Abstract
To investigate the molecular mechanism of assembly factor for spindle microtubules (ASPM) in the regulation of the malignant progression of hepatocellular carcinoma (HCC), bioinformatics analysis was utilized to analyze the role of ASPM in the malignant progression of HCC and its potential interaction with the kinesin family member 11 (KIF11) gene. The expression levels of ASPM and KIF11 were detected by reverse transcription-quantitative PCR and western blotting. Following knockdown of ASPM expression, Cell Counting Kit-8/colony formation assays were performed to detect cell viability and proliferation. Wound healing and Transwell assays were employed to detect cell migration and invasion. Additionally, a co-immunoprecipitation (CO-IP) assay was used to detect whether there was an interaction between ASPM and KIF11. KIF11 overexpression was performed to verify if ASPM exerted its effects via KIF11. ASPM was highly expressed in HCC tissues and cells, and was closely associated with a poor prognosis of patients with HCC. Interference with ASPM expression markedly inhibited the viability, proliferation, invasion and migration of HCC cells. Using a CO-IP assay, it was revealed that there was an interaction between ASPM and KIF11. Rescue experiments subsequently revealed the regulatory effects of ASPM on the activity, proliferation, invasion and migration of HCC cells via KIF11. Finally, western blot analysis demonstrated that ASPM in combination with KIF11 promoted the malignant progression of HCC by regulating the activity of the Wnt/β-catenin signaling pathway. Therefore, the present study demonstrated that ASPM may interact with KIF11 in HCC cells to promote the malignant progression of HCC via the Wnt/β-catenin signaling pathway.
Keywords: assembly factor for spindle microtubules, kinesin family member 11, Wnt/β-catenin, hepatocellular carcinoma
Introduction
Liver cancer is one of the most common causes of cancer-associated mortality worldwide, with the fifth highest incidence and mortality rates in the United States, and a continuously increasing mortality rate on an annual basis, despite the advances in medical technology (1). Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer, and its occurrence has been associated with hepatitis B virus, hepatitis C virus, smoking, alcohol consumption, obesity, diabetes and other factors (2,3). However, there is a lack of effective treatment strategies for HCC due to its high metastasis and recurrence rates (4). In recent years, with in-depth research on the molecular mechanism of HCC tumorigenesis, certain targeted drugs for patients with HCC have been developed (5). However, their efficacy is unsatisfactory due to the heterogeneity of tumors and the complexity of the molecular mechanisms involved (6). Therefore, it is urgent to further study the pathogenesis of HCC and to explore potential molecular targets to facilitate the development of treatments for the disease.
Assembly factor for spindle microtubules (ASPM) is an ASP homologous gene of Drosophila melanogaster expressed in the cytoplasm, which is crucial for the normal mitotic spindle function of embryonic neuroblasts (7,8). A number of studies have reported that ASPM serves an important role in the progression of various types of cancer. For instance, Pai et al (9) has reported that ASPM increases the stability of the upstream regulatory factor of the Wnt signaling pathway, enhances the Wnt-dishevelled segment polarity protein 3-β-catenin signaling pathway and promotes the malignant progression of prostate cancer. Different subtypes of ASPM serve different roles in pancreatic cancer, and ASPM-II mainly regulates Wnt signaling and tumor growth, while ASPM-III selectively regulates the cell cycle progression of pancreatic ductal adenocarcinoma cells (10). In addition, it has been demonstrated in a number of studies that ASPM can serve as a biomarker in bladder cancer (11,12), breast cancer (13), non-small cell lung cancer (14), prostate cancer (15) and ovarian cancer (16,17). Additionally, high ASPM expression is often associated with a poor prognosis (18). Lin et al (19) reported that ASPM expression is upregulated in HCC, and that upregulation of ASPM increases the invasion and metastasis of HCC. Furthermore, several studies have suggested ASPM as a prognostic biomarker for HCC (20,21), although the specific mechanism of action of ASPM in HCC remains elusive.
Kinesin family member 11 (KIF11), which encodes EG5, belongs to the kinesin-like protein family. KIF11 serves an essential role in cell mitosis (22) and the transport of secreted proteins (23). It has been demonstrated that KIF11 mutation causes autosomal dominant familial exudative vitreoretinopathy, as well as microcephaly, with or without choroidal retinopathy, lymphedema or hypophrenia (24,25). KIF11 acts as an oncogene in tumors and is associated with a poor prognosis in breast cancer (26). Additionally, it promotes cell invasion, proliferation and self-renewal in glioblastoma (27).
The present study aimed to investigate the expression levels of ASPM in HCC cells and to determine whether interference with ASPM expression could affect cell proliferation, invasion, migration and epithelial-to-mesenchymal transition (EMT). It was further investigated whether ASPM was associated with KIF11 to promote the malignant progression of HCC, as well as the involvement of the Wnt/β-catenin signaling pathway in this process. The aim of the present study was to provide a novel approach to the targeted therapy and clinical diagnosis of HCC in the future.
Materials and methods
Cell lines
The normal human liver cell line (THLE-2) and human HCC cell lines (SK-HEP-1, Huh7, HCC-LM3 and Hep3B) were purchased from National Infrastructure of Cell Line Resource. Each cell line was cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Thermo Fisher Scientific, Inc.) and 100 U/ml Penicillin/Streptomycin (Thermo Fisher Scientific, Inc.) with 5% CO2 at 37˚C.
Bioinformatics analysis
Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/) is a network server for gene expression analysis based on tumor and normal samples in The Cancer Genome Atlas and Genotype-Tissue Expression databases (28). GEPIA was used to detect the expression levels of ASPM and KIF11 in patients with liver hepatocellular carcinoma (LIHC), as well as their overall survival and disease-free survival rates, with |log2 fold change|>1 and P<0.01 used as cut-off values. The Human Protein Atlas (HPA; http://www.proteinatlas.org/) database is a data resource that integrates a tissue atlas, cell atlas, pathological atlas, brain atlas, blood atlas and metabolic atlas. The present study analyzed the association between prognosis and ASPM and KIF11 in HCC using the pathological atlas in the HPA database. The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; https://www.string-db.org/) database is an online database for the search of known protein-protein interactions. The interactive association between ASPM and KIF11 was explored through the multi-protein retrieval of the STRING database. The present study examined the correlation between ASPM and KIF11 expression using the pan-cancer analysis platform starBase (http://starbase.sysu.edu.cn/), which enables the analysis of interactions between multiple RNAs (29).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. cDNA was synthesized using a reverse transcription system kit (Invitrogen; Thermo Fisher Scientific, Inc.) as follows: Priming at 25˚C for 5 min, reverse transcription at 37˚C for 30 min and reverse transcription inactivation at 85˚C for 5 min. For qPCR, the MiScript SYBR-Green PCR kit (Qiagen GmbH) was used according to the manufacturer's protocol. The thermocycling condition was as follows: Initial denaturation at 95˚C for 3 min, 40 cycles of denaturation at 95˚C for 10 sec and annealing at 60˚C for 30 sec and extension at 60˚C for 30 sec. The normalization method was adopted with GADPH as the internal reference, and the results were presented as the differences in target gene expression quantity between the control group and the experimental group with the use of 2-ΔΔCq values (30). The primers used in the present study are listed in Table I.
Table I.
