Abstract
Severe acute respiratory syndrome coronavirus-2 causes coronavirus disease 2019, a pandemic which was originated from Wuhan city of China. The pandemic has affected millions of people worldwide. The pathogenesis of SARS-CoV-2 is characterized by a cytokine storm in the blood (cytokinemia) and tissues, especially the lungs. One of the major repercussions of this inflammatory process is the endothelial injury-causing intestinal bleeding, coagulopathy, and thromboembolism which result in various sudden and unexpected post-COVID complications including kidney failure, myocardial infarction, or multiorgan failure. In this review, we have summarized the immune responses, biochemical changes, and inflammatory responses in the human body after infection with the SARS-CoV-2 virus. The increased amount of inflammatory cytokines, chemokines, and involvement of complement proteins in inflammatory reaction increase the risk of occurrence of disease.
Keywords: SARS-CoV-2, Coronavirus, Coagulopathy, Inflammation, Cytokines storm, Interferons, Interleukins, Immune system
Graphical abstract
Contribution of interleukins, TNF-α and proinflammatory cytokines in the pathogenesis of COVID-19.
1. Introduction
Coronavirus (CoV) is positive-sense with single-stranded RNA (ss-RNA) virus and is zoonotic. The coronavirus disease of 2019 (or COVID-19) has killed millions of people and is counted as a major source for the initiation of many diseases. CoV belongs to the family of Coronaviridae while Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) belongs to the family of beta-coronavirus [1]. Its closest relative among human coronaviruses is Severe Acute Respiratory Syndrome Coronavirus-2 (or SARS-CoV-2). CoV comprises 4 genus namely alpha coronaviruses, beta coronaviruses, gamma coronaviruses, and delta coronaviruses [2]. These viruses cause respiratory ailments in humans and are responsible for causing gastrointestinal ailments in animals. In the year 2003, the universe was firstly attacked by the SARS-CoV, which results in an indefinite number of deaths. The virus was known to originate from the seafood market of Wuhan city in China. Earlier coronavirus was known by the name of WH-Human-1. During the genomic investigation of a virus, it was identified that virus is originated from bat coronavirus that allows the host cell to bind with human Angiotensin Converting Enzyme type 2 (ACE2) present on epithelial cells [3]. A respiratory disorder with unspecified cause was reported in December 2019 in the Wuhan city of China [4]. Later, the virus spread at a higher rate all over the world, and World Health Organization gives the virus an official name i.e. SARS-CoV-2 and COVID-19.
The contact of an individual with microdroplets and virus particles present in the contaminated inanimate objects helps in the transmission of viruses such as SARS-CoV-2 and others etc. [5]. The bronchial epithelium and type-II ACE2 pneumocytes cells of bronchial and alveolar epithelium are the main target cells for viruses [6]. Viral infection in bronchial epithelium progresses and can cause infections in other organs like liver, kidney, and lungs, etc. The division in basal membranes, initiation of autophagy, and reduction in the expression of ACE2 are results of SARS-CoV infections [7]. Production of type-I and III interferons (IFNs) are the prime function of the early defense mechanism played by the infected cell [8]. The CoV is effective to inhibit the induction of type-I and III IFNs because they are highly sensitive to their anti-viral effects [9]. The spread of infection from one cell to nearby cells and viremia occurred due to the release of virions in a large quantity [10]. All the abbreviations used in this manuscript have been enlisted in Table 1 . And, all the drugs studied in this manuscript have been enlisted in Table 2 along with their general, IUPAC names, their mechanism of action, and the respective PubChem IDs.
Table 1.
List of abbreviations
| S. No. | Abbreviation | Full Name |
|---|---|---|
| 1. | CoV | Coronavirus |
| 2. | COVID-19 | Coronavirus disease of 2019 |
| 3. | MERS-CoV | Middle East Respiratory Syndrome Coronavirus |
| 4. | SARS-CoV-2 | Severe Acute Respiratory Syndrome Coronavirus-2 |
| 5. | SARS-CoV | Severe Acute Respiratory Syndrome Coronavirus |
| 6. | ACE2 | Angiotensin Converting Enzyme 2 |
| 7. | IFNs | Interferons |
| 8. | ILs | Interleukins |
| 9. | ss-RNA | Single-stranded-RNA |
| 10. | ds-RNA | Double-stranded-RNA |
| 11. | NF-kB | Nuclear Factor-Kappa B |
| 12. | TMPRSS2 | Transmembrane Serine Protease 2 |
| 13. | TNF | Tumor Necrosis Factor |
| 14. | TLR | Toll Like Receptor |
| 15. | RBD | Receptor Binding Domain |
Table 2.
List of drugs with general, IUPAC names, PubChem IDs with mechanism.
| S. No. | Drug general name | IUPAC name | PubChem ID | Mechanism | Reference |
|---|---|---|---|---|---|
| 1. | Remdesivir | 2-ethylbutyl (2S)-2-[[[(2R,3S,4R,5R)-5-(4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-cyano-3,4-dihydroxyoxolan-2-yl]methoxy-phenoxyphosphoryl]amino]propanoate | 121304016 | Inhibition of RNA Polymerase | [170] |
| 2. | Baricitinib | 2-[1-ethylsulfonyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrazol-1-yl]azetidin-3-yl]acetonitrile | 44205240 | Inhibition of Janus Kinase 1 and 2 | [175] |
| 3. | Ruxolitinib | (3R)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrazol-1-yl]propanenitrile | 25126798 | Inhibition of Janus Kinase | [176] |
| 4. | Nafamostat mesylate | (6-carbamimidoylnaphthalen-2-yl) 4-(diaminomethylideneamino)benzoate;methanesulfonic acid | 5311180 | Inhibitor of TMPRSS2 | [178] |
| 5. | Camostat mesylate | [4-[2-[2-(dimethylamino)-2-oxoethoxy]-2-oxoethyl]phenyl] 4-(diaminomethylideneamino)benzoate;methanesulfonic acid | 5284360 | Inhibitor of TMPRSS2 | [178] |
| 6. | Warfarin | 4-hydroxy-3(3-oxo-1-phenylbutyl)-2H-chromen-2-one | 54678486 | Inhibition of blood clots | [180] |
| 7. | Aspirin | 2-acetyloxybenzoic acid | 2244 | Inhibition of initiation of inflammatory reactions and blood clots | [181] |
2. Structural organization of coronavirus
The genomic studies confirm that the virus is composed of ss-RNA, having 30Kb nucleotides [11]. Only four proteins are encoded by these nucleotides. These proteins are spike protein, membrane protein, envelope protein, and nucleocapsid protein and are abbreviated as S, M, E, and N proteins respectively [12]. The crown-like shape of a virus is due to club-shaped glycoproteins of the S-protein [13]. The genome of SARS-CoV-2 is 96% similar to bat coronavirus but this similarity is very less with SARS-CoV [14]. The single-stranded RNA is associated with the 5’ poly cap and poly-A tail. The presence of 3’-5’ exoribonucleases in coronavirus helps the virus to be different from other RNA viruses [15]. The virus is a betacoronavirus, found to affect the lower respiratory tract at a higher rate. Upon measurement, the diameter of spherical coronavirus is measured 80-120 nm [16]. ORF1a and ORF1b are the replicases responsible for coding pp1a and pp1b polyproteins [17]. This complex structure of coronavirus is responsible for its pathogenicity.
3. SARS-CoV-2 infection cycle and pathogenesis
Amongst S, E, M, and N proteins, only S-protein attain much interest towards researchers to investigate the SARS-CoV-2 pathology mechanism. Anatomically, Receptor-Binding-Domain (RDB) is present in the CoV, whose main function is the ligand binding on the cell membrane of the host [18]. T and B-cells address the epitopes on RDB signals the immune system to continue the production of neutralizing antibodies [19]. S1 contains the RDB and is a type I trimeric glycoproteins. The attachment of trimeric spike protein of the SARS-CoV-2 with ACE2 is considered as the first step for this viral pathogenicity [20]. The ACE2 enzyme is present in the epithelial cells of the lungs. The relationship between the SARS-CoV-2 and ACE2 expressions is unidentified yet. Upon binding of RDB and ACE2, S-protein is cleaved into S1 and S2 subunits once the cell undergoes conformational changes [21]. Transmembrane Serine Protease 2 (or TMPRSS2) is a serine protease which plays an important function in cleavage events of S-protein, by allowing the S2 subunit to mediate the fusion of the virus envelope with the cell membrane [22].
Once the spike protein binds to the ACE2 receptor in host, it undergoes molecular events like internalization and fusion [23]. These events help the virus to release its genomic RNA in the cytoplasm [24], which further helps in the translation of pp1a and pp1ab polyproteins. Polyprotein pp1a encodes the non-structural protein ranging from 1 to 11 and Polyprotein pp1ab encodes the proteins from 1 to 16 [25]. Upon cleavage of S-protein, the S2 subunit governs the passage of RNA of a virus into the cytoplasm of a target cell, and later polyproteins like pp1a and pp1b undergo the translation process if the viral RNA serves as a template for these polyproteins [26]. These polyproteins split into non-structural protein2, 3, 4, 5, 6, 7, 8, and 9. Splitting of these proteins signals the membrane to rearrange itself and forms the site where viral replication and transcription complexes are anchored. The rough endoplasmic reticulum is the site where these non-structural proteins are translated themselves [27]. And, the endoplasmic reticulum Golgi intermediate compartment (ERGIC) is the site where these non-structural proteins are assembled [28]. The assembly of these proteins allows the virions to move out of the cell by exocytosis through the secretory pathway [29], which later damages the epithelial and endothelial cells [30].
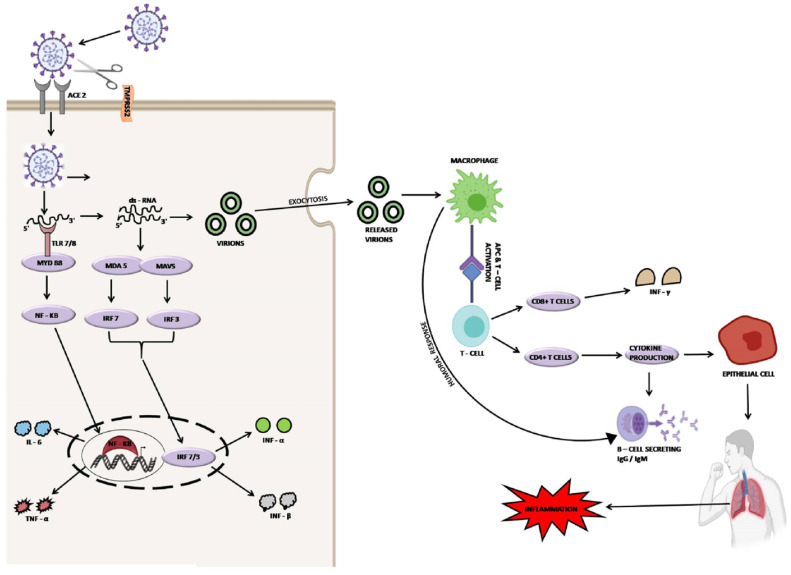
Transmission of virus and enhanced presentation of ACE2 on the epithelium, blood vessels, and fibroblasts increase the risk of spread of SARS-CoV-2 infection to the whole body. Other than ACE2, various proteins such as glucose-regulated proteins (GRP), alanyl aminopeptidase (ANPEP), dendritic cell-specific intercellular adhesion molecule-2-grabbing non-integrin-1 (DCSIGN1), dipeptidyl peptidase-4 (DPP4), heparin sulfate, neuropilin-1 (NRP-1), a cell adhesion molecule 5 (CEACAM5), angiotensin II type 2 receptor (AGTR2) and ganglioside GM1 are known to interact with the SARS-CoV-2 [31]. The furin cleaved substrates i.e. NRP-1 is known to be expressed abundantly in neurons, blood vessels, and respiratory tract and thus helps in the pathogenesis of SARS-CoV-2 infection [32]. The spike protein triggers the TLR4, ssRNA which further helps in the activation of TLR7 and double-stranded-RNA (dsRNA) [33]. The activation of dsRNA and TLR7 signals the IRFs and NF-kB-dependent signaling pathways to initiate the production and activation of pro-inflammatory cytokines and IFNs (type-I and III). The enhanced production of inflammatory cytokines results in the cytokine storm which directly damages the lungs and initiates the shortness of breath [34], (Fig. 1 ).
Fig. 1.
Life cycle of SARS-CoV-2.
4. The immune system in infection
The immune system is an essential component of a living organism for survival. Any abnormalities in the immune system provide access to bacteria, viruses, and parasites to enter the host cell. Proper functioning of the immune system prevents the host cell from the attack of pathogens [35]. The foremost function of the immune system is the differentiation of self and non-self. The innate immune system and adaptive immune system are two parts of the immune system which work together for elimination of pathogen [36]. The complement system, antibodies, bone marrow, white blood cells, thymus, spleen and inflammation are the major components of the immune system. All these components of the immune system play important role in regulating viral infections.
