Abstract
Background
Belamcanda chinensis (L.) DC. (BC) belongs to the family of Iridaceae and is widely cultivated and used in many Chinese patent medicine and Chinese medicinal formulae. However, due to the high similarities in appearance such as color and shape to Iris tectorum Maxim (ITM), another plant from the same family, BC is often confused or even misused with ITM.
Methods
Therefore, in order to distinguish the chemical constituents, qualities and biological activities of BC and ITM, multiple technologies including plant metabolomics, digital reference standard (DRS) analyzer and biological activities assay were employed to provide a sufficient basis for their practical applications.
Results
In plant metabolomics, the PCA and OPLS-DA score plot indicated the obvious differences in chemical profiling between BC and ITM and 6 compounds were successfully identified to contribute to the differences. In DRS study, the fingerprints of 10 and 8 compounds in BC and ITM were developed based on DRS analyzer, respectively, involving relative retention time (RRT) method and linear calibration using two reference substances (LCTRS) technique. The DRS analyzer also accurately identified 10 and 8 compounds from BC and ITM, respectively, by using only two reference standards. In biological activities assay, BC had a better anticancer effect than ITM due to the high abundance of irigenin, while ITM showed stronger hepatoprotective activity than BC because of the high abundance of tectoridin.
Conclusions
Therefore, due to the significant differences of B. chinensis and I. dichotoma in chemical composition and biological activities, the current studies strongly proved that these two medicinal plants could not be mixed in industrial production and clinical medication.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13020-021-00494-3.
Keywords: Belamcanda chinensis (L.) DC, Iris tectorum Maxim, Plant metabolomics, Digital reference standard, Biological activities
Background
Belamcanda chinensis (L.) DC. (BC), a perennial herbaceous plant whose rhizome is named as She-gan in a traditional Chinese medicine (TCM) belongs to the family of Iridaceae and is widely cultivated in China, Korea, Japan, India and eastern Russia as an economic medicinal plant. She-gan has been used in many Chinese patent medicine and Chinese medicinal formulae such as Xiaoer Qingre granules, Xiaoer Qingfei oral liquid, and Shengan Liyan oral liquid, etc., for the treatment coughing and pharyngitis.
However, Chuan-she-gan, the rhizome of Iris tectorum Maxim (ITM), another medicinal plant comes from the family of Iridacae mainly distributing in Sichuan of China, is also used for the treatment of asthma, cough, tonsillitis and pharyngitis. Actually, both of these two medicinal plants are rich in isoflavonoids, stilbenes, xanthones, and simple phenols [1–3]. Among them, isoflavones, such as tectoridin, iridin, tectorigenin, irigenin, irisflorentin and so on [3–5], are the major bioactive constituents of two medicines that have shown a wide range of biological activity, such as anticancer, hepatoprotective, antiatherosclerosis, antiosteoporosis and antihyperlipidemic, etc. [1, 6–8].
Consequently, in the medicinal market, as well as in the pharmaceutical industry, because of the high similarities in appearance such as color and shape between BC and ITM, they were often confused or even mixed with each other when used for the treatments of coughing and pharyngitis. Therefore, it is very necessary to distinguish these two plants from their chemical constituents and biological activities by multiple technologies and approaches, so as to provide a sufficient basis and guidance for their industrial production and clinical medication.
Metabolomics, genomics, and proteomics are important components of system biology. As an important branch of metabolomics, plant metabolomics describes the alterations in the content and composition of different plant phytochemicals [9]. Currently, the most promising technique in terms of metabolome coverage is ultra-high-performance liquid chromatography-high resolution tandem mass spectrometry (UHPLC-HRMS/MS), which is a powerful analytical tool for the analysis of the known compounds and elucidation of unknown compounds in herbal medicines. Here, UHPLC-Q Exactive-HRMS/MS based untargeted metabolomics techniques were used for qualitative studies to distinguish BC and ITM.
Quality control analysis of TCM is important for safe and effective use. The reference standard is the most important role for qualitative and quantitative analysis of TCM. There is an increasing demand for reference standards with the development of TCM quality control. At the same time, some TCM compounds are difficult to be extracted, isolated, and purified, led to a significant increase in the cost of TCM analysis. Linear calibration using two reference substances (LCTRS) approaches, a substitute reference standard method, can deal with the above problems effectively [10–12]. LCTRS is a method for the qualitative determination of several compounds to be measured by two reference standards by using several constant eigenvalues and algorithms. The principle of LCTRS is that there is a linear relationship between the retention time (tR) of the compounds on two different HPLC systems (including chromatographs and columns). The method has been successfully developed for the quality analysis of Salvia miltiorrhiza, Paris polyphylla, Rheum officinale [10–12]. Relative retention time (RRT) technique, a qualitative substitute reference standard method, can qualitative determination of several compounds to be measured by one reference standard. Finally, we introduced the concept of the digital reference standard (DRS), which supports the chromatographic algorithm methods of RRT and LCTRS. In the present study, quality control methods of fingerprint involving 10 compounds of BC and 8 compounds of ITM respectively were developed based on DRS method.
Recently, several studies on the quality control of BC were reported. Li et al. [13] evaluated the quality of BC by the establishment of chromatographic fingerprinting profile employing HPLC–DAD-MS method and simultaneous determination of seven phenol compounds. Chen et al. [14] reported the spatial chemical profiles of BC at different growth ages from various origins through qualitative and quantitative analyses by using UHPLC-Q/TOF–MS and UHPLC-QqQ-MS. Wen et al. [15] used a chemical profiling method to evaluate the quality of BC and compare the chemical compositions by HPLC analysis combining with multivariate data analysis. However, these methods only focused on one or several marker compounds and failed to distinguish these two plants from their chemical constituents and biological activities. Herein, the systematic studies on the chemical constituents, qualities and biological activities of BC and ITM were carried out by plant metabolomics, digital reference standard analyzer and biological activities assay.
Methods
Collection of plant materials
The rhizomes of two original medicinal plants of Belamcanda chinensis (L.) DC and Iris tectorum Maxim were collected from different habitats in China, and dried at room temperature. All of them were identified by Professor Yi Zhang. Their sample number, species, habitats and collection time were listed in Table 1.
Table 1.
The sorts of Belamcanda chinensis (L) DC and Iris tectorum Maxim
| No. | Species | Location | Collection time |
|---|---|---|---|
| BC01 | B. chineses | Anguo, Hebei | April 2020 |
| BC02 | B. chineses | Baoding, Hebei | April 2020 |
| BC03 | B. chineses | Yuncheng, Henan | April 2020 |
| BC04 | B. chineses | Qinhuangdao, Hebei | April 2020 |
| BC05 | B. chineses | Nangong, Hebei | May 2020 |
| BC06 | B. chineses | Yuncheng, Shanxi | May 2020 |
| ITM01 | I. tectorum | Chengdu, Sichuan | April 2020 |
| ITM02 | I. tectorum | Chengdu, Sichuan | April 2020 |
| ITM03 | I. tectorum | Chengdu, Sichuan | May 2020 |
| ITM04 | I. tectorum | Chengdu, Sichuan | May 2020 |
| ITM05 | I. tectorum | Baoding, Hebei | May 2020 |
| ITM06 | I. tectorum | Guiyang, Guizhou | April 2020 |
| ITM07 | I. tectorum | Bozhou, Anhui | April 2020 |
Chemicals and reagents
Chromatography-grade acetonitrile and methanol were purchased from Fisher Chemical (CA, USA). MS-grade ammonium acetate, formic acid, acetonitrile and methanol were obtained from Fisher Chemical (CA, USA). Chromatography grade phosphoric acid and ethanol were purchased from Sinopharm Chemical Reagent (Shanghai, China). Methyl-β-cyclodextrin (Me-β-CD) was obtained from Aladdin Co., Ltd (Shanghai, China). The water used was produced by Milli-Q system (Millipore, Bedford, MA, USA). Tectoridin, bicyclol and irisflorentine were purchased from National Institutes for Food and Drug Control (NIFDC, Beijing, China). Iristectorin A and iristectorigenin-A-7-glucoside were obtained from Nature Standard Technical Service Co., Ltd (Shanghai, China). Irigenin 7-glucoside was purchased from Chengdu Push Bio-technology Co., Ltd (Chengdu, China). Tectorigenin was obtained from Chengdu Puruifa Technology Co., Ltd (Chengdu, China). Iristectorigenin B, 3′,6-dimethoxy-4′,5,7-trihydroxyisoflavone, 5,7-dihydroxy-3-(3-hydroxy-4,5-dimethoxyphenyl)-6-methoxy-4-benzopyrone and 5,3-dihydroxy-4,5-dimethoxy-6,7-methylenedioxyisoflavone were purchased from Shanghai Tongtian Biotechnology Co., Ltd (Shanghai, China). Oxaliplatin was obtained from Oxaliplatin, Hospira Inc., (IL, Australia). d-galactosamine, curcumin, cell counting kit-8, RPMI1640 medium and DMEM high glucose culture medium were purchased from Meilun Biotechnology Co., Ltd (Dalian, China).
Standard solutions and sample preparation for LC‑HR/MS metabolomics and DRS study
The dry powder (0.1 g) of BC and ITM were extracted with 25 mL of 70% ethanol in an ultrasonic bath for 60 min. The supernatant was filtered through a 0.22 μm membrane filter before analysis. All reference chemicals were dissolved in 70% ethanol at 1.0 mg mL−1 as stock solutions. These stock solutions were stable for at least 1 week at room temperature. A defined amount of the above stock solutions were mixed and diluted to an appropriate concentration as the standard stock solution: these standards were stable at least for 2 weeks under 4 °C.
