Abstract
Translationally inactive histone mRNA is stored in frog oocytes, and translation is activated at oocyte maturation. The replication-dependent histone mRNAs are not polyadenylated and end in a conserved stem-loop structure. There are two proteins (SLBPs) which bind the 3′ end of histone mRNA in frog oocytes. SLBP1 participates in pre-mRNA processing in the nucleus. SLBP2 is oocyte specific, is present in the cytoplasm, and does not support pre-mRNA processing in vivo or in vitro. The stored histone mRNA is bound to SLBP2. As oocytes mature, SLBP2 is degraded and a larger fraction of the histone mRNA is bound to SLBP1. The mechanism of activation of translation of histone mRNAs may involve exchange of SLBPs associated with the 3′ end of histone mRNA.
In the early development of many organisms, the major level of gene regulation is translational. There is a “switch” from translation of maternal mRNAs that are necessary for oogenesis and maintenance of the oocyte to translation of stored maternal mRNAs that encode essential products for embryogenesis. At the time of translational activation there are a number of modifications to the stored mRNP (reviewed in reference 15). These modifications include cytoplasmic polyadenylation of selected mRNAs (35, 37, 38, 43, 47), which is determined by a sequence, the cytoplasmic polyadenylation element, in the 3′ untranslated region (UTR) (11); modification of the cap (19); and removal of proteins which inhibit translation (25, 28, 45). Some or all of these modifications are involved in the selective activation of translation of mRNAs at oocyte maturation in the frog. The lengthening of the poly(A) tail during oocyte maturation is an essential modification for translation of the c-mos (14, 37) and cyclin mRNAs (38). The poly(A) tail has been implicated in the recruitment of mRNAs by the ribosome (18, 30), through an interaction of the poly(A) binding protein with translation initiation factor eIF4G in yeast (40, 41), or by interaction with other proteins that may form a larger complex with the initiation factors (7).
Histone mRNAs are unique among metazoan mRNAs in that they lack a poly(A) tail, ending instead in a conserved stem-loop that fulfills many of the functions of the poly(A) tail on other mRNAs (23). The stem-loop interacts with a specific RNA-binding protein, termed the stem-loop binding protein (SLBP), which participates in pre-mRNA processing (22, 46) and remains associated with the mature histone mRNA as a component of the polyribosomal histone mRNP (8, 16). The stem-loop is essential for translation, presumably as a complex with SLBP (13, 39).
A common feature of all embryos that have a rapid cleavage stage is an extraordinary demand for histone proteins to incorporate into chromatin. In order to provide the necessary amount of histone protein during early embryogenesis, there is not the normal coupling of DNA replication and histone synthesis. Instead histone mRNAs are stable and histone proteins often are stored for later assembly into chromatin. The demand for histone proteins increases exponentially as cleavage proceeds. Thus, as much new histone must be incorporated into chromatin as the embryo cleaves from 1,000 to 2,000 cells as in the entire previous cleavage period. In the frog, all of the histone mRNA (and 75% of the histone protein) to support cleavage until the midblastula transition (MBT) (4,000 cells) is stored in the oocyte (51, 52), since there is no transcription until the MBT (31). After the MBT, there is active synthesis of histone mRNA to supply additional templates for the synthesis of new histones (42). Histone mRNA translation is activated at oocyte maturation to provide the remaining 25% of the histone protein needed prior to the MBT. The molecular basis of the activation of histone mRNA translation is not known, nor is it known how the histone mRNA is stored in an inactive form. It is known that some of the stored histone mRNAs have short oligo(A) tails (20, 34) that have been added to the stem-loop (4). These oligo(A) tails are removed at oocyte maturation (4).
We report here that Xenopus oocytes have two SLBPs. One, SLBP1, is the homologue of the mammalian SLBP present in somatic cells. The other, SLBP2, is an oocyte-specific form. SLBP2 is cytoplasmic, is present in high levels during early oogenesis when histone mRNA is stored, and is destroyed early in embryogenesis. In contrast, SLBP1 is present in low levels in early oogenesis and increases in amount during late oogenesis and early embryogenesis. It is likely that exchange of the SLBPs bound to histone mRNA is part of the mechanism of translational activation at oocyte maturation.
