Abstract
Iron regulatory protein 1 (IRP-1) binding to an iron-responsive element (IRE) located close to the cap structure of mRNAs represses translation by precluding the recruitment of the small ribosomal subunit to these mRNAs. This mechanism is position dependent; reporter mRNAs bearing IREs located further downstream exhibit diminished translational control in transfected mammalian cells. To investigate the underlying mechanism, we have recapitulated this position effect in a rabbit reticulocyte cell-free translation system. We show that the recruitment of the 43S preinitiation complex to the mRNA is unaffected when IRP-1 is bound to a cap-distal IRE. Following 43S complex recruitment, the translation initiation apparatus appears to stall, before linearly progressing to the initiation codon. The slow passive dissociation rate of IRP-1 from the cap-distal IRE suggests that the mammalian translation apparatus plays an active role in overcoming the cap-distal IRE–IRP-1 complex. In contrast, cap-distal IRE–IRP-1 complexes efficiently repress translation in wheat germ and yeast translation extracts. Since inhibition occurs subsequent to 43S complex recruitment, an efficient arrest of productive scanning may represent a second mechanism by which RNA-protein interactions within the 5′ untranslated region of an mRNA can regulate translation. In contrast to initiating ribosomes, elongating ribosomes from mammal, plant, and yeast cells are unaffected by IRE–IRP-1 complexes positioned within the open reading frame. These data shed light on a characteristic aspect of the IRE-IRP regulatory system and uncover properties of the initiation and elongation translation apparatus of eukaryotic cells.
The regulation of iron metabolism by the iron-responsive element (IRE)-iron regulatory protein (IRP) system represents an intensively studied example of translational control in higher eukaryotes. Several mRNAs encoding proteins involved in cellular iron metabolism harbor an IRE at a cap-proximal position of their 5′ untranslated regions (UTRs). The IRE is specifically recognized by IRP-1 and IRP-2, which bind to and repress the translation of IRE-containing mRNAs both in vivo and in vitro (17, 39). Translational control by specific mRNA-protein interactions is commonly enacted at the level of translation initiation (e.g., caudal [6, 38], 15-lipoxygenase [35, 36], and oskar [22, 43]). IRP binding to the IRE of ferritin mRNAs affects an early step of translation initiation: it prevents the recruitment of the 43S translation preinitiation complex (which includes the small ribosomal subunit) (13, 33). Transfection studies using mammalian tissue culture cells revealed a characteristic feature of this IRE-IRP regulatory mechanism: for IRP binding to efficiently block translation, the IRE must be located within <60 nucleotides from the m7GpppN-cap structure of the mRNA (9, 10). An IRE placed >60 nucleotides downstream from the cap structure mediates only partial translational inhibition by IRP binding. In keeping with this position effect, the cap-proximal location of mammalian IREs is phylogenetically conserved (10). The manifestation of the position effect displays cell-type-dependent quantitative differences (10), suggesting that the ability of the translation apparatus to overcome cap-distal IRE-IRP complexes can be affected by cellular determinants.
To investigate the mechanistic basis of the position effect, we have employed a rabbit reticulocyte cell-free translation system. Recapitulation of the position effect in this system provided the basis for biochemical analyses aimed at understanding the role of the mammalian translation apparatus in overcoming cap-distal IRE-IRP complexes. We show that sufficiently cap-distal IRE-IRP complexes permit the recruitment of the 43S complex, but temporarily slow its further progression. We demonstrate that initiating and elongating mammalian ribosomes progress through cap-distal IREs in a linear fashion in both the presence and absence of IRP-1 and suggest that the mammalian translation apparatus plays an active role in the displacement of such IRE-IRP complexes. Finally, we provide evidence that cap-distal IRE-IRP complexes can efficiently regulate translation in wheat germ extract (WGE) by arresting productive scanning.
MATERIALS AND METHODS
Plasmid construction.
Plasmid NOP1 is derived from pBluescript II SK +/−, and chloramphenicol acetyltransferase (CAT) plasmids were derived from pGEM-3Zf(−) and are cloned for transcription from the T7 RNA polymerase promoter. Plasmids NOP1 and IRE-mut have been previously described (11, 41). IRE.34 was created by insertion of a BamHI-XbaI fragment containing the IRE from F64 (10) into IRE-wt (11, 13) digested with BamHI-XbaI. IRE.66 was created from IRE-wt by digestion with BamHI and filling of the BamHI cohesive ends by using Klenow fragment, prior to digestion with XbaI and insertion of a StuI fragment from F64 containing the IRE and sequences 5′ of the IRE. IRE.100 was created by ligation of annealed phosphorylated oligodeoxyribonucleotides into IRE.66 after digestion with XhoI. The sequence of the sense-strand oligonucleotide is 5′-TCGAGATTTAACCTCTTCCA ACCCAAAGGC CTCT-3′. IRE.ORF was created from plasmid IRE-mut by introduction of annealed phosphorylated oligodeoxyribonucleotides containing the IRE sequence at position +135 (+1 is the T7 RNA polymerase transcription start site) by overlap extension by the PCR method described by Ho et al. (19). The sequence of the sense-strand oligonucleotide is 5′-GGATCCTGCT TCAACAGTGCTTGGACGGAT CTT-3′. IRE/AUG.34, IRE/AUG.66, and IRE/AUG.100 are identical to plasmids IRE.34, IRE.66, and IRE.100, respectively, except for the sequence of the IRE stem-loop and flanking nucleotides, which is 5′-GATCCATCGT TGCTTCAACA GTGCTTGGAC ACGATGGATC-3′ and which introduces five additional nucleotides between the cap structure and the IRE (Fig. 1A and C).
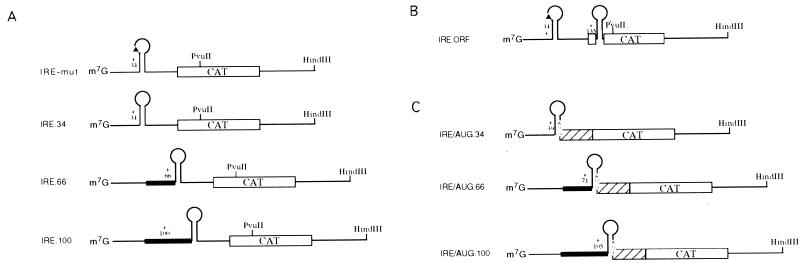
FIG. 1.
Reporter mRNAs used in this study. The open reading frame encoding CAT is denoted by an open box. The position of the IRE relative to the cap structure is indicated. The black triangle symbolizes an IRE which lacks the first nucleotide of the conserved loop. The restriction sites used to linearize the plasmids prior to transcription are indicated. (A) Diagrammatic representation of transcripts carrying an IRE at different positions in the 5′ UTR. The extension in the 5′ UTR is represented by the thick lines. The distances between the IRE and the initiator AUG are identical. (B) Insertion of an IRE into the CAT open reading frame. (C) Diagrammatic representation of transcripts carrying an AUG initiation codon within the lower part of the IRE stem. The N-terminal extension of the CAT open reading frame is represented by a striped box.
In vitro transcription.
Capped mRNAs were generated with T7 RNA polymerase from CAT plasmids after digestion with HindIII and NOP1 after BamHI digestion (16). Short 32P-labelled IRE.34, IRE.66, and IRE.ORF mRNAs (1.6 × 107 cpm/μg of RNA) were transcribed from the corresponding plasmids digested with PvuII as described above, with the exception that the final UTP concentration was reduced to 0.5 mM and 5 μM [32P]UTP was included. IRE short competitor RNA was transcribed from IRE-wt linearized with XbaI (37) and unlabelled IRE.34, IRE.66, IRE.100, IRE.ORF and IRE-mut competitor mRNAs from the corresponding plasmids digested with PvuII (12, 13). 32P-labelled IRE probe (2.9 × 107 cpm/μg of RNA) was transcribed from an XbaI-linearized plasmid (37).