Sequences of the primers used for reverse transcription-quantitative PCR.
| Gene | Sequence |
|---|---|
| KIF11 | F: 5'-GATGGACGTAAGGCAGCTCA-3' |
| R: 5'-TGTGGTGTCGTACCTGTTGG-3' | |
| ASPM | F: 5'-GGAGCGAGATCCCTCCAAAAT-3' |
| R: 5'-GGCTGTTGTCATACTTCTCATGG-3' | |
| Cadherin | F: 5'-CTGTGCCCAGCCTCCATGTTTT-3' |
| R: 5'-CTGGATAGCTGCCCATTGCAAGTTA-3' | |
| Cadherin | F: 5'-CCATCAAGCCTGTGGGAATC-3' |
| R: 5'-GCAGATCGGACCGGATACTG-3' | |
| Vimentin | F: 5'-TGTCCAAATCGATGTGGATGTTTC-3' |
| R: 5'-TTGTACCATTCTTCTGCCTCCTG-3' | |
| Ki-67 | F: 5'-CCACACTGTGTCGTCGTTTG-3' |
| R: 5'-CCGTGCGCTCATCCATTCA-3' | |
| PCNA | F: 5'-ATGTTTGAGGCACGCCTGATCCAG-3' |
| R: 5'-CTAAGATGCTTCCTCACTTCAATC-3' | |
| GAPDH | F: 5'-GGAGCGAGATCCCTCCAAAAT-3' |
| R: 5'-GGCTGTTGTCATACTTCTCATGG-3' |
F, forward; R, reverse; ASPM, assembly factor for spindle microtubules; KIF11, kinesin family member 11; PCNA, proliferating cell nuclear antigen.
Western blot analysis
The cells of each treatment group were collected and protein was extracted on ice with precooled RIPA lysis buffer (Beyotime Institute of Biotechnology) for 10 min. Protein was quantified using a BCA assay kit (Abcam) and the same amount of protein (25 µg/lane) was separated by 12% SDS-PAGE. Subsequently, the proteins were transferred to nitrocellulose membranes at 60 V for 120 min. The membranes were first blocked with 5% skimmed milk for 2 h at room temperature. The membranes were then incubated with primary antibodies at 4˚C overnight followed by HRP-conjugated goat anti-rabbit IgG secondary antibody at room temperature for 120 min. Finally, the chemiluminescence reaction was performed using an ECL kit (Beijing Solarbio Science & Technology Co., Ltd.), and images were captured to observe the blots. The densitometry was analyzed using the ImageJ software (v1.8; National Institutes of Health). Detailed information on the antibodies is shown in Table II. The experiment was repeated in triplicate.
Table II.
Information on the antibodies used for western blotting.
| Antibody | Dilution | Cat. no. | Specificity | Manufacturer |
|---|---|---|---|---|
| KIF11 | 1:1,000 | ab254298 | Rabbit monoclonal | Abcam |
| ASPM | 1:1,000 | ab238106 | Rabbit polyclonal | Abcam |
| PCNA | 1:1,000 | ab92552 | Rabbit monoclonal | Abcam |
| Ki-67 | 1:5,000 | ab16667 | Rabbit monoclonal | Abcam |
| MMP2 | 1:5,000 | ab92536 | Rabbit monoclonal | Abcam |
| MMP9 | 1:5,000 | ab76003 | Rabbit monoclonal | Abcam |
| N-cadherin | 1:5,000 | ab76011 | Rabbit monoclonal | Abcam |
| Vimentin | 1:5,000 | ab92547 | Rabbit monoclonal | Abcam |
| E-cadherin | 1:10,000 | ab133597 | Rabbit monoclonal | Abcam |
| β-catenin | 1:5,000 | ab32572 | Rabbit monoclonal | Abcam |
| p-GSK-3β | 1:1,000 | 9336 | Rabbit monoclonal | Cell Signaling Technology, Inc. |
| GSK-3β | 1:1,000 | 9325 | Rabbit monoclonal | Cell Signaling Technology, Inc. |
| GAPDH | 1:10,000 | ab181602 | Rabbit monoclonal | Abcam |
| IgG H&L (HRP) | 1:10,000 | ab6721 | Goat anti-rabbit HRP | Abcam |
ASPM, assembly factor for spindle microtubules; KIF11, kinesin family member 11; p, phosphorylated; PCNA, proliferating cell nuclear antigen.
Cell transfection
Silencer Select small interfering (si)RNA (si-ASPM; 5'-UGCCAUGGUGCAACUUGCU-3') and Silencer Select Negative Control No. 1 siRNA (si-NC; 5'-UUACCUCUAGUCGUCAUGU-3') were purchased from Thermo Fisher Scientific, Inc. pEGFP1-KIF11 and pEGFP-1 as negative control were purchased from Sangon Biotech Co., Ltd. A total of 0.8x106 cells were cultured in 35-mm culture dishes until the logarithmic growth phase, and Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfect 100 nM siRNA, 4 µg recombinant vector and their respective controls into Hep3B cells for 24 h at 37˚C, according to the product manual. Following transfection for 24 h, transfected Hep3B cells were utilized for subsequent experimentation.
Cell counting Kit-8 (CCK-8) assay
Transfected or non-tranfected Hep3B cells were inoculated into a 96-well plate at a density of 2,000 cells/well. Sterile CCK-8 solution was added after 0, 24, 48 and 72 h of cell culture. After 2 h of incubation at 37˚C, cell viability was detected. Absorbance at a wavelength of 450 nm was measured by a microplate reader (Molecular Devices, LLC).
Colony formation assay
Following digestion into a single-cell suspension using trypsin, the cells were inoculated into a 6-well plate at a density of 1,000 cells/well. After 1 week, each well was rinsed three times with PBS at room temperature. After fixation with 4% paraformaldehyde at room temperature for 15 min, 0.1% crystal violet (Thermo Fisher Scientific, Inc.) was added for staining at room temperature for 15 min. Subsequently, the clusters containing >50 cells were considered as a colony and counted under a light microscope (magnification, x40; Nikon Corporation).
Wound healing assay
Hep3B cells (5x105) were inoculated into a six-well plate. At 80% confluence, a gentle scrape on the well surface was performed with a pipette nozzle. Subsequently, the cells were washed three times with PBS to remove free cells. Following culture in serum-free medium at 37˚C for 24 h, cell migration was observed at 0 and 24 h and images were captured under a light microscope (magnification, x100; Nikon Corporation).
Transwell assay
Transwell inserts for 24-well plates (8 µm; Corning, Inc.) were coated with prediluted Matrigel (1:8; BD Biosciences) at 37˚C for 30 min. The cells were washed twice in PBS and then suspended in serum-free DMEM. A total of 5x105 cells in 200 µl medium were placed into the Matrigel-coated upper chamber and the lower chamber was filled with 400 µl DMEM with 10% FBS. Following incubation at 37˚C for 24 h, the non-invading cells in the upper chamber were gently removed. The cells on the submembrane surface were stained with 0.5% crystal violet for 15 min at room temperature and observed under a light microscope (magnification, x100; Nikon Corporation) and images were captured.