MATERIALS AND METHODS
Yeast three-hybrid screen.
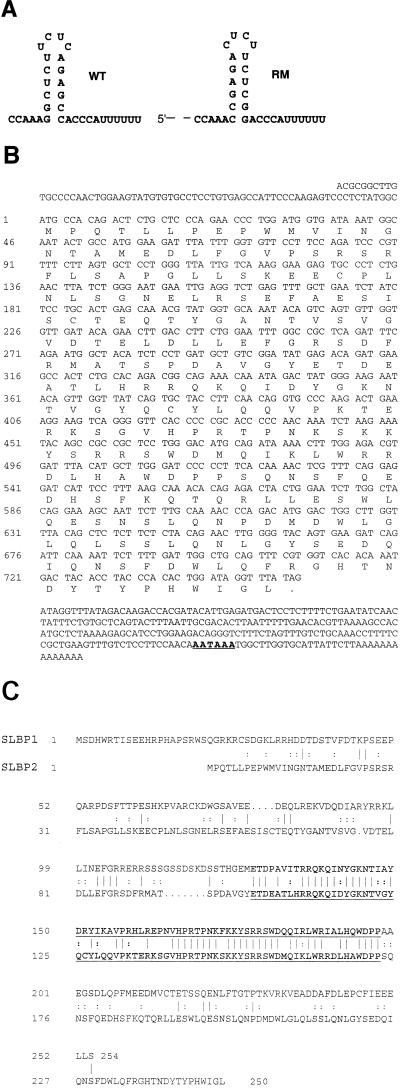
Four million transformants of Xenopus oocyte cDNA cloned into pGAD10 were screened as described previously with the stem loop sequence used as a bait (46). Plasmids were recovered from yeast that had activated both the his3 and lacZ genes and tested for the ability to activate these genes in yeast expressing a mutant stem-loop as the RNA bait (see Fig. 1A). Those plasmids that did not activate the mutant stem-loop were sequenced, and each encoded an SLBP. The complete sequence of the longest SLBP2 clone was obtained at the University of North Carolina DNA Sequencing Facility.
FIG. 1.
Isolation of SLBP2. (A) Wild-type (WT) and mutant (RM) stem-loop sequences used in the yeast three-hybrid selection and RNA binding experiments reported in this paper. (B) Complete nucleotide sequence of the longest SLBP2 clone. The putative polyadenylation signal is underlined. (C) Comparison of the protein sequences of SLBP1 and SLBP2. The RNA binding domains of the two SLBPs are shown in boldface type and underlined. Vertical lines indicate identical amino acids, and double dots indicate similar amino acids.
Treatment of oocytes with antisense oligonucleotides.
A series of 12 oligonucleotides (AX01 to AX12, each 15 nucleotides [nt]) were designed to hybridize to the SLBP1 mRNA every 80 nucleotides, with the first, AX01, complementary to a region in the 5′ UTR and the last, AX12, complementary to a sequence in the 3′ UTR. The oligonucleotides were dissolved in 10 mM Tris–1 mM EDTA, extracted with phenol-chloroform, precipitated with ethanol, and then resuspended in water at a concentration of 1 mg/ml. These oligonucleotides were tested both for the ability to destroy SLBP1 mRNA and for toxicity to the oocyte. Thirty nanoliters (30 ng) of each oligonucleotide was injected into the oocyte cytoplasm. Four hours later, RNA was prepared, resolved by gel electrophoresis, and assayed by Northern blotting with an SLBP1-specific probe. Injected oocytes were also harvested at various times and cell lysates were assayed for SLBP by mobility shift assay, using a radiolabeled stem-loop probe. The oligonucleotide that destroyed SLBP1 mRNA and showed the least toxicity was AX03, which was complementary to nt 457 to 471 of SLBP1 (accession no. U75681) (with the initiator methionine at nt 1) and had the following sequence: 5′-CATATCTTCCTTCCTT. Oocytes were injected with AX09 and allowed to recover for 16 h before subsequent injections with synthetic mRNAs. Injection of this oligonucleotide did not affect U7 snRNA levels (data not shown).
