Abstract
A number of previous studies have reported that dysregulated miR-184 expression is associated with the development of cancer. The aim of the present study was to investigate the role of miR-184 in prostate cancer (PC) and the mechanism underlying its effects. Data from human tumor tissue samples were collected from The CEancer Genome Atlas to determine the expression levels of miR-184 and DLX1. The miR-184 mimic and pcDNA3.1-DLX1 plasmid were utilized to induce overexpression of miR-184 and DLX1 in Du145 cells, respectively. Cell Counting Kit-8, wound healing and Transwell assays were performed to examine the effects of miR-184 on the aggressiveness of PC cells. Dual-luciferase reporter gene assay was used to investigate the association between miR-184 and DLX1, and reverse transcription-quantitative PCR and western blot analyses were utilized to determine the mRNA and protein levels. miR-184 expression was found to be downregulated whereas DLX1 was upregulated in PC tissues compared with normal prostate tissues. Cell propagation, migration and invasion were all inhibited by miR-184 upregulation in Du145 cells. Dual luciferase reporter assay confirmed the association between miR-184 and DLX1. The inhibitory effect of miR-184 mimic on cell behaviors was reversed by upregulation of DLX1. These findings suggest that miR-184 plays a beneficial role in suppressing the tumorigenesis of PC by directly targeting DLX1, and it may represent a potential therapeutic strategy for PC.
Keywords: microRNA-184, prostate cancer, distal-less homeobox 1, proliferation, migration, invasion
Introduction
Prostate cancer (PC) is the most frequent type of cancer in men worldwide, accounting for 9.5% of morbidity and mortality among all new cancer cases, and for 4.8% of all cancer-related deaths (1). Older men appear to be more susceptible to PC, as it has been reported that ~99% of patients with PC are aged >50 years (2). Similar to other malignancies, the timing of diagnosis is crucial for the survival of patients with PC. Current treatment strategies for PC include surgery, chemotherapy, radiotherapy and hormonal therapy (3-6); however, treatment at the molecular level has rarely been reported in PC. Therefore, the aim of the present study was to improve the outcome of PC by investigating a molecular treatment approach.
MicroRNAs (miRNAs) are a class of short non-coding RNAs, which have been found to be implicated in a number of human malignancies (7,8). miRNAs have been reported to affect tumor development by binding to the 3' untranslated region (3'UTR) of target genes (9,10). Accumulating evidence has demonstrated that miRNAs, including miR-216b-5p, miR-30d, miR-4638-5p and miR-141, are associated with the tumorigenesis of PC (11-14).
miR-184 is a single copy gene, evolutionarily conserved at the nucleotide level, that is located on chromosome 15(15). A strong association between miR-184 and protection from cancer has been reported by previous investigations. For example, the upregulation of miR-184 reduced nasopharyngeal carcinoma cell migration and invasion (16), and it was also shown to inhibit colorectal cancer cell viability and metastasis by targeting IGF-1R (17). In addition, lncRNA UCA1 has been reported to promote oral squamous cell carcinoma cell proliferation and cisplatin resistance through miR-184 suppression (18). However, the role of miR-184 in PC remains unclear.
In the present study, the expression levels of miR-184 and DLX1 were measured in PC tissues and cell lines. Functional tests were performed to evaluate the effect of miR-184 overexpression on PC cell lines in vitro. Further experiments were conducted to confirm whether DLX1 is a direct target of miR-184, and whether its expression could be reduced by miR-184 overexpression. Collectively, the results of the present study may provide a potential novel approach to the molecular treatment of PC.
Materials and methods
Patient specimens and cell lines
The expression profiles of miR-184 and DLX1 were downloaded from TCGA, including 52 normal human tissue samples and 499 PC samples. PC cell lines (C4-2, PC-3, Du145 and LNCaP) and a human prostate epithelial cell line (RWPE-2) were used in the present study. RWPE-2, C4-2 and Du145 cells were purchased from the American Type Culture Collection, whereas PC-3 and LNCaP cells were obtained from the cell bank of Type Culture Collection of the Chinese Academy of Sciences. All cells were incubated in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin at 37˚C with 5% CO2, and digested by 0.25% trypsin upon each passage.
Cell transfection
miR-184 mimic and pcDNA3.1-DLX1 were used to upregulate miR-184 and DLX1 expression, respectively. In addition, to examine the correlation of miR-184 and DLX1, pcDNA3.1-DLX1 (4 µg) and miR-184 mimic (0.1 nmol; designed and synthesized by Shanghai GenePharma Co., Ltd.) were used to co-transfect Du145 cells. All transfections were conducted using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Reverse transcription-quantitative PCR (RT-qPCR) analysis was utilized to verify transfection efficiency at 48 h following transfection.
