Abstract
Improved quality control and prolonged shelf life are important actions in preventing food waste. To get an overview of the bacterial diversity of fillets from live stored mature Atlantic cod, bacterial isolates were identified before and after storage (air and vacuum) and freezing/thawing. Based on the load of dominating bacteria, the effect of different packaging methods and a short freezing/thawing process on prolonged shelf-life was evaluated (total viable counts, bacteriota, sensory attributes, and volatile components). Hand filleted (strict hygiene) cod fillets had a low initial bacterial load dominated by the spoilage organism Photobacterium, whereas industrially produced fillets had higher bacterial loads and diversity (Pseudomonas, Arthrobacter, Psychrobacter, Shewanella). The identified bacteria after storage in vacuum or air were similar to the initially identified bacteria. Bacteriota analysis showed that a short time freezing/thawing process reduced Photobacterium while modified atmosphere packaging (MAP; 60% CO2/40% O2 or 60% CO2/40% N2) inhibited the growth of important spoilage bacteria (Photobacterium, Shewanella, Pseudomonas) and allowed the growth of Carnobacterium/Carnobacteriaceae and Acinetobacter. Despite being dominated by Photobacterium, fresh fillets stored in MAP 60% CO2/40% N2 demonstrated better sensory quality after 13 days of storage than fillets stored in MAP 60% CO2/40% O2 (dominated by Carnobacterium/Carnobacteriaceae). Carnobacterium spp. or other members of Carnobacteriaceae may therefore be potential spoilage organisms in cod when other spoilage bacteria are reduced or inhibited.
Keywords: microbiota, bacteriota, spoilage bacteria, Photobacterium, Carnobacterium, sensory analysis, volatile components, MAP, vacuum packaging, freezing
1. Introduction
One of the causes of food waste at the retail level is linked to a limited shelf life, and this is especially true for food with a relatively short shelf life such as fresh fish products [1,2,3]. It is also reported that fish has a high food loss in the EU (51%), of both edible and not fully utilized inedible parts of the fish [4]. In a circular economy, it is important to improve quality control and define best practices for a better utilization of fish raw materials. Atlantic cod (Gadus morhua L.) is an important species for the seafood industry connected to the Atlantic Ocean, and an improved knowledge of factors affecting shelf life is therefore important.
The quality deterioration of fish is primarily caused by bacterial growth and metabolism, autolytic changes, and chemical oxidation [5]. Such changes can lead to undesired development of volatile organic compounds (VOCs), resulting in off-flavors and off-odors. One of the most common VOC in cod is the pungent trimethylamine (TMA) caused by the reduction of trimethylamine oxide (TMAO) by specific spoilage bacteria such as Photobacterium [6,7]. There are, however, several different bacteria that can contribute to the fish spoilage through production of off-flavors and off-odors. Storage conditions such as temperature and packaging methods affect the bacterial composition, the lag phase and growth and hence also the presence of off-flavors and off-odors [7,8,9,10,11,12]. Although the fish muscle is considered sterile at the time of slaughtering or catch, bacteria from the skin, gastrointestinal tract, water, and processing environment can contaminate the fillets during processing [13]. Photobacterium spp. are the most dominant species in the intestinal microbiome of Atlantic cod and may contaminate the flesh during processing [14]. As far as we know, the bacteriota (bacterial composition) on cod processing machines has not been studied. However, in salmon processing plants, spoilage bacteria, such as Pseudomonas and Shewanella, were isolated from processing equipment after sanitation and Photobacterium were found in seawater and live fish, thereby enabling the cross contamination of fillets [15].
To extend shelf life, the processing and storage conditions should be optimized to prevent the growth of spoilage organisms present in fish. A common packaging method of gutted cod and cod fillets is aerobic storage in expanded polystyrene (EPS) boxes supplemented with ice to ensure a low chilling temperature [16]. Single packaged cod fillets and fillet products are also sold vacuum packaged or modified atmosphere packaged (MAP). Packaging with use of MAP is reported to preserve quality better than by vacuum packaging [6,11,12,17], and vacuum packaging is regarded as better to ensure quality and shelf life than aerobic storage [17,18]. However, a sufficient partial pressure of CO2 or headspace volume is required to obtain delayed bacterial growth compared to vacuum packaging [12,19,20,21].
A common gas mixture of MAP for fish fillets consists of carbon dioxide and nitrogen to suppress the growth of aerobic bacteria and bacteria sensitive to CO2, such as Pseudomonas [13]. The bacteriota at the end of the shelf life is reported to be dominated by Photobacterium phosphoreum which is relatively resistant to CO2 [12,22,23], or a mixed bacteriota (Pseudomonas, Shewanella, and Photobacterium) though under different storage conditions [24,25]. Some food packaging also include high levels of O2, which has shown to reduce the formation of TMAO to TMA in fish [23,24,26,27,28]. However, the bacteriota under high O2 storage has only been described in a few studies [16,23,24]. Studies also show that a mixture of bacteria presented during storage with a dominance of Photobacterium and Psychrobacter, followed by Moritella, Carnobacterium, Shewanella, and Vibrio as potential spoilage bacteria [29], and Carnobacterium spp. and Vagococcus spp. as part of the spoilage bacteriota of MAP sea bream [30].
A first indication of freezing as a technique to reduce luminous bacteria in fish came as early as in 1934 [31] and more recently studies showed that P. phosphoreum was not detectable in cod that had been stored frozen (6–12 weeks, −20 °C) and further refrigerated stored under modified atmosphere [22,32,33]. This indicated that a combination of a long-term freezing process to reduce P. phosphoreum and packaging with CO2 to inhibit Shewanella and Pseudomonas could extend the shelf life at refrigerated storage.
Even though many packaging studies of cod fillets have been performed, there is a lack of a full sensory descriptive analysis of the products, except for a description of heated samples [32,34]. Most of the studies where sensory analyses were included focused on overall freshness using the Quality Index Method (QIM) on raw whole fish [35], or evaluated the samples based on acceptance or rejection [6,36,37], or focussed on other species than cod [38].
Since total bacterial count does not necessarily correlate to spoilage due to the description of specific spoilage organisms (SSO) [13,39], it is important to consider the bacterial composition when deciding the shelf life and not only refer to the total viable count [29]. In order to better understand the diversity of spoilage bacterial communities, culture-independent analyses can provide additional information compared to traditional cultivation based methods [40]. Improved knowledge about how the bacteriota and sensory quality will respond to different packaging atmospheres and freezing strategies is therefore crucial, and it is an important step towards the reduction of food waste.
The aim of the present study was therefore to identify packaging concepts (different atmospheres combined with or without a freezing/thawing process) that can direct the bacteriota of cod fillets towards a composition with fewer detrimental sensory effects, thus increasing the shelf life. To obtain this, we identified bacteria on fillets from different processing plants (initially and after storage and a long-term freezing process) and investigated the effect of different packaging atmospheres on a range of potential spoilage organisms from raw materials and stored cod fillets. Furthermore, the effect of packaging atmospheres and a short-term freezing on the microbial growth, bacteriota, sensory attributes, and volatile compounds in cod filets during refrigerated storage were determined.
2. Materials and Methods
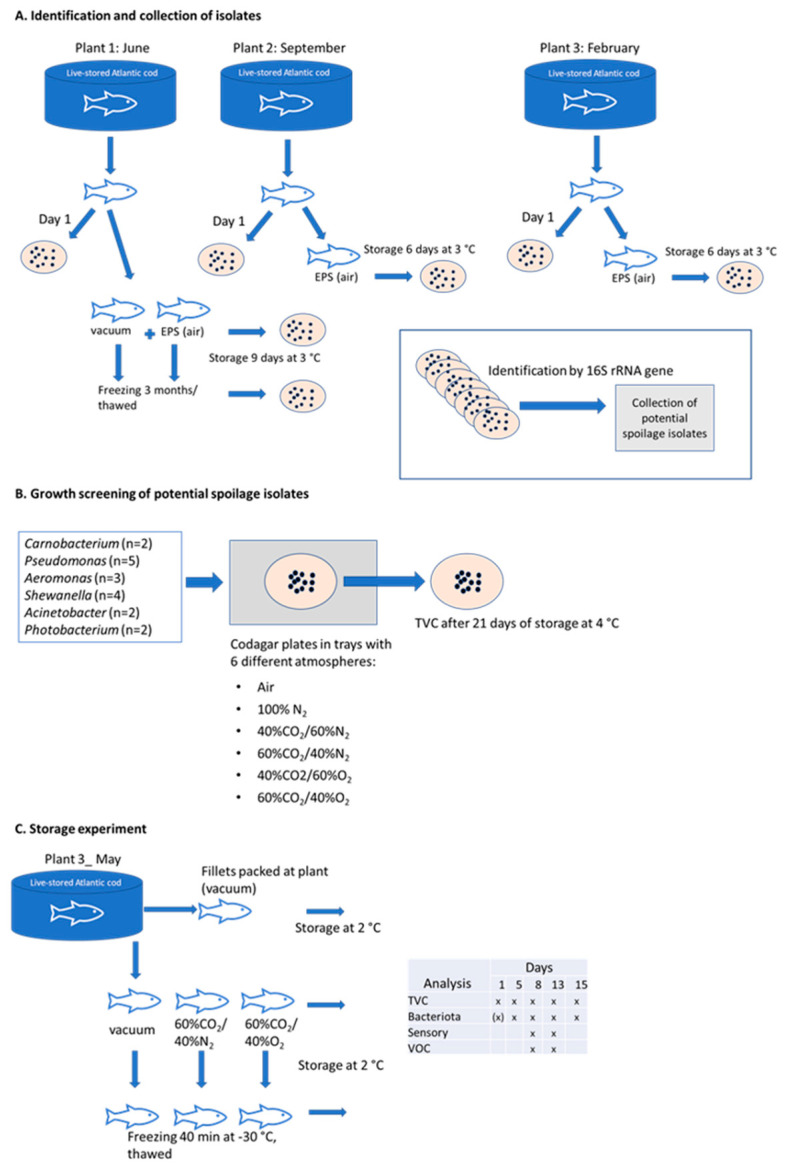
The presented work contains three parts: (1) Identification of bacterial strains from cod fillets (initial bacteriota and during a pre-storage test), (2) growth screening of selected bacterial strains on cod agar medium trays packaged with different gas mixtures, and (3) a storage experiment with packaging and short-term freezing. Figure 1 shows an overview of these three different parts.
Figure 1.
An overview of the three different parts (A–C) of the presented study. (A): Identification and collection of isolates. Cod fillets from three different plants were collected and stored at 3 °C in EPS boxes (air) for 6 (Plant 2 and 3) or 9 days (Plant 1). Fillets from Plant 1 were also frozen/thawed and stored in EPS boxes (air) and vacuum packaged for 9 days. Bacterial colonies were collected from agar plates at arrival (day 1) or after storage. The bacterial colonies were identified by partial 16S rRNA gene sequencing. (B): Growth screening of potential spoilage isolates. The growth of selected isolates from part (A) at 6 different atmospheres was tested on cod agar in trays. The total viable counts were registered after 21 days at 4 °C. (C): Storage experiment. Fresh fillets from Plant 3 were vacuum packaged (from commercial production) and packaged specific designed for the study by use of vacuum, and modified atmosphere (MAP) containing 60% CO2/40% N2 and 60% CO2/40% O2, all stored at 2 °C for up to 15 days. In addition, half of the packages were subjected to a short freezing process (40 min at −30 °C) before thawed and stored the same way as for the fresh fillets. Samples for TVC and bacteriota were collected after 1, 5, 8, 13 and 15 days, while samples for sensory- and volatile component (VOC) analysis were collected after 8 and 13 days. All fish samples used were of wild matured cod stored live in net pens for about two to four weeks prior to slaughtering (mean weight 5 kg) and pre-rigor filleting.
2.1. Characterization of Bacterial Strains from Cod Fillet
2.1.1. Fish Material
Cod fillets (Gadus morhua L.) were transported from three different processing plants in Norway within a period of one year, resulting in three different batches. Batch 1 was delivered in June (2014) and originated from a research station with a fish farm site (Plant 1). Batches 2 and 3 were delivered in September (2015) and February (2016), respectively, from two different commercial processing plants for fillet production (Plant 2 and 3, respectively). All samples were Atlantic cod (wild matured cod) stored live without feed supply in net pens for about two to four weeks prior to slaughtering (mean weight 5 kg). The current Norwegian regulations state that wild caught cod can be live stored in sea cages up to 12 weeks, and during the first four weeks feeding is not required [41], and the quality of fish stored live in net pens for about four weeks is reported to be unchanged compared to the initial quality [42]. Live storage of cod is a method to extend the fishing season and is used as a supplement during periods with bad weather conditions. The pre-rigor filleting (by hand at Plant 1) and packaging were performed at site the same day as slaughtering, and the fillets were packaged in EPS boxes wrapped with plastic sheets of polyethylene (OTR 5000 cm3 O2/m2/day) and added wet ice on top. The boxes were transported to the laboratory at Nofima (Ås, Norway) by airplane and truck (arrival the day after) for sampling at arrival (Day 1).
To map a broad range of microbes that can be present on cod after filleting and storage, the bacteriota on cod fillets one day after processing and after storage under different conditions relevant for the cod production chain was investigated.