Expression of proteins and characterization of antibodies.
Glutathione S-transferase (GST) fusion proteins were expressed after cloning of full-length SLBP1 and SLBP2 into pGEX-KG. The SLBP1 fusion protein was purified on glutathione-agarose. The SLBP2 fusion protein was present in inclusion bodies and was solubilized in sodium dodecyl sulfate (SDS) and resolved by preparative gel electrophoresis. The region containing SLBP2 was excised, and the gel was ground up and injected into rabbits. Full-length SLBP1 and SLBP2 were also expressed in baculovirus (Bac-to-Bac; GIBCO-BRL), and the resulting His-tagged proteins were purified by chromatography on Ni-agarose. The baculovirus-expressed SLBP2 protein was used to boost the rabbits that had been injected with the GST fusion protein. Baculovirus-expressed SLBP1 was injected into rabbits to prepare polyclonal antibodies. The SLBP2 antibodies were purified by resolving the GST-SLBP2 fusion protein expressed in bacteria by SDS-polyacrylamide gel electrophoresis, transferring the protein to nitrocellulose, and then incubating the serum with the nitrocellulose strip containing the GST-SLBP2 protein for 2 h at 4°C. The affinity-purified antibodies were eluted with 0.1 M glycine at pH 2.8 and immediately neutralized. The antiserum to the complete SLBP1 protein disrupted the RNA-protein complexes and also cross-reacted weakly with the SLBP2 protein on Western blots. To further purify these antibodies, a clone encoding an SLBP1-GST fusion protein with the RNA binding domain deleted was constructed and the SLBP1 antibody was purified by binding to this protein immobilized on nitrocellulose, followed by elution with 0.1 M glycine-HCl, pH 2.8. Each of the affinity-purified antibodies detected a single polypeptide by Western blotting and efficiently supershifted the appropriate RNA-SLBP complexes. Polyclonal antibodies against the C-terminal 15 amino acids of SLBP1 coupled to keyhole limpet hemocyanin were also prepared. These antibodies did not recognize SLBP1 on Western blots and were useful only for analyses of mobility shift assays.
Mobility shift assays.
Frog oocytes of different stages were homogenized in 5 to 10 μl of 15 mM Tris (pH 7.0)–0.1 mM PMSF (phenylmethylsulfonyl fluoride) per oocyte. Nuclei were manually dissected, and extracts were prepared from nuclei and cytoplasm in the same buffer. The homogenate was extracted with freon to remove the yolk (10). Similar results were obtained with extracts prepared without freon extraction, although not as much protein could be analyzed without affecting the resolution of the gel. Mobility shift and competition assays were performed essentially as previously described with 10% polyacrylamide gels (50).
Injection of oocytes.
The SLBP1 and SLBP2 proteins were expressed at high levels by injection of a synthetic capped mRNA transcribed from the respective SLBPs cloned into a modified version of pSP64T (Promega), which contains the 5′ and 3′ UTRs of the human β-globin mRNA and a poly(A) tail (a gift of Enrique Amaya and Doug Melton, Harvard University). To express histone mRNAs, the mouse histone H2a-614 gene (15 nl, 30 ng/μl) was injected into frog oocytes and RNA was prepared 18 h later (49). To assay for both processed and unprocessed RNAs, a probe starting at the Sau3A site 163 nt from the 3′ end of the mRNA and extending 33 nt past the stem-loop was constructed, labeled at the 3′ end with [α-32P]dCTP, and used for S1 nuclease mapping. The processed RNAs protect a 163-nt fragment, and the unprocessed RNAs protect a 196-nt fragment. The protected fragments were resolved by polyacrylamide gel electrophoresis and quantified on a PhosphorImager.