RT-qPCR
Total RNA was isolated from PC cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and then reverse-transcribed to cDNA (annealing at 25˚C, extension at 42˚C) using the GoScript® Reverse Transcription kit (Promega Corporation) in accordance with the protocol of the manufacturer. For the measurement of miRNA expression, the GoTaq® qPCR Master Mix (cat. no. A6001; Promega Corporation) was used with SYBR®-Green. The program was as follows: Initial denaturation at 95˚C for 10 min, followed by 40 cycles at 95˚C for 15 sec and at 60˚C for 1 min. The U6 small nuclear RNA was used as an internal reference for miR-184 expression, while GAPDH was used as internal control for DLX1 expression. The sequences of the primers were as follows: miR-184 forward, 5'-AGTGCAGTGGGGTTGGTCTA-3' and reverse, 5'-TCCAAATGGCTCATCTCCGAC-3'; U6 forward, 5'-CTCGCTTCGGCAGCACA-3' and reverse, 5'-AACGCTTCACGAATTTGCGT-3'; DLX1 forward, 5'-GCTACCCCTACGTCAACTCG-3' and reverse, 5'-CAGATCTTGACCTGGGTCCTC-3'; and GAPDH forward, 5'-GGAGCGAGATCCCTCCAAAAT-3' and reverse, 5'-GGCTGTTGTCATACTTCTCATGG-3'. The relative expression level was calculated using the 2-ΔΔCq method (19).
Cell Counting Kit-8 (CCK-8) assay
Du145 cells were seeded into 96-well plates at a density of 1,000 cells/well 24 h after transfection. The cells were then cultured in a CO2 incubator with 5% CO2 at 37˚C and examined every 24 h. For cell viability, 10 µl CCK-8 reagent was added to each well and incubated for 1.5 h at 37˚C. The OD value at 450 nm was subsequently determined using an automatic multi-well spectrophotometer (Bio-Rad Laboratories, Inc.), and a cell viability curve was drawn. All experiments were performed at least three times.
Cell migration and invasion assays
A wound healing assay was conducted to examine cell migration. Following transfection with specific vectors, cells suspended in RPMI-1640 medium supplemented with 10% FBS and 1% antibiotics (penicillin and streptomycin) were seeded into 6-well plates at a density of 5x105 cells/well overnight, and then incubated to >90% confluence. Subsequently, a 200-µl pipette tip was used to create a wound. The existing medium was then removed and the cells were rinsed with PBS before the addition of serum-free medium. Images of the wounds were captured 24 h following incubation using a light microscope and analyzed using ImageJ version 1.44p software (National Institutes of Health). The results were quantified using the following formulas: Wound closure rate=[area (0 h)-area (24 h)]/area (0 h); wound closure (%)=(wound closure rate in experimental group)/(wound closure rate in control group) x100.
Cell invasion was assessed using Transwell assay. Matrigel (BD Biosciences) was diluted in serum-free medium at 1:6 and placed in the upper chamber of transwell chamber at 37˚C for 4 h to polymerize Matrigel into a gel. All cells were seeded into the upper chambers of Transwell chambers (pore size, 8 µm; Corning Inc.) at a density of 2x105 cells/well. The upper chamber was filled with 200 µl serum-free DMEM, while the lower chamber was filled with 500 µl DMEM supplemented with 10% FBS. Cells were then cultured for a further 16 h, and cells invading into the lower chamber were fixed with 4% paraformaldehyde for 30 min at room temperature and stained with 0.1% crystal violet for 20 min at room temperature. The number of invading cells was subsequently calculated from five random fields of view using a light microscope.
Bioinformatics analysis
The bioinformatics software PicTar 21.0 (http://pictar.mdc-berlin.de) and TargetScan 7.2 (http://www.targetscan.org/vert_72/) (20) were used to predict potential target genes for hsa-miR-184.
