Abstract
Polycomb (Pc) is part of a Pc group (PcG) protein complex that is involved in repression of gene activity during Drosophila and vertebrate development. To identify proteins that interact with vertebrate Pc homologs, we performed two-hybrid screens with Xenopus Pc (XPc) and human Pc 2 (HPC2). We find that the C-terminal binding protein (CtBP) interacts with XPc and HPC2, that CtBP and HPC2 coimmunoprecipitate, and that CtBP and HPC2 partially colocalize in large PcG domains in interphase nuclei. CtBP is a protein with unknown function that binds to a conserved 6-amino-acid motif in the C terminus of the adenovirus E1A protein. Also, the Drosophila CtBP homolog interacts, through this conserved amino acid motif, with several segmentation proteins that act as repressors. Similarly, we find that CtBP binds with HPC2 and XPc through the conserved 6-amino-acid motif. Importantly, CtBP does not interact with another vertebrate Pc homolog, M33, which lacks this amino acid motif, indicating specificity among vertebrate Pc homologs. Finally, we show that CtBP is a transcriptional repressor. The results are discussed in terms of a model that brings together PcG-mediated repression and repression systems that require corepressors such as CtBP.
In Drosophila the Polycomb (Pc) group (PcG) genes have been identified as being part of a cellular memory system that is responsible for the stable and heritable repression of gene expression (3, 16). The PcG genes are required for maintenance of the repressed state of certain homeotic genes. Mutations in PcG genes result in derepression of these homeotic genes, which leads to homeotic transformations. In recent years vertebrate homologs of PcG genes have been identified and characterized. Mutations in these vertebrate PcG genes also lead to homeotic transformations, indicating that the vertebrate PcG genes have a function similar to that of their Drosophila homologs (reviewed in references 8 and 24).
Despite the extensive knowledge concerning the identity of Drosophila and vertebrate PcG genes, the molecular mechanism of how PcG proteins achieve inheritably stable transcriptional repression of target genes is not understood. Several models in which the PcG proteins can package target genes in a heterochromatin-like conformation or induce modifications of the nucleosomal organization have been considered (16). It also is not understood how PcG proteins interfere with transcription regulation. In theory, the PcG proteins might directly interact with enhancer proteins, with proteins of the basal transcription machinery, or with proteins that modify chromatin structure, such as histone deacetylases.
Insight into the molecular mechanisms underlying PcG action comes from observations indicating that PcG proteins function as large multimeric complexes. In Drosophila, several PcG proteins share 60 to 100 sites on polytene chromosomes of the salivary gland (18, 28), and coimmunoprecipitation experiments have shown that the Pc protein is present in a large protein complex that also includes the PcG protein Polyhomeotic (Ph) (6). The vertebrate PcG proteins also form multimeric protein complexes. Recently, we have shown that there are at least two distinct human PcG protein complexes (25). On the one hand, there is a complex which consists of human Pc 2 (HPC2), a human Pc homolog; a human homolog of the murine Pc protein M33 (21); HPH1 and HPH2, human homologs of the Drosophila PcG protein Ph; and BMI1, a human homolog of the Drosophila PcG protein Posterior sex combs (1, 9). This complex also contains the RING1 protein (20). All of these proteins coimmunoprecipitate with each other and colocalize in large nuclear domains termed PcG domains (9, 20, 21). On the other hand, Enx1/EZH2 and EED, mammalian homologs of the Drosophila PcG proteins Enhancer of zeste and Extra sex combs, respectively, appear to be part of a distinct PcG complex. Enx1/EZH2 and EED coimmunoprecipitate and colocalize with each other but not with the above-mentioned PcG proteins (25, 27).
To identify additional proteins that interact with the PcG complex, we screened two-hybrid cDNA libraries with vertebrate Pc homologs as targets. We found that a Xenopus homolog of C-terminal binding protein 1 (XCtBP1) (22) interacts with Xenopus Pc (XPc) (19) and that human CtBP2 (11) interacts with HPC2 (21). The CtBP1 protein has previously been identified as a protein that binds to the extreme C terminus of the adenovirus type 2 and 5 (Ad2/5) E1A protein, and CtBP1 attenuates transcriptional activation and tumorigenesis mediated by the E1A protein (2, 22, 26). We show that the CtBP proteins coimmunoprecipitate with HPC2, that the CtBP proteins partially colocalize in nuclear domains with HPC2, and, finally, that CtBP is able to repress gene activity. These findings are of particular interest since the recently identified Drosophila homolog of CtBP is able to interact with the Drosophila pair-rule segmentation protein Hairy (17) and the gap segmentation protein Knirps and the zinc finger protein Snail (14). Remarkably, all of these Drosophila CtBP-interacting proteins are, like HPC2 and XPc, repressors of gene activity. Our data suggest that HPC2-mediated repression involves an association with corepressors such as CtBP.
MATERIALS AND METHODS
Yeast two-hybrid screen.
The full-length coding regions of XPc (19) and HPC2 (21) were cloned into the pAS2 vector (5) (Clontech) and were used separately as targets to screen for interacting proteins (9, 20, 25). The other Pc and CtBP hybrids were derived by PCR (Expand; Boehringer) and were sequenced entirely. The pAS2-XPc plasmid was cotransformed with a Xenopus oocyte Matchmaker two-hybrid library (Clontech), and the pAS2-HPC2 plasmid was cotransformed with a human fetal brain Matchmaker two-hybrid library (Clontech), into Saccharomyces cerevisiae Y190. The transformants were plated on selective medium lacking the amino acids leucine, tryptophan, and histidine but containing 30 mM 3-amino-1,2,4-triazole (3-AT) (9, 20, 25). Potential interactions between different subclones of CtBP and HPC2 were tested. The transformants were plated on medium lacking the amino acids leucine, tryptophan, and histidine with or without 30 mM 3-AT. Cells with interactions that were scored as negative failed to grow in the presence of 30 mM 3-AT. Due to residual HIS3 promoter activity, however, they are able to grow on medium without 3-AT (9, 20, 25). Under these nonselective conditions, cells with negative interactions were β-galactosidase negative and the colony was white. Positive interactions meet the two criteria of growth in the presence of 3-AT and β-galactosidase positivity.
GST fusion proteins and in vitro binding assay.
The previously described (19) glutathione S-transferase–XPc (GST-XPc) (amino acids [aa] 1 to 521) and GST-XPc (aa 1 to 178) fusion proteins contain, respectively, the full-length XPc and the N-terminal first 178 aa of XPc, encompassing the chromodomain (19). Expression of the GST fusion proteins was induced for 3 h at 30°C with 0.4 mM isopropyl-β-d-thiogalactopyranoside as described previously (19). The cells were pelleted, resuspended in binding buffer (phosphate-buffered saline containing 1 mM EDTA, 1 mM dithiothreitol, 2 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, 10 μg of benzamidine per ml, 10 μg of trypsin inhibitor per ml, and 10 μg of aprotinin per ml), and sonicated. Triton X-100 was added to a final concentration of 1% (vol/vol), and the lysate was incubated for 30 min on ice. Cell debris was removed by centrifugation for 10 min at 14,000 × g, the supernatant was added to glutathione-Sepharose 4B, and the mixture was incubated for 30 min at 4°C. The beads were collected by centrifugation and washed extensively with binding buffer. Capped synthetic CtBP2 mRNA was made by in vitro transcription and translated at 20 μg/ml in a rabbit reticulocyte lysate in the presence of [35S]methionine (19). A 10-μl slurry of GST fusion protein (immobilized on glutathione-Sepharose) was preincubated for 30 min on ice in a final volume of 200 μl of binding buffer, containing 0.5% Nonidet P-40 and 1 mg of bovine serum albumin per ml. Subsequently, 3 μl of the reticulocyte lysate was added to the mixture and incubated for 30 min at 4°C with rotation. The beads were washed five times with 1 ml of ice-cold binding buffer. The complexes were separated on sodium dodecyl sulfate (SDS)-polyacrylamide gels, which were subjected to fluorography.
RNA analysis.
