Abstract
Mounting evidence suggests that eukaryotic RNA polymerases preassociate with multiple transcription factors in the absence of DNA, forming RNA polymerase holoenzyme complexes. We have purified an apparent RNA polymerase I (Pol I) holoenzyme from Xenopus laevis cells by sequential chromatography on five columns: DEAE-Sepharose, Biorex 70, Sephacryl S300, Mono Q, and DNA-cellulose. Single fractions from every column programmed accurate promoter-dependent transcription. Upon gel filtration chromatography, the Pol I holoenzyme elutes at a position overlapping the peak of Blue Dextran, suggesting a molecular mass in the range of ∼2 MDa. Consistent with its large mass, Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gels reveal approximately 55 proteins in fractions purified to near homogeneity. Western blotting shows that TATA-binding protein precisely copurifies with holoenzyme activity, whereas the abundant Pol I transactivator upstream binding factor does not. Also copurifying with the holoenzyme are casein kinase II and a histone acetyltransferase activity with a substrate preference for histone H3. These results extend to Pol I the suggestion that signal transduction and chromatin-modifying activities are associated with eukaryotic RNA polymerases.
In eukaryotes, the three multisubunit RNA polymerases utilize auxiliary transcription factors to recognize gene promoters and initiate transcription from defined start points (10, 21, 24, 36, 44, 48, 50, 59, 73). RNA polymerase II (Pol II) was the first eukaryotic RNA polymerase shown to be associated with general transcription factors in a holoenzyme complex (28, 29, 46). The Pol II holoenzyme was subsequently shown to include additional activities including the SWI/SNF chromatin-remodeling activity (71), the protein complex dubbed “mediator” which interacts with the C-terminal domain of the largest Pol II subunit (27, 38), multiple protein kinases (34), and enzymes involved in DNA repair (38). The list of proteins associated with the Pol II holoenzyme continues to grow as antibodies against suspected components are developed and tested.
Evidence for a Pol III holoenzyme was first reported a decade ago (72). Recently, a Pol III holoenzyme was characterized in greater detail by the Roeder laboratory and shown to contain the essential transcription factors TFIIIB and TFIIIC (70). The Pol III holoenzyme is self-sufficient for transcription of tRNAs and other class III genes which do not require TFIIIA. The latter gene-specific activator must be added to the holoenzyme for transcription of 5S rRNA genes. The Pol III holoenzyme was also shown to include at least one activity involved in the rapid down-regulation of Pol III transcription following treatment with cycloheximide or adenovirus infection (70). Therefore, it appears that the Pol III holoenzyme, like the Pol II holoenzyme, contains activities which are not strictly required for transcription but which probably link transcription with cellular signaling pathways.
Initial evidence for RNA Pol I holoenzymes has come from the extensive purification of a plant Pol I-containing complex self-sufficient for accurate, promoter-dependent transcription (53) and from functional studies of mouse complexes immunoprecipitated with antibodies against Pol I subunits (58). Plant Pol I transcription factors have not yet been characterized; thus, the identification of proteins in the putative Pol I holoenzyme of Brassica oleracea has been limited to several small subunits of the Pol I core enzyme for which antibodies are available (53). In the better-characterized Pol I transcription systems of vertebrates, namely, human, mouse, rat, and Xenopus systems, rRNA gene transcription is thought to require the transcription factor SL1 (also known as TIF-IB, Rib-1, TIF, and TFI-D), assisted by upstream binding factor (UBF) (3–5, 12, 22, 25, 39, 54, 61, 64, 66). SL1 is a complex of TATA-binding protein (TBP) and several associated factors (13, 52, 57). UBF is a homodimer that can bend and wrap DNA, presumably facilitating correct juxtaposition of SL1, polymerase, or other proteins essential for promoter and enhancer function (2, 49, 52). Though stimulatory, UBF is nonessential for basal-level in vitro transcription of rat and mouse rRNA gene templates (31, 60). Interestingly, immunoprecipitated mouse holoenzyme complexes contain both TBP and UBF but are not self-sufficient for promoter-dependent transcription. Instead, addition of another activity, TIF-IC, is needed (58). Thus far, a TIF-IC-like activity has been identified only in rodents.
In this paper, we show that Xenopus laevis cell extracts can be purified by DEAE (anion-exchange), Biorex (cation-exchange), Sephacryl (gel filtration), Mono Q (analytical anion-exchange), DNA-cellulose (affinity), and glycerol gradient sedimentation to yield single fractions that initiate accurate rRNA gene transcription in vitro. We show that TBP copurifies with the putative Pol I holoenzyme, whereas UBF does not. Protein kinase and histone acetyltransferase (HAT) activities also copurify with holoenzyme activity, extending the suggestion that all three nuclear polymerases associate with other protein complexes to integrate cellular signaling, chromatin modification, and transcription.
MATERIALS AND METHODS
Preparation of X. laevis S100 extracts.
X. laevis Xlk2 kidney cells were grown as monolayers in glass roller bottles in 50% L-15 medium (Sigma or Gibco) supplemented with 5% Nu serum (Collaborative Research), 5% fetal calf serum (Gibco), and 100 U each of penicillin and streptomycin per ml. Cell transcription extracts were made according to the method of McStay and Reeder (41). Briefly, cells were harvested at late log phase with phosphate-buffered saline supplemented with 1 mM EDTA (pH 8.0) and collected by low-speed centrifugation. After being washed in EDTA-free phosphate-buffered saline, cells were swollen in hypotonic buffer and ruptured with a Dounce homogenizer. After addition of KCl to a final concentration of 140 mM, extracts were subjected to centrifugation at 100,000 × g for 2 h at 4°C. The supernatant (S100) was dialyzed against column buffer (20% glycerol, 25 mM HEPES [pH 7.9], 1 mM dithiothreitol [DTT], and 0.1 mM EDTA) containing 100 mM KCl (CB100), frozen in liquid nitrogen, and stored at −80°C.
Glycerol gradient sedimentation.
For glycerol gradient sedimentation, 0.5 ml of transcriptionally active DE350 fraction was layered onto 11.5-ml, 20 to 40% glycerol gradients containing 100 mM KCl, 10 mM MgCl2, 0.2 mM EDTA, 0.2 mM EGTA, and 1 mM DTT. Gradients were subjected to centrifugation at 40,000 rpm in a Beckman SW41 rotor for 18 h at 4°C. Thirty ∼0.4-ml fractions were collected from the bottom of the tube. Aliquots were mixed with an equal volume of glycerol-free buffer and tested for both total Pol I activity (nicked calf thymus DNA template; 150 μg of α-amanitin per ml) and promoter-dependent transcription.
Assay for nonspecific RNA Pol I activity.
Promoter-independent (nonspecific) total RNA Pol I activity was measured as described in the work of Schwartz and Roeder (56). Fractions dialyzed against CB100 were added to reaction mixtures containing sheared calf thymus DNA (final concentration, 25 μg/ml), α-amanitin (150 to 50 μg/ml), nucleoside triphosphates (0.5 mM [each] ATP, GTP, and CTP and 0.04 mM UTP), [α-32P]UTP (0.05 μCi/μl; specific activity, 3,000 Ci/mmol), MnCl2 (2 mM), and bovine serum albumin (1 mg/ml). After 15 min at 30°C, reaction mixtures were spotted on DE81 filters and washed repeatedly with 0.5 M sodium phosphate to remove unincorporated UTP, followed by washing in 95% ethanol to speed drying. Incorporated [α-32P]UTP bound to filters was quantified by scintillation counting.
Assay for promoter-dependent RNA Pol I activity.
Promoter-dependent (specific) RNA Pol I transcription was performed in 30- to 40-μl reaction mixtures containing 10% glycerol, 25 mM HEPES (pH 7.9), 90 mM KCl, 6 mM MgCl2, 1 mM DTT, 100 μg of α-amanitin per ml, 0.5 mM (each) nucleoside triphosphates, and 200 to 400 ng of template DNA. Reaction mixtures were incubated for 2 to 3 h at 25°C. Transcription reactions were analyzed by S1 protection with end-labeled, single-stranded 65-nucleotide probes complementary to the RNA and spanning −15 to +50 or −21 to +44 relative to the transcription start site, +1. The labeled oligonucleotide and RNA were hybridized overnight at 65°C in 0.3 M NaCl–10 mM Tris-HCl (pH 7.5)–1 mM EDTA. S1 digestion was carried out at 37°C for 1 h in 5% glycerol–50 mM NaCl–30 mM sodium acetate (pH 4.5)–1 mM zinc sulfate–100 to 150 U of S1 nuclease per ml. Products were resolved on 8% polyacrylamide–8 M urea gels. Gels were vacuum dried onto filter paper and exposed to X-ray film.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
Peak fractions were analyzed on SDS–4.5 to 18% gradient polyacrylamide gels and stained with Coomassie brilliant blue R-250. For Western blotting, SDS–7.5 or 10% polyacrylamide gels were used. Proteins were transferred to nitrocellulose or polyvinylidene difluoride membranes (Amersham) with a semidry blotting apparatus (Sartorius) at 100 mA for 3 to 4 h. Membranes were incubated with several rabbit antisera: anti-TBP was a generous gift of Paul Labhart, anti-Drosophila casein kinase II (CKII) cross-reacting with the Xenopus β subunit was a generous gift of Neil Osheroff, and anti-human CKII α and α′ were purchased from Upstate Biotechnology. Anti-Xenopus UBF was raised in rabbits against the 328 N-terminal amino acids of UBF expressed in Escherichia coli. Secondary antibodies for Western blotting, coupled to horseradish peroxidase, were detected by enhanced chemiluminescence according to the directions of the manufacturer (Amersham). Alternatively, antibodies coupled to alkaline phosphatase were detected by the conventional colorimetric assay (Bio-Rad).
