Abstract
Pericytes and endothelial cells share membranous interdigitations called “peg-and-socket” interactions that facilitate their adhesion and biochemical crosstalk during vascular homeostasis. However, the morphology and distribution of these ultrastructures have remained elusive. Using a combination of 3D electron microscopy techniques, we examined peg-and-socket interactions in mouse brain capillaries. We found that pegs extending from pericytes to endothelial cells were morphologically diverse, exhibiting claw-like morphologies at the edge of the cell and bouton-shaped swellings away from the edge. Reciprocal endothelial pegs projecting into pericytes were less abundant and appeared as larger columnar protuberances. A large-scale 3D EM data set revealed enrichment of both pericyte and endothelial pegs around pericyte somata. The ratio of pericyte versus endothelial pegs was conserved among the pericytes examined, but total peg abundance was heterogeneous across cells. These data show considerable investment between pericytes and endothelial cells, and provide morphological evidence for pericyte somata as sites of enriched physical and biochemical interaction.
Keywords: Capillary, electron microscopy, endothelial cell, peg-and-socket, and pericyte
Introduction
Of all organs in the body, pericytes are most abundant in microvascular networks of the brain.1 Pericytes reside on the abluminal surface of endothelial cells and interact physically and biochemically with the endothelium to support numerous capillary functions,2 including angiogenesis and development of capillary network architecture,3 blood-brain barrier integrity4–6 and blood flow regulation.7–9 Although the basal lamina forms a physical division between pericytes and endothelial cells, this layer thins or becomes absent where the cells come in contact through a variety of subcellular structures, such as adhesion plaques, gap junctions, and peg-and-socket interactions.10–13 These structures are thought to be the loci of cell-cell communication and adherence. Their importance is highlighted by studies that disrupt pericyte-endothelial communication. For instance, mural cell differentiation is blocked in the absence of endothelial gap junctions.14,15 Impaired expression of N-cadherin, which is enriched in adhesion plaques, leads to loose pericyte attachment on the endothelium.16,17 While data on pericyte-endothelial interactions is increasing, precise 3D ultrastructural detail of these cellular interactions has been lacking.
Peg-and-socket connections are a defining feature of pericyte-endothelial interaction.18 Pegs are typically seen as protrusions from either pericytes or endothelial cells, resulting in indentations within the apposing cell type. In addition to anchoring the two cell types to each other, observations of enriched signaling protein expression and specialized vesicular structures at peg-and-sockets suggest their involvement in cellular cross-talk.19 However, despite decades of research, basic attributes of peg-and-socket structures including their 3D morphology, abundance, and distribution along capillary networks have remained uncharacterized. Their sub-micrometer size (tens to hundreds of nm) necessitates the use of electron microscopy for visualization, but the cross-sectional views provided by standard 2D transmission electron microscopy (TEM) capture only limited information on their morphology and distribution.20–23 Conventional scanning electron microscopy does not improve our understanding of peg-and-sockets because the cell-cell interface cannot be accessed.24 Serial section volume electron microscopy (3D EM) overcomes these limitations by providing EM resolution over 3D space with sequential sectioning and imaging. Two prior 3D EM studies described pericyte morphology in brain microvasculature. They showed how the abluminal surface of pericyte processes interacted with astrocytic endfeet,25 and how pericyte somata change shape in response to cerebral ischemia.26 However, these studies did not examine the pericyte-endothelial interface, where peg-and-socket interactions reside.
Acquiring 3D EM data from the capillary bed requires overcoming unique challenges in ultrastructural imaging. First, the microvasculature only occupies about 1% of the cortical tissue volume in mouse,27 making it necessary to first scan larger regions of tissue in search for vasculature. Second, pericyte somata, which have a larger endothelial interface to study, are only intermittently present along the capillary network.28 Third, the tortuosity of the capillary network makes it difficult to find capillaries cut in the cross-sectional orientation, which allow pericytes and endothelial interfaces to be more clearly resolved and segmented. Given these limitations, we used a two-step approach to locate pericyte somata. This involved using serial block-face scanning electron microscopy (SBF-SEM)29 to first survey larger regions of tissue, followed by focused ion beam scanning electron microscopy (FIB-SEM)30 to collect higher resolution 3D data for detailed morphological analysis. SBF-SEM is ideal for imaging larger fields of view (FOV), but results in thicker slices in the z-dimension (5 nm × 5 nm × 50 nm voxels). In contrast, FIB-SEM images at higher resolution in the z-dimension (isotropic voxels of 5 nm), but has a more restricted FOV. When used in combination, these complementary EM techniques are ideal for imaging the ultrastructure of rare events in tissue specimens.31
In addition to studying pericyte structure in a FIB-SEM data set, we further examined microvasculature in a large-scale SBF-SEM data set from mouse visual cortex publicly available through MICrONS Explorer, a consortium effort to map brain neural connectivity.32 This enabled us to examine peg-and-socket abundance and distribution across capillary networks, and also to compare SBF-SEM and FIB-SEM data sets. Collectively, our findings provide a unique view of the brain pericyte-endothelial interface and the reciprocal peg-and-socket interactions between these cell types. These data highlight the pericyte soma as a hub of pericyte-endothelial interaction in the adult mouse brain.
Methods
The Institutional Animal Care and Use Committee at the Medical University of South Carolina approved tissue collection procedures used for FIB-SEM imaging. The University has accreditation from the Association for Assessment and Accreditation of Laboratory Animal Care International, and all experiments were performed within its guidelines. All data were analyzed and reported according to ARRIVE guidelines.
Tissue preparation
For the FIB-SEM data set, a young adult male PDGFRβ-tdTomato mouse28 was transcardially perfused with modified Karnovsky’s fixative containing 2.5% EM grade glutaraldehyde, 2% paraformaldehyde (freshly made from powder), 0.025% CaCl2 in 0.15 M cacodylate buffer (pH 7.0-7.2). The perfusion pressure was carefully monitored to not exceed 120 mmHg using a specialized perfusion fixation system with sphygmomanometer to monitor pressure applied. After fixation, the brain was removed and sectioned into 1 mm thick slices on a brain matrix. The mouse S1 region of the somatosensory cortex was then excised with a scalpel blade, producing a roughly 1 mm3 piece of tissue that was left in fixative overnight. The following day, the tissue was washed in 0.15 M cacodylate buffer (pH7.4), and then treated with 1% osmium tetroxide, 1.5% potassium ferrocyanide in cacodylate buffer (0.15 M, pH 7.4) for 60 min. The tissues were then washed again in 0.15 M cacodylate buffer. Next, samples were immersed in 1% thiocarbohydrazide for 20 min at room temperature. For the following steps, until resin infiltration, we made use of a PELCO Biowave (Tedpella) for microwave processing.44 Washes in ultra-pure water (UPW) were followed by a second osmication of 2% osmium in UPW. After washing 5 × 3 min in UPW samples were placed in uranyl acetate replacement stain (Electron Microscopy Sciences) in water (1:3). During this incubation, Walton’s lead aspartate staining was prepared by freshly dissolving lead nitrate (20 mM) in 30 mM l-aspartic acid. This solution was adjusted to pH 5.5 with potassium hydroxide. After washing the samples in UPW, they were incubated in the fresh Walton’s lead at 65 °C for 30 min. After the final washes, the samples were dehydrated using a series of solutions of increasing EtOH concentration (50, 70, 90, and 2× 100%). Subsequent infiltration with resin (Spurr’s; EMS) was done by first incubating the samples for 2–4 h in 50% resin in EtOH, followed by an overnight incubation in 100% resin. The next day, after two more changes in fresh resin, the samples were flat-embedded between two microscopy slides in fresh resin and cured in the oven at 65°C for 72 h.45
SBF–SEM and FIB imaging
For SBF-SEM, the resin-embedded samples were mounted on an aluminum specimen pin (Melotte, Belgium) using conductive epoxy (Circuit Works).44 The specimens were trimmed into a pyramid shape using an ultramicrotome (UltraCut, 175 Reichert-Jung, Vienna, Austria). Next, samples were coated with 3 nm of platinum in a Quorum Q 150 T ES sputter coater (Quorum Technologies, Laughton, UK). The aluminum pins were placed in the Gatan 3View 2XP (Gatan Inc, Milton UK) in a Zeiss Merlin SEM (Carl Zeiss, Oberkochen Germany) for imaging at 2 kV with a Gatan Digiscan II ESB detector. The sample blocked was scanned for two capillaries close to each other. When found, these capillaries were followed by sectioning at 100 nm sections. Just before we expected the capillary branching point, the sample was removed from the SBF-SEM and transferred to the FIB-SEM
FIB-SEM imaging was performed using a Zeiss Crossbeam 540 system with Atlas5 software. The Focused Ion Beam (FIB) was set to remove 5 nm sections by propelling Gallium ions at the surface. The Atlas software was set-up to image an area of 22 µm × 7 µm at 5 nm pixels using an ESB (back-scattered electron) detector (grid voltage = 1200 V) with the electron beam at 1.5 kV and 1 nA.