Cell-free translations.
Cell-free translations were performed in the presence of [35S]methionine (10 mCi/ml). Reactions in rabbit reticulocyte lysate (RRL; 40% lysate [12 μl]) (Promega, Wis.) contained 0.7 or 2.1 mM Mg2+ (as stated in text and figure legends) and 63 mM K+. Reactions in WGE (50% extract [15 μl]) (Promega) contained 117 mM K+ and 1.9 or 2.1 mM Mg2+. Recombinant IRP-1 (16) was added to mRNAs on ice immediately prior to the addition of the translation extract. Translation reaction which did not receive recombinant protein were compensated with buffer N (150 mM KOAc, 24 mM HEPES [pH 7.6], 1.5 mM MgCl2, 5% glycerol). Short competitor transcripts were added to mRNAs on ice prior to addition of translation extract. Translation reaction mixtures were incubated at 30°C (RRL; except in Fig. 10) or 25°C (WGE) for 60 min. [35S]methionine-labelled products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and fluorography. Translation initiation assays and sucrose gradient analysis were performed as described previously (12, 13).
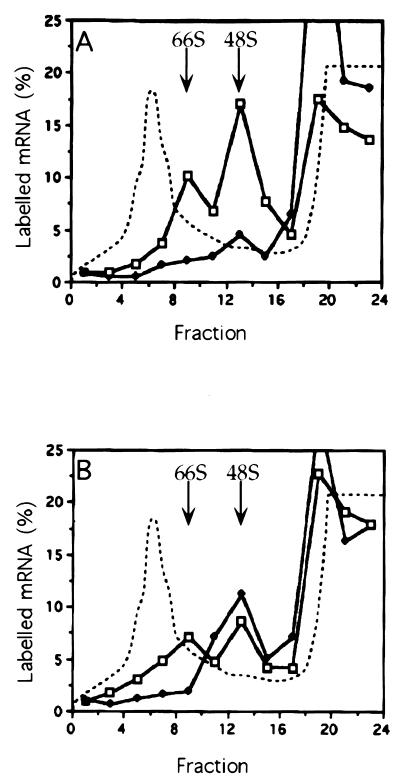
FIG. 10.
Ribosome assembly on IRE.34 and IRE.100 mRNAs in RRL (A and B) and WGE (C). Shortened mRNA transcripts of IRE.34 (A) and IRE.100 (B and C) were assayed in the absence of IRP-1 (C [open squares]) and the presence of either 45 ng of IRE competitor RNA (A and B [open squares]) or 125 ng of recombinant IRP-1 (A, B, and C [solid diamonds]). The samples were incubated for 30 min at 25°C in the presence of cycloheximide and analyzed on 5 to 25% linear sucrose gradients. The labelled mRNA in the fractions is expressed as a percentage of total counts recovered and is plotted against the fraction number. The dashed line denotes the A254 profile, which was unaltered by the addition of IRE competitor transcripts or IRP-1.
Gel retardation assays.
Protein samples were incubated with 1 ng of probe for 20 min at room temperature (unless otherwise stated), followed by a 10-min incubation with 20 U of RNase T1 (Boehringer, Mannheim, Germany) (except for the short ferritin IRE probe). Heparin (5 to 15 μg/ml) was added for an additional 10 min. Unlabelled competitor mRNAs were added together with the probe (unless otherwise stated). RNA-protein complexes were resolved on 4% nondenaturing polyacrylamide gels.
PhosphorImager quantitative analysis.
For quantification, the gels were exposed in a Storage Phosphor screen (Molecular Dynamics) that was subsequently scanned with a PhosphorImager (Molecular Dynamics). Quantification was done with the Image Quant program (Molecular Dynamics).
RESULTS
Recapitulation of the IRE-IRP position effect in a reticulocyte cell translation extract.
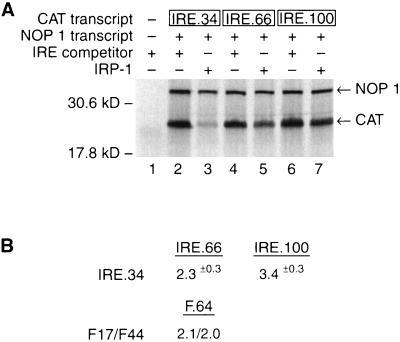
To examine the molecular basis of the IRE-IRP position effect by biochemical means, we aimed to recapitulate this effect in a cell-free translation extract. To this end, we designed the CAT reporter mRNAs IRE.34, IRE.66, and IRE.100, bearing IREs 34, 66, and 100 nucleotides, respectively, downstream from the cap structure (Fig. 1A). The sequences of the spacer inserts between the cap structure and the IRE were predicted not to form extensive secondary structure nor to disrupt the structure of the IRE itself. To examine the translation of these mRNAs in the absence of bound IRPs, short IRE competitor transcripts were added to sequester endogenous IRPs present in the extract. While all three CAT mRNAs are efficiently translated in the presence of the IRE competitor RNA (Fig. 2A, lanes 2, 4, and 6), IRE.34 mRNA is profoundly repressed by the addition of recombinant IRP-1 (lane 3). Increasing the distance between the cap structure and the IRE in IRE.66 and IRE.100 mRNAs leads to partial derepression of IRP-mediated translational control (compare lanes 5 and 7 with lane 3). Since the magnesium concentration can affect cell-free translation reactions (4, 27), the experiment was performed at low (0.7 mM [Fig. 2A]) and high (2.1 mM [data not shown]) magnesium concentrations. The position effect is clearly reflected at both magnesium concentrations. Importantly, the quantitative difference in translational repression between mRNAs bearing a cap-proximal IRE (IRE.34) and a more cap-distal IRE (IRE.66 [2.3-fold]) in vitro accurately reflects the quantitative difference seen in vivo (2.05-fold) (10) when similar reporter mRNAs are assayed (see Fig. 2 and Discussion). Further increases in the distance between the IRE and the cap structure (IRE.100) cause a further loss in translational repression by IRP binding (3.4-fold [Fig. 2]).
FIG. 2.
Translation of IRE.34, IRE.66, and IRE.100 mRNAs in RRL. (A) NOP1 (2 ng) (lanes 2 to 7) and 2.5 ng of IRE.34 (lanes 2 and 3), IRE.66 (lanes 4 and 5), or IRE.100 mRNA (lanes 6 and 7) were translated in vitro. Where indicated (+), 15 ng of IRE competitor transcript (lanes 1, 2, 4, and 6) or 150 ng of recombinant IRP-1 (lanes 3, 5, and 7) was added to the reactions. 35S-labelled translation products were analyzed by SDS-PAGE. The migration of the NOP1 and CAT translation products is indicated by arrows; molecular mass markers are shown on the left. (B) The quantative effects of moving the IRE are expressed (in fold differences) as the relative repression between IRE.34 and IRE.66 or IRE.100. Values are normalized for the NOP1 internal control. The data are derived from four experimental repetitions. The values for the position effect in cells are from a study by Goossen and Hentze (10).
The position of the IRE critically affects the recruitment of the small ribosomal subunit.