Plant metabolomics with liquid chromatography/high‑resolution mass spectrometry (LC‑HR/MS) analysis
The analyses were performed on an Ultimate 3000 UHPLC system coupled to Q Exactive MS (Thermo Fisher Scientific, CA, USA). UHPLC analyses were performed on a Waters Acquity UPLC BEH C18 column (2.1 × 100 mm, 1.7 μm; Waters Technologies, MA, USA) and Waters Van Guard BEH C18 column (2.1 × 5 mm, 1.7 μm; Waters Technologies, MA, USA). The mobile phase was consisted of (A) 0.1% formic acid–water and (B) acetonitrile, and the gradient program was optimized as follows: 0–1 min, 5% B; 1–9 min, 5–25% B; 9–19 min, 25–75% B; 19–25 min, 75–100% B; 25–26 min 100–5% B, 26–30 min 5% B. The column temperature was set at 45 °C. The injection volume was 1 μL. The flow rate was set at 0.4 mL min−1. The MS analysis of lipids was carried out on Q Exactive under the conditions: full MS/ddMS2 mode; evaporation temperature, 350 °C; capillary temperature, 320 °C; spray voltage, 3.0 kV for negative ion mode and 3.5 kV for positive ion mode; aux gas flow rate (arb), 10; sheath gas rate (arb), 35; mass range (m/z), 100–1500.
DRS study with instruments and chromatographic conditions
Chromatographic analysis was performed on Agilent 1260 high-performance liquid chromatography with a DAD detector (Agilent Technologies, CA, USA) and Waters e2695 high-performance liquid chromatography with a 2998 PDA detector (Waters Technologies, MA, USA). Twenty-four columns (Table 2) from mainstream manufacturers were randomly selected. DRS method research is recommended to use at least ten columns from three manufacturers.
Table 2.
Information of columns
| No. | Brand | Type | Specification |
|---|---|---|---|
| Column 1 | Osaka Soda | Capcell pak C18 | 250 × 4.6 mm, 5 μm |
| Column 2 | Dikma | Diamonsil C18 | 250 × 4.6 mm, 5 μm |
| Column 3 | Phenomenex | Luna C18 | 250 × 4.6 mm, 5 μm |
| Column 4 | TechMate | TechMate C18 | 250 × 4.6 mm, 5 μm |
| Column 5 | FLM | Titank C18 | 250 × 4.6 mm, 5 μm |
| Column 6 | Waters | Symmetry C18 | 250 × 4.6 mm, 5 μm |
| Column 7 | Waters | SunFire C18 | 250 × 4.6 mm, 5 μm |
| Column 8 | AkzoNobel | Kromasil 100-5-C18 | 250 × 4.6 mm, 5 μm |
| Column 9 | Agilent | ZORBAX Eclipse Plus C18 | 250 × 4.6 mm, 5 μm |
| Column 10 | SHIMADZU | Shim-pack GIST C18 | 250 × 4.6 mm, 5 μm |
| Column 11 | Exmere Ltd | Exsil Mono 100 C18 | 250 × 4.6 mm, 5 μm |
| Column 12 | SHIMADZU GL | Inertsil ODS-3 C18 | 250 × 4.6 mm, 5 μm |
| Column 13 | Dikma | Inspire C18 | 250 × 4.6 mm, 5 μm |
| Column 14 | Agilent | ZORBAX Eclipse XDB C18 | 250 × 4.6 mm, 5 μm |
| Column 15 | Agilent | ZORBAX SB C18 | 250 × 4.6 mm, 5 μm |
| Column 16 | Agilent | 5 HC C18 | 250 × 4.6 mm, 5 μm |
| Column 17 | Agilent | 5 TC C18 | 250 × 4.6 mm, 5 μm |
| Column 18 | YMC | Pack ODS-A | 250 × 4.6 mm, 5 μm |
| Column 19 | ZHONGPU | RP-C18 | 250 × 4.6 mm, 5 μm |
| Column 20 | SVEA | C18 Opal | 250 × 4.6 mm, 5 μm |
The main constituents of B. chinensis and I. tectorum are iristectorigenin A, iristectorigenin B, and irigenin. Because of the high similarity in their chemical constituents (Appendix Fig. 8), it is difficult to separate these three compounds by conventional HPLC method. Previous study found that methyl-β-cyclodextrin (Me-β-CD) could successfully resolve these problems and the resolution of the other components could also meet the content determination requirements [16]. Thus, Me-β-CD is used to the mobile phase additive. Mobile phase A was acetonitrile, mobile phase B was 0.1% phosphoric acid and 0.55% Me-β-CD-water and mobile phase C was 0.1% phosphoric acid–water. The elution procedure was shown in Table 3. The detection wavelength was 266 nm, and the UV–Vis absorption spectra (210–600 nm) were collected. The column temperature was 35 °C, the flow rate was 1 mL min−1, and the injection volume was 10 μL.
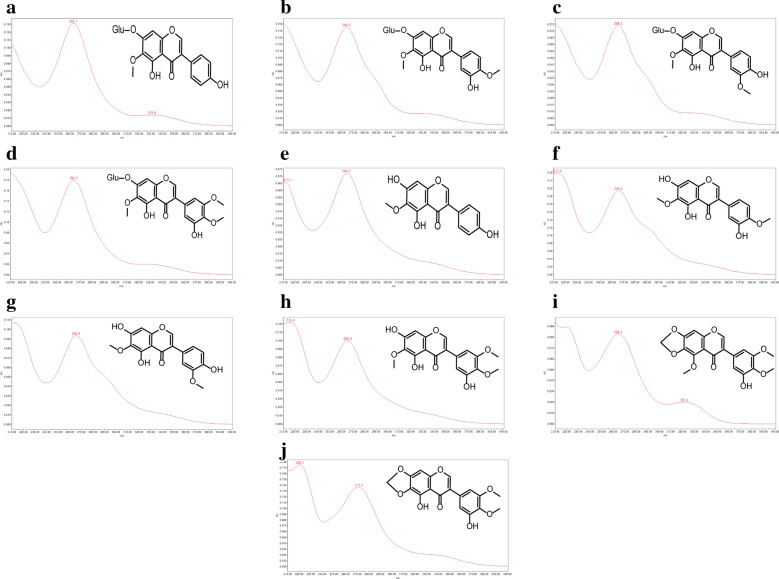
Fig. 8.
Chemical structures and UV spectra. a Tectoridin, b Iristectorin A, c Iristectorin B, d Iridin, e Tectorigenin, f Iristectorigenin B, g Iristectorigenin A, h Irigenin, i Irisflorentine, j Dichotomitin
Table 3.
The gradient elution conditions of the mobile phase
| Time (min) | Phase A (%) | Phase B (%) | Phase C (%) |
|---|---|---|---|
| 0–15 | 5–17 | 0–83 | 95–0 |
| 15–24 | 17–20 | 83–80 | 0 |
| 24–48 | 20–24 | 80–76 | 0 |
| 48–52 | 24–28 | 76–72 | 0 |
| 52–60 | 28–31 | 72–69 | 0 |
| 60–65 | 31–35 | 69–65 | 0 |
| 65–80 | 35–70 | 65–30 | 0 |
| 80–81 | 70–5 | 30–0 | 0–95 |
| 81–90 | 5 | 0 | 95 |
Data processing
The raw LC-HRMS data files (.raw) were uploaded to the XCMS web version platform (https://xcmsonline.scripps.edu/) for retention time alignment, peak picking, and annotation [17]. The chromatographic peak data were normalized uniformly, and the multidimensional data were further analyzed by the SIMCA-P software 14.1 (Umetrics, Umea, Sweden) for multivariate data analysis by principal components analysis (PCA), orthogonal partial least squares-discriminant analysis (OPLS-DA).
Evaluation of in vitro bioactivities
All compounds (tectoridin, iridin, tectorigenin, iristectorigenin B, iristectorigenin A, irigenin, irisflorentine, dichotomitin), BC 70% ethanol extract and ITM 70% ethanol extract were evaluated for their cytotoxicity against HepG2 and A549 cell lines using cell counting kit-8 (CCK-8) methods. Simultaneously, all standard compounds and ethanol extracts were evaluated for their hepatoprotective activities in BRL-3A and L02 cell lines as well as neuroprotective activities in BV2 cells in vitro.
Cell culture
HepG2, A549, BRL-3A, L02 and BV2 cell lines were obtained from Shenyang Pharmaceutical University. Cells were cultured in appropriate medium (DMEM or RPMI1640) supplemented with 10% FBS, 1% penicillin/streptomycin and 5% CO2 at 37 °C.
Toxicity measurements
The cytotoxicity against HepG2 and A549 cells of 8 compounds, BC and ITM ethanol extracts were determined by CCK-8 assay. Briefly, 1 × 104 cells were placed into each well of a 96-well plate and preincubated for 24 h in cell culture incubator (37 °C, 5% CO2). Secondly, cells were treated with different concentrations of 8 compounds (100–0.7813 μM), BC and ITM ethanol extracts (6400–25 μg mL−1) for 24 h, respectively. Then, cell viability was measured by cell counting kit-8 (CCK-8) assay (Dalian Meilun Biotechnology, Dalian, China) according to the manufacturer’s instructions. The absorbance values at 450 nm (OD450) were observed by Tecan Spark 10 K microplate reader (Tecan, Mannedorf, Switzerland). The viability of the control group is defined as 100%. Oxaliplatin was tested as a positive control.