Western blots.
Oocytes were homogenized in a solution of 0.3 M KCl, 2 mM MgCl2, 20 mM HEPES (pH 7.4), 0.5% Nonidet P-40 (NP-40), 20% glycerol, 2.5 mM dithiothreitol, and 0.1 mM PMSF (5 to 10 μl/oocyte). The homogenate was centrifuged for 10 min at 14,000 rpm, and the clear supernatant was mixed with an equal volume of 2% SDS–10% glycerol–0.06 M Tris (pH 6.8)–5% β-mercaptoethanol and boiled; proteins from one to three oocytes were resolved by electrophoresis on 12% SDS-polyacrylamide gels and transferred to nitrocellulose. The filter was incubated with the affinity-purified antibody, and the bound antibodies were detected by chemiluminescence with ECL technology (Amersham).
In vitro processing.
Nuclear extracts active in histone pre-mRNA processing were prepared from mouse myeloma cells, as previously described (8, 24). To remove the endogenous mouse SLBP, an antibody to the C-terminal peptide of SLBP was incubated with 0.5 to 1.0 ml of nuclear extract for 2 h at 4°C. Twenty microliters of protein A beads were added per 100 μl of extract. The beads were precoated by incubation with nuclear extract at 4°C for 30 min prior to use. The beads containing the bound antibody-SLBP complex were removed from the extract by centrifugation.
To complement processing, 100 to 200 ng of purified baculovirus-expressed protein encoding human SLBP, frog SLBP1, or frog SLBP2 was added to the extract. The substrate used was derived from the histone H2a-614 gene with the U7 snRNP binding site of the mouse histone H1t gene substituted for the U7 binding site of the H2a-614 gene (35a). Processing reaction mixtures were incubated for 1 h at 32°C, and the RNA was prepared and analyzed by gel electrophoresis.
Histone mRNA immunoprecipitation.
The histone mRNAs were precipitated by a modification of the method developed recently in our laboratory (46a). Fifty to 100 frog oocytes or eggs were homogenized (2 μl/oocyte) in a solution of 0.3 M KCl, 20 mM HEPES (pH 7.7), 2 mM MgCl2, 0.5% NP-40, 2.5 mM dithiothreitol, 1 mM cycloheximide, 100 units of the protease inhibitor ACE per ml, 1 mM PMSF, and 100 U of RNasin per ml. The homogenate was centrifuged for 10 min at 14,000 rpm in a microcentrifuge, and the clear supernatant was used for precipitation. An extract equivalent to five oocytes was incubated with affinity-purified antibody (2.5 μg) and 1 U of additional RNasin per μl for 4 h to overnight at 4°C on a rotator. Protein A beads (100 μl) were incubated with 200 μl of nuclear extract from blastula-stage sea urchin embryos (21). The precoated beads were washed with buffer D (0.1 M KCl, 10 mM HEPES [pH 7.4], 0.1% NP-40, 20% glycerol). Ten microliters of the precoated protein A beads and 30 μl of buffer D were added to the oocyte extracts, and the mixture was incubated for 2 h at 4°C on a rotator. The beads were collected by centrifugation and washed three times (5 min, 1 h, and 5 min) with 1 ml of buffer D. One hundred microliters of 7 M urea, 2% SDS, 0.35 M NaCl, 10 mM EDTA, and 10 mM Tris (pH 7.5) containing 10 μg of tRNA was added, the suspension was extracted once with phenol-chloroform, and the RNA was recovered by precipitation with ethanol. RNA was also prepared from the supernatant of the incubation with protein A beads. The Xenopus histone H2a mRNA was detected with an S1 nuclease protection assay using a Xenopus H2a gene from a major histone gene cluster (32) cloned from the oocyte cDNA library.
Northern blots.