We collected four data sets from one mouse. However, only one proved to be suitable for accurate segmentation, as images were obtained cross-sectionally from the capillary, allowing the basement membrane that separated the pericyte and endothelium to be clearly distinguished. In other data sets, imaging orientation was longitudinal to the capillary axis and the separation between pericyte and endothelium became harder to discern. Re-slicing the image volume at image analysis to obtain a cross-sectional orientation revealed jitter between physical slices that deteriorated the quality of the basement membrane signal.
Segmentation and 3D rendering of endothelial and pericyte structures
The FIB-SEM data set consisted of 2299 sections cut in series. The full data set was captured a 5 nm isotropic resolution. The data set was reduced in size by binning in the x-y plane to obtain 20 nm pixel dimensions. This binning reduced the size of the overall data set, and was necessary to facilitate reconstruction using available computing resources. The resolution of the z-axis was unchanged, and thus voxels in the analyzed data set were 20 × 20 × 5 nm resolution. Image segmentation was performed using Microscopy Image Browser (MIB) software.46 During this process, each plane in the image stack was manually traced, carefully defining the boundaries of the pericyte and endothelium. This manual analysis required ∼90 h to complete. After segmentation was completed the result was a Z-stack of raw traces for each structure that could be transported to Imaris (Bitplane) software for 3D rendering. Using the Imaris surface tool we generated 3D models of each structure by utilizing the automatic algorithm for surface detection and setting the threshold to the absolute intensity of raw data. The surfaces were smoothed out to a 40 nm detail level. Supplementary movies of the surface models were made in Imaris and edited in Adobe Premiere.
Peg analysis
To analyze the characteristics of pericyte and endothelial pegs, we individually segmented all endothelial pegs (n = 8) as well as a subset of pericyte pegs (n = 14). Creating segmentations for individual pegs allowed us to extract surface area and volume measurements from individual pegs in Imaris. The inclusion criteria used to define pegs, explained in Supplementary Fig. 2, was applied to analysis of both the FIB-SEM and MICrONS data sets.
Socket measurements
The width of both endothelial and pericyte sockets were measured on surface models utilizing the Measuring Point tool in Imaris. This tool allowed us to define two points directly on the 3D structure and calculate the distance between two points. We defined the socket width as the longest length across the whole socket. For sockets with amorphous shapes (neither circle or ellipse) we took multiple measurements of what appeared to be the longest width and reported the average of these values.
Navigation and annotation in MICrONS data set
The MICrONS data set consisted of a volume from layer 2/3 of a P36 male mouse visual cortex imaged at 3.58 × 3.58 × 40 nm resolution.32 A number of cell types were segmented and available for 3D viewing. For the purposes of this paper, only the capillary network was examined, as this encompassed both cells of interest (pericyte and endothelial) and the high-resolution 2D images allowed us to discern peg-and-socket locations with high confidence. However, pericytes and endothelial cells were not separately segmented in this online resource. First, pericyte and endothelial nuclei were located and annotated in the 2D view throughout the entire tissue volume. Second, using peg-and-socket criteria defined in Supplementary Fig. 2, all instances of pericyte and endothelial pegs were annotated throughout the capillary network (by SO). These annotations were then reviewed by two independent raters (VCS and SKB), with prior experience viewing vasculature in 2D EM images. If one reviewer disagreed or was unsure about the veracity of a peg, then that peg was omitted from the data set. Once all pegs were confirmed, we used the 3D coordinates from each peg annotation to calculate its distance from the nearest pericyte nucleus. To define the average width of the pericyte nucleus we located the start and end points of all pericyte nuclei throughout the EM data; omitting pericyte #2 as depicted in Figure 6 due to missing data. We then used the line-measuring tool in Neuromancer to calculate the distance between these two points and obtain the average width of a pericyte nucleus. Next, we defined the center point (midpoint of average pericyte nuclei width) to calculate the distance between all pegs and the pericyte nucleus using custom-code written in MATLAB. The locations of endothelial tight junctions and edges of pericyte processes were similarly annotated on 2D images on every 5–10 slices in the MICrONS dataset. Neuromancer annotations for all pericyte and endothelial peg locations, as well as endothelial tight junction positions examined within the MICrONS data set, can be viewed by web addresses provided in Supplementary Data.
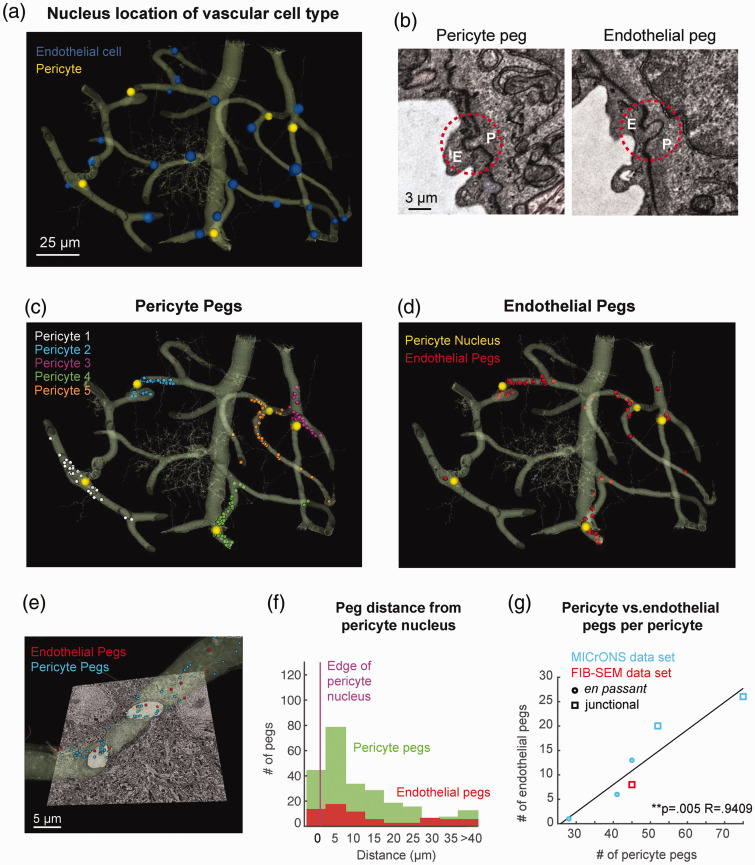
Figure 6.
Peg-and-socket enrichment at pericyte somata revealed in a large-scale EM data set: (a) 3D rendering of the capillary network surrounding a cortical ascending venules captured in the MICrONS data set, including 25 endothelial cells (blue) and 5 pericytes (yellow). A neuron is present within the vascular segmentation (cell with fine processes at center of image); (b) representative pericyte peg (left) and endothelial peg (right) seen in 2D slices, outlined with red-dashed circles; (c) distribution of pericyte pegs in the capillary network. Pegs are annotated in different colors based on the pericyte from which they originated; (d) distribution of endothelial pegs in the capillary network; (e) pericyte soma located at capillary bifurcation (Pericyte 2 from panel c). Endothelial pegs are annotated with red dots and pericyte pegs with blue dots; (f) histograms showing distance of both pericyte and endothelial pegs relative to nearest pericyte soma; (g) scatter plot of endothelial peg number versus pericyte pegs number for each pericyte soma, including both data from the MICrONS data set and the FIB-SEM example. A strong positive correlation is observed (Pearson’s correlation analysis, **p = 0.005; R = 0.9409).
Comparison of MICrONS and FIB-SEM data sets
Our analyses of the MICrONS data set spanned Z sections 34-2176 (or 1360–87,040 nm), giving a total Z examined range of 85.7 µm. The total size of the MICrONS data set is 250 × 140 × 90 µm. The dimensions of the FIB data set are 7 × 22 × 11 μm, as shown in Figure 2(a). The fixation procedures between data sets were slightly different. The MICrONS preparation used 2.5% paraformaldehyde and 1.25% glutaradehyde for perfusion, and the brain was then post-fixed for 16–72 h at 4°C. The FIB preparation used 2% paraformaldehyde and 2.5% glutaradehyde for perfusion, and small tissue sample (1 mm3) was post-fixed overnight at 4°C. The MICrONS post fixation was also different, using 2% osmium tetroxide and 2.5% potassium ferricyanide (90 min), as opposed to 1% osmium tetroxide and 1.5% potassium ferrocyanide (60 min) for the FIB data.