Hepatoprotective and neuroprotective assay
Eight compounds and 2 ethanol extracts were evaluated for their hepatoprotective activities against d-galactosamine induced L02 cell injury in vitro. The neuroprotective effects were determined by BV2 cells. The cells were maintained in an appropriate medium (DMEM or RPMI1640, Dalian Meilun Biotechnology, Dalian, China) in a humidified atmosphere of 5% CO2 at 37 °C. The cells were seeded into 96-well plates at a density of 1 × 105 cells mL−1. After attachment, the cells were pretreated with the test compounds and extracts in different concentrations for 2 h. d-galactosamine (25 mM) or LPS solution (10 μg mL−1) was subsequently added for incubating another 24 h. Then, cell viability was measured by cell counting kit-8 (CCK-8) assay (Dalian Meilun Biotechnology, Dalian, China) according to the manufacturer’s instructions. The absorbance values at 450 nm (OD450) were observed by Tecan Spark 10 K microplate reader (Tecan, Mannedorf, Switzerland). The viability of the control group is defined as 100%. Bicyclol and curcumin was tested as a positive control.
Statistical analysis
All the data are presented as mean ± SD. The level of significance between the two groups was analyzed by Students’ test, more than two groups were assessed by one-way or two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison’s test. Statistical analysis was performed using SPSS 19.0 (IBM, Chicago, IL, USA). All the results were considered statistically significant at P < 0.05.
Results
Phytochemical analysis by LC–MS/MS in BC and ITM
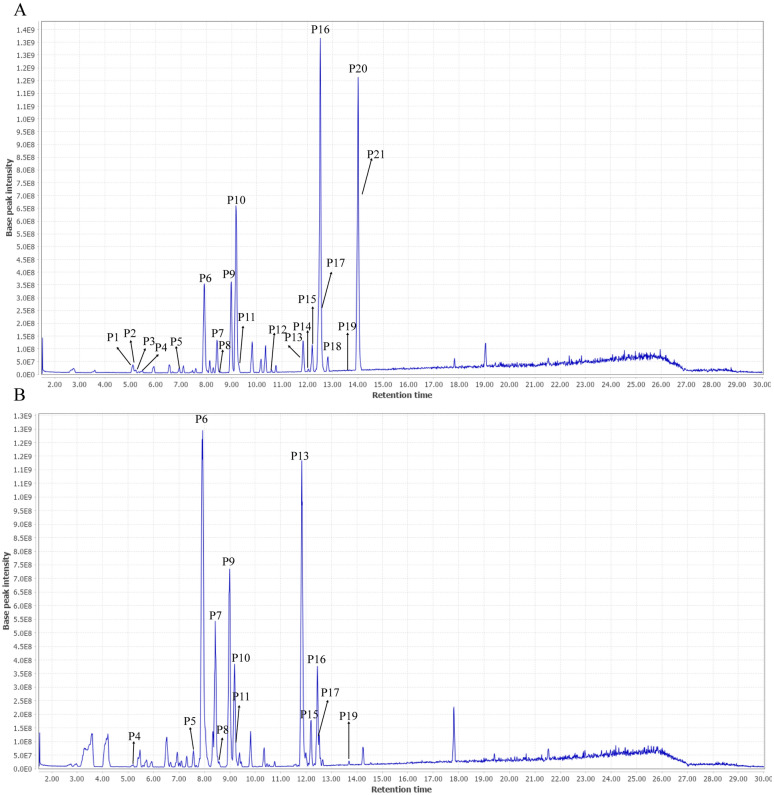
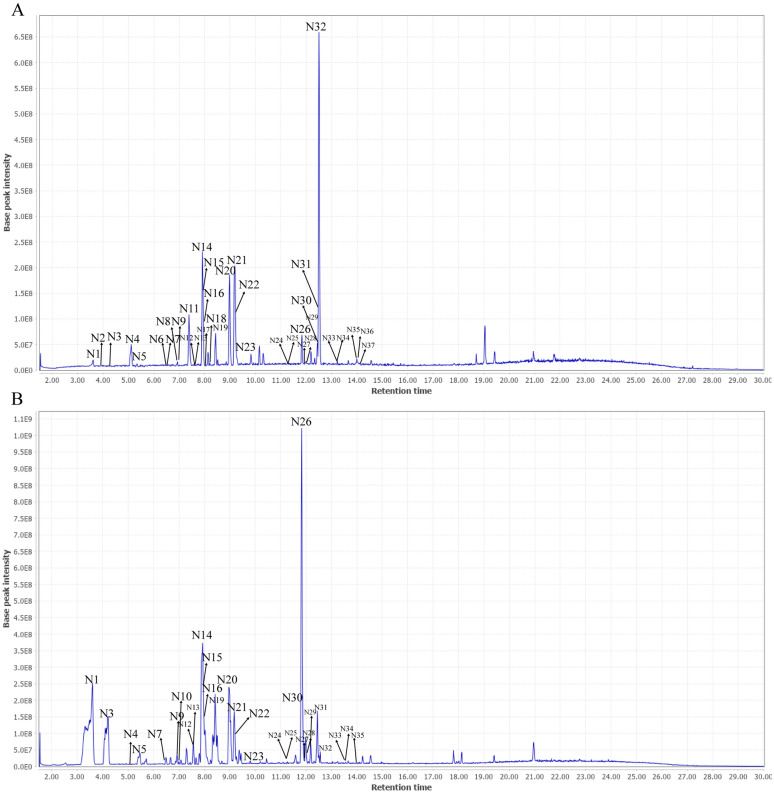
UHPLC-Q Exactive-MS/MS was conducted in both positive and negative ion mode to explore the phytochemical identification of BC and ITM extracts. The retention time, accurate molecular weight, quasi-molecular, formula and MS/MS fragments observed in the positive and negative mode were summarized in Tables 4 and 5. In present study, 40 compounds, including 25 isoflavone glycosides, 5 xanthones, 1 flavone and 9 other compounds were unambiguously or tentatively identified (Figs. 1, 2). By comparing with authentic standards and MS spectra, 10 compounds, including tectoridin, iristectorin A, iristectorin B, iridin, tectorigenin, iristectorigenin B, iristectorigenin A, irigenin, irisflorentine and dichotomitin were identified definitely, respectively (Additional file 1: Tables S1–S3).
Table 4.
Chemical characterization of B. chinensis (BC) and I. tectorum (ITM) by UHPLC-Q-exactive-MS/MS in positive ion
| Peaks | Retention time (min) | m/z Quasi-molecular [M + H]+ | m/z Calculated [M + H]+ | Error (ppm) | Formula | MS/MS fragments | Identification | Reference | Source |
|---|---|---|---|---|---|---|---|---|---|
| P1 | 5.13 | 423.0919 | 423.0922 | − 7.09 | C19H18O11 | 405, 333, 305, 303, 275 | Mangiferin | [14, 18–20] | BC, ITM |
| P2 | 5.35 | 423.0919 | 423.0922 | − 7.09 | C19H18O11 | 405, 333, 305, 303, 275 | Isomangiferin | [14, 18, 20] | BC, ITM |
| P3 | 5.35 | 437.1087 | 437.1078 | 20.59 | C20H20O11 | 315, 303, 301, 279, 227 | 7-O-Methylmangiferin | [14, 18] | BC |
| P4 | 5.49 | 625.1762 | 625.1763 | − 1.60 | C28H32O16 | 463, 340, 303, 279, 227 | Tectorigenin-7-O-glucosyl-4'-O-glucoside | [14, 18, 19, 21] | BC, ITM |
| P5 | 6.95 | 625.1760 | 625.1763 | − 4.80 | C28H32O16 | 540, 510, 463, 437, 315, 301 | Tectorigenin-7-O-β-glucosyl (1–6) glucoside | [14, 18, 20] | BC, ITM |
| P6 | 7.93 | 463.1232 | 463.1235 | − 6.48 | C22H22O11 | 301, 286 | Tectoridina | [14, 18–20] | BC, ITM |
| P7 | 8.45 | 493.1345 | 493.1341 | 8.11 | C23H24O12 | 463, 331, 316, 307, 229 | Iristectorin Aa | [14, 18, 20] | BC, ITM |
| P8 | 8.52 | 479.119 | 479.1184 | 12.52 | C22H22O12 | 331, 317, 287 | 3′-Hydroxytectoridin | [14, 18, 20] | BC, ITM |
| P9 | 8.99 | 493.1347 | 493.1341 | 12.17 | C23H24O12 | 331, 314, 279, 261, 199, 154 | Iristectorin Ba | [14, 18–20] | BC, ITM |
| P10 | 9.19 | 523.1381 | 523.1373 | 15.29 | C24H26O13 | 361, 340 | Iridina | [14, 18, 20] | BC, ITM |
| P11 | 9.19 | 523.1441 | 523.1446 | − 9.56 | C24H26O13 | 361, 340, 279, 227, 199, 154, 142 | Iridinisomer | [14, 18, 20] | BC, ITM |
| P12 | 10.6 | 535.1435 | 535.1446 | − 20.56 | C25H26O13 | 421, 377, 336, 315 | 3′,5′-Dimethoxyirisolone-4′-O-β-d-glucoside | [14, 18, 20] | BC |
| P13 | 11.84 | 301.0706 | 301.0707 | − 3.32 | C16H12O6 | 286, 231, 154, 142 | Tectorigenina | [14, 18, 20] | BC, ITM |
| P14 | 12.05 | 673.1777 | 673.1763 | 20.80 | C32H32O16 | 643, 615, 515, 361, 301 | 6′′-O-vanilloyliridin | [14, 18] | BC |
| P15 | 12.18 | 331.0798 | 331.0812 | − 42.29 | C17H14O7 | 316, 303, 279, 254, 234 | Iristectorigenin Ba | [14, 18–20] | BC, ITM |
| P16 | 12.43 | 331.0796 | 331.0812 | − 48.33 | C17H14O7 | 316, 303, 279, 254, 234 | Iristectorigenin Aa | [14, 18, 20] | BC, ITM |
| P17 | 12.51 | 361.0912 | 361.0918 | − 16.62 | C18H16O8 | 346, 331, 183 | Irigenina | [14, 18, 20] | BC, ITM |
| P18 | 12.82 | 373.0919 | 373.0918 | 2.68 | C19H16O8 | 361, 331, 301, 279, 226 | Noririsflorentin | [14, 18] | BC |
| P19 | 13.69 | 299.0549 | 299.055 | − 3.34 | C16H10O6 | 228, 199, 169, 154 | Irilone | [14, 18–19] | BC, ITM |
| P20 | 14.00 | 387.1071 | 387.1074 | − 7.75 | C20H18O8 | 359, 329, 262 | Irisflorentina | [14, 18–20] | BC |
| P21 | 14.06 | 359.0765 | 359.0761 | 11.14 | C18H14O8 | 329, 299, 271, 248, 223 | Dichotomitina | [14, 18–20] | BC |
aAuthentic standards
Table 5.