Establishment of histogram based on TCGA database
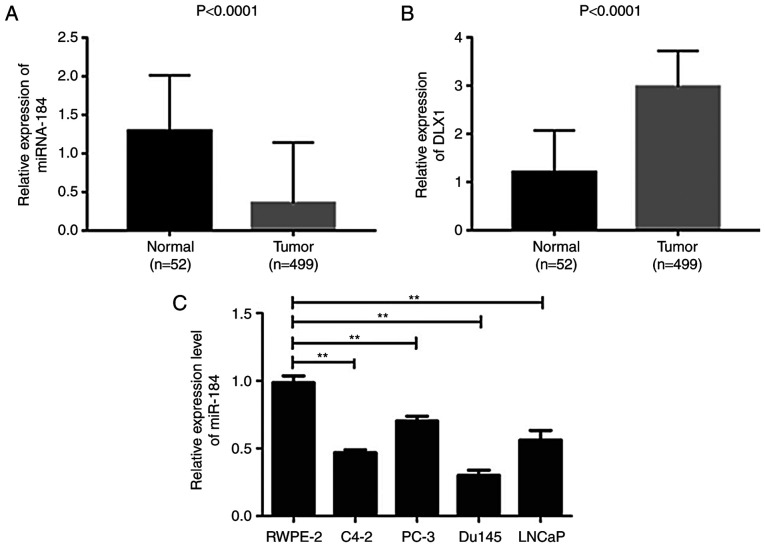
The present study used prostate cancer expression data from the TCGA database (https://cancergenome.nih.gov/). Prostate cancer gene expression profile data and clinical data were downloaded, and the first group of samples (499 samples, name Tumor) and the second group (52 samples, named normal), a total of 551 samples, were selected for analysis. The RNA-Seq data was preprocessed and the obtained data was used as an input file for subsequent methods. The method of preprocessing is as follows: First, the raw data was extracted from the matrix file. RNASeq data obtained from all 551 samples was processed and combined using the Perl programming language software package (version 5.88; Perl.org), and the matrix file was extracted to obtain the expression profile of the gene in all samples. Expression maps as shown in Fig. 1A and B were then generated using GraphPad Prism 5.0 (GraphPad Software, Inc.) based on this expression profile data. Among them, the height of the bar graph indicates the mean value of the relative expression of miR-184 in the sample, and the error line represents the maximum relative expression of miR-184 in the group.
Figure 1.
miR-184 is downregulated and DLX1 is upregulated in PC samples and cell lines. (A) miR-184 expression in PC tissues was lower compared with that in normal tissues. P<0.0001 vs. normal. (B) DLX1 expression was upregulated in PC tissues compared with that in normal tissues. P<0.0001 vs. normal. (C) miR-184 expression is higher in the RWPE-2 cell line compared with that in PC cell lines, as measured using reverse transcription-quantitative PCR. **P<0.01 vs. RWPE-2. DLX1, distal-less homeobox 1; miR, microRNA; PC, prostate cancer.
Dual luciferase reporter assays
A dual-luciferase reporter assay was carried out to confirm the prediction results. pMIR-DLX1-WT and pMIR-DLX1-MUT were synthesized by Shanghai GenePharma Co., Ltd. pMIR-DLX1-WT (0.2 µg) or pMIR-DLX1-MUT (0.2 µg) containing seven mutant nucleotides in the 3'UTR of DLX were co-transfected into Du145 cells (using the method described above) seeded in 96 well plates at a density of 1x104, along with miR-184 NC (0.05 nmol) or miR-184 mimic (0.05 nmol). Cells were subsequently incubated for 48 h at 37˚C with 5% CO2 before luciferase assays were performed. Luciferase activities in Du145 cells were determined using the Dual-Luciferase® reporter assay system (Promega Corporation). Firefly luciferase activity was normalized to Renilla luciferase activity. Each experiment was performed in triplicate.
Western blot analysis
Protein samples were extracted from the cells using RIPA lysis buffer (Beyotime Institute of Biotechnology) supplemented with protease inhibitors. The concentration of protein was quantified using a bicinchoninic acid assay kit (Beyotime Institute of Biotechnology). The lysates were then mixed with 5X loading buffer and boiled at 95˚C for 5 min. SDS-PAGE (12%) was performed to separate proteins (20 µg in each well) at 100 V for 1 h before being transferred to PVDF membranes at 200 mA for 1.5 h. The PVDF membrane was then incubated with 5% skimmed milk powder (diluted in 1X TBST buffer containing 0.1% Tween-20) at room temperature for 1 h prior to incubation with mouse anti-human DLX1 (1:1,000; cat. no. ab126054; Abcam) or mouse anti-human β-actin primary antibodies (1:1,000; cat. no. ab179467; Abcam) overnight at 4˚C. The membranes were subsequently rinsed once with PBS prior to incubation with horseradish peroxidase-conjugated secondary antibody (1:10,000; cat. no. ab205718; Abcam) for 2 h at room temperature. The protein bands were visualized using an ECL chromogenic kit and subsequently quantified using the Quantity One system (Version number 1708195; Bio-Rad Laboratories Inc.).