Half of the fillets from Plant 1 were packaged at Day 1 into vacuum prior to further storage, and half continued stored in EPS box. Furthermore, half of the EPS-samples and the vacuum samples were frozen at Day 1 for 3 months (−20 °C), as a pre-test of long-term frozen stored fillets, then thawed overnight at 1 °C before further storage. Both the thawed and the fresh samples from Plant 1 were sampled after 9 days of refrigerated storage (n = 6) (mean storage temperature 3 °C). The fillets from Plant 2 and 3 were sampled (n = 5) after 6 days of aerobic storage (mean storage temperature 3 °C). The 6 and 9 days were empirically chosen to achieve approximately similar levels of total bacterial count related to different initial count levels (Table 1), and at this time of storage, to obtain bacteriota that can be important for quality and shelf life.
Table 1.
Bacteria (%) identified (by 16S rRNA gene sequencing of colonies) from three different processing plants, one day after slaughtering and filleting. At Plant 1 samples were taken from both the skin and from the flesh (fillet). Plant 2 and 3 were industrial processing plants with sampling of the fillets.
| Plant 1 | Plant 2 | Plant 3 | Colonies (n), | ||
|---|---|---|---|---|---|
| Colonies of Bacterial Genus | Filet (n = 6) | Skin (n = 6) | Filet (n = 5) | Filet (n = 5) | Filet/Skin |
| Photobacterium | 92.6 | 78.9 | 3.8 | 88/30 | |
| Pseudomonas | 1.1 | 7.9 | 27.8 | 30.8 | 14/3 |
| Shewanella | 11.1 | 2/0 | |||
| Aeromonas | 5.6 | 1/0 | |||
| Psychrobacter | 1.1 | 5.3 | 11.1 | 19.2 | 8/2 |
| Arthrobacter | 2.6 | 26.9 | 7/1 | ||
| Acinetobacter | 22.2 | 3.8 | 5/0 | ||
| Janthinobacterium | 3.2 | 3/0 | |||
| Chryseobacterium | 2.6 | 3.8 | 1/1 | ||
| Stenotrophomonas | 1.1 | 1/0 | |||
| Sphingobacterium | 5.6 | 3.8 | 2/0 | ||
| Flavobacterium | 5.6 | 7.7 | 3/0 | ||
| Massilia | 1.1 | 2.6 | 1 /1 | ||
| Comamonas | 5.6 | 1/0 | |||
| Myroides | 5.6 | 1/0 | |||
| Colonies in sample (n) | 94 | 38 | 18 | 26 | Total: 138/38 |
| Total count, log10 CFU/g | 1.2 ± 1.0 | 2.0 ± 0.8 | 4.7 ± 0.1 | 3.6 ± 0.3 | |
2.1.2. Microbial Sampling
Microbial samples were taken from the fillets (skin and meat, separately) of Plant 1 and analyzed at Day 1 (n = 6) by use of sterile swabs (Cotton Tipped Applicator, single tip, OneMed, Helsinki, Finland) of a 5 × 20 cm surface of the fillet, suspended in 5 mL Pepton water and grown on Long & Hammer medium [43] at 15 °C for 5–7 days. The Long & Hammer agar contained 1% NaCl.
Additionally, bacterial plate count after 1 and 6/9 days of storage was performed by taking 3 × 3 cm and 1 cm depth (approximately 10 g) at the dorsal part of the fillets (without skin). The samples were homogenized in a stomacher for 60 s, and appropriate 10-fold dilutions were made and spread on Iron agar plates (Oxoid, Basingstoke, Hampshire, U.K.). Incubation was at 15 °C for about 5 days.
To identify the taxonomy of the cultivated bacteria from the three batches, 15 colonies (randomly selected from one sector per readable agar plate per fish fillet) for each sampling time were selected for DNA sequencing (16S rRNA gene).
2.1.3. 16S rRNA Gene Sequencing of Colonies
Colonies were identified from agar plates by partial 16S rRNA gene sequencing (universal primers [44] covering variable regions 3 and 4 in the 16S rRNA gene (V3 and V4) directly on colonies using a variation of a yeast colony PCR method as described before [45,46]. Amplification was performed using 0.25 µmol/l (each) primer, 5 µL of 5 Prime HotMastermix (AH Diagnostics, Oslo, Norway) to a total volume of 25 µL. The cycling conditions were: 94 °C 2 min, then 30 cycles of denaturing at 94 °C for 30 s, annealing at 60 °C for 30 s, extension at 72 °C for 30 s, and a final extension at 72 °C for 7 min. The PCR products were purified before sequencing, using ExoSap-IT (USB Corp., Cleveland, OH, USA). Sequencing PCR was carried out using the Big Dye® Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Dublin, Ireland) according to the manufacturer’s instructions with the forward universal 16S rRNA gene primer. The sequencing reactions were carried out in 25 cycles of 96 °C for 15 s and 60 °C for 4 min. A BigDye XTerminator Purification Kit (Applied Biosystems) was used according to the manufacturer’s recommendations to clean up the sequencing reactions and sequencing was performed on an ABI PRISM 3130xl Genetic Analyzer (Applied Biosystems). The taxonomy was identified using the RDP (Ribosomal Database Project) SeqMatch (http://rdp.cme.msu.edu/, accessed on 25 June 2014, 21 October 2015, 16 March 2016). The thresholds used in the RDP search were as follows: both type and non-type strains, both uncultured and isolates, only good sequences >1200 nt and KNN = 1.
2.1.4. Muscle pH
The pH was measured in triplicate directly in the cod fillets using a Knick pH meter (Knick GmbH & Co, Berlin, Germany) and muscle electrode S/N 5290739 (Mettler Toledo, Urdorf, Switzerland), only from Plant 1, at Day 1 and Day 6/9.
2.2. Growth Screening of Selected Bacteria Strains on Cod Agar Medium Trays
Based on abundance and knowledge of spoilage potential [13,15,24,40], 20 bacterial strains from the identified bacterial isolates and from culture collection, were selected for screening of growth (Table A1). Selections were performed to represent both bacteria that previous has been reported to cause spoilage, and bacteriota (initial and after storage) from the different processing plants.
To make an effective screening of the selected bacterial strains, a model system of cod agar plates were made, packaged with different gas mixtures (Table A2) and incubated at 4 °C. Cod agar was prepared as described in an earlier study [47], by use of 5 kg of fresh Atlantic cod fillets bought from a retailer.
The different bacterial cultures were incubated aerobically at 15 °C on Iron agar for about 5 days, colonies were picked and further cultivated in Long & Hammer broth (contained 1% NaCl) for 2 days at 15 °C, until reaching 8.1 ± 0.6 log10 CFU/mL. Each bacterial culture was diluted to achieve 5 log10 CFU/mL and an inoculate volume of 100 µL was used for the cod agar plates. There was one agar plate (without the lid) per packaged sample, and one agar plate per strain per gas mixture was included, which means 2–5 strains for each genus.
The packaging consisted of APET preformed tray (with 50 mm drawing depth), Biaxer topweb (65 XX FP AFM, Wipak, Oy, Finland) using a tray sealer (Multivac T200, Wolfertschwenden, Germany) with different pre-mixtures of gas (Nippon Gases Norge AS, Oslo, Norway).
After 13 days of storage, all packages were measured for the content of CO2 and O2 (Table A2) using a ChackMate 9900 O2/CO2 analyser (PBI Dansensor, Ringsted, Denmark). The samples were stored at 4 °C for 3 weeks.
To analyze the bacterial counts, the cells were rinsed off the agar plates using one ml sterile peptone water and a L-shaped spreader (VWR International, Oslo, Norway). The viable cell counts analysis was performed using an automated spiral plater (WASP Whitley automatic WB 05 SH, VWR International) on Iron agar plates, incubated at 15 °C for 3–5 days (read by ProtoCOL 2, Synbiosis (Cambrigde, United Kingdom)).
2.3. Storage Experiment of Pre-Rigor Processed Cod Loins
Based on results from the initial experiments and existing literature [23,24,27,28,48], it was hypothesized that a combination of CO2 and O2 (high levels of both O2 and CO2) would show a greater inhibitory effect on bacterial growth than CO2 and N2 (with high levels of CO2) and that vacuum packaging would allow for rapid bacterial growth. Furthermore, it was hypothesized that a short-term freezing process would eliminate a fraction of the initial bacteriota (e.g., Photobacterium) and delay microbial growth further. To test the hypotheses, industrially produced cod loins processed at Plant 3 were packed with different atmospheres as described in Section 2.3.1 (“Vacuum”, “CO2” and “O2/CO2”) and with and without being subjected to a short-term freezing after packaging, followed by a thawing process/refrigerated storage.
2.3.1. Fish material and Packaging
Atlantic cod was caught at the coast of northern Norway and stored live in cage (feed-deprived) for about four weeks (Senjahopen, Norway) before slaughter. Filleting was performed within two hours after slaughtering (mean weight was 4.8 kg; headed and gutted).
The fillets were cut into loins (dorsal part of the fillet) using a Valka Cutter water jet machine (Valka, Reykjavik, Iceland). Three different packaging methods were used: (1) MAP with 60% CO2 and 40% N2 and with a CO2-emitter pad (“CO2”), (2) MAP with 60% CO2 and 40% O2 (“O2/CO2”), and (3) vacuum packaging (“Vacuum”). In addition, an industrial processed sample of the ordinary production at the processing plant were used (line caught cod at the coast of northern Norway, delivered to the processing plant immediately after catch). The industrial processed samples (designated “Industry”) were filleted, cut into loins, and vacuum packed 2–3 h after processing of the other test samples.
A thermoformer packaging machine (Multivac R145, Multivac, Germany) was used for modified atmosphere packaging (“CO2”) and vacuum packaging at the processing plant. Drawing depth of vacuum packages was 35 mm (package volume 400 mL) and 40 mm for MAP (627 mL volume, gas/product volume ratio of 1/1). Film used for the bottom web was NICE XX 18 Black (PE/PA/EVOH/PA/PE), thickness of flat material 350 µm (Wipak Oy, Nastola, Finland). For the top web SC XX 3PA (PA/PP/PA/EVOH/PA/PE) with thickness of flat material 80 µm (Wipak Oy) was used. For the “Industry” samples SC 4 PA (PA/PP/PA/PE) was used both for top and bottom web (thickness of flat material 90 µm, Wipak Oy). The industrial processing was part of the processing plants regular production process of frozen single packed fillets.
For safety reasons, the O2/CO2 samples were packed at Nofima (packaging machine, Multivac T200, adjusted to high levels of O2): the fillets were vacuum packaged at the commercial processing plant, transported in EPS Air boxes added ice, and hygienically repackaged into MAP (40% O2 and 60% CO2) the day after filleting. Preformed trays (50 mm depth) of HDPE (600 mL volume) and a Biaxer (PET/PE/EVOH/PE) top web were used, with similar gas/product volume ratio as for the “CO2”. The gas mixture for the “O2/CO2” was measured to be 39.2% O2 and 55.3% CO2 in an empty package at the day of packaging.
In “CO2” packages, a CO2 emitter pad with 35 mL liquid absorption capacity and 230 mL CO2 emission capacity (XP-CO2-35-230-070175-Y, 70 × 175 mm,; Mc Airlaid’s GmbH, Steinfurt, Germany) was added to achieve an optimal CO2 availability. The CO2 emitter develop CO2 gas inside the package initiated by the liquid loss from the fish sample.
Liquid absorbent pad with 110 mL absorption capacity (MGS-110-070175-70, 70 × 175 mm; Mc Airlaid’s GmbH), was used for the O2/CO2 and the Vacuum packages.
The CO2 and O2 were analyzed at each sampling time by a CheckMate 9900 O2/CO2 analyser (PBI Dansensor, Ringsted, Denmark).
2.3.2. Freezing
Based on results from the pretest characterizing bacterial strains from fresh and thawed samples, half of the samples (CO2, O2/CO2 and Vacuum, not the Industry samples) were frozen by use of a Torry continuous air blast freezer (IQF; Individual Quick Freezing, Refrigeration Aberdeen LTD, Aberdeen, Scotland, UK) immediately after packaging. Freezing time was 40 min with air temperature monitored to be −30 °C. After freezing, the samples were placed in EPS boxes added wet ice for chill transport, in which the samples were thawed at arrival to the Nofima laboratory (Ås, Norway) the day after.
In the following, thawed samples will be designated as “CO2-T”, “O2/CO2-T” and “Vacuum-T”.
2.3.3. Temperature and Sampling
The mean temperature for the thawed samples and the ordinary chilled samples during transport were −2.3 ± 0.9 °C and 0.8 ± 0.6 °C, respectively (temperature loggers used: Kooltrak GmbH, Kiedrich, The Netherlands) and it took about 24 h for the thawed samples to reach the same temperature as the chilled samples (2 °C). Sampling was performed after 1, 5, 8, 13 and 15 days (n = 3 per sampling and treatment) at 2 °C storage (loggers used: Ecolog TN4-L, Elpro-Buchs AG, Buchs, Switzerland) for bacterial analyses. Day 0 was the time of processing and packaging at the industrial processing plant. Volatile components and sensory analysis were performed on day 8 and 13, based on current time of shelf life.
2.3.4. Culture-Dependent Analyses of Bacteria (Plate Count)
Analyses of total bacterial count were performed as earlier described (Section 2.1.2) of 3 × 3 cm cut from samples (Iron agar).
2.3.5. Culture Independent Analysis of Bacteria
Forty-five ml of the stomacher solutions were centrifuged at 13,000× g for 5 min. The pellets were frozen at −20 °C until DNA extraction using the Fast DNA-96 Soil Microbe kit (MP Biomedical) and following the manufacture’s MP-96 Inhibitor Removal Plate protocol.