Multitissue Northern blots containing approximately 2 μg of poly(A)+ RNA from different human tissues or human cell lines per lane were obtained commercially (Clontech). The U-2 OS osteosarcoma cell line was not present on the commercial Northern blot. Poly(A)+ RNA of U-2 OS was isolated and blotted, and the expression patterns of CtBP1 and CtBP2 were analyzed. To allow a comparison with the commercial Northern blot, poly(A)+ RNA of SW480 cells, which is also represented on the commercial blot and in which both the CtBP1 gene and the CtBP2 gene are strongly expressed, was blotted. We used part of the 3′ untranslated region (3′ UTR) of CtBP1 or CtBP2 as a probe. To obtain these probes, a PCR was performed on a human fetal brain Matchmaker two-hybrid library (Clontech). CtBP1 primers were 5′-CGCCAGTGACCAGTTGTAGC-3′ and 5′-CGTGATGATGCCGTCTTCA-3′, extending from bp 1324 to 1884. CtBP2 primers were 5′-TGCCAGAAGGTAATCAC-TCA-3′ and 5′-AATCCTATGCGTGCAGGTGT-3′, extending from bp 1365 to 1835. The blots were hybridized with [α−32P]dATP-labelled DNA probes, and the blots were autoradiographed with intensifying screens at −70°C with X-ray films.
Production of the CtBP polyclonal antibodies.
A fusion protein was made from the full-length cDNA of XCtBP1 encoding aa 1 to 440. The cDNA was cloned in frame into a pET-23 expression vector (Novagen). The fusion protein was produced in Escherichia coli BL21(DE), and the purified protein was injected into a rabbit. Serum was affinity purified over an antigen-coupled CNBr-Sepharose column (Pharmacia) to determine whether the rabbit anti-XCtBP1 polyclonal antibodies recognize both CtBP1 and CtBP2. T7-tagged CtBP1 and T7-tagged CtBP2 were expressed in E. coli BL21(DE). The bacterial cell lysates were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose. The blots were probed with a 1:10,000 dilution of either mouse monoclonal antibody against T7 (Novagen) or a 1:1,000 dilution of the rabbit polyclonal antibody against XCtBP1.
Immunoprecipitations and Western blotting.
COS-7 cells were transiently transfected with either T7-tagged HPC2 or T7-tagged CtBP2 or with both, using the calcium-phosphate transfection method (Gibco BRL). Both constructs were cloned in the pcDNA3 plasmid (Invitrogen). At 48 h after transfection, COS-7 cells were harvested and lysed in ELB lysis buffer (250 mM NaCl, 0.1% Nonidet P-40, 50 mM HEPES [pH 7.0], 5 mM EDTA) containing 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and the protease inhibitors leupeptin, benzamidine, and aprotinin. The cell lysate was sonicated three times with bursts of 15 s. The cell lysate was centrifuged at 14,000 × g at 4°C for 10 min, and the supernatant was subsequently aliquoted and stored at −70°C. Fifty microliters of the supernatant was incubated with the indicated antibodies for 4 h at 4°C. Goat anti-rabbit immunoglobulin G (IgG) antibodies or goat anti-chicken IgG antibodies (Jackson ImmunoResearch Laboratories) were added to the mixture and incubated for 1 h at 4°C. Protein A–Sepharose CL-4B (Pharmacia) and ELB lysis buffer with protease inhibitors were added up to 300 μl. The mixture was incubated for 1 h at 4°C with continuous mixing. Next, the mixture was centrifuged for 1 min at 1,500 × g at 4°C, the supernatant was transferred to a fresh tube, and the immunoprecipitate was washed with 1 ml of ice-cold ELB buffer without protease inhibitors. The mixture was then centrifuged for 1 min at 1,500 × g at 4°C. This washing procedure was repeated five times. After heating and removal of the protein A-Sepharose beads, the proteins were separated by SDS-PAGE and transferred to nitrocellulose. The blots were probed with a mouse monoclonal antibody against T7 (Novagen). The secondary alkaline phosphatase-conjugated goat antimouse antibodies or goat antichicken antibodies (Jackson ImmunoResearch Laboratories) were diluted 1:10,000, and nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (Boehringer) was used as substrate for detection.
Immunofluorescence labelling of tissue culture cells.
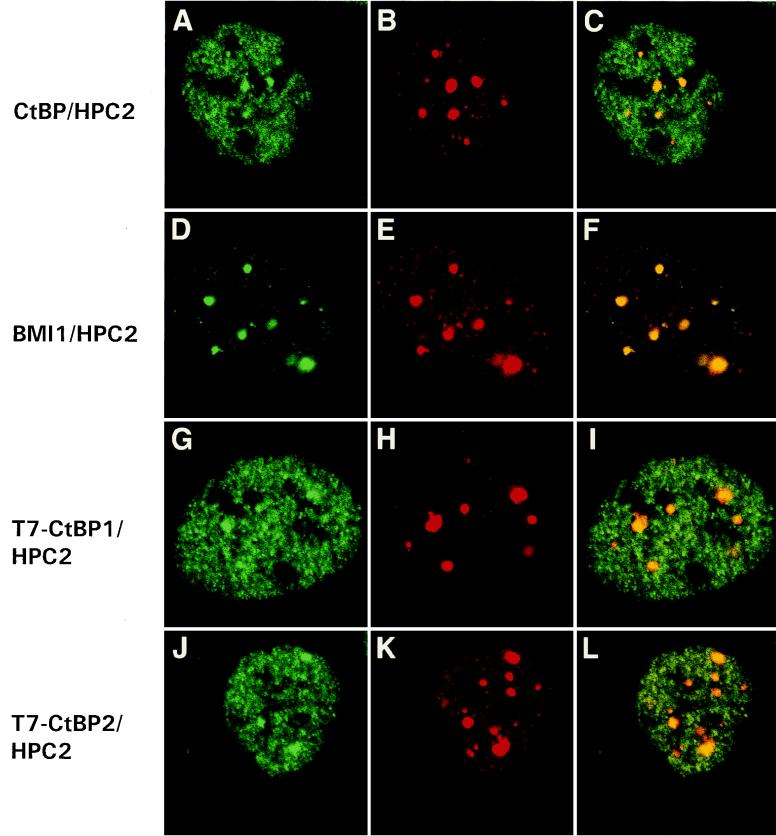
U-2 OS cells were cultured and labelled as described previously (9, 20, 21, 25). The labelling was analyzed by confocal laser scanning microscopy, and optical sections were made (see Fig. 8, where the first two pictures of each row represent the two different scanned channels for imaging the double labelling and the last picture in each row represents the reconstituted image). For labelling CtBP and BMI1, donkey anti-rabbit IgG coupled to Cy3 (Jackson Immunoresearch Laboratories) was used. For labelling HPC2, donkey anti-chicken IgG coupled to fluorescein isothiocyanate (Jackson Immunoresearch Laboratories) was used. To discriminate between the CtBP1 protein and the CtBP2 protein, U-2 OS cells were transiently transfected with either T7-tagged CtBP1 or T7-tagged CtBP2, and cells were double labelled with antibodies against HPC2 and mouse monoclonal antibodies against T7 (Novagen).
FIG. 8.
HPC2 and CtBP partially colocalize in nuclear domains of U-2 OS cells. Confocal single optical sections are shown. (A to C) Rabbit anti-XCtBP1 and chicken anti-HPC2 double labelling. CtBP (A) colocalizes with HPC2 (B) in large nuclear PcG domains (C; indicated by yellow), but CtBP is also abundantly expressed in a fine granular pattern throughout the nucleus (B and C). (D to F) Rabbit anti-BMI1 (D) and chicken anti-HPC2 (E) double labelling demonstrates colocalization (F) of BMI1 and HPC2 in large nuclear PcG domains. We transiently transfected U-2 OS cells with either T7-tagged CtBP1 (G) or T7-tagged CtBP2 (J). Double labelling was performed with a mouse monoclonal antibody against T7 (G and J) and the chicken anti-HPC2 antibody (H and K). We observed colocalization of HPC2 with either T7-CtBP1 (I) or T7-CtBP2 (L) in large nuclear PcG domains.
LexA fusion reporter gene-targeted repression assay.