Statistical analysis
All data were analyzed using SPSS 18.0 statistical software (IBM Corp.) and normal distribution was described as mean ± standard deviation. Student's t-test and ANOVA followed by Dunnett's or Bonferroni's post hoc test was used to compare the difference between two or multiple groups, respectively. Correlation between miR-184 and DLX1 was evaluated using Pearson's analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-184 expression is lower and DLX1 expression is higher in PC tissues and cell lines compared with normal tissues
It was previously reported that DLX1 modulates a number of signaling pathways during tumor progression by binding to β-catenin (21). Therefore, RNA-seq data from PC and normal prostate tissue samples was collected from the TCGA database to investigate the expression of miR-184 and DLX1, in order to establish whether there was a correlation between miR-184 and DLX1 expression and PC tumorigenesis. The expression level of miR-184 was found to be significantly lower in tumor tissues from the 499 PC patients compared with normal tissues (Fig. 1A; P<0.0001). By contrast, DLX1 expression was significantly higher in PC tissues compared with that in normal tissues (Fig. 1B; P<0.0001). The expression level of miR-184 was next measured in human PC cell lines (C4-2, PC-3, Du145 and LNCaP) and the normal human prostate epithelial cell line (RWPE-2) using RT-qPCR. miR-184 expression levels were found to be significantly lower in PC cell lines compared with that in the RWPE-2 cell line and the Du145 cell line exhibited the lowest level of expression compared with other PC cell lines (C4-2, PC-3 and LNCaP; Fig. 1C; P<0.01). To further investigate the mechanism underlying the potentially beneficial role of miR-184 in PC, subsequent experiments were conducted using the Du145 cell line.
miR-184 overexpression inhibits Du145 cell proliferation, migration and invasion
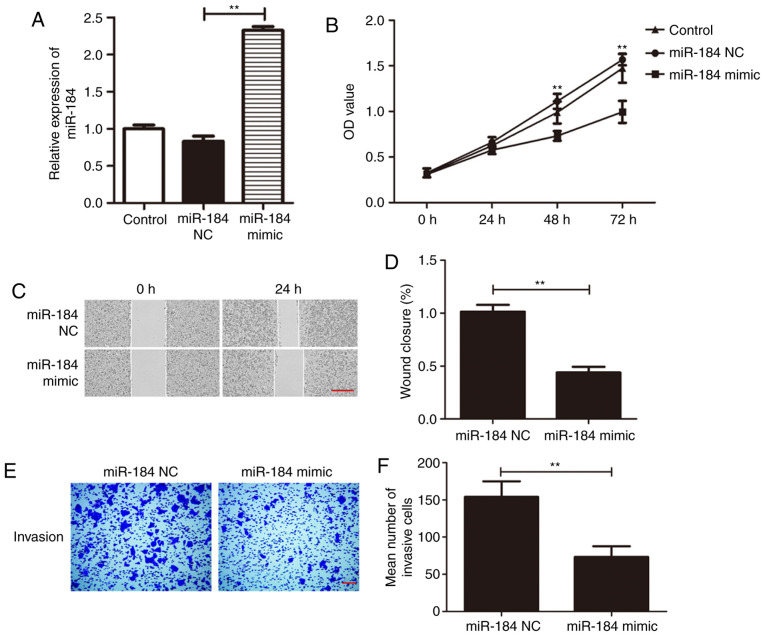
To investigate the specific effect of miR-184 on Du145 cells, a miR-184 mimic was used to overexpress miR-184. miR-184 expression was found to be significantly higher in cells transfected with miR-184 mimic compared with that in the control group (Fig. 2A; P<0.01). The CCK-8 assay was next applied to examine cell viability following transfection with miR-184 mimic. Du145 cell viability was significantly increased at 48 and 72 h following miR-184 overexpression (P<0.01; Fig. 2B). In addition, the migratory and invasive abilities of Du145 cells transfected with either miR-184 mimic or miR-184 NC were examined using wound healing and Transwell assays. Wound closure rate was significantly reduced in the miR-184 mimic group compared with the miR-NC group after 24 h of incubation (P<0.01; Fig. 2C and D). The results from the Transwell assay revealed that the number of invading cells in the miR-184 mimic group was significantly lower compared with that in the miR-NC group (P<0.01; Fig. 2E and F). These findings suggest that miR-184 overexpression prevents the proliferation, migration and invasion of PC cells.
Figure 2.
miR-184 overexpression inhibits Du145 cell proliferation, migration and invasion. (A) miR-184 upregulation following transfection with miR-184 mimic was confirmed using reverse transcription-quantitative PCR. **P<0.01 vs. miR-NC. (B) miR-184 overexpression inhibited Du145 cell viability compared with control and miR-NC. **P<0.01 vs. miR-NC. (C and D) Du145 cell migration was evaluated using wound healing assay. (C) Representative images of wounded Du145 cell monolayers at 0 and 24 h following transfection with miR-184 mimic or miR-NC. (D) miR-184 overexpression inhibited wound closure. Scale bar, 100 µm. **P<0.01 vs. miR NC. (E and F) Transwell assay was conducted to examine the role of miR-184 overexpression in Du145 cells by calculating the mean number of invading cells. (E) Representative images of invading Du145 cells transfected with miR-184 mimic or miR-NC. (F) miR-184 overexpression inhibited Du145 cell invasion. Scale bar, 200 µm. **P<0.01 vs. miR-184 NC. miR, microRNA; NC, negative control; OD, optical density.