4.1. Innate immune system in viral infections
Viral infections are mediated by the entry of viruses into the host cells. Generally, the entry of virus in cells that structurally contains the single-stranded RNA is first recognized by the certain receptors characterized as Pattern Recognition Receptors (PPRs) such as TLR7 and TLR8, NLR and RIG-I-like receptors (RLRs) [37]. These all receptors are expressed by alveolar macrophages and epithelial cells [38]. Some adaptor proteins are recruited by PPRs action and, lead to the further activation of transcription factors i.e. INF-regulatory factor, AP-1, and Nuclear-Factor Kappa-B (NF-kB) [39]. The generation of chemokines and type-I and type III antiviral interferons is an end product of the activation of these transcription factors [40]. The newly formed chemokines show attraction towards various immune cells involved in the innate response such as Monocytes, Dendritic cells, Natural Killer cells, and Polymorphonuclear leukocytes, etc. MCP-1, IP-10, and MIG are chemokines involved in recruiting the lymphocytes [41]. Later, they are identified by the viral antigens, and these antigens are presenting themselves in dendritic cells.
Chu and his co-authors had done the in-vitro study using human lung explants affected by SARS-CoV-2 and SARS-CoV. They found that both viruses can equally induce infections in alveolar macrophage and type-I and type-II pneumocytes. Additionally, they found, SARS-CoV has less potential to replicate itself in pulmonary tissues of the lung as compared to SARS-CoV-2 [42]. SARS-CoV-2 is not capable to regulate the expressions of immune mediators such as type-I, II, and III IFNs, but SARS-CoV can regulate the expressions of IFNs [43]. During infection, SARS-CoV-2 initiates the production of CXCL1, CXCL5, IL-6, IP-10, and MCP-1; however, SARS-CoV initiates the production of 11 cytokines [44]. Blanco and his co-authors worked on lung samples of COVID-19 patients to investigate the nature of transcriptional response generated in SARS-CoV-2 and SARS-CoV. They found that there was a sudden increase in the number of proinflammatory chemokines including reduced expression of IFN-I and III [45]. The enhanced level of interleukins (ILs) such as IL1RA, IL-1B, and IL-6 in the serum of SARS-CoV-2 affected individuals and, generation of cytokines involved in adaptive immunity makes SARS-CoV-2 different from SARS-CoV.
4.2. Role of adaptive immune response in SARS-CoV-2
The SARS-CoV-2 infection occurs due to the transformation between innate and adaptive responses of the immune system. CD4 cells help the B cells to generate the neutralizing antibodies however CD8 cells are cytotoxic but they help to remove the virus-infected cells [46]. It has been found that around 80% of infiltrating cells are CD8 cells in COVID-19 patients [47]. During infection, a deterioration response is generated in the body which does not remove the virus-infected cells and thus it can promote the replication of the virus. The uncontrolled replication of the virus develops the inflammatory response and cytokine storm, and is the main cause of severe acute respiratory distress syndrome (or ARDS) and disseminated intravascular coagulation [48]. This evidence is supported by the report of Clay and his co-authors by using a SARS-CoV primate model of infection. They found that, till the 10th day of post-infection, the virus was able to replicate itself in the lungs and cause lung inflammation. However, on the 14th day of infection, inflammation was found to be acute after the clearance of the virus and remains until the 28th day. This report gives a clear indication towards the dependency of the early phase on the replication of virus and immune dependent phase found to be intensifying the inflammation [49].
Dendritic cells have certain receptors such as DC-SIGN, which behave as a trans-receptor for SARS-CoV on dendritic cells [50]. CD26 is an aminopeptidase that helps in the activation of T-cells on binding with the spike protein of SARS-CoV-2 and generates the ineffectual T-cells [51]. CD147 is a protein that belongs to the immunoglobin family and involved in the T-cell activation. The main function of CD147 is to bind with the S1 domain of spike protein and thus allow the entry of the virus into host cells [52]. An event such as activation-induced cell death (or AICD) may be developed upon binding of SARS-CoV-2 to CD147 and CD26 [53] however; MERS-CoV has been reported to cause the apoptosis of T-cells [54]. These findings suggest the abnormal functioning of T-cells in COVID-19 patients.
4.3. Immune cells involved in SARS-CoV-2 infection
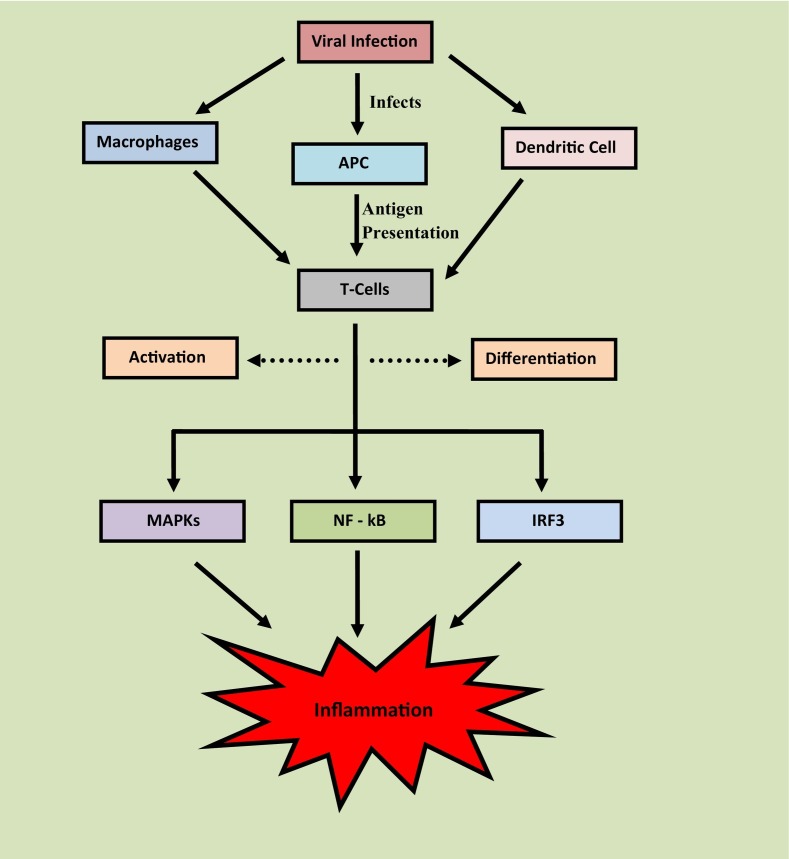
Mild respiratory infection with inflammatory responses was observed once the SARS-CoV-2 attacks on the respiratory tract [55]. The infection affects the functioning of a wide number of immune cells which includes macrophages, antigen-presenting cells, and dendritic cells [56]. The infection to these immune cells presents the antigens of SARS-CoV-2 to T-cells and laid the activation and differentiation of T-cell. The innate immune system of the host detects the infection by using Toll-like receptors (TLRs) and PPRs followed by the detection of pathogen-associated molecular patterns such as proteins, lipids, lipoproteins, and nucleic acid of the virus [57]. Moreover, the mitogen-activated protein kinase (MAPKs) pathway and factors like NF-kB; IRF3 gets activated which promotes the inflammation [58]. For example, SARS-CoV-2 shows a high binding affinity towards TLR which causes the release of pro-IL-1-β [59]. Later this IL undergoes cleavage through Caspase-1, which activates the inflammasome and enhances the generation of active mature IL-1β, as depicted in Fig. 2 .
Fig. 2.
Involvement of immune cells in development of inflammation.
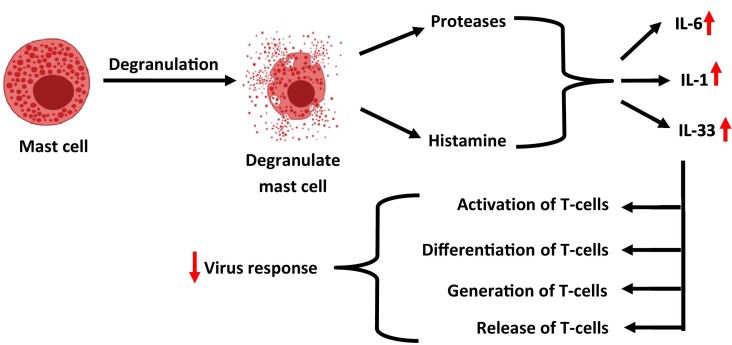
Mast cells are the immune cells present in the submucosa of the respiratory tract. They are activated in response to an attack of virus and their activation results in the release of histamine and proteases [60]. Additionally, their activation triggers the generation of ILs such as IL-1, IL-6, and IL-33 [61]. Activation of pro-inflammatory ILs further governs the activation, differentiation, generation, and release of T-cells and cytokines. Activated T-cells act as anti-viral cells by decreasing the risk of the virus [62], as depicted in Fig. 3 . The activation of T-dependent B cells guides the Helper T cells such as CD4 to generate the virus-specific antibodies. CD8+ is another type of T-cells, which is cytotoxic and causes damage to virus-infected cells. Pro-inflammatory chemokines and cytokines are produced from the T helper cells through the activation of the NF-kB signaling cascade. The signaling through the NF-kB cascade recruits the immune cells like monocytes and neutrophils to the site of infection [63].
Fig. 3.
Activity of interleukins in viral response.
Neutrophils are the important cells of immune system, also called as polymorphonuclear leukocytes. Whenever there is any injury to tissue or a pathogen is present in the body, a signal is generated that activates neutrophils. These activated neutrophils are released from the blood vessels and reach first at the site of infection [64]. Due to the increase in number of neutrophils in the COVID-19, a condition develops in the body which is called neutrophilia. Neutrophilia is also called neutrophil leukocytosis. Excessive inflammation is a major cause of neutrophilia. IL-17 plays an important role in inflammation by increasing the number of neutrophils. In addition to IL17, IL-1 is also responsible for promoting the production of neutrophils. This results in the formation of a positive loop by which neutrophils enrolls them. Thus by doing this, neutrophils produce IL-17 and attract the source of IL i.e. IL-17-producing-T-lymphocytes. This attraction enrolls the neutrophils in excess so that maximum numbers of neutrophils are produced and eventually the state of neutrophilia occurs in the body [65]. In COVID-19 patients, neutrophilia plays a negative role as the condition is believed to promote the respiration-related ailments by releasing the cytokines [66].
Pneumocytes are the specialized cells that are present in the alveoli of the lungs. These cells are known to be involved in the exchange of carbon-dioxide and oxygen. The damage to alveolar cells from COVID-19 causes the systemic hyper-inflammation, also known as the macrophage activation syndrome [67]. Furthermore, this virus enters macrophages by altering the function of pneumocytes cells. The release of damaged cell molecules and inflammatory cytokines is induced by the cell death that occurs due to the infection. Upon entry of viral genome, the inflammatory signal facilitates the activation of macrophages, which in turn leads to the production of inflammatory cytokines and chemokines. Macrophages are activated when T-cells produce inflammatory signals [68]. Macrophages are activated during the inflammatory response [69]; they trigger the release of cytokines and TNF-α. However, macrophages are the phagocytic immune cell that helps in the engulfment of pathogens but despite this, macrophages promote the release of high amounts of cytokines resulting in the formation of the IL-6 cascade. Additionally, a collaborative mechanism of inflammation and macrophage activation, greatly affects the major parts of the body, especially the lungs. Because, the hyperactivation of macrophages results in the formation of cytokine storm which acts on lungs and initiates the ailments related to respiration [70].
4.4. Inflammatory mediators in SARS-CoV-2
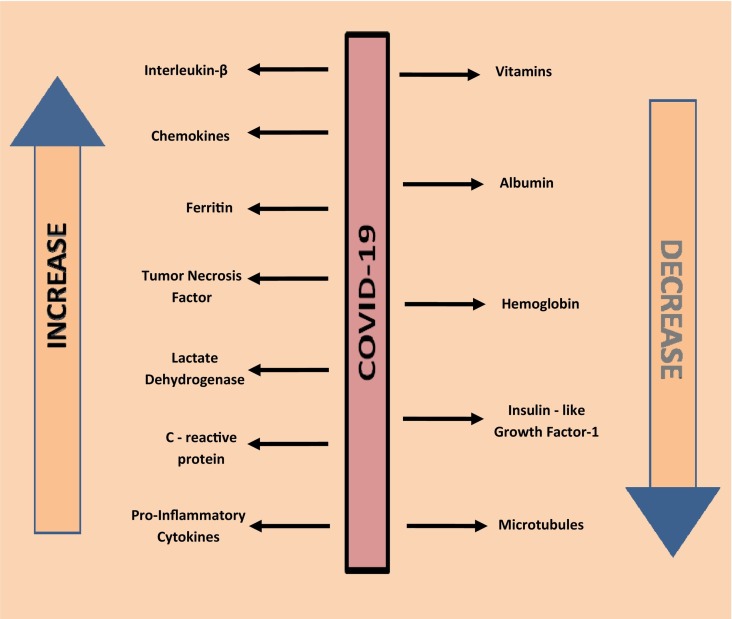
A large number of diseases such as inflammation, diabetes, cardiovascular diseases, obesity, and blood clotting are known to associate with the COVID-19 [71]. The levels of inflammatory markers such as chemokines, pro-inflammatory cytokines, ferritin, lactate dehydrogenase (LDH) enzyme, and C - reactive protein (CRP) are found to be high in COVID-19 suffered patients [72]. The elevated level of these markers initiates the inflammatory reactions in the body which regulates the inflammatory response. This indicates the relationship of CoV with inflammation. The level of albumin, vitamins, hemoglobin, insulin-like growth factor 1, and micronutrients are decreased with the rise in the level of pro-inflammatory mediators [73]. IL-1-beta, IL-6, tumor necrosis factor and C-reactive protein are some examples of pro-inflammatory mediators, causing a high rate of mortality in virus-infected individuals [74], (Fig. 4 ).