Protein kinase activity assay.
Mono Q peak fractions in CB100 (4 μl) were incubated for 30 min at 25°C in 10-μl reaction mixtures containing 3.5 μM ATP, 1 to 2 μCi of [γ-32P]ATP (6,000 Ci/mmol), and 7 mM MgCl2. Reaction mixtures were then loaded onto an SDS-polyacrylamide gel. Following electrophoresis, gels were fixed in methanol-acetic acid, dried, and exposed to X-ray film.
HAT assays.
HAT assays were performed as described by Brownell and Allis (8), with minor modifications. Ten micrograms of HeLa core histones (kindly provided by J. Workman) or 25 μg of total calf thymus histones (Worthington; fraction HLY) was incubated with 5 μl of S100 extract or dialyzed column fractions for 45 min at 37°C in 30- to 50-μl reaction mixtures. Buffer conditions were 50 mM Tris-HCl (pH 7.9), 1 mM DTT, 0.1 mM EDTA, 10% glycerol, 10 mM butyric acid, 0.25 μM acetyl coenzyme A (CoA), and 100 nCi of [3H] acetyl-CoA (26 Ci/mmol; Sigma). Reactions were stopped with 5 volumes of cold (−20°C) acetone. Precipitated proteins were washed with acetone, dissolved in SDS sample buffer, and subjected to electrophoresis on an SDS–15% polyacrylamide gel. Gels were treated with En3Hance cocktail (Kodak), and 3H-labeled proteins were detected by fluorography with Kodak XAR film.
RESULTS
Initial evidence for a Pol I holoenzyme.
Cell extracts of cultured X. laevis cells support accurate RNA Pol I transcription initiation from recombinant X. laevis rRNA minigenes (41). In this procedure, mechanically disrupted cells are adjusted to 140 mM KCl and subjected to centrifugation at 100,000 × g, and the high-speed supernatant (S100) is saved. The S100 fraction supports transcription from the rRNA gene promoter but contains only ∼15% of the nonspecific RNA Pol I activity (promoter-independent transcription on nicked DNA) that can be assayed. The remaining 85% of the Pol I activity that can be detected is what can be extracted from the cell pellet at high salt concentrations (0.4 to 0.8 M KCl); these fractions do not support promoter-dependent transcription (1). It is likely that additional Pol I remains in crude nuclear-chromatin pellets after high salt extraction and that histones and other chromatin proteins extracted at high salt inhibit the in vitro Pol I assay (56). Therefore, we suspect that the actual amount of Pol I extracted with 140 mM KCl is substantially less than 15% of the total Pol I and is probably enriched in free polymerase not yet engaged in transcription elongation.
S100 fractions were dialyzed against 100 mM KCl buffer and applied to a DEAE column. The column was then washed with 100 mM KCl column buffer (CB100) and sequentially eluted with column buffer containing 175 mM KCl (CB175), CB350, and CB1000 to yield fractions designated DE175, DE350, and DE1000, respectively. Following dialysis, the flowthrough and step-eluted fractions were tested alone and in all combinations for their ability to support α-amanitin-resistant transcription. Approximately 30% of the assayable Pol I activity flowed through the DEAE column, ∼10% was present in the DE175 fraction, and the remaining ∼60% was in the DE350 fraction. Only the DE350 fraction programmed accurate transcription initiation, as expected from prior studies (39). Though consistently less active than the starting S100 (compare lanes 1 and 2 in Fig. 1B), the promoter-dependent transcriptional activity of the DE350 fraction was not further stimulated by addition of any combination of the flowthrough, DE175, or DE1000 fractions (data not shown).
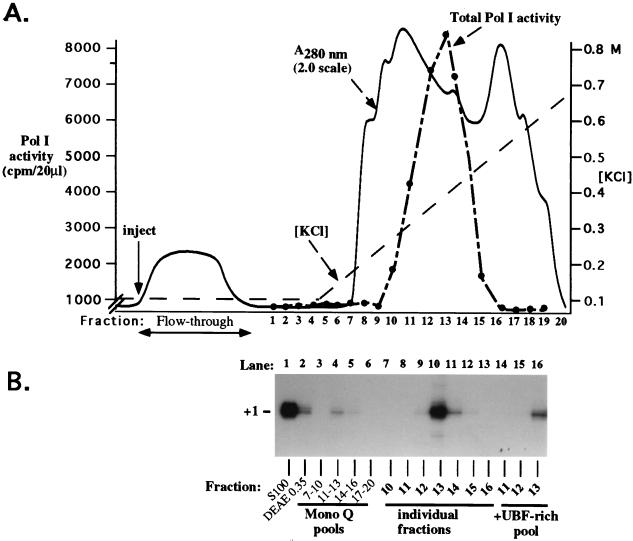
FIG. 1.
Coelution of all proteins required for RNA Pol I transcription in single Mono Q fractions. (A) Assay for total Pol I activity. Proteins eluted from DEAE-Sepharose with 350 mM KCl were subjected to chromatography on Mono Q by fast protein liquid chromatography. After being washed at 0.1 M KCl, the column was eluted with a linear gradient from 0.1 to 0.7 M KCl. Aliquots (20 μl) of individual fractions were tested for total Pol I activity on nicked calf thymus DNA in the presence of 150 μg of α-amanitin per ml. (B) Assay for promoter-dependent transcription. Equal aliquots of three to four Mono Q fractions were mixed to form pools and tested alone and in various combinations for their ability to support transcription from an X. laevis rRNA minigene promoter. Transcripts were detected by S1 nuclease protection. The pools of fractions 11 to 13 and 14 to 16 tested positive in this assay (panel B and data not shown). Individual Mono Q fractions from the positive pools were then tested (lanes 7 to 16) and compared to the activity of the pooled fractions (lanes 3 to 6). Fraction 13 corresponded to the peak of both total and promoter-dependent Pol I transcription. UBF peaked in fractions 14 and 15 (data not shown). Addition of the UBF-rich pool (fractions 14 to 16) to fractions 11 to 13 did not improve their promoter-dependent transcription activity.
The transcriptionally competent DE350 fraction was dialyzed to 0.1 M KCl, loaded onto a Mono Q fast protein liquid chromatography column, and eluted with a 10-column-volume linear gradient from 0.1 to 0.7 M KCl (Fig. 1A). Twenty fractions were collected, dialyzed, and tested for Pol I activity with nicked calf thymus template DNA. Pol I activity was detected in fractions 10 to 15, with the peak represented by fractions 12 to 14 (Fig. 1A). Equal aliquots from several successive fractions were next combined to form pools and tested alone and in all possible combinations for their ability to support promoter-dependent transcription from an rRNA minigene, assayed by the S1 nuclease protection assay (6). The pool of fractions 11 to 13 was transcriptionally competent (Fig. 1B, lane 4) and was not stimulated by addition of any other pool (data not shown). Pooled fractions 14 to 16 also had weak activity (Fig. 1B, lane 5). Column fractions contributing to the active pools were next tested individually. Fractions 12 to 14 were able to program promoter-dependent transcription, with fraction 13 representing the peak of both promoter-dependent and nonspecific Pol I activity (Fig. 1B, lanes 9 to 11). Interestingly, fractions 12 and 14 had nearly as much total Pol I activity as fraction 13 (Fig. 1A), yet directed only low levels of promoter-dependent transcription. DNase I footprinting on rRNA gene enhancer probes showed that UBF activity peaked in fractions 15 and 16 and overlapped the right flank of the polymerase peak (data not shown). UBF was not detectable on the left flank of the peak in fraction 11 or 12. This suggested the possibility that fraction 13 was better for promoter-dependent transcription due to an optimal ratio of polymerase to UBF. If so, adding UBF-enriched fractions from the right side of the polymerase peak to fractions on the left side of the Pol I peak was expected to stimulate promoter-dependent transcription. However, mixing the UBF-rich pool (fractions 14 to 16) with fractions 11 to 13 did not stimulate their promoter-dependent transcriptional activity but only diluted them (Fig. 1B, lanes 14 to 16).