Total RNA was prepared from Xenopus oocytes and embryos, as previously described (49). The RNA was treated with formaldehyde and resolved on a 0.8% agarose gel, transferred to nitrocellulose, and hybridized with a mixture of probes from SLBP1, SLBP2, and histone H3. The probes were prepared by random primer labeling with [α-32P]dCTP. The specific activity of the dCTP used to make the histone H3 probe was 1/20 of that of the dCTP used to make the histone H3 probe.
Nucleotide sequence accession number.
The complete nucleotide sequence of the longest SLBP2 clone obtained in this study has been assigned accession no. AF106799.
RESULTS
We previously reported the isolation of two orthologous SLBPs from human (HeLa cell) and Xenopus oocyte two-hybrid libraries (46) by using the yeast three-hybrid system (36). We screened for proteins which bound the wild-type stem-loop sequence in vivo but did not bind a mutant sequence with the stem-loop reversed (Fig. 1A). We obtained multiple isolates of the major SLBP from both human and Xenopus libraries (46). However, one of the eight initial clones selected from the Xenopus library encoded a protein different from the major SLBP. Further analysis of this clone confirmed that it passed the selection criteria (specific binding to the stem-loop in yeast). Sequencing of this clone revealed that it encodes a 250-amino-acid polypeptide (Fig. 1B), which we have termed SLBP2. SLBP2 has similarity to the major SLBP (46), which we have now termed SLBP1, only in the 73-amino-acid RNA binding domain (68% identity [Fig. 1C]). SLBP2, like SLBP1, is small, about 28 kDa, and has its RNA binding domain located in the central region of the protein.
Subsequent PCR analysis of the 30 clones we isolated from the frog oocyte library with the three-hybrid screen revealed that 23 encoded the homologue of the human protein (SLBP1) and 7 encoded SLBP2. Like the SLBP1 clones, many of the SLBP2 clones were fused to the Gal4 activation domain in the 5′ UTR, in frame with the rest of the protein. Since the baculovirus-expressed SLBP2 and in vitro-translated SLBP2 have electrophoretic mobilities similar to that of the SLBP2 we have detected in Xenopus oocytes by Western blotting (not shown), we are confident that we have identified the entire open reading frame. One of the clones we isolated had a poly(A) tail and polyadenylation signal, suggesting that it was a near-full-length clone. The size of this clone (1,066 nt) agrees well with the size of the SLBP2 mRNA measured by Northern blots (see Fig. 5A).
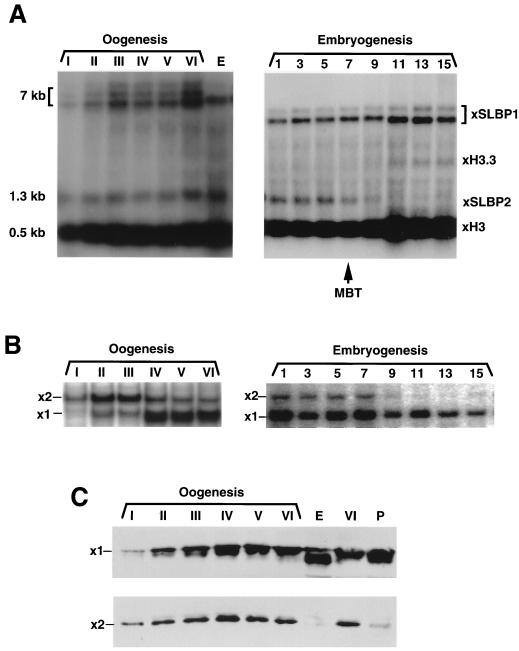
FIG. 5.
Expression of SLBP1 and SLBP2 during oogenesis and early embryogenesis. (A) RNA was prepared from oocytes and early embryos and resolved by gel electrophoresis on a 1% agarose gel. RNA from two oocytes or embryos was analyzed. The gel was transferred to nitrocellulose and hybridized with a mixture of probes to SLBP1, SLBP2, and histone mRNA. The histone probe was made at one-half of the specific activity of the other two probes. The identity of the bands was confirmed by hybridizing with single probes. (B) Extracts were prepared from oocytes and embryos and assayed for SLBP1 and SLBP2 by mobility shift assay. Each lane is analysis of one oocyte. (C) Protein from one oocyte (top) or four oocytes (bottom) was analyzed by Western blotting with either affinity-purified anti-SLBP1 (top) or affinity-purified anti-SLBP2 (bottom). Lanes P show oocytes matured by treatment with progesterone, and lanes E show eggs. The apparent higher mobility of SLBP1 in eggs in this experiment was not seen in other experiments.
