Abstract
Production of ribosomal protein S14 in Saccharomyces cerevisiae is coordinated with the rate of ribosome assembly by a feedback mechanism that represses expression of RPS14B. Three-hybrid assays in vivo and filter binding assays in vitro demonstrate that rpS14 directly binds to an RNA stem-loop structure in RPS14B pre-mRNA that is necessary for RPS14B regulation. Moreover, rpS14 binds to a conserved helix in 18S rRNA with approximately five- to sixfold-greater affinity. These results support the model that RPS14B regulation is mediated by direct binding of rpS14 either to its pre-mRNA or to rRNA. Investigation of these interactions with the three-hybrid system reveals two regions of rpS14 that are involved in RNA recognition. D52G and E55G mutations in rpS14 alter the specificity of rpS14 for RNA, as indicated by increased affinity for RPS14B RNA but reduced affinity for the rRNA target. Deletion of the C terminus of rpS14, where multiple antibiotic resistance mutations map, prevents binding of rpS14 to RNA and production of functional 40S subunits. The emetine-resistant protein, rpS14-EmRR, which contains two mutations near the C terminus of rpS14, does not bind either RNA target in the three-hybrid or in vitro assays. This is the first direct demonstration that an antibiotic resistance mutation alters binding of an r protein to rRNA and is consistent with the hypothesis that antibiotic resistance mutations can result from local alterations in rRNA structure.
The complexity and abundance of ribosomes necessitate the coordinate regulation of a large group of genes to avoid unnecessary investments of cellular energy in the production of excess components. In Saccharomyces cerevisiae, 78 different ribosomal proteins (r proteins) and 4 ribosomal RNAs are synthesized in nearly equimolar amounts (reviewed in reference 67). Because so much energy is invested in ribosome assembly, small adjustments to the rate of ribosome assembly or even the production of individual ribosomal components can be advantageous to the cell (31).
The coordinate regulation of ribosomal protein genes in Escherichia coli occurs by autogenous regulation (reviewed in references 11, 42, and 68). A subset of unassembled r proteins inhibits the expression of their own operons by exploiting their RNA binding capacity. It is generally assumed that these r proteins must bind preferentially to their rRNA target rather than to the corresponding mRNA binding site to allow repression only in the absence of assembly targets.
Fewer examples of feedback regulation are known in eukaryotes; only a few genes have been studied in detail (66). Three different yeast r protein genes are subjected to feedback control; L32 regulates the splicing and translation of its message (9, 15), rpL4 [L2] stimulates the degradation of its transcripts (46, 47), and rpS14 is thought to repress RPS14B [CRY2] expression posttranscriptionally (33). Homologs of two of these genes are also regulated in higher eukaryotes. The Xenopus laevis homolog of L4 is autogenously regulated at the level of splicing (3, 6), and the transcription of the mammalian RPS14 gene is repressed by unassembled protein (57). However, direct binding of the r protein to its messenger RNA target has been demonstrated in only two of these examples; yeast rpL32 binds directly to its pre-mRNA and mRNA (63), and mammalian S14 binds to its message and to antisense RNAs involved in its regulation (57). In neither of these cases has a direct interaction been demonstrated between the r protein and both mRNA and rRNA targets.
The RPS14B [CRY1] and RPS14B [CRY2] genes of S. cerevisiae are unlinked, duplicated genes that encode the essential 40S ribosomal subunit protein rpS14 (30, 43). Mutations in the last codon of either of these genes confer resistance to the translation inhibitor cryptopleurine (43). Similarly, mutations in two arginines at the C terminus of the mammalian homolog of RPS14 confer resistance to emetine (34). These inhibitors block protein synthesis by binding to a high-affinity site on the 40S ribosomal subunit and preventing the elongation factor EF-2-translocation step (5). In wild-type cells, RPS14A and RPS14B are expressed at a 10:1 ratio, respectively. A deficit of rpS14, caused by the deletion or inactivation of RPS14A, results in a the 10-fold derepression of RPS14B (43). Current evidence suggests that RPS14B is regulated posttranscriptionally by the recognition of an RNA stem-loop structure formed from sequences in the 5′ exon and first 62 nucleotides in the intron of RPS14B (33).
A fundamental prediction of this feedback model is that unassembled rpS14 interacts directly or indirectly with two different RNA targets—one in the ribosome and one in RPS14B pre-mRNA. Using the three-hybrid system (55) and a filter binding assay, we demonstrate that rpS14 directly interacts with RPS14B pre-mRNA and with a stem-loop in 18S rRNA. This is the first direct demonstration of the binding of a eukaryotic ribosomal protein to both rRNA and mRNA targets. Mutations in rpS14 that affect the affinity of the protein for both targets were generated to identify potential RNA binding domains of the protein. Interestingly, mutations that confer resistance to cryptopleurine or emetine altered the affinity of rpS14 for both RNAs in the three-hybrid assay. This result supports previous observations that antibiotic resistance mutations map to regions of bacterial r proteins predicted to bind to rRNA (reviewed in reference 50).
MATERIALS AND METHODS
Nomenclature of yeast r proteins and rpS14 mutant proteins.
The duplicated yeast genes encoding the 40S ribosomal protein rpS14 were originally designated CRY1 and CRY2 because mutations in either gene confer resistance to cryptopleurine (29, 43). Likewise, the CRY gene products were previously named rp59 with the original nomenclature of Gorenstein and Warner (19). Here we refer to CRY1 as RPS14A, CRY2 as RPS14B, and rp59 as rpS14 according to the new nomenclature for the ribosomal proteins of S. cerevisiae (36). In addition, other yeast r protein genes are referred to by their new names, with their old names in brackets. Alleles of RPS14B and the corresponding rpS14 proteins presented in this study are as follows: rps14b-4 (rpS14-CryR, L138stop); rps14b-5 (rpS14-EmRR, R133C, R134H); rps14b-6 (rpS14-E55G); rps14b-7 (rpS14-E55K); rps14b-8 (rpS14-P123L); and rps14b-9 (rpS14-ΔC127–138).
Three-hybrid assay.
The yeast strain L40 and plasmids pIII/MS2-1 and pACTII, used in the three-hybrid assay (55), were a gift from Marvin Wickens (University of Wisconsin). To facilitate cloning of fragments into the hybrid RNA vector, additional restriction sites were introduced into pIII/MS2-1 by ligating annealed oligonucleotides SWF20 5′-GGGAGATCTAAGCTTTACGTAATCGAT-3′ and SWF21 5′-ATCGATTACGTAAAGCTTAGATCTCCC-3′ into a unique SmaI site in the plasmid to generate p4130. DNA encoding the RPS14B regulatory element was amplified by PCR with oligonucleotides SWF16B 5′-GAAAGGCCTATTAAGAATGGCTAAGC-3′ and SWF18B 5′-AAGATCGATAAGAATAACTAAATGGT-3′, digested with StuI and ClaI, and ligated between the SmaI and ClaI sites of p4130. DNA encoding nucleotides 1515 to 1587 of S. cerevisiae 18S rRNA was amplified from p518 (a gift from Susan Liebman, University of Illinois, Chicago) with oligonucleotides S11up 5′-GAAAGGCCTGGGCATCAGGTATTCAATTG-3′ and S11dn 5′-AGGATCGATGGGCAAATGCTTTCGC-3′, digested with StuI and ClaI, and cloned into the SmaI and ClaI sites of p4130. The sequence of all recombinant DNAs was verified with the AmpliCycle sequencing kit (Perkin-Elmer).
The pACT-S14 hybrid protein vector was constructed by ligating a NruI-XhoI fragment from p4075 encoding the 3′ exon of RPS14B to pACTII digested with SmaI and XhoI. DNAs containing the C-terminal deletion mutation, a cryptopleurine resistance mutation, and the double emetine resistance mutation were amplified by PCR with SWF16B and SWF13BamHI 5′-CGGGATCCTCAGGTGGAGTCTGATGGGAC-3′, SWF37 5′-GGTAGAAGATGATGATTTCTTTTTTTTTTACTC-3′, or SWFEmR 5′-CGGGATCCTCATAAATGACAACCTCTTCTACCACCCTTCTTTC-3′, respectively, and subsequently cloned into pACTII.