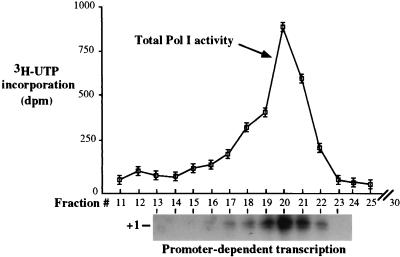
The finding that all necessary activities required for promoter-dependent Pol I transcription coeluted within single fractions on Mono Q suggested that they might be associated as a complex. Alternatively, coelution due to similar charge distributions was a possibility. As a further test, DE350 peak fractions were sedimented through 20 to 40% glycerol gradients, which were then fractionated. Six of the 30 glycerol gradient fractions (fractions 17 to 22) had significant total Pol I activity on nicked calf thymus DNA, which peaked in fraction 20 (Fig. 2; see graph). These same fractions were found to be capable of directing accurate transcription from the X. laevis rRNA gene promoter (Fig. 2, at bottom). Promoter-dependent transcription activity closely reflected the profile of total polymerase activity.
FIG. 2.
Individual fractions support accurate transcription initiation following sedimentation of DE350 fractions through glycerol gradients. Gradients (11.5 ml) were fractionated into 30 fractions and tested for total Pol I activity on nicked calf thymus DNA (top). Reactions were performed in triplicate, and the mean values for each fraction were plotted. Error bars represent the standard errors of the means. Fractions were also tested for their ability to program accurate, promoter-dependent Pol I transcription (autoradiogram at bottom). Transcripts were detected by S1 nuclease protection. Fraction 20 represented the peak in both assays.
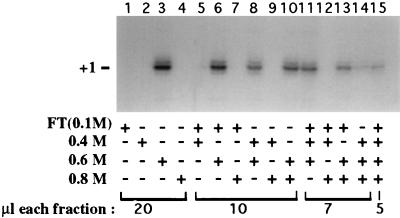
The data in Fig. 1 and 2 showed that the Pol I transcription machinery coeluted from a positively charged column matrix (DEAE) and also cosedimented when fractionated according to molecular mass. As a third test of the possible association of the Pol I transcription factors, we subjected the peak Mono Q fraction to chromatography on a negatively charged column matrix. Mono Q fraction 13 (Fig. 1), dialyzed to 100 mM KCl, was loaded onto a Biorex 70 column. The flowthrough was collected, and the column was washed prior to sequential step elutions with CB400, CB600, and CB800. These fractions were then dialyzed to 100 mM KCl and tested alone and in all possible combinations for their ability to direct transcription. The fraction eluting at 0.6 M KCl supported accurate, promoter-dependent transcription (Fig. 3, lane 3) and was not stimulated by addition of other fractions, but was diluted by them (lanes 6, 8, 10, 11, and 13 to 15).
FIG. 3.
Coelution of all activities essential for promoter-dependent Pol I transcription from Biorex 70. Mono Q fraction 13 (Fig. 1) in 0.1 M KCl buffer was applied to a 0.5-ml Biorex column. The flowthrough (FT) was collected, and the column was washed with CB100. Bound proteins were sequentially eluted with buffer containing 0.4, 0.6, and 0.8 M KCl. Individual fractions and combined fractions were tested for their ability to direct accurate transcription from the Xenopus minigene promoter. Transcripts were detected by S1 nuclease protection.
Purification of the putative Pol I holoenzyme to near homogeneity.
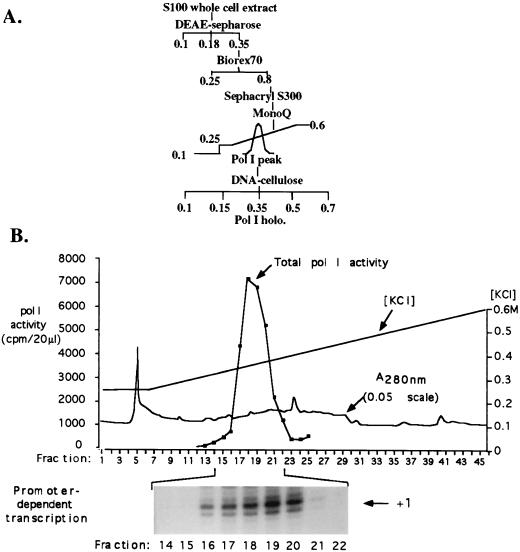
Based on the initial results in Fig. 1 to 3, we developed a scheme for purification of the putative Pol I holoenzyme that integrated DEAE-Sepharose, Biorex 70, and Mono Q ion-exchange chromatography; DNA-affinity chromatography (DNA-cellulose); and purification according to mass by gel filtration (Sephacryl S300) or glycerol gradient sedimentation. The protocol empirically found to provide the highest degree of purification while maintaining the greatest yield of promoter-dependent transcription activity was one that minimized dilution and dialysis (Fig. 4A). According to this purification scheme, crude S100 extract was initially fractionated on DEAE. Proteins eluted in CB350 were diluted slightly to 250 mM KCl and then loaded onto a Biorex column equilibrated in CB250. The Biorex column was eluted in a single step with CB800. Though a 600 mM elution from Biorex is sufficient to recover polymerase, as shown in Fig. 3, the 800 mM elution was found not to disrupt the complex and was expected to dissociate any contaminating nucleic acid or nonspecifically associated proteins prior to gel filtration. The Biorex eluate in 800 mM KCl buffer was next loaded onto a 195-ml Sephacryl S300 gel filtration column equilibrated and developed in CB100. The Sephacryl column served two purposes, facilitating the purification of the holoenzyme according to its mass and at the same time desalting the holoenzyme fractions. The holoenzyme peak eluting from the Sephacryl column was fractionated on Mono Q, by using a relatively shallow gradient from 250 to 600 mM KCl to optimize separation from other proteins. Finally, Mono Q peak fractions were subjected to chromatography on DNA-cellulose and eluted with steps of increasing salt concentration.
FIG. 4.
Extensive purification of the putative Xenopus holoenzyme. (A) Purification scheme. DNase-treated S100 extract (in CB100) was subjected to chromatography on DEAE-Sepharose. The 350 mM KCl fraction was diluted to 250 mM KCl and injected onto Biorex 70. Proteins eluting at 800 mM KCl were injected onto a 195-ml Sephacryl S300 gel filtration column equilibrated in CB100. The peak of total Pol I activity eluted near the Blue Dextran peak well in advance of thyroglobulin (669 kDa) and ferritin (450 kDa) molecular mass markers. The Sephacryl peak fractions were pooled and injected onto Mono Q, which was eluted with a gradient from 250 to 600 mM KCl. Peak fractions, eluting near 400 mM KCl, were dialyzed against CB100 and loaded onto a DNA-cellulose column. After being washed in CB100, fractions were eluted with CB150, CB350, CB500, and CB700. Pol I activity eluted in the 350 mM KCl fraction; other fractions had negligible activity. (B) Purification of the putative holoenzyme on the penultimate Mono Q column. The elution profile of total Pol I activity is shown in the graph. Individual fractions in the vicinity of the Pol I peak were then tested for their ability to direct accurate, promoter-dependent transcription (autoradiogram just below graph) by using 400 ng of supercoiled plasmid DNA containing the Xenopus rRNA minigene Ψ40.
By the purification scheme shown in Fig. 4A, the peak of Pol I activity eluted from the Sephacryl column after the void volume and overlapping the leading edge of the peak of Blue Dextran (Pharmacia), a polymer with an average molecular mass of ∼2 MDa. The lack of suitable mass standards in this size range and the nonlinear relationship between log10 mass and elution volume in this portion of the elution profile preclude a precise mass estimate for the complex. The peak of Pol I activity was followed by a broad shoulder of activity extending to ∼600 kDa, the approximate mass of the Pol I core enzyme. Sephacryl Pol I peak fractions (>1 MDa) were pooled and subjected to chromatography on Mono Q. Total Pol I activity, assayed with nicked calf thymus template DNA, eluted from Mono Q at approximately 0.35 to 0.38 M KCl, peaking in fractions 17 to 20 (Fig. 4B, graph). These same fractions were found to correspond to the peak holoenzyme fractions capable of promoter-dependent transcription (Fig. 4B, autoradiogram at bottom).
