Abstract
Since the outbreak at the end of 2019, SARS-CoV-2 has been spreading around the world for more than one year. Scientists have been intensely conducting research on this newly emerged coronavirus and the disease caused by it. Angiotensin-converting enzyme 2 (ACE2), as a receptor mediating the cellular entry of SARS-CoV-2, has become a hot spot for researchers. Here, we summarized the recent progresses on the function, expression and distribution characteristics of ACE2 in human body and among populations. We further discussed the interaction mechanism of ACE2 and SARS-CoV-2 S protein, focusing on key residues that effect interaction and binding ability of SARS-CoV-2 variants. This will facilitate researchers to better understand SARS-CoV-2 infection and transmission route, adaptation mechanism, and designing treatment strategies.
Keywords: SARS-CoV-2, ACE2, Expression pattern, Interaction
1. Introduction
Angiotensin-converting enzyme 2 (ACE2) gene belongs to the angiotensin converting enzyme family of dipeptidyl carboxypeptidase and has considerable homology with human angiotensin converting enzyme1. The ACE2 protein catalyzes the cleavage of angiotensin I into angiotensin 1–9, and cleavage of angiotensin II into the vasodilator angiotensin 1–7. Angiotensin 1–7 acts as a beneficial vasodilator and anti-proliferation agent, counterbalancing the actions of the vasoconstrictor angiotensin II [1], [2], [3], [4], [5]. rhACE2 (recombinant human ACE2) as the negative regulator of the renin-angiotensin system, has completed clinical trials and efficiently lowered or increased plasma angiotensin II and angiotensin 1–7 levels, respectively [6].
In 2004, scientists identified ACE2 protein as a functional receptor for the S protein of human coronavirus SARS-CoV [7]. A human coronavirus HCoV-NL63 that causes mild respiratory tract illness also employs ACE2 for cellular entry [8]. Due to the sequence similarity between SARS-CoV-2 and SARS-CoV, when SARS-COV-2 broke out, the researchers deduced that ACE2 may be a potential receptor for SARS-COV-2 [9], [10]. Zhou et al. conducted virus infectivity studies and found that exogenously expressed ACE2 is essential for SARS-CoV-2 to enter HeLa cells [11]. Later, scientists completed the construction of the crystal structure of SARS-CoV-2 S protein and receptor-binding domain (RBD) of the S protein and found that ACE2 binds to the SARS-CoV2 S protein extracellular domain/RBD with an affinity higher than the affinity of ACE2 for SARS-CoV S protein [12], [13]. Moreover, other than ACE2, SARS-CoV-2′s entry into target cell employs the cellular serine protease TMPRSS2 for S protein priming, which entails S protein cleavage at the S1/S2 and the S2′ site and allows fusion of viral and cellular membranes, a process driven by the S2 subunit [10].
2. Human tissue and Single-cell expression pattern of ACE2
The expression pattern of receptors determines the route of virus infection and transmission and is of great significance for understanding the pathogenesis and designing treatment strategies. Typical manifestations of coronavirus disease 2019 (COVID-19) are fever, fatigue, dry cough, pneumonia, and acute respiratory distress syndrome (ARDS) in severe cases [14], [15]. Other symptoms including sputum production, headache, diarrhea, anorexia, sore throat, chest pain, chills, and olfactory/taste disorders were also reported [16], [17], [18].
Through qRT-PCR research on 72 tissues of the human body, a study found that ACE2 mRNA is highly expressed in the kidney, heart, and gastrointestinal system [19]. This result is in line with what we learned from the human RNASeq database (Fig. 1 ). TMPRSS2 were dominantly expressed in the prostate, stomach and small intestine (Fig. 1). After the SARS-CoV outbreak, there were studies on the expression and distribution of ACE2 protein in humans. Through immunostaining of multiple tissues and organs of the human body, the researchers found that ACE2 protein is expressed in large amounts in type I (AT1) and type II alveolar epithelial cells (AT2), small intestinal epithelial cells [20].
Fig. 1.
Dotplot deciphering tissue expression pattern of ACE2 and TMPRSS2. RNA HPA tissue gene data, TPM (Transcripts Per Kilobase of exon model per Million mapped reads) represent transcript expression levels summarized per gene in 36 tissues based on RNA-Seq, https://www.proteinatlas.org/about/download.