Sucrose gradient analysis of initiation reactions had previously shown that cap-proximal IRE–IRP-1 complexes prevent the recruitment of 43S preinitiation complexes to the mRNA (13). To examine the effect of a cap-distal IRE–IRP-1 complex on the association of the small ribosomal subunit with the IRE.100 mRNA, we employed sucrose gradient analysis of initiation assays containing guanylyl-imidodiphosphate (GMP-PNP). This nonhydrolyzable GTP analog permits the recruitment of the small ribosomal subunit to the mRNA and its migration toward the translation initiation codon, but inhibits the GTP-dependent subsequent joining of the large ribosomal subunit leading to the accumulation of 48S complexes (43S preinitiation complex plus mRNA) (2, 18). In the absence of available IRP (i.e., when IRE competitor transcripts are added), both mRNAs accumulate in 48S and 66S complexes (Fig. 3), indicating the association of one or two small ribosomal subunits, respectively, with the mRNAs (13). The identity of these complexes was confirmed by parallel reactions with mixtures containing either cycloheximide or cap analog and by RNase treatment of GMP-PNP-induced complexes which resulted in a single peak at 48S (reference 13 and data not shown). Upon addition of IRP-1, the recruitment of 43S preinitiation complexes to IRE.34 mRNA is prevented, and the mRNA accumulates in the top fractions of the gradient in mRNPs (Fig. 3A). In contrast, IRE.100 mRNA redistributes into the 48S region of the gradient, indicating that single 43S complexes associate with the IRE.100 mRNA in the presence of IRP-1 (compare Fig. 3B and A). This result shows that the inhibition of 43S complex recruitment by a cap-proximal IRE-IRP complex is position dependent. Interestingly, the presence of a single 43S preinitiation complex (rather than multiple complexes) on IRE.100 mRNA may indicate that the IRE–IRP-1 complex stalls scanning along the 5′ UTR, leaving insufficient room for a second 43S complex to bind.
FIG. 3.
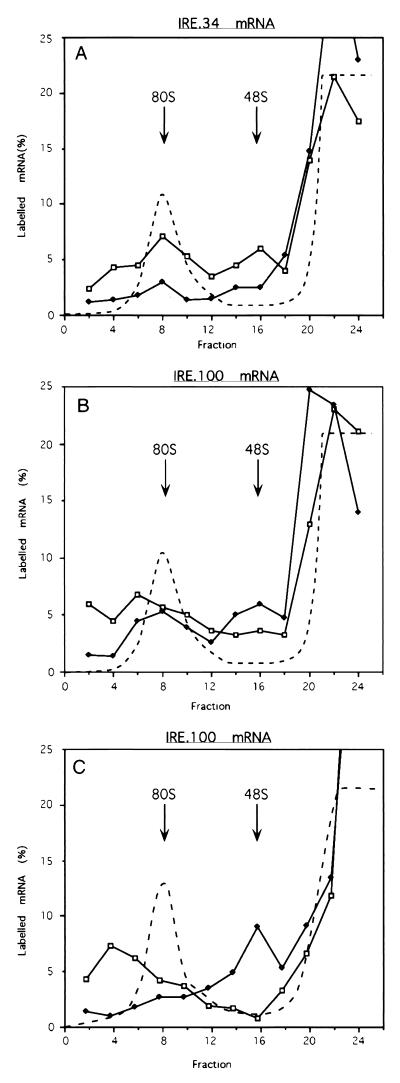
Recruitment of 43S complexes to IRE.34 and IRE.100 mRNAs in RRL. Shortened mRNA transcripts of IRE.34 (A) and IRE.100 (B) were incubated in RRL for 5 min in the presence of either 45 ng of IRE competitor RNA (open squares) or 125 ng of recombinant IRP-1 (solid diamonds) and fractionated after centrifugation through 5 to 25% linear sucrose gradients. The assays contained 0.5 mM GMP-PNP. The labelled mRNA in the fractions is expressed as a percentage of total counts recovered and is plotted against the fraction number. The dashed line denotes the A254 absorption profiles, which were identical for gradients with added IRE or IRP-1.
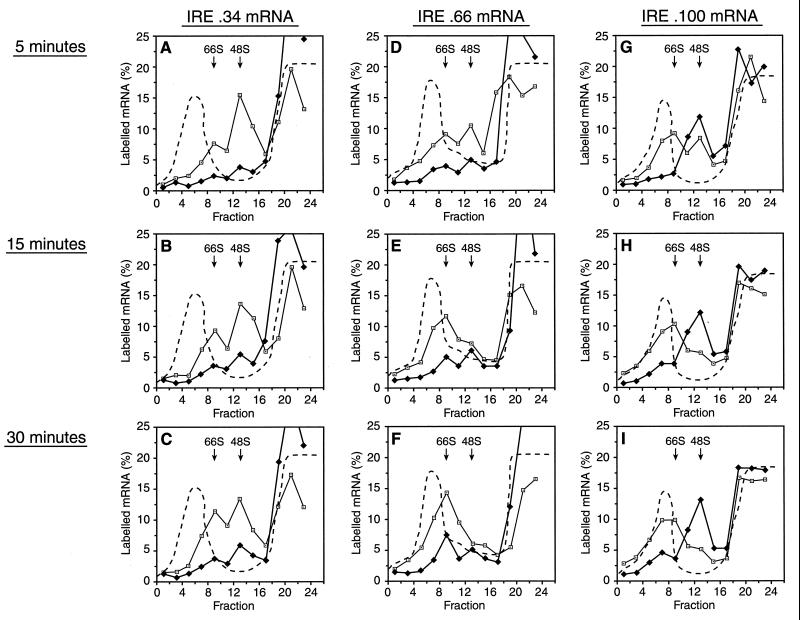
To further explore 43S complex recruitment to these mRNAs, kinetic experiments in the presence of GMP-PNP were undertaken (Fig. 4). These experiments were performed at high magnesium concentrations, where initiation proceeds more slowly (2), to increase the kinetic resolution. Analysis of IRE.34 mRNA revealed the expected large difference between ribosomal subunit recruitment in the presence and absence of IRP (Fig. 4A to C). The pattern of small ribosomal subunit recruitment in the absence of IRP does not significantly change over time, although a minor shift from 48S to 66S complexes occurs (Fig. 4A to C). Even at the longest time points, in the presence of IRP, there is not a significant accumulation of IRE.34 mRNA in 48S or 66S complexes (Fig. 4C). A greater proportion of IRE.100 mRNA is associated with 66S rather than 48S complexes compared to IRE.34 mRNA in the absence of IRP (Fig. 4G to I). This can be explained in terms of the longer 5′ UTR, which favors preloading (28). In the presence of IRP, small ribosomal subunits are already efficiently recruited to IRE.100 mRNA at 5 min (Fig. 4G), and there is not a noticeable difference in binding at 2 min (data not shown). However, even at 30 min, the majority of IRE.100 mRNAs are present in 48S rather than 66S complexes (Fig. 4I), strongly supporting the notion that the small ribosomal subunits have not migrated sufficiently far from the 5′ end of the mRNA to provide unimpeded access for subsequent subunits.
FIG. 4.
Kinetic analysis of the recruitment of small ribosomal subunits. Shortened transcripts of IRE.34 (A to C), IRE.66 (D to F), and IRE.100 (G to I) were assayed in the presence of either 45 ng of IRE competitor mRNA (open squares) or 125 ng of recombinant IRP-1 (solid diamonds) for 5 (A, D, and G), 15 (B, E, and H), or 30 (C, F, and I) min. The assays contained 0.5 mM GMP-PNP and were analyzed by using 5 to 25% linear sucrose gradients. The labelled mRNA in the fractions is expressed as a percentage of total counts recovered and is plotted against the fraction number. The dashed line denotes the A254 profiles, which were identical for gradients with added IRE or IRP-1.