PCR was performed in triplicate and paired end sequencing (2 × 150 bp) was performed using the protocol presented in Caporaso et al. [49] and as described before [50]. The library quantification and sequencing were performed at Nofima. The MiSeq Control Software (MCS) version used was RTA v1.18.54.
The forward and reverse reads were joined in QIIME version 1.9.1, and the barcodes corresponding to the reads that failed to assemble were removed. The sequences were then demultiplexed in QIIME allowing zero barcode errors and a quality score of 30 (Q30) using the QIIME toolkit [51]. The maximum and minimum number of sequences per sample was 171,601 and 64,940, respectively. Reads were assigned to their respective bacterial taxonomy using an openref Operational Taxonomic Unit (OTU) picking workflow. Reads that did not match a reference sequence were discarded resulting in 3779 OTUs with n > 2, each of these represents a phylotype and may be a representative of a bacterial species. The level 6 (genus) table derived from QIIME was used for further statistical analyses.
2.3.6. Volatile Organic Compounds (VOCs)
Dynamic headspace/GC-MS analyses of volatile organic compounds (VOCs) were performed on samples from the packages stored for 8 and 13 days. Samples were cut from the same loin as performed for the bacterial analyses and the sensory analysis. The content of volatiles was analyzed by dynamic headspace/GC-MS as described by [23,52] with small modifications to the method. The peaks were integrated, and compounds tentatively identified with MSD Chemstation software (E.02.02.1431) and NIST/EPA/NIH Mass Spectral Library (version2.0 g, 2012). Concentrations of the individual volatiles were calculated as µg/g sample based on an internal standard.
2.3.7. Sensory Analysis
To describe the objective perception of the various samples, a trained panel performed a Quality Descriptive Analysis (QDA), ISO 13299:2016 (E) of the samples [53]. The panel consisted of 10 subjects employed exclusively to work as sensory assessors at Nofima AS (Ås, Norway). Assessors are selected based on their sensory abilities and trained according to recommendations in ISO 8586-1:2012 (E) [54]. The sensory laboratory is designed according to guidelines in ISO 8589: 2007(E) [55] and electronic registration of data (EyeQuestion®, Logic8 BV, Utrecht, The Netherlands).
The assessors were trained and calibrated on two samples, fresh and stored cod, for the purpose to agree on the variation in attribute intensity. The samples were evaluated for the intensity of sensory odor attributes sour, seawater, cloying, fermented sour, yeast, chemical, sulfur, ammonia and pungent. Appearance was evaluated for the intensity of whiteness and glossiness (Table A3), evaluated after the odor attributes. In total, 21 loins were cut into ten pieces (one for each assessor) and served during five sessions with at least fifteen minutes break between each session. The coded samples (15 °C at time of evaluation) were served in blind trials randomized according to sample, assessor, and replicate. The samples were served to each assessor as raw samples in plastic cups with a three-digit code, with lid, with an approximate surface size of 3 × 3 cm (samples placed with skin side of the fillet towards the bottom of the cup). The intensity of each odor attribute was perceived by sniffing into the newly opened plastic cup.
The panelists recorded their results at an individual rate on a 15-cm non-structured continuous scale with the left side of the scale corresponding to the lowest intensity, and the right side corresponding to the highest intensity. The software transformed the responses into numbers between 1 (low intensity) and 9 (high intensity).
2.3.8. Statistical Analyses
Bacterial numbers were log10 transformed and mean values and standard error of the mean calculated in Minitab (Minitab 18.1, 2017, www.minitab.com, accessed on 29 May 2019). One-way ANOVA and Fisher least square of differences at 95% confidence were used to calculated differences between means.
Logarithmic change of the selected bacteria strains on cod agar medium trays, was calculated by subtracting the initial number of bacteria from the number after incubation for each strain. Statistical tests were done using variances between strains to calculate error.
Total counts (log10 transformed) were analysed by ANOVA using gas (CO2, O2/CO2, Vacuum), process (Fresh, Thawed), day, and second order interactions as effects. The industry sample was not included as a part of the ANOVA. Terms were considered significant for p-values below 0.05. Post hoc comparisons were done using contrasts for the expected marginal means, Tukey-HSD adjustment of p-values was performed for multiple testing adjustments.
The effects of experimental factors (gas, processing, days of storage) and their interactions on bacteriota and volatile compounds were tested using fifty-fifty MANOVA [56] on each of the datasets separately. Rotation tests [57] were applied for the false discovery rate (FDR) adjustment of p-values (considered significantly different when p-values below 0.05). Log transformation of the data were performed to improve normality conditions. Because there are many zeros in the data for volatile compounds, an offset corresponding to 1% of the minimum value was added to the data prior to log transformation. Bacteriota was analysed at level 6, i.e., the genus level, and only genera with an average abundance above 0.01 or a maximum of 0.1 were included in the analyses, hence the focus was on the most abundant genera, twelve genera passed the filter (Table A4).
The sensory data were analysed using a linear mixed model comprising the factors product (all seven different packaging variants), replicate and assessor and the second order interactions. Assessor and interactions involving assessors were considered random whereas the other factors were fixed.
Principal component analysis (PCA) was performed for each of the data sets from the experiment (bacteriota, sensory profiles and volatile compounds) to visualise and explore effects of the packaging concepts.
Multivariate analyses (fifty-fifty-MANOVA and PCA) were done in Matlab (Mathworks, R2018b), whereas ANOVA on total counts were done using R [58], and the sensory analyses were done with EyeOpenR (EyeQuestion Software 4.4.6, Logic8 BV, The Netherlands).
3. Results and Discussion
3.1. Bacterial Levels and Bacteriota in Cod Fillets from Three Different Producers before and after Storage at Different Conditions
3.1.1. Bacterial Levels and Diversity on Fillets One Day after Slaughter
Fresh cod fillets, processed immediately after slaughtering, were obtained from three different producers at different time points. All samples were of live stored cod (feed- deprived) that had previously been shown to keep stable fillet quality during the first four weeks of storage [42]. The pH of the fillets from Plant 1 was 6.9 ± 0.2 at Day 1, and 6.5 ± 0.3 and 6.6 ± 0.3 for the air and vacuum stored samples respectively (similar for fresh and thawed samples), which is in accordance with previous results on longline caught cod and farmed cod [23,59].
Large variations in the counts and the bacteriota on cod fillets one day after slaughter were found and reflected most likely differences in contamination during processing. Fillets from Plant 1, which were hand filleted under strict hygiene conditions, had a very low initial bacterial number with a clear dominance of Photobacterium (93% of the isolates from fillets and 79% from skin samples) (Table 1). To our knowledge bacteriota on fillet products of live stored wild Atlantic cod (northeast arctic cod) is not previously described. However, a similar bacteriota has been described for hand filleted, farmed Atlantic cod [60]. A dominance of Photobacterium is also typically found on the fillets of wild caught cod raw materials and dominating in the intestinal microbiome [14,37,61,62]. In Plant 1, the total viable count numbers were lower on skin compared to previously reported (3–6 log10 CFU/g) [63].
Industrially produced fillets from Plant 2 had the highest number of bacteria among the processing plants, including Pseudomonas (28%), Acinetobacter (22%), and Shewanella (11%), but no Photobacterium were detected. Plant 3 had also high total bacterial counts (1000 times higher than Plant 1) and the most dominant bacteria belonged to Pseudomonas (30%), Arthrobacter (26%), and Psychrobacter (19%). Although they represented only a small fraction (3.8%) of the total bacteriota, the level of Photobacterium on Plant 3 fillets was higher than for Plant 1 fillets. The fillets from Plant 2 and 3 contained higher bacterial numbers and the bacteriota was typical for what is found in the seafood processing environments [13,15,64]. A variation between batches was found, as shown in Table 1. Differences between lots and fish species are also reported by Zotta et al. [65]. In contrast, the bacteriota on fillets from Plant 1 (hand filleted) was more typical for the fish raw material [15]. Although the total number of colonies identified were 176, we cannot rule out that the diversity of the initial bacteriota could be higher given more colonies. However, it is likely that the initial bacteriota on cod industrially processed fillets is a combination of the bacteriota of the raw material and bacteria contaminating the fillets during processing.
3.1.2. Effect of Anaerobic and Aerobic Storage Conditions on the Bacterial Level and Bacteriota
Interestingly, fillets stored aerobically, or vacuum packed to a bacterial count of about 6 log10 CFU/g showed similar bacteriota as the initial bacteriota of the fillets (the fresh samples, Table 2). Photobacterium dominated in hand fileted cod from Plant 1 (92% and 98%, aerobically and vacuum packaged samples, respectively, from initially 93%) and was absent (<4.3% based on the number of isolates analyzed) in Plant 2 fillets, which was still dominated by Pseudomonas (39% from initially 28%), Shewanella (30% from initially 11%), and Acinetobacter (17% from initially 22%). In fillets from Plant 3, Pseudomonas still dominated, as also reported on vacuum packaged cod fillets [23], but the Photobacterium fraction had increased to 24% (initially 3%). Earlier reports have described that vacuum packaging of fresh fish can inhibit Pseudomonas, but it does not necessarily increase shelf life as Photobacterium or Shewanella (depending on initial numbers) will spoil the product [66,67,68]. Still, vacuum packaging is a common technology for fresh cod consumer packages, but the results of both the present and earlier studies show that it has no improved effect on the microbial shelf life compared to aerobic storage.
Table 2.
Bacteria (%) characterized (by 16S rRNA gene sequencing of colonies) from the same samples of fish fillets (n) after 6 (industrial processing plants) and 9 days (controlled facility) of storage (3 °C).
| Plant 1, Day 9 | Plant 1, Day 9 | Plant 2, Day 6 | Plant 3, Day 6 | ||||
|---|---|---|---|---|---|---|---|
| Colonies of Bacterial Genus | Air (n = 4) |
Air (Thawed) (n = 4) |
Vacuum (n = 4) | Vacuum (Thawed) (n = 4) | Air (n = 3) |
Air (n = 3) |
Colonies (n) |
| Photobacterium | 91.7 | 98.1 | 23.8 | 101 | |||
| Pseudomonas | 8.3 | 98.0 | 1.9 | 85.4 | 39.1 | 71.4 | 113 |
| Shewanella | 30.4 | 4.8 | 8 | ||||
| Aeromonas | 4.3 | 1 | |||||
| Psychrobacter | 9.8 | 4 | |||||
| Acinetobacter | 17.4 | 4 | |||||
| Myroides | 4.3 | 1 | |||||
| Pseudochrobactrum | 4.3 | 1 | |||||
| Carnobacterium | 2.0 | 4.9 | |||||
| Colonies in sample (n) | 48 | 50 | 53 | 41 | 21 | 21 | Total: 236 |
| Total count, log10 CFU/g | 6.3 ± 1.0 | 4.2 ± 0.8 | 5.7 ± 0.5 | 4.5 ± 1.2 | 6.3 ± 0.6 | 6.8 ± 0.5 | |
Most studies report bacterial content or bacteriota after a time of storage, and only a few studies refer to initial detection of bacteriota, like Hovda et al. [60] and Antunes-Rohling et al. [29]. They found Pseudomonas, Photobacterium and Shewanella both at Day 0 and after storage for hand filleted cod, similar as shown in the present study.
As expected, the storage time needed to reach bacterial levels associated with spoilage was longer when the initial contamination number was low. Thus, the shelf life of cod fillets stored aerobically and in vacuum seemed to be directly determined by the initial level and composition of the bacteriota.
3.1.3. The Effect of Freezing-Thawing and Vacuum Packaging on the Bacterial Level and Bacteriota
The combination of vacuum packaging and freezing could theoretically improve the microbial shelf life, combining the inhibition of Pseudomonas and lethal effect on Photobacterium after thawing. In the present study, long-term frozen storage (3 months at −20 °C) reduced the total bacterial count of the thawed, refrigerated storage fillets and eliminated Photobacterium of Plant 1 fillets. A lethal effect of long-term freezing on Photobacterium has previously been reported by others [22,32,33,69]. In addition, it allowed the growth of other bacteria, dominated by Pseudomonas, for both the thawed vacuum packaged fillets and the aerobically stored thawed fillets (85% and 98%, respectively) (Table 2).
3.2. Growth of Potential Spoilage Bacteria in a Cod Agar Model System under Different Atmospheres
Several studies have suggested that packaging with carbon dioxide gas combined with oxygen can extend the shelf life of cod [6,16,23,24,27,28,70]. Since the initial bacteriota varied considerably between processing plants, we screened a selection of potential spoilage bacteria isolated from cod fillets stored under different packaging atmospheres to investigate how different combinations of CO2, O2, and N2 inhibit growth (Table 3). Although we observed some strain differences within genera, we found that a combination of high concentrations of CO2 (60%) and O2 (40%) gas completely inhibited growth of Photobacterium, Shewanella, Aeromonas, and Acinetobacter, and partially Pseudomonas when grown on cod juice agar plates (Table 3). In general, the highest reduction or growth inhibition compared to aerobic storage was found for 60% CO2/40% O2, followed by 40% CO2/60% O2, 60% CO2/40% N2 and 40% CO2/60% N2. This corresponds to results obtained on Shewanella [71] and Photobacterium [72] showing highest inhibition of bacterial growth by the combination of CO2 and O2. The additive effect of oxygen together with CO2 on growth reduction was most notable for Photobacterium, Shewanella, and Aeromonas. Among the genera tested, Carnobacterium appeared to have highest tolerance to the different combinations of gas tested, including CO2 and O2.
