the AML1/CBFβ transcription factor complex in myeloid cell differentiation (21), underscoring a role for CBP/p300 as a hematopoietic transcriptional coactivator. Moreover, the AML1/CBFβ transcription factor complex is the most frequent target of chromosomal translocations in human leukemias. Thus, aberrant formation of this complex by AML1 or CBFβ fusion proteins may affect the role of CBP/p300 in myeloid cells.
Interaction of CBP with RNA polymerase II in vitro requires RNA helicase A, a nuclear 3′-5′ double-stranded DNA-RNA helicase containing a consensus DEAH box motif (32). The NUP98-DDX10 fusion protein in AML patients with the inv(11)(p15q22) chromosomal rearrangement contains the NUP98 portion that binds CBP and the DEAH box of the putative RNA helicase DDX10, which may have affinity for CBP as well (2). Thus, it will be of interest to determine whether NUP98-DDX10 promotes leukemogenesis by deregulation of CBP/p300 function. It is also tempting to speculate that NUP98 and CBP/p300 may interact at the NE, perhaps to promote transcription near the nuclear pores. However, in preliminary studies, we could not detect colocalization of NUP98 and CBP/p300 at nuclear pores or coimmunoprecipitate CBP/p300 with endogenous NUP98 by using NUP98-specific antibodies (8a). This suggests that nuclear relocation of the FG repeats and/or their fusion with a specific DNA binding domain may be essential for their interaction with CBP/p300.
ACKNOWLEDGMENTS
We thank Gerard Grosveld for plasmids pHA1triple-tag#10 and pUHD10S and support at the initiation of this work, L. C. Chan for the t(7;11) patient sample, Neal Copeland and Takuro Nakamura for the full-length HOXA9 plasmid, Hermann Bujard for HtTA cells, Chris Denny for NIH 3T3 cells, Martine Roussel for vector pSRαMSVtkCD8, and Albert Reynolds for the 12CA5 monoclonal antibody. We are grateful to Arjan Buijs, Ereke Bruce, Richard Moriggl, and Steve Morris for critical help and advice throughout this work and to Susan Baker, Jim Downing, Maarten Fornerod, and Tom Look for critically reading the manuscript.
These studies were supported by Cancer Center CORE grant CA-21765 and the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children’s Research Hospital. J.M.A.V.D., C.E.J.P., and L.H.K. were supported by NIH grant CA77262-01; M.L.C. is supported by NIH grant CA-42971; and C.A.S. is a fellow of the Leukemia Society of America and the Stanford Immunology Program (AI-07290).
REFERENCES
- 1.Adler H T, Nallaseth F S, Walter G, Tkachuk D C. HRX leukemic fusion proteins form a heterocomplex with the leukemia-associated protein SET and protein phosphatase 2A. J Biol Chem. 1997;272:28407–28414. doi: 10.1074/jbc.272.45.28407. [DOI] [PubMed] [Google Scholar]
- 2.Arai Y, Hosoda F, Kobayashi H, Arai K, Hayashi Y, Kamada N, Kaneko Y, Ohki M. The inv(11)(p15q22) chromosome translocation of de novo and therapy-related myeloid malignancies results in fusion of the nucleoporin gene, NUP98, with the putative RNA helicase gene, DDX10. Blood. 1997;89:3936–3944. [PubMed] [Google Scholar]
- 3.Bastos R, Lin A, Enarson M, Burke B. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J Cell Biol. 1996;134:1141–1156. doi: 10.1083/jcb.134.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bedigian H G, Johnson D A, Jenkins N A, Copeland N G, Evans R. Spontaneous and induced leukemias of myeloid origin in recombinant inbred BXH mice. J Virol. 1984;51:586–594. doi: 10.1128/jvi.51.3.586-594.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bedigian H G, Taylor B A, Meier H. Expression of murine leukemia viruses in the highly lymphomatous BXH-2 recombinant inbred mouse strain. J Virol. 1981;39:632–640. doi: 10.1128/jvi.39.2.632-640.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson G V, Nguyen T H, Maas R L. The expression pattern of the murine Hoxa-10 gene and the sequence recognition of its homeodomain reveal specific properties of Abdominal B-like genes. Mol Cell Biol. 1995;15:1591–1601. doi: 10.1128/mcb.15.3.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borrow J, Shearman A M, Stanton V P, Jr, Becher R, Collins T, Williams A J, Dube I, Katz F, Kwong Y L, Morris C, Ohyashiki K, Toyama K, Rowley J, Housman D E. The t(7;11)(p15;p15) translocation in acute myeloid leukaemia fuses the genes for nucleoporin NUP98 and class I homeoprotein HOXA9. Nat Genet. 1996;12:159–167. doi: 10.1038/ng0296-159. [DOI] [PubMed] [Google Scholar]
- 8.Borrow J, Stanton V P, Jr, Andresen J M, Becher R, Behm F G, Chaganti R S, Civin C I, Disteche C, Dube I, Frischauf A M, Horsman D, Mitelman F, Volinia S, Watmore A E, Housman D E. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 8a.Brindle, P., and J. van Deursen. Unpublished data.