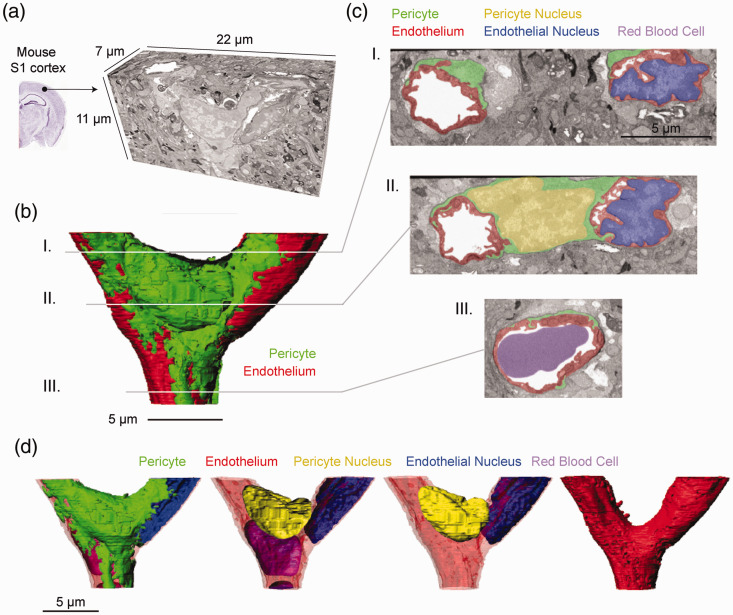
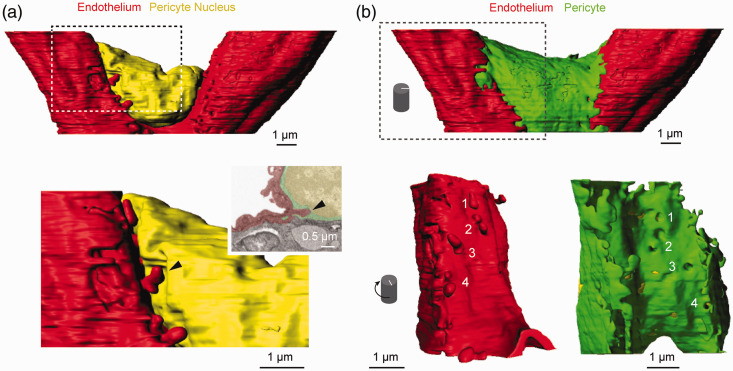
Figure 2.
Segmentation and surface modeling of a pericyte and endothelial cell: (a) representation of raw 3D FIB-SEM data used for segmentation. Total tissue volume of 1694 cubic micrometers was obtained from mouse S1 cortex; (b, c) 3D surface rendering of EM data displaying the capillary endothelium (red) and pericyte (green). Two-dimensional images from different planes in the data set are shown. Segmented components used to generate surfaces are each color coded according to structure; (d) 3D reconstructions of different sets of the five segmented components in the data set.
In vivo two-photon imaging
We generated chronic, skull-removed cranial windows for in vivo imaging, as previously described.47 The cranial window was created over the left hemisphere, and centered over 1.5 mm posterior and 3 mm lateral to bregma, which encompasses the S1 region of somatosensory cortex. Mice were imaged at least three weeks after cranial window construction. To label the vasculature, 50 µL of 5% (w/v in saline) 2 MDa FITC-dextran (Sigma-Aldrich; FD2000S) was injected through the retro-orbital vein under deep isoflurane anesthesia (4% MAC in medical air). Isoflurane was reduced to ∼1.5% MAC in medical air during imaging. The cortical microvasculature was imaged with a Bruker Investigator coupled to a Spectra-Physics Insight X3. Green and red fluorescence emission was collected through 515/30 nm and 615/60 nm bandpass filters, respectively, and detected by photomultiplier tubes. High-resolution image stacks were collected with a 20-X (1.0 NA) water-immersion objective (Olympus; XLUMPLFLN) and imaged with 975 nm excitation.
Analysis of pericyte locations in two-photon imaging data
In vivo two-photon images were acquired from three to six-month-old PDGFRβ-tdTomato mice (14 image stacks from eight mice). For all stacks, pericytes in a 200 × 200 × 200 μm volume were examined. Pericytes were defined as mural cells on vessels smaller than ∼6 µm in diameter with protruding ovoid cell bodies. These were identified as “junctional pericytes” if their somata were located directly on a capillary bifurcation. En passant pericytes were considered as cells with somata further than ∼10 µm from the nearest junction. Pericytes with somata within 10 µm of a junction but not directly at the bifurcation were excluded from the analysis, based on ambiguous positioning (∼10% of cells excluded, out of the 623 total somata identified).
Histology and super-resolution confocal imaging
Coronal sections of brain tissue 50 µm thick were obtained from transcardially perfused PDGFRβ-tdTomato mice (4% paraformaldehyde) using a vibratome. Antigen retrieval was performed with 0.5% trypsin (Sigma; T4049) in PBS at 37°C for 20 min. Tissue sections were then incubated overnight with anti-collagen IV (ab19808, 1:250 dilution, Abcam) in a solution of 2% TritonX-100 (v/v, Sigma-Aldrich; X100), 10% goat serum (v/v, Vector Laboratories; S1000), and 0.1% sodium azide (w/v, Sigma-Aldrich; S2002) in PBS at room temperature. The next day, sections were washed in PBS before being incubated with anti-rabbit Alexa 488 (A-11008, 1:1000 dilution, Invitrogen) for 2 h. A final PBS wash step was taken before mounting tissues on a glass slide with mounting medium (Fluoromount-G; ThermoFisher). Super-resolution confocal images were taken on a Zeiss LMS 880 confocal microscope with an Airyscan detector using a 63× oil immersion objective (Plan-Apochromat, Carl Zeiss, NA = 1.4, 190 µm working distance). For the en passant pericyte example, voxel size was 110 × 110 × 150 nm; 2 frame averaging; 1.6× zoom. For junctional pericyte example, voxel size was 132 × 132 × 150 nm; 2 frame averaging; 1.3× zoom). Images were taken with 561 nm excitation.
Statistical analyses
We conducted a Lilliefors test in MATLAB to determine normality for data on both pericyte and endothelial pegs. For data that followed a normal distribution we conducted unpaired t-tests to assess statistical significance. Non-parametric Mann Whitney U test was used for data that was not normally distributed. All statistical analyses were performed in Graphpad Prism software (ver. 8) or MATLAB. Results of analyses are reported in the figure legends.
Results
Pericyte somata are preferentially localized to capillary junctions
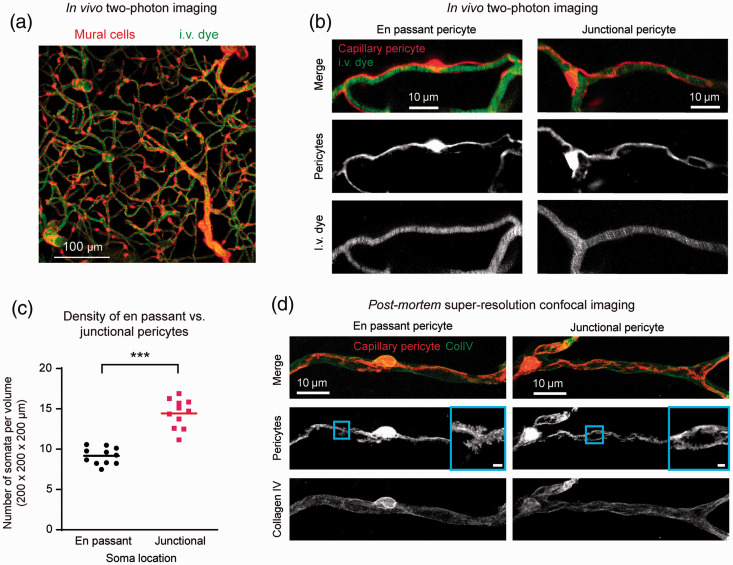
Prior studies have shown that pericyte somata are preferentially located at capillary junctions (i.e., junctional pericytes) compared to intervening capillary regions (i.e., en passant pericytes).28 We corroborated this finding using in vivo two-photon imaging to visualize pericyte soma location in cerebral cortex of PDGFRβ-tdTomato mice, which express tdTomato in all mural cells (Figure 1(a)). A ∼50% greater abundance of junctional pericytes was observed over en passant pericytes per tissue volume (Figure 1(b) and (c)). This demonstrates that locating capillary junctions during SBF-SEM improves chances of locating a pericyte soma.