Chemical characterization of B. chinensis (BC) and I. tectorum (ITM) by UHPLC-Q-exactive-MS/MS in negative ion
| Peaks | Retention time (min) | m/z Quasi-molecular [M-H]− | m/z Calculated [M-H]− | Error (ppm) | Formula | MS/MS fragments | Identification | Reference | Source |
|---|---|---|---|---|---|---|---|---|---|
| N1 | 3.6 | 535.166 | 535.1668 | − 14.95 | C21H30O13 | 525, 489, 323, 235, 215 | Tectoruside | [3, 19] | BC, ITM |
| N2 | 3.96 | 583.1314 | 583.1305 | 15.43 | C25H28O16 | 535, 489, 403, 352, 307, 273 | Neomangiferin | [3, 18, 19] | BC |
| N3 | 4.23 | 373.1141 | 373.114 | 2.68 | C15H20O8 | 363, 273, 235, 215 | Androsin | [3, 19] | BC, ITM |
| N4 | 5.12 | 421.078 | 421.0776 | 9.50 | C19H18O11 | 364, 327, 307, 273, 235, 215 | Mangiferin | [3, 18, 19] | BC, ITM |
| N5 | 5.35 | 421.0779 | 421.0776 | 7.12 | C19H18O11 | 395, 333, 307, 273, 255, 235, 215 | Isomangiferin | [3, 18] | BC, ITM |
| N6 | 6.40 | 435.0923 | 435.0933 | − 22.98 | C20H20O11 | 377, 339, 307, 275, 235, 215 | 7-O-methylmangiferin | [3, 18] | BC |
| N7 | 6.50 | 447.0926 | 447.0933 | − 15.66 | C21H20O11 | 327, 313, 285, 235, 215 | Luteolin-6-C-β-d-glucoside | [3, 22] | BC, ITM |
| N8 | 6.93 | 435.0925 | 435.0933 | − 18.39 | C20H20O11 | 391, 352, 313, 275, 235, 215 | 7-O-methylisomangiferin | [3, 18] | BC |
| N9 | 6.95 | 623.1622 | 623.1618 | 6.42 | C28H32O16 | 567, 537, 435, 313, 299, 284, 235,215 | Tectorigenin-7-O-glucosyl-4′-O-glucoside | [3, 23] | BC, ITM |
| N10 | 7.11 | 463.1246 | 463.1246 | 0.00 | C22H24O11 | 327, 273, 235, 215 | Dihydrokaempferol-7-O-glucoside | [3, 23] | ITM |
| N11 | 7.37 | 653.1712 | 653.1723 | − 16.84 | C29H34O17 | 595, 509, 403, 329, 243 | Iristectorigenin-A-7-O-β-glucosyl (1 → 6)-glucoside | [3, 18, 19] | BC |
| N12 | 7.57 | 431.0973 | 431.0984 | − 25.52 | C21H20O10 | 431, 269, 235, 215 | Saponaretin | [3, 22] | BC, ITM |
| N13 | 7.57 | 477.1029 | 477.1038 | − 18.86 | C21H20O10 | 431, 269, 235 | Genistein-7-O-glucoside | [3, 24] | BC, ITM |
| N14 | 7.92 | 461.1087 | 461.1089 | − 4.34 | C22H22O11 | 299 | Tectoridina | [3, 18] | BC, ITM |
| N15 | 7.92 | 461.1087 | 461.1089 | − 4.34 | C22H22O11 | 413, 352, 329, 299, 284, 255 | Tectorigenin-4′-O-β-d-glucoside | [3, 18] | BC, ITM |
| N16 | 7.92 | 507.1147 | 507.1144 | 5.92 | C22H22O11 | 461, 299 | Isotectorigenin-7-O-β-d-glucoside | [3, 18] | BC, ITM |
| N17 | 8.01 | 593.1499 | 593.1512 | − 21.92 | C27H30O15 | 507, 461, 299 | Genistein-7-Ogentiobioside | [3, 25] | BC |
| N18 | 8.21 | 447.0925 | 447.0933 | − 17.89 | C21H20O11 | 352, 317, 273, 235, 215 | Orobol-7-O-d-glucoside | [3, 24] | BC |
| N19 | 8.43 | 491.1202 | 491.1195 | 14.25 | C23H24O12 | 329, 314, 227, 215 | Iristectorin Aa | [3, 18, 19] | BC, ITM |
| N20 | 8.99 | 491.1201 | 491.1195 | 12.22 | C23H24O12 | 329, 314, 227, 215 | Iristectorin Ba | [3, 18, 19] | BC, ITM |
| N21 | 9.19 | 521.1293 | 521.1301 | − 15.35 | C24H26O13 | 427, 359, 344 | Iridina | [3, 18] | BC, ITM |
| N22 | 9.19 | 567.1348 | 567.1355 | − 12.34 | C24H26O13 | 521, 507, 359, 344 | Isoiridin | [3, 18] | BC, ITM |
| N23 | 9.83 | 257.0815 | 257.0819 | − 15.56 | C15H14O4 | 246, 230, 215 | Gnetucleistol D | [3] | BC, ITM |
| N24 | 11.57 | 653.1708 | 653.1723 | − 22.96 | C29H34O17 | 326, 299, 269 | Iristectorin-B-4′-O-glucoside | [3, 18] | ITM |
| N25 | 11.68 | 519.113 | 519.1144 | − 26.97 | C24H24O13 | 326, 268, 230 | Dichotomitin-3′-O-glucoside | [3, 18, 19] | BC, ITM |
| N26 | 11.84 | 299.0557 | 299.0561 | − 13.38 | C16H12O6 | 284, 268, 242, 230, 215, 195 | Tectorigenina | [3, 18, 19] | BC, ITM |
| N27 | 12.02 | 641.151 | 641.1512 | − 3.12 | C31H30O15 | 459, 299 | 6′′-O-phydroxybenzoyliridin | [3, 18] | BC, ITM |
| N28 | 12.04 | 671.1599 | 671.1618 | − 28.31 | C32H32O16 | 641, 613, 359, 326, 299 | 6′′-O-vanilloyliridin | [3, 18] | BC, ITM |
| N29 | 12.18 | 329.0667 | 329.0667 | 0.00 | C17H14O7 | 315, 286, 268, 242 | Iristectorigenin Ba | [3, 18, 19] | BC, ITM |
| N30 | 12.43 | 329.0667 | 329.0667 | 0.00 | C17H14O7 | 315, 286, 268, 242, 198 | Iristectorigenin Aa | [3, 18] | BC, ITM |
| N31 | 12.46 | 301.0715 | 301.0718 | − 9.96 | C16H14O6 | 280, 273, 242, 215 | Dihydrokaempferide | [3, 23] | BC, ITM |
| N32 | 12.51 | 359.0763 | 359.0772 | − 25.06 | C18H16O8 | 344, 329, 181 | Irigenina | [3, 18] | BC, ITM |
| N33 | 13.66 | 297.0406 | 297.0405 | 3.37 | C16H10O6 | 280, 258, 230, 215 | Irilone | [3, 18, 19] | BC, ITM |
| N34 | 13.68 | 343.0822 | 343.0823 | − 2.91 | C18H16O7 | 318, 297, 280 | Dalspinosin | [3] | BC, ITM |
| N35 | 13.94 | 327.0511 | 327.051 | 3.06 | C17H12O7 | 294, 258, 230 | Iriflogenin | [3] | BC, ITM |
| N36 | 13.94 | 373.0919 | 373.0929 | − 26.80 | C19H18O8 | 336, 294, 280, 258, 230, 215 | Junipegenin C | [3, 18] | BC |
| N37 | 14.04 | 357.0607 | 357.0616 | − 25.21 | C18H14O8 | 294, 230 | Dichotomitina | [3, 18, 19] | BC |
aAuthentic standards
Fig. 1.
UHPLC-Q-Exactive MS base peak intensity (BPI) chromatogram of BC (A) and ITM (B) in positive ion mode
Fig. 2.