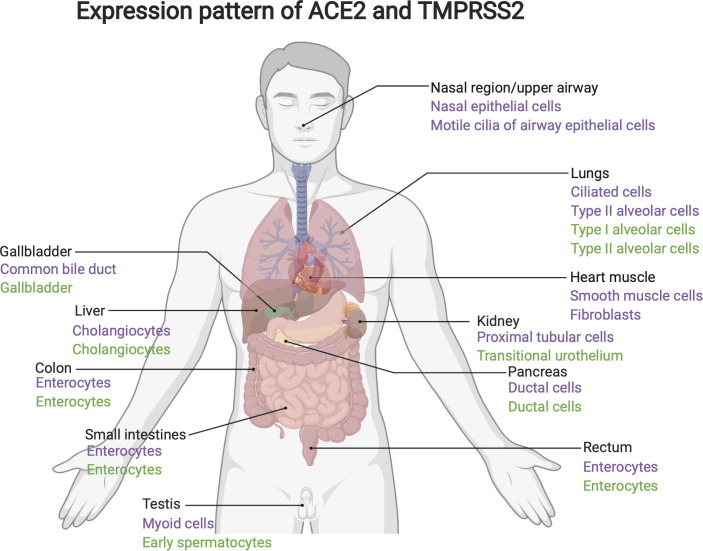
Since the SARS-CoV-2 outbreak, many studies have used public single-cell RNASeq databases to analyze the expression and distribution of ACE2 at the cell level in different organs of human. In lungs, studies found that ACE2 mainly expressed in AT2 and ciliated cells. While TMPRSS2 are mainly expressed in AT2, AT1 and club cells, and the expression level is higher than ACE2 [21], [22], [23]. In the nasal region/upper airway, ACE2 were found predominantly expressed in nasal epithelial cells and the motile cilia of airway epithelial cells, which likely represents the initial site of SARS-CoV-2 viral entry [22], [24], [25]. In the gastrointestinal system, ACE2 and TMPRSS2 were both highly expressed in enterocytes of ileum, colon and rectum [22], [26]. In the liver, ACE2 were specifically expressed in cholangiocytes [22], [27]. In the kidney, ACE2 is abundantly expressed in all subtypes of renal proximal tubule cells [22], [28]. Moreover, in the pancreas, by scRNA-Seq and ex vivo analyses, Kusmartseva and colleagues demonstrated prominent expression of ACE2 in ductal epithelium and microvasculature [29]. Based on data from above references and data from single cell transcriptomic datasets (https://www.proteinatlas.org/about/assays+annotation#normalization_rna), we concluded major cell types from different organs that expressing ACE2 and TMPRSS2 (Fig. 2 ). The tissue expression pattern of ACE2 and TMPRSS2 in non-human primates (NHP) are similar with that in human. In NHP, ACE2 was found expressing in Type II Pneumocytes (AT2) of lungs, and ACE2/TMPRSS2 were found co-expressing in absorptive enterocytes of ileum [21].
Fig. 2.
Major cell types from different organs/tissues that expressing ACE2 and TMPRSS2. Cell types expressing ACE2 were marked in purple. Cell types expressing TMPRSS2 were marked in green. This figure is created with Biorender.com.
3. ACE2 expression divergence among different populations and during virus infection
Due to the different infection rates of SARS-CoV-2 in different populations, the researchers suspect that the human ACE2 (hACE2) has different expression patterns in different populations. Old age and male sex are significant risk factors for severe SARS-CoV-2 infections [14], [30]. Cigarette smoking is also strongly associated with adverse outcomes from COVID-19 [31], [32]. By analyzing RNA data from different databases, Smith et al. found hACE2 expression was equivalent between men and women and between age groups (>70 years old versus <29 years old). Using single-cell sequencing data, they demonstrated that hACE2 was expressed in a subset of secretory cells in the respiratory tract. Chronic smoke exposure triggers the expansion of this cell population and a concomitant increase in hACE2 expression. In contrast, quitting smoking decreases the abundance of these secretory cells and reduces hACE2 levels [33]. Another study also found that significantly higher hACE2 gene expression in former smoker’s lung compared to non-smoker’s lung, while proved no significant difference in ACE2 gene expression between ethnic groups (Asian and Caucasian), age groups (>60 years old versus <60 years old), or gender groups (male versus female) [34]. Moreover, prior research has proved that hACE2 was upregulated in the population that suffered failing heart or myocardial infarction [35], [36].