Two complexes which bind the 3′ end of histone mRNA are present in Xenopus oocytes.
To study the complexes containing the different SLBPs which are present in Xenopus oocytes, polyclonal antibodies were raised against both recombinant SLBP1 and SLBP2 and were affinity purified to remove any cross-reacting antibodies. Synthetic mRNAs encoding SLBP1 and SLBP2 were translated in a rabbit reticulocyte lysate. SLBP1 has mobility similar to mammalian SLBP on SDS-polyacrylamide gels (45 kDa), and SLBP2 has mobility of 35 kDa, close to the predicted molecular weight of 28 kDa (Fig. 2A). Specific complexes were formed with each in vitro translation product (Fig. 2B). The complex formed with SLBP1 has mobility similar to the complex formed with the mammalian SLBP (not shown), and the complex was supershifted by the antibody against SLBP1 but not against SLBP2 (Fig. 2B, lanes 5 to 7). The SLBP2 in vitro translation product also bound the stem-loop specifically (Fig. 2B, lanes 11 to 13) and reacted only with the antibody against SLBP2 (Fig. 2B, lanes 9 and 10). The complex formed with SLBP2 has lower mobility than the complex formed with SLBP1 in both the reticulocyte lysate and in frog oocyte extracts (see below).
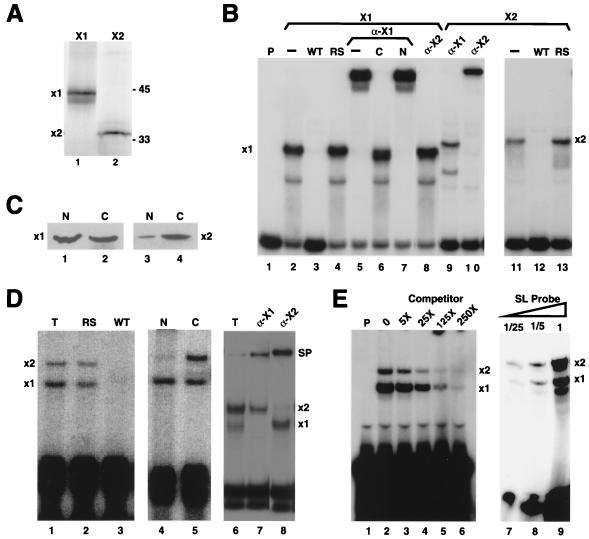
FIG. 2.