Figure 1.
Pericyte somata are found preferentially at capillary bifurcations: (a) in vivo two-photon imaging of cortical microvasculature in PDGFRβ-tdTomato mice; (b) pericytes were categorized as en passant (not at a capillary bifurcation) or junctional (at a capillary bifurcation); (c) capillary pericytes were found with greater numbers at capillary bifurcations (***p = 0.0005, paired t-test; n = 14 image stacks examined from eight mice); (d) super-resolution confocal imaging of capillary pericytes reveals substantial substructure of pericytes, including a serrated pattern at the lamellar edges (scale bar for inset is 1 μm).
To understand the extent to which optical imaging can be used to evaluate subcellular pericyte morphology, we also imaged tdTomato-positive pericytes using super-resolution confocal microscopy in fixed tissues (Figure 1(d)). This revealed fine lamellae with serrated edges that extended from the soma and processes, similar to that previously reported by confocal microscopy.33 However, whether these features corresponded with peg-and-socket interactions could not be resolved, necessitating detailed ultrastructural examination.
3-D ultrastructural imaging of a junctional pericyte
To collect 3D-EM data from a junctional pericyte, we prepared a block of tissue obtained from adult mouse cerebral cortex (primary sensory cortex). SBF-SEM was first used to scan large fields for converging vessels indicative of capillary junctions. Once potential junctions were located, the tissue block was transitioned to a FIB-SEM for collection of serial high-resolution images through the junction. This process allowed us to capture a 7 × 22 × 11 μm image volume centered on a capillary junction (Figure 2(a)). Data was collected at 5 nm isotropic voxel resolution but then binned in the x-y dimension to 20 ×20 × 5 nm to facilitate data handling and analysis. A pericyte soma was located at the junction, and an endothelial cell nucleus was observed on one of the three capillaries forming the junction. We segmented both the pericyte and endothelial cells, including their nuclei and cytosolic space (Figure 2(b) to (d); Supplementary Movie 1). A red blood cell within the capillary lumen was also segmented. Much of the pericyte volume was composed of its nucleus. However, lamellae extending from the soma and its emerging processes covered a significant portion of the endothelial surface. These lamellae were roughly 200–300 nm in thickness and their edges exhibited the same serrated pattern as observed with optical imaging (Figure 2(b)). Cross-sectional views of the capillary wall revealed significant interlocking between the pericyte and endothelial cell, some of which represented peg-and-socket interactions between the two cell types (Figure 2(c)). The individual cytoplasmic spaces of the pericyte and endothelium could be clearly distinguished, because they were separated by an electron-dense basement membrane. The basement membrane was also seen on the abluminal surface of the pericyte, and thus surrounded the cell completely, as expected for a bonafide capillary pericyte. The basement membrane appeared to become less dense at some peg-and-socket interactions. However, the images lacked the resolution to confidently measure basement membrane thickness, or to segment the layer as a separate compartment. Additionally, smaller structures such as adherens junctions or endothelial tight junctions were not reliably resolved, and thus were not quantified.
Appearance and arrangement of pericyte pegs
We examined the separately rendered surface volumes of the pericyte and endothelium in detail using Imaris, to study the morphology of pericyte protrusions and their corresponding sockets and grooves within the endothelium (Figure 3(a); Supplementary Movie 2). The resolution of the surface models was set to best capture the morphologies of the peg-and-sockets. As such, occasional small “islands” of pericyte were observed in the model because their thin links to the cell body were not well resolved.
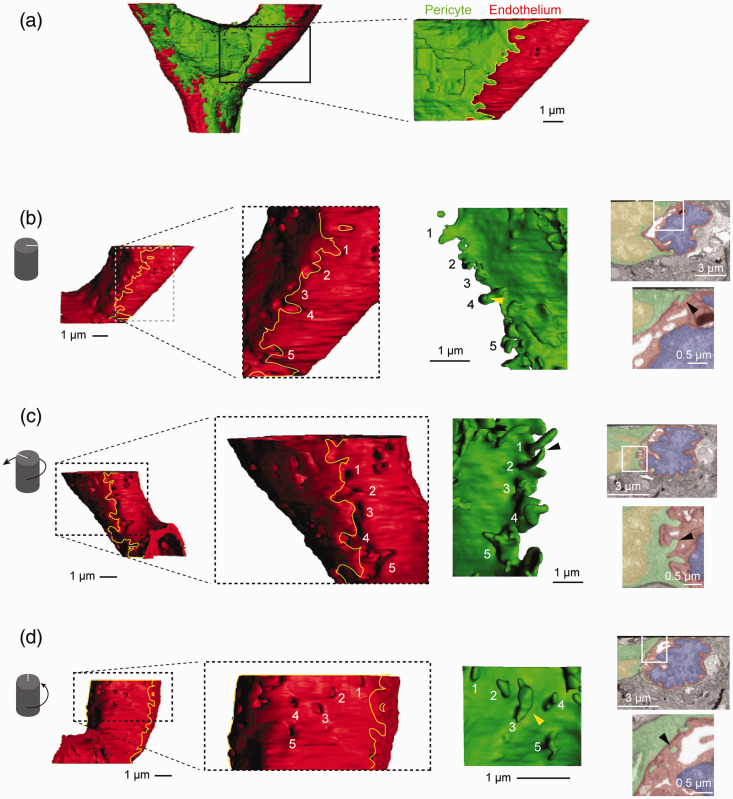
Figure 3.
Pericyte peg-and-socket interactions at the pericyte nucleus: (a) a cropped region of the data set was taken for detailed examination of the pericyte-endothelial interface. This region includes part of the pericyte nucleus and endothelial nucleus; (b) surface model of the endothelium alone with yellow outline showing region of pericyte overlap. Magnified view (dashed box) shows endothelium sockets and shallower indentations created by the lateral edge of the pericyte. The under-surface of the pericyte region in contact with the endothelium is shown (green), with pegs numbered to match their corresponding endothelial sockets. In a single image from the raw 2D data (boxed area enlarged) the appearance of one pericyte peg (#2) (yellow arrowhead in reconstruction) is marked (black arrowhead); (c) surface model of the endothelium rotated ∼120° from its position in panel B to show the other lateral side of the pericyte. The magnified view (dashed box) displays deep sockets and indentations generated by finger-like pegs on the opposite edge of the pericyte (green). In a single image from the raw 2D data (boxed area enlarged), one finger-like peg (#2) is shown (arrowhead); (d) surface model of the endothelium oriented to show the undersurface of the pericyte. Magnified view (dashed box) shows sockets made by pegs found directly beneath the pericyte nucleus. The under-surface of the pericyte reveals pegs with bouton-like shapes (green). In a single image from the raw 2D data (boxed area enlarged), the appearance of one peg (#3) is shown (yellow arrowhead). Note how the 2D slice (right) fails to capture the neck of the peg, and it appears completely contained within the endothelium.
We observed an enrichment of pegs that embedded into the endothelium at the edge of pericyte lamellae (Figure 3(b) and (c)). Some of these pegs penetrated deeply into the endothelium and took on diverse morphologies, such as hook (Figure 3(b), arrowhead) and finger-like protrusions (Figure 3(c), arrowhead). The corresponding impressions made in the endothelium by the pericyte were quite diverse with deep sockets created by the longer pericyte pegs, as well as widespread shallow grooves created by more superficial interactions. Pericyte pegs were also observed directly beneath the pericyte nucleus, away from the edges of the cell (Figure 3(d)). The endothelial sockets created by these more medially localized pegs were circular or elliptical in shape at the entrance. Their corresponding pericyte pegs often swelled in volume beyond the socket entry-point to create bouton-like shapes (Figure 3(d), arrowhead). Thus, pericyte somata are securely anchored to the endothelium by pegs at their lateral edges and medial surfaces. Pericyte pegs may create additional surface area at the pericyte-endothelial interface for direct cell-cell communication.