- 9.Chan S K, Jaffe L, Capovilla M, Botas J, Mann R S. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell. 1994;78:603–615. doi: 10.1016/0092-8674(94)90525-8. [DOI] [PubMed] [Google Scholar]
- 10.Chang C P, Brocchieri L, Shen W F, Largman C, Cleary M L. Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol Cell Biol. 1996;16:1734–1745. doi: 10.1128/mcb.16.4.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang C P, Shen W F, Rozenfeld S, Lawrence H J, Largman C, Cleary M L. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 12.Dalrymple M A, McGeoch D J, Davison A J, Preston C M. DNA sequence of the herpes simplex virus type 1 gene whose product is responsible for transcriptional activation of immediate early promoters. Nucleic Acids Res. 1985;13:7865–7879. doi: 10.1093/nar/13.21.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fornerod M, Boer J, van Baal S, Jaegle M, von Lindern M, Murti K G, Davis D, Bonten J, Buijs A, Grosveld G. Relocation of the carboxyterminal part of CAN from the nuclear envelope to the nucleus as a result of leukemia-specific chromosome rearrangements. Oncogene. 1995;10:1739–1748. [PubMed] [Google Scholar]
- 14.Fu G K, Grosveld G, Markovitz D M. DEK, an autoantigen involved in a chromosomal translocation in acute myelogenous leukemia, binds to the HIV-2 enhancer. Proc Natl Acad Sci USA. 1997;94:1811–1815. doi: 10.1073/pnas.94.5.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuda M, Gotoh Y, Nishida E. Interaction of MAP kinase with MAP kinase kinase: its possible role in the control of nucleocytoplasmic transport of MAP kinase. EMBO J. 1997;16:1901–1908. doi: 10.1093/emboj/16.8.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirai H, Roussel M F, Kato J Y, Ashmun R A, Sherr C J. Novel INK4 proteins, p19 and p18, are specific inhibitors of the cyclin D-dependent kinases CDK4 and CDK6. Mol Cell Biol. 1995;15:2672–2681. doi: 10.1128/mcb.15.5.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang S Y, Tang J L, Liang Y J, Wang C H, Chen Y C, Tien H F. Clinical, haematological and molecular studies in patients with chromosome translocation t(7;11): a study of four Chinese patients in Taiwan. Br J Haematol. 1997;96:682–687. doi: 10.1046/j.1365-2141.1997.d01-2100.x. [DOI] [PubMed] [Google Scholar]
- 19.Ida K, Kitabayashi I, Taki T, Taniwaki M, Noro K, Yamamoto M, Ohki M, Hayashi Y. Adenoviral E1A-associated protein p300 is involved in acute myeloid leukemia with t(11;22)(q23;q13) Blood. 1997;90:4699–4704. [PubMed] [Google Scholar]
- 20.Janknecht R, Hunter T. Transcription. A growing coactivator network. Nature. 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- 21.Kitabayashi I, Ida K, Morohoshi F, Yokoyama A, Mitsuhashi N, Shimizu K, Nomura N, Hayashi Y, Ohki M. The AML1-MTG8 leukemic fusion protein forms a complex with a novel member of the MTG8(ETO/CDR) family, MTGR1. Mol Cell Biol. 1998;18:846–858. doi: 10.1128/mcb.18.2.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraemer D, Wozniak R W, Blobel G, Radu A. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc Natl Acad Sci USA. 1994;91:1519–1523. doi: 10.1073/pnas.91.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg A M, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavau C, Szilvassy S J, Slany R, Cleary M L. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence H J, Helgason C D, Sauvageau G, Fong S, Izon D J, Humphries R K, Largman C. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood. 1997;89:1922–1930. [PubMed] [Google Scholar]
- 26.Lu Q, Kamps M P. Heterodimerization of Hox proteins with Pbx1 and oncoprotein E2a-Pbx1 generates unique DNA-binding specificities at nucleotides predicted to contact the N-terminal arm of the Hox homeodomain—demonstration of Hox-dependent targeting of E2a-Pbx1 in vivo. Oncogene. 1997;14:75–83. doi: 10.1038/sj.onc.1200799. [DOI] [PubMed] [Google Scholar]
- 27.Martin K J, Lillie J W, Green M R. Evidence for interaction of different eukaryotic transcriptional activators with distinct cellular targets. Nature. 1990;346:147–152. doi: 10.1038/346147a0. [DOI] [PubMed] [Google Scholar]