Other than cigarette smoking, researchers found hACE2 expression in airway epithelial cells could be upregulated by various viral infections including influenza, respiratory syncytial virus, SARS-CoV, and MERS-CoV. They further proved that dsRNA mimic poly(I:C) and interferons could both induce upregulation of hACE2. Therefore, researchers conclude that hACE2 is an interferon-stimulated gene (ISG), and inflammation stimulation leads to its up-regulation and further promotes SARS-CoV-2 infection [34]. Another study found that in cells expressing hACE2, the expression of ISGs was often upregulated. By treating primary human upper airway basal cells with IFN-a, they also found upregulation of hACE2 expression [21]. However, recent research identified a novel, transcriptionally independent truncated isoform of ACE2 (dACE2), but not hACE2, as an ISG. dACE2, which lacks 356 amino-terminal amino acids, was non-functional in binding the SARS-CoV-2 spike protein and as a carboxypeptidase [37]. Therefore, whether hACE2 could act as an ISG needs to be further verified.
4. ACE2 interaction with SARS-CoV-2 S protein
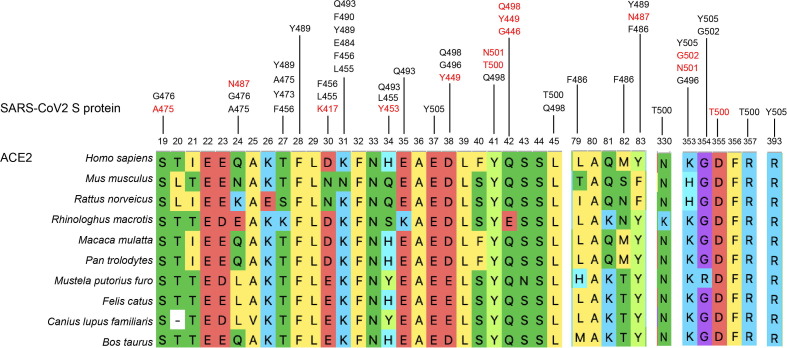
SARS-CoV-2 RBD (amino acids 319–541 of S protein) is the key region in SARS-CoV-2 S protein that interacts with the hACE2 receptor [38], [39], [40]. The structures of the hACE2 receptor in complex with the SARS-CoV-2 RBD/ the C-terminal domain of the spike protein have been constructed to reveal the mechanisms of binding of SARS-CoV-2. The major RBD interaction regions in ACE2 include helix H1 (Q24–Q42), a loop in a beta-sheet (K353–R357), and the end of helix H2 (L79–Y83). A series of hydrophilic residues located along the interface was found to form a solid network of H-bond and salt bridge interactions (Fig. 3 ). These strong polar contacts include the SARS-CoV-2-RBD residue A475 interacting with hACE2 residue S19, N487 with Q24, and Y453 with H34. Residue K417 was shown to contribute ionic interactions with hACE2 D30. G446, Y449, G496, Q498, T500, and G502 are in close proximity with hACE2 amino acids D38, Y41, Q42, K353, and D355. SARS-CoV-2-RBD Y489 and F486 pack against hACE2 residues F28, L79, M82, and Y83, forming a small patch of hydrophobic interactions at the interface. Overall, the virus-receptor engagement is dominated by polar contacts mediated by the hydrophilic residues [41], [42]. Moreover, scientists found that the glycosylation of the hACE2 contributes substantially to the binding of the virus. Both the ACE2 receptor and the spike protein are heavily glycosylated, including at sites near their binding interface [43], [44]. Atomistic molecular dynamics simulations showed contrasting effects of ACE2 glycosylation, weakening the binding of SARS- CoV-2 spike in the case of the N90 glycan, strengthening the binding in case of the N322 glycan, and being neutral in case of other sites [45].
Fig. 3.
Segments where hACE2 interacts with SARS-COV-2 S protein, and sequence alignment of these segments in other putative host animals. The amino acids marked in red indicate they have H-bond or salt bridge interactions with hACE2. The listed animals are Homo sapiens (human), Mus musculus (mouse), Rattus norveicus (rat), Rhinologhus macrotis (bat), Macaca mulatta (monkey), Pan trolodytes (orangutan), Mustela putorius furo (ferret), Felis catus (cat), Canius lupus familiaris (dog) and Bos taurus (cow).