Transformants containing plasmids expressing the hybrid RNAs and the hybrid proteins were selected on media lacking uracil and leucine. Multiple transformants were then assayed for the ability to grow on selective media containing 5, 10, 15, or 20 mM 3-amino-triazole (3-AT) or on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) indicator plates. Direct measurement of beta-galactosidase activity was determined as described by Kippert (24). Enzymatic activity values represent the averages of three independent experiments. Expression of the hybrid proteins in yeast was verified by Western analysis with rabbit polyclonal antibodies generated against glutathione S-transferase (GST)–S14 fusion protein (32).
Analysis of rpS14 mutants in vivo.
To assay the function of rpS14 mutant proteins in vivo independently of the three-hybrid system, mutations were site directed into a vector containing an EcoRI-ClaRI RPS14B fragment from which the intron of RPS14B was previously removed (33). A stop codon was engineered at codon 127 by site-directed mutagenesis with oligonucleotide SWF34ΔC 5′-CCATCAGACTCCAACTAGTAGAAGGGTGGTAG-3′ (27). The double emetine resistance mutation and the E55G, E55K, and P123L mutations were also site directed with oligonucleotides SWF35Em 5′-GGTAGAAGAGGTTGTCATTTATGATTTCTTTT-3′, SWF31 5′-CCGACAGAGACAAATCATCTCCATAC-3′, SWF32 5′-CCGACAGAGACGGATCATCTCCATAC-3′, or WSF33 5′-GTTACTCCAGTCCTATCAGACTCCACC-3′. The plasmids were subsequently transformed into JWY3245 (rps14A-Δ RPS14B RPS14B-lacZ) or JWY1884 (rps14A-Δ rps14B-Δ pRPS14A) to assay the ability of rpS14 mutants to repress RPS14B expression or assemble into ribosomes, respectively (33, 39).
In vitro transcription.
Templates for transcription of RPS14B RNAs were generated as follows. A BglII-BamHI fragment containing RPS14B was inserted at the BamHI site in Bluescript Ks+ (Stratagene) with a BglII site created in a previous study at nucleotide +1 (33). A functional BglII site in RPS14B was then introduced by site-directed mutagenesis (27) at nucleotide +105 to generate plasmid p4051. Linearized DNA suitable for transcription of RPS14B nucleotides +1 to +105 was prepared by digestion with BglII followed by purification from 1% agarose-TAE gels. Linearized plasmids were purified from gels as described by Zhen and Swank (70) and suspended in diethyl pyrocarbonate-treated water at a final concentration of 0.5 to 1.0 mg/ml. DNA including nucleotides 1515 to 1587 of S. cerevisiae 18S rRNA was inserted into Bluescript Ks− for use in transcription reactions as follows. The ribosomal DNA (rDNA) fragment was amplified by PCR with oligonucleotides S11up and S11dn and cloned between the SmaI and ClaI restriction sites of Bluescript Ks−. This plasmid was subsequently linearized with ClaI and purified as described above. Template for the transcription of ferritin L chain iron-responsive element (IRE) was a gift from Chuck Allerson, National Institutes of Health.
Radiolabeled RNA was synthesized by run-off transcription from either the plasmids described above or PCR-generated templates with the Ambion T7-MEGAshortscript transcription kit. Reactions were performed according to the manufacturer’s instructions. For filter binding experiments, RNA was uniformly labeled by including 20 μCi of [α-32P]UTP (3,000 Ci/mmol; Amersham) in the reaction. Full-length transcripts were purified from 6% polyacrylamide (19:1 acrylamide:bis-acrylamide)–5 M urea gels by soaking gel slices in elution buffer (0.3 M NaOAc, 1 mM EDTA) overnight at 4°C. The eluted transcripts were extracted once with phenol-chloroform, precipitated with ethanol, and suspended in an appropriate volume of Tris-EDTA.
MBP-S14 purification.
The MBP-S14 fusion was constructed as follows. Following the precise removal of the RPS14B intron from a plasmid containing RPS14B on an EcoRI-ClaI fragment (33), a unique StuI site was introduced just before the initiator ATG of RPS14B by site-directed mutagenesis with oligonucleotide SWF7 5′-GGTCGTTAGCCATAGGCCTCTTAATTGTTATTGGG-3′. The entire RPS14B coding sequence was then fused in-frame to malE in the pMalc vector (BioLabs) to generate p4078. The fusion plasmid was expressed in E. coli BL21 by induction with 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2.5 h. Induced cells were lysed by sonication and applied to an amylose column as described in the manufacturer’s instruction manual (ProFusion kit; BioLabs). Protein concentration was estimated by a micro-Bradford assay and concentrated with Microcon 10 concentrators (Millipore) as necessary. Purified protein was stored in column elution buffer (CEB) at 4°C for several weeks or at −80°C for longer periods of time.
Filter binding.
RNA probes were diluted in renaturation buffer containing 30 mM Tris-HCl (pH 8.0), 350 mM KCl, and 10 mM MgCl2. The RNA was heated at 60°C for 5 min and immediately placed on ice for 10 min. Typical binding reactions consisted of 5 μl of RNA (10 to 15 nM), 43 μl of binding buffer (30 mM Tris-HCl [pH 8.0], 150 mM KCl, 2 mM MgCl2, and 60 mg of E. coli tRNA per ml), and 0 to 20 μg of MBP-S14 diluted in CEB as necessary. Binding reactions were incubated at 25°C for 30 min and then applied directly onto nitrocellulose filters (HAWP 024; Millipore) under gentle vacuum. Before application of the binding reactions, 1 ml of binding buffer (without tRNA) was used to equilibrate each filter. Subsequent to sample filtration, the filters were rinsed with 100 μl of binding buffer (without tRNA) to reduce background radioactivity. Binding was quantified by scintillation counting, and binding isotherms were plotted with Kaleidagraph 3.0.
Selection of gain-of-function mutants.
To generate a library of randomly mutated plasmids, pACT-S14 was transformed into E. coli XL1-Red (Stratagene) according to the manufacturer’s suggestions. Colonies from multiple independent transformations were scraped from transformation plates and used to inoculate 5 ml of overnight cultures in Luria broth plus ampicillin for subsequent plasmid extraction.
Mutagenized pACT-S14 plasmids were transformed into the three-hybrid yeast strain carrying the wild-type MS2-RPS14B hybrid RNA vector. Transformants were grown on minimal media lacking uracil and leucine for 3 days at 30°C. Subsequently, the transformation plates were replica plated to minimal media lacking uracil, leucine, and histidine and containing 20 mM 3-AT. Resistant colonies were chosen after 5 to 7 days and restreaked onto selective plates without 3-AT. Plasmids conferring 3-AT resistance were shuttled through E. coli and into yeast again to confirm that the resistance phenotype was associated with the plasmid. Mutations in rpS14 were identified by DNA sequencing, and expressing of mutant hybrid proteins was checked by Western analysis.
Analysis of yeast ribosomal subunits.
Ribosomal subunits were extracted and analyzed as described in Tsay et al. (59) with the following modifications. Yeast cells were grown to early log phase in 100 ml of yeast extract-peptone-dextrose at 30°C. Forty optical density units (ODs) of cell extract was loaded onto 35-ml 7% to 47% linear sucrose gradients. The gradients were centrifuged at 27,000 rpm in a SW28 swinging bucket rotor for 4 h at 4°C.
RESULTS
rpS14 and RPS14B RNA interact in the yeast three-hybrid system.