DLX1 is a target gene of miR-184
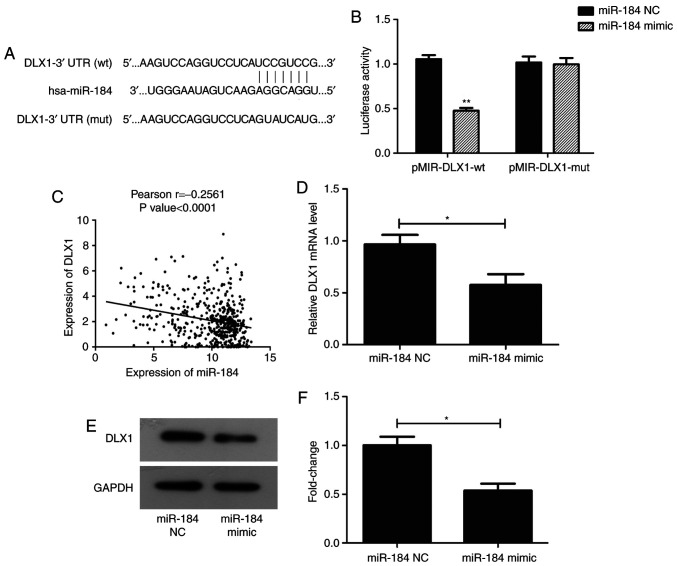
TargetScan 7.2 was used to confirm whether DLX1 is directly targeted by miR-184. DLX1 has a seven-nucleotide miR-184-binding site in its 3'UTR (Fig. 3A). Luciferase activity was significantly repressed in PC cells co-transfected with pMIR-DLX1-WT and miR-184 mimic (P<0.01), but not in those co-transfected with pMIR-DLX1-mut and miR-184 mimic (Fig. 3B). In addition, Pearson's analysis revealed that the expression levels of miR-184 and DLX1 were negatively correlated using the clinical data obtained from TCGA (P<0.0001; Fig. 3C). RT-qPCR and western blotting data also revealed that, compared with GAPDH, the expression of DLX1 was reduced at the mRNA and protein levels following transfection with miR-184 mimic (Fig. 3D-F; P<0.05).
Figure 3.
DLX1 is a direct target of miR-184. (A) Sequence of the miR-184-binding site in DLX1 3'UTR. (B) Relative luciferase activity in Du145 cells co-transfected with miR-184 mimic or miR-NC and DLX1-3'UTR (wt) or DLX1-3'UTR (mut) and corresponding quantification. **P<0.01 vs. miR-NC. (C) Pearson's analysis between miR-184 and DLX1 expression based on the PC tissue data from TCGA database. P<0.0001. (D) Reverse transcription-quantitative PCR measurement of DLX1 expression following miR-184 overexpression, *P<0.05. (E and F) Expression of DLX1 protein was measured in Du145 cells using western blot analysis following transfection with miR-184 mimic or miR-NC; the results were normalized to GAPDH. (E) Representative western blot images showing DLX1 and GAPDH expression. (F) Quantified data from. (E) *P<0.05. DLX1, distal-less homeobox 1; miR, microRNA; 3'UTR, 3' untranslated region; wt, wild-type; mut, mutant; NC, negative control.
The effects of miR-184 overexpression on Du145 cells are reversed by DLX1 overexpression
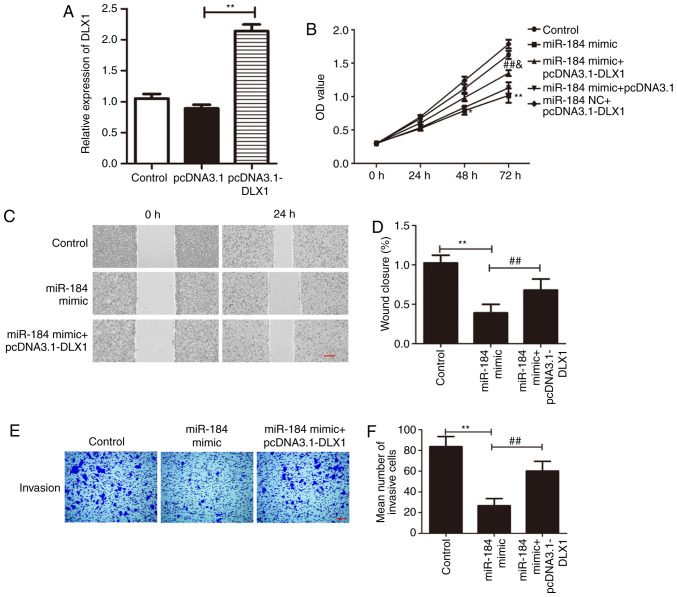
To further elucidate the molecular mechanisms underlying the anti-proliferative and anti-migratory effects of miR-184, the role of DLX1 was further studied. Given the negative correlation between miR-184 expression and DLX1 expression, it was hypothesized that the anti-proliferative and anti-migratory effects of miR-184 mimic in Du145 cells might be reversed by overexpression of DLX1. RT-qPCR showed that DLX1 expression was significantly increased in Du145 cells transfected with pcDNA3.1-DLX when compared with cells transfected with empty vector (Fig. 4A; P<0.01). It was subsequently found that the overexpression of DLX1 reversed the effects of miR-184 mimic on Du145 cells. The results of the CCK-8 assay suggested that the anti-proliferative effects of miR-184 mimic overexpression in Du145 cells were partially reversed following co-transfection with plasmids expressing DLX1 (P<0.05; F=0.37; Fig. 4B). Similarly, the wound healing and Transwell assays showed similar patterns. Although the overexpression of DLX1 did not restore the migration and invasiveness of Du145 cells to levels comparable to the control group, it partially alleviated the inhibitory effects of miR-184 overexpression on Du145 cells (P<0.01; Fig. 4C-F). Taken together, these findings indicate that DLX1 is a direct target gene of miR-184, and its overexpression can partially reverse the effects of miR-184 overexpression.