Co-immunoprecipitation (CO-IP) assay
The cells were separated, washed twice with PBS, and lysed in precooled cell lysis buffer for IP (cat. no. P0013; Beyotime Institute of Biotechnology) containing protease inhibitors. The supernatant was collected after centrifugation at 13,000 x g for 10 min at 4˚C. The supernatant of cell lysate (500 µg) were incubated at 4˚C overnight after the addition of 1 µg KIF11 (cat. no. ab254298; Abcam) or IgG antibody (cat. no. 2729; Cell Signaling Technology, Inc.). The pretreated 50 µg protein A agarose beads were added to the cell lysis buffer incubated with the antibody overnight for a slow shake, followed by incubation at 4˚C for 4 h, so that the antibody was coupled with protein A agarose beads. After the IP reaction, agarose beads were centrifuged at 1,000 x g for 3 min at 4˚C to the bottom of the tube. The supernatant was then carefully absorbed, and the agarose beads were washed three times with 1 ml lysis buffer. A total of 15 µl 2X SDS sample buffer was finally added for boiling at 100˚C for 5 min, followed by western blotting, as aforementioned.
Statistical analysis
All data were processed using GraphPad Prism 8.0 (GraphPad Software, Inc.). Measurement data are presented as the mean ± SD. Comparisons between two groups were conducted using a unpaired Student's t-test, and one-way ANOVA with Tukey's test was used to compare multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
ASPM is highly expressed in HCC tissues and cell lines
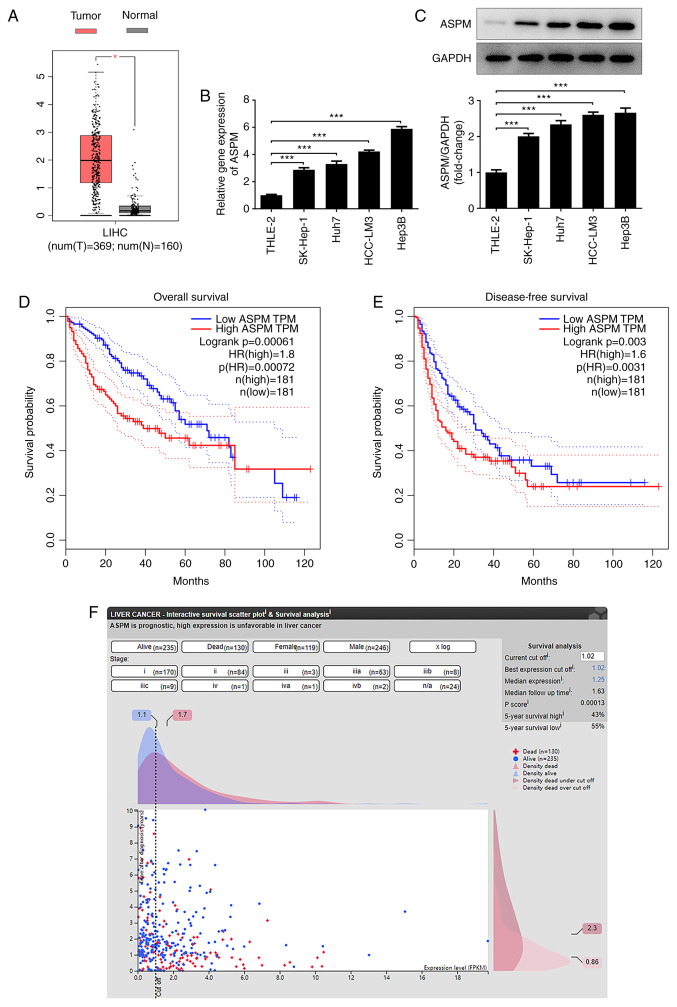
To explore the expression levels of ASPM in HCC and its clinical significance, the GEPIA database was used for the detection of the expression levels of ASPM in HCC and normal liver tissues, and to analyze the overall survival and disease-free survival rates. A total of 160 normal samples and 369 tumor samples were obtained from the LIHC dataset of GEPIA2, with |Log2 fold change|>1 and P<0.01 as cut-off values. Higher ASPM expression was observed in HCC tissues compared with in normal tissues (Fig. 1A). In addition, the expression levels of ASPM in normal hepatocytes and HCC cell lines were detected by RT-qPCR and western blotting, and the results revealed significantly higher expression levels of ASPM in HCC cell lines compared with in normal hepatocytes (Fig. 1B and C). Furthermore, Hep3B cells were selected for further experiments, as they exhibited the highest expression levels of ASPM. To evaluate of the significance of ASPM in clinical studies of HCC, the present study revealed through GEPIA database analysis that the overall survival rate was markedly different between the low and high ASPM groups before 80 months, while there was no difference observed after 80 weeks. In terms of the disease-free survival rate, there was no difference at ~50 months. In general, the overall and disease-free survival of patients with high ASPM expression were markedly lower than those of patients with low ASPM expression (Fig. 1D and E). Furthermore, in HPA database analysis, most patients with low ASPM expression were alive within 10 years after diagnosis. In patients with high ASPM expression, a large proportion of deaths had already occurred within 1 year after diagnosis. This indicated that ASPM was closely associated with prognosis (Fig. 1F), revealing that ASPM may serve as an independent prognostic factor for patients with HCC.
Figure 1.
ASPM is highly expressed in HCC tissues and cell lines. (A) ASPM expression in HCC and normal liver tissues was examined using the GEPIA database. *P<0.05. ASPM expression in normal hepatocytes and HCC cell lines was detected by (B) reverse transcription-quantitative PCR and (C) western blotting. (D) 5-year overall survival and (E) disease-free survival of patients were analyzed using the GEPIA database. (F) Association between ASPM and the prognosis of patients with HCC was analyzed using the Human Protein Atlas database. The plus-shaped dots on the graph represent the deaths, the circular dots represent the patients alive, the x-axis represents ASPM expression and the y-axis represents years after diagnosis. ***P<0.001; n≥3. ASPM, assembly factor for spindle microtubules; HCC, hepatocellular carcinoma; GEPIA, Gene Expression Profiling Interactive Analysis.
Interference with ASPM inhibits the proliferation of HCC cells
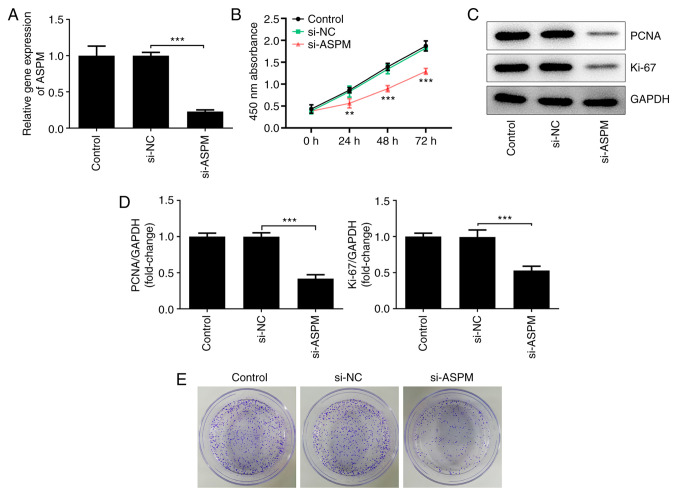
To further investigate the biological functions of ASPM in HCC cell lines, the present study interfered with ASPM expression in Hep3B cells and detected the expression levels of ASPM after interference using RT-qPCR (Fig. 2A). Cell viability was detected using a CCK-8 assay, and it was revealed that the number of viable Hep3B cells was decreased significantly following ASPM-knockdown compared with the si-NC group (Fig. 2B). Western blotting was used to detect the expression levels of proliferation-associated proteins, namely proliferating cell nuclear antigen (PCNA) and Ki-67. Compared with the si-NC group, the expression levels of PCNA and Ki-67 were decreased significantly after interference with ASPM expression (Fig. 2C and D). In addition, using a colony formation assay, it was demonstrated that interference with ASPM expression markedly inhibited the colony formation ability of Hep3B cells (Fig. 2E). The aforementioned experiments collectively indicated that interference with ASPM expression inhibited the proliferation of HCC cells.