The kinetics of small ribosomal subunit association with IRE.66 mRNA was also examined (Fig. 4D to F). When IRP is not available, IRE.66 mRNA is present in a higher proportion of 66S rather than 48S complexes compared to IRE.34 mRNA (Fig. 4D to F). At the shortest time point, IRE.66 mRNA association with small ribosomal subunits is strongly inhibited by IRP-1 and appears similar to IRE.34 mRNA (Fig. 4D). However, the effect of IRP-1 on small ribosomal subunit recruitment is much less profound after 30 min than for IRE.34 mRNA (Fig. 4F). It therefore seems that the recruitment of small ribosomal subunits to IRE.66 mRNA is kinetically delayed by IRP-1.
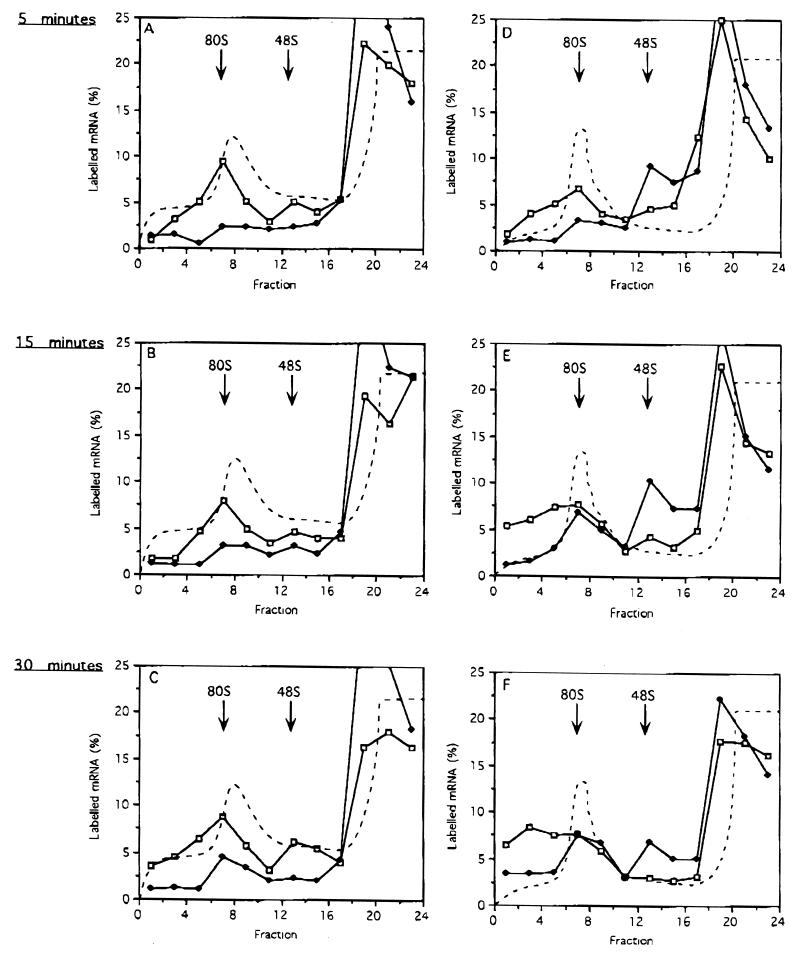
We next analyzed the kinetics of 80S ribosome formation on IRE.34 and IRE.100 mRNAs. In agreement with the previous analysis, Fig. 5D to F show that the presence of IRP-1 does not retard 48S complex formation with IRE.100 mRNA compared to in its absence. Furthermore, comparison of Fig. 5D to F with Fig. 5A to C reveals the temporary accumulation of 48S complexes on IRE.100 mRNA even in the absence of GTP analogs, indicative of stalled scanning. In contrast to analysis in the presence of GMP-PNP, in its absence, small ribosomal subunits eventually overcome the cap-distal IRE-IRP complexes, as the formation of 80S ribosomes can be observed (Fig. 5E to F). The formation of 80S complexes on IRE.100 mRNA at the later time points does not reflect an inactivation or passive dissociation of IRP-1, because IRE.34 mRNA remains repressed even at the latest time point (Fig. 5C). Compatible results were obtained at low (0.7 mM) magnesium concentrations (percent mRNA in 80S or heavier complexes in the presence of IRP: IRE.34, 6%; IRE.100, 20%). Thus the sucrose gradient analysis of IRE.100 mRNA suggests that its partial translational repression cannot be attributed to a slower recruitment of 43S complexes to the mRNA, but more likely is due to the kinetic effect of pausing scanning ribosomal subunits.
FIG. 5.
Kinetic analysis of 80S ribosome assembly on IRE.34 and IRE.100 mRNAs. Shortened transcripts of IRE.34 (A to C) and IRE.100 (D to F) were assayed in the presence of either 45 ng of IRE competitor RNA (open squares) or 125 ng of recombinant IRP-1 (solid diamonds) for 5 (A and D), 15 (B and E), or 30 (C and F) min. The assays contained 0.1 mM cycloheximide and were analyzed by using 5 to 25% linear sucrose gradients. The labelled mRNA in the fractions is expressed as a percentage of total counts recovered and is plotted against the fraction number. The dashed line denotes the A254 profiles, which were identical for gradients with added IRE or IRP-1.
In conclusion, this analysis reveals that two mechanisms exist to impede translation by RNA-protein complexes. In the first, cap-proximal RNA-protein complexes inhibit ribosomal association with the mRNA (IRE.34). In the second, cap-distal RNA-protein complexes delay productive scanning towards 80S complex formation (IRE.100).
Elongating ribosomes linearly transgress IRE–IRP-1 complexes located within the open reading frame.
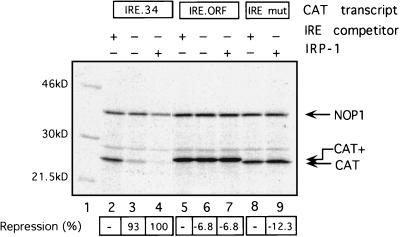
To examine if an IRE–IRP-1 complex located within the open reading frame of an mRNA can affect translation, an IRE was inserted into the CAT coding region, 32 nucleotides downstream of the A of the AUG codon, to create IRE.ORF (Fig. 1B). This insertion preserved and extended the CAT open reading frame. IRE.34 mRNA and IRE.ORF mRNA were translated in the RRL cell-free system (Fig. 6), including NOP1 mRNA as an internal control and IRE-mut mRNA as a negative control with a point-mutated IRE that cannot bind IRP-1. In the presence of competitor IRE transcripts to titrate the endogenous IRPs, all mRNAs are translated with similar efficiencies (lanes 2, 5, and 8). The translation product of ORF.CAT mRNA displays a slower mobility (labelled CAT+ [lane 5]), consistent with the insertion of the IRE sequence. When the competitor IRE is omitted (lanes 3 and 6) or when saturating quantities of recombinant IRP-1 are added (lanes 4, 7, and 9), only IRE.34 mRNA translation is repressed, while the translation of IRE.ORF mRNA is entirely unaffected and yields the extended CAT+ translation product (lanes 6 and 7). Thus, elongating ribosomes possess the capacity to linearly translate regions of the mRNA that are bound by high-affinity RNA binding proteins. The impediment imposed by such RNA-protein complexes is not sufficient to significantly impinge on the yield of translation products.