The LexA repression assay was performed as described previously (20, 21, 25). U-2 OS cells were cultured in a 25-cm2 flask and cotransfected with 2 μg of the heat shock factor (HSF)-inducible luciferase (LUC) reporter plasmid (20, 21), 4 μg of the LexA fusion constructs, and 2 μg of the pSV/β-Gal construct (Promega), using the calcium phosphate transfection method. The HSF-inducible LUC reporter plasmid was activated by exposure of the cells at 43°C for 1 h, followed by a 6-h recovery at 37°C. LUC activity was normalized to β-galactosidase activity. The LUC activity in cells transfected with only the LUC reporter plasmid was therefore set at 100%, and LUC activities in cells cotransfected with the indicated plasmids were expressed as percentages of this control value. The degree of repression by LexA fusion proteins is expressed as the mean ± standard error of the mean. All experiments were performed seven times independently, including the transfections.
Nucleotide sequence accession numbers.
The GenBank accession numbers for XCtBP1 and CtBP1 are AF091554 and AF091555, respectively.
RESULTS
Identification of CtBP1 and CtBP2 as proteins that interact with the vertebrate Pc homologs XPc and HPC2.
To identify genes encoding proteins that interact with HPC2 and XPc, both of which are vertebrate homologs of the Drosophila PcG protein Pc, we performed two-hybrid screens. The full-length coding regions for XPc (19) and HPC2 (21) were cloned into the pAS2 vector (5). The plasmids pAS2-XPc and pAS2-HPC2 were cotransformed with, respectively, a Xenopus oocyte and a human fetal brain two-hybrid library. Approximately 106 independent clones were obtained for each screen. One hundred thirty-six growing colonies were obtained from the two-hybrid screen with XPc. Twelve colonies, of which eight colonies contained similar cDNA inserts, remained histidine and β-galactosidase positive after DNA isolation and rescreening. From the two-hybrid screen with HPC2, 100 growing colonies were obtained, of which 3 colonies remained histidine and β-galactosidase positive after DNA isolation and rescreening. A 1,519-bp cDNA clone that we isolated from the two-hybrid screen with HPC2 was identical to CtBP2 (11). The isolated CtBP2 clone encodes aa 1 to 445 of the 445-aa CtBP2 protein. A 1,414-bp cDNA clone obtained from the XPc screen was homologous to CtBP1 and CtBP2 (11, 22). The predicted 440-aa protein is 85% identical to CtBP1 based on the encoding sequence published by Schaeper et al. (22) and is 78% identical to CtBP2 (11) (Fig. 1). However, comparison of the open reading frames of XCtBP and CtBP1 revealed potential frameshifts in the reading frame. We therefore searched for different EST clones in the database of CtBP1 and compared these with CtBP1 and XCtBP. Comparison of different EST clones (accession no. H46860, AA282011, and AA312167) indeed revealed that there are several frameshifts in the published sequence of CtBP1. We confirmed these differences by sequencing the CtBP1 cDNA, which we obtained by PCR. When the corrections are taken into account, the XCtBP protein is 96% identical to CtBP1 instead of 85%. Based on the extensive homology between CtBP1 and XCtBP we therefore named the novel Xenopus protein XCtBP1.
FIG. 1.
Comparison of the XCtBP1 and the human CtBP1 and CtBP2 proteins. Identical amino acids are indicated as black boxes.
In conclusion, a two-hybrid screen with XPc as a target resulted in the isolation of XCtBP1, a Xenopus homolog of CtBP1. A two-hybrid screen with HPC2 as a target resulted in the isolation of the CtBP2 protein.
A specific 6-amino acid motif in HPC2 is crucial for binding of CtBP.
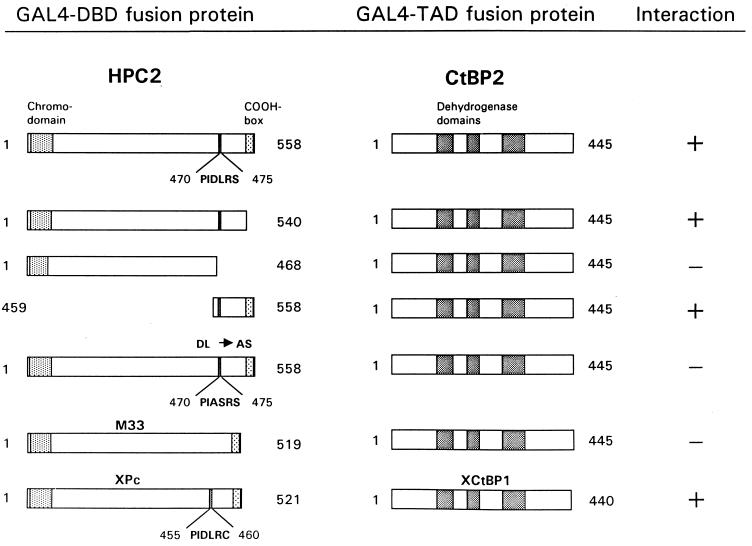
To define domains that are responsible for the interaction of the Pc proteins and the CtBP proteins, we cloned different parts of HPC2 in frame with the GAL4 DNA binding domain (GAL4 DBD) and tested whether these proteins could still interact with full-length CtBP2 (Fig. 2). HPC2 comprises two functional domains. The first domain is the N-terminal chromodomain, which is essential for binding of the Pc protein to chromatin (12). The other domain is the C-terminal COOH box (aa 540 to 558). This COOH box is necessary for the repression of gene activity (4, 13, 21) and is also the domain to which the RING1 protein binds (20, 23). We found that an HPC2 mutant (aa 1 to 540) which lacks the COOH box is still able to interact with CtBP2 (Fig. 2). In contrast, a smaller portion of the HPC2 protein (aa 1 to 468) does not interact with CtBP2, whereas a C-terminal fragment (aa 459 to 558) is able to interact with CtBP2. Thus, CtBP2 interacts with a part of the C terminus of HPC2 but not with the extreme C-terminal COOH box (aa 540 to 558), which is involved in gene repression and RING1 binding.
FIG. 2.
Mapping of the CtBP2 interaction domain in the HPC2 protein and specificity among vertebrate Pc homologs for binding CtBP. The indicated portions of HPC2 were fused to the GAL4 DBD. The HPC2 regions include the shaded chromodomain (aa 6 to 58), a 6-aa motif (PIDLRS) (aa 470 to 475), and the shaded COOH box (aa 540 to 554). The mutation from DL to AS within the 6-aa motif is indicated. The full-length vertebrate Pc proteins M33 and XPc were also fused to the GAL4 DBD. The conserved 6-aa motif (PIDLRC) in the XPc protein is indicated. The three dehydrogenase homology domains within CtBP2 and XCtBP1 are shaded. Constructs that encompass different portions of the HPC2 protein are indicated. The plasmids were cotransformed with full-length CtBP2 (aa 1 to 445) or XCtBP1 (aa 1 to 440), which is fused to the GAL4 TAD. Interactions were positive when cells grew on selective medium lacking histidine and when they were also β-galactosidase positive. When a negative interaction is indicated, no β-galactosidase activity was detected.
Within the C-terminal fragment to which CtBP2 binds, we observed a 6-aa motif (PIDLRS) (aa 470 to 475) (Fig. 2) which is very similar to a 6-aa motif (PLDLSC) present in the extreme C terminus of the Ad2/5 E1A protein. This motif is essential for the interaction between E1A and CtBP1 (22). Mutations within the first four amino acids of the E1A motif completely abolish the interaction between E1A and CtBP1 (22). We created a similar mutation within this corresponding 6-aa motif of HPC2 by changing the motif from PIDLRS to PIASRS, using PCR primers which contained the specific mutations. Subsequently, we tested whether the HPC2(DL→AS) mutant protein is still able to interact with CtBP2. We found that the DL-to-AS mutation in the HPC2 protein completely abolishes the interaction with CtBP2 in the two-hybrid system. Importantly, the mutation within the 6-aa motif leaves intact the C-terminal COOH box of the HPC2 protein to which the RING1 protein binds (20). We therefore tested whether the RING1 protein is still able to interact with the HPC2(DL→AS) mutant protein. We observed no loss of interaction between this mutant HPC2 protein and RING1 (data not shown), underlining the specificity of the interaction between CtBP2 and the conserved 6-aa motif in HPC2.