Fig. 4.
Variations in inflammatory mediators and proteins during COVID-19 infection.
4.4.1. INFs in SARS-CoV-2
IFNs are signaling proteins, known to secrete from host cells in response to viral attacks [75]. They protect the host cell from the attack of virus and hence named anti-viral proteins. Type I, II, and Type III IFNs are known to involve in the innate immunity to bacteria or viruses and hence act as central cytokines. All nucleated cells are a prime source of type I and III IFNs [76]. Virus-infected cells such as leukocytes are the major site for the generation of IFN-α [77]. On the contrary, virus-infected cells such as fibroblasts are the main site for the generation of IFN-β [78]. The initiation of viral response or intracellular bacterial infections from the natural killer cells signals the macrophages to produce the type II IFNs. During the antigen-specific immunity, the T helper CD4 and CD8 cytotoxic T lymphocyte cells activate the effector T cells which enhance the production of IFN-γ [79].
The attachment of INFAR1 or INFAR2 with type I IFNs and IFN-γR1 or IFN-γR2 with type II IFNs and attachment of IL-28R or IL-10Rβ with type III IFNs result in the activation of a meshwork of downstream signaling which specifically shifts the transcription factors to an activated state [80]. Moreover, levels of IFN-γ are reported to increase in the COVID-19 patients. The continuous decline in the number of lymphocytes and rapid increase in the infiltration of neutrophils in the alveoli of lungs were observed in the COVID-19 patients [81]. The increased levels of IFN-γ are primarily associated with the generation of inflammation in the pulmonary area of the lungs which causes damage to lungs in SARS-CoV-1 [82] and MERS-CoV [83]. CD4 TH cells are the chief producers of IFN-γ [84]. The production of IFN-γ significantly promotes the differentiating mechanism of CD8 T cells [85]. Granulocytes and monocytes colony-stimulating factors are produced from CD4 TH cells which extensively activate the promotion of differentiation of various monocytes such as CD16+, CD14+, and CD15+ in blood [86]. The increased levels of IFNs in COVID-19 can be managed using the corticosteroids [87].
4.4.2. ILs in SARS-CoV-2
ILs are known for the generation of multiple immune cells and are involved in the interaction of leucocytes-to-leucocytes hence they are termed ILs. They are meant for stimulating the immune response. The IL-1 is involved in the immunity derived from the T cells [88]. IL-1 is responsible for the expression of receptors present on the surface of IL-2 [89]. Moreover, they governed the homeostasis of T-cells by secreting the IL-2. The proliferation and activation of secondary cytokines including epithelial cells are increased by the action of IL-1α and IL-1-β [90]. Additionally, they play an important function by increasing the signaling in the acute phase and carry the immune cells to the site of infection. The IL-1 has been found to exhibit the maximum affinity for the IL-1-receptor1 (IL-1-R1) and the receptor is profoundly known to express on the surface of TH2 cells [91]. Recent studies confirm the decline in allergic symptoms in the model of hypersensitivity by diminishing the levels of IL-4 and IL-5 [92].
The signaling events from IL-1 receptors significantly increase the levels of IgM antibodies which enhance the survival rate of mice infected with the influenza virus. However, this signaling event is responsible for acute lung immunopathology [93]. Immunological analysis signifies that the level of IL-1-β, IL-7, IL-8, and IL-9 is rapidly increased in the plasma of COVID-19 patients [94]. Damaged tissue or cells are the major producers of these cytokines and hence these are considered as prime immune drivers to generate the immune response in COVID-affected persons. Levels of IL-2 and IL-7 are gradually increased in the COVID-affected patients admitted in ICU and non-ICU patients [95]. Antigen-presenting cells are responsible for the secretion of IL-10. IL-10 acts as a mediator for the differentiation and activation of CD8 T cells and TH immune cells [96]. Later this IL represents itself as a stable immune modulator in COVID-19 patients and hence it is considered a vital sign of immune delinquency [97]. To reduce the ILs levels in COVID-19, Anakinra can be used as it is an antagonist of the IL-1-receptor [98].
4.4.3. Tumor Necrosis Factor (TNF) in SARS-CoV-2
Amongst all the superfamilies of TNF, only 19 members of this family contain the type II membrane protein [99]. This membrane protein behaves as a cytokine once they are secreted out from the cell following the extracellular proteolytic cleavage mechanism. Cells such as activated lymphocytes, monocytes, astrocytes, macrophages, adipocytes, neutrophils, and smooth muscle cells are responsible for the generation of TNF-α [100]. TNFR1 is a primary receptor of TNF-α, known to exert the pleiotropic action of cytokines. TNF-α plays a crucial function in the cytokine storm and hence mediates the expression of cytokines, transcription factors, etc [101]. However, they also mediate the expression of those genes and receptors which are responsible for the growth. The levels of IL-6 and soluble IL-2 receptors were increased in COVID infection [102]. The induction of HA-synthetase-2 in EpCAM+ lung alveolar epithelium was observed in the lungs of COVID infected patients [103]. Hyaluronan is responsible for the influx of fluid in the alveoli of the lungs and becomes the main cause of deoxygenation [104]. However, it has been observed that epithelial cells present in the lungs do not secrete the TNF-α in H5N1 influenza infection [105]. Anti-TNF drugs such as infliximab are recommended to combat the inflammation in COVID-19.
5. Molecular, biochemical and immunological changes in SARS-CoV-2
During the CoV attack, the human body undergoes many molecular biochemical and immunological changes. These changes include the virus attachment to host cell, increase in cytokine level, changes in circulating cells and inflammatory events, etc.
5.1. Changes in circulating cells during SARS-CoV-2 infection
The changes in the immune system are characterized with the help of abnormal functioning of neutrophils, decrease in the activity of T-cells to any new antigens, decrease in the expression of MHC-II, alternation in the productive mechanism of cytokines including alternation in signaling and expression of TLR, reduction in the cytotoxic potential of natural killer cells, elevation in senescent T-cells, decrease in phagocytic activity of macrophages, and rapid decrease in the count of monocytes, are observed in the COVID-19 patients having chronic low-grade inflammation [106]. During the infection of SARS-CoV-2, T cells including CD4 and CD8 cells undergo the modification process [107]. Lymphopenia is considered as most prominent in the SARS-CoV-2. It is proven to harm the regulatory T (Tregs) cells, CD4 Th1 and CD8 cells [108]. Studies highlights that, there is a gradual decrease in the number of memory T-cells but the number of naive T-cells increased. Various phenotypes manifest during the circulation of CD8 in the SARS-CoV-2 [109].
Perforin is present in NK cells and cytotoxic-T-Lymphocytes which are known to create pores in the cell-membrane during degranulation [110]. Continuous rise in the amount of circulating myeloid-derived suppressor cells shows the presence of negative relationship between perforin content of CD8+ and NK-cells and; serum levels of IL-6 and IL-8 [111]. Numerous activation markers i.e. HLA-DR, Th17 CD4+CCR6+, CD38, CD44, and CD69, are expressed due to a reduction in the number of CD4 cells [112,113]. Monocytopenia occurs when the level of monocytes continuously falls to an average level [114]. This decrease in the level of blood monocyte is considered a serious threat in SARS-CoV-19 patients [115]. In Monocytopenia, monocytes play an important role by circulating themselves and they have a relation with the CD14+CD16+ inflammatory monocytes subset [116]. These changes in circulating cells increase the complications in SARS-CoV-19 patients.
5.2. Occurrence of chronological events in SARS-CoV-2 infection
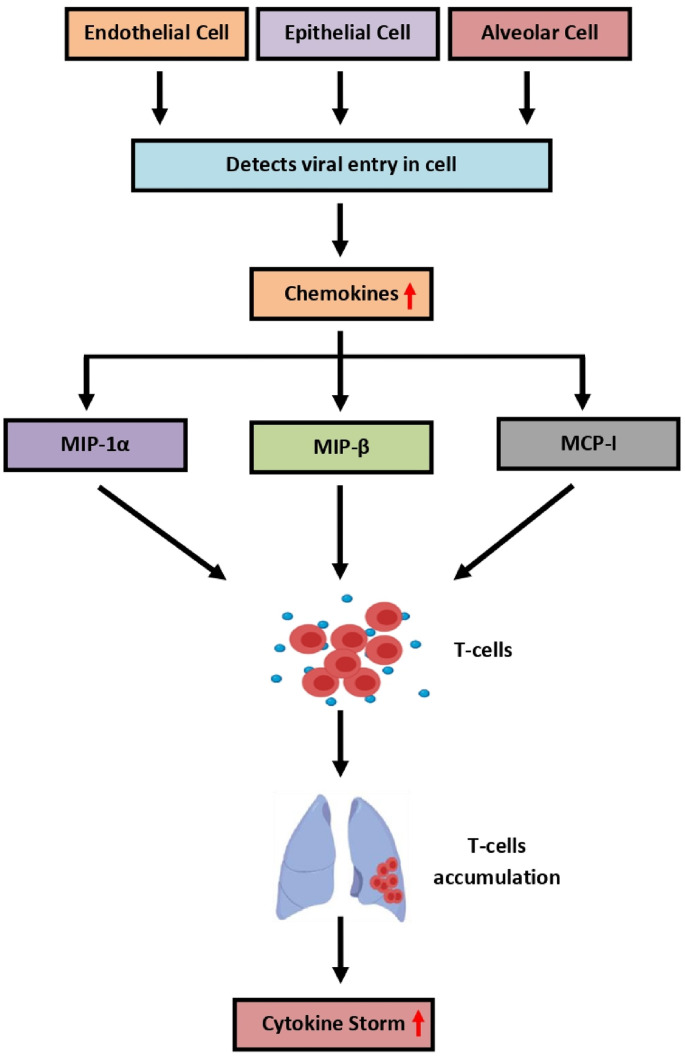
The infection cycle of SARS-CoV-2 initiates with the binding of ACE2 with the spike protein of SARS-CoV-2. The attachment of spike to ACE2 allows the virus molecule to enter the host cell. Upon entry, the RNA of the virus gradually starts to replicate itself and prepare multiple copies [117]. These changes are easily identified by surrounding cells like endothelial, epithelial, and alveolar cells and thus enhance the productivity of chemokines and pro-inflammatory cytokines in the targeted organ. These chemokines include the various macrophage inflammatory proteins (MIP) i.e. MIP-1α, MIP-β, and MCP-1. These MIPs supports the inflammation by migrating the T-cells, monocytes, and macrophages towards the infection site. The migration of these cells causes the development of a feedback loop that includes the pro-inflammatory cytokines [118]. The pro-inflammatory feedback loop mainly affects the lungs by accumulating the immune cells. Their accumulation in the lungs results in the over-production of pro-inflammatory cytokines, which provides damage to the lungs by inducing the cytokine storm event [119]. However, only lungs are not affected due to cytokine storm, many other body organs are affected, resulting in the damage of multiple organs, as depicted in Fig. 5 .
Fig. 5.
T-cells in Cytokine storm.
5.3. Cytokine storm in SARS-CoV-2
The continuous cytokine storm causes macrophage activation syndrome (MAS) and secondary hemophagocytic lymphohistiocytosis (HLH) which fail the organ system [120]. The increased levels of CD25 and hepatosplenomegaly alternations in fibrinogen are observed in MAS and HLH ailments however, these findings are not observed in COVID-19 [121]. The shortage in the count of lymphocytes could be a possible factor for misbalancing the innate and acquired immune responses. Moreover, this misbalancing slows down the system which causes a delay in some immunological events like clearance of virus, abnormal regulation of INFs and hyperstimulated macrophages and neutrophils, etc. The conditions such as excessive activation of CD4+ T-cells and exhaustion of CD8+ T-cells including lack of responses generated from B and T cells demonstrates the severe conditions of COVID-19 [122].