We next examined a region further from the pericyte nucleus that had the structure of pericyte processes (Supplementary Fig. 1A; Supplementary Movie 3). The endothelial interaction in this region was similar to that of edges near the pericyte nucleus. The pericyte created shallow grooves throughout the endothelial surface, and more sparsely distributed pegs near the edge created deeper penetrations into the endothelium (Supplementary Fig. 1B–D). A diversity of structural features could be observed at the edge of the pericyte, including claw-like projections (Supplementary Fig. 1C, arrowhead), and small loops (Supplementary Fig. 1D, arrowhead). However, this region lacked the medially located bouton-shaped pegs seen closer to the pericyte soma.
Appearance and arrangement of endothelial pegs
We also examined the reciprocal interaction where endothelial pegs inserted into sockets on the pericyte surface. Endothelial pegs appeared larger in size and fewer in number compared to pericyte pegs (8 endothelial pegs vs. 14 pericyte pegs in the total pericyte-endothelial surface examined) (Figure 4(a) and (b)). In this data set, they were only observed directly beneath the pericyte nucleus on one branch of the capillary bifurcation that had greater association with the pericyte nucleus. The endothelial pegs came within very close contact to the pericyte nucleus, in some cases creating indentations in the nuclear surface (Figure 4(a)).
Figure 4.
Characteristics of endothelial pegs: (a) image shows endothelium and pericyte nucleus. Several large endothelial pegs (one denoted with arrowhead; bottom panel) project toward the pericyte nucleus (yellow) and create indentations in the nuclear surface. An accompanying 2D image from the raw segmented data shows the same peg coming in close proximity with the pericyte nucleus (black arrowhead); (b) cropped section of endothelium and pericyte rotated ∼45° to the left from panel A to show frontal view of the endothelial pegs (bottom right). The under-surface of the pericyte (green) shows corresponding large sockets created by the endothelial pegs (bottom left).
Pericyte pegs are prevalent and exhibit greater diversity in morphology
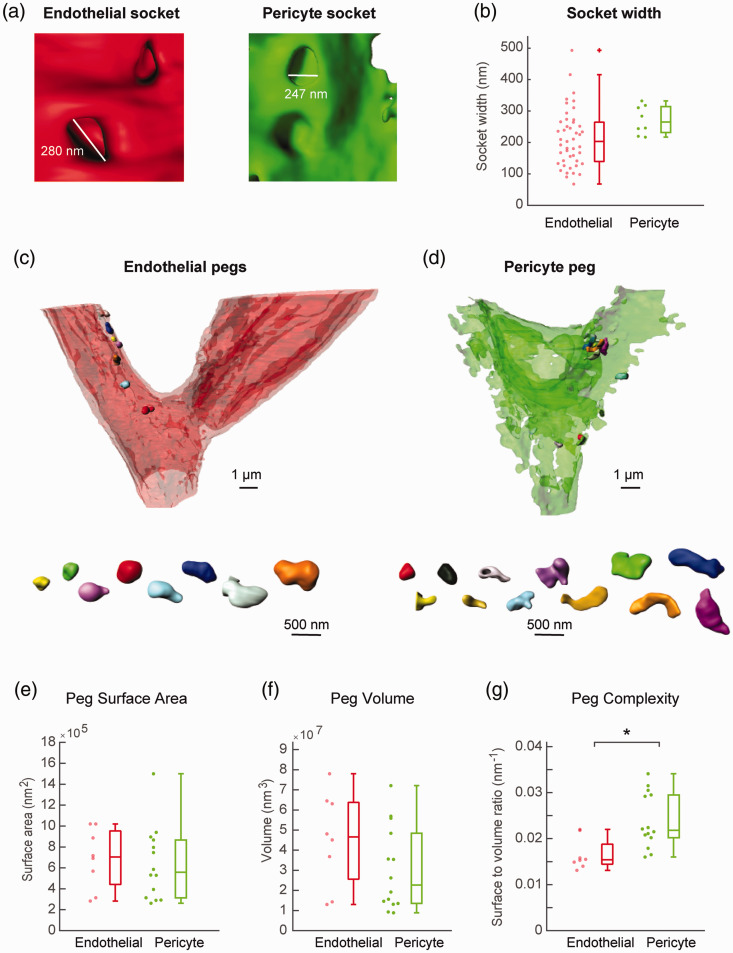
We next quantified differences between pericyte peg-and-socket structures versus reciprocal endothelial peg-and-socket structures. We first compared the width of sockets at their entry-point in which pericyte or endothelial pegs first created membrane indentations. Since the entryway was not always round in shape, we took measurements at the widest distances across the socket (Figure 5(a)). As a population, sockets created by endothelial pegs were ∼25% larger in width than sockets for pericyte pegs, and this difference trended toward significance (Figure 5(b)). Sockets for pericyte pegs were highly varied in width, exhibiting nearly an order of magnitude in range (from ∼50 to 500 nm). In contrast, sockets in pericytes created by endothelial pegs were more consistent in size, ranging between 200 and 300 nm. There was a trend toward significantly higher variance in socket width for pericyte pegs (p = 0.062, F-test of equality of variance).
Figure 5.
Diversity in peg-and-socket morphologies: (a) examples of endothelial and pericyte sockets with measurements of their widths; (b) box plot comparing the socket widths between endothelial cells and pericytes (p = 0.6818, unpaired t-test). A trend toward higher variance of endothelial sockets created by pericyte pegs was detected (p = 0.062; F-test of equality of variance); (c) endothelium shown at reduced opacity for clear visualization of individually segmented and rendered endothelial pegs, displayed in different colors. Below, individual pegs are displayed side-by-side at the same scale; (d) individually segmented and rendered pericyte pegs shown within the pericyte surface model and displayed side-by-side at the same scale below; (e–g) box plot of peg surface area (e), volume (f), and complexity (surface-to-volume ratio) (g). Complexity is significantly higher with pericyte pegs (*p = 0.0046; Mann–Whitney U test). The complexity ratio of pericyte pegs trended towards higher variance than that of endothelial pegs (p = 0.15; F-test of equality of variance). Peg surface area, and volume were not significantly different (p = 0.916 and p = 0.360, unpaired t-test).
We then examined the shape of pericyte and endothelial pegs. A number of individual pegs were segmented such that their morphological characteristics could be assessed (Figure 5(c) and (d); Supplementary Movie 4,5). We considered peg surface area, volume, and the surface area/volume ratio (as a measure of complexity) (Figure 5(e) to (g)). The surface area of pericyte and endothelial pegs was not markedly different, but the volume of endothelial pegs tended to be larger though area was not statistically significant between groups. However, the complexity of pericyte pegs (surface area to volume ratio), was significantly greater than that of endothelial pegs. There was a trend toward greater variance in complexity of pericyte pegs (p = 0.15; F-test of equality of variance). This indicates that pericyte and endothelial pegs have a similar surface area through which to make contact and communicate with the apposing cell, but pericyte pegs appear to take less volume to achieve this.
Peg-and-socket interactions are enriched at the pericyte soma
While the high-resolution FIB-SEM data set permitted detailed renderings of the pericyte-endothelial interface, we sought to further understand the broader distribution of peg-and-socket interactions throughout a capillary network. We therefore made use of a larger SBF-SEM data set made publicly available through MICrONS Explorer (https://microns-explorer.org/). This data set encompasses a 250 × 140 × 90 µm volume (with voxel dimensions of 3.58 × 3.58 × 40 nm) from layer 2/3 of a P36 male mouse visual cortex, and included a portion of an ascending venule and its surrounding capillary networks. While neuronal connectivity has been the primary focus of recent work with this dataset,32 (the vasculature as a whole has already been segmented) but the arduous task of separating pericytes from endothelial cells had not yet been undertaken. However, this data provided us the framework to navigate and manually annotate the positions of peg-and-socket interactions throughout the microvascular network, albeit not at the resolution level of the FIB-SEM dataset.
We focused our analyses only on the capillaries, as venular mural cells are distinct from capillary pericytes in morphological and physiological properties.28 In total, 5 pericyte somata (yellow) and 25 endothelial nuclei (blue) were found in the data set (Figure 6(a)). Criteria used to identify pegs in the prior FIB-SEM data set were applied (see Methods and Supplementary Fig. 2), and each peg was verified by three independent raters to create a consensus data set (Figure 6(b)). We annotated the resulting pericyte peg locations throughout the capillary network, with each annotation color-coded to mark the pericyte from which it originated (Figure 6(c) and (e)). This revealed that pericyte pegs were concentrated around pericyte somata. When identifying endothelial pegs, we found that these were also concentrated near the pericyte somata (Figure 6(d) and (e)).