The ability of rpS14 to interact with RPS14B pre-mRNA was tested by using the yeast three-hybrid system (55). Analogous to the two-hybrid system, the three-hybrid system depends upon the interaction of RNA and protein components to bring together an array of factors required to activate reporter gene expression in yeast. A number of specific RNA-protein interactions have already been demonstrated in this system, including IRE/IRP1, TAR/Tat (55), histone mRNA/HBF or SLBP (37, 65), and fem-3 PME/FBF (69).
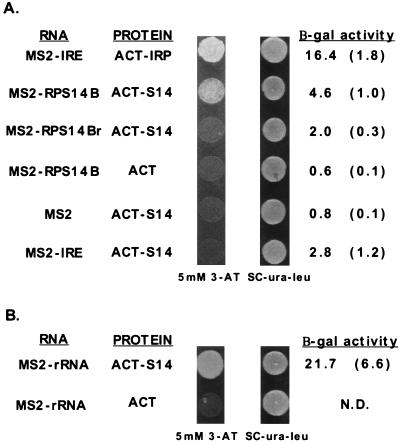
To determine whether rpS14 interacts with RPS14B pre-mRNA in the three-hybrid system, RPS14B encoding rpS14 was fused in-frame to the GAL4 transcriptional activation domain in pACTII, and the RPS14B regulatory stem-loop sequence (Fig. 1) was cloned upstream of the MS2 coat protein binding site in p4130 derived from pIII/MS2-1. When both plasmids were transformed together into a yeast strain expressing the LexA-MS2 coat protein hybrid (55), the resulting colonies exhibited increased reporter gene expression as indicated by growth on plates containing 5 mM 3-AT and increased beta-galactosidase activity (Fig. 2A). This three-hybrid interaction between RPS14B RNA and rpS14 was reproducible but weak compared to the IRE/IRP positive control. The positive response was dependent upon both components, since substitution of either with the vector plasmid did not activate the reporters. Moreover, both an antisense RPS14B sequence and an IRE RNA failed to interact positively with rpS14 in three-hybrid assays. These three-hybrid experiments demonstrate a link between unassembled rpS14 and RPS14B pre-mRNA.
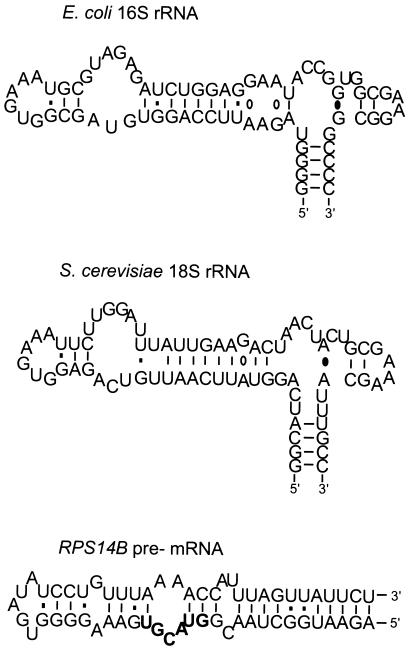
FIG. 1.
Predicted secondary structure of helix 23 from E. coli 16S rRNA (nucleotides 668 to 738) and S. cerevisiae 18S rRNA (nucleotides 876 to 948) and the RPS14B regulatory stem-loop (nucleotides 31 to 89). The RPS14B sequence was previously defined by extensive mutational analysis as necessary for feedback regulation (33). The 5′ splice site of RPS14B is shown in bold-faced letters. The structure of this RNA was predicted by using the University of Wisconsin FOLD program as described in reference 33. Ribosomal RNA structures are adapted from Gutell (21) and are available at http://pundit.colorado.edu:8080/RNA/16S/16s.html.
FIG. 2.
Three-hybrid assay of interactions between rpS14 and the regulatory stem-loop in RPS14B pre-mRNA or helix 23 of 18S rRNA. The three-hybrid yeast strain L40 (55), containing an integrated copy of the gene encoding the LexA:MS2 coat binding fusion protein, was transformed with plasmids carrying different GAL4 activation domain fusions and MS2 RNA fusions. Transformants were selected on SC-Ura-Leu medium and subsequently tested for expression of the HIS3 and lacZ reporter genes. A positive three-hybrid interaction is indicated by growth on medium supplemented with 5 mM 3-AT and elevated beta-galactosidase (β-gal) activity. (A) MS2-IRE and ACT-IRP hybrid plasmids encode the IRE and the iron-responsive protein fused to the MS2 hairpin and the GAL4 activation domain, respectively (55). The RPS14B regulatory RNA (MS2-RPS14B) and rpS14 (ACT-S14) interact positively in this assay. Antisense RPS14B (MS2-RPS14r), the IRE (MS2-IRE), MS2 alone (MS2), and the GAL4 activation domain alone all failed to interact with the appropriate protein or RNA target. The averages of three independent measurements of beta-galactosidase activity are shown, with standard deviations in parentheses. (B) Helix 23 of S. cerevisiae 18S rRNA (MS2-rRNA) interacts positively with S14. N.D., not determined.
rpS14 also interacts with 18S rRNA.
Since most r proteins are thought to interact with rRNA during the processing of rRNA and its assembly into ribosomes (56), it is likely that rpS14 also recognizes an rRNA target. A possible rRNA target for rpS14 is suggested by experiments with the bacterial homolog of rpS14, designated rpS11. Nucleotides 668 to 738 in helix 23 of the E. coli 16S rRNA can be cross-linked to rpS11 in vitro (20) and protected by rpS11 in hydroxyl-radical structure probing experiments (45). Given that rpS11 and rpS14 have 37% identity (30) and that helix 23 of rRNA is also conserved (Fig. 1), it seems likely that both proteins recognize the same region of rRNA in their respective organisms. To test this hypothesis, we introduced the region of yeast 18S rRNA corresponding to helix 23 in bacterial rRNA (nucleotides 876 to 948) into the three-hybrid system. As shown in Fig. 2B, the rRNA-rpS14 interaction led to 4.5-fold-greater reporter gene expression than that for the RPS14B-rpS14 combination. In contrast, no interaction was observed between rpS14 and the ITS2 sequence of rRNA (data not shown). The observation that rpS14 binds to both RPS14B pre-mRNA and, with higher affinity, to rRNA is consistent with a model in which competition between the RPS14B pre-mRNA and rRNA binding sites dictates the relative expression of RPS14B. Thus, when rpS14 is in excess of its ribosomal assembly partners, it binds to its pre-mRNA and prevents its expression.
S14 directly interacts with both RNA targets.
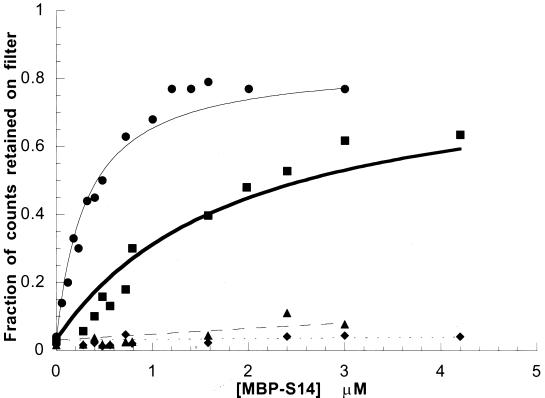
To determine whether rpS14 could interact directly with RNA, we tested the ability of purified MBP-S14 fusion protein to bind to RPS14B pre-mRNA and rRNA targets in vitro in a filter binding assay (Fig. 3). MBP-S14 bound directly to both RNAs as determined by the retention of increasing amounts of radiolabeled RNAs with increasing concentrations of fusion protein. Consistent with the three-hybrid result, rpS14 exhibited greater affinity for the rRNA target (Kd ≅ 3 μM−1) compared to the RPS14B pre-mRNA (Kd ≅ 0.5 μM−1). The specificity of this interaction was verified by the inability of MBP-S14 to recognize the IRE RNA in a similar filter binding experiment. These data not only validate the three-hybrid interaction but also demonstrate a direct interaction between S14 and its two RNA targets.
FIG. 3.