Sequence differences of ACE2 among host animals may determine the adaptability of SARS-CoV-2 to different hosts. Bats are considered as the reservoir host animals of SARS-CoV-2 [11], [46]. Liu and colleagues determined the complex structure of SARS-CoV-2 RBD and bat ACE2 from Rhinolophus macrotis (bACE2-Rm). SARS-CoV-2 RBD binds to bACE2-Rm with a lower affinity than that to hACE2. Mutational analysis revealed that the Y41 and E42 of bACE2-Rm (Y41 and Q42 in hACE2), which contains variations in many bat species, play central roles in the interaction with SARS-CoV-2 RBD [47]. Two recent studies show that New World monkey, koala, and mouse ACE2 cannot serve as functional receptors to support SARS-CoV-2 entry. They verified residues H41/E42 in New World monkey ACE2, T31 in koala ACE2, H353 in mouse ACE2 restrict SARS-CoV-2′s entry. K353 in human ACE2 can hydrogen bond with G502 of the SARS-CoV-2 spike protein, stabilizing the ACE2-spike complex [48], [49].
Since sequence variants of ACE2 among populations may influence the spreading of SARS-CoV-2, several studies focused on the binding of the proteins encoded by different human ACE2 allelic variants [38], [50], [51], [52], [53], [54]. A study compared the structural and functional consequences of 17 ACE2 allelic variants and found rs73635825 (S19P) and rs143936283 (E329G) showed noticeable variations in their intermolecular interactions with the SARS-CoV-2 protein [51]. Another study found putative missense ACE2 variants including rs147311723 (L731F, AF (allele frequency) = 0.01 in African), rs372272603 (R219C, AF = 7 × 10−4 in European), rs373025684 (S547C, AF = 4 × 10−4 in European) and rs142984500 (H378R, AF = 2 × 10−4 in European). They also found rs142984500 (H378R, AF = 2 × 10−4 in European) could directly weaken the binding of catalytic metal atoms to decrease ACE2 catalytic activity, and rs73635825 (S19P, AF = 3 × 10−3 in African) could distort the most important helix to interact with the S protein [52].
5. ACE2 interaction with SARS-CoV-2 variants
With the global pandemic of SARS-CoV-2, many variants have emerged. Variants including B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), and B.1.617.2 (Delta) have become the main strains currently circulating. Studies have proved B.1.1.7, B.1.351, P.1, and B.1.617.2 showed significantly increased transmission [55], [56]. Compared with the original epidemic strains, they all have amino acid mutations at the RBD region. All RBD substitutions of these variants are concentrated at 417, 452, 478, 484, 490, and 501 (Table 1 ). Changes in these sites that affect interaction with hACE2 will greatly affect the spread of the virus. N501Y that shared by B.1.1.7, B.1.351, and P.1 enhances the binding affinity to hACE2 [57], [58], [59]. A single K417N substantially reduced hACE2 binding, while E484K slightly reduced binding affinity to hACE2, but this reduction will be restored when combined with N501Y. The triple mutant K417N/E484K/N501Y (as in B.1.351) had a similar binding ability to wild type [60]. L452R mutation (in B.1.617 and C.37) reinforces affinity toward hACE2 [61]. Interestingly, B.1.351 and P.1 acquired much higher binding ability to mouse and mink ACE2 receptors [62]. Mutations in the SARS-CoV-2 protein can lead to the cross‐species transmission of the virus. A mouse-adapted SARS-CoV-2 virus MASCp36 which bears N501Y, Q493H, and K417N mutations at RBD of S protein showed increased infectivity in mouse lungs. The structure of mouse ACE2 and MASCp36 RBD complex showed these mutations contributed to the tight binding of the MASCp36 to mouse ACE2 [63], [64]. A recent study found T372A which is present in all human SARS-CoV-2 RBD sequences but not in closely related viruses from bats and pangolins bound hACE2 with higher affinity in experimental binding assays. This mutation likely contributed to SARS-CoV-2′s emergence from animal reservoirs to humans [65].
Table 1.