Two complexes are formed on the stem-loop in Xenopus oocytes. (A) Clones encoding SLBP1 (lane 1) and SLBP2 (lane 2) were transcribed in vitro and then translated in a rabbit reticulocyte lysate in the presence of [35S]methionine. The products were resolved by electrophoresis on an SDS–12% polyacrylamide gel and detected by radioautography. (B) Reticulocyte lysates programmed with either SLBP1 (lanes 2 to 8) or SLBP2 (lanes 9 to 13) mRNAs were incubated with the radiolabeled probe containing the stem-loop, and the complexes were resolved by gel electrophoresis. The antibody to the C-terminal peptide of SLBP1 was added in lanes 5 to 7 and lane 9. The SLBP1 antibody was preincubated with the antigenic peptide in lane 6 and with control peptide from the N terminus of SLBP1 in lane 7. The affinity-purified antibody to SLBP2 was added in lanes 8 and 10. A 100-fold excess of competitor unlabeled RNAs containing the stem-loop (WT) (lanes 3 and 12) or the reverse-stem (RS) (lanes 4 and 13) sequence was added to some reaction mixtures. (C) Oocytes were fractionated into nuclei and cytoplasm, and proteins from three oocytes were resolved by SDS-polyacrylamide gel electrophoresis. The gel was transferred to nitrocellulose and probed with affinity-purified anti-SLBP1 (lanes 1 and 2) or anti-SLBP2 (lanes 2 and 3) antibodies. (D) Total extracts from frog oocytes were incubated with the wild-type stem-loop probe (lanes 1 to 3 and 6 to 8). A 100-fold excess of unlabeled reverse-stem RNA (RS) (lane 2) or wild-type RNA (WT) (lane 3) was included as competitor. The oocytes were fractionated into nuclei (lane 4) and cytoplasm (lane 5). Affinity-purified polyclonal antibodies to SLBP1 (α-X1) (lane 7) or SLBP2 (α-X2) (lane 8) were added to the reaction mixture. The complexes formed with SLBP1 and SLBP2 are denoted x1 and x2, respectively, and the supershifted complexes are denoted SP (lanes 7 and 8). (E) An extract from stage IV oocytes was incubated with the radiolabeled probe and increasing molar amounts of stem-loop competitor (lanes 2 to 6), and the complexes were resolved by gel electrophoresis. A similar extract from stage III oocytes was incubated with increasing amounts of radiolabeled probe (lanes 7 to 9), and the complexes were resolved by gel electrophoresis.
Extracts were prepared from stage VI Xenopus oocytes, incubated with a 30-nt radiolabeled stem-loop RNA, and then analyzed by electrophoresis under native conditions. Two complexes were detected in extracts from the whole oocyte (Fig. 2D, lane 1). Formation of both complexes was competed by the wild-type stem-loop but not by the reverse-stem RNA (Fig. 2D, lanes 2 and 3). To determine the subcellular localization of the two activities, the oocytes were manually fractionated into nucleus and cytoplasm. Complex x1 was present in both the nuclear and the cytoplasmic fractions, but complex x2 was found exclusively in the cytoplasm (Fig. 2D, lanes 4 and 5). Antibodies to SLBP1 specifically supershifted complex x1, and antibodies to SLBP2 supershifted complex x2 (Fig. 2D, lanes 7 and 8).
We also measured the total amount (as compared to free active SLBP) of each SLBP protein in the nucleus and cytoplasm by Western blotting with affinity-purified SLBP1 and SLBP2 antibodies. SLBP1 was found in both the nucleus and the cytoplasm (Fig. 2C, lanes 1 and 2), while SLBP2 was found almost exclusively in the cytoplasm (Fig. 2C, lanes 3 and 4). The small amount of SLBP2 in the nuclear fraction probably results from slight contamination with cytoplasm.
To assess the relative affinities of the two SLBPs for the stem-loop, we performed competition experiments with frog extracts that contained both SLBP1 and SLBP2 binding activity. Two types of assays were performed (Fig. 2E). When the assay was performed in probe excess and varying amounts of the wild-type competitor were added to the extract, formation of both complexes was competed to a similar extent at all concentrations of competitor (Fig. 2E, lanes 2 to 6). In the second assay, increasing amounts of probe were added to the extract to determine whether one SLBP bound preferentially to limiting amounts of probe. As increasing amounts of probe were added, the amounts of complex x1 and x2 increased in parallel (Fig. 2E, lanes 7 to 9). Taken together, these results demonstrate that SLBP1 and SLBP2 have similar affinities for the stem-loop.
SLBP1, not SLBP2, functions in histone pre-mRNA processing.