MBP-S14 binds to the RPS14B regulatory stem-loop or helix 23 of 18S rRNA in vitro. Filter binding isotherms for MBP-S14 and its two RNA targets are shown. MBP-S14 and helix 23 of 18S rRNA ( ); MBP-S14 and RPS14B regulatory stem-loop (
); MBP-S14 and RPS14B regulatory stem-loop ( ); MBP-S14 and IRE RNA (—♦—); MBP-S14ΔC and RPS14B regulatory stem-loop (
); MBP-S14 and IRE RNA (—♦—); MBP-S14ΔC and RPS14B regulatory stem-loop ( ).
).
The C terminus of S14 is required for RNA binding.
Like many ribosomal proteins, the amino acid sequence of rpS14 does not contain a discernible RNA recognition domain. However, analyses of bacterial r proteins suggest that conserved, basic amino acids, particularly those located in loops or turns in the protein structure, conserved solvent-exposed hydrophobic residues, and amino acids mutable to drug resistance phenotypes are hallmarks of RNA binding domains in r proteins (60, 61; reviewed in references 49 and 50). These observations suggest that the C terminus of rpS14 might be involved in RNA recognition because it is rich in highly conserved, basic residues (30) that are predicted (by the Chou-Fasman algorithm) to fold into a loop-turn structure. In addition, resistance to the translational inhibitors cryptopleurine and emetine maps to the last three residues of rpS14 (Fig. 4). Mutations that confer resistance to cryptopleurine map to the last codon of yeast rpS14, changing leucine 138 to a serine or a stop codon (29, 43). Likewise, emetine resistance mutations in the mammalian RPS14 gene change two highly conserved arginines, residues 136 and 137 (52). Based upon these criteria, the C terminus of rpS14 is a good candidate for an RNA binding domain.
FIG. 4.
Amino acid sequence of S14 encoded by the RPS14B gene. The locations of mutations that confer resistance to emetine (underline) and cryptopleurine (asterisk) are indicated. Amino acids that can be mutated to increase the three-hybrid interaction between S14 and the regulatory stem-loop of RPS14B are indicated by arrowheads. An alignment of E. coli rpS11, yeast, and human rpS14, shown in Larkin et al. (30), illustrates that the rpS14 sequence is highly conserved.
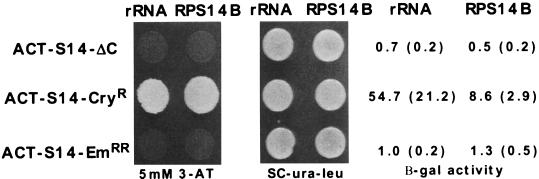
To test the importance of the C terminus in RNA binding, the ability of a truncated version of rpS14 (11-amino-acid C-terminal truncation, designated rpS14-ΔC) to interact with RPS14B pre-mRNA and rRNA was examined by using the three-hybrid system (Fig. 5) and by filter binding (Fig. 3 and data not shown). The truncated protein failed to interact with either RNA target in both assays. Western blot analysis indicated that steady-state levels of the rpS14-ΔC mutant protein were comparable to the wild-type protein in the three-hybrid yeast strain (data not shown). Thus, the inability of rpS14-ΔC to interact with RNA in the three-hybrid system cannot be attributed to protein instability. The effect of this mutation was further examined by determining whether the truncated protein could assemble into functional ribosomes in vivo. A plasmid shuffle experiment was used to demonstrate that the truncated protein could not complement the lethal phenotype of a rps14A::TRP1 rps14b::LEU2 double-knockout strain (data not shown). We have previously shown that wild-type rpS14 is present in 40S ribosomal subunits and that it is necessary for the assembly of stable functional 40S subunits. In the absence of rpS14, no stable 40S subunits assemble (39). Therefore, the failure of rpS14-ΔC to complement the lethality of rps14a::TRP1 rps14b::LEU2 suggests that this truncated protein does not assemble into functional 40S subunits. Taken together, these data indicate that the C terminus of rpS14 is necessary for RNA recognition in vivo and in vitro.
FIG. 5.
Altered interactions of antibiotic-resistant S14 proteins or C-terminally truncated S14 with RPS14B RNA or rRNA. ACT-rpS14 three-hybrid constructs containing the C-terminal truncation of 11 amino acids (ACT-S14-ΔC), the CryR mutation (ACT-S14-CryR), or the EmRR mutation (ACT-S14-EmRR) were assayed with the RPS14B regulatory stem-loop or with helix 23 or 18S rRNA. Results are presented as in Fig. 2. β-gal, beta-galactosidase.
Antibiotic resistance mutations alter the affinity of rpS14 for RNA.
To further investigate the role of the C terminus of rpS14 in RNA recognition and to explore the link between antibiotic resistance and RNA binding, the ability of rpS14 containing the R136C R137H emetine resistance double mutation (rpS14-EmRR) or the L138stop cryptopleurine resistance mutation (rpS14-CryR) to bind RPS14B and rRNA targets was tested in the three-hybrid system. Cells expressing rpS14-EmRR and either RNA target did not grow on 3-AT plates and exhibited low levels of beta-galactosidase activity. Furthermore, rpS14-EmRR did not bind to either target in the in vitro filter binding assay (data not shown). In contrast, rpS14-CryR interacted more strongly with both RNAs than did the wild-type rpS14 in the three-hybrid assay (Fig. 5). Thus, the two drug resistance mutations appear to affect the affinity of rpS14 for RNA in different ways. Despite this difference, these data are nonetheless consistent with the hypothesis that antibiotic resistance mutations in r proteins confer their effect by altering the r protein’s interaction with rRNA.
rpS14-EmRR fails to repress RPS14B but assembles into ribosomes.
The ability of rpS14-EmRR to recognize the two natural, full-length RNA targets in vivo was examined by determining (i) whether rpS14-EmRR could function as a repressor of RPS14B expression and (ii) whether the mutant protein could assemble into functional ribosomes. The ability of rpS14-EmRR to repress RPS14B was assessed by comparing beta-galactosidase levels in a rps14a::TRP1 RPS14B RPS14B-lacZ yeast strain transformed with either RPS14A, RPS14B, or the emetine-resistant allele of RPS14B (33). As expected, wild-type rpS14 encoded by either RPS14A or RPS14B can repress RPS14B in vivo; expression of RPS14B-lacZ decreases from 504 to 73 U/OD when RPS14A is introduced into the strain or to 90 U/OD upon transformation with RPS14B. However, expression of the RPS14B-lacZ reporter was not significantly repressed when the emetine-resistant allele of RPS14B was introduced; 381 U of beta-galactosidase activity per OD was observed. It is not clear whether the modest repression of RPS14B-lacZ expression in this case results from the ability of rpS14-EmRR to directly function as a repressor, albeit less efficiently than wild-type rpS14, or if rpS14-EmRR indirectly represses RPS14B-lacZ by competing with RPS14B-encoded rpS14 for assembly into ribosomes. If the latter is true, the decrease in RPS14B-lacZ expression might result from a slight excess of wild-type unassembled rpS14.
The ability of rpS14-EmRR to assemble into functional ribosomes in yeast was tested by a plasmid shuffle experiment. rps14a-Δ rps14b-Δ cells containing a plasmid encoding the emetine-resistant allele of RPS14B as the only source of S14 were viable but exhibited a slow-growth phenotype (data not shown). This result indicates that rpS14-EmRR can assemble into functional ribosomes but suggests that the assembly and/or functionality of the 40S subunits is aberrant.
Our observation that rpS14-EmRR assembles into ribosomes but does not bind RNA in vitro is not unprecedented. Mutations in yeast rpL25 and rpL32 that weaken or eliminate binding to RNA in vitro do not prevent assembly of these proteins into ribosomes in vivo and are not lethal (25, 54, 64). These results suggest that other factors, such as protein-protein or additional RNA-protein interactions, stabilize the association of rpS14 with the assembling ribosome. The incorporation of rpS14-EmRR into ribosomes might involve interaction with not only helix 23 but also helix 24 of 18S rRNA. In addition to helix 23, nucleotides in helix 24 of E. coli rRNA were protected from hydroxyl radical attack by rpS11 (45). Hence, an interaction between rpS14 and helix 24, in the absence of strong interactions with helix 23, might be sufficient to permit the assembly of functional, albeit less stable, 40S subunits.