5.4. Activation of monocyte-derived macrophages in COVID-19
The cytopathic effects were found to be increased with the slowdown in productivity of type-1 IFNs. With the increase in cytopathic effects, monocyte chemoattractants were also secreted at high concentrations from alveolar epithelial cells. These sudden secretions cause the sustained recruitment of blood monocyte in the lungs [123]. The monocytes are differentiated into pro-inflammatory macrophages [124], and the Janus kinase signal transducer helps in the differentiation of monocytes [125]. Monocytes that are derived from macrophages are recruited and activated with the production and activation of tumor necrosis factor, granulocyte-macrophage colony-stimulating factor (GM-CSF), IFN-γ, and natural killer cells, etc [126]. The accumulation of oxidized phospholipids in the lungs of virus-infected patients signals the Toll-like receptor 4 (TLR4)-TRAF6-NF-kB pathway to activate the monocyte-derived macrophages [127]. The recognition of single-stranded RNA helps the virus sensing and trigger the activation of TLR7 [128].
Some theories indicate that viruses can enter the cytoplasm of macrophages only if the type I IFN mediates the expression of receptors responsible for their entry [129]. Once the virus entered the cytoplasmic region, it activates the NLRP3 inflammasome [130]. Their activation further causes the release of IL-1β or IL-18 (mature ILs). Monocytes-derived macrophages are activated through the action of IL-1β. It can either use the paracrine mode or autocrine mode for the activation of derived monocytes [131]. Additionally, the productivity mechanism of type I IFNs in infected lungs can be diminished with IL-1β [132]. An increase in the activation of monocyte-derived macrophages supports the generation of cytokine storm in SARS-CoV-2 [133]. Uncontrolled release of pro-inflammatory cytokines from virus-infected lungs results in the cytokine storm [134].
6. Targets for vaccine development against SARS-CoV-2
As of now, there is no vaccine available in the market that can completely inhibit the infection of coronavirus. B-cell and T-cells are considered the most prominent antibody targets. The immunological response generated from B and T-cells can be considered as a potential antibody target for vaccine development against COVID-19 [135] and have been discussed in the following sections.
6.1. B-Cells as antibody target in SARS-CoV-2
Conformational changes are induced during the attachment of the virus with the cell and are needed for the fusion of the cell membrane with the virus. Viruses neutralizing antibodies block the conformational changes and thus restrict the virus to attach to the cell membrane and protect the cell from the attack of the virus [136]. Spike protein of the virus is governed by the furin and serine protease such as TMPRSS2 and TMPRSS4 which collectively enable the binding of the virus with the cell membrane. Interaction of spike protein to ACE2 continues on a domain called RBD [137]. Thus, targeting the RBD with antibodies is counted as one of the best examples to slow down the effect of various types of coronaviruses. Generally, the RBD shows post-transcriptional modifications like glycosylation and methylation due to which they are not able to reproduce in vaccines [138]. A receptor interaction site (RIS) is a site where ACE2 binds with the receptor [139]. The RIS is found to be non-glycosylated and thus it would be a prominent target for vaccine development [140]. IgG is the most preferred isotype to be induced in the SARS-CoV-2 vaccine [141]. The widest targeted isotype by a virus is IgA because IgA reduces the risk of infection to cells present in the respiratory tract i.e. epithelial cell or mucosal cell [142]. Normally adjuvants are used at a high scale for triggering the production of IgA [143]. Moreover, TLR7/8 and TLR9 ligands are helpful to boost the immunological response generated by IgA and thus considered as potential candidates against the virus [144].
6.2. T-cells as antibody target in SARS-CoV-2
The major function of immune cells is to diminish the risk of disease by protecting the immune system. Initially, CD8 and CD4 T-cells identify the antigens and continue their reaction with SARS-CoV-2 antigens [145]. T-cells alone are not considered as a strong candidate for the prevention of ailments; however neutralizing antibodies have a high potential to prevent the risk of ailments [146]. The use of anti-viral vaccines available on market is highly recommended because they have antibodies that protect the body. These anti-viral vaccines work by two mechanisms, they either block the entry of virus in a cell or either signals the host cell to prevent itself from the attack of virus. These vaccines specifically act in the extracellular space where the viral particles are neutralized by the antibodies. CD4 T-helper cells help the B-cell to enhance the production of antibodies [147]. HLA molecule represents the antigens of T-cells on the outer surface of infected and antigen-presenting cells. Few factors with T-cells are responsible for creating the difference in HLA molecules. These factors include the viral recognition and genetic polymorphism in HLA molecules etc [148]. The T-cell specificity for antigenic peptides is different in every individual which allows it to bind with their HLA molecules [149]. The presentation of HLA molecules by their antigenic peptide is necessary for every individual [150].
Short antigenic peptides and mini-genes are present in the vaccines. But still, these vaccines cannot work efficiently for some individuals which are not capable to present their HLA molecule [151]. The presence of either DNA/RNA or full-length viral proteins/long peptides in vaccines can be used as an alternate strategy for those who are not capable to present their HLA molecule [152]. Infected cells are a prime source for the generation of viral peptides and these are recognized by CD8 cytotoxic T-cells. Some antigenic proteins are collected from extracellular space, are not able to represent themselves in front of CD8 T-cells [153]. Cross-presentation is one major factor responsible for the inefficient behavior of full-length protein vaccines [154]. The event of cross-presentation in vaccines does not cause the immunological responses that are mediated by CD8 T-cells. After vaccination, the generation of T-cell response for a long time is always being considered a difficult task [155]. This hinders the development of vaccine against SARS-CoV-2.
7. Proficiency of antibodies in SARS-CoV-2
The use of antibodies against the spike protein of the virus helps the immune system by blocking the binding of spike protein with ACE2. The protective mechanisms of antibodies are still a mystery for researchers that how these antibodies work against viral proteins and their mechanism of cross-reactivity against alpha coronavirus and beta coronavirus [156]. These show the maximum cross-reactivity for beta-coronavirus as compared to alpha-coronavirus. Immunological study of HCoV-OC43 addresses that, anti-N-antibodies have high reactivity for SARS-CoV [157]. Serological studies explain the time duration to detect the antibodies in serum during infection. For example, IgG antibodies can be easily recognized on the 14th day of infection [158], and IgA and IgM antibodies can be recognized during the 1st week of infection [159]. Respiratory mucosa behaves as a mediator for the entry of virus (SARS-CoV-2) into the body [160]. Secretory Immunoglobulin A (SIgA) is an immunoglobulin that plays an important function in the defense mechanisms of the mucosa. SIgA performs the anti-viral neutralizing bioactivity in respiratory tract mucosa of COVID-19 patients [161]. However, the presence of neutralizing IgA in bronchoalveolar lavages of COVID-19 patients gives proof of their anti-viral bioactivity [162].
The main aim of using coronavirus-neutralizing antibodies is to target the spike protein [163]. S-protein is composed of two subunits (or domains) i.e. S1 and S2. Structurally, S1A, S1B, S1C, and S1D collectively formed the S1 subunit. The main function of the S1 subunit is to initiate the attachment of virus with cell [164] meanwhile; S2 subunit mediates the fusion of viral and cellular membrane [165]. In CoV, the irreversible conformational changes are caused due to receptor interactions which enable the fusion of membrane [166]. In-silico studies reveal that the primary amino acid sequences of 1273 residue of the spike protein of SARS-CoV-2 and 1255 residue of SARS-CoV are structurally 77.5% similar in nature [167]. The similarities in the amino acid sequence of these residues mediate the attachment of ACE2 protein with the spike protein of the virus through the S1 subunit [168]. Thus, protein neutralizing antibodies are continuously used to target the receptor interactions.
Upon investigation, Gruber and his co-authors found that neutralizing antibodies are present in the MIS-C patients, which can be used in the treatment of COVID-19. Later these antibodies are known to be associated with activated immune cells and ILs such as monocytes, lymphocytes, natural killer cells, myeloid chemotaxis, and IL-18 or IL-16 [169]. The upregulation of Fc-γ receptor-1 on macrophage and intercellular adhesion molecule-1 on neutrophils collectively increase the immunological response mediated by the Fc- receptors and antigen presentation [170]. The activation of host immune cells through viral superantigens and production of autoantibodies by recognizing the T-cell these antibodies cause the attenuation of virus SARS-CoV-2 [171]. Additionally, infected cells are the site for viral antigens to increase the expression of T-cells and antibodies, which are considered as a prime mechanism to attenuate the pathogenesis of SARS-CoV-2 [172].
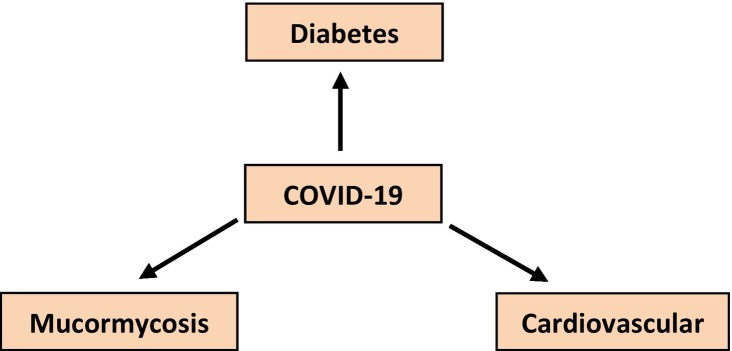
8. Complications of COVID-19
In medical sciences, complications can be defined as the abrupt conditions that arise either during the progression of the disease or after treatment. People suffering from diabetes, asthma, and cancer are at major risk of this viral disease [173]. However, many diseases have been known to associate with the pathogenesis of coronavirus. Lungs are not only targeted; other organs such as the heart, kidney, blood vessels, gut, and brain are also at great risk [174]. To the present date, a number of complications have been noted in chronic and viral diseases. For example, COVID-19 is also associated with many complications such as cardiovascular disease; diabetes and mucormycosis etc (Fig. 6 ) are explained as below:
Fig. 6.
Complications of COVID-19.
8.1. Cardiovascular disease
Cardiovascular diseases are one of the major comorbidities of COVID-19. Complications of cardiovascular disease have been reported in COVID patients who have no history of heart disease [175]. Cardiovascular disease can be defined a disease that damages the blood vessels or heart. Cardiovascular diseases can be classified into four types i.e. stroke, aortic disease, peripheral arterial disease and coronary heart disease etc. Blood clotting plays an important role in the development of stroke. Small clots in small vessels were seen in COVID infected people. These tiny clots may form in multiple places of the body and brain and bring some severe complications including heart attack or stroke [176]. People infected with diabetes or high blood pressures are more likely to form clots in the body. The accumulated blood clot constricts the area of blood vessels and prevents the flow of oxygen and blood which causes serious conditions like cardiac arrest. There are many antiplatelet drugs available in the market to combat this condition which can effectively reduce the risk of disease [177]. For example, Warfarin [178] and aspirin are major two anti-platelet drugs used for the treatment. Higher doses of aspirin and Warfarin are found to be toxic, however; at lower doses, they reduce the risk of heart attack and stroke [179].
8.2. Diabetes
A metabolic disorder which is caused due to the presence of excess blood sugar in the body is called diabetes. It is known to affect the multiple organs of the body such as kidney, nerve and heart etc. Type 2 diabetes is considered a major comorbidity associated with COVID-19 that occurs when the body does not accept the insulin [180]. Hence, Chinese researchers conducted a study on COVID-19 patients. Eventually, they found that people who did not even have a history of diabetes were found to have Ketosis or Ketoacidosis, indicating diabetes as a confirmed concomitant of COVID-19 [181]. The management of ketosis and ketoacidosis involves the correction of dehydration, hyperglycemia and electrolyte imbalance.
8.3. Mucormycosis
It is an unknown fungal infection which is also called as the black fungus. The disease is being detected in patients who have been recovered or have been cured of the COVID-19. The risk of mucormycosis is quite higher in COVID-19 patients with diabetic complications [182]. However, the disease can be managed by taking the Amphotericin B deoxycholate [183].
9. Therapeutic strategies against COVID-19
Various preventive measures can be used to avoid the risk of viral infections including COVID-19. These therapeutic strategies include the use of antiviral drugs, inhibitors of enzymes and therapies, etc.
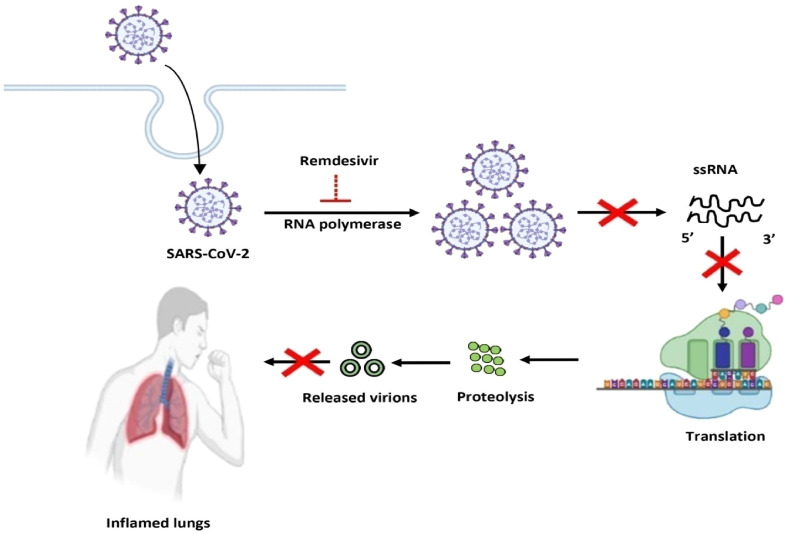
9.1. Remdesivir as an anti-viral drug in COVID-19
Various anti-viral drugs are available in the national and international market that inhibits the action of death-causing viruses. Remdesivir is one such example that can be used against the infection caused by SARS-CoV-2. Remdesivir is an analog of nucleoside whose prime function is the inhibition of the viral RNA polymerase [184]. During viral infection, the transcription mechanism is continuously active to form more viral RNA polymerase copies [185]. But the inhibition of RNA polymerase by remdesivir restricts the continuous production of viral RNA which causes the decrease in the death rate of COVID-19 patients [186]. The therapeutic action of remdesivir increased its market value at the international level. The risk of abnormalities like diabetes, non-alcoholic fatty liver, and diabetic nephropathy is increased due to consumption of remdesivir [187]. However, remdesivir is found to be effective in severe forms of SARS-CoV-2 infection, as the mechanism is depicted in Fig. 7 .