Figure 2.
ASPM-knockdown inhibits the proliferation of hepatocellular carcinoma cells. (A) ASPM expression following its knockdown was detected by reverse transcription-quantitative PCR. (B) Cell viability was detected using a Cell Counting Kit-8 assay. (C and D) Expression levels of proliferation-associated proteins PCNA and Ki-67 were detected by western blotting. (E) Colony formation ability of Hep3B cells was detected using a colony formation assay. **P<0.01 and ***P<0.001 as indicated or vs. si-NC; n≥3. ASPM, assembly factor for spindle microtubules; PCNA, proliferating cell nuclear antigen; si, small interfering RNA; NC, negative control.
Interference with ASPM inhibits the migration, invasion and EMT of HCC cells
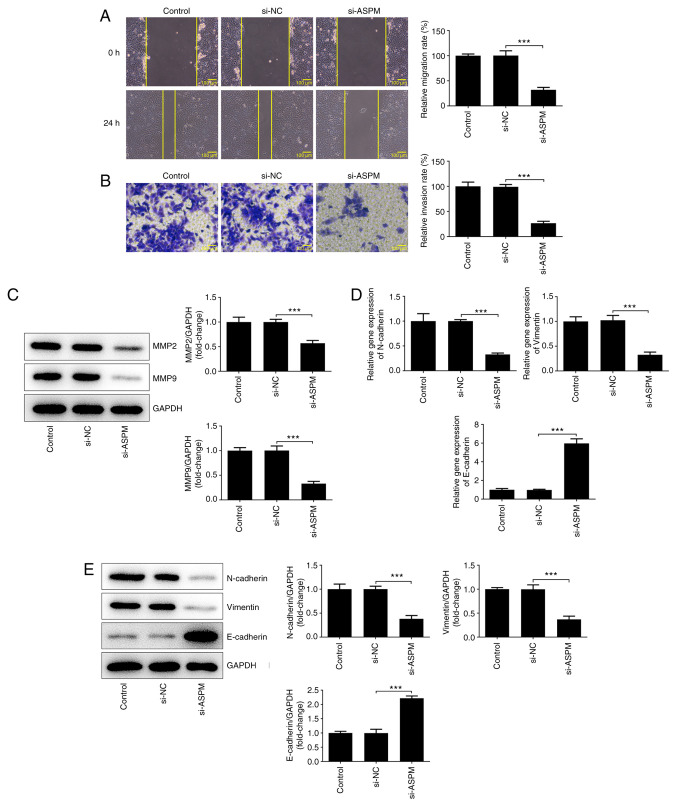
The high metastatic propensity of HCC serves a key role in the malignant progression of HCC (19). Therefore, the effects of ASPM on the migration, invasion and EMT of HCC cells were examined. The results of the wound healing assay revealed a significant decrease in the migratory ability of the si-ASPM group compared with that of the si-NC group (Fig. 3A). The Transwell assay demonstrated a significant decrease in the invasive ability of HCC cells after interference with ASPM expression (Fig. 3B). MMP2 and MMP9 both serve an important role in the invasion and metastasis of tumor cells (31). Therefore, their protein expression levels were detected by western blotting, and it was revealed that the expression levels of these two proteins were significantly downregulated after ASPM-knockdown (Fig. 3C). The invasion and metastasis of tumors are often accompanied by the occurrence of EMT. Therefore, the present study examined the expression levels of N-cadherin/vimentin and E-cadherin using RT-qPCR and western blotting, and the results revealed that the expression levels of N-cadherin/vimentin were decreased, while those of E-cadherin were elevated following ASPM-knockdown (Fig. 3D and E). The aforementioned experimental results indicated that interference with ASPM expression inhibited the migration, invasion and EMT of HCC cells.
Figure 3.
ASPM-knockdown inhibits the migration, invasion and epithelial-to-mesenchymal transition of hepatocellular carcinoma cells. (A) Migration was detected using a wound healing assay; magnification, x100. (B) Invasion was detected using a Transwell assay; magnification, x100. (C) Protein expression levels of MMP2 and MMP9 were detected by western blotting. Expression levels of N-cadherin, vimentin and E-cadherin were detected by (D) reverse transcription-quantitative PCR and (E) western blotting. ***P<0.001; n≥3. ASPM, assembly factor for spindle microtubules; si, small interfering RNA; NC, negative control.
Interference with ASPM inhibits KIF11 expression in HCC cells
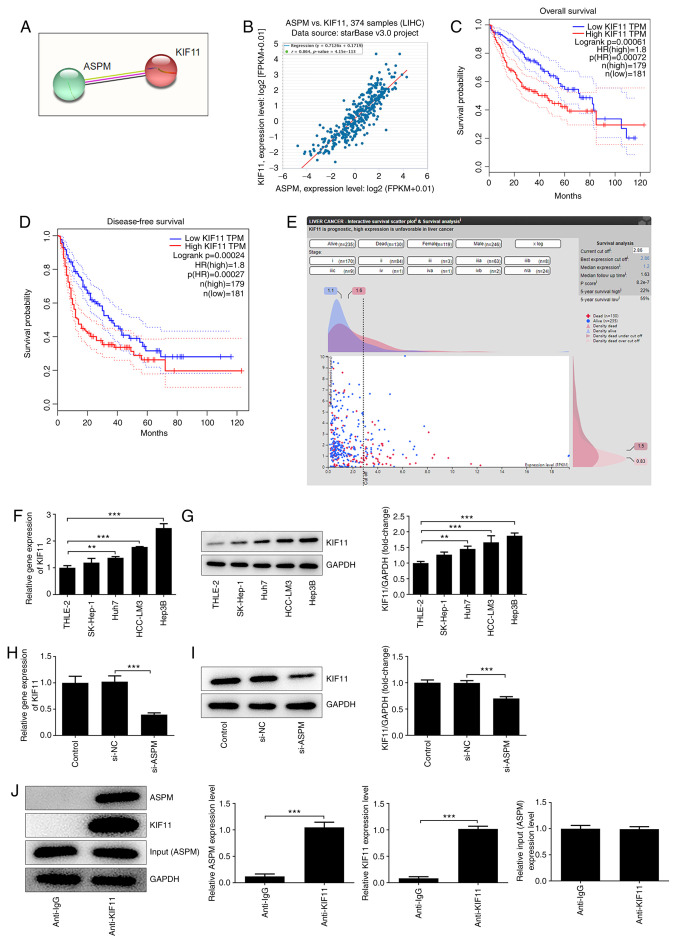
To further explore the mechanism of ASPM acting on HCC, STRING and starBase were used to predict the possible interaction between ASPM and KIF11 (Fig. 4A and B). The starBase database indicated an R value of 0.864 between ASPM and KIF11, suggesting a distinct positive correlation between the two (Fig. 4B). Additionally, GEPIA database analysis suggested that the overall and disease-free survival of patients with high KIF11 expression were significantly lower than those of patients with low KIF11 expression (Fig. 4C and D). Additionally, the HPA database analysis revealed that KIF11 expression was closely associated with the prognosis of patients (Fig. 4E), and may serve as an independent prognostic factor analogous to ASPM for patients with HCC. These results indicated that there exists a strong association between ASPM and KIF11, both of which are closely associated with the development of HCC. To further study the association between ASPM and KIF11, RT-qPCR and western blotting were performed to detect the expression levels of KIF11 in normal liver cells and HCC cells after interference with ASPM expression. The results demonstrated that KIF11 expression was significantly higher in HCC cell lines compared with in normal liver cells (Fig. 4F and G), whereas it was significantly lower after interference with ASPM expression (Fig. 4H and I). CO-IP was conducted to verify whether ASPM and KIF11 interacted with each other, and the results revealed that endogenous ASPM and endogenous KIF11 co-precipitated in Hep3B cells (Fig. 4J). The aforementioned experiments indicated an interaction between ASPM and KIF11, and an inhibitory effect of ASPM-knockdown on KIF11 expression in HCC cell lines.