To quantify the distance between pegs and pericyte soma location, we plotted peg prevalence as a function of distance to the wall of the pericyte nucleus. This revealed that 64% of the pericyte pegs were within 15 µm from the nucleus (Figure 6(f), green). In contrast, 5% of the total pericyte pegs were located at distances >40 µm from the pericyte nucleus. This distribution was mimicked by endothelial pegs, despite their lower abundance (Figure 6(f), red).
In total, 241 pericyte pegs and 66 endothelial pegs were identified throughout the MICrONS data set. Individual pericyte somata exhibited a broad range of pegs, from 28 to 75 pericyte pegs and 1 to 27 endothelial pegs per pericyte, suggesting heterogeneity in the degree of pericyte-endothelial interaction even in this relatively small network. Interestingly, pericytes extending larger numbers of pegs also received a larger number of endothelial pegs, suggesting that a balance of reciprocal interaction is maintained between pericyte and endothelial cell. A plot of pericyte peg versus endothelial peg number, per pericyte soma, revealed a strong positive correlation (Figure 6(g)). Each pericyte had a baseline of 30 pegs, but above this, a factor of ∼3 pericyte pegs to 1 endothelial peg was maintained. The pericyte imaged in our separate FIB-SEM data set fit well into this correlation. In this limited data set, junctional pericytes tended to have a higher number of pegs than en passant pericytes.
Pericyte processes occasionally track along the endothelial tight junction
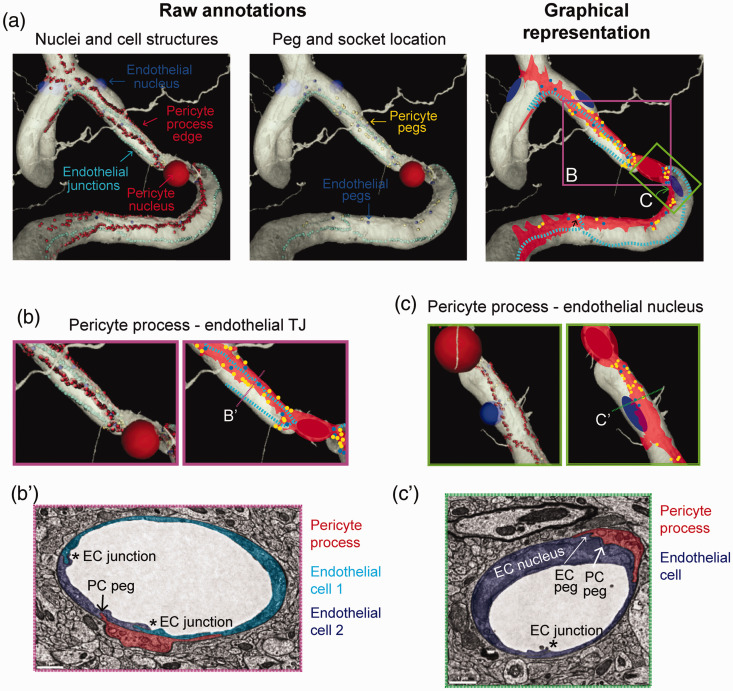
To gain a broader view of the interactions between pericytes and the endothelium, we mapped the location of pericyte processes throughout the MICrONs data set. We found that four out of the five total pericytes identified were of thin-strand pericyte morphology (Figure 7(a)). Thin-strand pericytes extend long and thin processes.34 The borders of their processes were annotated in raw 2D EM data, such that their positions could be visualized in the 3D rendered view alongside annotations for positions of endothelial tight junctions and pericyte and endothelial pegs (as described above). We then created a graphical representation of the pericyte processes in relation to the endothelial features. This showed that pericyte processes tend to track along many, but not all, endothelial junctions (Figure 7(a) and (b)). In regions in proximity to pericyte somata, both pericyte and endothelial pegs flanked alongside the endothelial junctions (arrows; Figure 7(b)). Occasionally, the thin-strand processes would extend a secondary process to move toward endothelial cell nuclei, as they were encountered (Figure 7(c)). This created a greater pericyte-endothelial interface at the endothelial nucleus, suggesting an additional structure for cell-cell communication, though these regions were not enriched in peg-and-socket interactions as seen at pericyte somata. The final pericyte out of the five exhibited more complex morphology consistent with a mesh pericyte (Supplementary Fig. 3). Interestingly, despite its complexity, the process of the mesh pericyte still associated with the underlying endothelial tight junction, and secondary processes could also be seen extending toward the endothelial nucleus. This interaction of pericyte processes with the endothelial nucleus could also be observed by light microscopy in super-resolution confocal images (Supplementary Fig. 4)
Figure 7.
Pericyte processes associate with endothelial tight junctions and nuclei: (a) raw annotations and graphical representation of pericyte and endothelial ultrastructures from the 3D rendered MICrONs data set showing a thin-strand pericyte (red) along the brain vasculature. (Left panel) Pericyte process edges annotations (red dots) outline the boundary of the pericyte processes. Light blue dots demarcate the position of endothelial tight junctions. (Middle panel) Pericyte peg (yellow dots) and endothelial peg (dark blue dots) positions, as described in Figure 7. (Right panel) Graphical representation of all annotated structures with the areas covered by pericyte processes shaded in red; (b) raw segmented data and graphical representation highlight the tendency for pericyte processes to track the endothelial tight junction, and for pericyte pegs (yellow) and endothelial pegs (dark blue) to flank along the endothelial junctions (light blue); (b’) corresponding 2D electron microscopic image from the dataset, taken at location shown in panel B; (c) raw segmented data and graphical representation demonstrate how a pericyte process sends secondary extensions to increase contact with the vasculature around an endothelial cell nucleus (blue); (c’) corresponding 2D electron microscopic image from the dataset, taken at location shown in panel c.
Despite having sufficient image quality to observe endothelial substructures, including caveolae and tight junctions, pericyte pegs did not appear to be consistently apposed to any endothelial structure or increase in electron dense intracellular material.
Discussion
This study has employed 3D volume EM technologies to document the morphology of the pericyte-endothelial interface in brain microvasculature. The results show substantial, reciprocal peg-and-socket interactions between capillary pericytes and endothelial cells of cerebral cortex. These interactions are enriched at pericyte somata and could represent “hotspots” for pericyte-endothelial communication in the expansive capillary network. We have further identified key distinctions in the number, distribution, and individual morphologies of pericyte and endothelial pegs. Pericyte pegs are more numerous, diverse in structure, and broadly distributed across the pericyte-endothelial interface. They line the edges of pericyte processes, appear to provide anchoring support for maintenance of endothelial coverage and suggest a structural basis for the conveyance of mechanical force. Endothelial pegs are fewer in number, but are larger and mirror the distribution of pericyte pegs. Further, our findings suggest a balanced interaction between pericyte and endothelial cells. Above a baseline of 30 pericyte pegs per soma, a conserved ∼3:1 ratio of pericyte and endothelial pegs was observed for each pericyte examined.
With respect to roles in vascular mechanics, it has remained unclear how capillary pericytes that only partially cover the endothelium exert force on capillary diameter to affect blood flow.8 Past studies have alluded to presence of adhesion proteins such as fibronectin and other dense plaques specifically located at the pericyte-endothelial interface.20 The presence of adhesive proteins, along with the multitude of pegs along the edges of the pericyte, suggests that pericytes are securely anchored to the endothelium in a way that would allow them to maintain endothelial coverage and translate force to the underlying endothelium during contraction or relaxation. Force applied circumferentially along the capillary axis “cinch” up regions of the endothelial wall that they contact, and this occurrence has been captured in prior TEM studies.35,36 Beyond local mechanics, peg-and-socket interactions may be a site of aggregation for gap junctions, ion channels and receptors involved in vascular conduction of electrical signals during neurovascular coupling.37,38 However, immunohistochemical studies using APEX, or another electron dense tracer, combined with 3D-EM studies are still needed to define the true subcellular distribution of pericyte-endothelial signaling proteins.
In numerous diseases of the central nervous system, pericyte investment in the endothelium is diminished. This is often attributed to pericyte death, as measured by loss of pericyte-expressing proteins such as PDGFRβ. However, other factors that affect pericyte investment, such as altered pericyte-endothelial signaling, thickening of the vascular basement membrane, or pericyte detachment and migration, can occur in the absence of pericyte death, and may emerge earlier than overt pericyte loss.7,39,40 This pathology is more challenging to characterize and has relied upon ultrastructural imaging. An evaluation of peg-and-socket interactions would be logical in these cases, as the structure and density of pegs, or their ability to make cell-cell contact may change in disease conditions.35,41 While we have shown that 3D EM is ideal to study these interactions in detail, and is a logical complement to conventional 2D EM, the approach remains a bottleneck in studies of pericyte biology due to the significant investment in time and money to acquire and process 3D EM data.