UHPLC-Q-Exactive MS BPI chromatogram of BC (A) and ITM (B) in negative ion mode
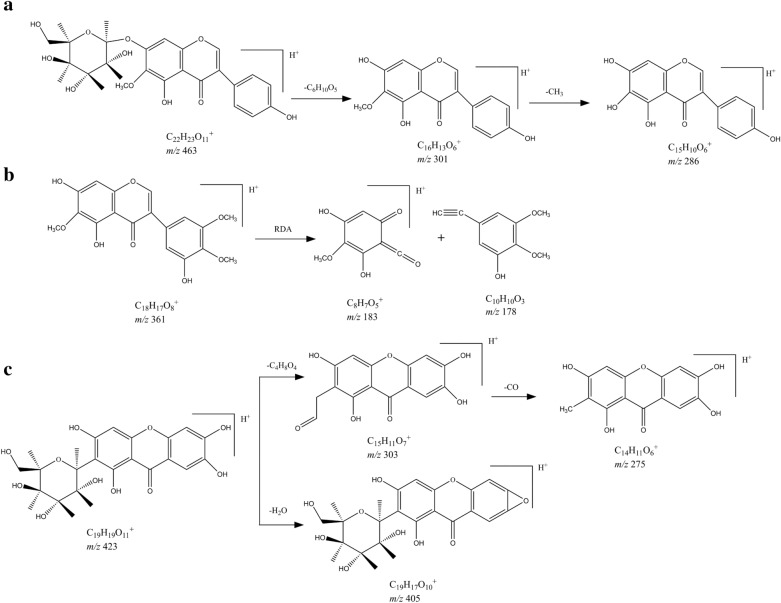
For the MS spectra of isoflavonoid glycosides, the neutral loss of 162 Da was demonstrated as the characteristic [M + H − 162]+ ion and [M − H − 162]− of O-glycosides such as P4/N9, P5, P6/N14, P7/N19, P8, P9/N20, P10/N21, N11, N15, N16, N22, N24 (Appendix Fig. 9a). Tectorigenin-7-O-β-glucosyl (1–6) glucoside and tectorigenin-7-O-glucosyl-4′-O-glucoside showed loss of 324 Da (-2glycosides), and generated m/z 301 or 299 ions corresponding to the [M + H − 2glycosyl]+ or [M − H − 2glycosyl]− fragments which was the same as [M + H]+ or [M − H]− ion of tectorigenin [3]. MS/MS spectra of irigenin showed successive loss of 15 Da, 30 Da and 178 Da, and generated m/z 346, 331 and 183 ions corresponding to the [M + H − CH3]+, [M + H − 2CH3]+ and [M + H − C10H10O3]+ fragments. The fragment ion at m/z 183 derived from irigenin Retro–Diels–Alder (RDA) fragmentation, which were observed in all the isoflavones and the most characteristic ion for these isoflavonoids [13]. This RDA fragmentation was shown in Appendix Fig. 9b. By the similar method, other isoflavone aglycones were also characterized.
Fig. 9.
The proposed fragmentation pathways. a Tectoridin fragmentation, b Irigenin Retro–Diels–Alder (RDA) fragmentation (m/z 361→183), c Xanthone mangiferin fragmentation
In this work, five xanthones including mangiferin, isomangiferin, 7-O-methylmangiferin, neomangiferin, 7-O-methylisomangiferin were identified in the extracts of BC and ITM. Xanthones showed [M + H − 120]+ and [M − H − 120]− fragment ions in MS/MS spectra, which were typical of xanthone C-glucosides [18]. MS/MS spectra of mangiferin showed successive loss of 18 Da and 120 Da, and generated m/z 405 and 303 ions corresponding to the [M + H − H2O]+ and [M + H − C4H8O4]+ fragments (Appendix Fig. 9c). P2 was tentatively assigned as an isomer of mangiferin, isomangiferin. By the similar method, neomangiferin were characterized. Compounds 7-O-methylmangiferin and 7-O-methylisomangiferin showed similar fragmentation behaviors as that of mangiferin, thus they could be deduced as derivatives of mangiferin. The [M − H]− ions of 7-O-methylmangiferin and 7-O-methylisomangiferin were both observed at m/z 435, 14 Da more than that of mangiferin. It could be presumed that a hydroxyl group was replaced by a methoxy group in structures of 7-O-methylmangiferin and 7-O-methylisomangiferin [18]. This fragmentation was shown in Appendix Fig. 9.
BC and ITM in LC‑HR/MS metabolomics
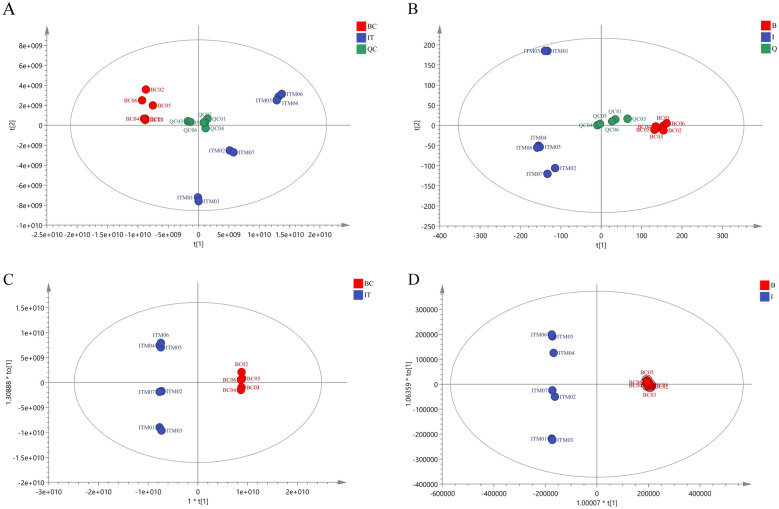
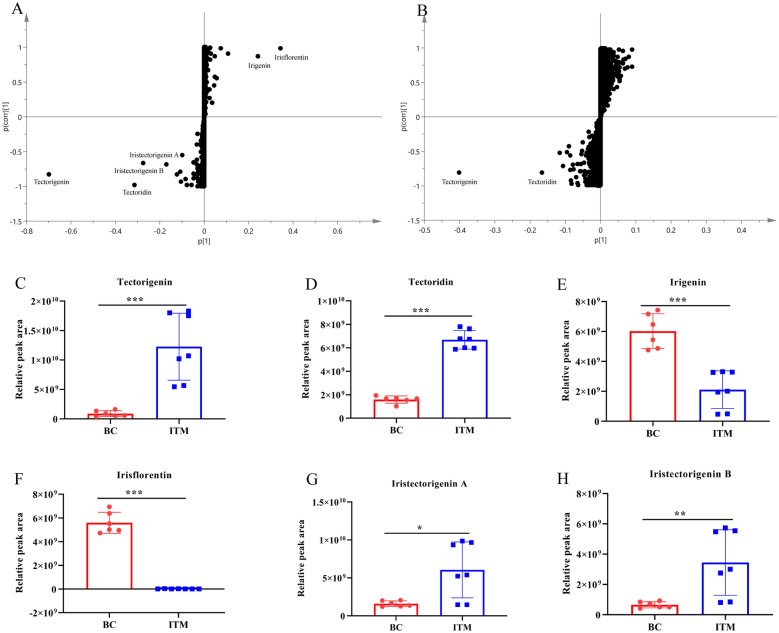
PCA was carried out using the resultant data matrix. BC (red circles) and ITM (blue circles) samples were roughly separated on the PC1 (70.2%) vs PC2 (11.5%) plane (Fig. 3A) in positive ion and the PC1 (27.6%) vs PC2 (10.3%) plane (Fig. 3B) in negative ion. Simultaneously, we performed OPLS-DA to search for components that could help to distinguish BC from ITM. The OPLS-DA projection models had a clearer separation of the BC and ITM samples in positive and negative ion (Fig. 3C–D). Furthermore, the s-plot indicated the following characteristic peaks: m/z 301.0706 (tectorigenin), m/z 463.1232 (tectoridin), m/z 361.0912 (irigenin), m/z 387.1071 (irisflorentin), m/z 331.0798 (iristectorigenin A) and m/z 331.0796 (iristectorigenin B) (Fig. 4A, B). These remarkable variables of chemical constitute were major phytochemical-marker between BC and ITM (Fig. 4C–H). These components were at the edge of the vertical and horizontal axes, and were explanatory variables that strongly contributed to the objective variable. The multivariate chemometric analysis demonstrated that the remarkable differences of chemical constitute between BC and ITM.
Fig. 3.
Score plots of PCA in positive ion (A) and negative ion (B). Score plots of OPLS-DA in positive ion (C) and negative ion (D) for LC-HR/MS metabolomics. Red circles: B. chineses; blue circles: I. tectorum; green circles: QC sample
Fig. 4.