Figure 4.
ASPM-knockdown inhibits KIF11 expression in HCC cells. Interaction between ASPM and KIF11 was predicted using (A) Search Tool for the Retrieval of Interacting Genes/Proteins and (B) StarBase. (C) 5-year overall survival and (D) disease-free survival of patients were analyzed using the Gene Expression Profiling Interactive Analysis database. (E) Association between KIF11 and prognosis of patients was analyzed using the Human Protein Atlas database. The plus shaped dots on the graph represent the deaths, the circular dots represent the patients alive, the x-axis represents KIF11 expression and the y-axis represents years after diagnosis. Expression levels of KIF11 in normal hepatocytes and HCC cell lines were detected using (F) RT-qPCR and (G) western blotting. Expression levels of KIF11 in Hep3B cells were detected by (H) RT-qPCR and (I) western blotting. (J) Interaction between ASPM and KIF11 verified by co-immunoprecipitation. **P<0.01; ***P<0.001; n≥3. ASPM, assembly factor for spindle microtubules; KIF11, kinesin family member 11; HCC, hepatocellular carcinoma; RT-qPCR, reverse transcription-quantitative PCR; si, small interfering RNA; NC, negative control; LIHC, liver hepatocellular carcinoma.
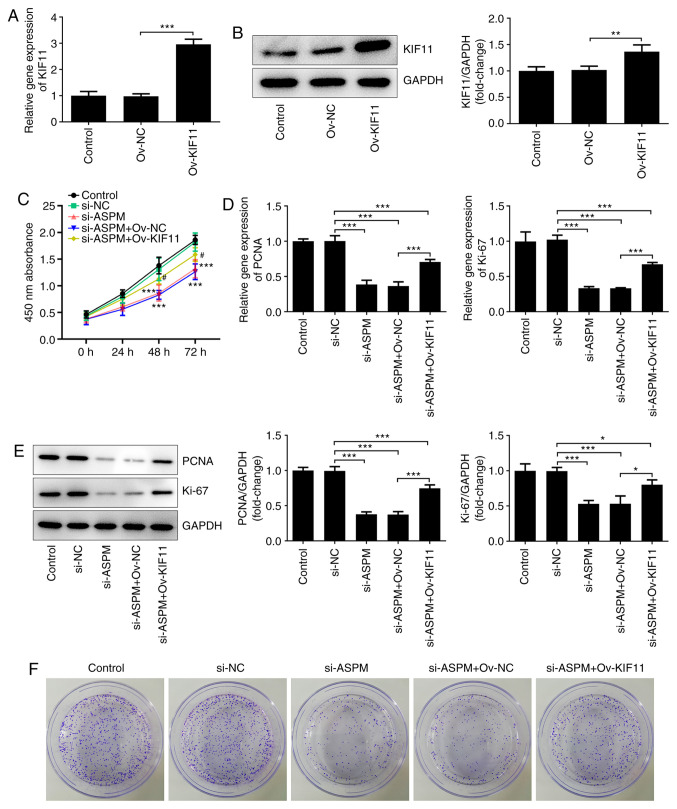
Interference with ASPM inhibits the proliferation of HCC cells via KIF11
To verify that ASPM exerted an effect via KIF11, the KIF11 overexpression vector (Ov-KIF11) was first constructed. The results of RT-qPCR and western blotting demonstrated that the mRNA and protein expression levels of KIF11 were significantly increased after transfection with the KIF11 overexpression vector (Fig. 5A and B). In addition, si-ASPM, Ov-KIF11 and their respective NCs were co-transfected into Hep3B cells, and their proliferation levels were then detected using a CCK-8 assay. The results revealed that interference with ASPM expression significantly inhibited the proliferative ability of HCC cells, while this effect was reversed by simultaneous transfection of si-ASPM and Ov-KIF11 (Fig. 5C). RT-qPCR and western blotting revealed that the simultaneous transfection of si-ASPM and Ov-KIF11 restored the mRNA and protein expression levels of proliferation-associated genes PCNA and Ki-67 (Fig. 5D and E), and this trend was consistent with the results of the CCK-8 assay. Furthermore, the colony formation assay demonstrated that simultaneous transfection of si-ASPM and Ov-KIF11 reversed the inhibitory effect of si-ASPM on the colony formation capacity of Hep3B cells (Fig. 5F). Overall, the aforementioned experimental results indicated that ASPM-knockdown inhibited the proliferation and colony formation ability of HCC cells via KIF11.
Figure 5.
ASPM-knockdown inhibits the proliferation of hepatocellular carcinoma cells via KIF11. Expression levels of KIF11 were detected by (A) RT-qPCR and (B) western blotting. (C) Proliferation in the si-ASPM and Ov-KIF11 groups was detected using a Cell Counting Kit-8 assay. Expression levels of PCNA and Ki67 were detected by (D) RT-qPCR and (E) western blotting. (F) Colony formation ability was detected using a colony formation assay. *P<0.05; **P<0.01; ***P<0.001 as indicated or vs. si-NC; #P<0.05 vs. si-ASPM + Ov-NC; n≥3. ASPM, assembly factor for spindle microtubules; KIF11, kinesin family member 11; RT-qPCR, reverse transcription-quantitative PCR; si, small interfering RNA; Ov, overexpression; NC, negative control; PCNA, proliferating cell nuclear antigen.