Despite challenges in cost and throughput, data from volume EM studies can begin to inform the application of more conventional laboratory approaches. For instance, our EM findings offer guidance toward how immunohistochemical analyses and use of fluorescent transgenic mice might be further refined to document changes in pericyte-endothelial interaction. First, focus could be given to changes in protein expression at and around pericyte somata, especially as it pertains to proteins enriched at peg-and sockets, such as Ang1-Tie2 and perhaps gap junctional proteins. Second, our data suggests that the serrated patterns along the edge of pericyte, particularly near the cell soma, correspond to peg-and-socket interactions, and that these structures are partially resolvable with high-resolution confocal imaging of transgenic lines such as PDGFRβ-tdTomato (Figure 1(d)). The serrated patterns unfortunately are not yet resolvable by in vivo two-photon imaging, leaving open the question of whether pericyte pegs exhibit any structural plasticity. Third, our volume EM studies have identified some spatial correspondence between thin-strand pericyte processes and inter-endothelial tight junctions, as well as secondary pericyte processes that associate with endothelial nuclei. Whether these features deviate from the normal state during pathology will be worth examining, as they may reflect alterations in pericyte-endothelial adherence and signaling. This possibility needs to be further examined in various disease models involving pericyte dysfunction or loss.
SBF-SEM is the approach of choice for collection of large-scale volume EM data. Our findings show that, despite thicker slices in the Z-dimension, there is sufficient resolution to identify most peg-and-socket interactions in SBF-SEM data. The width of endothelial cell pegs, as defined by their sockets size, is well above typical slice thickness, and the majority of pericyte pegs are also larger. However, it is likely that smaller pericyte pegs ∼ 100 nm (two slices per peg) are under-sampled and potentially overlooked. The 3D rendering of pegs would also be less accurate using SBF-SEM data, compared to FIB-SEM. Nevertheless, our data indicate that resolutions achieved by SBF-SEM are sufficient to identify many peg-and-sockets, allowing us to discover conserved ratios between pericyte and endothelial peg number per pericyte soma, and heterogeneity in peg-and-socket interaction within local pericyte populations. Data from our separate FIB-SEM sampled pericyte fell within the range of this heterogeneity. Thus a combination of volume EM approaches, as used here, would be useful to further fully document changes in structural interactions in neuropathological conditions.
Finally, recent studies also indicate that pericyte-endothelial communication differs in capillaries closer to the parenchymal arterioles, and are important to promote upstream conductivity of depolarizing and hyperpolarization signals during neurovascular coupling in brain and retina.38,42 The physical basis of this signaling directionality was shown to be specialized gap junctional structures in capillaries near arterioles.43 As such, it will be important to use 3D EM to investigate pericyte-endothelial interaction along different zones of the microvascular network, ideally with immuno-labeling of proteins involved in pericyte-endothelial signaling to better link structure and function. The growing availability of automated 3D EM technologies will undoubtedly create more impressive datasets for examination of neurovascular ultrastructure. However, advances in automated segmentation of pericytes from endothelial cells will be essential to mine these rich data sets. Despite the effort involved, 3D volume EM studies provide the only way to study the interaction of cells at nanometer resolution, and we believe that this study is an important proof of principle of the value of such work.
Supplemental Material
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X211012836 for Three-dimensional ultrastructure of the brain pericyte-endothelial interface by Sharon Ornelas, Andrée-Anne Berthiaume, Stephanie K Bonney, Vanessa Coelho-Santos, Robert G Underly, Anna Kremer, Christopher J Guérin, Saskia Lippens and Andy Y Shih in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-pdf-2-jcb-10.1177_0271678X211012836 for Three-dimensional ultrastructure of the brain pericyte-endothelial interface by Sharon Ornelas, Andrée-Anne Berthiaume, Stephanie K Bonney, Vanessa Coelho-Santos, Robert G Underly, Anna Kremer, Christopher J Guérin, Saskia Lippens and Andy Y Shih in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-pdf-3-jcb-10.1177_0271678X211012836 for Three-dimensional ultrastructure of the brain pericyte-endothelial interface by Sharon Ornelas, Andrée-Anne Berthiaume, Stephanie K Bonney, Vanessa Coelho-Santos, Robert G Underly, Anna Kremer, Christopher J Guérin, Saskia Lippens and Andy Y Shih in Journal of Cerebral Blood Flow & Metabolism
Acknowledgements
We appreciate the helpful comments and discussion of Stefan Stamenkovic, Mark Takeno and Nuno Maçarico da Costa. We thank Patrick J Mulholland for use of a Zeiss LSM 880 with Airyscan for super-resolution imaging of pericytes (NIH S10 OD021532).
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Our work is supported by grants to AYS from the NIH (NS106138, AG063031, NS097775). AAB is funded by a scholarship from the American Federation for Aging Research. VCS is supported by awards American Heart Association (20POST35160001) and Luso-American Development Foundation (2017/165). SKB is supported by an NIH/NINDS F32 fellowship (1F32NS117649-01). The purchase of the volume EM equipment was funded by a grant from the Flemish Government.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions: The FIB-SEM data set was collected by RGU, AK, SL, CJG. Two-photon and super-resolution confocal imaging data was collected by AAB. Image segmentation and analyses were performed by SO. Peg verification and evaluation of MICrONS data was aided by SO, SKB and VCS. Detailed annotation of pericyte structure and endothelial tight junctions was performed by SKB. Statistics were performed by SO. The manuscript was written by SO and AYS with contributions from all authors.
Supplementary material: Supplementary material for this paper can be found at: https://journals.sagepub.com/home/jcb
ORCID iD: Vanessa Coelho-Santos https://orcid.org/0000-0002-9450-6103
References
- 1.Winkler EA, Bell RD, Zlokovic B.Central nervous system pericytes in health and disease. Nat Neurosci 2011; 14: 1398–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng Z, Chopp M, Chen J.Multifaceted roles of pericytes in central nervous system homeostasis and disease. J Cereb Blood Flow Metab 2020; 40: 1381–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerhardt H, Betsholtz C.Endothelial-pericyte interactions in angiogenesis. Cell Tissue Res 2003; 314: 15–23. [DOI] [PubMed] [Google Scholar]
- 4.Daneman R, Zhou L, Kebede AA, et al. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature 2010; 468: 562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armulik A, Genové G, Mäe M, et al. Pericytes regulate the blood-brain barrier. Nature 2010; 468: 557–561. [DOI] [PubMed] [Google Scholar]
- 6.Bell RD, Winkler EA, Sagare AP, et al. Pericytes control key neurovascular functions and neuronal phenotype in the adult brain and during brain aging. Neuron 2010; 68: 409–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall CN, Reynell C, Gesslein B, et al. Capillary pericytes regulate cerebral blood flow in health and disease. Nature 2014; 508: 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartmann DA, Berthiaume AA, Grant RI, et al. Brain capillary pericytes exert a substantial but slow influence on blood flow. Nat Neurosci 2020; online ahead of print. [DOI] [PMC free article] [PubMed]
- 9.Watson AN, Berthiaume AA, Faino AV, et al. Mild pericyte deficiency is associated with aberrant brain microvascular flow in aged PDGFRβ+/– mice. J Cereb Blood Flow Metab 2020; 40: 2387–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuevas P, Gutierrez-Diaz JA, Reimers D, et al. Pericyte endothelial gap junctions in human cerebral capillaries. Anat Embryol (Berl) 1984; 170: 155–159. [DOI] [PubMed] [Google Scholar]
- 11.Leeson TS.Rat retinal blood vessels. Can^ J Opthalmol 1979; 14: 21–28. [PubMed] [Google Scholar]
- 12.Dore-Duffy P, Cleary K.Morphology and properties of pericytes. Methods Mol Biol 2011; 686: 49–68. [DOI] [PubMed] [Google Scholar]