We have identified the XCtBP1 protein in a two-hybrid screen with the XPc protein as the target. The XPc protein encompasses a specific 6-aa motif, PIDLRC, related to the 6-aa motif in HPC2 which is crucial for binding CtBP (Fig. 2). We also tested whether the CtBP protein could interact with another murine homolog, M33, which is more homologous to the human Pc homolog, CBX2/HPC1, than to HPC2 (7, 15, 21). Surprisingly, we observed no interaction between M33 and CtBP2 in the two-hybrid system (Fig. 2) or between M33 and CtBP1 (data not shown). Importantly, M33 does not encompass the conserved 6-amino-acid motif that is present in HPC2 and that is crucial for the interaction with CtBP. It is therefore likely that the lack of this conserved 6-aa motif in M33 is responsible for the lack of interaction between M33 and CtBP. This result is the first indication that, despite the high degree of homology in the chromodomain and the COOH box, there is specificity among different vertebrate Pc proteins, particularly in their ability to interact with other proteins.
In conclusion, the highly homologous proteins CtBP1, CtBP2, and XCtBP1 interact with HPC2 and XPc. Strikingly, no interaction could be observed between CtBP and M33, a murine Pc homolog, indicating specificity among the different vertebrate Pc proteins.
CtBP1 and CtBP2 are able to homo- and heterodimerize, and the interaction domain differs from the domain responsible for interaction with HPC2.
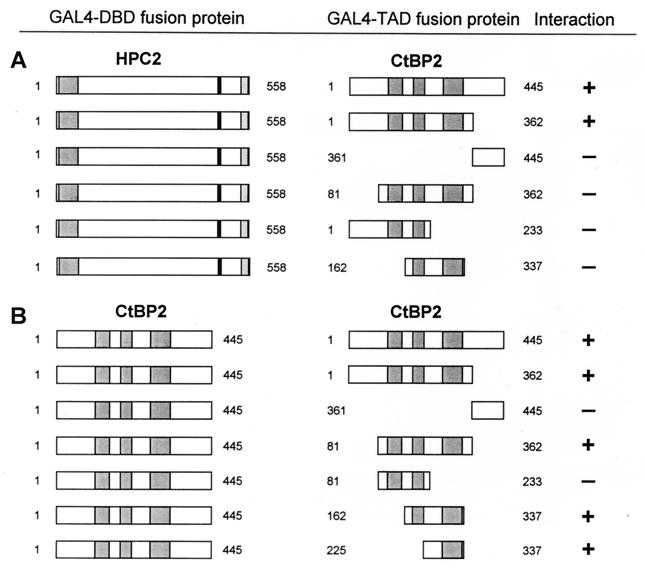
To determine which part of the CtBP proteins is responsible for the interaction with HPC2, we subcloned different protein fragments of CtBP2 in frame with the GAL4 transactivating domain (GAL4 TAD) (Fig. 3A). The C-terminal region of CtBP2 encompassing aa 361 to 445 is not capable of interaction with HPC2, whereas the N-terminal region containing aa 1 to 362 is still able to interact with HPC2. This region encompasses three domains which have strong homology with various NAD-dependent D-isomer-specific 2-hydroxy acid dehydrogenases (11, 22). To analyze whether these dehydrogenase homology domains are responsible for the interaction with HPC2, we made three constructs containing different sets of these dehydrogenase homology domains.
FIG. 3.
Mapping of domains of interaction of CtBP2 with HPC2 (A) and CtBP2 (B). (A) The indicated portions of CtBP2 were fused to the GAL4 TAD. These CtBP2 regions include three dehydrogenase homology domains. Plasmids were cotransformed with full-length HPC2 which was fused to the GAL4 DBD. (B) Full-length CtBP2 which was fused to the GAL4 DBD was tested for interaction against the indicated portions of CtBP2. When a negative interaction is indicated, no β-galactoctosidase activity was detected.
We found that a region of CtBP2 encompassing aa 81 to 362, which contains all three dehydrogenase homology domains, is not able to interact with HPC2. Also, a CtBP2 region (aa 1 to 233) encompassing the N terminus and the first two dehydrogenase homology domains and a CtBP2 region (aa 162 to 337) encompassing the second and the third dehydrogenase homology domains are not able to interact with HPC2. These results indicate that a large region of CtBP2 (aa 1 to 362), which encompasses both the extreme N-terminal part and the dehydrogenase homology domains, is responsible for the interaction with HPC2.
The HPC2 protein (20, 21) is part of a complex which constitutes the mammalian homologs of the Drosophila Ph protein, HPH1 and HPH2. These two proteins are able to homo- and heterodimerize with each other (9). To address the question of whether this is also true for CtBP1 and CtBP2, we cloned the full-length coding region for CtBP2 in frame with the GAL4 DBD and tested whether CtBP2 could interact with itself or CtBP1. Both CtBP1 (data not shown) and CtBP2 (Fig. 3B) are able to interact with CtBP2 in the two-hybrid system, indicating that these proteins are able to homodimerize and to heterodimerize.
To define the domains that are responsible for the interaction between CtBP2 and CtBP2, we subcloned different parts of CtBP2 in frame with the GAL4 TAD and tested whether these domains are still able to interact with full-length CtBP2. The C-terminal region of CtBP2 encompassing aa 361 to 445 is not able to interact with CtBP2, whereas the N-terminal region containing aa 1 to 362 is still able to interact with CtBP2 (Fig. 3B). A region containing only the three dehydrogenase homology domains (aa 81 to 361) still interacts with CtBP2. Detailed analysis of this region showed that CtBP2 aa 81 to 233, encompassing the first two dehydrogenase homology domains, exhibits no interaction with CtBP2. In contrast, CtBP2 aa 162 to 337, containing dehydrogenase homology domains two and three, still interacts with CtBP2. Also, CtBP2 aa 225 to 337, containing only the third dehydrogenase homology domain, is still able to interact with CtBP2 (Fig. 3B). These data indicate that a region in CtBP2 encompassing the third dehydrogenase homology domain is sufficient for the interaction with full-length CtBP2. Interestingly, this relatively small interaction domain, which is necessary to convey homodimerization between CtBP2 and CtBP2, differs from the domain for interaction with HPC2. Above we showed that a much larger region of CtBP2, containing the N terminus as well as all three dehydrogenase domains, is necessary for the interaction with HPC2 (Fig. 3A).
In summary, the CtBP1 protein and the CtBP2 protein each can interact with itself, and they are also able to interact with each other. The domain responsible for this interaction is a region encompassing the third dehydrogenase homology domain. This interaction domain differs from the domain that is responsible for the interaction with HPC2, which involves the N terminus and all three dehydrogenase homology domains.
The XPc and CtBP2 proteins interact directly in vitro.
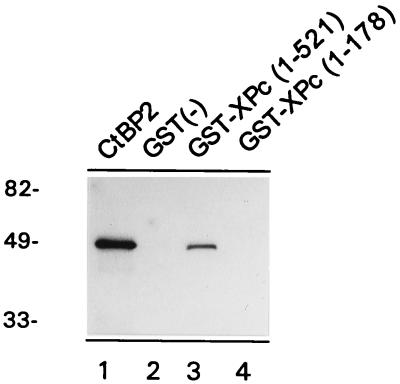
To determine whether the interaction between the vertebrate Pc homologs and CtBP is a direct interaction, we employed an in vitro pull-down assay. The previous described (19) fusion protein of GST and full-length XPc (aa 1 to 521) was expressed in bacteria. The affinity-purified protein was subsequently immobilized on GST-Sepharose and incubated with [35S]methionine-labelled, in vitro-translated CtBP2. After extensive washing, the [35S]methionine-labelled proteins bound to GST-XPc were analyzed by SDS-PAGE. The in vitro-translated full-length CtBP2 protein of 48 kDa (Fig. 4, lane 1) was able to bind to the immobilized GST-XPc (lane 3) but did not bind to the immobilized GST alone (lane 2). We also tested whether CtBP interacted with another GST-XPc (aa 1 to 178) fusion protein (19). This portion of the XPc protein encompasses the chromodomain of XPc but lacks the entire C-terminal domain that contains the 6-amino-acid motif to which CtBP binds. CtBP does not bind to such a C-terminal deletion HPC2 mutant in the two-hybrid assay (Fig. 2). Importantly, we found that the GST-XPc aa 1 to 178 protein does not interact with CtBP2 (Fig. 4, lane 4). These results confirm the two-hybrid assay data (Fig. 2) and underline the specificity of the in vitro pull-down assay.