FIG. 6.
IRE–IRP-1 complexes within the open reading frame do not affect mRNA translation. NOP1 (5 ng) (lanes 2 to 9) and 2.5 ng of IRE.34 (lanes 2 to 4), IRE.ORF (lanes 5 to 7), or IRE-mut (lanes 8 and 9) were translated in RRL. Where indicated, 15 ng of IRE competitor transcript (lanes 2, 5, and 8) or 200 ng of recombinant IRP-1 (lanes 4, 7, and 9) was added to the reactions. 35S-labelled translation products were analyzed by SDS-PAGE. [35S]Met incorporation into translation products was determined by phosphoimaging, and the relative repression of CAT synthesis in the presence of recombinant IRP-1 normalized for the NOP1 internal control is shown below each lane, with repression of IRE.34 set as 100%. The migrations of the NOP1 and CAT and longer CAT+ translation products are indicated by arrows; molecular mass markers are shown on the left.
Linear scanning through cap-distal IRE-IRP complexes during translation initiation.
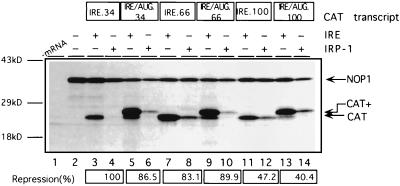
To overcome cap-distal IRE-IRP complexes subsequent to the recruitment of the small ribosomal subunit to IRE.100 mRNA, the initiation apparatus might either traverse the entire 5′ UTR in a linear fashion or bypass the IRE–IRP-1 complex by a jumping or shunting mechanism, as shown for the initiation of translation of cauliflower mosaic virus 35S RNA (7) or adenovirus late mRNAs (44). To distinguish between these possibilities, an in-frame AUG codon was introduced into the IREs of IRE.34, IRE.66, and IRE.100 mRNAs to create IRE/AUG.34, IRE/AUG.66, and IRE/AUG.100 mRNAs, respectively (Fig. 1C). If the translation machinery were to hop or shunt past the IRE–IRP-1 complex, initiation should occur at the downstream CAT initiation codon. However, if the entire 5′ UTR is linearly scanned, protein synthesis should start from the AUG within the IRE and yield an extended protein product. When endogenous IRPs are titrated by competitor IREs, translation of the three different IRE/AUG mRNAs predominantly yields the N-terminally extended CAT polypeptide, indicating that the AUG codon within the IRE is efficiently recognized in the absence of IRP-1 (Fig. 7, lanes 5, 9, and 13). Interestingly, leaky scanning, which leads to the synthesis of the smaller CAT polypeptide, decreases as the distance between the 5′ end of the mRNA and the initiation codon is increased (compare IRE/AUG.34 with IRE/AUG.100). All three IRE/AUG mRNAs display the same response to IRP-1 as their non-IRE/AUG counterparts (compare lanes 5 and 6 with lanes 3 and 4, lanes 9 and 10 with lanes 7 and 8, and lanes 13 and 14 with lanes 11 and 12, respectively), demonstrating that the position effect is preserved following the insertion of the AUGs. Importantly, IRE/AUG.66 and IRE/AUG.100 mRNAs yield the N-terminally extended CAT polypeptides, even in the presence of IRP-1. Thus, the entire 5′ UTR including the IRE is scanned in a linear fashion during initiation of translation, inconsistent with the notion that the IRE-IRP complex is bypassed by a hopping or shunting mechanism.
FIG. 7.
Cap-distal IRE-IRP complexes are not bypassed during initiation of translation. NOP1 (5 ng) (lanes 2 to 14) and 2.5 ng of IRE.34 (lanes 3 and 4), IRE/AUG.34 (lanes 5 and 6), IRE.66 (lanes 7 and 8), IRE/AUG.66 (lanes 9 and 10), IRE.100 (lanes 11 and 12), or IRE/AUG.100 (lanes 13 and 14) were translated in RRL, in the presence of 15 ng of IRE competitor RNA or 200 ng of recombinant IRP-1. 35S-labelled translation products were analyzed by SDS-PAGE. [35S]Met incorporation into translation products was determined by phosphoimaging, and the relative repression of CAT synthesis in the presence of recombinant IRP-1 normalized for the NOP1 internal control is shown below, with repression of IRE.34 set as 100%. The migrations of the NOP1 and CAT and longer CAT+ translation products are indicated by arrows; molecular mass markers are shown on the left.
Active IRP-1 displacement from cap-distal IREs by the mammalian translation apparatus.
The possibility that the observed IRE position effect could result from different affinities of the respective IREs for IRP-1 was excluded by competition gel retardation assays (data not shown). This is consistent with previous results (10).
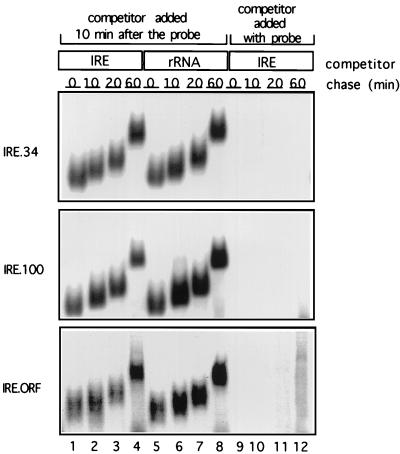
Another series of competition gel retardation assays were then performed to estimate the passive dissociation rate of IRP-1 in the cell-free-translation extract (Fig. 8). The RRL was pretreated with an m7GpppG cap analog to prevent the recruitment of the translation apparatus to the mRNAs, which might cause active IRP-1 displacement. Capped radiolabelled IRE.34, IRE.100, and IRE.ORF mRNAs were subsequently incubated in this extract with 50 ng of recombinant IRP-1 to allow formation of IRE–IRP-1 complexes. After 10 min, 1 μg of IRE competitor or rRNA as a nonspecific competitor was added, and the incubation was continued to assess the passive dissociation of IRP-1 from the mRNAs. Aliquots taken at 0, 10, 20, and 60 min after the addition of competitor RNA were loaded on a running, native polyacrylamide gel, resulting in the shorter migration distances of IRE-IRP complexes of samples loaded at later time points (Fig. 8). Addition of the IRE competitor simultaneously with the probe completely abolishes complex formation (lanes 9 to 12), demonstrating that the competitor is in excess. Complexes formed between the IRE.34, IRE.100, and IRE.ORF IREs and IRP-1 appear to be equally stable, and the majority of the mRNAs are still associated with IRP-1 after 60 min. Due to the stability of the IRE–IRP-1 complexes, their half-lives (t1/2s) exceed the duration of the assay; the t1/2s were thus extrapolated to range between 66 and 73 min (Fig. 8). The slow passive dissociation of IRP-1 from IRE.34, IRE.100, and IRE.ORF RNAs suggests that both the initiation apparatus and the elongation translation apparatus play an active role in displacing IRP-1 from cap-distal IREs.
FIG. 8.
Stability of the IRE–IRP-1 complexes. 32P-labelled IRE.34, IRE.100, or IRE.ORF probes (2 ng) were incubated with 100 ng of recombinant IRP-1 in 40% RRL for 10 min at 30°C, to allow formation of IRE-IRP complexes, before addition of IRE competitor (1 μg) (lanes 1 to 4) or 1 μg of rRNA (lanes 5 to 8). Aliquots were taken after 0, 10, 20, and 60 min and analyzed on a running nondenaturing gel. In control reactions, the IRE competitor RNA was added simultaneously with the probe (lanes 9 to 12).