Mutations that alter the specificity of rpS14 binding to RPS14B pre-mRNA and rRNA.
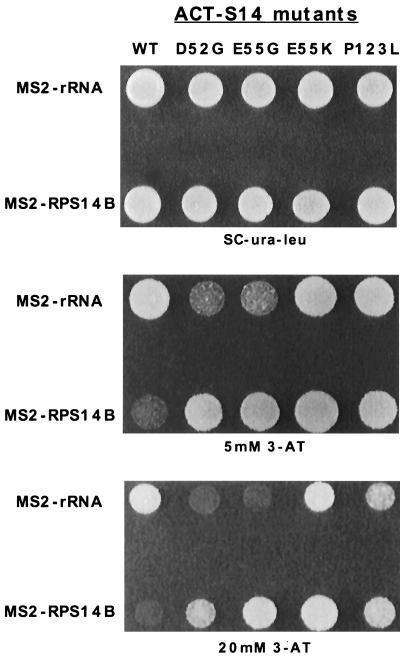
To define other regions of rpS14 that are important for RNA recognition, mutations in this protein that increased the interaction between RPS14B pre-mRNA and rpS14 were selected by using the three-hybrid system. Unlike the interaction between rpS14 and 18S rRNA, the weak interactions between wild-type rpS14 and RPS14B pre-mRNA did not allow growth on 20 mM 3-AT plates. Since mutations that increased the binding of rpS14 to this target might be expected in regions of the protein involved in RNA recognition, we transformed a library of randomly mutagenized pACT-S14 plasmids into the three-hybrid system and selected for strong RNA-protein interactions on plates containing 20 mM 3-AT. Ninety 3-AT-resistant colonies were recovered out of approximately 34,000 transformants. The 3-AT-resistant phenotype proved to be plasmid borne for only 13 of these 90 strains. These 13 pACTII-S14 plasmids were sequenced, and each was found to contain a single mutation that altered one of three codons in rpS14 (Fig. 4 and 7). The codons affected were D52G (1), E55G (6), E55K (5), and P123L (1). These mutations pinpoint another region of rpS14 that might play a role in RNA binding.
The ability of these mutant proteins to bind nonspecifically to RNA was assayed by using several different RNAs in the three-hybrid system, including the IRE RNA, an antisense RPS14B RNA, and an empty MS2 vector with no other additional RNA sequence. All four mutant proteins demonstrated increased nonspecific binding to these different RNAs (data not shown). When assayed with the 18S rRNA target, however, two of the four mutant rpS14 proteins, D52G and E55G, exhibited weaker interactions (Fig. 6 and Table 1). This change in specificity, namely increased binding to RPS14B pre-mRNA and decreased binding to 18S rRNA in the three-hybrid assay, was not tested in vitro. Nevertheless, these data suggest that this region of rpS14 is involved in discriminating between the two RNA targets. Interestingly, both rpS14-E55G and rpS14-E55K were able to complement the rps14a-Δ rps14b-Δ double knockout without any obvious effect on the growth rate of the cells (data not shown). Hence, the reduced affinity of rpS14-E55G for rRNA and the promiscuous RNA binding behavior of rpS14-E55K and rpS14-E55G were not overtly deleterious to the cell. The identification of this region in rpS14 that is important for RNA specificity highlights the utility of the three-hybrid system for detailed analysis of RNA-protein interactions.
FIG. 6.
Mutations in RPS14 that alter the binding specificity of rpS14 to the RPS14B stem-loop regulatory RNA or helix 23 of 18S rRNA. rpS14 mutants with increased affinity for RPS14B regulatory RNA were selected by using the 3-ATR phenotype of the three-hybrid assay. Mutant proteins were subsequently screened for altered interactions with helix 23 of 18S rRNA. The plates shown were incubated only for 2 days to accentuate differences in the three-hybrid interactions; a 3-day incubation is necessary to clearly see the interaction between wild-type (WT) rpS14 and the RPS14B RNA target as shown in Fig. 2.
TABLE 1.
Beta-galactosidase activities for rpS14 mutants and RPS14B pre-mRNA or rRNA in the three-hybrid assay
| RNA | Protein | U/OD | SD |
|---|---|---|---|
| RPS14B | Wild type | 4.60 | 1.61 |
| RPS14B | D52G | 20.39 | 5.69 |
| RPS14B | E55G | 39.24 | 18.16 |
| RPS14B | E55K | 160.87 | 54.15 |
| RPS14B | P123L | 15.24 | 8.01 |
| rRNA | Wild type | 21.69 | 6.55 |
| rRNA | D52G | 6.44 | 2.19 |
| rRNA | E55G | 12.93 | 1.15 |
| rRNA | E55K | 163.04 | 54.97 |
| rRNA | P123L | 16.11 | 4.47 |
DISCUSSION
We have demonstrated that rpS14 binds directly to the regulatory sequence in RPS14B pre-mRNA (nucleotides 39 to 89) and to a conserved helix in 18S rRNA. rpS14 is the first eukaryotic r protein for which both mRNA and rRNA targets are known; this finding strengthens our model that RPS14B regulation results from differences in the affinity of rpS14 for RPS14B pre-mRNA and 18S rRNA. Second, we have used the three-hybrid system to begin to identify residues to rpS14 important for binding these RNAs. This is the first example of the use of the three-hybrid system to map RNA binding domains and thus demonstrates the utility of this in vivo genetic method to study RNA-protein interactions in more detail. Third, we have established that two different drug resistance mutations alter the binding of rpS14 to RNA. This finding supports the model that antibiotic resistance is mediated by alterations in rRNA structure, in this case through changes in r protein-rRNA interactions.
Model for the autogenous control of RPS14B expression.
Our results support the model that the autogenous regulation of RPS14B expression is governed by a competition between two RNA binding sites, the RPS14B regulatory stem-loop and helix 23 in 18S rRNA. Experiments with the three-hybrid system and in vitro filter binding demonstrate that rpS14 binds directly to both RNAs. Moreover, the interaction with rRNA is about fivefold stronger than the one with RPS14B RNA. These observations are consistent with a model for the autogenous control of RPS14B expression in which rpS14 is preferentially consumed by ribosome assembly. Only when rpS14 accumulates in excess of its assembly partners is it available to interact with RPS14B pre-mRNA and prevent expression of the gene.
In E. coli, almost all of the ribosomal proteins involved in autogenous regulation are primary binding proteins (reviewed in reference 68). While it was once thought that only primary binding proteins were involved in direct interactions with rRNA, more recent evidence suggests that most, if not all, r proteins recognize rRNA and influence its structure throughout the course of ribosome assembly and function (40, 45, 56). S11, the E. coli homolog of yeast rpS14, is assembled into the ribosome as a tertiary binding protein (41). Since the assembly order of yeast r proteins has not yet been established in much detail (26, 58), it is not clear when rpS14 interacts with the assembling yeast ribosome.
The rRNA binding sites for two other yeast r proteins were identified by phylogenetic comparison to their bacterial homologs. Yeast L25 and E. coli EL23 as well as yeast rpL12 [rpL15] and E. coli EL1 recognize each other’s binding site in the respective organisms (13, 14). In both examples, the rRNA binding sites are conserved. The interaction between rpS14 and 18S rRNA provides the third example of a conserved rRNA-ribosomal protein interaction and thus supports the prediction that many other interactions in the ribosome also are conserved. Furthermore, our detection of an interaction between rpS14 and its rRNA ligand in the three-hybrid system demonstrates the utility of this method for mapping eukaryotic ribosomal protein-rRNA interactions.
One protein binding to two different RNAs.