FIG. 4.
XPc and CtBP2 interact directly in vitro. [35S]methionine-labelled CtBP2 protein (lane 1) was incubated with GST-Sepharose alone (lane 2), GST-XPc aa 1 to 521 (lane 3), or GST-XPc aa 1 to 178 (lane 4). The GST-XPc aa 1 to 521 but not the GST-XPc aa 1 to 178 fusion protein is able to interact with in vitro-translated [35S]methionine-labelled CtBP2 protein. Molecular weights in thousands are indicated on the left.
Expression of CtBP1 and CtBP2 in human tissues and human cancer cell lines.
To investigate the expression patterns of CtBP1 and CtBP2, we needed unique cDNA fragments in order to avoid cross-hybridization between CtBP1 and CtBP2 mRNA species. Since there is no homology between the UTRs of CtBP1 and CtBP2, we used a 560-bp fragment of the 3′ UTR of CtBP1 and a 470-bp fragment of the 3′ UTR of CtBP2 as probes. These probes were hybridized to Northern blots containing poly(A)+ mRNAs from different human cancer cell lines or human tissues (Clontech). We detected single transcripts of approximately 2.4 kb for CtBP1 and approximately 3.0 kb for CtBP2. In all human tissues present on the commercial Northern blot (Fig. 5A), CtBP1 was expressed at approximately the same level as CtBP2, with the exception of the thymus and peripheral blood leukocytes. In these two tissues, the CtBP2 transcript was hardly detectable (Fig. 5A, lanes 2 and 8).
FIG. 5.
Expression patterns of CtBP1 and CtBP2 in human tissues (A) and in human cancer cell lines (B). (A) Expression levels in spleen (lane 1), thymus (lane 2), prostate (lane 3), testis (lane 4), ovary (lane 5), small intestine (lane 6), colon (lane 7), and peripheral blood leukocytes (lane 8). (B) Expression levels in promyelocytic leukemia HL-60 (lane 1), HeLa S3 (lane 2), chronic myelogenous leukemia K-562 (lane 3), lymphoblastic leukemia MOLT-4 (lane 4), Burkitt’s lymphoma Raji (lane 5), colorectal adenocarcinoma SW480 (lane 6), lung carcinoma A549 (lane 7), and melanoma G361 (lane 8) cell lines. Lanes 1 to 8, commercially obtained Northern blot. We also isolated and blotted poly(A)+ RNA from U-2 OS cells (lane 10) and SW480 cells (lane 9), the latter to allow comparison with the commercial multiple-tissue Northern blot. To verify the loading of RNA in each lane, the blots were hybridized with a probe for glyceraldehyde-3-phosphate dehydrogenase (GAPDM).
In human cancer cell lines, differences in expression of either CtBP1 or CtBP2 were more pronounced than in normal tissues. In the case of CtBP1, high expression of the commercial blot was detected in HL-60 cells (Fig. 5B, lane 1) and in the adenocarcinoma SW480 cell line (lane 6). Expression of CtBP1 was still well pronounced in HeLa S3 cells (Fig. 5B, lane 2), K-562 cells (lane 3), MOLT-4 cells (lane 4), and U-2 OS cells (lane 10). Low expression of CtBP1 was detected in Raji cells (lane 5) and G361 cells (lane 8), whereas almost no CtBP1 expression was found in A549 cells. In the case of CtBP2, high expression was detected in HeLa S3 cells (Fig. 5B, lane 2) and SW480 cells (lane 6), whereas significantly lower expression was detected in HL-60 (lane 1), G361 (lane 8), and U-2 OS (lane 10) cells. A very low level of CtBP2 expression was found in A549 cells (lane 7), but no detectable CtBP2 transcript could be observed in K-562 (lane 3), MOLT-4 (lane 4), and Raji (lane 5) cells. Interestingly, CtBP2 was highly expressed in the spleen (Fig. 5A, lane 1), whereas no expression could be observed in a B-cell-derived cell line, Raji (Fig. 5B, lane 5). Strikingly, in all tissues or cell lines either one or two CtBP transcripts could be detected, with the exception of lung carcinoma cells (lane 7), in which both CtBP transcripts were hardly detectable.
A polyclonal antibody raised against XCtBP1 recognizes both CtBP1 and CtBP2.
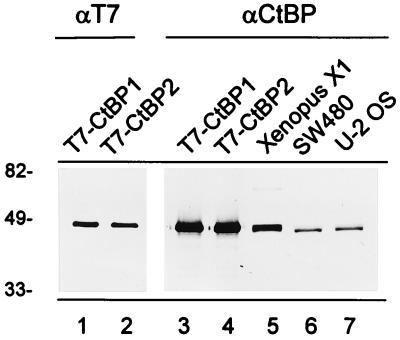
To determine the distribution of the CtBP proteins in the cell nucleus and to be able to detect CtBP proteins in immunoprecipitates, we raised a polyclonal antibody against full-length XCtBP1. To test whether the polyclonal antibody also recognizes both CtBP1 and CtBP2, we created constructs containing the full-length coding region for either CtBP1 or CtBP2, with a T7 tag at the N terminus. Fusion proteins were produced in E. coli BL21(DE), and the bacterial cell lysates were subsequently separated by SDS-PAGE and transferred to nitrocellulose. The blots were probed with either a mouse monoclonal antibody against T7 (Fig. 6, lanes 1 and 2) or our rabbit polyclonal antibody against XCtBP1 (lanes 3 to 7). The T7 antibody recognizes both the 48-kDa T7-tagged CtBP1 (lane 1) and T7-tagged CtBP2 (lane 2) proteins. Also, the anti-XCtBP1 polyclonal antibody recognizes the 48-kDa T7-tagged CtBP1 (lane 3) and T7-tagged CtBP2 (lane 4) proteins, indicating that both CtBP1 and CtBP2 are recognized by the polyclonal antibody raised against XCtBP1. We further analyzed cell extracts of Xenopus X1 cells (Fig. 6, lane 5), SW480 cells (lane 6), and U-2 OS cells (lane 7). In all three cell extracts a doublet protein band of approximately 48 kDa was observed. We conclude that the antibody against XCtBP1 recognizes both the CtBP1 and CtBP2 proteins.
FIG. 6.
A rabbit polyclonal antibody recognizes XCtBP1, CtBP1, and CtBP2. T7-tagged CtBP1 (lanes 1 and 3) and T7-tagged CtBP2 (lanes 2 and 4) were expressed in E. coli. Cell lysates were analyzed by Western blotting and probed with either a mouse monoclonal antibody against T7 (αT7) (lanes 1 and 2) or the polyclonal antibody against CtBP (αCtBP) (lanes 3 and 4). In cell lysates of Xenopus X1 cells (lane 5), colorectal adenocarcinoma SW 480 cells (lane 6), and osteosarcoma U-2 OS cells (lane 7), the polyclonal antibody against CtBP recognizes a doublet of 48 kDa. Molecular weights in thousands are indicated on the left.
An in vivo interaction between CtBP2 and HPC2.
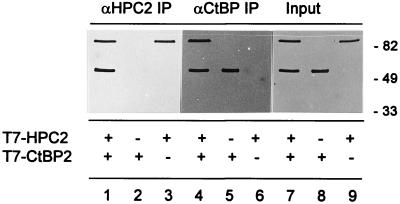
To determine whether the interaction between CtBP proteins and HPC2 also exists in vivo, we performed coimmunoprecipitation experiments. We transiently transfected COS-7 cells with T7-tagged HPC2 and T7-tagged CtBP2. We used polyclonal rabbit antibodies directed against XCtBP1 and HPC2 for the immunoprecipitations and a mouse monoclonal antibody against T7 to detect either the 82-kDa T7-HPC2 (21) or the 48-kDa T7-CtBP2 protein.