Table 3.
Growth screening of bacterial strains. Two–five strains (N) per genus were tested. Logarithmic change in bacterial numbers after 21 days of 4 °C storage on cod agar trays inoculated with selected bacteria strains under different atmospheric conditions. Mean values for 2–5 strains for each genus (from Table A1) are given. Positive numbers indicate growth. For each row, entries with the same letter attached to them are not significantly different at the 0.05 level.
| Genus | Air | 100% N2 | 40% CO2, 60% N2 | 60% CO2, 40% N2 | 60% O2, 40% CO2 | 40% O2, 60% CO2 | N (Number of Strains) |
|---|---|---|---|---|---|---|---|
| Carnobacterium | 6.7 a | 6.7 a | 5.1 b | 4.3 b | 5.0 c | 3.7 d | 2 |
| Pseudomonas | 7.9 a | 7.6 a | 2.7 b | 1.5 c | 1.3 c | 0.6 c | 5 |
| Aeromonas | 7.6 a | 7.5 a | 2.1 b | 2.2 b | 0.4 c | 0.3 c | 3 |
| Shewanella | 7.5 a | 6.5 a | 3.9 b | 2.0 c | 0.1 d | 0.1 d | 4 |
| Acinetobacter | 6.4 a | 3.1 a,b | 1.8 a,b | 0.3 b | 0.4 b | 0.0 b | 2 |
| Photobacterium | 3.5 a | 3.8 a | 2.2 a | 2.5 a | −1.3 b | −1.3 b | 2 |
Restricted oxygen conditions (100% N2, 0.02 ± 0.0% O2, Table A2) had an overall insignificant effect on the bacterial growth compared to aerobic storage (Table 3). This result supported the findings in the storage tests described above (Section 3.1.3), that virtually all specific spoilage bacteria in cod may grow relatively fast under oxygen restriction (with no CO2), and the final dominating genera may be dependent on the initial contamination and probably also storage temperature.
Immediately after packaging, the gas mixtures were measured to be 37.7% CO2, 60.0% O2 and 57.9% CO2, 40.1% O2 for the 40% CO2/60% O2 and the 60% CO2/40% O2 mixtures, respectively (measurements only on two of the gas mixtures), showing good compliance to the pre-mixture of the packaging gases.
3.3. Storage Experiment of Pre-Rigor Processed Cod Fillets
Based on the mapping of microbial levels before and after storage of cod under different conditions and the growth potential of bacteria associated with cod, we designed an experiment combining both freezing/thawing and the MAP conditions that showed the most promising results in the growth screening to measure the effect of TVC, bacteriota, VOCs and sensory quality on industrial processed cod fillets. A short-time freezing (40 min at −30 °C) was chosen instead of the previous long-term freezing process as a bacteria inhibitory treatment after the packaging, allowing short-time occupying of freezing capacity at the processing plant prior transport to the market. Vacuum was also tested as this is a commonly used packaging method of cod fillets in Norway. In addition, vacuum packaged fillets of traditional wild catch of Atlantic cod were included as a reference (called “Industry”).
3.3.1. The Effect of Packaging Atmosphere on Development of the Bacteriota during Storage
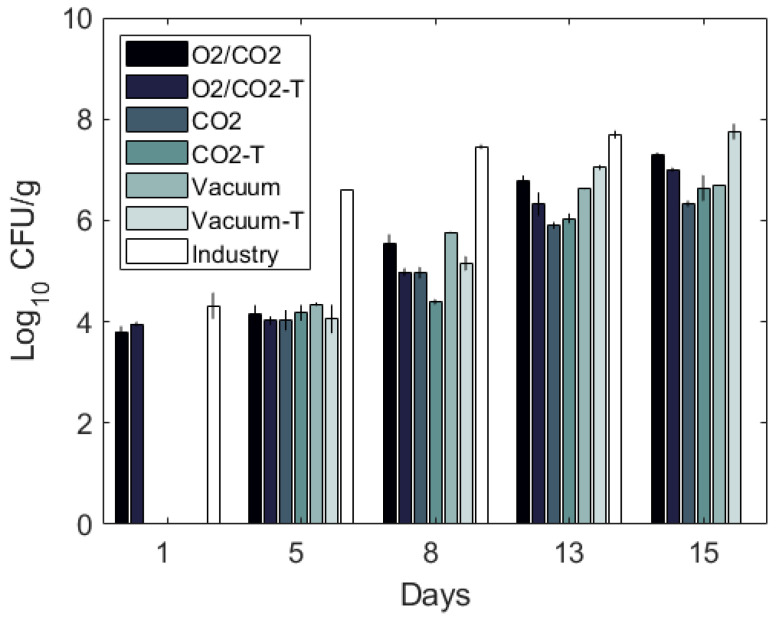
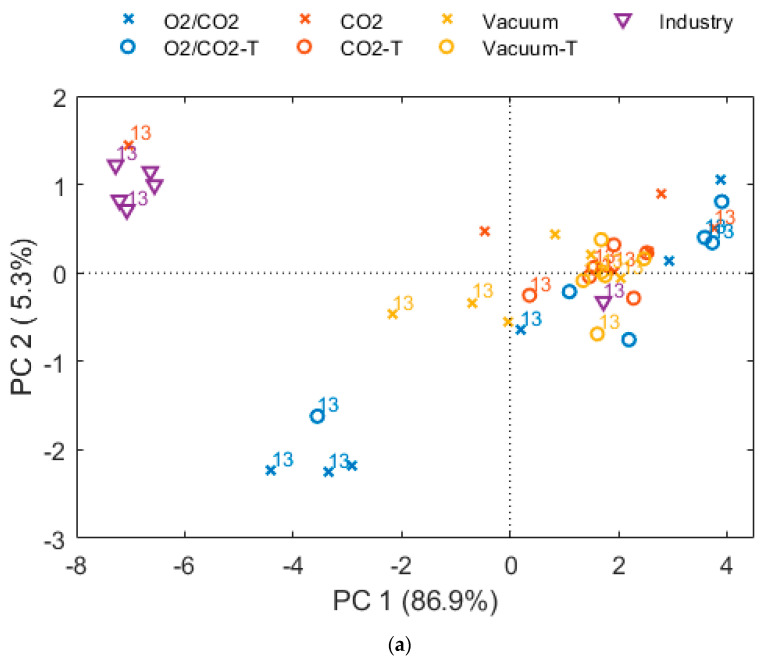
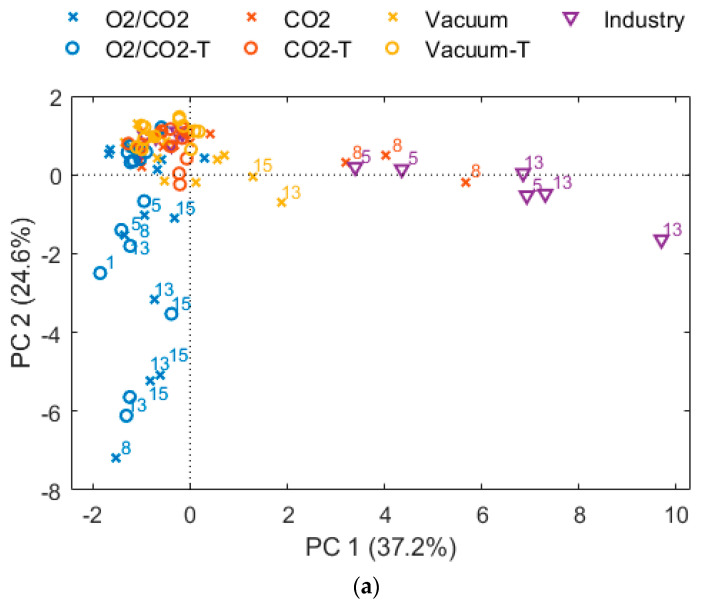
Variations in the bacterial growth rates were found between packaging atmospheres for fresh cod fillets during storage (Figure 2). The lowest level of bacteria after 15 days of storage was found in cod packed with 60% CO2 and highest in cod packed with O2/CO2.
Figure 2.
Total bacterial count (log10 CFU/g) (mean ± standard error of the mean, SEM) per packaging method of cod loins during storage (2 °C). The “CO2” samples are packaged with 60% CO2, 40% N2 and a CO2 emitter, and “O2/CO2” is packaged with 60% CO2 and 40% O2. “Industry” samples and “Vacuum” samples are packaged with vacuum. The samples marked with “T” are thawed samples, and the “CO2”, “O2/CO2”, “Vacuum” and “Industry” are fresh packaged fillets.
The content of CO2 in package headspace during storage can partial explain this. It was measured to be 53.0 ± 1.5% and 35.3 ± 3.2 for the 60% CO2/40% N2 and 60% CO2/40% O2 mixtures, respectively (no differences between fresh and thawed) and 59.7 ± 2.3% O2 during storage for the fresh and thawed O2/CO2 packages.
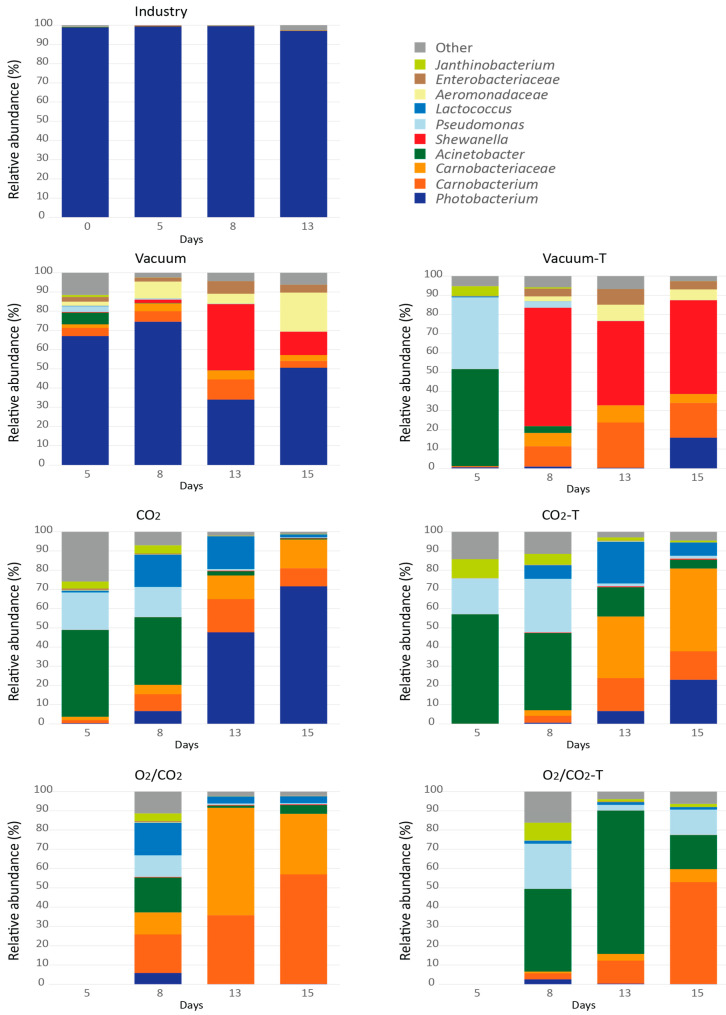
Profound differences were also found for the bacteriota between the packaging atmospheres (Supplementary Table S1 and Figure 2 and Figure 3). A rapid growth of Photobacterium was observed in vacuum packaged samples. After 13 days, the total growth plateaued to some degree, and the bacteriota was dominated by Photobacterium, Shewanella and Aeromonadaceae. As expected, 60% CO2 appeared to inhibit growth of a wide range of bacteria naturally present on cod fillets, including Shewanella and Aeromonadaceae, but to a less degree Photobacterium and Carnobacterium/Carnobacteriaceae, which dominated at the end of storage. In O2/CO2 atmosphere, there was a rapid growth and dominance of Carnobacterium/Carnobacteriaceae. This was not seen by Hovda et al. [24], either for the CO2/N2 or the CO2/O2 atmospheres, although they detected Carnobacterium at Day 0. However, Antunes-Rohling et al. [29] found that Carnobacterium can contribute to spoilage of hake, in addition to Photobacterium under CO2/N2 atmosphere.
Figure 3.
The dominating bacteriota
(relative abundance) during storage with different packaging and processing. Each point is represented as an average of one to three parallels. Taxa with abundance above 1% across all samples is represented, the rest is presented as “Other”. Taxa is represented at either genus (g) or family (f) level by different colors:  Photobacterium;
Photobacterium;  Carnobacterium;
Carnobacterium;  Carnobacteriaceae;
Carnobacteriaceae;  Acinetobacter;
Acinetobacter;  Shewanella;
Shewanella;  Pseudomonas;
Pseudomonas;  Lactococcus;
Lactococcus;  Aeromonadaceae;
Aeromonadaceae;  Enterobacteriaceae;
Enterobacteriaceae;  Jantinobacterium;
Jantinobacterium;  Other. Day 5 samples from O2/CO2 are missing from the MiSeq analysis and are marked with grey shading in the figure.
Other. Day 5 samples from O2/CO2 are missing from the MiSeq analysis and are marked with grey shading in the figure.