Fig. 7.
Role of remdesivir in suppression of COVID-19.
9.2. Inhibitors of TMPRSS2 or ACE2 in SARS-CoV-2 infection
The SARS-CoV-2 treatment can be achieved either by blocking the TMPRSS2 or ACE2 receptor [188]. Many inhibitor compounds are clinically being tested and some are approved for the COVID-19 treatment. In rheumatoid arthritis, the ACE2 mediated endocytosis can be inhibited by using the Janus kinase inhibitor i.e. baricitinib. So the use of baricitinib against SARS-CoV-2 could show the same effect as rheumatoid arthritis [189]. Ruxolitinib is another inhibitor, used for the inhibition of Janus kinase. It is now being clinically tested in the laboratories to get the appropriate COVID-19 treatment [190]. However, the entry of the virus can be restricted by increasing the amount of soluble form of ACE2. APN01 is the recombinant form of ACE2 that is prepared by the APEIRON. The strategy of using APN01 with soluble ACE2 is clinically tested [191]. In COVID-19 treatment, the use of TMPRS2 inhibitors is under consideration. Two inhibitors of TMPRSS2 i.e. Nafamostat mesylate and Camostat mesylate are available in the market and are clinically approved [192]. But these inhibitors are not used against SARS-CoV-2 due to lack of knowledge. Further validation of these inhibitors on COVID-19 patients provides new insights into their mechanism of action.
9.3. Convalescent plasma therapy
Many infectious diseases can be cured with convalescent plasma therapy. The use of convalescent plasma therapy gives a positive output with a high efficacy rate against SARS, MERS, and H1N1 [193]. Due to the lack of neutralizing antibodies in convalescent plasma, the therapy does not found to be effective against the Ebola virus. It has been proposed that SARS-CoV-2, MERS, and SARS are sharing the same virological features, so convalescent plasma therapy is recommended by physicians for treatment [194]. If the virus-infected patients are benefited after plasma therapy then the plasma of those patients can be used for the treatment of others. COVID-19 recovered people donate their convalescent plasma to COVID-19 suffered patients. Only those people are allowed to donate their plasma that tests as seronegative for Human Immuno virus or Hepatitis C virus [195]. Recently it was found that convalescent plasma therapy does not show a good result in SARS-CoV-2 patients. It has been found that neutralizing antibodies were present in the convalescent plasma of COVID-19 patients, which are found to diminish the inflammatory response generated in COVID-19. These neutralizing antibodies restrict the attachment of the virus with the ACE2 enzyme and thus helps in the treatment of COVID-19 suffered patients [196]. The lifespan of this antibody is only 2 years so it can be easily detected even after 2 years.
9.4. TNF-alpha as a model target for SARS-CoV-2 treatment
An enzyme responsible for the secretion of TNF into blood circulation is called the TNF-converting enzyme or TACE [197]. The activity of the TNF-converting enzyme (TACE) increased upon binding of ACE2 to spike protein. An interaction of ACE2 and TACE signals the host cell to mediate the entry of the virus. The release of TACE in circulation suggests the role of TNF in an early phase of infection. Anti-TNF drugs or antagonists will be of great interest to avoid the release of TNF in SARS-CoV-2 [198]. So targeting the TNF will inhibit the spread of infection at the early phase and helps the COVID-19 patients to fight against it.
10. Conclusion
SARS-CoV-2 is a novel virus responsible for the initiation of inflammatory reactions in the body. The pathogenesis of the virus results in kidney failure, heart attack, and generation of inflammatory reactions including lungs. The use of neutralizing antibodies is the best strategy to reduce the risk of SARS-CoV-2. In addition to this, anti-TNF therapies effectively work against SARS-CoV-2 by decreasing the level of inflammatory cytokines thus inhibiting the inflammatory mechanism. Therefore, adopting therapeutic strategies to improve the immune response can serve as the best treatment against COVID and its associated diseases.
Credit author’s statement
JG has conceptualized and designed the manuscript. SSG has done the literature review, diagrammatical work, manuscript writing, and referencing work. JG and AS have edited the manuscript.
Declaration of Competing Interest
We wish to confirm that there is no conflict of interest associated with this publication. Further, there has been no significant financial support for this work that could have influenced its outcome.
Acknowledgments
The authors thank the Division of Research and Development (DRD), Lovely Professional University, Phagwara, India for providing us space, time and atmosphere to bridge this collaborative work.
References
- 1.Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin. Microbiol. Infect. 2020;26:729–734. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pal M., Berhanu G., Desalegn C., Kandi V. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): An update. Cureus. 2020;12:7423. doi: 10.7759/cureus.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ni W., Yang X., Yang D., Bao J., Li R., Xiao Y., Hou C., Wang H., Liu J., Yang D., Xu Y., Cao Z., Gao Z. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care. 2020;24:422. doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alzoughool F., Alanagreh L. Coronavirus drugs: Using plasma from recovered patients as a treatment for COVID-19. Int J Risk Saf Med. 2020;31:47–51. doi: 10.3233/JRS-201017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson N.M., Norton A., Young F.P., Collins D.W. Airborne transmission of severe acute respiratory syndrome coronavirus-2 to healthcare workers: a narrative review. Anaesthesia. 2020;75:1086–1095. doi: 10.1111/anae.15093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salamanna F., Maglio M., Landini M.P., Fini M. Body localization o ACE-2: On the trail of the keyhole of SARS-CoV-2. Front Med (Lausanne) 2020;7:594495. doi: 10.3389/fmed.2020.594495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fung T.S., Liu D.X. Human coronavirus: Host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 8.Hendrickson C.M., Matthay M.A. Viral pathogens and acute lung injury: investigations inspired by the SARS epidemic and the 2009 H1N1 influenza pandemic. Semin Respir Crit Care Med. 2013;34:475–486. doi: 10.1055/s-0033-1351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park A., Iwasaki A. Type I and Type III Interferons- induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia L.F. Immune response, inflammation, and the clinical spectrum of COVD-19. Front. Immunol. 2020;11:1441. doi: 10.3389/fimmu.2020.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: rna synthesis, proofreading and final capping. Cells. 2020;9:1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan Y.-J., Lim S.G., Hong W. Characterization of viral proteins encoded by the SARS-coronavirus genome. Antivir. Res. 2005;65:69–78. doi: 10.1016/j.antiviral.2004.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malik Y.A. Properties of coronavirus and SARS-CoV-2. Malays J Pathol. 2020;42:3–11. [PubMed] [Google Scholar]
- 14.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogando N.S., Ferron F., Decroly E., Canard B., Posthuma C.C., Snijder E.J. The curious case of the nidovirus exoribonuclease: Its role in RNA synthesis and replication fidelity. Front. Microbiol. 2019;10:1813. doi: 10.3389/fmicb.2019.01813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guy J.S., Breslin J.J., Vivrette S., Smith L.G. Characterization of a coronavirus isolated from a diarrheic foal. J. Clin. Microbiol. 2000;38:4523–4526. doi: 10.1128/jcm.38.12.4523-4526.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. basis Dis. 1866;2020:165878. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giron C.C., Laaksonen A., da Silva F.L.B. On the interactions of the receptor-binding domain of SARS-CoV-1 and SARS-CoV-2 spike proteins with monoclonal antibodies and the receptor ACE2. Virus Res. 2020;285:198021. doi: 10.1016/j.virusres.2020.198021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez-Trincado J.L., Gomez-Perosanz M., Reche P.A. Fundamentals and Methods for T- and B-Cell Epitope Prediction. J Immunol Res. 2017:1–14. doi: 10.1155/2017/2680160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., Zhou Y., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implications for development of RBD proteins as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antivir. Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanism of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medina-Enriquez M.M., Lopez-Leon S., Carlos-Escalante J.A., Aponte-Torres Z., Cuapio A., Wegman-Ostrosky T. ACE2: the molecular doorway to SARS-Cov-2. Cell Biosci. 2020;10:148. doi: 10.1186/s13578-020-00519-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakagawa K., Lokugamage K.G., Makino S. Viral and Cellular mRNA Translation in Coronavirus-Infected Cells. Adv. Virus Res. 2016;96:165–192. doi: 10.1016/bs.aivir.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yadav R., Chaudhary J.K., Jain N., Chaudhary P.K., Khanra S., Dhamija P., Sharma A., Kumar A., Handu S. Role of structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells. 2021;10:821. doi: 10.3390/cells10040821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pascarella G., Strumia A., Piliego C., Bruno F., Buono R.D., Costa F., Scarlata S., Agro F.E. COVID-19 diagnosis and management: a comprehensive review. J. Intern. Med. 2020;288:192–206. doi: 10.1111/joim.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Risco C., Rodriguez J.R., Lopez-Lglesias C., Carrascosa J.L., Esteban M., Rodriguez D. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J. Virol. 2002;76:1839–1855. doi: 10.1128/JVI.76.4.1839-1855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villanueva R.A., Rouille Y., Dubuisson J. Interactions between virus proteins and host cell membranes during the viral life cycle. Int. Rev. Cytol. 2005;245:171–244. doi: 10.1016/S0074-7696(05)45006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deinhardt-Emmer S., Bottcher S., Haring C., Giebeler L., Henke A., Zell R., Jungwirth J., Jordan P.M., Werz O., Hornung F., Brandt C., Marquet M., Mosig A.S., Pletz M.W., Schacke M., Rodel J., Heller R., Nietzsche S., Loffler B., Ehrhardt C. SARS-CoV-2 causes severe epithelial inflammation and barrier dysfunction. J. Virol. 2021;95 doi: 10.1128/JVI.00110-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karmakar D., Lahiri B., Ranjan P., Chatterjee J., Lahiri P., Sengupta S. Road Map to Understanding SARS-CoV-2 Clinico-Immunopathology and COVID-19 Disease Severity. Pathogens. 2020;10:5. doi: 10.3390/pathogens10010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., Meer F.V.D., Kallio K., Kaya T., Anastasina M., Smura T., Levanov L., Szirovicza L., Tobi A., Kallio-Kokko H., Osterlund P., Joensuu M., Meunier F.A., Butcher S.J., Winkler M.S., Mollenhauer B., Helenius A., Gokce O., Teesalu T., Hepojoki J., Vapalahti O., Stadelmann C., Balisteri G., Simons M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khanmohammadi S., Rezaei N. Role of Toll-like receptors in the pathogenesis of COVID-19. J. Med. Virol. 2021;93:2735–2739. doi: 10.1002/jmv.26826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ragab D., Eldin H.S., Taeimah M., Khattab R., Salem R. The COVID-19 Cytokine Storm; What We Know So Far. Front. Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaplin D.D. Overview of the immune response. J. Allergy Clin. Immunol. 2010;125 doi: 10.1016/j.jaci.2009.12.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marshall J.S., Warrington R., Watson W., Kim H.L. An introduction to immunology and immunopathology. Allergy, Asthma Clin. Immunol. 2018;14:49. doi: 10.1186/s13223-018-0278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson M.R., Kaminski J.J., Kurt-Jones E.A., Fitzgerald K.A. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 2011;3:920–940. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorley A.J., Grandolfo D., Lim E., Goldstraw P., Young A., Tetley T.D. Innate immune response to bacterial ligands in the peripheral human lung-role of alveolar epithelial TLR expression and signaling. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayden M.S., Ghosh S. Shared principles in NF-Kappa B signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 40.Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engelhardt E., Toksov A., Goebeler M., Debus S., Brocker E.B., Gillitzer R. Chemokines IL-8, GROalpha, MCP-1, IP-10, and Mig are sequentially and differentially expressed during phase-specific infiltration of leukocyte subsets in human wound healing. Am. J. Pathol. 1998;153:1849–1860. doi: 10.1016/s0002-9440(10)65699-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu H., Chan J.F.-W., Wang Y., Yuen T.T.-T., Chai Y., Hou Y., Shuai H., Yang D., Huang X., Zhang X., Cai J.-P., Zhou J., Yuan S., Kok K.-H., K. K.-W. To, Chan I.H.-Y., Zhang A.J., Sit K.-Y., Au W.-K., Yuen K.