- 28.Moriggl R, Berchtold S, Friedrich K, Standke G J, Kammer W, Heim M, Wissler M, Stocklin E, Gouilleux F, Groner B. Comparison of the transactivation domains of Stat5 and Stat6 in lymphoid cells and mammary epithelial cells. Mol Cell Biol. 1997;17:3663–3678. doi: 10.1128/mcb.17.7.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moskow J J, Bullrich F, Huebner K, Daar I O, Buchberg A M. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller A J, Young J C, Pendergast A M, Pondel M, Landau N R, Littman D R, Witte O N. BCR first exon sequences specifically activate the BCR/ABL tyrosine kinase oncogene of Philadelphia chromosome-positive human leukemias. Mol Cell Biol. 1991;11:1785–1792. doi: 10.1128/mcb.11.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagata K, Kawase H, Handa H, Yano K, Yamasaki M, Ishimi Y, Okuda A, Kikuchi A, Matsumoto K. Replication factor encoded by a putative oncogene, set, associated with myeloid leukemogenesis. Proc Natl Acad Sci USA. 1995;92:4279–4283. doi: 10.1073/pnas.92.10.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura T, Largaespada D A, Lee M P, Johnson L A, Ohyashiki K, Toyama K, Chen S J, Willman C L, Chen I M, Feinberg A P, Jenkins N A, Copeland N G, Shaughnessy J D., Jr Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet. 1996;12:154–158. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura T, Largaespada D A, Shaughnessy J D, Jr, Jenkins N A, Copeland N G. Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukaemias. Nat Genet. 1996;12:149–153. doi: 10.1038/ng0296-149. [DOI] [PubMed] [Google Scholar]
- 35.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 36.Ohno M, Fornerod M, Mattaj I W. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- 37.Phelan M L, Rambaldi I, Featherstone M S. Cooperative interactions between HOX and PBX proteins mediated by a conserved peptide motif. Mol Cell Biol. 1995;15:3989–3997. doi: 10.1128/mcb.15.8.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Popperl H, Bienz M, Studer M, Chan S K, Aparicio S, Brenner S, Mann R S, Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- 39.Radu A, Moore M S, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 40.Rauskolb C, Wieschaus E. Coordinate regulation of downstream genes by extradenticle and the homeotic selector proteins. EMBO J. 1994;13:3561–3569. doi: 10.1002/j.1460-2075.1994.tb06663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowley J D, Reshmi S, Sobulo O, Musvee T, Anastasi J, Raimondi S, Schneider N R, Barredo J C, Cantu E S, Schlegelberger B, Behm F, Doggett N A, Borrow J, Zeleznik-Le N. All patients with the T(11;16)(q23;p13.3) that involves MLL and CBP have treatment-related hematologic disorders. Blood. 1997;90:535–541. [PubMed] [Google Scholar]
- 42.Sauvageau G, Lansdorp P M, Eaves C J, Hogge D E, Dragowska W H, Reid D S, Largman C, Lawrence H J, Humphries R K. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc Natl Acad Sci USA. 1994;91:12223–12227. doi: 10.1073/pnas.91.25.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schnabel C A, Abate-Shen C. Repression by HoxA7 is mediated by the homeodomain and the modulatory action of its N-terminal-arm residues. Mol Cell Biol. 1996;16:2678–2688. doi: 10.1128/mcb.16.6.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen W F, Montgomery J C, Rozenfeld S, Moskow J J, Lawrence H J, Buchberg A M, Largman C. AbdB-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol Cell Biol. 1997;17:6448–6458. doi: 10.1128/mcb.17.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen W F, Rozenfeld S, Lawrence H J, Largman C. The Abd-B-like Hox homeodomain proteins can be subdivided by the ability to form complexes with Pbx1a on a novel DNA target. J Biol Chem. 1997;272:8198–8206. doi: 10.1074/jbc.272.13.8198. [DOI] [PubMed] [Google Scholar]
- 46.Taki T, Sako M, Tsuchida M, Hayashi Y. The t(11;16)(q23;p13) translocation in myelodysplastic syndrome fuses the MLL gene to the CBP gene. Blood. 1997;89:3945–3950. [PubMed] [Google Scholar]
- 47.van Dijk M A, Murre C. Extradenticle raises the DNA binding specificity of homeotic selector gene products. Cell. 1994;78:617–624. doi: 10.1016/0092-8674(94)90526-6. [DOI] [PubMed] [Google Scholar]
- 48.van Dijk M A, Peltenburg L T, Murre C. Hox gene products modulate the DNA binding activity of Pbx1 and Pbx2. Mech Dev. 1995;52:99–108. doi: 10.1016/0925-4773(95)00394-g. [DOI] [PubMed] [Google Scholar]
- 49.von Lindern M, Fornerod M, Soekarman N, van Baal S, Jaegle M, Hagemeijer A, Bootsma D, Grosveld G. Translocation t(6;9) in acute nonlymphocytic leukaemia results in the formation of a DEK-CAN fusion gene. Baillieres Clin Haematol. 1992;5:857–879. doi: 10.1016/s0950-3536(11)80049-1. [DOI] [PubMed] [Google Scholar]
- 50.von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol Cell Biol. 1992;12:3346–3355. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 52.Yang C, Shapiro L H, Rivera M, Kumar A, Brindle P K. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol Cell Biol. 1998;18:2218–2229. doi: 10.1128/mcb.18.4.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu B D, Hess J L, Horning S E, Brown G A, Korsmeyer S J. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]