The bacterial growth in vacuum packaged cod fillets that were directly delivered from a fishing boat to the fish factory at the same day (termed “Industry”), differed significantly from the fillets produced from live stored cod (Figure 2, Figure 3 and Figure 4). No lag phase and a rapid growth of Photobacterium up to almost 8 log10 CFU/g after 13 days was observed (7.5 log10 CFU/g after 8 days). The origin of the fillets (wild caught vs live stored with a different initial contamination and/or fish flesh properties) and time of processing (later in the day) could have been the cause of the large differences.
Figure 4.
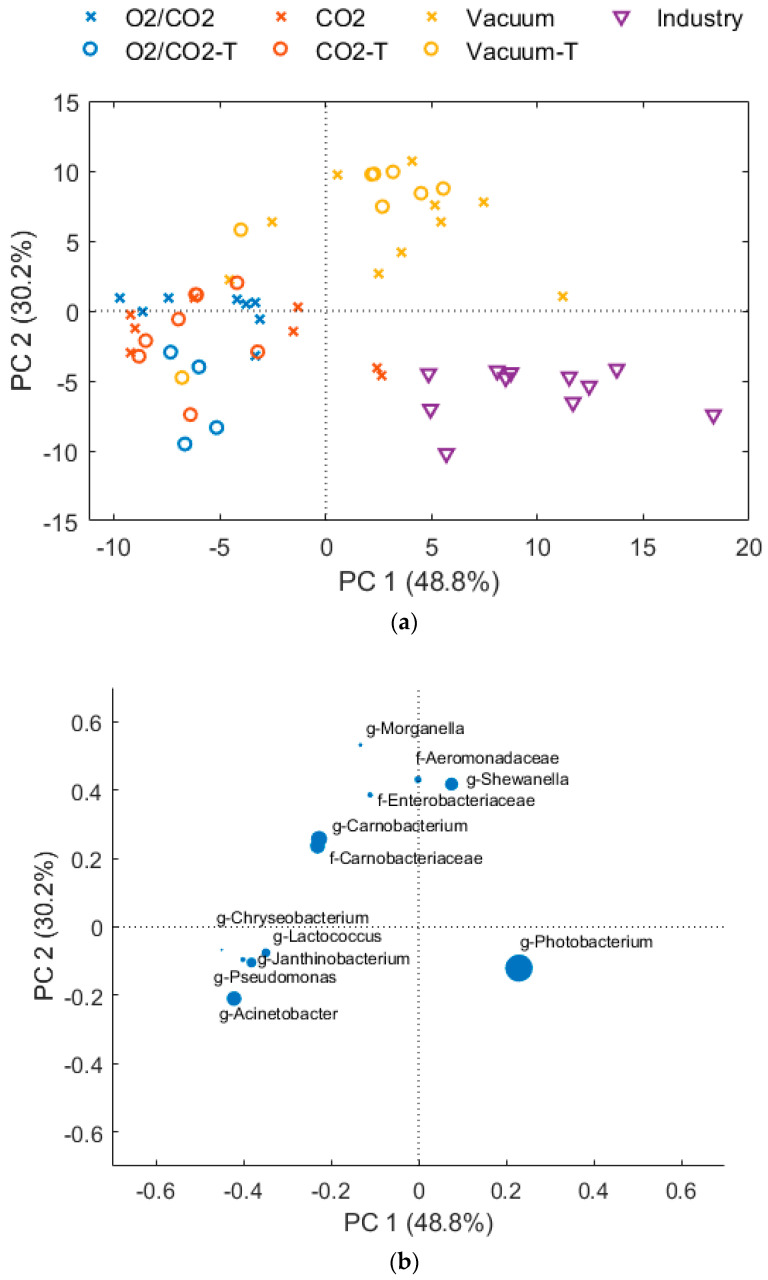
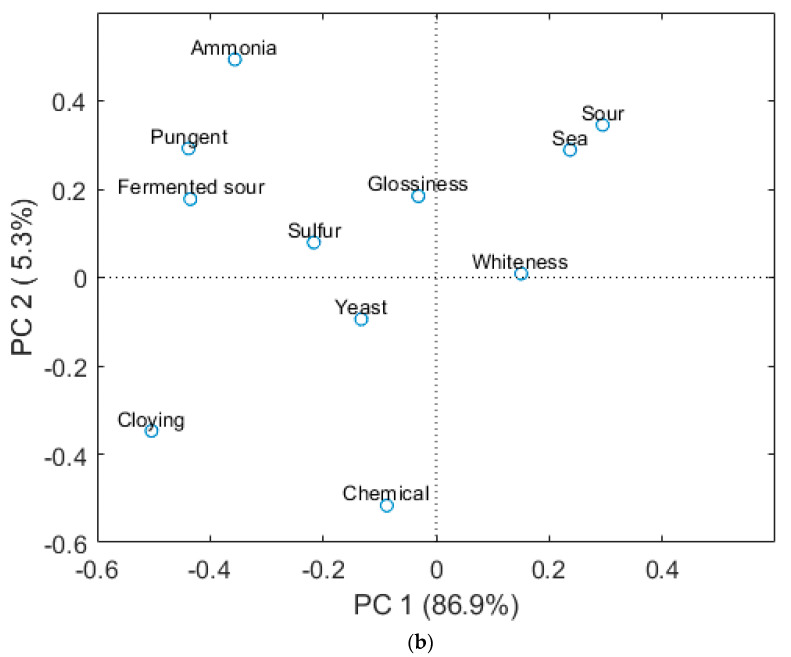
PCA of bacteriota after 1, 5, 8, 13 and 15 days of storage of fresh (cross symbol) and thawed (circle symbol) samples. The “CO2” samples (red color) are packaged with 60% CO2, 40% N2 and a CO2 emitter, and “O2/CO2” (blue color) is packaged with 60% CO2 and 40% O2. “Industry” samples (purple triangle symbol) and “Vacuum” samples (yellow color) are packaged with vacuum. (a) Scores plot, days of storage are indicated by label, (b) loading plot, dot size corresponds to relative abundance.
Overall, the results were in accordance with previous literature that identify a shift in the dominant populations during storage of cod as an effect of storage conditions [8,10]. We have also in an earlier study found a dominance of Carnobacterium during storage of 40% O2/60% CO2 packaged cod [23]. However, Rudi et al. [8] detected a strong association of coalfish with Photobacterium, but salmon was associated with Carnobacterium and Brochothrix (both species packaged with CO2/N2). Similar to the present study, Hovda et al. [24] did not find any difference in total bacterial count between cod packed in CO2 with and without high-O2. However, in contrast to our findings, Pseudomonas dominated, not Carnobacterium in cod packaged with O2/CO2 gas. Another study has reported lower bacterial growth rates for cod packaged with high-O2/CO2 compared to CO2/N2 [28]. Differences between studies may reflect differences between raw materials used and the initial contamination, as shown in the present study where both counts and composition of the contamination of fresh cod varied significantly between three factories. Difference in bacterial composition between different batches is also reported on hake [29].
3.3.2. The Effect of Short-Term Freezing on Development of the Bacteriota during Refrigerated Storage
Depending on packaging gas, the short-term freezing and thawing affected both the total growth rate (Figure 2 and Table A1) and the bacteriota (Supplementary Table S1 and Figure 3 and Figure 4) (p = 0.002 and p < 0.001, respectively). Overall, it seemed like the freezing process eliminated or strongly reduced the relative abundance of Photobacterium in all packaging types until the end of storage and increased the relative abundance of Pseudomonas and Acinetobacter early in the storage period. For thawed, vacuum packed cod, a rapid growth of Shewanella and Carnobacterium/Carnobacteriaceae were observed, resulting nearly 8 log10 CFU/g numbers after 15 days of storage. As also found for fresh samples, CO2 inhibited Shewanella, and when Photobacterium is reduced by freezing/thawing Carnobacterium/Carnobacteriaceae dominated. Moreover, in cod packaged with O2/CO2, there was a rapid growth of Carnobacterium/Carnobacteriaceae, accompanied by Acinetobacter and Pseudomonas in the absence of Photobacterium (for details see Supplementary Table S1). A similar effect of freezing on the microbial composition has also been reported by Emborg et al. [73] and Bøknæs et al. [33]. The present study and other studies imply that important spoilage bacteria, such as Photobacterium, Shewanella and Pseudomonas can be inhibited by a combination of packaging gas and freezing-thawing processes. In the present study, they were replaced by Carnobacterium/Carnobacteriaceae. Carnobacterium has been detected as the prevalent bacteria in tuna, shrimps, swordfish, and cod packaged in high-oxygen MAP (80% O2, 20% N2, and 60% CO2, 40% O2) [16,74]. It has been reported that Carnobacterium is less competitive compared to Photobacterium under CO2/N2 atmosphere, but grows well together with Shewanella baltica [67,75,76].
Although the effects of CO2 and O2 on the bacteriota from the experiment with cod fillets were partly in agreement with the model experiment with agar plates, it was obvious that much less inhibitory effect were found with a food matrix and that growth rates in cod were different from the cod based nutrient agar. For example, Photobacterium showed rapid growth in cod, but very slow growth on agar. There are probably several reasons for this, such as different strains, mixed bacterial communities, better exposure to the gas, and differences between nutrients between the two systems. However, growth on fish is the most relevant as weakness of using model systems may occur.
3.3.3. Effect of Packaging Atmospheres and Short-Term Freezing on Sensory Profiles and Volatile Compounds after Storage
The different packaging concepts, both the type of packaging gas and whether the cod had been frozen and thawed, affected the sensory profiles and level of metabolites (Table A5 and Table A6, Figure 5 and Figure 6). Nevertheless, the “Industry” fillets were deviant compared to the other ones with higher intensity scores of the attributes cloying, fermented sour, ammonia, and pungent compared to the other packaging variants (Table 4 and Figure 5). These attributes can be related to the presence and metabolism of Photobacterium, as this bacterium totally dominated during the refrigerated storage.
Figure 5.
PCA analysis of sensory attributes evaluated after 8 days and 13 days of storage for the fresh (cross symbol) and thawed (circle symbol) samples. The “CO2” samples (red color) are packaged with 60% CO2, 40% N2 and a CO2 emitter, and “O2/CO2” (blue color) is packaged with 60% CO2 and 40% O2. “Industry” samples (purple triangle symbol) and “Vacuum” samples (yellow color) are packaged with vacuum. (a) Scores plot (13 days of storage are indicated by label), (b) loading plot: odor attributes.
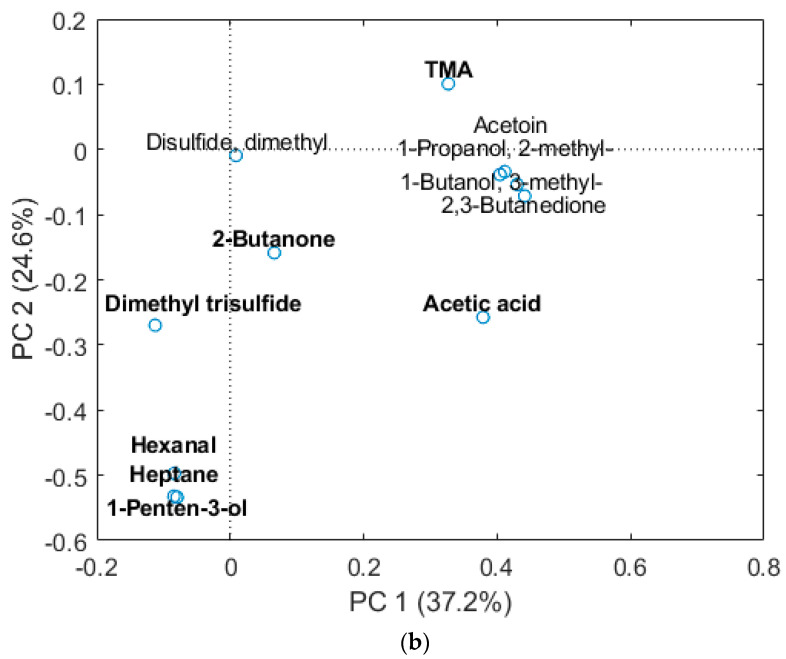
Figure 6.
PCA of volatile components after 8 and 13 days of storage for fresh (cross symbol) and thawed (circle symbol) samples, Table 2. samples (red color) are packaged with 60% CO2, 40% N2 and a CO2 emitter, and “O2/CO2” (blue color) is packaged with 60% CO2 and 40% O2. “Industry” samples (purple triangle symbol) and “Vacuum” samples (yellow color) are packaged with vacuum. (a) Score plot and (b) loading plot. Days of storage are indicated by label for those with score PC1 > 1 or score PC2 < 1. Loading plot: Compounds with significant effect of packaging gas (FDR < 0.05) from the 5050 manova/rotation test for design samples are in bold letters.
Table 4.