We found that CtBP2 and HPC2 coimmunoprecipitate with each other (Fig. 7). The anti-HPC2 antibody coimmunoprecipitated both T7-CtBP2 and T7-HPC2 (Fig. 7, lane 1) from cells expressing both T7-HPC2 and T7-CtBP2 (lane 7), as was detected with the anti-T7 monoclonal antibody. No T7-CtBP2 could be detected in the anti-HPC2-immunoprecipitated material (lane 2) when T7-CtBP2 but not T7-HPC2 was expressed (lane 8). Also, no T7-CtBP2 could be detected in the anti-HPC2 immunoprecipitated material (lane 3) when T7-HPC2 but not T7-CtBP2 was expressed (lane 9).
FIG. 7.
In vivo interaction between HPC2 and CtBP2. Immunoprecipitation (IP) was performed with polyclonal rabbit antibodies against HPC2 (αHPC2) (lanes 1 to 3) or polyclonal rabbit antibodies against XCtBP1 (αCtBP) (lanes 4 to 6). The resulting immunoprecipitates were Western blotted and analyzed with mouse monoclonal antibodies against T7. The total cell extracts (Input) are shown in lanes 7 to 9. COS-7 cells were transiently transfected with both pcDNA3-T7-HPC2 and pcDNA3-T7-CtBP2 (lanes 1, 4, and 7) or with either pcDNA3-T7-CtBP2 (lanes 2, 5, and 8) or pcDNA3-T7-HPC2 (lanes 3, 6, and 9). Molecular weights in thousands are indicated on the right.
Similarly, the anti-CtBP antibody immunoprecipitated both T7-HPC2 and T7-CtBP2 (Fig. 7, lane 4) from cells expressing both T7-HPC2 and T7-CtBP2 (lane 7). No T7-HPC2 could be detected in the anti-CtBP-immunoprecipitated material (lane 5) when T7-CtBP2 but not T7-HPC2 was expressed (lane 8). Finally, no T7-HPC2 could be detected in the anti-CtBP-immunoprecipitated material (lane 6) when T7-HPC2 but not T7-CtBP2 was expressed (lane 9).
Also, in extracts of SW480 cells, in which the PcG proteins are highly expressed (9, 21) and in which the CtBP proteins are expressed, we observed coimmunoprecipitation of either HPC2 and CtBP or BMI1 and CtBP (data not shown). However, in both cases the recovery of the proteins in the immunoprecipitations was approximately 20% of the input. This result further strengthens the notion that an interaction between CtBP and HPC2 exists in vivo. The low recovery might indicate that the interaction between CtBP and the PcG complex is of a transient nature.
In conclusion, we show that CtBP2 and HPC2 coimmunoprecipitated with each other from extracts of COS-7 cells in which we overexpressed CtBP2 and HPC2. These findings indicate that CtBP2 and HPC2 interact with each other in vivo.
CtBP1 and CtBP2 partially colocalize with HPC2 in nuclei of U-2 OS cells.
To determine the subcellular distribution of the CtBP1 protein and the CtBP2 protein in relation to the HPC2 protein, we performed immunofluorescence labelling experiments. Previously we have shown that the HPC2 protein colocalizes in large nuclear domains, termed PcG domains, with BMI1, HPH1, HPH2, and RING1 (9, 20, 21). To compare the distributions of the CtBP proteins relative to the distribution of HPC2, we performed double-labelling experiments with the rabbit anti-XCtBP1 antibody, which recognizes both CtBP1 and CtBP2 (Fig. 6), and a chicken anti-HPC2 antibody (20, 21). We found that the CtBP proteins are abundantly present in nuclei of U-2 OS cells in a fine granular pattern but also in larger nuclear domains (Fig. 8A). Within these larger nuclear domains, the CtBP proteins colocalize with HPC2 (Fig. 8B and C). However, the colocalization within these domains differs slightly from the colocalization of the BMI1 protein (Fig. 8D) with the HPC2 protein (Fig. 8E and F). The BMI1 and HPC2 proteins completely colocalize in bright, sharply edged PcG domains (Fig. 8F). This specific labelling pattern has also been observed with antibodies against the human PcG homologs HPH1 and HPH2 (9) and the RING1 protein (20). The nuclear domains that are detected by the anti-XCtBP1 antibody and that colocalize with the more sharply edged PcG domains have a more diffuse shape (Fig. 8A, B, and C). Another difference between the CtBP and PcG labelling patterns is that most of the BMI1 and HPC2 proteins appear to be concentrated within the large PcG domains (Fig. 8D, E, and F). In contrast, most of the CtBP labelling is detected in the smaller domains throughout the nucleoplasm and not in the larger domains that colocalize with the PcG domains. This fine granular pattern is too complex to allow analysis of any systematic colocalization.
Since the rabbit anti-CtBP antibody recognizes both the CtBP1 protein and the CtBP2 protein, it is not possible to directly test for differences in nuclear localization of the CtBP1 protein and the CtBP2 protein. In order to distinguish between the distributions of the CtBP1 protein and the CtBP2 protein, we transiently transfected U-2 OS cells with either the T7-tagged CtBP1 protein (Fig. 8G) or the T7-tagged CtBP2 protein (Fig. 8J). Double labelling was performed with a mouse monoclonal antibody against T7 and the affinity-purified chicken antibody against HPC2. We found that T7-tagged CtBP1 (Fig. 8G) colocalizes with HPC2 (Fig. 8H) in the large PcG domains (Fig. 8I). Also, T7-tagged CtBP2 (Fig. 8J) colocalizes with HPC2 (Fig. 8K) within these large PcG domains (Fig. 8L). These results indicate that there are no major detectable differences in the localizations of CtBP1 and CtBP2 and that both proteins are present in the same PcG domains.
CtBP acts as a transcriptional repressor when targeted to a reporter gene.
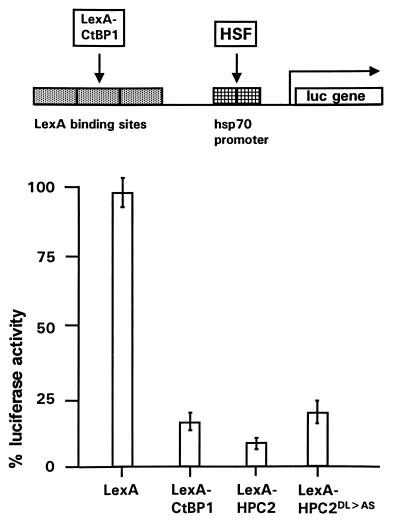
The PcG proteins are involved in the repression of gene expression, but the identified PcG proteins do not bind directly to DNA. Nevertheless, the ability of the PcG proteins to repress gene activity can be tested by targeting LexA fusion proteins to a reporter gene (20, 21, 25). Previously, we have shown that LexA-HPC2 was able to repress gene activity (20, 21). We asked whether this is also true for the CtBP proteins. We therefore tested whether LexA-CtBP1 was able to repress gene expression when targeted to a reporter gene. U-2 OS human osteosarcoma cells were transfected with a construct containing a tandem of four LexA operators, binding sites for the HSF transcriptional activator, and the hsp70 TATA promoter region, immediately upstream of the LUC reporter gene. The endogenous HSF was used as transcriptional activator. In absence of this activator, no LUC expression could be measured (data not shown). In the presence of the HSF, expression was maximal and was set at 100%. Cotransfection with LexA alone had no significant effect on LUC expression (Fig. 9) (97% ± 6% [n = 7]). We found that LexA-CtBP1 was able to repress LUC expression significantly (16% ± 4% [n = 7]). This degree of LUC repression was also found for LexA-CtBP2 (data not shown). In the same experiment we found that LexA-HPC2 could repress LUC activity most efficiently (9% ± 3% [n = 7]). Previously, we have shown that a LexA-HPC2 mutant which lacks the C-terminal domain, to which the RING1 protein binds, was no longer able to repress LUC expression (21). We tested whether the HPC2(DL→AS) mutant also has lost the ability to repress LUC expression. In this HPC2 mutant the specific 6-aa motif is mutated, which leads to abolishment of the interaction with CtBP (Fig. 2). We observed a slight but significant decrease in the ability of the HPC2 protein to repress gene activity when the DL→AS mutation is introduced. However, the LexA-HPC2(DL→AS) mutant still represses LUC activity significantly (20% ± 5% [n = 7]).