Ribosomal protein S14 is among a unique group of proteins that bind multiple, specific RNA targets (reviewed in references 12, 62, and 68). Most, but not all, of the other known proteins in this class are ribosomal proteins that recognize mRNA and rRNA targets, including the E. coli proteins S4, S7, S15, L10/L12, and S8. The molecular basis for recognition of two RNAs by one protein is still not fully understood. In some cases, the two RNA targets of ribosomal proteins contain sequence and structural similarities that suggest a common mode of recognition by the protein. In other examples, the mRNA and rRNA ligands bear little resemblance to each other. It is possible that the r proteins that recognize these seemingly distinct targets do so by using two separate RNA binding domains. In support of this hypothesis, structural studies indicate that several r proteins do contain at least two RNA binding domains. These domains could interact independently with two RNA binding sites on the same RNA molecular (e.g., two distinct sites on rRNA) or on two different RNA molecules (e.g., mRNA and rRNA). More detailed analysis of the structures of the two RNA targets of rpS14 should reveal whether there are any common features necessary for binding or any unique elements responsible for the different affinities of rpS14 for these RNAs.
Utility of the yeast three-hybrid system.
In addition to their role in ribosome assembly and function, RNA-protein interactions are instrumental to many other biological activities. Despite this importance, very little is known about the intricacies of how proteins recognize specific RNA targets. The recent development of genetic systems, however, should greatly facilitate endeavors to investigate these interactions by providing tools to rapidly and randomly survey both RNA and protein molecules for important residues that contribute to binding (23, 28, 55).
The yeast three-hybrid system was originally developed and tested by using well-established RNA-protein interactions. The binding constants for these interactions as estimated from in vitro binding experiments range from 0.01 to 10 nM. Here we report that weak interactions which are in the micromolar range in vitro can be detected and studied in this system in vivo. We also demonstrate that the three-hybrid system provides a tool for analyzing established RNA-protein interactions by providing a means to rapidly survey a protein for regions involved in the interaction. This is particularly useful for proteins like rpS14 that do not contain known RNA recognition motifs.
Selection for rpS14 variants with greater affinity for the RPS14B regulatory stem-loop uncovered four mutations in three different codons. These mutations highlight two regions of the protein that may be important for RNA recognition and specificity. All four mutations reduced the ability of rpS14 to discriminate among different RNA targets. However, two of the mutations, D52G and E55G, also reduced the protein’s affinity for its rRNA target. This change in specificity, increased affinity for RPS14B pre-mRNA and decreased affinity for rRNA, suggests that these two residues are crucial for the recognition of rRNA in this assay and implies that this region of rpS14 is involved in establishing the specificity of RNA binding. The C terminus of rpS14 is required for interaction with both RNA targets. The effects of the emetine resistance double mutation, the P123L mutation, and the C-terminal truncation collectively indicate that the architecture and sequence of this region are important for RNA recognition. That the structure of the C terminus is important for RNA recognition is suggested by the P123L mutation; changing the proline at position 123 to leucine might eliminate a beta turn that provides the rigidity to this region necessary for specific binding. It seems probable that the conserved, basic residues in this C-terminal loop contribute to the stability of the RNA-protein interactions by participating in electrostatic interaction with the phosphate backbone of RNA.
Because the three-hybrid assay takes place in vivo, its output potentially reflects both direct as well as indirect effects. It is notable, however, that in each case in which we have performed complementary in vitro binding assays, the results agree with those observed in the three-hybrid system. While additional experiments are clearly necessary for an understanding of how the protein interacts with its two RNA targets, our experiments with the three-hybrid system provide a launching pad for further investigation of these interactions.
Antibiotic resistance mutations and rRNA structure.
Antibiotic resistance mutations in rRNA or r proteins provide powerful genetic tools for studying the structure and function of the ribosome. Translational antibiotics appear to function by binding directly to rRNA and altering rRNA tertiary structures that are important for ribosome function (1, 4, 17, 38, 48, 53). Ribosomal proteins are thought to direct folding of rRNA in assembling ribosomes and to maintain rRNA structure necessary for the function of mature ribosomes. Mutations in rRNA or in r proteins that confer antibiotic resistance are thought to perturb or occlude the antibiotic binding site in the ribosome by locally altering rRNA structure. Results with mutations in E. coli r protein S12 are consistent with this view. The conformation of rRNA in ribosomes containing a streptomycin-resistant or -dependent mutant S12 protein is altered compared to that of rRNA in wild-type cells (1). As mentioned previously, antibiotic resistance mutations in several r proteins are located in or near amino acids that can be cross-linked to rRNA (8, 10, 18, 49, 51, 60, 61). The observation that drug resistance mutations in rpS14 alter the interaction of this protein with its two RNA targets demonstrates that resistance mutations reside in RNA binding domains of r proteins. It is currently not clear why rpS14-EmRR fails to interact with RNA in the three-hybrid system, while rpS14-CryR binds to RNA better than the wild-type protein in this assay. Future experiments may reveal that the two resistant proteins affect the conformation of rRNA in different ways.
That rpS14 plays an important role in translation is foreshadowed by several studies in which E. coli S11 was among a subset of r proteins that cross-linked to an AUG analog (44), initiation factor IF-2 (2), and initiation factor IF-3 (22). In addition, rpS11 is thought to be involved in establishing the binding site for messenger RNA (7) and transfer RNA in the 30S subunit (16). Future experiments that investigate the interactions between rpS14 and 18S rRNA as well as cryptopleurine and emetine with 18S rRNA should provide valuable insight into both the mechanism of antibiotic resistance and the function of rpS14 in ribosome assembly and function.
ACKNOWLEDGMENTS
We thank Rachel Green, Javier Lopez, Jon Warner, Josep Vilardell, and colleagues in our laboratory for critical reading of the manuscript. We are grateful to Dhruba SenGupta, Beilin Zhang, and Marvin Wickens for sharing the three-hybrid system and for advice on using the system. We also thank Chuck Allerson and Susan Liebman for providing the templates for transcription of the IRE and yeast 18S rRNA, respectively, and Robin Gutell for 16S and 18S rRNA structure predictions.
This work was supported by U.S. Public Health Service research grant GM28301 to J.L.W.