SARS-CoV-2 variants with amino acid substitutions on RBD.
| WHO label | Pango lineage | RBD substitutions | Earliest documented samples |
|---|---|---|---|
| Alpha | B.1.1.7 | N501Y | United Kingdom, Sep-2020 |
| Beta | B.1.351 | K417N E484K N501Y | South Africa, May-2020 |
| Gamma | P.1 | K417T E484K N501Y | Brazil, Nov-2020 |
| Delta | B.1.617.2 | L452R T478K | India, Oct-2020 |
| Eta | B.1.525 | E484K | Multiple countries, Dec-2020 |
| Lota | B.1.526 | E484K | United States of America, Nov-2020 |
| Kappa | B.1.617.1 | L452R E484Q | India, Oct-2020 |
| Lambda | C.37 | L452Q F490S | Peru, Dec-2020 |
6. Conclusion and perspectives
Here, we reviewed the function, expression pattern in the human body and population, interaction with SARS-CoV-2 S protein of ACE2. In the human body, ACE2 is abundantly expressed in the gastrointestinal system, kidney, testis, and heart. In most of the detectable tissues, ACE2 tends to express in epidermal cells. In the lungs, ACE2 is mainly expressed in AT2 and ciliated cells. In the nasal region/upper airway, ACE2 was found predominantly expressed in nasal epithelial cells and the motile cilia of airway epithelial cells. The expression level of ACE2 is upregulated in smokers, but there is no significant difference between different ages and genders. ACE2 interacts with SARS-CoV-2 RBD, and some of the residues including Y41, Q42, K353 that form polar contacts with RBD are essential for binding. Sequence differences of ACE2 in these residues among host animals determine the adaptability of SARS-CoV-2 to different hosts. Moreover, sequence variants of ACE2 among populations may influence the spreading of SARS-CoV-2. An ACE2 allelic variant rs73635825 (S19P) whose allele frequency is 3 × 10−3 in African could distort the most important helix to interact with the S protein. Other than sequence and expression pattern, glycosylation on ACE2 amino acids can also affect virus entry. On the other hand, mutations in the SARS-CoV-2 protein can lead to the cross‐species transmission of the virus. Newly emerged SARS-CoV-2 variants with mutations like N501Y on RBD have stronger interaction with hACE2, some gained the ability to infect mice. The study of ACE2 is of great significance for finding the intermediate host and potential host of SARS-CoV-2 and predicting the transmission ability of the mutant virus.
There are some questions that need to be answered in the future. Although some studies have proved that SARS-CoV-2 can use coreceptors like NRP1 for host cell entry, other ACE2-independent receptors need to be further studied [66], [67], [68]. Moreover, whether the high expression of ACE2 in certain organs is related to the sequelae of COVID-19 needs to be further followed up and studied. It is still controversial that whether ACE2 could act as an ISG to facilitate infection. Will other newly emerged SARS-CoV-2 with mutations on RBD have stronger interaction with ACE2? Except for glycosylation, whether other post-translational modifications on ACE2 amino acids could affect virus entry. Moreover, soluble ACE2 as a promising therapeutic candidate that neutralizes SARS-CoV-2 infection needs to be further studied.
Conflict of interest statement
The authors declare that there are no conflicts of interests.
Acknowledgements
This work was supported by the China Postdoctoral Science Fund (No. 2019M664012 and No. 2020T130135ZX).
Author contributions
Ruiting Li: Data Curation, Writing - Original Draft. Chengfeng Qin: Conceptualization, Writing - Review & Editing.
References
- 1.Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., Breitbart R.E., Acton S. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ. Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 2.Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 3.Vickers C., Hales P., Kaushik V., Dick L., Gavin J., Tang J., Godbout K., Parsons T., Baronas E., Hsieh F., Acton S., Patane M., Nichols A., Tummino P. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J. Biol. Chem. 2002;277:14838–14843. doi: 10.1074/jbc.M200581200. [DOI] [PubMed] [Google Scholar]
- 4.Rushworth C.A., Guy J.L., Turner A.J. Residues affecting the chloride regulation and substrate selectivity of the angiotensin-converting enzymes (ACE and ACE2) identified by site-directed mutagenesis. FEBS J. 2008;275:6033–6042. doi: 10.1111/j.1742-4658.2008.06733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zisman L.S., Keller R.S., Weaver B., Lin Q., Speth R., Bristow M.R., Canver C.C. Increased angiotensin-(1–7)-forming activity in failing human heart ventricles: evidence for upregulation of the angiotensin-converting enzyme Homologue ACE2. Circulation. 2003;108:1707–1712. doi: 10.1161/01.CIR.0000094734.67990.99. [DOI] [PubMed] [Google Scholar]