Sensory scores of odor attributes (1 = low intensity, 9 = high intensity) after 13 days of storage (2 °C). For each column, entries with the same letter attached to them are not significantly different at the 0.05 level.
| Sour Odor 1 | Sea Odor 1 | Cloying | Fermented Sour | Yeast | Chemi-Cal | Sulfur | Ammonia | Pungent | White-Ness | Glossiness | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CO2 | 2.78 a | 2.44 ab | 3.71 b | 3.26 bcd | 1.49 abc | 1.72 b | 2.40 ab | 2.96 ab | 3.01 b | 5.59 ab | 4.85 a |
| CO2-T | 3.02 a | 2.75 a | 3.17 b | 2.32 cd | 1.22 c | 1.75 b | 2.74 ab | 2.22 b | 1.92 bc | 5.58 ab | 3.35 b |
| Vacuum | 2.09 ab | 2.17 abc | 3.64 b | 3.51 bc | 1.32 c | 1.96 b | 2.55 ab | 2.65 b | 2.04 bc | 5.39 b | 3.99 ab |
| Vacuum-T | 2.98 a | 2.77 a | 3.07 b | 2.66 bcd | 1.14 c | 1.99 b | 1.85 b | 1.79 b | 1.57 c | 6.10 a | 4.61 a |
| O2/CO2 | 1.55 b | 1.37 c | 5.71 a | 3.91 ab | 2.06 a | 3.10 a | 2.62 ab | 2.58 b | 3.06 b | 5.16 b | 4.15 ab |
| O2/CO2-T | 3.10 a | 2.63 a | 3.12 b | 2.22 d | 1.36 bc | 2.22 ab | 2.29 b | 1.89 b | 2.23 bc | 6.07 a | 4.15 ab |
| Industry | 1.68 b | 1.61 bc | 5.90 a | 4.84 a | 1.99 ab | 1.84 b | 3.43 a | 4.27 a | 4.54 a | 5.42 b | 4.42 a |
1 positive associated odor attributes; Green color means that scores are significant different to “Industry” (p = 0.05), and orange color means score different to both “Industry” and the other packaging methods.
In the fresh cod fillets of “CO2” and “Vacuum” the bacteriota was dominated by Photobacterium, and Shewanella and Photobacterium, respectively. The bacterial levels were probably too low (<7 log10 CFU/g) to cause significant sensory changes, especially for the level of Shewanella (sensory impact at levels above 8 log10 CFU/g, according to Dalgaard et al. [39]). In general, the intensities of negatively associated sensory attributes were low after 13 days for all cod fillets packaged in CO2 or vacuum (Table 4). Although Photobacterium dominated in CO2 packaged fillets, it seemed to have a slower growth compared to what was found for the vacuum packaged fillets. A difference in odor intensity scores between vacuum and CO2 packaged could possibly been detected if a further time of storage had been conducted, as differences in odor attributes is previously found between similar packaging methods for cod fillets [12].
After 13 days of storage, fresh cod fillets packed in O2/CO2 showed significantly higher intensities for cloying and chemical odor compared to fillets packaged with “CO2” or in vacuum. The difference in bacteriota for the fresh samples were clear; O2/CO2 samples were dominated by Carnobacterium/Carnobacteriaceae whereas all other samples were dominated by Photobacterium and Photobacterium/Shewanella. Higher intensities of cloying and chemical odor may therefore be caused by Carnobacterium or other members of Carnobacteriaceae, however this must be investigated further both to identify the specific species and to show any causal relationship.
The thawed fillets were dominated by Acinetobacter and Carnobacterium. The growth of Acinetobacter was in contradiction to what was found in the screening test, which showed almost completed inhibition of Acinetobacter under different modified atmospheres. Acinetobacter has not been reported as a spoilage bacterium and seems to be outcompeted by Carnobacterium during further storage (Figure 2).
Laursen et al. [77] reported development of off-flavors like chlorine, chemical, malty, and sour with growth of Carnobacterium on chilled MAP Nordic shrimps. The attribute chlorine was not included in the sensory attribute list in our study but was a part of the description of the attribute chemical (Table A3). Additionally, chlorine was mentioned by the sensory panelist for O2/CO2 packaged fillets, but also for fillets packed with vacuum and CO2. At the same time, the presence of 1-penten-3-ol, heptane, and hexanal were significant effected by packaging gas with higher levels for the cod packaged with O2/CO2 (Figure 6). These components are associated with oxidation of lipids facilitated by the high-oxygen gas mixture [23,78]. The chemical attribute might therefore also be related to oxidation of the fillets, but probably not very likely because of the low lipid content in cod fillet, though this must be further investigated.
Samples that had gone through the freezing/thawing process had lower intensities of fermented sour, and higher sour odor and sea odor after 13 days storage (Table 4). The effect of freezing/thawing depended on packaging gas, and significant interactions between the freezing thawing process and packaging gas was observed for ammonia, cloying, glossiness, pungent, and fermented sour odor (Table A5). Results therefore indicate that freezing/thawing may have a positive effect on the product quality, which correspond to findings by Guldager et al. [32].
Several volatile components of alcohols, esters, ketones, aldehydes, and acids were detected, with a selection of the relevant components shown in Figure 6 and Table A6. TMA is commonly known as a bacterial metabolite of Photobacterium [7,38,39], with an estimation of 30 times higher TMA production by Photobacterium compared to Shewanella [39]. Cod fillets dominated by Carnobacterium have not shown any development of TMA as demonstrated on fillets dominated by Photobacterium [23]. In addition, an association between acetic acid and Photobacterium has been reported [79]. Our results showed that the levels of TMA and acetic acid were affected by packaging gas and process, in addition to 2-butanone and dimethyl trisulfide that were also affected by packaging (Table A6), and as shown in Figure 6. The contents of TMA and acetic acid were also especially pronounced for ordinary industrial vacuum packaged cod fillets (“Industry”) and some of the fillets packaged with CO2 that were dominated by Photobacterium (Figure 6). After eight days of storage, the “Industry” samples had reached high bacterial levels (7.5 log10 CFU/g) and a higher intensity of pungent, cloying, sour fermented, ammonia, and sulfur compared to the other samples (data not shown). A more pronounced difference in the quality of the different packaging concepts would probably have been detected if sensory analyses had been performed after the 15 days of storage.
4. Conclusions
A number of strategies have been proposed to extend the shelf life of cod, such as strict production hygiene, optimization of packaging gases, long-term and short-term freezing and thawing, and extreme cooling regimes. The present work illustrates how technologies that have been reported to inhibit or reduce known spoilage bacteria may not always extend the shelf life. For example, the bacterial growth under vacuum packaging and aerobic storage proved similar for fresh cod. However, combined with freezing/thawing, this packaging technology may improve sensory quality due to a shift in dominating bacteriota from Photobacterium (high spoilage potential) to Shewanella/Carnobacterium.
The present study supports previous studies showing that storage under different modified atmospheres (CO2 and N2 or O2) can be used to control the dominating bacteriota, including specific spoilage organisms, such as Shewanella, Photobacterium, and Pseudomonas. However, when these bacteria were inhibited, a rapid growth of Carnobacterium was found. The dominance of Carnobacterium was linked to the negatively associated attributes of cloying and chemical odor, and possibly also chlorine odor. Carnobacterium spp. may thereby be defined as a specific spoilage organism in cod when other spoilage bacteria are reduced or inhibited.
Altogether, this study presents results that are important to achieve improved quality control of fish fillet products and ultimately inhibit food waste in the value chain.
Acknowledgments
We would like to thank the project leader Geir Sogn-Grundvåg, Nofima. We would also like to thank the skillful project partners; Nergård, Multivac and Tommen Gram, and the staff at the laboratories of Nofima at Ås; microbial and chemical labs, and the sensory panel and packaging facilities.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/foods10081754/s1, Table S1: Bacteriota relative abundance table (Excel file).
Appendix A
Table A1.
Bacterial strains (20) used in growth screening on cod agar plates at different packaging gas mixtures. The bacterial strains were selected from the fillets from Plant 2 and 3 (aerobically stored fillets), which were the commercial processing plants, but similar strains were also detected from Plant 1. Two strains were from vacuum packaged fillets of Plant 1. Three strains were chosen from former industrial studies (*).
| Bacterial Taxa Bacterial Taxa | Source | Strain Number |
|---|---|---|
| Pseudomonas putida | Plant 2, Day 1 | MF 7459 |
| Pseudomonas syringae | Plant 2, Day 6 | MF 7464 |
| Pseudomonas psychrophila | Plant 2, Day 6 | MF 7465 |
| Pseudomonas fluorescens | Plant 3, Day 6 | MF 7470 |
| Pseudomonas fragi | Plant 3, Day 6 | MF 7471 |
| Photobacterium phosphoreum | Plant 3, Day 1 | MF 7469 |
| Photobacterium sp. | Plant 3, Day 6 | MF 7473 |
| Shewanella sp. | Plant 2, Day 1 | MF 7460 |
| Shewanella sp. | Plant 3, Day 6 | MF 7472 |
| Shewanella baltica | From salmon-industry * | MF 5509 |
| Shewanella putrefaciens | From salmon-industry * | MF 5496 |
| Aeromonas sp. | Plant 2, Day 1 | MF 7461 |
| Aeromonas rivuli | Plant 2, Day 6 | MF 7466 |
| Aeromonas salmonicida | From salmon-industry * | MF 5413 |
| Acinetobacter sp. | Plant 2, Day 1 | MF 7462 |
| Acinetobacter sp. | Plant 2, Day 6 | MF 7463 |
| Arthrobacter sp. | Plant 3, Day 1 | MF 7467 |
| Psychrobacter glacincola | Plant 3, Day 1 | MF 7468 |
| Carnobacterium sp. | Plant 1, Day 9 (vacuum, thawed) | MF 7458 |
| Carnobacterium maltaromaticum | Plant 1, Day 9 (vacuum, thawed) | MF 7457 |
Table A2.
The different packaging gas mixtures used for packaging of cod agar plates for growth screening, and the measured levels (%) of CO2 and O2 after 13 days of refrigerated 4 °C storage of at total storage period of three weeks.
| Premix Packaging Gas | Gas Measured Day 13 |
|---|---|
| 60% CO2, 40% N2 | 53.1 ± 0.5% CO2, 0.5 ± 0.3% O2 |
| 40% CO2, 60% N2 | 34.8 ± 0.4% CO2, 0.3 ± 0.4% O2 |
| 60% CO2, 40% O2 | 51.4 ± 0.2% CO2, 44.9 ± 0.2% O2 |
| 40% CO2, 60% O2 | 32.3 ± 0.0% CO2, 63.7 ± 0.0% O2 |
| 100% N2 | 1.0 ± 0.2% CO2, 0.02 ± 0.0% O2 |
| Air | 0.3 ± 0.1% CO2, 20.3 ± 0.0% O2 |
Table A3.
Sensory vocabulary for raw samples of cod fillets (Gadus morhua) of odor attributes and for color/appearance. No intensity = no presence of the specific attributes, evident intensity = evident presence of the specific attributes.
| Sensory Attributes | Description of Attribute |
|---|---|
| Odor | |
| Seawater | Fresh salty, sea, ocean |
| Sour | Related to a fresh, balanced odor due to the presence of organic acids |
| Fermented sour | Fermented sour odor, tainted |
| Cloying | An unfresh and sickening odor |
| Yeast | Odor of yeast |
| Chemical | Related to odors like diesel, burnt, scorched, rubber, chlorine |
| Sulfur | Sulfur odor, boiled egg, cabbage, onion |
| Ammonia | odor of ammonia |
| Pungent | Relates to a sharp and stinging odor |
| Color/Appearance | |
| Whiteness | Paleness, lack of pure color or black |
| Glossiness | overall impression |
Table A4.
5050 manova results for bacteriota. The table shows the false discovery rate (FDR) adjusted p-values and the explained variance for each of the terms included in the model. The "*" is showing the interactions.
| OTUlabel | Day | Gas | Process | Day * Gas | Day * Process | Gas * Process |
|---|---|---|---|---|---|---|
| Explained variance (%) | 13.4 | 50.1 | 4.3 | 3.2 | 1.5 | 4.9 |
| g-Chryseobacterium | 0.000 | 0.000 | 0.090 | 0.968 | 0.654 | 0.468 |
| f-Carnobacteriaceae; Other | 0.054 | 0.001 | 0.889 | 0.461 | 0.654 | 0.009 |
| g-Carnobacterium | 0.066 | 0.002 | 0.316 | 0.461 | 0.654 | 0.031 |
| g-Lactococcus | 0.028 | 0.000 | 0.346 | 0.671 | 0.654 | 0.070 |
| g-Janthinobacterium | 0.000 | 0.000 | 0.000 | 0.968 | 0.654 | 0.666 |
| f-Aeromonadaceae | 0.099 | 0.000 | 0.501 | 0.461 | 0.654 | 0.070 |
| g-Shewanella | 0.230 | 0.000 | 0.289 | 0.597 | 0.654 | 0.024 |
| f-Enterobacteriaceae | 0.054 | 0.000 | 0.316 | 0.461 | 0.654 | 0.439 |
| g-Morganella | 0.256 | 0.000 | 0.056 | 0.461 | 0.483 | 0.024 |
| g-Acinetobacter | 0.000 | 0.000 | 0.003 | 0.461 | 0.654 | 0.148 |
| g-Pseudomonas | 0.000 | 0.000 | 0.001 | 0.671 | 0.882 | 0.666 |
| g-Photobacterium | 0.401 | 0.002 | 0.021 | 0.540 | 0.745 | 0.148 |
Table A5.