S-plots of OPLS-DA in positive ion (A) and negative ion (B) for LC-HR/MS metabolomics. (C-H) The remarkable variable of chemical constitutes
Fingerprint analysis of BC and ITM by DRS study
Optimization of HPLC conditions and method validation
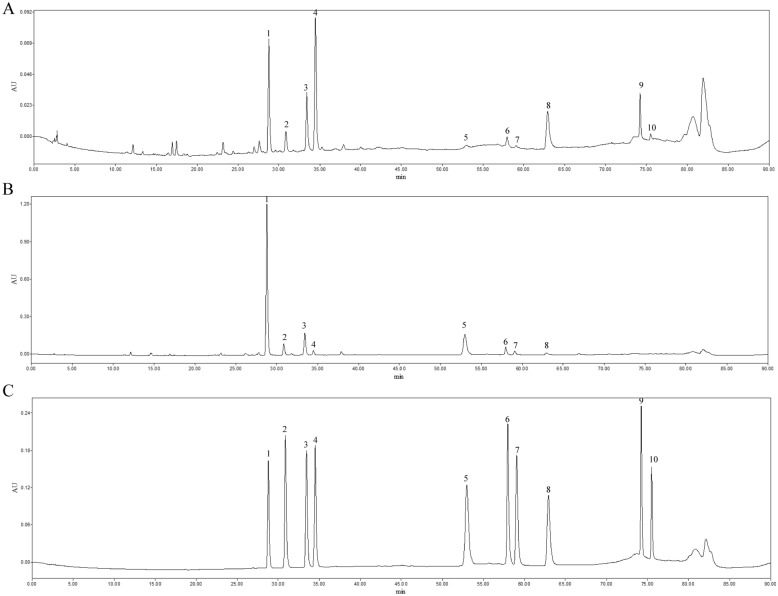
The mobile phase, gradient elution procedures, detection wavelength and flow rates were optimized. The selected chromatographic conditions had satisfactory peak shape and resolution between peaks. Representative chromatograms and spectra were shown in Fig. 5. The peaks were identified by UV spectra and retention time (Appendix Fig. 8).
Fig. 5.
Representative HPLC chromatogram of sample on Column 12 (Exsil Mono 100 C18). A BC sample, B ITM sample, C Chromatograms of mixed standard. Tectoridin (1), Iristectorin A (2), Iristectorin B (3), Iridin (4), Tectorigenin (5), Iristectorigenin B (6), Iristectorigenin A (7), Irigenin (8), Irisflorentine (9), Dichotomitin (10)
Methodological validation experiments were performed on column 14 (Inertsil ODS-3 C18). The precision (n = 6), stability (48 h, n = 9), and repeatability (n = 6) were tested. The results showed that RSD of the peaks tR and peak areas were both less than 2%, thus meeting the requirements of fingerprint analysis.
Initialization for the DRS method
Details of the operating principle and applications of LCTRS were well documented in previous our literature [10–12]. In brief, the LCTRS method consisted of several steps, including data importing, peak assignment, setting the qualitative chromatographic method.
Optimization and evaluation of DRS method
Retention time prediction by LCTRS method
In our study, the StR values compounds, as a reference value for retention time prediction, were determined by the arithmetic average of the retention times on nineteen columns [10–12]. In B. chineses, we had identified 10 compounds by standard, including tectoridin, iristectorin A, iristectorin B, iridin, tectorigenin, iristectorigenin B, iristectorigenin A, irigenin, irisflorentine and dichotomitin. Reference compounds selection is very important for the qualitative analysis in the substitute methods. According to DRS method principle, the tectoridin and dichotomitin were selected as two reference compounds for LCTRS method. The actual retention times of the 10 compounds on different columns and chromatographic instruments showed good linear relationships with their StR (Table 6). Meanwhile, in I. tectorum, we also had identified 8 compounds by standard, including tectoridin, iristectorin A, iristectorin B, iridin, tectorigenin, iristectorigenin B, iristectorigenin A and irigenin. The tectoridin and irigenin were selected as two reference compounds for LCTRS method in I. tectorum, and the linear fitting results were shown in Table 6.
Table 6.
Linear fitting results of actual retention times by LCTRS method for BC and ITM
| No. | BC | ITM | ||
|---|---|---|---|---|
| Calibration curve | R2 | Calibration curve | R2 | |
| Column 1 | Y = 1.0425X − 3.9978 | 0.9995 | Y = 1.0285X − 3.4495 | 0.9991 |
| Column 2 | Y = 0.9660X + 2.0604 | 0.9995 | Y = 0.9480X + 2.6624 | 0.9996 |
| Column 3 | Y = 0.9967X − 0.9580 | 0.9994 | Y = 0.9740X + 0.0858 | 0.9999 |
| Column 4 | Y = 0.9985X − 1.4591 | 0.9989 | Y = 0.9686X − 0.3842 | 0.9994 |
| Column 5 | Y = 1.0189X − 4.5116 | 0.9945 | Y = 0.9522X − 1.7815 | 0.9968 |
| Column 6 | Y = 0.9961X + 4.0136 | 0.9931 | Y = 1.0885X + 0.7441 | 0.9954 |
| Column 7 | Y = 1.0128X − 0.3540 | 0.9997 | Y = 1.0260X-0.9166 | 0.9997 |
| Column 8 | Y = 0.9829X + 1.1290 | 0.9998 | Y = 0.9740X + 1.4514 | 0.9996 |
| Column 9 | Y = 0.9851X + 3.4759 | 0.9972 | Y = 1.0386X + 1.5402 | 0.9983 |
| Column 10 | Y = 1.0075X − 1.0370 | 0.9999 | Y = 0.9978X − 0.6472 | 0.9999 |
| Column 11 | Y = 0.9711X-1.1506 | 0.9929 | Y = 0.8989X + 1.6232 | 0.9960 |
| Column 12 | Y = 1.0107X − 4.1726 | 0.9939 | Y = 0.93393X − 1.2438 | 0.9971 |
| Column 13 | Y = 0.9632X + 3.0443 | 0.9997 | Y = 0.9688X + 2.8581 | 0.9994 |
| Column 14 | Y = 0.9707X + 5.8023 | 0.9929 | Y = 1.0602X + 2.8936 | 0.9959 |
| Column 15 | Y = 0.9838X + 4.0523 | 0.9952 | Y = 1.0649X + 1.4692 | 0.9964 |
| Column 16 | Y = 1.0038X + 0.5351 | 0.9985 | Y = 1.0407X − 0.8421 | 0.9987 |
| Column 17 | Y = 1.0233X − 2.1767 | 0.9999 | Y = 1.0165X − 2.0256 | 0.9998 |
| Column 18 | Y = 0.9947X + 0.7155 | 0.9996 | Y = 1.0065X + 0.2546 | 0.9994 |
| Column 19 | Y = 1.0340X − 3.6999 | 0.9995 | Y = 1.0217X − 3.1698 | 0.9991 |
Retention time prediction by RRT method
The RRT method was used the single standard to identify chromatographic peaks. In B. chineses, we selected irigenin as the reference compound because of its appropriate retention time and availability. Meanwhile, in I. tectorum, we chose iristectorigenin B as the reference compound. The RRT of ten (BC) and eight (ITM) analytes relative to irigenin and iristectorigenin B were calculated by the arithmetic average of the RRTs on nineteen columns.
Comparison between LCTRS and RRT method
To evaluate the advantages and disadvantages of LCTRS and RRT method, we then calculated the absolute deviations (ΔtR) of the actual retention time and predicted retention time on nineteen columns (Additional file 1: Tables S4–S7). As shown in Table 7, the RRT method had a larger average deviation, a lower identification rate and available column amount than LCTRS. The above results showed that LCTRS had a precise, feasible, and superior to identify peaks than RRT.
Table 7.
Comparison of different methods (19 columns for method establishment)
| Method | Average tR deviation/min | Identification rate/% (ΔtR ≤ 1.5 min) | Available column amount (ΔtR ≤ 1.5 min) |
|---|---|---|---|
| RRT (BC) | 1.07 | 75.44 | 8 |
| LCTRS (BC) | 0.77 | 82.89 | 11 |
| RRT (ITM) | 0.72 | 84.21 | 10 |
| LCTRS (ITM) | 0.45 | 94.74 | 13 |
Sample tests
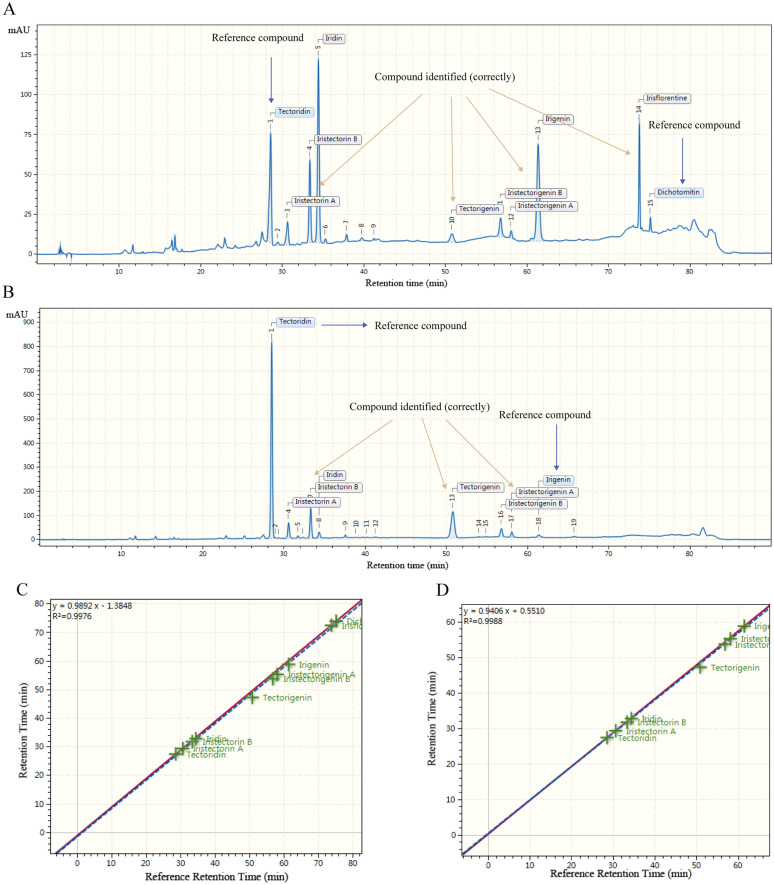
To further verify the reliability of our method LCTRS, we chose SVEA C18 Opal column for sample testing. The detailed procedure of LCTRS method was described in our previous literature [10–12]. In brief, the LCTRS sample tests consisted of three steps, including data integrated, reference compounds assigned and the results obtained. The sample test results were exhibited in Fig. 6, which included the qualitative results of peaks and linear fitting results (Fig. 6).