REFERENCES
- 1.Allen P N, Noller H F. Mutations in ribosomal proteins S4 and S12 influence the higher order structure of 16S ribosomal RNA. J Mol Biol. 1989;208:457–468. doi: 10.1016/0022-2836(89)90509-3. [DOI] [PubMed] [Google Scholar]
- 2.Bollen A, Heimark R L, Cozzone A, Traut R R, Hershey J W B. Cross-linking of initiation factor IF-2 to Escherichia coli 30S ribosomal proteins with dimethylsuberimidate. J Biol Chem. 1975;250:4310–4314. [PubMed] [Google Scholar]
- 3.Bozzoni I, Fragapane P, Annesi F, Pierandrei-Amaldi P, Amaldi F, Beccari E. Expression of two Xenopus laevis ribosomal protein genes in injected frog oocytes: a specific splicing block interferes with the L1 RNA maturation. J Mol Biol. 1984;180:987–1005. doi: 10.1016/0022-2836(84)90267-5. [DOI] [PubMed] [Google Scholar]
- 4.Brink M F, Brink G, Verbeet M P, de Boer H A. Spectinomycin interacts specifically with the residues G1064 and C1192 in 16S rRNA, thereby potentially freezing this molecule into an inactive conformation. Nucleic Acids Res. 1994;22:325–331. doi: 10.1093/nar/22.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bucher K, Skogerson L. Cryptopleurine—an inhibitor of translocation. Biochemistry. 1976;15:4755–4759. doi: 10.1021/bi00667a001. [DOI] [PubMed] [Google Scholar]
- 6.Caffarelli E, Fragapane P, Gehring C, Bozzoni I. The accumulation of mature RNA for the X. laevis ribosomal protein L1 is controlled at the level of splicing and turnover of the precursor RNA. EMBO J. 1987;6:3493–3498. doi: 10.1002/j.1460-2075.1987.tb02674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang C, Craven G R. Identification of several proteins involved in the messenger RNA binding site of the 30S ribosome by inactivation with 2-methoxy-5-nitrotropone. J Mol Biol. 1977;117:401–418. doi: 10.1016/0022-2836(77)90135-8. [DOI] [PubMed] [Google Scholar]
- 8.Chittum H S, Champney W S. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J Bacteriol. 1994;176:6192–6198. doi: 10.1128/jb.176.20.6192-6198.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dabeva M D, Warner J R. Ribosomal protein L32 of Saccharomyces cerevisiae regulates both splicing and translation of its own transcript. J Biol Chem. 1993;268:19669–19674. [PubMed] [Google Scholar]
- 10.Davies C, Ramakrishnan V, White S W. Structural evidence for specific S8-RNA and S8-protein interactions within the 30S subunit: ribosomal protein S8 from Bacillus stearothermophilus at 1.9 Å resolution. Structure. 1996;4:1093–1104. doi: 10.1016/s0969-2126(96)00115-3. [DOI] [PubMed] [Google Scholar]
- 11.Draper D E. RNA-protein interactions in ribosomes. In: Nagai K, Mattaj I W, editors. RNA-protein interactions. Oxford, United Kingdom: Oxford University Press; 1994. pp. 82–102. [Google Scholar]
- 12.Draper D E. Ribosomal protein-RNA interactions. In: Zimmerman R A, Dahlberg A E, editors. Ribosomal RNA: structure, evolution, processing, and function in protein biosynthesis. Boca Raton, Fla: CRC Press, Inc.; 1996. pp. 171–198. [Google Scholar]
- 13.El-Baradi T T A L, Raué H A, de Regt V C H F, Verbree E C, Planta R J. Yeast ribosomal protein L25 binds to an evolutionary conserved site on yeast 26S and E. coli 23S rRNA. EMBO J. 1985;4:2101–2107. doi: 10.1002/j.1460-2075.1985.tb03898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Baradi T T A L, de Regt V C H F, Einerhand S W C, Teixido J, Planta R J, Ballesta J P G, Raué H A. Ribosomal proteins EL11 from Escherichia coli and L15 from Saccharomyces cerevisiae bind to the same site in both yeast 26S and mouse 28S rRNA. J Mol Biol. 1987;195:909–917. doi: 10.1016/0022-2836(87)90494-3. [DOI] [PubMed] [Google Scholar]
- 15.Eng F J, Warner J R. Structural basis for the regulation of splicing of a yeast messenger RNA. Cell. 1991;65:797–804. doi: 10.1016/0092-8674(91)90387-e. [DOI] [PubMed] [Google Scholar]
- 16.Fanning T G, Cantrell M, Shih C Y, Craven G R. Evidence that proteins S1, S11, and S21 directly participate in the binding of transfer RNA to the 30S ribosome. Nucleic Acids Res. 1978;5:933–950. doi: 10.1093/nar/5.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fourmy D, Recht M I, Blanchard S C, Puglisi J D. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996;274:1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- 18.Golden B L, Hoffman D W, Ramakrishnan V, White S W. Ribosomal protein S17: characterization of the three-dimensional structure by 1H and 15N NMR. Biochemistry. 1993;32:12812–12820. doi: 10.1021/bi00210a033. [DOI] [PubMed] [Google Scholar]
- 19.Gorenstein C, Warner J R. Coordinate regulation of the synthesis of eukaryotic ribosomal proteins. Proc Natl Acad Sci USA. 1976;73:1547–1551. doi: 10.1073/pnas.73.5.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greuer B, Osswald M, Brimacombe R, Stöffler G. RNA-protein cross-linking in Escherichia coli 30S ribosomal subunits: determination of sites on 16S rRNA that are cross-linked to proteins S3, S4, S7, S9, S10, S11, S17, S18 and S21 by treatment with bis-(2-chloroethyl)-methylamine. Nucleic Acids Res. 1987;15:3241–3255. doi: 10.1093/nar/15.8.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutell R R. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures. Nucleic Acids Res. 1994;22:3502–3507. doi: 10.1093/nar/22.17.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heimark R L, Kahan L, Johnston K, Hershey J W B, Tract R R. Cross-linking of initiation factor IF3 to proteins of the Escherichia coli 30S ribosomal subunit. J Mol Biol. 1976;105:219–230. doi: 10.1016/0022-2836(76)90108-x. [DOI] [PubMed] [Google Scholar]
- 23.Jain C, Belasco J G. A structural model for the HIV-1 Rev-RRE complex deduced from altered-specificity Rev variants isolated by a rapid genetic strategy. Cell. 1996;87:115–125. doi: 10.1016/s0092-8674(00)81328-8. [DOI] [PubMed] [Google Scholar]
- 24.Kippert F. A rapid permeabilization procedure for accurate quantitative determination of beta-galactosidase activity in yeast cells. FEMS Microbiol Lett. 1995;128:201–206. doi: 10.1111/j.1574-6968.1995.tb07523.x. [DOI] [PubMed] [Google Scholar]
- 25.Kooi E A, Rutgers C A, Kleijmeer M J, van’t Riet J, Venema J, Raué H A. Mutational analysis of the C-terminal region of Saccharomyces cerevisiae ribosomal protein L25 in vitro and in vivo demonstrates the presence of two distinct functional elements. J Mol Biol. 1994;240:243–255. doi: 10.1006/jmbi.1994.1438. [DOI] [PubMed] [Google Scholar]
- 26.Kruiswijk T, Planta R J, Krop J M. The course of the assembly of ribosomal subunits in yeast. Biochim Biophys Acta. 1978;517:378–389. doi: 10.1016/0005-2787(78)90204-6. [DOI] [PubMed] [Google Scholar]
- 27.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 28.Laird-Offringa I A, Belasco J G. Analysis of RNA-binding proteins by in vitro genetic selection: identification of an amino acid residue important for locking U1A onto its RNA target. Proc Natl Acad Sci USA. 1995;92:11859–11863. doi: 10.1073/pnas.92.25.11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larkin J C, Woolford J L., Jr Molecular cloning and analysis of the CRY1 gene: a yeast ribosomal protein gene. Nucleic Acids Res. 1983;11:403–420. doi: 10.1093/nar/11.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larkin J C, Thompson J R, Woolford J L., Jr Structure and expression of the Saccharomyces cerevisiae CRY1 gene: a highly conserved ribosomal protein gene. Mol Cell Biol. 1987;7:1764–1775. doi: 10.1128/mcb.7.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B, Vilardell J, Warner J R. An RNA structure involved in feedback regulation of splicing and of translation is critical for biological fitness. Proc Natl Acad Sci USA. 1996;93:1596–1600. doi: 10.1073/pnas.93.4.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z. Feedback regulation of CRY2, a yeast ribosomal protein gene. Ph.D. thesis. Pittsburgh, Pa: Carnegie Mellon University; 1995. [Google Scholar]