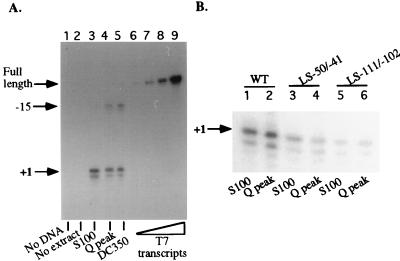
Peak Mono Q fractions were pooled, dialyzed to 100 mM KCl, and subjected to chromatography on double-stranded DNA-cellulose (Sigma). After the flowthrough was collected and washed with CB100, the column was step eluted with CB150, CB350, CB500, and CB700. Nonspecific Pol I activity bound to the column and eluted at 350 mM KCl (DC350 fraction), with only trace amounts of polymerase detected in other fractions. The peak DC350 fraction directed promoter-dependent Pol I transcription from the correct start site, +1 (Fig. 5A, lane 5). Interestingly, a signal mapping to −15 was also prevalent in transcription reactions with Mono Q (Fig. 5A, lane 4) or DC350 (lane 5) fractions, whereas this signal was much weaker (but detectable) with S100 extract (lane 3). Neither +1 nor −15 signals were obtained in control reactions (lanes 1 and 2), as expected. McStay and Reeder showed that an activity required for Pol I termination flows through DEAE, whereas the Pol I transcription initiation factors bind to the column and elute in the DE350 fraction (40). This activity should be missing from our purified fractions. This suggested that the −15 signal might result from internal S1 cleavage of readthrough transcripts initiated at +1 and going around the entire circular plasmid construct and traversing the promoter region complementary to the probe. Alternatively, transcription initiation sites elsewhere on the plasmid could be a source of readthrough transcripts. A third possibility is that the −15 signal represents an alternative transcription initiation site. The possibility that the −15 or +1 signals could be S1 artifacts resulting from internal digestion of readthrough transcripts seemed unlikely given that neither site is particularly AT rich. Nonetheless, a simple control experiment was performed to directly test this possibility. For this experiment, rRNA minigene sequences from −245 to +350 were transcribed into RNA with the T7 promoter in the pBluescript plasmid located adjacent to the upstream border of the cloned minigene sequences (−245). These transcripts would be identical to any putative readthrough transcripts traversing the minigene. Increasing amounts of these T7 transcripts were hybridized to a 65-mer probe spanning −21 to +44. RNA-probe hybrids were then subjected to S1 nuclease digestion in reactions performed side by side with those using transcripts generated with the S100, Mono Q, or DC350 fractions (lanes 3 to 5). Importantly, T7 transcripts were only digested to a size corresponding to the full-length probe (Fig. 5A, lanes 6 to 9). No fragments whose 5′ ends mapped to −15 or +1 were generated, indicating that the latter are not S1 artifacts of readthrough transcripts. This suggests that the −15 signal probably represents an alternative initiation site upstream of +1. A possibility is that the increased use of this site by proteins in Mono Q peak and DC350 fractions compared to starting S100 extracts may reflect the modification or loss of an activity that helps restrict transcription initiation to +1. Interestingly, we have noted that in fractions that have been stored for many months, the ability to transcribe from +1 is labile and progressively lost over time. In such “dead” fractions, transcription signals mapping to −15 can still be detected, also suggesting a time-dependent decay in a specificity factor (data not shown).
FIG. 5.
Accuracy and promoter specificity of highly purified holoenzyme fractions. (A) In lanes 3 to 5, S100, Mono Q, and DNA-cellulose (DC350) peak fractions were compared for their ability to program accurate transcription initiation from the Ψ40 minigene (400 ng). A reaction containing S100 but without added template (lane 1) and a reaction containing template but no added protein (lane 2) were run as controls. Lanes 6 to 9 are controls in which in vitro transcripts corresponding to the RNA strand of the complete minigene were generated by T7 polymerase and subjected to S1 nuclease protection alongside the other reactions. A single protected product, corresponding to the size of the full-length probe, was generated in proportion to the amount of input RNA. No −15 or +1 products were generated, suggesting that the latter are not artifacts due to cleavage of readthrough transcripts. (B) Comparison of S100 and Mono Q peak fractions for their sensitivity to promoter mutations. The wild-type (WT) Ψ40 minigene (500 ng) was used as the template in lanes 1 and 2. Two different linker scanner mutants of Ψ40, LS-50/-41 (lanes 3 and 4) and LS-111/-102 (lanes 5 and 6), were also tested.
As another test of promoter specificity, we compared Mono Q peak holoenzyme fractions to S100 fractions for their ability to initiate transcription from wild-type or linker scanner mutant promoters. Using promoter mutants shown previously to support diminished levels of transcription in vitro (with crude S100 fractions) or in vivo (by oocyte injection) (51), we found that S100 and holoenzyme fractions had similarly reduced activity on these templates. An example is shown in Fig. 5B with the strong linker scanner mutants LS-50/-41 and LS-111/-102. With a template concentration optimal for the Mono Q fractions (though higher than optimal for S100 fractions), the S100 and Mono Q fractions programmed similar levels of transcription from the wild-type minigene Ψ40 (lanes 1 and 2). Transcription was reduced significantly with LS-50/-41 (lanes 3 and 4) and was further reduced with LS-111/-102 (lanes 5 and 6), in agreement with prior studies (51).
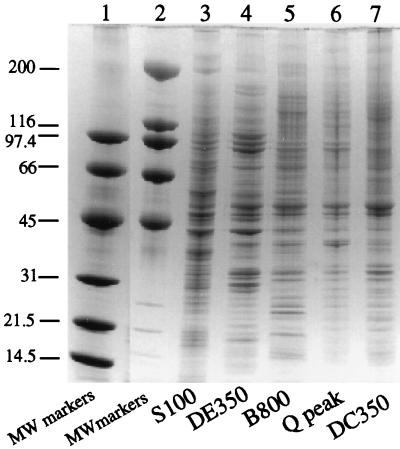
The protein compositions of peak fractions throughout the purification scheme were compared following gradient SDS-PAGE and Coomassie blue staining (Fig. 6). The protein compositions of the peak fractions from the final two columns, Mono Q and DNA-cellulose, are qualitatively similar. This suggests that purification of the complex is approaching apparent homogeneity, defined as the point at which no further reduction in complexity is achieved by additional steps. Approximately 55 distinct bands are visible in the DC350 fraction, and with the exception of prominent bands at ∼50 and ∼33 kDa, most have similar staining intensities.
FIG. 6.
Polypeptide composition of peak fractions throughout the purification scheme. Equal amounts (on a mass basis) of S100, DEAE, Biorex, Mono Q, or DNA-cellulose peak fractions were subjected to SDS-PAGE on a 4.5 to 18% gradient gel (lanes 3 to 7, respectively). Following electrophoresis, the gel was stained with Coomassie blue. Two different-size classes of molecular mass (MW) markers (Bio-Rad) were run on the same gel (lanes 1 and 2); their sizes in kilodaltons are indicated to the left of the figure.
TBP, but not UBF, copurifies with the Pol I holoenzyme.
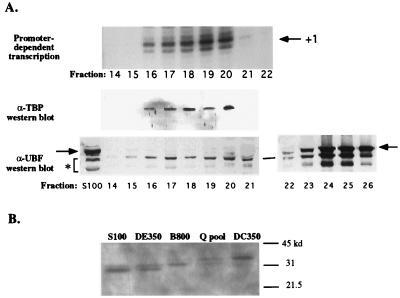
Mono Q and DNA-cellulose peak fractions were tested for activities suspected to be associated with the Pol I complex and for which antibodies or biochemical assays were available. With a polyclonal antiserum raised against recombinant Xenopus TBP (generously provided by Paul Labhart), TBP was readily detected in Mono Q fractions 16 to 20, precisely cofractionating with the fractions competent for promoter-dependent transcription (Fig. 7A). TBP was also detected in the DNA-cellulose peak fraction (Fig. 7B, last lane). Interestingly, comparison of peak fractions throughout the purification scheme reveals the progressive enrichment of a TBP isoform with decreased SDS-PAGE gel mobility relative to the starting S100 (Fig. 7B), most likely due to phosphorylation or some other posttranslational modification. Preimmune serum did not cross-react with any Xenopus proteins of the expected size for TBP (data not shown).
FIG. 7.
TBP copurifies with Pol I holoenzyme activity. (A) Mono Q fractions including the peak of Pol I holoenzyme activity (same column run as that shown in Fig. 4) were subjected to Western blotting with polyclonal antibodies directed against Xenopus TBP or UBF. The blots are aligned with an autoradiogram revealing the transcriptionally active fractions. Antibody-antigen complexes were detected by enhanced chemiluminescence. Full-length UBF is denoted by arrows; smaller proteins thought to be UBF degradation products are denoted by an asterisk. (B) Detection of TBP in peak fractions throughout the Pol I holoenzyme purification scheme reveals that an isoform with reduced gel mobility is enriched during purification.