Fig. 6.
Results of sample tests on column 20 [SVEA C18 Opal]. A The result of the LCTRS method of BC. B The result of the LCTRS method of ITM. C The result of peaks and linear fitting results of BC. D The result of peaks and linear fitting results of ITM
Toxicity and protective analysis
The cytotoxicity of 8 compounds (0.78–100 μM) and the extracts (25–6400 μg mL−1) was evaluated against HepG2 and A549 cells, respectively (Table 8). Among them, irigenin showed significantly inhibiting activity on the HepG2 cells with IC50 values of 18.66 µM and iristectorigenin B showed significantly inhibiting activity on the A549 cells with IC50 values of 35.21 µM (Table 8). Compared with BC extracts, ITM extracts showed strongly inhibitory activity against HepG2 and A549 cells with IC50 values of 31.41 μg mL−1 and 147.40 μg mL−1, respectively (Table 8).
Table 8.
The toxicity of 8 compounds, BC extracts, ITM extracts and positive drug in HepG2 and A549 cells
| Concentration (μM/μg mL−1) | HepG2 (IC50 ± SD) | A549(IC50 ± SD) |
|---|---|---|
| Oxaliplatin | 6.02 ± 0.23 | 13.13 ± 0.16 |
| Tectoridin | 135.80 ± 0.63 | 83.98 ± 0.02 |
| Iridin | 126.20 ± 0.07 | 102.90 ± 0.50 |
| Tectorigenin | 60.28 ± 0.34 | 68.44 ± 0.33 |
| Iristectorigenin B | 38.68 ± 0.28 | 35.21 ± 0.22 |
| Iristectorigenin A | 44.12 ± 0.66 | 42.06 ± 0.02 |
| Irigenin | 18.66 ± 0.11 | 47.63 ± 0.44 |
| Irisflorentine | 155.70 ± 0.17 | 37.23 ± 0.28 |
| Dichotomitin | 161.80 ± 0.63 | 62.01 ± 0.11 |
| BC extracts | 31.41 ± 0.48 | 147.40 ± 0.57 |
| ITM extracts | 68.94 ± 0.12 | 366.60 ± 0.56 |
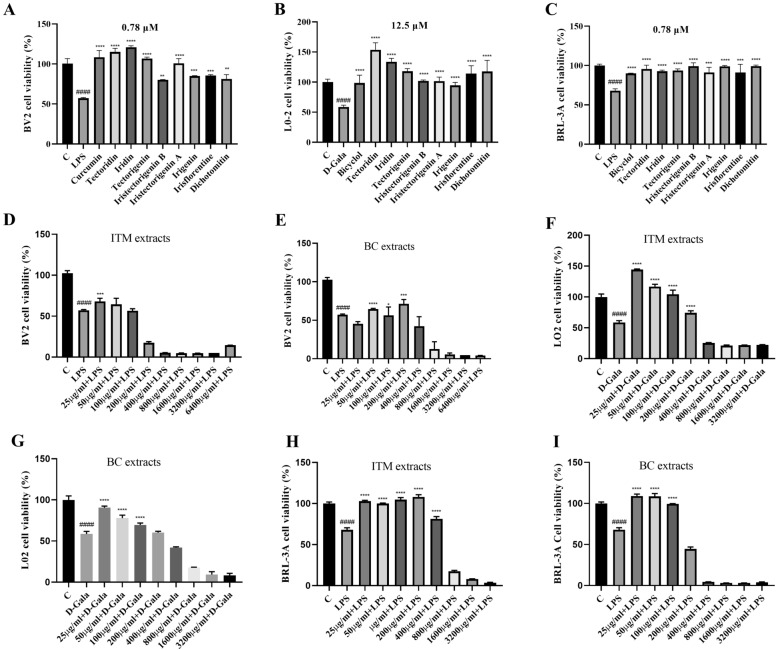
To further investigate their neuroprotective and hepatoprotective, BV2 microglial cells, L02 and BRL-3A cells were treated with 8 compounds and 2 extracts (Additional file 1: Table S8). Results showed that Tectoridin and iridin at concentration of 0.75 μM exhibited higher neuroprotective activity than the positive control, curcumin (Fig. 7A, Additional file 1: Table S8). However, BC extracts and ITM extracts showed no neuroprotective effects on BV2 cells (Fig. 7D–E). Tectoridin and iridin at concentration of 12.5 μM showed more potent hepatoprotective activity than bicyclol (positive control) on L02 cells (Fig. 7B, Additional file 1: Table S8). Compared with BC extracts, ITM extracts showed more potent neuroprotective activity on L02 cells (Fig. 7F, G). Iristectorigenin B, tectoridin and irigenin showed more potent hepatoprotective activity than bicyclol on BRL-3A cells (Fig. 7C, Additional file 1: Table S8). ITM extracts showed more potent hepatoprotective activity on BRL-3A cells than BC extracts (Fig. 7H, I).
Fig. 7.
The neuroprotective and hepatoprotective of 8 compounds, BC extracts, ITM extracts and positive drug. A–C The neuroprotective and hepatoprotective of 8 compounds. D–E The neuroprotective of ITM and BC extracts in BV2 cell. F, G The hepatoprotective of ITM and BC extracts in L02 cell. H, I The hepatoprotective of ITM and BC extracts in BRL-3A cell. Cell viability was measured with a CCK-8 assay. The data are expressed as the percentage of the relative untreated control cells. All values are expressed as the mean ± SD. ####P < 0.0001 versus the control group. **P < 0.01, ***P < 0.001, ****P < 0.0001 versus the model group
Discussions
In our work, the plant metabolomics, digital reference standard, as well as in vitro activity assay strategy were used to evaluate the chemical constituents, qualities and biological activities of BC and ITM. In the multivariate analysis, the PCA and OPLS-DA score plot indicated the obvious differences in chemical profiling between BC and ITM. Moreover, it showed that 6 principal compounds were successfully identified to contribute to the differences in chemical profiling between BC and ITM.
In digital reference standard study, a series of quality control methods of fingerprints in BC and ITM were developed based on the DRS analyzer, involving the RRT method, LCTRS method. In BC, the tectoridin and dichotomitin were selected as two reference compounds for LCTRS method. In ITM, the tectoridin and irigenin were selected as two reference compounds for LCTRS method. The digital reference standard strategy significantly reduced the analysis cost, saved time and improved the multicomponent analysis efficiency of the analysis method.
In biological activities assay, BC has better anticancer activity than ITM due to its high abundance of irigenin. In contrast, the hepatoprotective activity of ITM was higher than that of BC because of the high abundance of tectoridin. Unfortunately, neither BC nor ITM showed good neuroprotective activity in BV2 cells.
Conclusions
In summary, based on multidimensional strategy, it was indicated that B. chinensis and I. dichotoma were significantly different in their chemical constituents and biological activities. The results not only showed that these two medicinal plants could not be mixed in clinical medication, but also provided a novel multidimensional strategy for the identification of easily confused, easily adulterated, and even counterfeit medicinal materials.
Supplementary Information
Additional file 1: Table S1. Structures of all isoflavones found in Belamcanda chinensis (L) DC and Iris tectorum Maxim. Me: CH3, Glu: glucose. Table S2. Structures of isoflavones from Belamcanda chinensis (L) DC and Iris tectorum Maximwith additional dioxolane ring. Me: CH3, Glc: glucose. Table S3. Structures of xanthonesfrom Belamcanda chinensis (L) DC and Iris tectorum Maximwith. Me: CH3, Glu: glucose. Table S4. The absolute deviations (ΔtR) of the actual retention time and predicted retention time on nineteen columns in BC LCTRS method. Table S5. The absolute deviations (ΔtR) of the actual retention time and predicted retention time on nineteen columns in BC RRT method. Table S6. The absolute deviations (ΔtR) of the actual retention time and predicted retention time on nineteen columns in ITM LCTRS method. Table S7. The absolute deviations (ΔtR) of the actual retention time and predicted retention time on nineteen columns in ITM RRT method. Table S8. Neuroprotective and hepatoprotective effects of compounds and positive drug.
Acknowledgements
Not applicable.
Abbreviations
- BC
Belamcanda chinensis (L.) DC
- ITM
Iris tectorum Maxim
- DRS
Digital reference standard
- RRT
Relative retention time
- LCTRS
Linear calibration using two reference substances
- TCM
Traditional Chinese medicine
- UHPLC-HRMS/MS
Ultra-high-performance liquid chromatography-high resolution tandem mass spectrometry
- Me-β-CD
Methyl-β-cyclodextrin
- PCA
Principal components analysis
- OPLS-DA
Orthogonal partial least squares-discriminant analysis
- CCK-8
Cell counting kit-8
Appendix
Authors’ contributions
HXZ, DLM, YZ, SCM, LS conceived and designed the experiments. HXZ, LL, JZ, JMZ, HJS, HL performed the experiments. HXZ, LL analyzed and interpreted the data. HXZ, DLM wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work is supported by the National Natural Science Foundation of China (Grant No. 81573694), and the National Drug Standard Improvement Project in 2021 (General Technology) (Grant No. 2021Z01). The project is also sponsored by “Liaoning BaiQianWan Talents Program in 2018”.