- 13.Moffat DB.The fine structure of the blood vessels of the renal medulla with particular reference to the control of the medullary circulation. J Utrastruct Res 1967; 19: 532–545. [DOI] [PubMed] [Google Scholar]
- 14.Antonelli-Orlidge A, Saunders KB, Smith SR, et al. An activated form of transforming growth factor beta is produced by cocultures of endothelial cells and pericytes. Proc Natl Acad Sci USA 1989; 86: 45544–44548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirschi KK, Burt JM, Hirschi KD, et al. Gap junction communication mediates transforming growth factor-beta activation and endothelial-induced mural cell differentiation. Circ Res 2003; 93: 429–437. [DOI] [PubMed] [Google Scholar]
- 16.Gerhardt H, Wolburg H, Redies C.N-cadherin mediates pericytic-endothelial interaction during brain angiogenesis in the chicken. Dev Dyn 2000; 218: 472–479. [DOI] [PubMed] [Google Scholar]
- 17.Li F, Lan Y, Wang Y, et al. Endothelial Smad4 maintains cerebrovascular integrity by activating N-cadherin through cooperation with notch. Dev Cell 2011; 20: 291–302. [DOI] [PubMed] [Google Scholar]
- 18.Armulik A, Genove G, Betsholtz C.Pericytes: developmental, physiological, and pathological perspectives, problems and promises. Dev Cell 2011; 21: 193–215. [DOI] [PubMed] [Google Scholar]
- 19.Wakui S, Yokoo K, Muto T, et al. Localization of ang-1, -2, tie-2, and VEGF expression at endothelial-pericyte interdigitation in rat angiogenesis. Lab Invest 2006; 86: 1172–1184. [DOI] [PubMed] [Google Scholar]
- 20.Courtoy PJ, Boyles J.Fibronectin in the microvasculature: localization in the pericyte-endothelial interstitium. J Utrastruct Res 1983; 83: 258–273. [DOI] [PubMed] [Google Scholar]
- 21.Díaz-Flores L, Gutiérrez R, Madrid JF, et al. Pericytes. Morphofunction, interactions and pathology in a quiescent and activated mesenchymal cell niche. Histol Histopathol 2009; 24: 909–969. [DOI] [PubMed] [Google Scholar]
- 22.Allsopp G, Gamble HJ.An electron microscopic study of the pericytes of the developing capillaries in human fetal brain and muscle. J Anat 1979; 128: 155–168. [PMC free article] [PubMed] [Google Scholar]
- 23.Tilton RG, Kilo C, Williamson JR.Pericyte-endothelial relationships in cardiac and skeletal muscle capillaries. Microvasc Res 1979; 18: 325–335. [DOI] [PubMed] [Google Scholar]
- 24.Fujiwara T, Uehara Y.The cytoarchitecture of the wall and the innervation pattern of the microvessels in the rat mammary gland: a scanning electron microscopic observation. Am J Anat 1984; 170: 39–54. [DOI] [PubMed] [Google Scholar]
- 25.Mathiisen TM, Lehre KP, Danbolt NC, et al. The perivascular astroglial sheath provides a complete covering of the brain microvessels: an electron microscopic 3D reconstruction. Glia 2010; 58: 1094–1103. [DOI] [PubMed] [Google Scholar]
- 26.Haley M, J, Lawrence CB.The blood-brain barrier after stroke: structural studies and the role of transcytotic vesicles. J Cereb Blood Flow Metab 2017; 37: 456–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai PS, Kaufhold J, Blinder P, et al. Correlations of neuronal and microvascular densities in murine cortex revealed by direct counting and colocalization of cell nuclei and microvessels. J Neurosci 2009; 29: 14553–14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartmann DA, Underly RG, Grant RI, et al. Pericyte structure and distribution in the cerebral cortex revealed by high-resolution imaging of transgenic mice. Neurophotonics 2015; 2: 041402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denk W, Horstmann H.Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. Plos Biol 2004; 2: e329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heymann JA, Hayles M, Gestmann I, et al. Site-specific 3D imaging of cells and tissues with a dual beam microscope. J Struct Biol 2006; 155: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guérin CJ, Kremer A, Borghgraef P, et al. Combining serial block face and focused ion beam scanning electron microscopy for 3D studies of rare events. Methods Cell Biol 2019; 12: 69–85. [DOI] [PubMed] [Google Scholar]
- 32.Schneider-Mizell C, Bodor AL, Collman F, et al. Chandelier cell anatomy and function suggest a variably distributed but common signal. bioRxiv. Epub ahead of print 1 April 2020. DOI: 10.1101/2020.03.31.018952v1.
- 33.Hill RA, Tong L, Yuan P, et al. Regional blood flow in the normal and ischemic brain is controlled by arteriolar smooth muscle cell contractility and not by capillary pericytes. Neuron 2015; 87: 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grant RI, Hartmann DA, Underly RG, et al. Organizational hierarchy and structural diversity of microvascular pericytes in adult mouse cortex. J Cereb Blood Flow Metab 2019; 39: 411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nahirney PC, Reeson P, Brown CE.Ultrastructural analysis of blood-brain barrier breakdown in the peri-infarct zone in young and aged mice. J Cereb Blood Flow Metab 2016; 36: 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakano M, Atobe Y, Goris RC, et al. Ultrastructure of the capillary pericytes and the expression of smooth muscle alpha-actin and desmin in the snake infrared sensory organs. Anat Rec 2000; 260: 299–307. [DOI] [PubMed] [Google Scholar]
- 37.Hariharan A, Weir N, Robertson C, et al. The ion channel and GPCR toolkit of brain capillary pericytes. Front Cell Neurosci 2020; 14: 601324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kovacs-Oller T, Ivanova E, Bianchimano P, et al. The pericyte connectome: spatial precision of neurovascular coupling is driven by selective connectivity maps of pericytes and endothelial cells and is disrupted in diabetes. Cell Discov 2020; 6: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sagare AP, Bell RD, Zhao Z, et al. Pericyte loss influences Alzheimer-like neurodegeneration in mice. Nat Commun 2013; 4: 2932. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Dore-Duffy P, Owen C, Balabanov R, et al. Pericyte migration from the vascular wall in response to traumatic brain injury. Microvasc Res 2000; 60: 55–69. [DOI] [PubMed] [Google Scholar]
- 41.Braverman IM, Sibley J, Keh A.Ultrastructural analysis of the endothelial-pericyte relationship in diabetic cutaneous vessels. J Invest Dermatol 1990; 95: 147–153. [DOI] [PubMed] [Google Scholar]
- 42.Longden TA, Dabertrand F, Koide M, et al. Capillary K+-sensing initiates retrograde hyperpolarization to increase local cerebral blood flow. Nat Neurosci 2017; 20: 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ivanova E, Kovacs-Oller T, Sagdullaev BT.Domain-specific distribution of gap junctions defines cellular coupling to establish a vascular relay in the retina. J Comp Neurol 2019; 527: 2675–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guérin CJ, Kremer A, Borghgraef P, et al. Targeted studies using serial block face and focused ion beam scan electron microscopy. J Vis Exp 2019; 150: e59480. [DOI] [PubMed]
- 45.Lippens S, Kremer A, Borghgraef P, et al. Serial block face-scanning electron microscopy for volume electron microscopy. Methods Cell Biol 2019; 152: 68–85. [DOI] [PubMed] [Google Scholar]
- 46.Belevich I, Joensuu M, Kumar D, et al. Microscopy image browser: a platform for segmentation and analysis of multidimensional datasets. PLOS Biol 2016; 14: e1002340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berthiaume AA, Grant RI, McDowell KP, et al. Dynamic remodeling of pericytes in vivo maintains capillary coverage in the adult mouse brain. Cell Rep 2018; 22: 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jcb-10.1177_0271678X211012836 for Three-dimensional ultrastructure of the brain pericyte-endothelial interface by Sharon Ornelas, Andrée-Anne Berthiaume, Stephanie K Bonney, Vanessa Coelho-Santos, Robert G Underly, Anna Kremer, Christopher J Guérin, Saskia Lippens and Andy Y Shih in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-pdf-2-jcb-10.1177_0271678X211012836 for Three-dimensional ultrastructure of the brain pericyte-endothelial interface by Sharon Ornelas, Andrée-Anne Berthiaume, Stephanie K Bonney, Vanessa Coelho-Santos, Robert G Underly, Anna Kremer, Christopher J Guérin, Saskia Lippens and Andy Y Shih in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-pdf-3-jcb-10.1177_0271678X211012836 for Three-dimensional ultrastructure of the brain pericyte-endothelial interface by Sharon Ornelas, Andrée-Anne Berthiaume, Stephanie K Bonney, Vanessa Coelho-Santos, Robert G Underly, Anna Kremer, Christopher J Guérin, Saskia Lippens and Andy Y Shih in Journal of Cerebral Blood Flow & Metabolism