We tested whether the abundant Pol I transcription factor UBF cofractionated with the holoenzyme by Western blotting with a polyclonal antiserum raised against the amino-terminal 328 amino acids of Xenopus UBF. This antiserum (but not preimmune serum) cross-reacts with the full-length UBF species of 82 and 85 kDa (denoted by the arrow in Fig. 7A) as well as a series of smaller doublets suspected to be UBF turnover products missing various portions of the C terminus (see S100 control lane). These smaller proteins are also observed if growing Xenopus cells are quickly suspended in SDS sample buffer and boiled and the resulting cell lysates are subjected to SDS-PAGE and Western blotting, suggesting that these putative turnover products are present in the cell (data not shown). As discussed previously, UBF footprinting activity elutes after the Pol I peak on Mono Q. Consistent with these footprinting data, Western blotting shows that the vast majority of full-length UBF elutes after the Pol I peak, being most abundant in fractions 23 to 27 (Fig. 7A, bottom right). However, a trace of the full-length UBF doublet can be detected in peak holoenzyme fractions. A more abundant protein doublet ∼10 kDa smaller than full-length UBF is detected in Mono Q fractions 14 to 26, possibly corresponding to UBF degradation products. Upon subsequent chromatography on DNA-cellulose, neither the full-length nor these smaller UBF-related proteins coelute with the holoenzyme in the DC350 fraction. Instead, these UBF-related proteins eluted in the DC500 fraction (data not shown).
One could argue that UBF must associate with the Pol I holoenzyme to copurify up until the Mono Q column and might be separated from the holoenzyme only as a consequence of the elution procedure. Interpretation is complicated by the abundance of UBF. On the Sephacryl column prior to Mono Q, full-length UBF was found in virtually all fractions by Western blotting, beginning with the void volume and extending to ∼85 kDa, the approximate size of the UBF monomer (data not shown). UBF’s unusually broad elution profile could be due to aggregation or participation in a variety of distinct protein complexes. Regardless, UBF’s presence in Sephacryl fractions containing the polymerase holoenzyme peak may be fortuitous, thus explaining UBF’s failure to precisely coelute with the holoenzyme on Mono Q or DNA-cellulose.
Protein kinase activity copurifies with the Pol I holoenzyme.
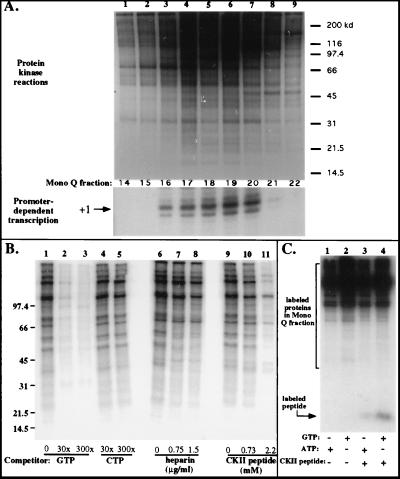
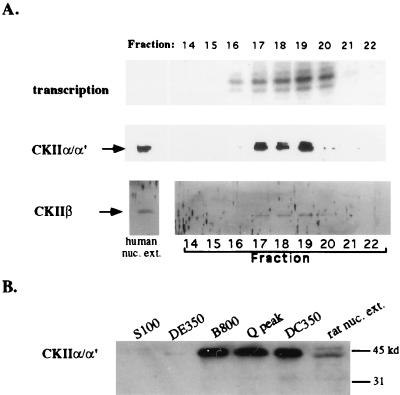
We tested for protein kinase(s) by incubating Mono Q fractions flanking and including the holoenzyme peak with [γ-32P]ATP in a magnesium-containing buffer, followed by SDS-PAGE and autoradiography. As shown in Fig. 8, one or more protein kinases were able to phosphorylate many of the proteins in Mono Q fractions 14 to 22, with labeling activity highest in fractions 17 to 20, closely corresponding to the peak holoenzyme fractions. A test of the nucleotide specificities of the kinase(s) revealed that labeling with ATP could be competed with excess unlabeled GTP but not CTP (Fig. 8B, compare lanes 2 to 5 to lane 1), consistent with the known properties of CKII. Labeling of some, though not all, protein bands in peak holoenzyme fractions was inhibited by heparin, a known inhibitor of CKII (20) (Fig. 8B, compare lanes 7 and 8 to lane 6). Furthermore, a peptide containing a consensus CKII phosphorylation site (15) was a competitor for the labeling of most proteins phosphorylated by the endogenous kinase within the peak Mono Q fraction (Fig. 8B, lane 11). This same peptide could be phosphorylated when mixed with the peak holoenzyme fraction and supplied with either [γ-32P]ATP or [γ-32P]GTP as the phosphate donor (Fig. 8C). Confirmation that CKII is present in Mono Q peak holoenzyme fractions was obtained by Western blotting with a polyclonal antiserum raised against Drosophila CKII (generously provided by Neil Osheroff) and a commercial antiserum raised against human α and α′ CKII subunits, both of which cross-react with the appropriate subunits of Xenopus CKII. With these antibodies, CKII was detected in Mono Q fractions 17 to 20, corresponding closely with the Pol I holoenzyme peak (Fig. 9A). CKII was also present in the peak DNA-cellulose fraction (Fig. 9B).
FIG. 8.
Protein kinase activity coelutes with Pol I holoenzyme activity. (A) Aliquots (4 μl) of Mono Q fractions 14 to 22 (same column run as that shown in Fig. 4) were incubated for 30 min in a buffer containing MgCl2 and [γ-32P]ATP and then subjected to SDS-PAGE (8% polyacrylamide, Tris-Tricine buffer) and autoradiography. Positions of molecular mass markers (in kilodaltons) are shown on the right. The lanes of the SDS-PAGE gel are aligned with the transcription reactions to highlight the correspondence between the kinase and transcriptionally active fractions. (B) Biochemical characterization of holoenzyme-associated kinase activity. Kinase activity of Mono Q fraction 20 was tested in the presence of various competitors or inhibitors, which were added to the reactions prior to the addition of [γ-32P]ATP. Reaction mixtures subjected to electrophoresis in lanes 1, 6, and 9 were controls to which no competitors were added. Nonradioactive GTP or CTP was added in a 30- or 300-fold excess to the reaction mixtures subjected to electrophoresis in lanes 2 to 5. Heparin was added in two concentrations to reaction mixtures in lanes 7 and 8. In lanes 10 and 11, a synthetic peptide containing a consensus CKII phosphorylation site was added in two concentrations. Numbers at left indicate molecular mass in kilodaltons. (C) Mono Q peak fraction 20 will direct phosphorylation of a synthetic peptide (1.5 mM) containing a consensus CKII phosphorylation site with either [γ-32P]ATP (lane 3) or [γ-32P]GTP (lane 4) as the phosphate donor. Lanes 1 and 2 are controls to which no peptide was added. Reaction mixtures were subjected to electrophoresis on a 16.5% SDS–Tris-Tricine gel.
FIG. 9.
Detection of CKII in peak Pol I holoenzyme fractions by Western blotting. (A) Mono Q fractions (60 μl) were trichloroacetic acid precipitated and loaded on SDS–12% PAGE gels. Western blots were probed with an antiserum raised against Drosophila CKII which cross-reacts with the Xenopus β subunit or with an antibody raised against human CKII α and α′ subunits. In the far left lane of each gel, 2 μl of human nuclear extract was run as a control. Antigen-antibody complexes were detected by enhanced chemiluminescence on X-ray film (CKII α subunits) or by colorimetric reaction (β subunit). The Western blots are aligned with the gel showing the transcription reaction products to allow easy comparison. (B) Relative abundance of CKII in peak fractions throughout the purification scheme. A sample of rat nuclear extract was run as a positive control in the rightmost lane.
Unlike Pol II transcription initiation (9, 65), Pol I initiation does not require an ATPase activity to promote open-complex formation (30, 37). Therefore, one can substitute AMP-PNP and GMP-PNP for ATP and GTP, respectively, as substrates for Pol I transcription. These nucleotide analogs are not functional as phosphate donors for protein kinases; thus, we used them in place of ATP and GTP to determine if transcription would be reduced by inhibiting CKII and/or other associated kinases. We were disappointed to find that neither AMP-PNP nor GMP-PNP, alone or in combination, had a significant effect on the holoenzyme’s transcriptional activity (data not shown). Thus, the functional significance (if any) of the holoenzyme-associated protein kinase activity is unknown.
Holoenzyme fractions contain HAT activity.