-Y. Comparative Replication and Immune Activation Profiles of SARS-CoV-2 and SARS-CoV in Human Lungs: An Ex Vivo Study With Implications for the Pathogenesis of COVID-19. Clin. Infect. Dis. 2020;71:1400–1409. doi: 10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim Y.-M., Shin E.-C. Type I and III interferon responses in SARS-CoV-2infection. Exp. Mol. Med. 2021;53:750–760. doi: 10.1038/s12276-021-00592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mohammad M.H.S. Immune response scenario and vaccine development for SARS-Cov-2 infection. Int. Immunopharmacol. 2021;94:107439. doi: 10.1016/j.intimp.2021.107439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hogland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., Tenoever B.R. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;28:1036–1045. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tay M.Z., Poh C.M., Renia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li G., Fan Y., Han T., Li Z., Zhou P., Pan P., Wang W., Hu D., Liu X., Zhang Q., Wu J. Coronavirus infections and immune responses. J. Med. Virol. 2020;92:424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walls A.C., Park Y.-J., Tororici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clay C.C., Donart N., Fomukong N., Knight J.B., Lei W., Prince L., Hahn F., Westriene J.V., Harrod K.S. Primary severe acute respiratory syndrome coronavirus infection limits replication but not lung inflammation upon homologous rechallenge. J. Virol. 2012;86:4234–4244. doi: 10.1128/JVI.06791-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Z.-Y., Huang Y., Ganesh L., Leung K., Kong W.-P., Schwartz O., Subbarao K., Nabel G.J. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 2004;78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bailey S.R., Nelson M.H., Majchrzak K., Bowers J.S., Wyatt M.M., Smith A.S., Neal L.R., Shirai K., Carpenito C., June C.H., Zilliox M.J., Paulos C.M. Human CD26 high T cells elicit tumor immunity against multiple malignancies via enhanced migration and persistence. Nat. Commun. 2017;8:1961. doi: 10.1038/s41467-017-01867-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang K., Chen W., Zhang Z., Deng Y., Lian J.-Q., Du P., Wei D., Zhang Y., Sun X.-X., Gong L., Yang X., He L., Zhang L., Yang Z., Geng J.-J., Chen R., Zhang H., Wang B., Zhu Y.-M., Nan G., Jiang J.-L., Li L., Wu J., Lin P., Huang W., Xie L., Zheng Z.-H., Zhang K., Miao J.-L., Cui H.-Y., Huang M., Zhang J., Fu L., Yang X.-M., Zhao Z., Sun S., Gu H., Wang Z., Wang C.-F., Lu Y., Liu Y.-Y., Wang Q.-Y., Bian H., Zhu P., Chen Z.-N. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther. 2020;5:283. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang I.-C., Bailey C.C., Weyer J.L., Radoshitzky S.R., Becker M.M., Chiang J.J., Brass A.L., Ahmed A.A., Chi X., Dong L., Longobardi L.E., Boltz D., Kuhn J.H., Elledge S.J., Bavari S., Denison M.R., Choe H., Farzan M. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ying T., Li W., Dimitrov D.S. Discovery of T-Cell Infection and Apoptosis by Middle East Respiratory Syndrome Coronavirus. J. Infect. Dis. 2016;213:877–879. doi: 10.1093/infdis/jiv381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brosnahan S.B., Jonkman A.H., Kugler M.C., Munger J.S., Kaufman D.A. COVID-19 and Respiratory System Disorders: Current Knowledge, Future Clinical and Translational Research Questions. Arterioscler. Thromb. Vasc. Biol. 2020;40:2586–2597. doi: 10.1161/ATVBAHA.120.314515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kubicka-Sierszen A., Grzegorczyk J. The influence of infectious factors on dendritic cell apoptosis. Arch. Med. Sci. 2015;11:1044–1051. doi: 10.5114/aoms.2015.54860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amarante-Mendes G.P., Adjemian S., Branco L.M., Zanetti L.C., Weinlich R., Bortoluci K.R. Pattern recognition receptors and the host cell death molecular machinery. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.02379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu H., Lin L., Zhang Z., Zhang H., Hu H. Targeting NF-kB pathway for the therapy of diseases: mechanism and clinical study. Signal Transduct Target Ther. 2020;5:209. doi: 10.1038/s41392-020-00312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Conti P., Ronconi G., Caraffa A., Gallenga C., Ross R., Frydas I., Kritas S. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVID-19 or SARS-CoV-2): anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents. 2020;34:327–331. doi: 10.23812/CONTI-E. [DOI] [PubMed] [Google Scholar]
- 60.Thangam E.B., Jemima E.A., Singh H., Baig M.S., Khan M., Mathias C.B., Church M.K., Saluja R. The Role of Histamine and Histamine Receptors in Mast Cell-Mediated Allergy and Inflammation: The Hunt for New Therapeutic Targets. Front. Immunol. 2018;9:1873. doi: 10.3389/fimmu.2018.01873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jang T.Y., Kim Y.H. Interleukin-33 and Mast Cells Bridge Innate and Adaptive Immunity: From the Allergologist’s Perspective. Int Neurourol. 2015;19:142–150. doi: 10.5213/inj.2015.19.3.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whitmire J.K. Induction and function of virus-specific CD4+ T cell responses. Virology. 2011;411:216–228. doi: 10.1016/j.virol.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun S.-C. The non-canonical NF-kB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017;17:545–558. doi: 10.1038/nri.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Teng T.-S., Ji A.-L., Ji X.-Y. Li, Neutrophils and Immunity; From Bactericidal Action to Being Conquered, J Immunol Res.: 2017. Y.-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosales C. Neutrophil: A Cell with Many Roles in Inflammation or Several Cell Types? Front. Physiol. 2018;9:113. doi: 10.3389/fphys.2018.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang J., Li Q., Yin Y., Zhang Y., Cao Y., Lin X., Huang L., Hoffmann D., Lu M., Qiu Y. Excessive Neutrophils and Neutrophil Extracellular Traps in COVID-19. Front. Immunol. 2020;11:2063. doi: 10.3389/fimmu.2020.02063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Savelli G., Bonacina M., Rizzo A., Zaniboni A. Activated macrophages are the main inflammatory cell in COVID-19 interstitial pneumonia infiltrates. Is it possible to show their metabolic activity and thus the grade of inflammatory burden with 18F-Fluorocholine PET/CT? Med. Hypotheses. 2020;144:109885. doi: 10.1016/j.mehy.2020.109885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Otsuka R., Seino K.I. Macrophage activation syndrome and COVID-19. Inflamm Regen. 2020;40:19. doi: 10.1186/s41232-020-00131-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fujiwara N., Kobayashi K. Macrophages in inflammation. Curr Drug Targets Inflamm Allergy. 2005;4:281–286. doi: 10.2174/1568010054022024. [DOI] [PubMed] [Google Scholar]
- 70.Rowaiye A.B., Okpalefe O.A., Adejoke O.O., Ogidigo J.O., Oladipo O.H., Ogu A.C., Oli A.N., Olofinase S., Onyekwere O., Abubakar A.R., Jahan D., Islam S., Dutta S., Haque M. Attenuating the Effects of Novel COVID-19 (SARS-CoV-2) Infection-Induced Cytokine Storm and the Implications. J. Inflamm. Res. 2021;14:1487–1510. doi: 10.2147/JIR.S301784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lim S., Bae J.H., Kwon H.-S., Nauck M.A. COVID-19 and diabetes mellitus: from pathophysiology to clinical management. Nat. Rev. Endocrinol. 2021;17:11–30. doi: 10.1038/s41574-020-00435-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Poggiali E., Zaino D., Immovilli P., Rovero L., Losi G., Dacrema A., Nuccetelli M., Vadacca G.B., Guidetti D., Vercelli A., Magnacavallo A., Bernardini S., Terracciano C. Lactate dehydrogenase and C-reactive protein as predictors of respiratory failure in COVID-19 patients. Clin. Chim. Acta. 2020;509:135–138. doi: 10.1016/j.cca.2020.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chiappetta S., Sharma A.M., Bottino V., Stier C. COVID-19 and the role of chronic inflammation in patients with obesity. JEPHR. 2020;44:1790–1792. doi: 10.1038/s41366-020-0597-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kany S., Vollrath J.T., Relja B. Cytokines in Inflammatory Disease. Int. J. Mol. Sci. 2019;20:6008. doi: 10.3390/ijms20236008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Samuel C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ali S., Mann-Nuttel R., Schulze A., Richter L., Alferink J., Scheu S. Sources of Type I Interferons in Infectious Immunity: Plasmacytoid Dendritic Cells Not Always in the Driver’s Seat. Front. Immunol. 2019;10:778. doi: 10.3389/fimmu.2019.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McNab F., Mayer-Barber K., Sher A., Wack A., O’Garra A. Type I Interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Erlandsson L., Blumenthal R., Eloranta M.L., Engel H., Alm G., Weiss S., Leanderson T. Interferon-beta is required for interferon-alpha production in mouse fibroblasts. Curr. Biol. 1998;8:223–226. doi: 10.1016/s0960-9822(98)70086-7. [DOI] [PubMed] [Google Scholar]
- 79.Parkin J., Cohen B. An overview of the immune system. Lancet. 2001;357:1777–1789. doi: 10.1016/S0140-6736(00)04904-7. [DOI] [PubMed] [Google Scholar]
- 80.Fara A., Mitrev Z., Rosalia R.A., Assas B.M. Cytokine storm and COVID-19:a chronicle of pro-inflammatory cytokines. Open Biol. 2020;10:200160. doi: 10.1098/rsob.200160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cavalcante-Silva L.H.A., Carvalho D.C.M., de Almeida Lima E., Galvao J.G.F.M., de Franca da Silva J.S., de Sales-Neto J.M., Rodrigues-Mascarenhas S. Neutrophils and COVID-19: The road so far. Int. Immunopharmacol. 2021;90:107233. doi: 10.1016/j.intimp.2020.107233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ye Q., Wang B., Mao J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Inf. Secur. 2020;80:607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mubarak A., Alrfaei B., Aljurayyan A., Alqafil M.M., Farrag M.A., Hamed M.E., Alosaimi B., Almajhdi F., Alturaiki W. In vivo and in vitro Evaluation of Cytokine Expression Profiles During Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Infection. J. Inflamm. Res. 2021;14:2121–2131. doi: 10.2147/JIR.S312337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Swanson M.A., Lee W.T., Sanders V.M. IFN-gamma production by Th1 cells generated from naïve CD4+ T cells exposed to norepinephrine. J. Immunol. 2001;166:232–240. doi: 10.4049/jimmunol.166.1.232. [DOI] [PubMed] [Google Scholar]
- 85.Bhat P., Leggatt G., Waterhouse N., Frazer I.H. Interferon-γ derived from cytotoxic lymphocytes directly enhances their motility and cytotoxicity. Cell Death Dis. 2017;8:2836. doi: 10.1038/cddis.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ganguly D., Paul K., Bagchi J., Rakshit S., Mandal L., Bandyopadhyay G., Bandyopadhyay S. Granulocyte-macrophage colony-stimulating factor derives monocytes to CD14 low CD38+ DCSIGN-interleukin-10-producing myeloid cells with differential effects on T-cell subsets. Immunology. 2007;121:499–507. doi: 10.1111/j.1365-2567.2007.02596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heimfarth L., Serafini M.R., Martins-Filho P.R., de Souza Siqueria Quintans J., Quintans-Junior L.J. Drug repurposing and cytokine management in response to COVID-19: A review. Int. Immunopharmacol. 2020;88:106947. doi: 10.1016/j.intimp.2020.106947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Santarlasci V., Cosmi L., Maggi L., Liotta F., Annunziato F. IL-1 and T Helper Immune Responses. Front. Immunol. 2013;4:182. doi: 10.3389/fimmu.2013.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oelmann E., Stein H., Berdel W.E., Herbst H. Expression of Interleukin-1 and Interleukin-1 Receptors Type 1 and Type 2 in Hodgkin Lymphoma. PLoS One. 2015;10 doi: 10.1371/journal.pone.0138747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dinarello C.A. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol. Rev. 2018;281:8–27. doi: 10.1111/imr.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kaneko N., Kurata M., Yamamoto T., Morikawa S., Masumoto J. The role of interleukin-1 in general pathology. Inflamm Regen. 2019;39:12. doi: 10.1186/s41232-019-0101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Galli S.J., Tsai M., Piliponsky A.M. The development of allergic inflammation. Nature. 2008;454:445–454. doi: 10.1038/nature07204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schmitz N., Kurrer M., Bachmann M.F., Kopf M. Interleukin-1 is responsible for acute lung immunopathology but increases survival of respiratory influenza virus infection. J. Virol. 2005;79:6441–6448. doi: 10.1128/JVI.79.10.6441-6448.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang L., Liu S., Liu J., Zhang Z., Wan X., Huang B., Chen Y., Zhang Y. COVID-19: immunopathogenesis and immunotherapeutics. Signal Transduct Target Ther. 2020;5:128. doi: 10.1038/s41392-020-00243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Haljasmagi L., Salumets A., Rumm A.P., Jurgenson M., Krassohhina E., Remm A., Sein H., Kareinen L., Vapalathi O., Sironen T., Peterson H., Milani L., Tamm A., Hayday A., Kisand K., Peterson P. Longitudinal proteomic profiling reveals increased early inflammation and sustained apoptosis proteins in severe COVID-19. Sci. Rep. 2020;10:20533. doi: 10.1038/s41598-020-77525-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Iyer S.S., Cheng G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit. Rev. Immunol. 2012;32:22–63. doi: 10.1615/critrevimmunol.v32.i1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu L., Zhang H., Dauphars D.J., He Y.-W. A Potential Role of Interleukin 10 in COVID-19 Pathogenesis. Trends Immunol. 2021;42:3–5. doi: 10.1016/j.it.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dimopoulos G., de Mast Q., Markou N., Theodorakopoulou M., Komnos A., Mouktaroudi M., Netea M.G., Spyridopoulos T., Verheggen R.J., Hoogerwerf A. Lachana, Van de Veerdonk F.L., Giamarellos-Bourboulis E.J. Favorable anakinra responses in severe Covid-19 patients with secondary hemophagocytic lymphohistiocytosis. Cell Host Microbe. 2020;28:117–123. doi: 10.1016/j.chom.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Horiuchi T., Mitoma H., Harashima S.-I., Tsukamoto H., Shimoda T. Transmembrane TNF-alpha: structure, function and interaction with anti-TNF agents. Rheumatology. 2020;49:1215–1228. doi: 10.1093/rheumatology/keq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Long S.S. Diagnosis and management of undifferentiated fever in children. J. Inf. Secur. 2016;72:S68–S76. doi: 10.1016/j.jinf.2016.04.025. [DOI] [PubMed] [Google Scholar]