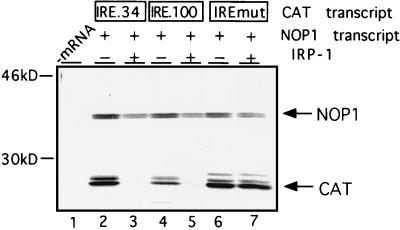
The position effect is species restricted.
IRP-1-mediated repression has previously been reconstituted in WGE (13, 16, 42), which, unlike mammalian cells and extracts, does not contain endogenous IRP (16, 42). To test whether IRP-1-mediated repression in this system is position dependent, the translation of IRE.34 and IRE.100 was examined in WGE by using IRE-mut and NOP mRNAs as negative and internal controls, respectively. In the absence of exogenous IRP-1, all three test transcripts are translated with comparable efficiencies (Fig. 9, lanes 2, 4, and 6). The translation of both IRE.34 and IRE.100 mRNAs is repressed specifically and to a comparable extent with saturating (lanes 3 and 5) and subsaturating (data not shown) amounts of exogenously added IRP-1. We also evaluated the effect of IRP-1 in a yeast-free-cell translation extract, which, like wheat germ, is devoid of endogenous IRE-binding activity (34). As in the wheat germ system, IRP-mediated translational repression does not exhibit a significant position dependence in yeast (25) or in a yeast-free-cell translation extract (data not shown). This apparent difference between RRL and the wheat germ and yeast extracts was further investigated with the wheat germ system. Control experiments showed that the observed differences were not due to different buffer conditions or incubation temperatures (data not shown). Direct comparison of ribosome assembly on IRE.100 mRNA in the presence of IRP-1 shows that both 48S and 80S complexes assemble in RRL after 30 min, whereas only 48S complexes assemble in WGE (compare Fig. 10B and C [Fig. 10A is a control for IRP-1 activity]). This result is best explained by extended stalling of the scanning subunits by the cap-distal IRE–IRP-1 complexes in WGE. These results also show that the position effect observed in mammalian cells and in the cell-free-translation extract derived from mammalian cells is not universal, but species restricted. They suggest that mammalian cells possess an activity with which they can overcome cap-distal high-affinity RNA-protein complexes.
FIG. 9.
Position-independent translational repression by IRP-1 in WGE. NOP1 (10 ng) (lanes 2 to 7) and 5 ng of IRE.34 (lanes 2 and 3), IRE.100 (lanes 4 and 5), or IRE-mut (lanes 6 and 7) mRNAs were translated in WGE in the absence (−) (lanes 2, 4, and 6) or presence (+) of 200 ng of recombinant IRP-1 (lanes 3, 5, and 7). 35S-labelled translation products were analyzed by SDS-PAGE. The migrations of NOP1 and CAT translation products are indicated by arrows; molecular mass markers are shown on the left.
DISCUSSION
This study addresses a long-standing question that was raised when the positions of IREs in various mRNAs and from different species were found to be phylogenetically conserved and functionally important (9, 10). Subsequent studies added further examples of cellular mRNAs that appear to obey the position effect (15, 32) and also uncovered possible exceptions (15, 23 [see below]). The mechanism by which cap-proximal IRE-IRP complexes regulate translation initiation is increasingly well understood. In this report, we provide a molecular analysis of the function of cap-distal IRE-IRP complexes. These findings have shed light on the understanding of the biology of the IRE-IRP regulatory system in particular and the understanding of the functional interplay between ribosomes and mRNA binding proteins in general.
The experimental system.
The position effect was recapitulated in a well-characterized mammalian cell-free translation system, RRL (20). Using reporter mRNAs with the same 5′ UTR spacer sequences as in previous transfection experiments with murine B6 fibroblasts and human HeLa cells (10), we could directly compare the qualitative and quantitative effects of IRP binding to IREs located in different positions of the mRNA. IRE.34 mRNA in this study and F44 mRNA in the transfection experiments (10) served as positive controls and points of reference for the profound repressive effect of IRP binding to a cap-proximal IRE. IRE.66 mRNA (this study) and F64 mRNA (in the transfection experiments [10]) allowed a comparative assessment of the role of IRE-IRP complexes formed more than 60 nucleotides downstream from the cap structure. Our results show that the RRL system reflects the roughly twofold (50%) difference between F44 and F64 mRNAs in vivo (10) and IRE.34 and IRE.66 mRNAs in vitro (Fig. 2) with reassuring accuracy. In addition to the IRE.34 and IRE.66 mRNAs, we designed the two constructs IRE.100 and IRE.ORF. The former displays a stronger 5′ UTR position effect than F64 and IRE.66, respectively, by placing the IRE 100 nucleotides downstream from the cap structure (Fig. 2). The latter allowed the analysis of the effect of IRE-IRP complexes on elongating ribosomes (Fig. 6).
Function of cap-distal IRE-IRP complexes in the 5′ UTRs of mammalian and nonmammalian mRNAs.
In contrast to cap-proximal IRE–IRP-1 complexes that preclude the recruitment of the 43S translation preinitiation complex (including the 40S ribosomal subunit) (Fig. 3A) (13, 33), 43S complex recruitment to IRE.100 mRNA is not impeded (Fig. 3B and Fig. 4G to I). Kinetic analyses employing IRE.66 and IRE.100 mRNAs revealed that IRP-1 binding to IRE.100 mRNA does not even delay 43S complex recruitment (Fig. 4), whereas this complex associates more slowly and less efficiently with IRE.66 mRNA in the presence of IRP-1 than in its absence (Fig. 4D to F). This kinetic delay contributes to the stronger repressive effect of IRP-1 on IRE.66 than that on IRE.100 mRNA. It is unclear from the analysis of IRE.66 whether small ribosomal subunit scanning is also delayed.
Subsequent to 43S complex recruitment, the small ribosomal subunit appears to be stalled by the IRE-IRP complex of IRE.100 mRNA. This is suggested by the reduced translational efficiency of IRE.100 mRNA in the presence of IRP-1 compared to that in its absence (despite the unimpeded initial association of the 43S complex) and the redistribution of IRE.100 mRNA from 66S complexes (indicative of two 43S complexes) to 48S complexes (with a single 43S complex) in the presence of IRP-1 (Fig. 3B and Fig. 4G to I). This redistribution indicates that the temporary stalling of a 43S complex which occupies ∼35 to 80 nucleotides of the 5′ UTR (29) prevents the binding of a second complex.
After the temporary scanning arrest, the translation machinery linearly transgresses the remainder of the 5′ UTR to the translation initiation codon (Fig. 5). Our findings with the IRE/AUG.66 and IRE/AUG.100 mRNAs (Fig. 7) show that the translation apparatus does not negotiate the IRE-IRP complex by a nonlinear hopping or shunting mechanism (7, 40, 44). Ribosomal scanning through the region of the IRE will likely require the absence of IRP-1, although this cannot be formally concluded from our experiments. This interpretation begs the question of whether the stalled preinitiation complex “sits and waits” for the passive dissociation of IRP-1 or whether it actively participates in dislodging the bound protein. We favor the latter hypothesis. In contrast to IRE.34 mRNA, IRE.100 mRNA displays only a 26 to 35% repression (Fig. 2 [27%]), yet both mRNAs bind IRP-1 with equal affinities (data not shown) and comparable dissociation rates (Fig. 8). While this difference may suggest that a preloaded ribosomal complex can actively contribute to IRP-1 displacement, the more efficient translation of IRE.100 mRNA could be alternatively explained by a faster rate at which a preloaded and stalled preinitiation complex scans through the IRE region before IRP-1 rebinding, compared to the less rapid recruitment of a 43S complex to IRE.34 mRNA. However, we favor an active contribution of the mammalian translational apparatus, since this is consistent with the very slow passive dissociation rate of IRP-1 (Fig. 8) and the species specificity of the position effect (Fig. 9). Assuming that the translation apparatus actively contributes to IRP-1 displacement, the displacement activity would be predicted to be recruited to the mRNA with or after the 43S preinitiation complex. This activity may represent an RNA helicase, which could indirectly displace IRP by disrupting the IRE, or a novel protein-displacing factor.