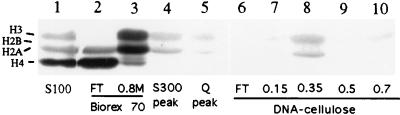
Our studies of uniparental rRNA gene silencing in interspecies hybrids (nucleolar dominance) have suggested that rRNA gene activity can be controlled via histone acetylation and associated chromatin modifications (11). We tested for HAT activity by assaying the ability of Pol I holoenzyme fractions to catalyze the labeling of purified histones with [3H]acetyl-CoA (Fig. 10). In starting S100 whole-cell extracts, HAT activity was readily detected, with histone H4 being the predominant substrate (lane 1). Different HAT activities were fractionated at the Biorex purification step; the flowthrough was enriched in H4 HAT activity (lane 2), whereas the 0.8 M KCl step was enriched for an activity with a substrate preference for histones H3 and H2A (lane 3). The latter HAT activity copurified with the holoenzyme peak on Sephacryl S300 (lane 4), Mono Q (lane 5), and DNA-cellulose (lane 8). These data indicate that HAT activity is closely associated with, or intrinsic to, the Pol I holoenzyme. Future studies using chromatin-assembled minigenes will be needed to test whether the associated HAT activity assists in the transcription of these templates.
FIG. 10.
Detection of HAT activity in X. laevis S100 extract and purified holoenzyme fractions. Core histones (10 μg) were incubated in reaction mixtures containing [3H]acetyl-CoA and 5 μl of the following: S100 extract (lane 1), Biorex flowthrough (FT) (lane 2), the Biorex 0.8 M holoenzyme-containing fraction (lane 3), the Sephacryl S300 Pol I holoenzyme peak (lane 4), the Mono Q holoenzyme peak (lane 5), or all five fractions from the DNA-cellulose column (lanes 6 to 10), including the holoenzyme peak (lane 8). Proteins were then subjected to SDS–15% PAGE and fluorography to detect labeled proteins. Coomassie blue staining allowed the positions of the different core histones to be determined.
DISCUSSION
Prior studies have shown that the Xenopus Pol I transcription system can be split by heparin chromatography into multiple transcription factors that must be added back together to reconstitute transcription (39). However, in our scheme, which purposely omits the heparin column, all activities essential for accurate, promoter-dependent transcription initiation copurify with Pol I on at least five columns. The order of the columns does not appear to be critical. For instance, the Biorex column can precede or follow the Mono Q column. Likewise, the purification scheme can be modified such that the Sephacryl gel filtration column can be omitted, instead being replaced by glycerol gradient sedimentation following DEAE, Biorex, and Mono Q chromatography (14a).
The degree to which we have purified the holoenzyme is frustratingly difficult to estimate based on conventional biochemical criteria. As we have discussed in detail previously (53), the major problem is the extreme lability of Pol I activity. Only a portion of the total Pol I activity loaded on each column can be recovered and accounted for among the subsequent fractions, and mixing fractions does not reconstitute lost activity. Therefore, specific activity is not a useful measure of purity because the continual loss of activity leads to large and compounded errors. On a protein mass basis, final DNA-cellulose peak fractions contain approximately 4,000-fold less protein than the starting amount of S100 protein.
The possibility that the Pol I transcription machinery copurifies by virtue of being bound to DNA seems unlikely. First, purification of the functional holoenzyme was unaffected by treatment of starting S100 extracts with DNase. Second, though the crude extract and DE350 fractions contain considerable amounts of nucleic acid (both RNA and DNA), virtually all contaminating nucleic acid flows through the Biorex column based on ethidium bromide staining and fluorometry (following Hoechst dye binding) and none can be detected following Mono Q chromatography. The fact that the holoenzyme binds to DNA-cellulose and elutes at moderate salt suggests that DNA-binding sites within the complex are unoccupied. One could argue that the interaction with DNA-cellulose is simply an ionic interaction rather than a DNA affinity interaction. However, we have evidence that the Pol I holoenzyme binds promoter DNA in vitro in a gel mobility shift assay and thus must be free of associated DNA or must be able to readily exchange onto a promoter probe, apparently as an intact complex (unpublished data). Probably the strongest argument is that the 800 mM KCl elution from the Biorex column is loaded without dialysis onto the Sephacryl gel filtration column; ionic interactions between the holoenzyme complex and DNA should be disrupted at such high salt concentrations and are unlikely to occur again during chromatography.
In yeast, in vitro transcription studies have suggested that neither TBP nor upstream activation factor is essential for basal-level Pol I transcription from the core promoter. However, TBP is required for full promoter activity in vitro, apparently by facilitating an interaction between upstream activation factor and core factor (62). TBP is also required for yeast Pol I transcription in vivo (14, 55). In vertebrates and Acanthamoeba, all studies to date have suggested that TBP is essential for Pol I transcription (44, 48); thus, one would predict that a Pol I holoenzyme capable of promoter-dependent transcription should include TBP. Indeed, TBP precisely cofractionates with Xenopus holoenzyme fractions that support transcription from the rRNA gene promoter, supporting this prediction. It is interesting that TBP should be so stably associated with the holoenzyme, even at salt concentrations as high as 0.8 M KCl, whereas it can apparently dissociate readily from the Xenopus Rib-1–SL1 complex unless stabilized by UBF (7). We speculate that protein-protein interactions within the putative holoenzyme complex prevent dissociation of TBP (and presumably all of Rib-1), perhaps in a manner analogous to the way in which UBF can stabilize TBP within the Rib1 complex (7).
The role of UBF in Pol I transcription is controversial. Due to its abundance and ability to bend and wrap naked DNA, we and others have proposed that UBF is likely to serve a structural role, helping to organize rRNA genes into a transcriptionally competent format (2, 49, 52). UBF’s ability to counteract transcriptional repression caused by histone H1 or Ku protein on naked DNA templates (31, 32) and its ability to displace linker histone from fully assembled nucleosomes (26) are consistent with a defining role in chromatin structure. However, there is disagreement concerning the need for UBF in the activation of Pol I transcription. In mouse and rat cells, UBF has been shown to be stimulatory but not essential for basal-level transcription. In contrast, available evidence has suggested that UBF is required for transcription from the Xenopus and human promoters (3, 4, 7, 13, 39). One possibility is that the requirement for UBF depends on the purity of the system, with crude fractions requiring UBF as an antirepressor. Experiments by Kuhn and Grummt support the latter interpretation (31).
Based on the current study, we suggest that UBF is nonessential for basal-level transcription in vitro from the Xenopus promoter, in agreement with the conclusions reached by using rodent systems. In apparent contradiction to our demonstration that UBF and the Xenopus holoenzyme do not precisely copurify, immunoprecipitation studies have suggested a UBF-Pol I holoenzyme association in mouse cell extracts (57). A possibility is that UBF-holoenzyme interactions occur but are transient, such that a small fraction of total UBF can be immunoprecipitated with the holoenzyme in crude fractions. Given the vast excess of UBF over holoenzyme in the cell, transient UBF-holoenzyme interactions would seem to be necessary to prevent the holoenzyme from being sequestered at nonpromoter sites bound by UBF. Importantly, our finding that UBF does not copurify with the holoenzyme suggests that UBF is not integral to the complex but does not rule out its ability to interact with the holoenzyme.
Another difference between the Xenopus and mouse holoenzymes is that the Xenopus complex is self-sufficient for promoter-dependent transcription, whereas the mouse holoenzyme requires supplemental TIF-IC (57). In agreement with the results reported here, highly purified Pol I holoenzyme fractions from the plant B. oleracea are also self-sufficient for transcription (53). One possibility is that TIF-IC is easily displaced from the mouse polymerase but is tightly associated with polymerase in other species. It is noteworthy that similar controversies exist concerning the composition of RNA Pol II holoenzymes in vertebrates and yeast (18). For instance, some groups have purified the Pol II holoenzyme in a form containing all essential transcription factors and self-sufficient for promoter-dependent transcription (46, 47) or in a form associated with the chromatin-remodeling complex SWI/SNF (71). Other groups have isolated the Pol II holoenzyme as a complex missing essential transcription factors or SWI/SNF (18, 28, 33).
CKII is a ubiquitous kinase known to be important for cell cycle control and signaling pathways regulating cell proliferation (1a, 35, 42). CKII has been implicated in the control of rRNA gene expression for some time (21), with good evidence that it is involved in the phosphorylation of the UBF acidic tail, the C terminus rich in serine and acidic amino acids (45, 67, 68). Our finding that CKII is associated with the Pol I holoenzyme suggests that UBF-holoenzyme interactions might result in UBF phosphorylation. The DNA-binding activity of UBF does not appear to be affected by its growth-dependent phosphorylation state (68), suggesting that phosphorylation affects another, currently undefined, step. We suspect that a role for CKII may become apparent under UBF-dependent transcription conditions. CKII was recently shown to be associated with immunoprecipitated rat RNA Pol I (19); therefore, it seems likely that CKII is associated with mammalian Pol I holoenzymes as it is in Xenopus. The significance of this association remains to be demonstrated.