- 101.Chen G., Wu D., Guo W., Cao Y., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pedersen S.F., Ho Y.-C. SARS-Cov-2: a storm is raging. J. Clin. Invest. 2020;130:2202–2205. doi: 10.1172/JCI137647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., Bucci E., Piacentini M., Ippolito G., Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chan M.C.W., Cheung C.Y., Chui W.H., Tsao S.W., Nicholls J.M., Chan Y.O., Chan R.W.Y., Long H.T., Poon L.L.M., Guan Y., Peiris J.S.M. Proinflammatory cytokine responses induced by influenza A (H5N1) viruses in primary human alveolar and bronchial epithelial cells. Respir. Res. 2005;6:135. doi: 10.1186/1465-9921-6-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Herold S., Becker C., Ridge K.M., Budinger G.R.S. Influenza virus-induced lung injury: pathogenesis and implications for treatment. Eur. Respir. J. 2015;45:1463–1478. doi: 10.1183/09031936.00186214. [DOI] [PubMed] [Google Scholar]
- 106.Buicu A.-L., Cernea S., Benedek I., Buicu C.-F., Benedek T. Systematic Inflammation and COVID-19 Mortality in Patients with Major Noncommunicable Diseases: Chronic Coronary Syndromes. Diabetes and Obesity, J Clin Med. 2021;10:1545. doi: 10.3390/jcm10081545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen Z., Wherry E.J. T cell responses in patients with COVID-19. Nat. Rev. Immunol. 2020;20:529–536. doi: 10.1038/s41577-020-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tavakolpour S., Rakshandehroo T., Wei E.X., Rashidian M. Lymphopenia during the COVID-19 infection: What it shows and what can be learned. Immunol. Lett. 2020;225:31–32. doi: 10.1016/j.imlet.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.de Candia P., Prattichizzo F., Garavelli S., Matarese G. T Cells: Warriors of SARS-CoV-2 Infection. Trends Immunol. 2021;42:18–30. doi: 10.1016/j.it.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Osinska I., Popko K., Demkow U. Perforin: an important player in immune response. Cent Eur J Immunol. 2014;39:109–115. doi: 10.5114/ceji.2014.42135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bordoni V., Sacchi A., Cimini E., Notari S., Grassi G., Tartaglia E., Casetti R., Giancola M.L., Bevilacqua N., Maeurer M., Zumla A., Locatelli F., De Benedetti F., Palmieri F., Marchioni L., Capobianchi M.R., D’Offizi G., Petrosillo N., Antinori A., Nicastri E., Ippolito G., Agrati C. An inflammatory Profile Correlates With Decreased Frequency of Cytotoxic Cells in Coronavirus Disease. Clin. Infect. Dis. 2019;71(2020):2272–2275. doi: 10.1093/cid/ciaa577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lei L., Qian H., Yang X., Zhang X., Zhang D., Dai T., Guo R., Shi L., Cheng Y., Zhang B., Zhou X., Hu J., Guo Y. The phenotypic changes of γδ T cells in COVID-19 patients. J. Cell. Mol. Med. 2020;24:11603–11606. doi: 10.1111/jcmm.15620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexpected individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nockher W.A., Wiemer J., Scherberich J.E. Haemodialysis monocytopenia: differential sesquestration kinetics of CD14+CD16+ and CD14++ blood monocyte subset. Clin. Exp. Immunol. 2001;123:49–55. doi: 10.1046/j.1365-2249.2001.01436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Martinez F.O., Combes T.W., Orsenigo F., Gordon S. Monocyte activation in systematic Covid-19 infection: Assay and rationale. EBioMedicine. 2020;59:102964. doi: 10.1016/j.ebiom.2020.102964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sampath P., Moideen K., Ranganathan U.D., Bethunaickan R. Monocyte Subsets: Phenotypes and Function in Tuberculosis Infection. Front. Immunol. 2018;9:1726. doi: 10.3389/fimmu.2018.01726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Galeotti C., Bayry J. Autoimmune and inflammatory diseases following COVID-19. Nat. Rev. Rheumatol. 2020;16:413–414. doi: 10.1038/s41584-020-0448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Castelli V., Cimini A., Ferri C. Cytokine Storm in COVID-19:When You Come Out of the Storm, You Won’t Be the Same Person Who Walked in. Front. Immunol. 2020;11:2132. doi: 10.3389/fimmu.2020.02132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schulert G.S., Grom A.A. Macrophage activation syndrome and cytokine-directed therapies. Best Pract. Res. Clin. Rheumatol. 2014;28:277–292. doi: 10.1016/j.berh.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lerkvaleekul B., Vilaiyuk S. Macrophage activation syndrome: early diagnosis is key. Open Access Rheumatol. 2018;10:117–128. doi: 10.2147/OARRR.S151013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., Yuan Z., Feng Z., Zhang Y., Wu Y., Chen Y. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yang J., Zhang L., Yu C., Yang X.-F., Wang H. Monocyte and macrophage differentiation: circulatory inflammatory monocyte as biomarker for inflammatory diseases. Biomark Res. 2014;2:1. doi: 10.1186/2050-7771-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ghoreschi K., Laurence A., O’Shea J. Janus Kinases in immune cell signaling. Immunol. Rev. 2009;228:273–287. doi: 10.1111/j.1600-065X.2008.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Petrina M., Martin J., Basta S. Granulocyte macrophage colony-stimulating factor has come of age: From a vaccine adjuvant to antiviral immunotherapy. Cytokine Growth Factor Rev. 2021;59:101–110. doi: 10.1016/j.cytogfr.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Imai Y., Kuba K., Neely G.G., Yaghubian-Malhami R., Perkmann T., Loo G.V., Ermolaeva M., Veldhuizen R., Leung Y.H.C., Wan H., Liu H., Sun Y., Pasparakis M., Kopf M., Mech C., Bavari S., Peiris J.S.M., Slutsky A.S., Akira S., Hultqvist M., Holmdahl R., Nicholls J., Jiang C., Binder C.J., Penninger J.M. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008;133:235–249. doi: 10.1016/j.cell.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lund J.M., Alexopoulou L., Sato A., Karow M., Adams N.C., Gale N.W., Iwasaki A., Flavell R.A. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. U. S. A. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li S.-F., Gong M.-J., Zhao F.-R., Shao J.-J., Xie Y.-L., Zhang Y.-G., Chang H.-Y. Type I Interferons: Distinct Biological Activities and Current Applications for Viral Infection. Cell. Physiol. Biochem. 2018;51:2377–2396. doi: 10.1159/000495897. [DOI] [PubMed] [Google Scholar]
- 130.Kelley N., Jeltema D., Duan Y., He Y. The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation. Int. J. Mol. Sci. 2019;20:3328. doi: 10.3390/ijms20133328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Duque G.A., Descoteaux A. Macrophage cytokines: involvement in immunity and infectious disease. Front. Immunol. 2014;5:491. doi: 10.3389/fimmu.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Pere H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pene F., Marin N., Roche N., Szwebel T.-A., Merkling S.H., Treluyer J.-M., Veyer D., Mouthon L., Blanc C., Tharaux P.-L., Rozenberg F., Fischer A., Duffy D., Rieux-Laucat F., Kerneis S., Terrier B. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tang Y., Liu J., Zhang D., Xu Z., Ji J., Wen C. Cytokine Storm in COVID-19: The Current Evidence and Treatment Strategies. Front. Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sun X., Wang T., Cai D., Hu Z., Chen J., Liao H., Zhi L., Wei H., Zhang Z., Qiu Y., Wang J., Wang A. Cytokine storm intervention in the early stages of COVID-19 pneumonia. Cytokine Growth Factor Rev. 2020;53:38–52. doi: 10.1016/j.cytogfr.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bettini E., Locci M. SARS-CoV-2 mRNA vaccines: immunological mechanism and beyond. Vaccines (Basel) 2021;9:147. doi: 10.3390/vaccines9020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ali M.G., Zhang Z., Gao Q., Pan M., Rowan E.G., Zhang J. Recent advances in therapeutic applications of neutralizing antibodies for virus infection: an overview. Immunol. Res. 2020;68:325–339. doi: 10.1007/s12026-020-09159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Speiser D.E., Bachmann M.F. COVID-19: Mechanism of Vaccination and Immunity. Vaccines (Basel) 2020;8:404. doi: 10.3390/vaccines8030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bian J., Li Z. Angiotensin-converting enzyme 2 (ACE2): SARS-CoV-2 receptor and RAS modulator. Acta Pharm. Sin. B. 2021;11:1–12. doi: 10.1016/j.apsb.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shin M.D., Shukla S., Chung Y.H., Beiss V., Chan S.K., Ortega-Rivera O.A., Wirth D.M., Chen A., Sack M., Pokorski J.K., Steinmetz N.F. COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 2020;15:646–655. doi: 10.1038/s41565-020-0737-y. [DOI] [PubMed] [Google Scholar]
- 141.Noval M.G., Kaczmarek M.E., Koide A., Rodriguez-Rodriguez B.A., Louie P., Tada T., Hattori T., Panchenko T., Romero L.A., Teng K.W., Bazley A., de Vries M., Samanovic M.I., Weiser J.N., Aifantis I., Cangiarella J., Mulligan M.J., Desvignes L., Dittmann M., Landau N.R., Aguero-Rosenfeld M., Koide S., Stapleford K.A. Antibody isotype diversity against SARS-CoV-2 is associated with differential serum neutralizing capacities. Sci. Rep. 2021;11:5538. doi: 10.1038/s41598-021-84913-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chao Y.X., Rotzschke O., Tan E.-K. The role of IgA in COVID-19. Brain Behav. Immun. 2020;87:182–183. doi: 10.1016/j.bbi.2020.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mohan T., Verma P., Rao D.N. Novel adjuvants & delivery vehicles for vaccines development: a road ahead. Indian J. Med. Res. 2013;138:779–795. [PMC free article] [PubMed] [Google Scholar]
- 144.Duthie M.S., Windish H.P., Fox C.B., Reed S.G. Use of defined TLR ligands as adjuvants within human vaccines. Immunol. Rev. 2011;239:179–196. doi: 10.1111/j.1600-065X.2010.00978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., Lopez-Camacho C., Slon-Campos J., Zhao Y., Stuart D.I., Paesen G.C., Grimes J.M., Antson A.A., Bayfield O.W., Hawkins D.E.D.P., Ker D.S., Wang B., Turtle L., Subramaniam K., Thomson P., Zhang P., Dold C., Ratcliff J., Simmonds P., de Silva T., Sopp P., Wellington D., Rajapaksa U., Chen Y.-L., Salio M., Napolitani G., Paes W., Borrow P., Kessler B.M., Fry J.W., Schwabe N.F., Semple M.G., Baillie J.K., Moore S.C., Openshaw P.J.M., Ansari M.A., Dunachie S., Barnes E., Farter J., Kerr G., Goulder P., Lockett T., Levin R., Zhang Y., Jing R., Ho L.-P. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lipsitch M., Grda Y.H., Sette A., Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat. Rev. Immunol. 2020;20:709–713. doi: 10.1038/s41577-020-00460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Swain S.L., McKinstry K.K., Sturtt T.M. Expanding roles for CD4+ T cells in immunity to viruses. Nat. Rev. Immunol. 2012;12:136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jin P., Wang E. Polymorphism in clinical immunology-From HLA typing to immunogenetic profiling. J. Transl. Med. 2003;1:8. doi: 10.1186/1479-5876-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wieczorek M., Abualrous E.T., Sticht J., Alvaro-Benito M., Stolzenberg S., Noe F., Freund C. Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front. Immunol. 2017;8:292. doi: 10.3389/fimmu.2017.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chen B., Khodadoust M.S., Olsson N., Wagar L.E., Fast E., Liu C.L., Muftuoglu Y., Sworder B.J., Diehn M., Levy R., Davis M.M., Elias J.E., Altman R.B., Alizadeh A.A. Predicting HLA class II antigen presentation through integrated deep learning. Nat. Biotechnol. 2019;37:1332–1343. doi: 10.1038/s41587-019-0280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Aurisicchio L., Fridman A., Bagchi A., Scarselli E., La Monica N., Ciliberto G. A novel minigene scaffold for therapeutic cancer vaccines. Oncoimmunology. 2014;3:27529. doi: 10.4161/onci.27529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dong Y., Dai T., Wei Y., Zhang L., Zheng M., Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct Target Ther. 2020;5:237. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.L. J. Sigal, Activation of CD8 T Lymphocytes during Viral Infections, Encyclopedia of Immunobiology. 286-290.