ANOVA of sensory descriptive analysis on odors for packaging concepts in storage experiment without industrial samples. Mixed model with effect of assessor, gas, process (freezing/thawing), replicate and two-way interactions. Assessor and interactions with assessors are random effects, other fixed. The "*" is showing the interactions.
| Attribute | Gas | Process | Interaction | Replicate | Gas * Replicate | Process * Replicate |
|---|---|---|---|---|---|---|
| Ammonia odor | 0.015 | 0.000 | 0.009 | 0.005 | 0.000 | 0.000 |
| Cloying odor | 0.059 | 0.000 | 0.004 | 0.148 | 0.000 | 0.000 |
| Yeast odor | 0.044 | 0.000 | 0.144 | 0.715 | 0.000 | 0.061 |
| Chemical odor | 0.063 | 0.091 | 0.147 | 0.157 | 0.100 | 0.086 |
| Seawater odor | 0.188 | 0.001 | 0.230 | 0.821 | 0.000 | 0.000 |
| Pungent odor | 0.004 | 0.000 | 0.000 | 0.105 | 0.000 | 0.000 |
| Fermented sour odor | 0.356 | 0.000 | 0.010 | 0.036 | 0.000 | 0.000 |
| Sulfur odor | 0.088 | 0.010 | 0.610 | 0.257 | 0.000 | 0.000 |
| Sour odor | 0.669 | 0.000 | 0.217 | 0.642 | 0.000 | 0.000 |
| Glossiness | 0.619 | 0.136 | 0.000 | 0.734 | 0.130 | 0.000 |
| Whiteness | 0.292 | 0.000 | 0.105 | 0.003 | 0.000 | 0.002 |
Table A6.
5050 manova results for selected volatile organic compounds. The table shows the false discovery rate (FDR) adjusted p-values for each of the terms included in the model. The "*" is showing the interactions.
| Compound | Day | Gas | Process | Day * Gas | Day * Process | Gas * Process |
|---|---|---|---|---|---|---|
| Explained variance (%) | 18.7 | 14.7 | 2.7 | 12.1 | 3.7 | 3.1 |
| Trimethylamine | 0.0001 | 0.0001 | 0.0421 | 0.00165 | 0.63499 | 0.86033 |
| Dimethyl disulfide | 0.29786 | 0.52467 | 0.13338 | 0.10174 | 0.39613 | 0.0776 |
| Dimethyl trisulfide | 0.47753 | 0.02586 | 0.40159 | 0.89936 | 0.59946 | 0.1146 |
| 3-methyl-1-Butanol | 0.0001 | 0.1159 | 0.12875 | 0.0001 | 0.63499 | 0.1091 |
| 2-methyl-1-Propanol | 0.03457 | 0.40634 | 0.10637 | 0.00233 | 0.30765 | 0.1091 |
| 1-Penten-3-ol | 0.66168 | 0.0001 | 0.83773 | 0.97357 | 0.39613 | 0.96596 |
| Heptane | 0.00887 | 0.00017 | 0.83773 | 0.89936 | 0.65986 | 0.57603 |
| Hexanal | 0.29786 | 0.00025 | 0.96558 | 0.30384 | 0.6959 | 0.99812 |
| 2-Butanone | 0.0001 | 0.03025 | 0.37694 | 0.11575 | 0.60465 | 0.7585 |
| 2,3-Butanedione | 0.0001 | 0.59535 | 0.44496 | 0.30384 | 0.65986 | 0.86033 |
| Acetoin | 0.0001 | 0.64372 | 0.21098 | 0.00347 | 0.63499 | 0.57603 |
| Acetic acid | 0.04599 | 0.04577 | 0.0077 | 0.27464 | 0.2337 | 0.95961 |
Author Contributions
Conceptualization, methodology, writing—review and editing, resources, A.Å.H., B.M. and S.L.; software, formal analysis, investigation, validation, data curation, writing—original draft preparation, visualization, A.Å.H., B.M., S.L., M.Ø.G. and I.B.; project administration, funding acquisition, A.Å.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Norwegian Research Council, grant number 233751 as a part of the CATCH project and as part of the “FoodPilot Plant”, grant number 296083.
Data Availability Statement
The data presented in this study are available on request to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lee P., Osborn S., Whitehead P. Reducing food waste by extending product life. [(accessed on 26 March 2021)];WRAP. 2015 1:44. Available online: wrap.org.uk. [Google Scholar]
- 2.Møller H., Hagtvedt T., Lødrup N., Andersen J.K., Madsen P.L., Werge M., Aare A.K., Reinikainen A., Rosengren Å., Kjellén J., et al. Food Waste and Date Labelling. Issues Affecting the Durability. Nordic Council of Ministers; Copenhagen, Denmark: 2016. p. 100. [Google Scholar]
- 3.FAO . The State of Food and Agriculture. Moving Forward on Food Loss and Waste Reduction. Food and Agricultrue Organization of the United Nations; Rome, Italy: 2019. p. 182. [Google Scholar]
- 4.Caldeira C., De Laurentiis V., Sala S. Assessment of Food Waste Prevention Actions. Publications Office of the European Union; Luxembourg: 2019. p. 175. [Google Scholar]
- 5.Olafsdóttir G., Martinsdóttir E., Oehlenschläger J., Dalgaard P., Jensen B., Undeland I., Mackie I.M., Henehan G., Nielsen J., Nilsen H. Methods to evaluate fish freshness in research and industry. Trends Food Sci. Technol. 1997;8:258–265. doi: 10.1016/S0924-2244(97)01049-2. [DOI] [Google Scholar]
- 6.Dalgaard P., Gram L., Huss H.H. Spoilage and Shelf-Life of Cod Fillets Packed in Vacuum or Modified Atmospheres. Int. J. Food Microbiol. 1993;19:283–294. doi: 10.1016/0168-1605(93)90020-H. [DOI] [PubMed] [Google Scholar]
- 7.Sørensen J.S., Bøknæs N., Mejlholm O., Dalgaard P. Superchilling in combination with modified atmosphere packaging resulted in long shelf-life and limited microbial growth in Atlantic cod (Gadus morhua L.) from capture-based-aquaculture in Greenland. Food Microbiol. 2020;88:1–11. doi: 10.1016/j.fm.2019.103405. [DOI] [PubMed] [Google Scholar]
- 8.Rudi K., Maugsten T., Hannevik S., Nissen H. Explorative multivariate analyses of 16S rDNA microbial community data from modified atmosphere-packed (MAP) salmon and coalfish. Appl. Environ. Microbiol. 2004;70:5010–5018. doi: 10.1128/AEM.70.8.5010-5018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gram L., Ravn L., Rasch M., Bruhn J.B., Christensen A.B., Givskov M. Food spoilage—interactions between food spoilage bacteria. Int. J. Food Microbiol. 2002;78:79–97. doi: 10.1016/S0168-1605(02)00233-7. [DOI] [PubMed] [Google Scholar]
- 10.Macé S., Cornet J., Chevalier F., Cardinal M., Pilet M.-F., Dousset X., Joffraud J.-J. Characterisation of the spoilage microbiota in raw salmon (Salmo salar) steaks stored under vacuum or modified atmosphere packaging combining conventional methods and PCR-TTGE. Food Microbiol. 2012;30:164–172. doi: 10.1016/j.fm.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 11.Hansen A.Å., Mørkøre T., Rudi K., Rødbotten M., Bjerke F., Eie T. Quality changes of pre-rigor filleted Atlantic salmon (Salmo salar L.) packaged in modified atmosphere using CO2 emitter, traditional MAP, and vacuum. J. Food Sci. 2009;74:M242–M249. doi: 10.1111/j.1750-3841.2009.01233.x. [DOI] [PubMed] [Google Scholar]
- 12.Hansen A.Å., Moen B., Rødbotten M., Berget I., Pettersen M.K. Effect of vacuum or modified atmosphere packaging (MAP) in combination with a CO2 emitter on quality parameters of cod loins (Gadus morhua) Food Packag. Shelf Life. 2016;9:29–37. doi: 10.1016/j.fpsl.2016.05.005. [DOI] [Google Scholar]
- 13.Gram L., Huss H.H. Microbiological spoilage of fish and fish products. Int. J. Food Microbiol. 1996;33:121–137. doi: 10.1016/0168-1605(96)01134-8. [DOI] [PubMed] [Google Scholar]
- 14.Riiser E.S., Haverkamp T.H.A., Varadharajan S., Borgan Ø., Jakobsen K.S., Jentoft S., Star B. Switching on the light: Using metagenomic shotgun sequencing to characterize the intestinal microbiome of Atlantic cod. Environ. Microbiol. 2019;21:2576–2594. doi: 10.1111/1462-2920.14652. [DOI] [PubMed] [Google Scholar]
- 15.Møretrø T., Moen B., Heir E., Hansen A.Å., Langsrud S. Contamination of salmon fillets and processing plants with spoilage bacteria. Int. J. Food Microbiol. 2016;237:98–108. doi: 10.1016/j.ijfoodmicro.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 16.Hansen A.Å., Rødbotten M., Lea P., Rotabakk B.T., Birkeland S., Pettersen M.K. Effect of transport packaging and repackaging into modified atmosphere on shelf life and quality of thawed Atlantic cod loins. Packag. Technol. Sci. 2015;28:925–938. doi: 10.1002/pts.2139. [DOI] [Google Scholar]
- 17.Pantazi D., Papavergou A., Pournis N., Kontominas M.G., Savvaidis I.N. Shelf-life of chilled fresh Mediterranean swordfish (Xiphias gladius) stored under various packaging conditions: Microbiological, biochemical and sensory attributes. Food Microbiol. 2008;25:136–143. doi: 10.1016/j.fm.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Pennacchia C., Ercolini D., Vallani F. Spoilage-related microbiota associated with chilled beef stored in air or vacuum pack. Food Microbiol. 2011;28:84–93. doi: 10.1016/j.fm.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Gill C.O., Penney N. The Effect of the Initial Gas Volume to Meat Weight Ratio on the Storage Life of Chilled Beef Packaged under Carbon-Dioxide. Meat Sci. 1988;22:53–63. doi: 10.1016/0309-1740(88)90026-5. [DOI] [PubMed] [Google Scholar]
- 20.Randell K., Ahvenainen R., Hattula T. Effect of the Gas/Product ratio and CO2 concentration on the shelf-life of MA packed fish. Packag. Technol. Sci. 1995;8:205–218. doi: 10.1002/pts.2770080404. [DOI] [Google Scholar]
- 21.Dixon N.M., Kell D.B. The Inhibition by CO2 of the Growth and Metabolism of Microorganisms. J. Appl. Bacteriol. 1989;67:109–136. doi: 10.1111/j.1365-2672.1989.tb03387.x. [DOI] [PubMed] [Google Scholar]
- 22.Sørensen J.S., Ørnfeld-jensen O., Bøknæs N., Mejlholm O., Jessen F., Dalgaard P. Thawed and chilled Atlantic cod (Gadus morhua L.) from Greenland—options for improved distribution. LWT Food Sci. Technol. 2020;131:109473. doi: 10.1016/j.lwt.2020.109473. [DOI] [Google Scholar]
- 23.Hansen A.Å., Mørkøre T., Rudi K., Olsen E., Eie T. Quality changes during refrigerated storage of MA-Packaged pre-rigor fillets of farmed Atlantic cod (Gadus morhua L.) using traditional MAP, CO2-emitter, and vacuum. J. Food Sci. 2007;72:M423–M430. doi: 10.1111/j.1750-3841.2007.00561.x. [DOI] [PubMed] [Google Scholar]
- 24.Hovda M.B., Lunestad B.T., Sivertsvik M., Rosnes J.T. Characterisation of the bacterial flora of modified atmosphere packaged farmed Atlantic cod (Gadus morhua) by PCR-DGGE of conserved 16S rRNA gene regions. Int. J. Food Microbiol. 2007;117:68–75. doi: 10.1016/j.ijfoodmicro.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 25.Kontominas M.G., Badeka A.V., Kosma I.S., Nathanailides C.I. Recent developments in seafood packaging technologies. Foods. 2021;10:940. doi: 10.3390/foods10050940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boskou G., Debevere J. Reduction of trimethylamine oxide by Shewanella spp. under modified atmospheres in vitro. Food Microbiol. 1997;14:543–553. doi: 10.1006/fmic.1997.0132. [DOI] [Google Scholar]
- 27.Debevere J., Boskou G. Effect of modified atmosphere packaging on the TVB/TMA-producing microflora of cod fillets. Int. J. Food Microbiol. 1996;31:221–229. doi: 10.1016/0168-1605(96)01001-X. [DOI] [PubMed] [Google Scholar]
- 28.Sivertsvik M. The optimized modified atmosphere for packaging of pre-rigor filleted farmed cod (Gadus morhua) is 63 mL/100 mL oxygen and 37 mL/100 mL carbon dioxide. LWT Food Sci. Technol. 2007;40:430–438. doi: 10.1016/j.lwt.2005.12.010. [DOI] [Google Scholar]
- 29.Antunes-Rohling A., Calero S., Halaihel N., Marquina P., Raso J., Calanche J., Beltran J.A., Alvarez I., Cebrian G. Characterization of the spoilage microbiota of Hake fillets packaged under a modified atmosphere (MAP) rich in CO2 (50% CO2/50% N2) and stored at different temperatures. Foods. 2019;8:489. doi: 10.3390/foods8100489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parlapani F.F., Kormas K.A., Boziaris I.S. Microbiological changes, shelf life and identification of initial and spoilage microbiota of sea bream fillets stored under various conditions using 16S rRNA gene anlaysis. J. Sci Food Agric. 2015;95:2386–2394. doi: 10.1002/jsfa.6957. [DOI] [PubMed] [Google Scholar]
- 31.Stewart M.M. The effect of exposure to low temperatures on the numbers of bacteria in fish’s muscle. J. Soc. Chem. Ind. 1934;24:273–278. [Google Scholar]
- 32.Guldager H.S., Bøknæs N., Osterberg C., Nielsen J., Dalgaard P. Thawed cod fillets spoil less rapidly than unfrozen fillets when stored under modified atmosphere at 2 degrees C. J. Food Prot. 1998;61:1129–1136. doi: 10.4315/0362-028X-61.9.1129. [DOI] [PubMed] [Google Scholar]