Availability of data and materials
All data are fully available without restriction.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hongxu Zhou and Yi Zhang contributed equally
Contributor Information
Shuangcheng Ma, Email: masc@nifdc.org.cn.
Dali Meng, Email: mengdali@syphu.edu.cn.
References
- 1.Woźniak D, Matkowski A. Belamcandae chinensis rhizome—a review of phytochemistry and bioactivity. Fitoterapia. 2015;107:1–14. doi: 10.1016/j.fitote.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y, Wu CM, Dai RJ, Li L, Yu YH, Li Y, Meng WW, Zhang L, Zhang Y, Deng YL. Combination of HPLC chromatogram and hypoglycemic effect identifies isoflavones as the principal active fraction of Belamcanda chinensis leaf extract in diabetes treatment. J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879:371–378. doi: 10.1016/j.jchromb.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 3.Xie GY, Zhu Y, Shu P, Qin XY, Wu G, Wang Q, Qin MJ. Phenolic metabolite profiles and antioxidants assay of three Iridaceae medicinal plants for traditional Chinese medicine “She-gan” by on-line HPLC-DAD coupled with chemiluminescence (CL) and ESI-Q-TOF-MS/MS. J Pharm Biomed Anal. 2014;98:40–51. doi: 10.1016/j.jpba.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Wu C, Li Y, Chen Y, Lao X, Sheng L, Dai R, Meng W, Deng Y. Hypoglycemic effect of Belamcanda chinensis leaf extract in normal and STZ-induced diabetic rats and its potential active faction. Phytomedicine. 2011;18:292–297. doi: 10.1016/j.phymed.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Ito H, Onoue S, Yoshida T. Isoflavonoids from Belamcanda chinensis. Chem Pharm Bull. 2001;49:1229–1231. doi: 10.1248/cpb.49.1229. [DOI] [PubMed] [Google Scholar]
- 6.Morrissey C, Bektic J, Spengler B, Galvin D, Christoffel V, Klocker H, Fitzpatrick JM, Watson RW. Phytoestrogens derived from Belamcanda chinensis have an antiproliferative effect on prostate cancer cells in vitro. J Urol. 2004;172:2426–2433. doi: 10.1097/01.ju.0000143537.86596.66. [DOI] [PubMed] [Google Scholar]
- 7.Lee HU, Bae EA, Kim DH. Hepatoprotective effect of tectoridin and tectorigenin on tert-butyl hyperoxide-induced liver injury. J Pharmacol Sci. 2005;97:541–544. doi: 10.1254/jphs.SCZ040467. [DOI] [PubMed] [Google Scholar]
- 8.Xiong Y, Yang Y, Yang J, Chai H, Li Y, Yang J, Jia Z, Wang Z. Tectoridin, an isoflavone glycoside from the flower of Pueraria lobata, prevents acute ethanol-induced liver steatosis in mice. Toxicology. 2010;276:64–72. doi: 10.1016/j.tox.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 9.PerezdeSouza L, Alseekh S, Naake T, Fernie A. Mass spectrometry-based untargeted plant metabolomics. Curr Protoc Plant Biol. 2019;19(4):e20100. doi: 10.1002/cppb.20100. [DOI] [PubMed] [Google Scholar]
- 10.Wang Q, Yu X, Sun L, Tian R, He H, Wang S, Ma S. Fingerprint analysis of phenolic acid extract of Salvia miltiorrhiza by digital reference standard analyzer with one or two reference standards. Chin Med. 2021;16:8. doi: 10.1186/s13020-020-00408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun L, Jin HY, Tian RT, Wang MJ, Liu LN, Ye LP, Zuo TT, Ma SC. A simple method for HPLC retention time prediction: linear calibration using two reference substances. Chin Med. 2017;12:16–27. doi: 10.1186/s13020-017-0137-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen AZ, Sun L, Yuan H, Wu AY, Lu JG, Ma SC. Simultaneous qualitative and quantitative analysis of 11 active compounds in rhubarb using two reference substances by UHPLC. J Sep Sci. 2018;41:3686–3696. doi: 10.1002/jssc.201800479. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Li WZ, Huang W, Cheung AW, Bi CW, Duan R, Guo AJ, Dong TT, Tsim KW. Quality evaluation of Rhizoma Belamcandae (Belamcanda chinensis (L.) DC.) by using high-performance liquid chromatography coupled with diode array detector and mass spectrometry. J Chromatogr A. 2009;1216:2071–2078. doi: 10.1016/j.chroma.2008.05.082. [DOI] [PubMed] [Google Scholar]
- 14.Chen YJ, Liang ZT, Zhu Y, Xie GY, Tian M, Zhao ZZ, Qin MJ. Tissue-specific metabolites profiling and quantitative analyses of flavonoids in the rhizome of Belamcanda chinensis by combining laser-microdissection with UHPLC-Q/TOF-MS and UHPLC-QqQ-MS. Talanta. 2014;130:585–597. doi: 10.1016/j.talanta.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Wen Y, He L, Peng R, Lin Y, Zhao L, Li X, Ye L, Yang J. A novel strategy to evaluate the quality of herbal products based on the chemical profiling, efficacy evaluation and pharmacokinetics. J Pharm Biomed Anal. 2018;161:326–335. doi: 10.1016/j.jpba.2018.08.047. [DOI] [PubMed] [Google Scholar]
- 16.Jiang H, Yue QI, Zou G, Guoxin LI. Separation of isomer and similar structure compounds in Belamcandae Rhizoma by methyl-β-CD chiral mobile phase additive HPLC. Chin J Mod Appl Pharm. 2017;34:1–3. [Google Scholar]
- 17.Forsberg EM, Huan T, Rinehart D, Benton HP, Warth B, Hilmers B, Siuzdak G. Data processing, multi-omic pathway mapping, and metabolite activity analysis using XCMS online. Nat Protoc. 2018;13:633–651. doi: 10.1038/nprot.2017.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang YY, Wang Q, Qi LW, Qin XY, Qin MJ. Characterization and determination of the major constituents in Belamcandae Rhizoma by HPLC-DAD-ESI-MS(n) J Pharm Biomed Anal. 2011;56:304–314. doi: 10.1016/j.jpba.2011.05.040. [DOI] [PubMed] [Google Scholar]
- 19.Wei Y, Shu P, Hong J, Qin M. Qualitative and quantitative evaluation of phenolic compounds in Iris dichotoma Pall. Phytochem Anal. 2012;23:197–207. doi: 10.1002/pca.1343. [DOI] [PubMed] [Google Scholar]
- 20.Ito H, Nishitani E, Konoshima T, Takasaki M, Kozuka M, Yoshida T. Flavonoid and benzophenone glycosides from Coleogyne ramosissima. Phytochemistry. 2000;4:695–700. doi: 10.1016/S0031-9422(00)00192-8. [DOI] [PubMed] [Google Scholar]
- 21.Ablajan K. A study of characteristic fragmentation of isoflavonoids by using negative ion ESI-MSn. J Mass Spectrom. 2011;46:77–84. doi: 10.1002/jms.1867. [DOI] [PubMed] [Google Scholar]
- 22.Liu S, Yan J, Xing J, Song F, Liu Z, Liu S. Characterization of compounds and potential neuraminidase inhibitors from the n-butanol extract of compound Indigowoad Root Granule using ultrafiltration and liquid chromatography-tandem mass spectrometry. J Pharm Biomed Anal. 2012;59:96–101. doi: 10.1016/j.jpba.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 23.Shu P, Hong JL, Gang WU, Yang YB, Qin MJ. Analysis of flavonoids and phenolic acids in Iris tectorum by HPLC-DAD-ESI-MS~n. Chin J Nat Med. 2010;8:202–207. doi: 10.3724/SP.J.1009.2010.00202. [DOI] [Google Scholar]
- 24.Duenas M, Hernandez T, Estrella I, Fernandez D. Germination as a process to increase the polyphenol content and antioxidant activity of lupin seeds (Lupinus angustifolius L.) Food Chem. 2010;117:599–607. doi: 10.1016/j.foodchem.2009.04.051. [DOI] [Google Scholar]
- 25.Kachlicki P, Einhorn J, Muth D, Kerhoas L, Stobiecki M. Evaluation of glycosylation and malonylation patterns in flavonoid glycosides during LC/MS/MS metabolite profiling. J Mass Spectrom. 2008;43:572–586. doi: 10.1002/jms.1344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Structures of all isoflavones found in Belamcanda chinensis (L) DC and Iris tectorum Maxim. Me: CH3, Glu: glucose. Table S2. Structures of isoflavones from Belamcanda chinensis (L) DC and Iris tectorum Maximwith additional dioxolane ring. Me: CH3, Glc: glucose. Table S3. Structures of xanthonesfrom Belamcanda chinensis (L) DC and Iris tectorum Maximwith. Me: CH3, Glu: glucose. Table S4. The absolute deviations (ΔtR) of the actual retention time and predicted retention time on nineteen columns in BC LCTRS method. Table S5. The absolute deviations (ΔtR) of the actual retention time and predicted retention time on nineteen columns in BC RRT method. Table S6. The absolute deviations (ΔtR) of the actual retention time and predicted retention time on nineteen columns in ITM LCTRS method. Table S7. The absolute deviations (ΔtR) of the actual retention time and predicted retention time on nineteen columns in ITM RRT method. Table S8. Neuroprotective and hepatoprotective effects of compounds and positive drug.
Data Availability Statement
All data are fully available without restriction.