FIG. 9.
Repression of HSF-induced LUC gene activity by CtBP. Activation of LUC expression is maximally induced by endogenous HSF in the absence of any LexA fusion protein. This LUC activity was set at 100%. LUC activities in cells cotransfected with the indicated plasmids were expressed as percentages of this control value. Bars represent the average degree of repression by LexA, LexA-CtBP1, LexA-HPC2, or LexA-HPC2(DL→AS) in seven independent experiments (means ± standard errors of the means).
We conclude that CtBP is able to repress gene activity when targeted to a reporter gene, almost as efficiently as HPC2. Furthermore, mutating the specific 6-aa motif within HPC2 which is crucial for CtBP binding has a significant but small effect on the ability of HPC2 to repress gene activity.
DISCUSSION
An interaction between CtBP and vertebrate Pc homologs.
The Pc protein is part of a multimeric PcG protein complex which is involved in the stable and heritable repression of gene activity during Drosophila and vertebrate development. To identify proteins that interact with vertebrate Pc proteins, we employed two-hybrid screens with a Xenopus Pc homolog, XPc, and a human Pc homolog, HPC2. Here, we describe the identification of two closely related proteins, the Xenopus homolog of CtBP1, XCtBP1, and CtBP2, which interact with XPc and HPC2. This interaction also exists in vivo, since the proteins coimmunoprecipitate with each other and partially colocalize in large PcG domains in interphase nuclei. However, our data also indicate that the interactions between CtBP and HPC2 differ substantially from the interaction between human PcG proteins that we previously described. The human PcG homologs BMI1, HPH1, HPH2, and HPC2, as well as the RING1 protein, almost quantitatively coimmunoprecipitate with each other from extracts of SW480 and U-2 OS cells (9, 20, 21). Furthermore, BMI1, HPH1, HPH2, and HPC2 completely colocalize within large nuclear domains of interphase cells termed PcG domains. The in vivo interaction between CtBP and HPC2 differs in both aspects. Only a small amount of the endogenous CtBP and HPC2 proteins coimmunoprecipitate from cell extracts. This may indicate that the interaction between CtBP and HPC2 is of a transient nature, whereas BMI1, HPH1, HPH2, HPC2, and RING1 form a more stable protein complex. Also, the partial colocalization between the CtBP proteins and HPC2 points towards differences. First of all, the CtBP proteins are more abundantly distributed than the PcG proteins outside the PcG domains in a fine granular pattern throughout the nucleoplasm. Further, even within the large PcG domains the CtBP proteins only partly colocalize with HPC2, since the large CtBP domains have a more diffuse shape than the sharply edged PcG domains. Therefore, although our data indicate that the CtBP proteins interact with HPC2, the differences in colocalization and the only partial coimmunoprecipitation of the endogenous proteins point towards a broader range of CtBP function. This notion is supported by the fact that the Drosophila homolog of CtBP, dCtBP, has been found to interact with repressors such as Hairy and Knirps (14, 17).
The conserved amino acid motif that is crucial for binding CtBP determines specificity of structurally related proteins to interact with CtBP.
Within the extreme C terminus of the Ad2/5 E1A protein, a specific 6-aa motif which is crucial for binding CtBP is present (2, 22). We find that within the C terminus of the vertebrate Pc homologs HPC2 and XPc, a similar 6-aa motif that is crucial for binding CtBP is present. Mutation of this 6-aa motif completely abolished the interaction with CtBP. Interestingly, the interaction between CtBP and its interacting proteins seems to be evolutionarily conserved through this 6-aa motif. A conserved amino acid motif is crucial for binding the Drosophila homolog of CtBP, dCtBP. This amino acid motif within the Drosophila repressors Knirps (P-DLS-K) and Snail (P-DLS-K) (14) and Hairy (PLSLV) (17) is similar to the 6-aa motif found within HPC2 (PIDLRS), XPc (PIDLRC), and E1A (PLDLSC) and is crucial for binding dCtBP.
Remarkably, another vertebrate Pc homolog, M33, which is very homologous to the human CBX2/HPC1 protein (7, 21), is not able to interact with CtBP. A likely explanation for this lack of interaction between CtBP and M33 is that the M33 protein does not encompass a conserved 6-aa motif that is found in HPC2 or XPc. Notably, this is the first indication that despite the high degree of homology between the different vertebrate Pc homologs, there is specificity among these proteins, particularly in their ability to interact with other proteins. This difference in their ability to interact with CtBP is not of a general nature, since previously it has been shown that the HPC2, XPc, and M33 proteins are all able to interact with the RING1 protein (20, 23).
The specificity of the CtBP interaction raises the question of whether there exists an interaction between dCtBP and Drosophila Pc. The fact that the Drosophila Pc protein does not encompass a conserved 6-aa motif suggests that the interaction between dCtBP and Pc does not exists in Drosophila. If this is true, then the interaction with CtBP is restricted to a particular class of vertebrate Pc homologs. However, it is still possible that a slightly degenerated amino acid sequence is present in Drosophila Pc, which could be responsible for a potential interaction with dCtBP.
Interestingly, a similar kind of specificity has been observed for the interaction between dCtBP and members of the Hairy/Enhancer of split [E(spl)]/Deadpan protein class (17). These proteins are structurally related basic helix-loop-helix protein and are all required as transcriptional repressors of genes necessary for processes such as sex determination, segmentation, and neurogenesis. At least seven members of the E(spl) basic helix-loop-helix class have been identified. However, of this class only the E(spl) mδ/C protein is able to interact with dCtBP, whereas all proteins are able to interact with Groucho (17). All of these data suggest a high degree of selectivity in the interactions of CtBP proteins with specific members of larger protein families.
Involvement of CtBP in HPC2-mediated gene repression.
We have identified vertebrate CtBP proteins that interact with a specific class of vertebrate Pc proteins, which are involved in repression of gene activity. It is not clear from our results to what degree CtBP proteins are involved in mediating the repressing abilities of these vertebrate Pc proteins. A mutation within the 6-aa motif that mediates the binding between CtBP and HPC2 results in a significant but only small decrease in the repressing abilities of HPC2 (Fig. 9). This result is in agreement with previous findings by us and others (4, 13, 21) showing that the main domain that mediates repression resides in the conserved, extreme C-terminal 30 aa of Pc proteins. Such a mutant, which we previously termed ΔHPC2, loses approximately 80% of its repressing ability, while it still retains the 6-aa motif to which CtBP binds. We are tempted to conclude that although our results indicate that CtBP contributes to the repressing ability of the HPC2 protein, this contribution is small compared to the contribution of the extreme C-terminal COOH box.
Alternatively, the significance of the interaction may be a targeting function of CtBP for the PcG complex. The recent finding that the dCtBP protein interacts with the repressors Knirps and Snail (14) and Hairy (17) supports this notion. These Drosophila repressors are all sequence-specific DNA binding proteins. It is conceivable that CtBP proteins target HPC2, and thereby the PcG complex, to particular loci in the chromatin that contain binding sites for specific repressors such as human homologs of Knirps and Hairy. The result would be a complex between these repressors and the PcG complex, with CtBP as a bridging protein. Such a model would not be feasible when CtBP proteins act as monomers, since HPC2 and these repressors interact through the same interaction domain within CtBP. This in turn would result in competition between HPC2 and these repressors. However, since the CtBP proteins have the ability to homo- and heterodimerize, both HPC2 and other CtBP-interacting repressor proteins could simultaneously bind to a CtBP homo- or heterodimer. This scenario permits enormous flexibility in the range of PcG action. For instance, the specificity of the interaction between CtBP and only a subclass of vertebrate Pc homologs allows targeting of distinct PcG complexes. Inclusion of HPC2 in the complex would permit recruitment to a CtBP-repressor target site, whereas inclusion of the M33 Pc homolog excludes such a recruitment.