- 6.Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.-C., Turner A.J., Raizada M.K., Grant M.B., Oudit G.Y. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ. Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pohlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W.u., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 Cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Yan B., Zhan F.X., Wang Y.Y., Xiao sG.F., Shi Z.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shang J., Ye G., Shi K.e., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan W.J., Ni Z.Y., Hu Y.U., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y.i., Zhang L.i., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L.i., Xie J., Wang G., Jiang R., Gao Z., Jin Q.i., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 18.Giacomelli A., Pezzati L., Conti F., Bernacchia D., Siano M., Oreni L., Rusconi S., Gervasoni C., Ridolfo A.L., Rizzardini G., et al. Self-reported olfactory and taste disorders in patients with severe acute respiratory coronavirus 2 infection: A cross-sectional study. Clin. Infect. Dis. 2020;71:889–890. doi: 10.1093/cid/ciaa330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harmer D., Gilbert M., Borman R., Clark K.L. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532:107–110. doi: 10.1016/s0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- 20.Hamming I., Timens W., Bulthuis MLC, Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:e1019. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sungnak W., Huang N.I., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., Worlock K.B., Yoshida M., Barnes J.L. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y., Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020;202:756–759. doi: 10.1164/rccm.202001-0179LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.C. Wu, S. Zheng, Y. Chen, M. Zheng, Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCoV, in the nasal tissue [Preprint.] medRxiv (2020), 2020.2002.2011.20022228, 10.1101/2020.01.26.919985. [DOI]
- 25.Lee I.T., Nakayama T., Wu C.T., Goltsev Y., Jiang S., Gall P.A., Liao C.K., Shih L.C., Schurch C.M., McIlwain D.R., et al. ACE2 localizes to the respiratory cilia and is not increased by ACE inhibitors or ARBs. Nat. Commun. 2020;11:5453. doi: 10.1038/s41467-020-19145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.H. Zhang, Z. Kang, H. Gong, D. Xu, J. Wang, Z. Li, X. Cui, J. Xiao, T. Meng, W. Zhou, et al. (2020). The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes [Preprint]. bioRxiv, 2020.2001.2030.927806, 10.1101/2020.01.30.927806. [DOI]
- 27.X. Chai, L. Hu, Y. Zhang, W. Han, Z. Lu, A. Ke, J. Zhou, G. Shi, N. Fang, J. Fan, et al., Specific ACE2 expression in cholangiocytes may cause liver damage after 2019-nCoV infection [Preprint]. bioRxiv (2020), 2020.2002.2003.931766, 10.1101/2020.02.03.931766. [DOI]
- 28.W. Lin, L. Hu, Y. Zhang, J.D. Ooi, T. Meng, P. Jin, X. Ding, L. Peng, L. Song, Z. Xiao, et al., Single-cell analysis of ACE2 expression in human kidneys and bladders reveals a potential route of 2019-nCoV infection [Preprint], bioRxiv. (2020), 2020.2002.2008.939892, 10.1101/2020.02.08.939892. [DOI] [PMC free article] [PubMed]
- 29.Kusmartseva I., Wu W., Syed F., Van Der Heide V., Jorgensen M., Joseph P., Tang X., Candelario-Jalil E., Yang C., Nick H., et al. Expression of SARS-CoV-2 entry factors in the pancreas of normal organ donors and individuals with COVID-19. Cell Metab. 2020;32:e1046. doi: 10.1016/j.cmet.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.C. Ma, J. Gu, P. Hou, L. Zhang, Y. Bai, Z. Guo, H. Wu, B. Zhang, P. Li, X. Zhao, Incidence, clinical characteristics and prognostic factor of patients with COVID-19: a systematic review and meta-analysis [Preprint], medRxiv (2020), 2020.2003.2017.20037572, 10.1101/2020.03.17.20037572. [DOI]