- 154.Montfoort N.V., der Aa E.V., Woltman A.M. Understanding MHC class I presentation of viral antigens by human dendritic cells as a basis for rational design of therapeutic vaccines. Front. Immunol. 2014;5:182. doi: 10.3389/fimmu.2014.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gilbert S.C. T-cell-inducing vaccines- what’s the future. Immunology. 2012;135:19–26. doi: 10.1111/j.1365-2567.2011.03517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yaqinuddin A. Cross-immunity between respiratory coronaviruses may limit COVID-19 fatalities. Med. Hypotheses. 2020;144:110049. doi: 10.1016/j.mehy.2020.110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Guo L., Wang Y., Kang L., Hu Y., Wang L., Zhong J., Chen H., Ren L., Gu X., Wang G., Wang C., Dong X., Wu C., Han L., Wang Y., Fan G., Zou X., Li H., Xu J., Jin Q., Cao B., Wang J. Cross-reactive antibody against human coronavirus OC43 spike protein correlates with disease severity in COVID-19 patients: a retrospective study. Emerg Microbes Infect. 2021;10:664–676. doi: 10.1080/22221751.2021.1905488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jacofsky D., Jacofsky E.M., Jacofsky M. Understanding Antibody Testing for COVID-19. J. Arthroplast. 2020;37:S74–S81. doi: 10.1016/j.arth.2020.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ma H., Zeng W., He H., Zhao D., Jiang D., Zhou P., Cheng L., Li Y., Ma X., Jin T. Serum IgA, IgM, and IgG responses in COVID-19. Cell. Mol. Immunol. 2020;17:773–775. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Subbarao K., Mahanty S. Respiratory Virus Infections: Understanding COVID-19. Immunity. 2020;52:905–909. doi: 10.1016/j.immuni.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Varadhachary A., Chtterjee D., Garza J., Garr R.P., Foley C., Letkeman A.F., Dean J., Haug D., Breeze J., Traylor R., Malek A., Nath R., Linbeck L. Salivary anti-SARS-CoV-2 IgA as an accessible biomarker of mucosal immunity against COVID-19. MedRxiv. 2020 doi: 10.1101/2020.08.07.20170258. [DOI] [Google Scholar]
- 162.Sterlin D., Mathian A., Miyara M., Mohr A., Anna F., Clear L., Quentric P., Fadlallah J., Devilliers H., Ghillani P., Gunn C., Hockett R., Mudumba S., Guihot A., Luyt C.E., Mayaux J., Beurton A., Fourati S., Bruel T., Schwartz O., Lacorte J.M., Yssel H., Parizot C., Dorgham K., Charneau P., Amoura Z., Gorochov G. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xiaojie S., Yu L., Lei Y., Guang Y., Min Q. Neutralizing antibodies targeting SARS-CoV-2 spike protein. Stem Cell Res. 2020;50:102125. doi: 10.1016/j.scr.2020.102125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Maginnis M.S. Virus-Receptor Interactions: The Key to Cellular Invasion. J. Mol. Biol. 2018;430:2590–2611. doi: 10.1016/j.jmb.2018.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Xu Y., Liu Y., Lou Z., Qin L., Li X., Bai Z., Pang H., Tien P., Gao G.F., Rao Z. Structural basis for coronavirus-mediated membrane fusion. Crystal structure of mouse hepatitis virus spike protein fusion core. J. Biol. Chem. 2004;279:30514–30522. doi: 10.1074/jbc.M403760200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sepahvandi A., Ghaffari M., Bahmanpour A.H., Moztarzadeh F., Zarrintaj P., Uludag H., Mozafari M. COVID-19: insights into virus-receptor interactions. Molecular Biomedicine. 2021;2 doi: 10.1186/s43556-021-00033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fedry J., Hurdiss D.L., Wang C., Li W., Obal G., Drulyte I., Du W., Howes S.C., Van Kuppeveld F.J.M., Forster F., Bosch B.-J. Structural insights into the cross-neutralization of SARS-CoV and SARS-CoV-2 by the human monoclonal antibody 47D11. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abf5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yang J., Petitjean S.J.L., Koehler M., Zhang Q., Dumitru A.C., Chen W., Derclaye S., Vincet S.P., Soumillion P., Alsteens D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020;11:4541. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang C., Li W., Drabek D., Okba N.M.A., Haperen R.V., Osterhaus A.D.M.E., Van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang Y., Jonsson F. Expression, Role, and Regulation of Neutrophil Fcγ Receptors. Front. Immunol. 2019;10:1958. doi: 10.3389/fimmu.2019.01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cheng M.H., Zhang S., Porritt R.A., Rivas M.N., Paschold L., Wollscher E., Binder M., Arditi M., Bahar I. Superantigenic character of an insert unique to SARS-CoV-2 spike supported by skewed TCR repertoire in patients with hyperinflammation. Proc. Natl. Acad. Sci. U. S. A. 2020;117:25254–25262. doi: 10.1073/pnas.2010722117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Poland G.A., Ovsyannikova I.G., Kennedy R.B. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020;396:1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ehrlich S.F., Quesenberry C.P., Jr., Van Den Eeden S.K., Shan J., Ferrara A. Patients diagnosed with diabetes are at increased risk for asthma, chronic obstructive pulmonary disease, pulmonary fibrosis, and pneumonia but not lung cancer. Diabetes Care. 2010;33:55–60. doi: 10.2337/dc09-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zabetakis I., Lordan R., Norton C., Tsoupras A. COVID-19: The Inflammation Link and the Role of Nutrition in Potential Mitigation. Nutrients. 2020;12:1466. doi: 10.3390/nu12051466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Callender L.A., Curran M., Bates S.M., Mairesse M., Weigandt J., Betts C.J. The impact of pre-existing comorbidities and therapeutic interventions on COVID-19. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.01991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tang N., Li D., Wang X., Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020;18:844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Biswas S., Thakur V., Kaur P., Khan A., Kulshrestha S., Kumar P. Blood clots in COVID-19 patients: Simplifying the curious mystery. Med. Hypotheses. 2021;146:110371. doi: 10.1016/j.mehy.2020.110371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Garg S.S., Gupta J., Sharma S., Sahu D. An insight into the therapeutic applications of coumarin compounds and their mechanism of action. Eur. J. Pharm. Sci. 2020;152:105424. doi: 10.1016/j.ejps.2020.105424. [DOI] [PubMed] [Google Scholar]
- 179.Nishikawa M. Antiplatelet drugs. Nihon Rinsho. 2014;72:1248–1253. [PubMed] [Google Scholar]
- 180.Demeulemeester F., Punder K., Van Heijningen M., Van Doesburg F. Obesity as a Risk Factor for Severe COVID-19 and Complications: A Review. Cells. 2021;10:933. doi: 10.3390/cells10040933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Li J., Wang X., Chen J., Zuo X., Zhang H., Deng A. COVID-19 infection may cause ketoacidosis. Diabetes Obes. Metab. 2020;22:1935–1941. doi: 10.1111/dom.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Singh A.K., Singh R., Joshi S.R., Misra A. Mucormycosis in COVID-19: A systematic review of case reported worldwide and in India. Diabetes Metab Syndr. 2021;15:102146. doi: 10.1016/j.dsx.2021.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Spellberg B., Walsh T.J., Kontoyiannis D.P., Edwards J., Jr., Ibrahim A.S. Recent advances in the management of mucormycosis: from bench to bedside. Clin. Infect. Dis. 2009;48:1743–1751. doi: 10.1086/599105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Frediansyah A., Nainu F., Dhama K., Mudatsir M., Harapan H. Remdesivir and its antiviral activity against COVID-19: A systematic review. Clin Epidemiol Glob Health. 2021;9:123–127. doi: 10.1016/j.cegh.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hi Y., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F.G., Horby P.W., Cao B., Wang C. Inhibition of blood clotsRemdesivir in adults with severe COVID-19: a randomized, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ceriello A., Prattichizzo F. Pharmacological management of COVID-19 in type 2 diabetes. J. Diabetes Complicat. 2021;35:107927. doi: 10.1016/j.jdiacomp.2021.107927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sharma P., Kumar A., Anikhindi S., Bansal N., Singla V., Shivam K., Arora A. Effect of COVID-19 on Pre-existing Liver disease: What Hepatologist Should Know? J Clin Exp Hepatol. 2021;11:484–493. doi: 10.1016/j.jceh.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.McKee D.L., Sternberg A., Stange U., Laufer S., Naujokat C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol. Res. 2020;157:104859. doi: 10.1016/j.phrs.2020.104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Rodriguez-Garcia J.L., Sanchez-Nievas G., Arevalo-Serrano J., Garcia-Gomez C., Jimenez-Vizuete J.M., Martinez-Alfaro E. Baricitinib improves respiratory function in patients treated with corticosteroids for SARS-CoV-2 pneumonia: an observational cohort study. Rheumatology. 2021;60:399–407. doi: 10.1093/rheumatology/keaa587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Y. Furumoto, M. Gadina, The arrival of JAK inhibitors: advancing the treatment of immune and hematologic disorders, BioDrugs. 27 (201) 431-438. [DOI] [PMC free article] [PubMed]
- 191.Zoufaly A., Poglitsch M., Aberle J.H., Hoepler W., Seitz T., Traugott M., Grieb A., Pawelka E., Laferl H., Wenisch C., Neuhold S., Haider D., Stiasny K., Bergthaler A., Puchhammer-Stoeckl E., Mirazimi A., Montserrat N., Zhang H., Slutsky A.S., Penninger J.M. Human recombinant soluble ACE2 in severe COVID-19. Lancet Respir. Med. 2020;8:1154–1158. doi: 10.1016/S2213-2600(20)30418-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hoffmann M., Schroeder S., Kleine-Weber H., Muller M.A., Drosten C., Pohlmann S. Nafamostat Mesylate Blocks Activation of SARS-CoV-2: New Treatment Option for COVID-19. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mair-Jenkins J., Saavedra-Campos M., Baillie J.K., Cleary P., Khaw F.-M., Lim W.S., Makki S., Rooney K.D., Nguyen-Van-Tam J.S., Beck C.R. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J. Infect. Dis. 2015;211:80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sharma A., Tiwari S., Deb M.K., Marty J.L. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): a global pandemic and treatment strategies. Int. J. Antimicrob. Agents. 2020;56:106054. doi: 10.1016/j.ijantimicag.2020.106054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ye M., Fu D., Ren Y., Wang F., Wang D., Zhang F., Xia X., Lv T. Treatment with convalescent plasma for COVID-19 patients in Wuhan. China, J Med Virol. 2020;92:1890–1901. doi: 10.1002/jmv.25882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mostafa-Hedeab G. ACE2 as Drug Target of COVID-19 Virus Treatment, Simplified Updated Review. Rep Biochem Mol Biol. 2020;9 doi: 10.29252/rbmb.9.1.97. 97-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Brynskov J., Foegh P., Pedersen G., Ellervik C., Kirkegaard T., Bingham A., Saermark T. Tumour necrosis factor alpha converting enzyme (TACE) activity in the colonic mucosa of patients with inflammatory bowel disease. Gut. 2002;51:37–43. doi: 10.1136/gut.51.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Robinson P.C., Liew D.F.L., Liew J.W., Monaco C., Richards D., Shivakumar S., Tanner H.L., Feldmann M. The Potential for Repurposing Anti-TNF as a Therapy for the Treatment of COVID-19. Med (NY) 2020;1:90–102. doi: 10.1016/j.medj.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]