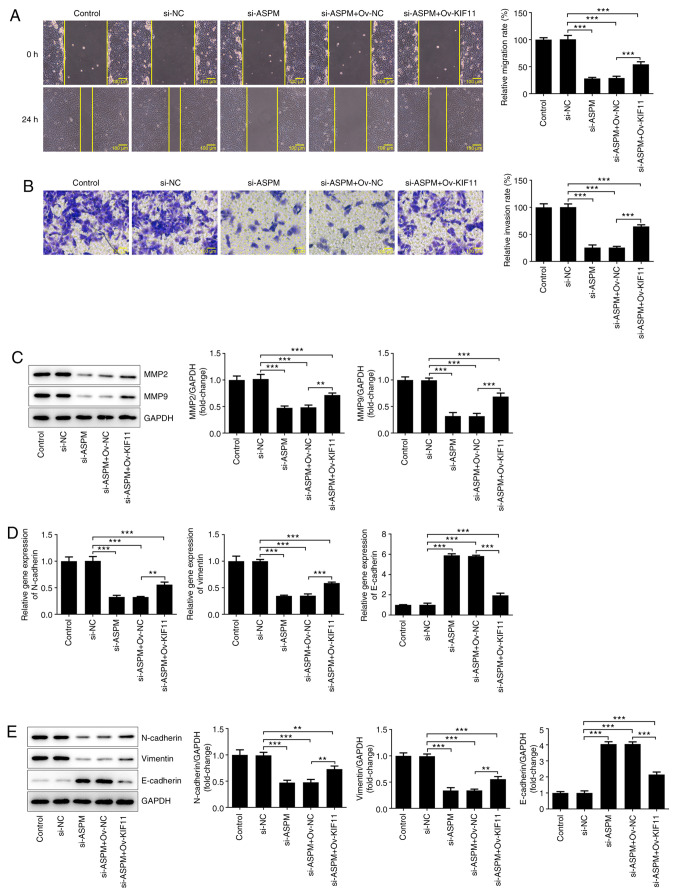
Interference with ASPM inhibits the migration, invasion and EMT of HCC cells via KIF11
To further determine the mechanism, si-ASPM, Ov-KIF11 and their respective NCs were co-transfected into Hep3B cells. Wound healing and Transwell assays were utilized to detect the changes in cell invasion and migration. The results demonstrated that simultaneous transfection of si-ASPM and Ov-KIF11 reversed the inhibitory effect of si-ASPM on the migration and invasion of Hep3B cells (Fig. 6A and B). The detection of MMP2 and MMP9 expression by western blotting demonstrated a significant decrease in the expression levels of MMP2 and MMP9 after interference with ASPM expression, whereas these expression levels were partly restored after simultaneous transfection of si-ASPM and Ov-KIF11 (Fig. 6C). The expression levels of EMT-associated proteins were detected by RT-qPCR and western blotting, revealing that, compared with the si-ASPM group, simultaneous transfection of si-ASPM and Ov-KIF11 restored the expression levels of N-cadherin and vimentin to some degree, in addition to inhibiting E-cadherin expression (Fig. 6D and E). These experiments suggested that interference with ASPM inhibited the migration, invasion and EMT of HCC cells via KIF11.
Figure 6.
ASPM-knockdown inhibits the migration, invasion and epithelial-to-mesenchymal transition of hepatocellular cancer cells via KIF11. (A) Migration was detected using a wound healing assay; magnification, x100. (B) Invasion was detected using a Transwell assay; magnification, x100. (C) Protein expression levels of MMP2 and MMP9 were detected by western blotting. Expression levels of N-cadherin, vimentin and E-cadherin were detected by (D) reverse transcription-quantitative PCR and (E) western blotting. **P<0.01; ***P<0.001; n≥3. ASPM, assembly factor for spindle microtubules; KIF11, kinesin family member 11; si, small interfering RNA; Ov, overexpression; NC, negative control.
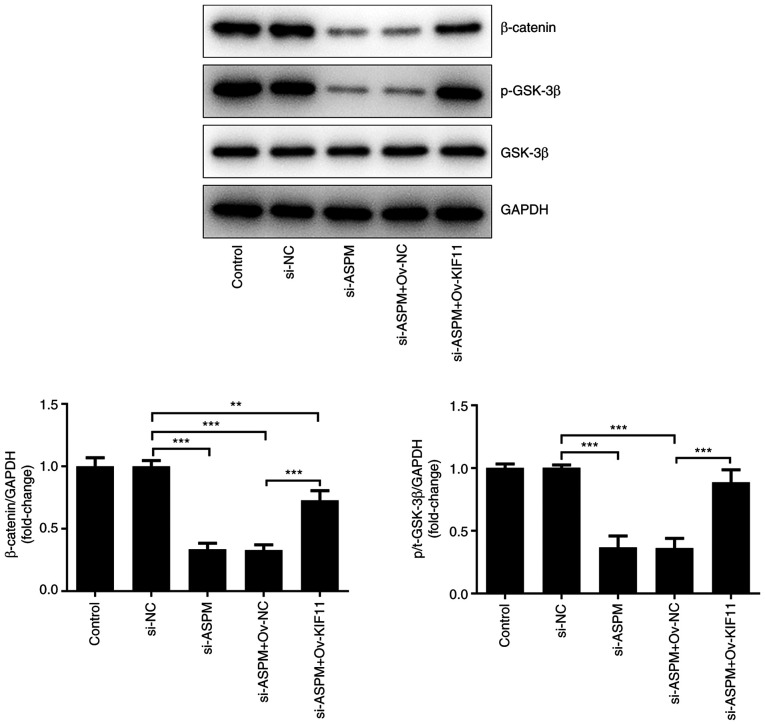
ASPM is involved in the Wnt/β-catenin signaling pathway by regulating KIF11
A previous study has reported the ability of ASPM to promote glioma growth through the Wnt/β-catenin signaling pathway (32). In addition, Pei et al (33) revealed that silencing of KIF11 markedly decreased the self-renewal ability of breast cancer cells by β-catenin. Based on these findings, it was hypothesized that ASPM could regulate the occurrence and development of HCC via the KIF11-mediated Wnt/β-catenin signaling pathway. To verify the aforementioned hypothesis, the expression levels of proteins associated with the Wnt/β-catenin signaling pathway, namely β-catenin, phosphorylated (p-)GSK-3β and GSK-3β, were detected after co-transfection of si-ASPM, Ov-KIF11 and their respective NCs into Hep3B cells. The results demonstrated that the levels of β-catenin and p-GSK-3 were significantly decreased after interference with ASPM, indicating that interference with ASPM expression silenced the Wnt/β-catenin signaling pathway. Additionally, the levels of β-catenin and p-GSK-3β were restored compared with the si-ASPM + Ov-NC group after simultaneous transfection of si-ASPM and Ov-KIF11 (Fig. 7). The aforementioned experiments indicated that ASPM may be involved in the Wnt/β-catenin signaling pathway through the regulation of KIF11 and thereby may regulate the malignant progression of HCC.
Figure 7.
ASPM participates in the Wnt/β-catenin signaling pathway by regulating KIF11. Expression levels of proteins associated with the Wnt/β-catenin signaling pathway were detected by western blotting. **P<0.01; ***P<0.001; n≥3. ASPM, assembly factor for spindle microtubules; KIF11, kinesin family member 11; si, small interfering RNA; Ov, overexpression; NC, negative control; p, phosphorylated.
Discussion
HCC ranks among the top five most common malignant tumors worldwide (34). Due to the insidiousness of this disease, patients are often diagnosed with advanced HCC at their initial examination in the hospital, resulting in a poor prognosis (35). At present, chemotherapy and immunotherapy are comparatively the best options for the treatment of HCC (36). Therefore, it is particularly important to perform more in-depth research on the pathogenesis of HCC, with the aim of facilitating the development of immunotherapy for the treatment of HCC.
ASPM is highly expressed in various types of cancer, including breast (13), bladder (12) and ovarian cancer (16). The present study revealed that ASPM is highly expressed in liver cancer tissues and cells, which was consistent with the findings of other studies (19-21). In vitro experiments revealed that interference with ASPM expression inhibited the proliferation, migration, invasion and EMT of HCC cells, suggesting that the upregulation of ASPM in HCC may promote the malignant progression of HCC. In addition, the present study further revealed that KIF11 and ASPM are associated using STRING and starBase database analysis, suggesting that KIF11 and ASPM may interact directly.