Several well-known Pol II coactivator and transcription factor complexes have HAT activity, including the GCN5-containing SAGA complex (16, 69) and TFIID (43). In addition, ATP-dependent chromatin-remodeling activities such as SWI/SNF have been found in association with Pol II holoenzymes (71). These results suggest that assembly of transcription preinitiation complexes involves activators, general transcription factors, and holoenzymes that not only bind to DNA and one another but also reposition nucleosomes and hyperacetylate their histones in the vicinity of gene promoters (17, 23, 63). Thus, gene activation increasingly appears to be dependent on chromatin modifications that facilitate gene derepression. Association of HAT activity with the Pol I holoenzyme is consistent with a genome-wide role for chromatin modifications in gene regulation and is consistent with our observation that histone hyperacetylation is correlated with rRNA gene activation in the epigenetic phenomenon of nucleolar dominance (11).
ACKNOWLEDGMENTS
We thank Liang Annie Shen for excellent technical assistance, Jerry Workman (Pennsylvania State University) for the gift of purified HeLa histones, Neil Osheroff (Vanderbilt University) for the gift of antibodies against Drosophila CKII, and Paul Labhart (Scripps Research Institute) and Brian McStay (University of Dundee) for gifts of antibodies against Xenopus TBP. We are grateful to Larry Rothblum (Geisinger Clinic) for helpful discussions and for sharing unpublished results and materials.
This work was supported by NIH grant 5-RO1-GM50910 to C.S.P. Annie-Claude Albert was supported, in part, by a W. M. Keck Fellowship awarded through the Washington University School of Medicine.
REFERENCES
- 1.Albert, A.-C. Unpublished data.
- 1a.Allende J E, Allende C C. Protein kinases. 4. Protein kinase CK2: an enzyme with multiple substrates and a puzzling regulation. FASEB J. 1995;9:313–323. doi: 10.1096/fasebj.9.5.7896000. [DOI] [PubMed] [Google Scholar]
- 2.Bazett-Jones D, Leblanc B, Herfort M, Moss T. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science. 1994;264:1134–1137. doi: 10.1126/science.8178172. [DOI] [PubMed] [Google Scholar]
- 3.Bell S P, Jantzen H M, Tjian R. Assembly of alternative multiprotein complexes directs rRNA promoter selectivity. Genes Dev. 1990;4:943–954. doi: 10.1101/gad.4.6.943. [DOI] [PubMed] [Google Scholar]
- 4.Bell S P, Learned R M, Jantzen H M, Tjian R. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science. 1988;241:1192–1197. doi: 10.1126/science.3413483. [DOI] [PubMed] [Google Scholar]
- 5.Bell S P, Pikaard C S, Reeder R H, Tjian R. Molecular mechanisms governing species-specific transcription of ribosomal RNA. Cell. 1989;59:489–497. doi: 10.1016/0092-8674(89)90032-9. [DOI] [PubMed] [Google Scholar]
- 6.Berk A J, Sharp P A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977;12:721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- 7.Bodeker M, Cairns C, McStay B. Upstream binding factor stabilizes Rib 1, the TATA-binding-protein-containing Xenopus laevis RNA polymerase I transcription factor, by multiple protein interactions in a DNA-independent manner. Mol Cell Biol. 1996;16:5572–5578. doi: 10.1128/mcb.16.10.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brownell J E, Allis C D. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bunick D, Zandomeni R, Ackerman S, Weinmann R. Mechanism of RNA polymerase II-specific initiation of transcription in vitro: ATP requirement and uncapped runoff transcripts. Cell. 1982;29:877–886. doi: 10.1016/0092-8674(82)90449-4. [DOI] [PubMed] [Google Scholar]
- 10.Buratowski S. The basics of basal transcription by RNA polymerase II. Cell. 1994;77:1–3. doi: 10.1016/0092-8674(94)90226-7. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z J, Pikaard C S. Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 1997;11:2124–2136. doi: 10.1101/gad.11.16.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clos J, Buttgereit D, Grummt I. A purified transcription factor (TIF-IB) binds to essential sequences of the mouse rDNA promoter. Proc Natl Acad Sci USA. 1986;83:604–608. doi: 10.1073/pnas.83.3.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comai L, Zomerdijk J C B M, Beckmann H, Zhou S, Admon A, Tjian R. Reconstitution of transcription factor SL1: exclusive binding of TBP by SL1 or TFIID subunits. Science. 1994;266:1966–1972. doi: 10.1126/science.7801123. [DOI] [PubMed] [Google Scholar]
- 14.Cormack B P, Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- 14a.Denton, M., and C. S. Pikaard. Unpublished data.
- 15.Gatica M, Jedlicki A, Allende C C, Allende J E. Activity of the E75E76 mutant of the alpha subunit of casein kinase II from Xenopus laevis. FEBS Lett. 1994;339:93–96. doi: 10.1016/0014-5793(94)80392-7. [DOI] [PubMed] [Google Scholar]
- 16.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 17.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 18.Halle J-P, Meisterernst M. Gene expression: increasing evidence for a transcriptosome. Trends Genet. 1996;12:161–163. doi: 10.1016/0168-9525(96)30035-8. [DOI] [PubMed] [Google Scholar]
- 19.Hannan R D, Hempel W M, Cavanaugh A, Arino T, Dimitrov S I, Moss T, Rothblum L I. Affinity purification of mammalian RNA polymerase I. J Biol Chem. 1998;273:1257–1267. doi: 10.1074/jbc.273.2.1257. [DOI] [PubMed] [Google Scholar]
- 20.Hathaway G M, Lubben T H, Traugh J A. Inhibition of casein kinase II by heparin. J Biol Chem. 1980;255:8038–8041. [PubMed] [Google Scholar]
- 21.Jacob S T. Regulation of ribosomal gene transcription. Biochem J. 1995;306:617–626. doi: 10.1042/bj3060617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jantzen H-M, Chow A M, King D S, Tjian R. Multiple domains of the RNA polymerase I activator hUBF interact with the TATA-binding protein complex hSL1 to mediate transcription. Genes Dev. 1992;6:1950–1963. doi: 10.1101/gad.6.10.1950. [DOI] [PubMed] [Google Scholar]
- 23.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 24.Kassavetis G A, Bardeleben C, Bartholomew B, Braun B R, Joazeiro C A P, Pisano M, Geiduschek E P. Transcription by RNA polymerase III. In: Conaway R C, Conaway J W, editors. Transcription mechanisms and regulation. New York, N.Y: Raven Press; 1994. pp. 107–126. [Google Scholar]
- 25.Kato H, Nagamine M, Kominami R, Muramatsu M. Formation of the transcription initiation complex on mammalian rDNA. Mol Cell Biol. 1986;6:3418–3427. doi: 10.1128/mcb.6.10.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kermekchiev M, Workman J L, Pikaard C S. Nucleosome binding by the polymerase I transactivator upstream binding factor displaces linker histone H1. Mol Cell Biol. 1997;17:5833–5842. doi: 10.1128/mcb.17.10.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim Y J, Björklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 28.Koleske A J, Young R A. The RNA polymerase II holoenzyme and its implications for gene regulation. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 29.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 30.Kuhn A, Gottlieb T M, Jackson S P, Grummt I. DNA-dependent protein kinase: a potent inhibitor of transcription by RNA polymerase I. Genes Dev. 1995;9:193–203. doi: 10.1101/gad.9.2.193. [DOI] [PubMed] [Google Scholar]
- 31.Kuhn A, Grummt I. Dual role of the nucleolar transcription factor UBF: trans-activator and antirepressor. Proc Natl Acad Sci USA. 1992;89:7340–7344. doi: 10.1073/pnas.89.16.7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhn A, Stefanovsky V, Grummt I. The nucleolar transcription activator UBF relieves Ku antigen-mediated repression of mouse ribosomal gene transcription. Nucleic Acids Res. 1993;21:2057–2063. doi: 10.1093/nar/21.9.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Bjorklund S, Kim Y J, Kornberg R D. Yeast RNA polymerase II holoenzyme. Methods Enzymol. 1996;273:172–175. doi: 10.1016/s0076-6879(96)73017-3. [DOI] [PubMed] [Google Scholar]
- 34.Liao S M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, vanVuuren H J, Young R A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 35.Litchfield D W, Luscher B. Casein kinase II in signal transduction and cell cycle regulation. Mol Cell Biochem. 1993;127/128:187–199. doi: 10.1007/BF01076770. [DOI] [PubMed] [Google Scholar]