Figure 4.
The effects of miR-184 mimic on Du145 cells were rescued by DLX1 overexpression. (A) The transfection efficiency of pcDNA3.1-DLX1 in Du145 cells was tested using reverse transcription-quantitative PCR. **P<0.01 vs. pcDNA3.1. (B) The inhibitory effects of miR-84 mimics on Du145 cell viability were reversed by co-transfection with pcDNA3.1-DLX1. *P<0.05 and **P<0.01 vs. control, ##P<0.01 vs. miR-184 mimic and &P<0.05 vs. miR NC + pcDNA3.1-DLX1. (C and D) The inhibitory effects of miR-184 mimics on Du145 cell migration were reversed by co-transfection with pcDNA3.1-DLX1. (C) Representative images of the Du145 cell monolayer following transfection with miR-184 mimic or co-transfection with miR-184 mimic and pcDNA3.1-DLX1. (D) Quantified results from. (C) Scale bar, 100 µm. **P<0.01 vs. control and ##P<0.01 vs. miR-184 mimic. (E and F) The inhibitory effects of miR-184 mimic on Du145 cell invasion were reversed by co-transfection with pcDNA3.1-DLX1. (E) Representative images of invading Du145 cells following transfection with miR-184 mimic or co-transfection with miR-184 mimic and pcDNA3.1-DLX1. (F) Quantified results from. (E) Scale bar, 200 µm. **P<0.01 vs. control and ##P<0.01 vs. miR-184 mimic. DLX1, distal-less homeobox 1; miR, microRNA; OD, optical density.
Discussion
Previous studies have demonstrated that miRNAs can modulate a diverse number of cellular functions in several diseases, rendering it meaningful to investigate the regulatory network of miRNAs (22). Indeed, a variety of biological functions have been found to be associated with the aberrant expression of miRNAs, including cell apoptosis, proliferation, differentiation and immune regulation, such as pathogen recognition (23-26). In particular, dysregulated miR-184 has been reported to be associated with a number of malignancies (16-18). The aim of the present study was to investigate the role of miR-184 in PC.
Accumulating evidence supports an association between miR-184 expression and the promotion or prevention of tumor development. miR-184 may impair cancer progression, as reported by Su et al (27), who found that the overexpression of miR-184 exerted an inhibitory effect on renal cell carcinoma cell proliferation. Feng et al (28) suggested that miR-184 expression was decreased in glioblastoma (GBM) tissues and suppressed the development of GBM by binding to stanniocalcin-2; similarly, an inhibitory effect was also reported in breast cancer (29). By contrast, miR-184 was found to promote cell proliferation in tongue squamous cell carcinoma by targeting SOX7(30). These findings collectively raise the possibility that miR-184 serves a dual function in tumorigenesis, namely as a tumor suppressor as well as tumor promoter, although it may be hypothesized that this largely depends on the tumor type and a diverse number of potential target genes. In the present study, the results were consistent with previous investigations (25,26), supporting the hypothesis of an inhibitory role of miR-184 in PC.
DLX1 is a binding protein of β-catenin, which can increase cancer cell viability and migration by activating β-catenin/TCF signaling (21). In fact, abnormal activation of β-catenin/TCF signaling is considered to be a signature of PC (31). Therefore, DLX1 appears to be a potential carcinogenic factor. DLX1 has been reported to promote ovarian cancer progression by interacting with Forkhead box protein M1, and to also contribute to the development of oral clefts (32,33). DLX1 has also been implicated in PC (34,35). In the present study, the results of the dual luciferase reporter assay and the analysis of cell proliferation, migration and invasion in vitro indicated that DLX1 is a downstream target gene of miR-184. In addition, the overexpression of DLX1 partially reversed the inhibitory effects of miR-184 overexpression on PC.