KIF11 acts as an oncogene in tumors and is associated with a poor prognosis in breast cancer (26). Additionally, it promotes cell invasion, proliferation and self-renewal in glioblastoma (27). A number of studies have reported that KIF11 may serve as a biological diagnostic and prognostic marker for numerous types of tumor, including oral (37), non-small cell lung (14,38), ovarian (39) and bladder cancer (40). Liu et al (41) observed high KIF11 expression in HCC tissues, which is strongly associated with liver cirrhosis and tumor stages, and can be considered as a biomarker for poor prognosis of HCC. The present study revealed that KIF11 was highly expressed in HCC through bioinformatics analysis, and that it was associated with HCC development, which was consistent with the study by Liu et al (41). The CO-IP assay demonstrated that ASPM could directly bind to KIF11, and overexpression of KIF11 reversed the inhibitory effect of ASPM-knockdown on proliferation, migration, invasion and EMT of HCC cells to some extent.
The Wnt/β-catenin signaling pathway is a highly conserved and tightly controlled molecular mechanism, which has been identified to be abnormally activated in HCC and to exert a major influence over the occurrence and development of this type of cancer (42,43). β-catenin is an important protein molecule in the Wnt signaling pathway, and its expression levels in HCC tissues directly affect the activation degree of the Wnt/β-catenin signaling pathway (44). At present, a number of studies have validated the effectiveness of Wnt/β-catenin antagonists in dampening the malignant progression of HCC (45,46). The development of novel drugs targeting the Wnt/β-catenin signaling pathway has now become one of the main focuses of research for the treatment of HCC. It was revealed that the levels of β-catenin and p-GSK-3β were decreased after interference with ASPM expression, silencing the Wnt/β-catenin signaling pathway. However, when KIF11 was overexpressed at the same time, the β-catenin and p-GSK-3β levels were restored, which indicated that KIF11 was involved in the Wnt/β-catenin signaling pathway. Additionally, the current results demonstrated that ASPM regulated the involvement of KIF11 in the Wnt/β-catenin signaling pathway to regulate the malignant progression of HCC.
In conclusion, the present study identified that ASPM was highly expressed in HCC tissues, and that ASPM-knockdown inhibited the proliferation, migration, invasion and EMT of HCC cells. Additionally, ASPM was involved in the Wnt/β-catenin signaling pathway through KIF11, thereby promoting the proliferation, migration, invasion and EMT of HCC cells.
Acknowledgements
Not applicable.
Funding Statement
Funding: The present study was supported by The National Natural Science Foundation of China (grant no. 81871260).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
BW designed and performed the experiments, and made considerable contributions to the manuscript writing. CH performed the experiments and analyzed the data. LK conceived the study, guided the experiments and revised the manuscript. BW and CH confirmed the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel R, Miller K, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Anwanwan D, Singh S, Singh S, Saikam V, Singh R. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer. 2020;1873(188314) doi: 10.1016/j.bbcan.2019.188314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Craig AJ, von Felden J, Garcia-Lezana T, Sarcognato S, Villanueva A. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2020;17:139–152. doi: 10.1038/s41575-019-0229-4. [DOI] [PubMed] [Google Scholar]
- 4.Nie J, Lin B, Zhou M, Wu L, Zheng T. Role of ferroptosis in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2018;144:2329–2337. doi: 10.1007/s00432-018-2740-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greten TF, Lai CW, Li G, Staveley-O'Carroll KF. Targeted and immune-based therapies for hepatocellular carcinoma. Gastroenterology. 2019;156:510–524. doi: 10.1053/j.gastro.2018.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang F, Wang BR, Wang YG. Role of autophagy in tumorigenesis, metastasis, targeted therapy and drug resistance of hepatocellular carcinoma. World J Gastroenterol. 2018;24:4643–4651. doi: 10.3748/wjg.v24.i41.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar A, Blanton S, Babu M, Markandaya M, Girimaji S. Genetic analysis of primary microcephaly in Indian families: novel ASPM mutations. Clin Genet. 2004;66:341–348. doi: 10.1111/j.1399-0004.2004.00304.x. [DOI] [PubMed] [Google Scholar]
- 8.Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, Rodionov V, Han DK. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal. 2009;2(ra46) doi: 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- 9.Pai VC, Hsu CC, Chan TS, Liao WY, Chuu CP, Chen WY, Li CR, Lin CY, Huang SP, Chen LT, Tsai KK. ASPM promotes prostate cancer stemness and progression by augmenting Wnt-Dvl-3-β-catenin signaling. Oncogene. 2019;38:1340–1353. doi: 10.1038/s41388-018-0497-4. [DOI] [PubMed] [Google Scholar]
- 10.Hsu CC, Liao WY, Chan TS, Chen WY, Lee CT, Shan YS, Huang PJ, Hou YC, Li CR, Tsai KK. The differential distributions of ASPM isoforms and their roles in Wnt signaling, cell cycle progression, and pancreatic cancer prognosis. J Pathol. 2019;249:498–508. doi: 10.1002/path.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Q, Hu J, Deng J, Fu B, Guo J. Bioinformatics analysis identified key molecular changes in bladder cancer development and recurrence. Biomed Res Int. 2019;2019(3917982) doi: 10.1155/2019/3917982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Z, Zhang Q, Luh F, Jin B, Liu X. Overexpression of the ASPM gene is associated with aggressiveness and poor outcome in bladder cancer. Oncol Lett. 2019;17:1865–1876. doi: 10.3892/ol.2018.9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang J, Lu M, Cui Q, Zhang D, Kong D, Liao X, Ren J, Gong Y, Wu G. Overexpression of ASPM, CDC20, and TTK confer a poorer prognosis in breast cancer identified by gene co-expression network analysis. Front Oncol. 2019;9(310) doi: 10.3389/fonc.2019.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Liu X, Li J, Ren F. Identification and integrated analysis of key biomarkers for diagnosis and prognosis of non-small cell lung cancer. Med Sci Monit. 2019;25:9280–9289. doi: 10.12659/MSM.918620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie JJ, Zhuo YJ, Zheng Y, Mo RJ, Liu ZZ, Li BW, Cai ZD, Zhu XJ, Liang YX, He HC, Zhong WD. High expression of ASPM correlates with tumor progression and predicts poor outcome in patients with prostate cancer. Int Urol Nephrol. 2017;49:817–823. doi: 10.1007/s11255-017-1545-7. [DOI] [PubMed] [Google Scholar]
- 16.Alsiary R, Brüning-Richardson A, Bond J, Morrison EE, Wilkinson N, Bell SM. Deregulation of microcephalin and ASPM expression are correlated with epithelial ovarian cancer progression. PLoS One. 2014;9(e97059) doi: 10.1371/journal.pone.0097059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brüning-Richardson A, Bond J, Alsiary R, Richardson J, Cairns DA, McCormack L, Hutson R, Burns P, Wilkinson N, Hall GD, et al. ASPM and microcephalin expression in epithelial ovarian cancer correlates with tumour grade and survival. Br J Cancer. 2011;104:1602–1610. doi: 10.1038/bjc.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao ZY, Yu F, Jia HX, Ye Z, Yao SJ. ASPM predicts poor prognosis and regulates cell proliferation in bladder cancer. Kaohsiung J Med Sci. 2020;36:1021–1029. doi: 10.1002/kjm2.12284. [DOI] [PubMed] [Google Scholar]