- 36.Lobo S M, Hernandez N T. Transcription of snRNA genes by RNA polymerases II and III. In: Conaway R C, Conaway J W, editors. Transcription mechanisms and regulation. New York, N.Y: Raven Press; 1994. pp. 127–160. [Google Scholar]
- 37.Lofquist A K, Li H, Imboden M A, Paule M R. Promoter opening (melting) and transcription initiation by RNA polymerase I requires neither nucleotide beta,gamma hydrolysis nor protein phosphorylation. Nucleic Acids Res. 1993;21:3233–3238. doi: 10.1093/nar/21.14.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 39.McStay B, Hu C H, Pikaard C S, Reeder R H. xUBF and Rib 1 are both required for formation of a stable polymerase I promoter complex in X. laevis. EMBO J. 1991;10:2297–2303. doi: 10.1002/j.1460-2075.1991.tb07766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McStay B, Reeder R H. A DNA-binding protein is required for termination of transcription by RNA polymerase I in Xenopus laevis. Mol Cell Biol. 1990;10:2793–2800. doi: 10.1128/mcb.10.6.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McStay B, Reeder R H. A termination site for Xenopus RNA polymerase I also acts as an element of an adjacent promoter. Cell. 1986;47:913–920. doi: 10.1016/0092-8674(86)90806-8. [DOI] [PubMed] [Google Scholar]
- 42.Meisner H, Czech M P. Phosphorylation of transcriptional factors and cell-cycle-dependent proteins by casein kinase II. Curr Opin Cell Biol. 1991;3:474–483. doi: 10.1016/0955-0674(91)90076-b. [DOI] [PubMed] [Google Scholar]
- 43.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 44.Moss T, Stefanovsky V Y. Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase I. Prog Nucleic Acids Res Mol Biol. 1995;50:25–66. doi: 10.1016/s0079-6603(08)60810-7. [DOI] [PubMed] [Google Scholar]
- 45.O’Mahony D J, Smith S D, Xie W, Rothblum L I. Analysis of the phosphorylation, DNA-binding and dimerization properties of the RNA polymerase I transcription factors UBF1 and UBF2. Nucleic Acids Res. 1992;20:1301–1308. doi: 10.1093/nar/20.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ossipow V, Tassan J P, Nigg E A, Schibler U. A mammalian RNA polymerase II holoenzyme containing all components required for promoter-specific transcription initiation. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 47.Pan G, Aso T, Greenblatt J. Interaction of elongation factors TFIIS and elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J Biol Chem. 1997;272:24563–24571. doi: 10.1074/jbc.272.39.24563. [DOI] [PubMed] [Google Scholar]
- 48.Paule M R. Transcription of ribosomal RNA by eukaryotic RNA polymerase I. In: Conaway R C, Conaway J W, editors. Transcription mechanisms and regulation. New York, N.Y: Raven Press; 1994. pp. 83–106. [Google Scholar]
- 49.Putnam C D, Copenhaver G P, Denton M L, Pikaard C S. The RNA polymerase I transactivator upstream binding factor requires its dimerization domain and high-mobility group (HMG) box 1 to bend, wrap, and positively supercoil enhancer DNA. Mol Cell Biol. 1994;14:6476–6488. doi: 10.1128/mcb.14.10.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reeder R H, editor. Regulation of transcription by RNA polymerase I. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 51.Reeder R H, Pennock D, McStay B, Roan J, Tolentino E, Walker P. Linker scanner mutagenesis of the Xenopus laevis ribosomal gene promoter. Nucleic Acids Res. 1987;15:7429–7441. doi: 10.1093/nar/15.18.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reeder R H, Pikaard C S, McStay B. UBF, an architectural element for RNA polymerase I promoters. Nucleic Acids Mol Biol. 1995;9:251–263. [Google Scholar]
- 53.Saez-Vasquez J, Pikaard C S. Extensive purification of a putative RNA polymerase I holoenzyme from plants that accurately initiates rRNA gene transcription in vitro. Proc Natl Acad Sci USA. 1997;94:11869–11874. doi: 10.1073/pnas.94.22.11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schnapp A, Grummt I. Transcription complex formation at the mouse rDNA promoter involves the stepwise association of four transcription factors and RNA polymerase I. J Biol Chem. 1991;266:24588–24595. [PubMed] [Google Scholar]
- 55.Schultz M C, Reeder R H, Hahn S. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II, and III promoters. Cell. 1992;69:697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- 56.Schwartz L B, Roeder R G. Purification and subunit structure of deoxyribonucleic acid-dependent ribonucleic acid polymerase I from mouse myeloma, MOPC 315. J Biol Chem. 1974;249:5898–5906. [PubMed] [Google Scholar]
- 57.Seither P, Grummt I. Molecular cloning of RPA2, the gene encoding the second largest subunit of mouse RNA polymerase I. Genomics. 1996;37:135–139. doi: 10.1006/geno.1996.0531. [DOI] [PubMed] [Google Scholar]
- 58.Seither P, Iben S, Grummt I. Mammalian RNA polymerase I exists as a holoenzyme with associated basal transcription factors. J Mol Biol. 1998;275:43–53. doi: 10.1006/jmbi.1997.1434. [DOI] [PubMed] [Google Scholar]
- 59.Serizawa H, Conaway J W, Conaway R C. Transcription initiation by mammalian RNA polymerase II. In: Conaway R C, Conaway J W, editors. Transcription mechanisms and regulation. New York, N.Y: Raven Press; 1994. pp. 27–44. [Google Scholar]
- 60.Smith S D, O’Mahony D J, Kinsella B T, Rothblum L I. Transcription from the rat 45S ribosomal DNA promoter does not require the factor UBF. Gene Expr. 1993;3:229–236. [PMC free article] [PubMed] [Google Scholar]
- 61.Smith S D, Oriahi E, Lowe D, Yang Y, O’Mahony D, Rose K, Chen K, Rothblum L I. Characterization of factors that direct transcription of rat ribosomal DNA. Mol Cell Biol. 1990;10:3105–3116. doi: 10.1128/mcb.10.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steffan J S, Keys D A, Dodd J A, Nomura M. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev. 1996;10:2551–2563. doi: 10.1101/gad.10.20.2551. [DOI] [PubMed] [Google Scholar]
- 63.Steger D J, Workman J L. Remodeling chromatin structures for transcription: what happens to the histones? Bioessays. 1996;18:875–884. doi: 10.1002/bies.950181106. [DOI] [PubMed] [Google Scholar]
- 64.Tanaka N, Kato H, Ishikawa Y, Hisatake K, Tashiro K, Kominami R, Muramatsu M. Sequence-specific binding of a transcription factor TFID to the promoter region of mouse ribosomal RNA gene. J Biol Chem. 1990;265:13836–13842. [PubMed] [Google Scholar]
- 65.Tantin D, Carey M. A heteroduplex template circumvents the energetic requirement for ATP during activated transcription by RNA polymerase II. J Biol Chem. 1994;269:17397–17400. [PubMed] [Google Scholar]
- 66.Tower J, Culotta V C, Sollner-Webb B. Factors and nucleotide sequences that direct ribosomal DNA transcription and their relationship to the stable transcription complex. Mol Cell Biol. 1986;6:3451–3462. doi: 10.1128/mcb.6.10.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Voit R, Kuhn A, Sander E E, Grummt I. Activation of mammalian ribosomal gene transcription requires phosphorylation of the nucleolar transcription factor UBF. Nucleic Acids Res. 1995;23:2593–2599. doi: 10.1093/nar/23.14.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Voit R, Schnapp A, Kuhn A, Rosenbauer H, Hirschmann P, Stunnenberg H G, Grummt I. The nucleolar transcription factor mUBF is phosphorylated by casein kinase II in the C-terminal hyperacidic tail which is essential for transactivation. EMBO J. 1992;11:2211–2218. doi: 10.1002/j.1460-2075.1992.tb05280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wade P A, Wolffe A P. Histone acetyltransferases in control. Curr Biol. 1997;7:82–84. doi: 10.1016/s0960-9822(06)00042-x. [DOI] [PubMed] [Google Scholar]
- 70.Wang Z, Luo T, Roeder R G. Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactivated during protein synthesis inhibition. Genes Dev. 1997;11:2371–2382. doi: 10.1101/gad.11.18.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 72.Wingender E, Jahn D, Seifart K H. Association of RNA polymerase III with transcription factors in the absence of DNA. J Biol Chem. 1986;261:1409–1413. [PubMed] [Google Scholar]
- 73.Zawel L, Reinberg D. Initiation of transcription by RNA polymerase II: a multi-step process. Prog Nucleic Acids Res Mol Biol. 1993;44:67–108. doi: 10.1016/s0079-6603(08)60217-2. [DOI] [PubMed] [Google Scholar]