Function of IRE-IRP complexes in the open reading frame of mRNAs.
When an IRE–IRP-1 complex is encountered by elongating rather than initiating ribosomes, translation proceeds without noticeable impediment. The translation of IRE.ORF mRNA (which bears an IRE 32 nucleotides downstream from the initiation codon) is equally efficient in the presence of IRP-1 as in its absence in RRL (Fig. 6) and yeast and wheat germ translation extracts (data not shown). Thus, this property of elongating ribosomes appears to be a more general one. These data are also consistent with other reports which have shown that impediments imposed by RNA secondary structures within the open reading frame are more easily overcome than those within the 5′ UTR (30, 31). Nonetheless, previous data indicate that even translation elongation can be affected by IRE-IRP complexes under special circumstances: the human immunodeficiency virus Gag-Pol ribosomal frameshift requires a so-called “heptanucleotide slippery sequence,” which induces a low level of basal frameshifting and a downstream RNA structure as an enhancer of this process. IRE-IRP complexes can functionally substitute for the downstream RNA structure and mediate regulated ribosomal frameshifting (24).
System-specific and general biological implications of the position effect.
An expanding number of mRNAs have been found to be translationally regulated by the IRE-IRP system (15, 17, 23). In mammalian cells, all of the examples that have been identified to date are regulated by cap-proximal IREs located <50 nucleotides downstream from the cap structure. Since IRE-IRP complexes that are located further downstream in the 5′ UTR still display translational control by IRP binding, albeit less efficiently (this study and reference 10), variation of the distance of the IRE from the transcription start site could be employed as a means of fine-tuning the regulatory effects of IRP binding.
In contrast to mammalian cells and RRL, we have found that plant and yeast translation extracts do not display a position effect. The lack of a position effect in yeast translation extracts correlates well with a recent study that suggests that yeast cells do not exhibit a position effect, at least when the IRE is moved approximately 60 nucleotides from the cap (25). Our results suggest that increasing this distance further would not lead to position dependence in yeast cells. The relative insensitivity of yeast cells and extracts to the positions of RNA-protein complexes within their 5′ UTR is consistent with the observation that the translation of mRNAs in yeast is particularly sensitive to the presence of secondary structures (1). No such observation has been made with WGE, where moderately stable structures have been shown not to inhibit the progression of the translational machinery (26). While neither yeast nor plants appear to regulate their iron metabolism by the IRE-IRP system, the succinate dehydrogenase (Ip subunit) mRNA from Drosophila melanogaster was found to be translationally regulated by IRP (15, 23). Dependent on the transcription start site, the IRE in this mRNA is located in either a cap-proximal or a cap-distal position. It is currently unknown whether Drosophila is mammal-like or wheat germ- or yeast-like with regard to the position effect. Nonetheless, our data (Fig. 3B, 4G to I, 5D to F, and 10) would suggest that, at least in part, regulation may be achieved by stalling the progression of the scanning preinitiation complex.
More generally, other RNA-protein interactions have been identified that regulate translation by means of 5′ UTR binding sites (14). The data in this report show that there are at least two different mechanisms by which translation initiation can be regulated by 5′ UTR mRNA binding proteins: inhibition of 43S complex recruitment (as in IRE.34 mRNA) and stalled scanning without inhibited 43S complex recruitment (as in IRE.100 mRNA). The newly identified mechanism of stalled scanning by cap-distal RNA-protein complexes could potentially explain known examples of translational control, such as the autoregulation of poly(A)-binding protein mRNA by poly(A)-binding protein (5) or the regulation of Drosophila msl-2 mRNA by the Sex-lethal protein (3, 8, 21). The regulation of F64 and IRE.66 mRNAs may result from a composite of the two mechanisms described above. Stalled scanning could represent a very efficient means of translational regulation in yeast and plant systems, so that the dissociation rate of the binding protein becomes a key determinant for control. In mammals, the position effect offers an opportunity for tissue-specific or developmental regulation by changing the concentration or activity of factors which assist the initiating ribosomes in overcoming cap-distal RNA-protein complexes or by altering the transcription start site.
ACKNOWLEDGMENTS
We thank Thomas Preiss for the gift of yeast translation extract. N.K.G. thanks Jeremy Brock for encouragement and advice during the course of this work. We also thank Thomas Preiss and George Simos for critical reading of the manuscript.
E.P. and N.K.G. were supported through grants by the European Commission and the Deutsche Forschungsgemeinschaft, respectively, to M.W.H.
E.P. and N.K.G. contributed equally to this work.
REFERENCES
- 1.Altmann M, Trachsel H. The yeast Saccharomyces cerevisiae system: a powerful tool to study the mechanism of protein synthesis initiation in eukaryotes. Biochimie. 1994;76:853–861. doi: 10.1016/0300-9084(94)90187-2. [DOI] [PubMed] [Google Scholar]
- 2.Anthony D D, Merrick W C. Analysis of 40 S and 80 S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J Biol Chem. 1992;267:1554–1562. [PubMed] [Google Scholar]
- 3.Bashaw G J, Baker B S. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell. 1997;89:789–798. doi: 10.1016/s0092-8674(00)80262-7. [DOI] [PubMed] [Google Scholar]
- 4.Dasso M C, Jackson R J. On the fidelity of mRNA translation in the nuclease-treated rabbit reticulocyte lysate system. Nucleic Acids Res. 1989;17:3129–3144. doi: 10.1093/nar/17.8.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Melo Neto O P, Standart N, Martins de Sa C. Autoregulation of poly(A)-binding protein synthesis in vitro. Nucleic Acids Res. 1995;23:2198–2205. doi: 10.1093/nar/23.12.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubnau J, Struhl G. RNA recognition and translational regulation by a homeodomain protein. Nature. 1996;379:694–699. doi: 10.1038/379694a0. [DOI] [PubMed] [Google Scholar]
- 7.Fütterer J, Kiss-Laszlo Z, Hohn T. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell. 1993;73:789–802. doi: 10.1016/0092-8674(93)90257-q. [DOI] [PubMed] [Google Scholar]
- 8.Gebauer F, Meredino L, Hentze M W, Valcarcel J. The Drosophila splicing regulator Sex-lethal directly inhibits translation of male-specific-lethal-2 mRNA. RNA. 1998;4:142–150. [PMC free article] [PubMed] [Google Scholar]
- 9.Goossen B, Caughman S W, Harford J B, Klausner R D, Hentze M W. Translational repression by a complex between the iron-responsive element of ferritin mRNA and its specific cytoplasmic binding protein is position-dependent in vivo. EMBO J. 1990;9:4127–4133. doi: 10.1002/j.1460-2075.1990.tb07635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goossen B, Hentze M W. Position is the critical determinant for function of iron-responsive elements as translational regulators. Mol Cell Biol. 1992;12:1959–1966. doi: 10.1128/mcb.12.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray N K. The mechanism of translational repression by iron regulatory protein. Ph.D. thesis. Glasgow, Scotland: University of Glasgow; 1994. [Google Scholar]