- 31.Patanavanich R., Glantz S.A. Smoking is associated with COVID-19 progression: A meta-analysis. Nicotine Tobacco Res. 2020;22:1653–1656. doi: 10.1093/ntr/ntaa082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Q., Meng M., Kumar R., Wu Y., Huang J., Lian N., Deng Y., Lin S.u. The impact of COPD and smoking history on the severity of COVID-19: A systemic review and meta-analysis. J. Med. Virol. 2020;92:1915–1921. doi: 10.1002/jmv.25889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith J.C., Sausville E.L., Girish V., Yuan M.L., Vasudevan A., John K.M., Sheltzer J.M. Cigarette smoke exposure and inflammatory signaling increase the expression of the SARS-CoV-2 receptor ACE2 in the respiratory tract. Dev. Cell. 2020;53:514–529.e3. doi: 10.1016/j.devcel.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.G. Cai, Bulk and single-cell transcriptomics identify tobacco-use disparity in lung gene expression of ACE2, the receptor of 2019-nCov [Preprint], medRxiv, 2020.2002.2005.20020107, 10.1101/2020.02.05.20020107. [DOI]
- 35.Goulter A.B., Goddard M.J., Allen J.C., Clark K.L. ACE2 gene expression is up-regulated in the human failing heart. BMC Med. 2004;2:19. doi: 10.1186/1741-7015-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.L.M. Burrell, J. Risvanis, E. Kubota, R.G. Dean, P.S. MacDonald, S. Lu, C. Tikellis, S.L. Grant, R.A. Lew, A.I. Smith, et al., Myocardial infarction increases ACE2 expression in rat and humans. Eur. Heart J. 26 (2005), 369-375; discussion 322-364, 10.1093/eurheartj/ehi114. [DOI] [PubMed]
- 37.Onabajo O.O., Banday A.R., Stanifer M.L., Yan W., Obajemu A., Santer D.M., Florez-Vargas O., Piontkivska H., Vargas J.M., Ring T.J., Kee C., Doldan P., Tyrrell D.L., Mendoza J.L., Boulant S., Prokunina-Olsson L. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat. Genet. 2020;52:1283–1293. doi: 10.1038/s41588-020-00731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Othman H., Bouslama Z., Brandenburg J.-T., da Rocha J., Hamdi Y., Ghedira K., Srairi-Abid N., Hazelhurst S. Interaction of the spike protein RBD from SARS-CoV-2 with ACE2: Similarity with SARS-CoV, hot-spot analysis and effect of the receptor polymorphism. Biochem. Biophys. Res. Commun. 2020;527:702–708. doi: 10.1016/j.bbrc.2020.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song W., Gui M., Wang X., Xiang Y.e., Heise M.T. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:e1007236. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi C., Sun X., Ye J., Ding L., Liu M., Yang Z., Lu X., Zhang Y., Ma L., Gu W., Qu A., Xu J., Shi Z., Ling Z., Sun B. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell. Mol. Immunol. 2020;17:621–630. doi: 10.1038/s41423-020-0458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:e899. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q.i., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 43.Shajahan A., Archer-Hartmann S., Supekar N.T., Gleinich A.S., Heiss C., Azadi P. Comprehensive characterization of N- and O- glycosylation of SARS-CoV-2 human receptor angiotensin converting enzyme 2. Glycobiology. 2021;31:410–424. doi: 10.1093/glycob/cwaa101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mehdipour A.R., Hummer G. Dual nature of human ACE2 glycosylation in binding to SARS-CoV-2 spike. Proc. Natl. Acad. Sci. U. S. A. 2021;1182100425118 doi: 10.1073/pnas.2100425118. e2100425118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Wang P., Liu D., Yang J., Holmes E.C., et al. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr. Biol. 2020;30:e2193. doi: 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu, K., Tan, S., Niu, S., Wang, J., Wu, L., Sun, H., Zhang, Y., Pan, X., Qu, X., Du, P., et al. Cross-species recognition of SARS-CoV-2 to bat ACE2. Proc. Natl. Acad. Sci. U. S. A. 118(2021), e2020216118. 10.1073/pnas.2020216118. [DOI] [PMC free article] [PubMed]
- 48.Liu Y., Hu G., Wang Y., Ren W., Zhao X., Ji F., Zhu Y., Feng F., Gong M., Ju X., et al. Functional and genetic analysis of viral receptor ACE2 orthologs reveals a broad potential host range of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2025373118. e2025373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren W., Zhu Y., Wang Y., Shi H., Yu Y., Hu G., Feng F., Zhao X., Lan J., Wu J., Kenney D.J., Douam F., Tong Y., Zhong J., Xie Y., Wang X., Yuan Z., Zhou D., Zhang R., Ding Q., Wang D. Comparative analysis reveals the species-specific genetic determinants of ACE2 required for SARS-CoV-2 entry. PLoS Pathog. 2021;17:e1009392. doi: 10.1371/journal.ppat.1009392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao Y., Li L., Feng Z., Wan S., Huang P., Sun X., Wen F., Huang X., Ning G., Wang W. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020;6:11. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hussain M., Jabeen N., Raza F., Shabbir S., Baig A.A., Amanullah A., Aziz B. Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J. Med. Virol. 2020;92:1580–1586. doi: 10.1002/jmv.25832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo X., Chen Z., Xia Y., Lin W., Li H. Investigation of the genetic variation in ACE2 on the structural recognition by the novel coronavirus (SARS-CoV-2) J. Transl. Med. 2020;18:321. doi: 10.1186/s12967-020-02486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Calcagnile M., Forgez P., Iannelli A., Bucci C., Alifano M., Alifano P. Molecular docking simulation reveals ACE2 polymorphisms that may increase the affinity of ACE2 with the SARS-CoV-2 Spike protein. Biochimie. 2021;180:143–148. doi: 10.1016/j.biochi.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darbani B. The expression and polymorphism of entry machinery for COVID-19 in human: juxtaposing population groups, gender, and different tissues. Int. J. Environ. Res. Public Health. 2020;17:3433. doi: 10.3390/ijerph17103433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., Sillitoe J., Kwiatkowski D.P., Flaxman S., Ratmann O., Bhatt S., Hopkins S., et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 56.Campbell F., Archer B., Laurenson-Schafer H., Jinnai Y., Konings F., Batra N., Pavlin B., Vandemaele K., Van Kerkhove M.D., Jombart T., et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill. 2021;26:2100509. doi: 10.2807/1560-7917.ES.2021.26.24.2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laffeber C., de Koning K., Kanaar R., Lebbink J.H.G. Experimental evidence for enhanced receptor binding by rapidly spreading SARS-CoV-2 variants. J. Mol. Biol. 2021;433:167058. doi: 10.1016/j.jmb.2021.167058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:e1220. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu X., Mannar D., Srivastava S.S., Berezuk A.M., Demers J.-P., Saville J.W., Leopold K., Li W., Dimitrov D.S., Tuttle K.S., Zhou S., Chittori S., Subramaniam S., Bhella D. Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies. PLoS Biol. 2021;19:e3001237. doi: 10.1371/journal.pbio.3001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuan M., Huang D., Lee C.D., Wu N.C., Jackson A.M., Zhu X., Liu H., Peng L., van Gils M.J., Sanders R.W., et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science. 2021;373:818–823. doi: 10.1126/science.abh1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Motozono C., Toyoda M., Zahradnik J., Saito A., Nasser H., Tan T.S., Ngare I., Kimura I., Ikeda T., Nakagawa S.O., Ueno T., Sato U., et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29:1124–1136.e11. doi: 10.1016/j.chom.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang R., Zhang Q., Ge J., Ren W., Zhang R., Lan J., Ju B., Su B., Yu F., Chen P., Liao H., Feng Y., Li X., Shi X., Zhang Z., Zhang F., Ding Q., Zhang T., Wang X., Zhang L. Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity. 2021;54:1611–1621.e5. doi: 10.1016/j.immuni.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gu H., Chen Q.i., Yang G., He L., Fan H., Zhou X., Geng S., Sheng X., Jiang S., Sun S., Qin C.F., Zhou Y., et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.S. Sun, H. Gu, L. Cao, Q. Chen, G. Yang, R.T. Li, H. Fan, Q. Ye, Y.Q. Deng, X. Song, et al., Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2 [Preprint], bioRxiv (2020), 2020.2011.2010.377333, 10.1101/2020.11.10.377333. [DOI] [PMC free article] [PubMed]
- 65.Kang L., He G., Sharp A.K., Wang X., Brown A.M., Michalak P., Weger-Lucarelli J. A selective sweep in the Spike gene has driven SARS-CoV-2 human adaptation. Cell. 2021;184:4392–4400.e4. doi: 10.1016/j.cell.2021.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Teesalu T., Hepojoki J., Vapalahti O., Stadelmann C., Balistreri G., Simons M., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Daly J.L., Simonetti B., Klein K., Chen K.-E., Williamson M.K., Antón-Plágaro C., Shoemark D.K., Simón-Gracia L., Bauer M., Hollandi R., Greber U.F., Horvath P., Sessions R.B., Helenius A., Hiscox J.A., Teesalu T., Matthews D.A., Davidson A.D., Collins B.M., Cullen P.J., Yamauchi Y. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zamorano Cuervo N., Grandvaux N. ACE2: Evidence of role as entry receptor for SARS-CoV-2 and implications in comorbidities. eLife. 2020;9:e61390. doi: 10.7554/eLife.61390. [DOI] [PMC free article] [PubMed] [Google Scholar]