In conclusion, miR-184 and DLX1 exhibited opposite trends in PC tissues; namely, miR-184 expression was suppressed, whereas DLX1 expression was increased. Although further mechanistic studies are required, including chromatin immunoprecipitation, immunohistochemical analyses, use of animal models and recording of the clinical characteristics of patients with PC during follow-up, the findings of the present study suggest, to a certain extent, that miR-184 can affect PC progression by suppressing DLX1 expression. Therefore, miR-184 may be of value as a potential treatment target of PC in the future.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
GGT and CX analyzed the data and wrote the manuscript. WKZ and CYW revised the manuscript. CYW, GGT, CX and WKZ collected the data and designed the study.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1. Cancer Stat Facts: Prostate Cancer in 2018; Available from: https://seer.cancer.gov/statfacts/html/prost.html. [Google Scholar]
- 2. World Cancer Report 2014. World Health Organization. 2014. pp. Chapter 5.11. [Google Scholar]
- 3.Caini S, Gandini S, Dudas M, Bremer V, Severi E, Gherasim A. Sexually transmitted infections and prostate cancer risk: A systematic review and meta-analysis. Cancer Epidemiol. 2014;38:329–338. doi: 10.1016/j.canep.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Miller DC, Hafez KS, Stewart A, Montie JE, Wei JT. Prostate carcinoma presentation, diagnosis, and staging: An update form the national cancer data base. Cancer. 2003;98:1169–1178. doi: 10.1002/cncr.11635. [DOI] [PubMed] [Google Scholar]
- 5.Redman JM, Gulley JL, Madan RA. Combining immunotherapies for the treatment of prostate cancer. Urol Oncol. 2017;35:694–700. doi: 10.1016/j.urolonc.2017.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jang TL, Kim IY, Scardino PT, Eastham JA. Reply to effectiveness of radical prostatectomy with adjuvant radiotherapy versus radiotherapy plus androgen deprivation therapy for men with advanced prostate cancer: Do we have certainties today? Cancer. 2019;125:2318–2320. doi: 10.1002/cncr.32054. [DOI] [PubMed] [Google Scholar]
- 7.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 9.O'Hara SP, Mott JL, Splinter PL, Gores GJ, LaRusso NF. MicroRNAs: Key modulators of posttranscriptional gene expression. Gastroenterology. 2009;136:17–25. doi: 10.1053/j.gastro.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang XJ, Reyes JL, Chua NH, Gaasterland T. Prediction and identification of arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5(R65) doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He J, Sun M, Geng H, Tian S. Long non-coding RNA Linc00518 promotes paclitaxel resistance of the human prostate cancer by sequestering miR-216b-5p. Biol Cell. 2019;111:39–50. doi: 10.1111/boc.201800054. [DOI] [PubMed] [Google Scholar]
- 12.Liang YK, Han ZD, Lu JM, Liu ZZ, Zhuo YJ, Zhu XJ, Chen JX, Ye JH, Liang YX, He HC, Zhong WD. Downregulation of ARID4A and ARID4B promote tumor progression and directly regulated by microRNA-30d in patient with prostate cancer. J Cell Biochem. 2018;119:7245–7255. doi: 10.1002/jcb.26913. [DOI] [PubMed] [Google Scholar]
- 13.Jiang W, Gao Y, Wang Z, Gong C, Hu C, Ding X, Qiang L, Gao S, Ren F. Co-delivery of miR-4638-5p and docetaxel based on redox-sensitive polypeptide micelles as an improved strategy for the treatment of castration-resistant prostate cancer. Mol Pharm. 2019;16:437–447. doi: 10.1021/acs.molpharmaceut.8b01074. [DOI] [PubMed] [Google Scholar]
- 14.Khorasani M, Teimoori-Toolabi L, Farivar TN, Asgari M, Abolhasani M, Shahrokh H, Afgar A, Kalantari E, Peymani A, Mahdian R. Aberrant expression of miR-141 and nuclear receptor small heterodimer partner in clinical samples of prostate cancer. Cancer Biomark. 2018;22:19–28. doi: 10.3233/CBM-170696. [DOI] [PubMed] [Google Scholar]
- 15.Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci USA. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu X, Ding X, Ding Z, Jia P. Total flavonoids from leaves of carya cathayensis ameliorate renal fibrosis via the miR-21/Smad7 signaling pathway. Cell Physiol Biochem. 2018;49:1551–1563. doi: 10.1159/000493458. [DOI] [PubMed] [Google Scholar]