- 12.Gray N K. Translational control by repressor proteins binding to the 5′UTR of mRNAs. Methods Mol Biol. 1998;77:379–397. doi: 10.1385/0-89603-397-X:379. [DOI] [PubMed] [Google Scholar]
- 13.Gray N K, Hentze M W. Iron regulatory protein prevents binding of the 43S translation pre-initiation complex to ferritin and eALAS mRNAs. EMBO J. 1994;13:3882–3891. doi: 10.1002/j.1460-2075.1994.tb06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray N K, Hentze M W. Regulation of protein synthesis by mRNA structure. Mol Biol Rep. 1994;19:195–200. doi: 10.1007/BF00986961. [DOI] [PubMed] [Google Scholar]
- 15.Gray N K, Pantopoulos K, Dandekar T, Ackrell B A C, Hentze M W. Translational regulation of mammalian and Drosophila Krebs cycle enzymes via iron-responsive elements. Proc Natl Acad Sci USA. 1996;93:4925–4930. doi: 10.1073/pnas.93.10.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray N K, Quick S, Goossen B, Constable A, Hirling H, Kühn L C, Hentze M W. Recombinant iron regulatory factor functions as an iron-responsive element-binding protein, a translational repressor and an aconitase. A functional assay for translational repression and direct demonstration of the iron switch. Eur J Biochem. 1993;218:657–667. doi: 10.1111/j.1432-1033.1993.tb18420.x. [DOI] [PubMed] [Google Scholar]
- 17.Hentze M W, Kühn L C. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide and oxidative stress. Proc Natl Acad Sci USA. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hershey J W B, Monro R E. A competitive inhibitor of the GTP reaction in protein synthesis. J Mol Biol. 1966;18:68–76. doi: 10.1016/s0022-2836(66)80077-3. [DOI] [PubMed] [Google Scholar]
- 19.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 20.Jackson R J, Hunt T. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- 21.Kelley R L, Wang J, Bell L, Kuroda M I. Sex-lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature. 1997;387:195–199. doi: 10.1038/387195a0. [DOI] [PubMed] [Google Scholar]
- 22.Kim-Ha J, Kerr K, Macdonald P M. Translational regulation of oskar mRNA by Bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- 23.Kohler S A, Henderson B R, Kuhn L C. Succinate dehydrogenase b mRNA of Drosophila melanogaster has a functional iron-responsive element in its 5′-untranslated region. J Biol Chem. 1995;270:30781–30789. doi: 10.1074/jbc.270.51.30781. [DOI] [PubMed] [Google Scholar]
- 24.Kollmus H, Hentze M W, Hauser H. Regulated ribosomal frameshifting by an RNA-protein interaction. RNA. 1996;2:316–323. [PMC free article] [PubMed] [Google Scholar]
- 25.Koloteva N, Mueller P P, McCarthy J E G. The position dependence of translational regulation via RNA-RNA and RNA-protein interactions in the 5′-untranslated region of eukaryotic mRNA is a function of the thermodynamic competence of 40S ribosomes in translational initiation. J Biol Chem. 1997;272:16531–16539. doi: 10.1074/jbc.272.26.16531. [DOI] [PubMed] [Google Scholar]
- 26.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozak M. Evaluation of the fidelity of initiation of translation in reticulocyte lysates from commercial sources. Nucleic Acids Res. 1990;18:2828. doi: 10.1093/nar/18.9.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak M. Effects of long 5′ leader sequences on initiation by eukaryotic ribosomes in vitro. Gene Expr. 1991;1:117–125. [PMC free article] [PubMed] [Google Scholar]
- 29.Kozak M, Shatkin A J. Sequences of two 5′-terminal ribosome-protected fragments from reovirus messenger RNAs. J Mol Biol. 1977;112:75–96. doi: 10.1016/s0022-2836(77)80157-5. [DOI] [PubMed] [Google Scholar]
- 30.Liebhaber S A, Cash F E, Shakin S H. Translationally associated helix-destabilizing activity in rabbit reticulocyte lysate. J Biol Chem. 1984;259:15597–15602. [PubMed] [Google Scholar]
- 31.Lingelbach K, Dobberstein B. An extended RNA/RNA duplex structure within the coding region of mRNA does not block translational elongation. Nucleic Acids Res. 1988;16:3405–3414. doi: 10.1093/nar/16.8.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melefors Ö, Goossen B, Johansson H E, Stripecke R, Gray N K, Hentze M W. Translational control of 5-aminolevulinate synthase mRNA by iron-responsive elements in erythroid cells. J Biol Chem. 1993;268:5974–5978. [PubMed] [Google Scholar]
- 33.Muckenthaler M, Gray N K, Hentze M W. IRP-1 binding to ferritin mRNA disrupts the bridging interaction between cap-binding complex and the small ribosomal subunit. Mol Cell. 1998;2:383–388. doi: 10.1016/s1097-2765(00)80282-8. [DOI] [PubMed] [Google Scholar]
- 34.Oliveira C C, Goossen B, Zanchin N I T, McCarthy J E G, Hentze M W, Stripecke R. Translational repression by the human iron-regulatory factor (IRF) in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:5316–5322. doi: 10.1093/nar/21.23.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ostareck D H, Ostareck-Lederer A, Wilm M, Thiele B J, Mann M, Hentze M W. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell. 1997;16:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- 36.Ostareck-Lederer A, Ostareck D H, Standart N, Thiele B. Translation of 15-lipoxygenase mRNA is inhibited by a protein that binds to a repeated sequence in the 3′ untranslated region. EMBO J. 1994;13:1476–1481. doi: 10.1002/j.1460-2075.1994.tb06402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pantopoulos K, Hentze M W. Nitric oxide signaling to iron-regulatory protein (IRP): direct control of ferritin mRNA translation and transferrin receptor mRNA stability in transfected fibroblasts. Proc Natl Acad Sci USA. 1995;92:1267–1271. doi: 10.1073/pnas.92.5.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rivera-Pomar R, Niessing D, Schmidt-Ott U, Gehring W J, Jaeckle H. RNA binding and translational suppression by bicoid. Nature. 1996;379:746–749. doi: 10.1038/379746a0. [DOI] [PubMed] [Google Scholar]
- 39.Rouault T A, Klausner R D, Harford J B. Translational control of ferritin. In: Hershey J W B, Matthews M B, Sonnenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 335–363. [Google Scholar]
- 40.Schneider R J. Adenovirus and vaccinia virus translational control. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 575–606. [Google Scholar]
- 41.Stripecke R, Hentze M W. Bacteriophage and spliceosomal proteins function as position-dependent cis/trans repressors of mRNA translation in vitro. Nucleic Acids Res. 1992;20:5555–5564. doi: 10.1093/nar/20.21.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walden W E, Patino M M, Gaffield L. Purification of a specific repressor of ferritin mRNA translation from rabbit liver. J Biol Chem. 1989;264:13765–13769. [PubMed] [Google Scholar]
- 43.Webster P J, Liang L, Berg C A, Lasko P, Macdonald P M. Translational repressor bruno plays multiple roles in development and is widely conserved. Genes Dev. 1997;11:2510–2521. doi: 10.1101/gad.11.19.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yueh A, Schneider R J. Selective translation initiation by ribosome jumping in adenovirus-infected and heat shocked cells. Genes Dev. 1996;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]