- 19.Lin SY, Pan HW, Liu SH, Jeng YM, Hu FC, Peng SY, Lai PL, Hsu HC. ASPM is a novel marker for vascular invasion, early recurrence, and poor prognosis of hepatocellular carcinoma. Clin Cancer Res. 2008;14:4814–4820. doi: 10.1158/1078-0432.CCR-07-5262. [DOI] [PubMed] [Google Scholar]
- 20.Xue JM, Liu Y, Wan LH, Zhu YX. Comprehensive analysis of differential gene expression to identify common gene signatures in multiple cancers. Med Sci Monit. 2020;26(e919953) doi: 10.12659/MSM.919953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Z, Li Y, Hao H, Wang Y, Zhou Z, Wang Z, Chu X. Screening Hub genes as prognostic biomarkers of hepatocellular carcinoma by bioinformatics analysis. Cell Transplant. 2019;28 (Suppl 1):76S–86S. doi: 10.1177/0963689719893950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rapley J, Nicolàs M, Groen A, Regué L, Bertran MT, Caelles C, Avruch J, Roig J. The NIMA-family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. J Cell Sci. 2008;121:3912–3921. doi: 10.1242/jcs.035360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wakana Y, Villeneuve J, van Galen J, Cruz-Garcia D, Tagaya M, Malhotra V. Kinesin-5/Eg5 is important for transport of CARTS from the trans-Golgi network to the cell surface. J Cell Biol. 2013;202:241–250. doi: 10.1083/jcb.201303163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Güneş N, Taşdemir E, Jeffery H, Yetik H, Ostergaard P, Tüysüz B. A novel mutation of KIF11 in a Child with 22q11.2 deletion syndrome associated with MCLMR. Mol Syndromol. 2019;9:266–270. doi: 10.1159/000491568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu H, Xiao X, Li S, Jia X, Guo X, Zhang Q. KIF11 mutations are a common cause of autosomal dominant familial exudative vitreoretinopathy. Br J Ophthalmol. 2016;100:278–283. doi: 10.1136/bjophthalmol-2015-306878. [DOI] [PubMed] [Google Scholar]
- 26.Zhou J, Chen WR, Yang LC, Wang J, Sun JY, Zhang WW, He ZY, Wu SG. KIF11 functions as an oncogene and is associated with poor outcomes from breast cancer. Cancer Res Treat. 2019;51:1207–1221. doi: 10.4143/crt.2018.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venere M, Horbinski C, Crish JF, Jin X, Vasanji A, Major J, Burrows AC, Chang C, Prokop J, Wu Q, et al. The mitotic kinesin KIF11 is a driver of invasion, proliferation, and self-renewal in glioblastoma. Sci Transl Med. 2015;7(304ra143) doi: 10.1126/scitranslmed.aac6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Qin L, Liao L, Redmond A, Young L, Yuan Y, Chen H, O'Malley BW, Xu J. The AIB1 oncogene promotes breast cancer metastasis by activation of PEA3-mediated matrix metalloproteinase 2 (MMP2) and MMP9 expression. Mol Cell Biol. 2008;28:5937–5950. doi: 10.1128/MCB.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Huang L, Yang Y, Chen S, Sun J, Ma C, Xie J, Song Y, Yang J. ASPM promotes glioblastoma growth by regulating G1 restriction point progression and Wnt-β-catenin signaling. Aging (Albany NY) 2020;12:224–241. doi: 10.18632/aging.102612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pei YY, Li GC, Ran J, Wan XH, Wei FX, Wang L. Kinesin family member 11 enhances the self-renewal ability of breast cancer cells by participating in the Wnt/β-catenin pathway. J Breast Cancer. 2019;22:522–532. doi: 10.4048/jbc.2019.22.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Stefano F, Chacon E, Turcios L, Marti F, Gedaly R. Novel biomarkers in hepatocellular carcinoma. Dig Liver Dis. 2018;50:1115–1123. doi: 10.1016/j.dld.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 35.Xie QL, Liu Y, Zhu Y. Chromosome region maintenance 1 expression and its association with clinical pathological features in primary carcinoma of the liver. Exp Ther Med. 2016;12:59–68. doi: 10.3892/etm.2016.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, Cao J, Wang P, He X. NT5DC2 is a novel prognostic marker in human hepatocellular carcinoma. Oncol Lett. 2020;20(70) doi: 10.3892/ol.2020.11931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daigo K, Takano A, Thang PM, Yoshitake Y, Shinohara M, Tohnai I, Murakami Y, Maegawa J, Daigo Y. Characterization of KIF11 as a novel prognostic biomarker and therapeutic target for oral cancer. Int J Oncol. 2018;52:155–165. doi: 10.3892/ijo.2017.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider MA, Christopoulos P, Muley T, Warth A, Klingmueller U, Thomas M, Herth FJ, Dienemann H, Mueller NS, Theis F, Meister M. AURKA, DLGAP5, TPX2, KIF11 and CKAP5: Five specific mitosis-associated genes correlate with poor prognosis for non-small cell lung cancer patients. Int J Oncol. 2017;50:365–372. doi: 10.3892/ijo.2017.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Z, Zhou Y, Cao Y, Dinh TL, Wan J, Zhao M. Identification of candidate biomarkers and analysis of prognostic values in ovarian cancer by integrated bioinformatics analysis. Med Oncol. 2016;33(130) doi: 10.1007/s12032-016-0840-y. [DOI] [PubMed] [Google Scholar]
- 40.Pan S, Zhan Y, Chen X, Wu B, Liu B. Identification of biomarkers for controlling cancer stem cell characteristics in bladder cancer by network analysis of transcriptome data stemness indices. Front Oncol. 2019;9(613) doi: 10.3389/fonc.2019.00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu C, Zhou N, Li J, Kong J, Guan X, Wang X. Eg5 overexpression is predictive of poor prognosis in hepatocellular carcinoma patients. Dis Markers. 2017;2017(2176460) doi: 10.1155/2017/2176460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perugorria MJ, Olaizola P, Labiano I, Esparza-Baquer A, Marzioni M, Marin JJG, Bujanda L, Banales JM. Wnt-β-catenin signalling in liver development, health and disease. Nat Rev Gastroenterol Hepatol. 2019;16:121–136. doi: 10.1038/s41575-018-0075-9. [DOI] [PubMed] [Google Scholar]
- 43.Wang W, Smits R, Hao H, He C. Wnt/β-catenin signaling in liver cancers. Cancers (Basel) 2019;11(926) doi: 10.3390/cancers11070926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang G, Shen T, Yi X, Zhang Z, Tang C, Wang L, Zhou Y, Zhou W. Crosstalk between long non-coding RNAs and Wnt/β-catenin signalling in cancer. J Cell Mol Med. 2018;22:2062–2070. doi: 10.1111/jcmm.13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi XD, Yu XH, Wu WR, Xu XL, Wang JY, Xu LB, Zhang R, Liu C. Dickkopf-1 expression is associated with tumorigenity and lymphatic metastasis in human hilar cholangiocarcinoma. Oncotarget. 2016;7:70378–70387. doi: 10.18632/oncotarget.11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu B, Yang X, Xu Y, Yao G, Shu H, Lin B, Hood L, Wang H, Yang S, Gu J, et al. Elevated expression of DKK1 is associated with cytoplasmic/nuclear beta-catenin accumulation and poor prognosis in hepatocellular carcinomas. J Hepatol. 2009;50:948–957. doi: 10.1016/j.jhep.2008.11.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.