- 17.Wu G, Liu J, Wu Z, Wu X, Yao X. MicroRNA-184 inhibits cell proliferation and metastasis in human colorectal cancer by directly targeting IGF-1R. Oncol Lett. 2017;14:3215–3222. doi: 10.3892/ol.2017.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang Z, Zhao J, Xie W, Sun Q, Wang H, Qiao B. LncRNA UCA1 promotes proliferation and cisplatin resistance of oral squamous cell carcinoma by suppressing miR-184 expression. Cancer Med. 2017;6:2897–2908. doi: 10.1002/cam4.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Qadir M, Faheem A. mRNA: A diagnostic and therapeutic tool for pancreatic cnacer. Crit Rev Eukaryot Gene Expr. 2017;27:197–204. doi: 10.1615/CritRevEukaryotGeneExpr.2017019494. [DOI] [PubMed] [Google Scholar]
- 21.Liang M, Sun Y, Yang HL, Zhang B, Wen J, Shi BK. DLX1, a binding protein of beta-catenin, promoted the growth and migration of prostate cancer cells. Exp Cell Res. 2018;363:26–32. doi: 10.1016/j.yexcr.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Li JY, Wei X, Sun Q, Zhao XQ, Zheng CY, Bai CX, Du J, Zhang Z, Zhu LG, Jia YS. MicroRNA-449b-5p promotes the progression of osteoporosis by inhibiting osteogenic differentiation of BMSCs via targeting Satb2. Eur Rev Med Pharmacol Sci. 2019;23:6394–6403. doi: 10.26355/eurrev_201908_18519. [DOI] [PubMed] [Google Scholar]
- 23.Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, Garcia S, Nowak J, Yeung ML, Jeang KT, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104:16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akao Y, Nakagawa Y, Naoe T. MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol Rep. 2006;16:845–850. [PubMed] [Google Scholar]
- 25.Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578–589. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen XM, Splinter PL, O'Hara SP, LaRusso NF. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against cryptosporidium parvum infection. J Biol Chem. 2007;282:28929–28938. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su Z, Chen D, Li Y, Zhang E, Yu Z, Chen T, Jiang Z, Ni L, Yang S, Gui Y, et al. microRNA-184 functions as tumor suppressor in renal cell carcinoma. Exp Ther Med. 2015;9:961–966. doi: 10.3892/etm.2015.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng L, Ma J, Ji H, Liu Y, Hu W. MiR-184 retarded the proliferation, invasiveness and migration of glioblastoma cells by repressing stanniocalcin-2. Pathol Oncol Res. 2018;24:853–860. doi: 10.1007/s12253-017-0298-z. [DOI] [PubMed] [Google Scholar]
- 29.Phua YW, Nguyen A, Roden DL, Elsworth B, Deng N, Nikolic I, Yang J, Mcfarland A, Russell R, Kaplan W, et al. MicroRNA profiling of the pubertal mouse mammary gland identifies miR-184 as a candidate breast tumour suppressor gene. Breast Cancer Res. 2015;17(83) doi: 10.1186/s13058-015-0593-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen D, Li J, Li S, Han P, Li N, Wang Y, Du S. miR-184 promotes cell proliferation in tongue squamous cell carcinoma by targeting SOX7. Oncol Lett. 2018;16:2221–2228. doi: 10.3892/ol.2018.8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider JA, Logan SK. Revisiting the role of Wnt/β-catenin signaling in prostate cancer. Mol Cell Endocrinol. 2018;462:3–8. doi: 10.1016/j.mce.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan DW, Hui WW, Wang JJ, Yung MMH, Hui LMN, Qin Y, Liang RR, Leung THY, Xu D, Chan KKL, et al. DLX1 acts as a crucial target of FOXM1 to promote ovarian cancer aggressiveness by enhancing TGF-β/SMAD4 signaling. Oncogene. 2017;36:1404–1416. doi: 10.1038/onc.2016.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabóia TM, Reis MF, Martins AM, Romanos HF, Tannure PN, Granjeiro JM, Vieira AR, Antunes LS, Küchler EC, Costa MC. DLX1 and MMP3 contribute to oral clefts with and without positive family history of cancer. Arch Oral Biol. 2015;60:223–228. doi: 10.1016/j.archoralbio.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Leyten GH, Hessels D, Smit FP, Jannink SA, de Jong H, Melchers WJ, Cornel EB, de Reijke TM, Vergunst H, Kil P, et al. Identification of a candidate gene panel for the early diagnosis of prostate cancer. Clin Cancer Res. 2015;21:3061–3070. doi: 10.1158/1078-0432.CCR-14-3334. [DOI] [PubMed] [Google Scholar]
- 35.Rhie SK, Guo Y, Tak YG, Yao L, Shen H, Coetzee GA, Laird PW, Farnham PJ. Identification of activated enhancers and linked transcription factors in breast, prostate, and kidney tumors by tracing enhancer networks using epigenetic traits. Epigenetics Chromatin. 2016;9(50) doi: 10.1186/s13072-016-0102-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.