Although the Ad E1A protein is involved in transcriptional activation and repression of several viral and cellular promoters, the E1A protein is not able to bind DNA by itself. The known transforming and transcriptional activities appear to be related to the ability of the E1A protein to interact with various cellular proteins (reviewed in reference 10). It is tempting to speculate that in vivo, the E1A protein disturbs the interaction between CtBP and the PcG complex by disrupting the interaction between CtBP and the HPC2 protein. Particularly, since the interaction between E1A and CtBP is stronger than the interaction between the vertebrate Pc homologs and CtBP (data not shown), E1A might be a strong competitor for binding with CtBP. A significant feature of the interference of E1A with the transcription machinery of the infected cell may involve interference with PcG-mediated repression, through the disruption of the CtBP-PcG interaction.
ACKNOWLEDGMENTS
R.G.A.B.S. and M.J.G. contributed equally to this work.
We thank Roel van Driel for critically reading the manuscript, Karien Hamer and Jan den Blaauwen for technical assistance, and Thijs Hendrix for raising rabbit antibodies.
This work was supported in part by grants from the Netherlands Organization for Scientific Research (NWO) to R.G.A.B.S.
REFERENCES
- 1.Alkema M J, Bronk M, Verhoeven E, Otte A P, Van ’t Veer L J, Berns A, Van Lohuizen M. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian Polycomb complex. Genes Dev. 1997;11:226–240. doi: 10.1101/gad.11.2.226. [DOI] [PubMed] [Google Scholar]
- 2.Boyd J M, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G. A region in the C-terminus of adenovirus 2/5 E1A protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 1993;12:469–478. doi: 10.1002/j.1460-2075.1993.tb05679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breiling A, Paro R. Mechanisms of heritable gene silencing during development of Drosophila. In: Van Driel R, Otte A P, editors. Nuclear organization, chromatin structure, and gene expression. Oxford, United Kingdom: Oxford University Press; 1997. pp. 236–249. [Google Scholar]
- 4.Bunker C A, Kingston R E. Transcription repression by Drosophila and mammalian Polycomb group proteins in transfected mammalian cells. Mol Cell Biol. 1994;14:1721–1732. doi: 10.1128/mcb.14.3.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. The retinoblastoma protein associates with the protein phophatase type I catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 6.Franke A, DeCamillis M, Zink D, Cheng N, Brock H W, Paro R. Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J. 1992;11:2941–2950. doi: 10.1002/j.1460-2075.1992.tb05364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gecz J, Gaunt J S, Passage E, Burton R D, Cudrey C, Pearce J J, Fontes M. Assignment of a Polycomb-like chromobox gene (CBX2) to human chromosome 17q25. Genomics. 1995;26:130–133. doi: 10.1016/0888-7543(95)80091-y. [DOI] [PubMed] [Google Scholar]
- 8.Gould A. Functions of mammalian Polycomb group and trithorax group related genes. Curr Opin Genet Dev. 1997;7:488–494. doi: 10.1016/s0959-437x(97)80075-5. [DOI] [PubMed] [Google Scholar]
- 9.Gunster M J, Satijn D P E, Hamer K M, den Blaauwen J L, De Bruijn D, Alkema M J, Van Lohuizen M, van Driel R, Otte A P. Identification and characterization of interactions between the vertebrate polycomb-group protein BMI1 and human homologs of polyhomeotic. Mol Cell Biol. 1997;17:2326–2335. doi: 10.1128/mcb.17.4.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones N. Transcriptional modulation by the adenovirus E1A gene. Curr Top Microbiol Immunol. 1995;199:113–130. doi: 10.1007/978-3-642-79586-2_4. [DOI] [PubMed] [Google Scholar]
- 11.Katsanis N, Fisher E M C. A novel C-terminal binding protein (CtBP2) is closely related to CtBP1, an adenovirus E1A-binding protein, and maps to human chromosome 21q21.3. Genomics. 1998;47:294–299. doi: 10.1006/geno.1997.5115. [DOI] [PubMed] [Google Scholar]
- 12.Messmer S, Franke A, Paro R. Analysis of the functional role of the Polycomb chromo domain in Drosophila melanogaster. Genes Dev. 1992;6:1241–1254. doi: 10.1101/gad.6.7.1241. [DOI] [PubMed] [Google Scholar]
- 13.Müller J. Transcriptional silencing by the Polycomb protein in Drosophila embryos. EMBO J. 1995;14:1209–1220. doi: 10.1002/j.1460-2075.1995.tb07104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nibu Y, Zhang H, Levine M. Interaction of short-range repressors with Drosophila CtBP in the embryo. Science. 1998;280:101–104. doi: 10.1126/science.280.5360.101. [DOI] [PubMed] [Google Scholar]
- 15.Pearce J J H, Singh P B, Gaunt S J. The mouse has a Polycomb-like chromobox gene. Development. 1992;114:921–929. doi: 10.1242/dev.114.4.921. [DOI] [PubMed] [Google Scholar]
- 16.Pirrotta V. PcG complexes and chromatin silencing. Curr Opin Genet Dev. 1997;7:249–258. doi: 10.1016/s0959-437x(97)80135-9. [DOI] [PubMed] [Google Scholar]
- 17.Poortinga G, Watanabe M, Parkhurst S M. Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rastelli L, Chan C S, Pirrotta V. Related chromosome binding sites for zeste, suppressors of zeste and polycomb group proteins in Drosophila and their dependence on Enhancer of zeste function. EMBO J. 1993;12:1513–1522. doi: 10.1002/j.1460-2075.1993.tb05795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reijnen M J, Hamer K M, den Blaauwen J L, Lambrechts C, Schoneveld I, van Driel R, Otte A P. Polycomb and Bmi-1 homologs are expressed in overlapping patterns in Xenopus embryos and are able to interact with each other. Mech Dev. 1995;53:35–46. doi: 10.1016/0925-4773(95)00422-x. [DOI] [PubMed] [Google Scholar]
- 20.Satijn D P E, Gunster M J, van der Vlag J, Hamer K M, Schul W, Alkema M J, Saurin A J, Freemont P S, van Driel R, Otte A P. RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol Cell Biol. 1997;17:4105–4113. doi: 10.1128/mcb.17.7.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satijn D P E, Olson D J, van der Vlag J, Hamer K M, Lambrechts C, Masselink H, Gunster M J, Sewalt R G A B, van Driel R, Otte A P. Interference with the expression of a novel human polycomb protein, HPC2, results in cellular transformation and apoptosis. Mol Cell Biol. 1997;17:6076–6086. doi: 10.1128/mcb.17.10.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaeper U, Boyd J M, Verma S, Uhlmann E, Subramanian T, Chinnadurai G. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc Natl Acad Sci USA. 1995;92:10467–10471. doi: 10.1073/pnas.92.23.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoorlemmer J, Marcos C, Were F, Martinez R, Garcia E, Satijn D P E, Otte A P, Vidal M. RING1A is a transcriptional repressor that interacts with the Polycomb-M33 protein and is expressed at rhombomere boundaries in the mouse hindbrain. EMBO J. 1997;16:5930–5942. doi: 10.1093/emboj/16.19.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schumacher A, Magnuson T. Murine Polycomb- and trithorax-group genes regulate homeotic pathways and beyond. Trends Genet. 1997;13:167–170. [PubMed] [Google Scholar]
- 25.Sewalt R G A B, van der Vlag J, Gunster M J, Hamer K M, den Blaauwen J L, Satijn D P E, Hendrix T, van Driel R, Otte A P. Characterization of interactions between the mammalian Polycomb-group proteins Enx1/EZH2 and EED suggests the existence of different mammalian Polycomb-group protein complexes. Mol Cell Biol. 1998;18:3586–3595. doi: 10.1128/mcb.18.6.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sollerbrant K, Chinnadurai G, Svensson C. The CtBP binding domain in the adenovirus E1A protein controls CR1-dependent transactivation. Nucleic Acids Res. 1996;24:2578–2584. doi: 10.1093/nar/24.13.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Lohuizen M, Tijms M, Voncken J W, Schumacher A, Magnuson T, Wientjens E. Interaction of mouse Polycomb-group (Pc-G) proteins Enx1 and Enx2 with Eed: indication for separate Pc-G complexes. Mol Cell Biol. 1998;18:3572–3579. doi: 10.1128/mcb.18.6.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zink B, Paro R. In vivo binding pattern of a transregulator of homeotic genes in Drosophila melanogaster. Nature. 1989;337:468–471. doi: 10.1038/337468a0. [DOI] [PubMed] [Google Scholar]