- 33.Bøknæs N., Østerberg C., Nielsen J., Dalgaard P. Influence of freshness and frozen storage temperature on quality of thawed cod fillets stored in modified atmosphere packaging. LWT Food Sci. Technol. 2000;33:244–248. doi: 10.1006/fstl.2000.0634. [DOI] [Google Scholar]
- 34.Bøknæs N., Jensen K.N., Guldager H.S., Østerberg C., Nielsen J., Dalgaard P. Thawed chilled Barents Sea cod fillets in modified atmosphere packaging-application of multivariate data analysis to select key parameters in good manufacturing practice. LWT Food Sci. Technol. 2002;35:436–443. doi: 10.1006/fstl.2001.0876. [DOI] [Google Scholar]
- 35.Warm K., Bøknæs N., Nielsen J. Development of Quality Index Methods for evaluation of frozen cod (Gadus morhua) and cod fillets. J. Aquat. Food Prod. Technol. 1998;7:45–59. doi: 10.1300/J030v07n01_04. [DOI] [Google Scholar]
- 36.Nuin M., Alfaro B., Cruz Z., Argarate N., George S., La Marc Y., Olley J., Pin C. Modelling spoilage of fresh turbot and evaluaton of time-temperature integrator (TTI) label under fluctuating temperature. Int. J. Food Microbiol. 2008;127:193–199. doi: 10.1016/j.ijfoodmicro.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 37.Dalgaard P., Mejholm O., Christiansen T.J., Huss H.H. Importance of Photobacterium phosphoreum in relation to spoilage of modified atmosphere-packed fish products. Lett. Appl. Microbiol. 1997;24:373–378. doi: 10.1046/j.1472-765X.1997.00152.x. [DOI] [Google Scholar]
- 38.Odeyemi O.A., Burke C.M., Bolch C.C.J., Stanley R. Seafood spoilage microbiota and associated volatile organic compounds at different storage temperatures and packaging conditions. Int. J. Food Microbiol. 2018;280:87–99. doi: 10.1016/j.ijfoodmicro.2017.12.029. [DOI] [PubMed] [Google Scholar]
- 39.Dalgaard P. Qualitative and Quantitative Characterization of Spoilage Bacteria from Packed Fish. Int. J. Food Microbiol. 1995;26:319–333. doi: 10.1016/0168-1605(94)00137-U. [DOI] [PubMed] [Google Scholar]
- 40.Chaillou S., Chaulot-Talmon A., Caekebeke H., Cardinal M., Christieans S., Denis C., Desmonts M.H., Dousset X., Feurer C., Hamon E., et al. Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. ISME J. 2015;9:1105–1118. doi: 10.1038/ismej.2014.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.FOR-2004-12-22-1878 . Norwegian Regulations (Forskrift om utøvelse av fisket i sjøen) Ministry of Trade, Industry and Fisheries; Oslo, Norway: [(accessed on 27 November 2020)]. Available online: https://lovdata.no/dokument/SF/forskrift/2004-12-22-1878#KAPITTEL_20. [Google Scholar]
- 42.Ageeva T.N., Olsen R.L., Joensen S., Esaiassen M. Quality aspects of fillets, loin and tail products made from live-stored feed-deprived Atlantic cod (Gadus morhua L.) at different times post mortem. LWT Food Sci. Technol. 2018;97:656–661. doi: 10.1016/j.lwt.2018.06.031. [DOI] [Google Scholar]
- 43.Van Spreekens K.J.A. The sustainability of a modification of Long and Hammer’s medium for the enumeration of more fastidious bacteria from fresh fishery products. Arch. Lebensm. 1974;25:213–219. [Google Scholar]
- 44.Nadkarni M.A., Martin F.E., Jacques N.A., Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- 45.McClean M.N. Colony PCR (Yeast) [(accessed on 14 January 2019)]; Available online: https://openwetware.org/wiki/McClean:_Colony_PCR_(Yeast)
- 46.Bjerke G.A., Rudi K., Avershina E., Moen B., Axelsson L. Exploring the brine mirobiota of a traditional norwegian fermented fish produtc (rakfisk) from six different producers during two consecutive seasonal productions. Foods. 2019;8:72. doi: 10.3390/foods8020072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chandrasekaran M., Lakshmanaperumalsamy P., Chandramohan D. Fish flesh agar medium—A suitable experimental medium for the detection of spoilage bacteria. Antonie Leeuwenhoek. 1985;51:219–225. doi: 10.1007/BF02310014. [DOI] [PubMed] [Google Scholar]
- 48.Boskou G., Debevere J. In vitro study of TMAO reduction by Shewanella putrefaciens isolated from cod fillets packed in modified atmosphere. Food Addit. Contam. 1998;15:229–236. doi: 10.1080/02652039809374634. [DOI] [PubMed] [Google Scholar]
- 49.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N., Owens S.M., Betley J., Fraser L., Bauer M., et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansen A.Å., Moen B., Storø P.G., Pettersen M.K. Improved microbial control of CO2 packaged salmon fillets compared to whole, gutted salmon. Adv. Food Process. Technol. 2018;10:1–11. doi: 10.29011/AFPT-106.100006. [DOI] [Google Scholar]
- 51.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushmn F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olsen E., Vogt G., Saarem K., Greibrokk T., Nilsson A. Autoxidation of cod liver oil with tocopherol and ascorbyl palmitate. J. Am. Oil Chem. Soc. 2005;82:97–103. doi: 10.1007/s11746-005-1049-6. [DOI] [Google Scholar]
- 53.ISO 13229:2016 . Sensory Analysis—General Guidance for Establishing a Sensory Profile. International Organization for Standardization; Geneva, Switzerland: 2016. [Google Scholar]
- 54.ISO 13229:2012 . Sensory Analysis—General Guidance for Selection, Training and Monitoring of Selected Sensory Assessors and Expert Assessors. International Organization for Standardization; Geneva, Switzerland: 2012. [Google Scholar]
- 55.ISO 8589:2007 . Sensory Analysis—General Guidance for the Design of Test Rooms. International Organization for Standardization; Geneva, Switzerland: 2007. [Google Scholar]
- 56.Langsrud Ø. 50-50 multivariate analysis of variance for collinear responses. J. R. Stat. Soc. Ser. D Stat. 2002;51:305–317. doi: 10.1111/1467-9884.00320. [DOI] [Google Scholar]
- 57.Langsrud Ø. Rotation tests. Stat. Comput. 2005;15:53–60. doi: 10.1007/s11222-005-4789-5. [DOI] [Google Scholar]
- 58.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: [(accessed on 21 August 2020)]. Available online: https://wwwR-project.org/ [Google Scholar]
- 59.Olafsdottir G., Lauzon H.L., Martinsdottir E. Evaluation of shelf life of superchilled cod (Gadus morhua) fillets and the influence of temperature fluctuations during storage on microbial and chemical quality indicators. J. Food Sci. 2006;71:S97–S109. doi: 10.1111/j.1365-2621.2006.tb08928.x. [DOI] [Google Scholar]
- 60.Hovda M.B., Sivertsvik M., Lunestad B.T., Lorentzen G., Rosnes J.T. Characterisation of the dominant bacterial population in modified atmosphere packaged farmed halibut (Hippoglossus hippoglossus) based on 16S rDNA-DGGE. Food Microbiol. 2007;24:362–371. doi: 10.1016/j.fm.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 61.Wilson B., Danilowicz B.S., Meijer W.G. The diversity of bacterial communities associated with Atlantic Cod Gadus morhua. Microb. Ecol. 2008;55:425–434. doi: 10.1007/s00248-007-9288-0. [DOI] [PubMed] [Google Scholar]
- 62.Reynisson E., Lauzon H.L., Magnússon H., Jónsdóttir R., Ólafsdóttir G., Marteinsson V., Hreggviðsson G.O. Bacterial composition and succession during storage of North-Atlantic cod (Gadus morhua) at superchilled temperatures. BMC Microbiol. 2009;9:250. doi: 10.1186/1471-2180-9-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Georgala D.L. The bacterial flora of the skin of North Sea cod. J. Gen. Microbiol. 1958;18:84–91. doi: 10.1099/00221287-18-1-84. [DOI] [PubMed] [Google Scholar]
- 64.Bagge-Ravn D., Ng Y., Hjelm M., Christiansen J.N., Johansen C., Gram L. The microbial ecology of processing equipment in different fish industries—analysis of the microflora during processing and following cleaning and disinfection. Int. J. Food Microbiol. 2003;87:239–250. doi: 10.1016/S0168-1605(03)00067-9. [DOI] [PubMed] [Google Scholar]
- 65.Zotta T., Parente E., Ianniello R.G., De Filippis F., Ricciardi A. Dynamics of bacterial communities and interaction networks in thawed fish fillets during chilled storage in air. Int. J. Food Microbiol. 2019;293:102–113. doi: 10.1016/j.ijfoodmicro.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 66.Jørgensen B.R., Huss H.H. Growth and activity of Shewanella putrefaciens isolated from spoiling fish. Int. J. Food Microbiol. 1989;9:51–62. doi: 10.1016/0168-1605(89)90037-8. [DOI] [PubMed] [Google Scholar]
- 67.Leroi F. Occurence and role of lactic acid bacteria in seafood products. Food Microbiol. 2010;27:698–709. doi: 10.1016/j.fm.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 68.Dalgaard P., Gram L., Huss H.H. The effect of anaerobic conditions and carbon dioxide. In: Huss H.H., editor. Quality and Quality Changes in Fresh Fish. FAO; Rome, Italy: 1995. pp. 65–69. [Google Scholar]
- 69.Fujii T., Kurihara K., Okuzumi M. Viability and histidine decarboxylase activity of halophilic histamine-forming bacteria during frozen storage. J. Food Prot. 1994;57:611–613. doi: 10.4315/0362-028X-57.7.611. [DOI] [PubMed] [Google Scholar]
- 70.Stenström I.M. Microbiological flora of cod fillets (Gadus morhua) stored at 2 °C in different mixtures of carbon dioxide and nitrogen/oxygen. J. Food Prot. 1985;48:585–589. doi: 10.4315/0362-028X-48.7.585. [DOI] [PubMed] [Google Scholar]
- 71.López-Caballero M.E., Sánchez-Fernández J.A., Moral A. Growth and metabolic activity of Shewanella putrefaciens maintained under different CO2 and O2 concentrations. Int. J. Food Microbiol. 2001;64:277–287. doi: 10.1016/S0168-1605(00)00473-6. [DOI] [PubMed] [Google Scholar]
- 72.López-Caballero M.E., Torres M.D.A., Sánchez-Fernández J.A., Moral A. Photobacterium phosphoreum isolated as a luminescent colony from spoiled fish, cultured in model system under controlled atmospheres. Eur. Food Res. Technol. 2002;215:390–395. doi: 10.1007/s00217-002-0575-1. [DOI] [Google Scholar]
- 73.Emborg J., Laursen B.G., Rathjen T., Dalgaard P. Microbial spoilage and formation of biogenic amines in fresh and thawed modified atmosphere-packed salmon (Salmo salar) at 2 degrees C. J. Appl. Microbiol. 2002;92:790–799. doi: 10.1046/j.1365-2672.2002.01588.x. [DOI] [PubMed] [Google Scholar]
- 74.Franzetti L., Scarpellini M., Mora D., Galli A. Carnobacterium spp. in seafood packaged in modified atmosphere. Ann. Microbiol. 2003;53:189–198. [Google Scholar]
- 75.Macé S., Cardinal M., Jaffrès E., Cornet J., Lalanne V., Chevalier F., Sérot T., Pilet M.-F., Dousset X., Joffraud J.J. Evaluation of the spoilage potential of bacteria isolated from spoiled cooked whole tropical shrimp (Penaeus vannamei) stored under modified atmosphere packaging. Food Microbiol. 2014;40:9–17. doi: 10.1016/j.fm.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 76.Leisner J.J., Laursen B.G., Prévost H., Drider D., Dalgaard P. Carnobacterium: Positive and negative effects in the environment and in foods. FEMS Microbiol. Rev. 2007;31:592–613. doi: 10.1111/j.1574-6976.2007.00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Laursen B.G., Leisner J.J., Dalgaard P. Carnobacterium species: Effect of metabolic activity and interaction with Brochotrix thermosphacta on sensory characteristics of modified atmosphere packed shrimp. J. Agric. Food Chem. 2006;54:3604–3611. doi: 10.1021/jf053017f. [DOI] [PubMed] [Google Scholar]
- 78.Nieminen T.T., Dalgaard P., Bjorkroth J. Volatile organic compounds and Photobacterium phosphoreum associated with spoilage of modified atmosphere packaged raw pork. Int. J. Food Microbiol. 2016;218:86–95. doi: 10.1016/j.ijfoodmicro.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 79.Macé S., Mamlouk K., Chipchakova S., Prévost H., Joffraud J.J., Dalgaard P., Pilet M.-F., Dousset X. Development of a rapid Real-time PCR method as a tool to quantify viable Photobacterium phosphoreum bacteria in salmon (Salmo salar) steaks. Appl. Environ. Microbiol. 2013;79:2612–2619. doi: 10.1128/AEM.03677-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request to the corresponding author.