- 33.Li Z, Paulovich A G, Woolford J L., Jr Feedback inhibition of the yeast ribosomal protein gene CRY2 is mediated by the nucleotide sequence and secondary structure of CRY2 pre-mRNA. Mol Cell Biol. 1995;15:6454–6464. doi: 10.1128/mcb.15.11.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madjar J-J, Nielson-Smith K, Frahm M, Roufa D J. Emetine resistance in Chinese hamster ovary cells is associated with an altered ribosomal protein S14 mRNA. Proc Natl Acad Sci USA. 1982;79:1003–1007. doi: 10.1073/pnas.79.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madjar J-J, Frahm M, McGill S, Roufa D J. Ribosomal protein S14 is altered by two-step emetine resistance in Chinese hamster cells. Mol Cell Biol. 1983;3:190–197. doi: 10.1128/mcb.3.2.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mager W H, Planta R J, Ballesta J-P G, Lee J C, Mizuta K, Suzuki K, Warner J R, Woolford J. A new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25:4872–4875. doi: 10.1093/nar/25.24.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin F, Schaller A, Eglite S, Schümperli D, Müller B. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997;16:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moazed D, Noller H F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 39.Moritz M, Paulovich A G, Tsay Y-F, Woolford J L., Jr Depletion of yeast ribosomal proteins L16 or rp59 disrupts ribosome assembly. J Cell Biol. 1990;111:2261–2274. doi: 10.1083/jcb.111.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mueller F, Brimacombe R. A new model for the three-dimensional folding of Escherichia coli 16S ribosomal RNA. II. The RNA-protein interaction data. J Mol Biol. 1997;271:545–565. doi: 10.1006/jmbi.1997.1211. [DOI] [PubMed] [Google Scholar]
- 41.Nomura M, Held W A. Reconstitution of ribosomes: studies of ribosome structure, function and assembly. In: Nomura M, Tissières A, Lengyel P, editors. Ribosomes. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1974. pp. 193–223. [Google Scholar]
- 42.Nomura M. History of ribosome research: a personal account. In: Hill W, et al., editors. The ribosome: structure, function, and evolution. Washington, D.C: American Society for Microbiology; 1990. pp. 3–55. [Google Scholar]
- 43.Paulovich A G, Thompson J R, Larkin J C, Li Z, Woolford J L., Jr Molecular genetics of cryptopleurine resistance in Saccharomyces cerevisiae: expression of a ribosomal protein gene family. Genetics. 1993;135:719–730. doi: 10.1093/genetics/135.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pongs O, Stöffler G, Lanka E. The codon binding site of the Escherichia coli ribosomes as studied with a chemically reactive A-U-G analog. J Mol Biol. 1975;99:301–315. doi: 10.1016/s0022-2836(75)80148-3. [DOI] [PubMed] [Google Scholar]
- 45.Powers T, Noller H G. Hydroxyl radical footprinting of ribosomal proteins on 16S rRNA. RNA. 1995;1:194–209. [PMC free article] [PubMed] [Google Scholar]
- 46.Presutti C, Caifre S-A, Bozzoni I. The ribosomal protein L2 in S. cerevisiae controls the level of accumulation of its own mRNA. EMBO J. 1991;10:2215–2221. doi: 10.1002/j.1460-2075.1991.tb07757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Presutti C, Villa T, Hall D, Pertica C, Bozzoni I. Identification of the cis-acting elements mediating the autogenous control of ribosomal protein L2 mRNA stability in yeast. EMBO J. 1995;14:101–109. doi: 10.1002/j.1460-2075.1995.tb00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puglisi E V, Puglisi J D. Nuclear magnetic resonance spectroscopy of RNA. In: Simons R W, Grunberg-Manago M, editors. RNA structure and function. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1998. pp. 117–146. [Google Scholar]
- 49.Ramakrishnan V, White S W. The structure of ribosomal protein S5 reveals sites of interaction with 16S rRNA. Nature. 1992;358:768–771. doi: 10.1038/358768a0. [DOI] [PubMed] [Google Scholar]
- 50.Ramakrishnan V, White S W. Ribosomal protein structures: insights into the architecture, machinery and evolution of the ribosome. Trends Biochem Sci. 1998;23:208–212. doi: 10.1016/s0968-0004(98)01214-6. [DOI] [PubMed] [Google Scholar]
- 51.Ramakrishnan V, Davies C, Gerchman S E, Golden B L, Hoffman D W, Jaishree T N, Kycia J H, Porter S, White S W. Structures of prokaryotic ribosomal proteins: implications for RNA binding and evolution. Biochem Cell Biol. 1995;73:979–986. doi: 10.1139/o95-105. [DOI] [PubMed] [Google Scholar]
- 52.Rhoads D D, Roufa D J. Emetine resistance of Chinese hamster cells: structures of wild-type and mutant ribosomal protein S14 mRNAs. Mol Cell Biol. 1985;5:1655–1659. doi: 10.1128/mcb.5.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosendahl G, Douthwaite S. The antibiotics micrococcin and thiostrepton interact directly with 23S rRNA mucleotides 1067A and 1095A. Nucleic Acids Res. 1995;22:357–363. doi: 10.1093/nar/22.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rutgers C A, Rientjes J M J, van’t Riet J, Raué H A. rRNA binding domain of yeast ribosomal protein L25: identification of its borders and a key leucine residue. J Mol Biol. 1991;218:375–385. doi: 10.1016/0022-2836(91)90719-m. [DOI] [PubMed] [Google Scholar]
- 55.SenGupta D J, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stern S, Powers T, Changchien L-M, Noller H F. RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science. 1989;244:783–790. doi: 10.1126/science.2658053. [DOI] [PubMed] [Google Scholar]
- 57.Tasheva E S, Roufa D J. Regulation of human RPS14 transcription by intronic antisense RNAs and ribosomal protein S14. Genes Dev. 1995;9:304–316. doi: 10.1101/gad.9.3.304. [DOI] [PubMed] [Google Scholar]
- 58.Trapman J, Retel J, Planta R J. Ribosomal precursor particles from yeast. Exp Cell Res. 1975;90:95–104. doi: 10.1016/0014-4827(75)90361-4. [DOI] [PubMed] [Google Scholar]
- 59.Tsay Y F, Thompson J R, Rotenberg M O, Larkin J C, Woolford J L., Jr Ribosomal protein synthesis is not regulated at the translational level in Saccharomyces cerevisiae: balanced accumulation of ribosomal proteins L16 and rp59 is mediated by turnover of excess protein. Genes Dev. 1988;2:664–676. doi: 10.1101/gad.2.6.664. [DOI] [PubMed] [Google Scholar]
- 60.Urlaub H, Kruft V, Bischof O, Müller E-C, Wittmann-Liebold B. Protein-rRNA binding features and their structural and functional implications in ribosomes as determined by cross-linking studies. EMBO J. 1995;14:4578–4588. doi: 10.1002/j.1460-2075.1995.tb00137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Urlaub H, Thiede B, Miller E-C, Brimacombe R, Wittmann-Liebold B. Identification and sequence analysis of contact sites between ribosomal proteins and rRNA in Escherichia coli 30S subunits by a new approach using matrix-assisted laser desorption/ionization-mass spectrometry combined with N-terminal microsequencing. J Biol Chem. 1997;272:14547–14555. doi: 10.1074/jbc.272.23.14547. [DOI] [PubMed] [Google Scholar]
- 62.Varani G, Nagai K. RNA recognition by RNP proteins during RNA processing. Annu Rev Biophys Biomol Struct. 1998;27:407–445. doi: 10.1146/annurev.biophys.27.1.407. [DOI] [PubMed] [Google Scholar]
- 63.Vilardell J, Warner J R. Regulation of splicing at an intermediate step in the formation of the splicesome. Genes Dev. 1994;8:211–220. doi: 10.1101/gad.8.2.211. [DOI] [PubMed] [Google Scholar]
- 64.Vilardell J, Warner J R. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol Cell Biol. 1997;17:1959–1965. doi: 10.1128/mcb.17.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Z F, Whitfield M L, Ingledue III T C, Dominski Z, Marzluff W F. The protein that binds the 3′ end of the histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- 66.Warner J R, Mitra G, Schwindinger W F, Studeny M, Fried H M. Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol Cell Biol. 1985;5:1512–1521. doi: 10.1128/mcb.5.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woolford J L. The structure and biogenesis of yeast ribosomes. Adv Genet. 1991;29:63–118. doi: 10.1016/s0065-2660(08)60107-8. [DOI] [PubMed] [Google Scholar]
- 68.Zengel J M, Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog, Nucleic Acids Res. Mol Biol. 1994;47:331–370. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]
- 69.Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens M P. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- 70.Zhen L, Swank R T. A simple and high yield method for recovering DNA from agarose gels. BioTechniques. 1993;14:894–898. [PubMed] [Google Scholar]