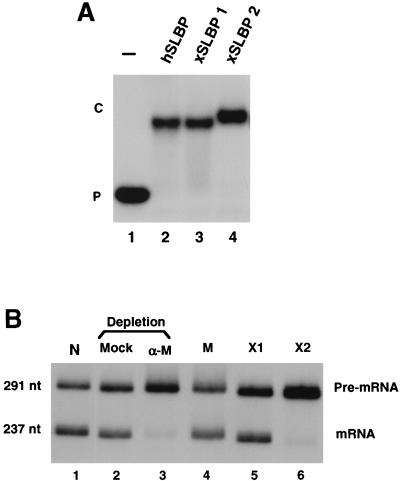
To determine whether both SLBP1 and SLBP2 can function in histone pre-mRNA processing, we expressed both SLBP1 and SLBP2 from baculovirus vectors, and tested the ability of the recombinant proteins to participate in pre-mRNA processing in vitro. Both of the baculovirus proteins bound to the stem-loop with similar affinity (Fig. 3A). A nuclear extract highly active in pre-mRNA processing was prepared from mouse myeloma cell nuclei, as previously described (8, 24). The mouse SLBP was removed from this extract by using an antibody against the C-terminal peptide and protein A-agarose. The depleted extract is inactive in pre-mRNA processing (Fig. 3B, lane 3), while a mock-depleted extract retained most of its initial activity (Fig. 3B, lanes 1 and 2). Recombinant mouse SLBP, frog SLBP1, and frog SLBP2 expressed in baculovirus were added back to the extract. Mouse SLBP and frog SLBP1 restored processing to normal levels (Fig. 3B, lanes 4 and 5), demonstrating that the only component necessary for processing depleted by the antibody was SLBP. Frog SLBP2 was totally inactive in pre-mRNA processing (Fig. 3B, lane 6) even though it bound the stem-loop (Fig. 3A, lane 4).
FIG. 3.
SLBP1, but not SLBP2, functions in histone pre-mRNA processing in vitro. (A) SLBP1, SLBP2, and human SLBP (hSLBP) were expressed in baculovirus, and 200 ng of protein was analyzed for binding to the stem-loop by mobility shift assay. (B) A 291-nt substrate was synthesized with T7 RNA polymerase from the mouse histone H1t gene. A nuclear extract prepared from mouse myeloma cells was very active in histone pre-mRNA processing (lane 1). The extract was depleted by using an antibody to the C terminus of mouse SLBP (lane 3), or was treated with immunoglobulin G purified from preimmune serum (lane 2). An equal amount (200 ng) of recombinant baculovirus-expressed mouse SLBP (M) (lane 4), frog SLBP1 (X1) (lane 5), or SLBP2 (X2) (lane 6) was added to the depleted extract. The extracts were incubated for 30 min at 27°C, and RNA was prepared and analyzed by electrophoresis on 8% polyacrylamide–7M urea gels.
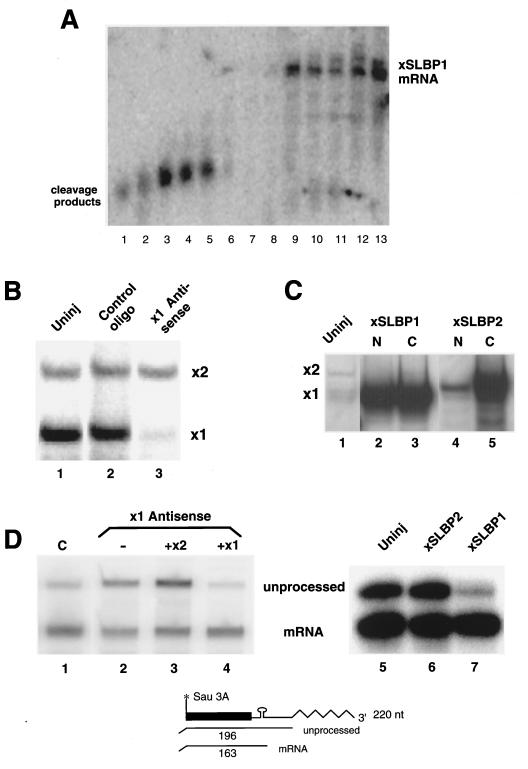
We also tested the ability of the two Xenopus SLBPs to complement histone pre-mRNA processing in vivo. Injection of antisense oligonucleotides into Xenopus oocytes results in degradation of the target mRNAs by RNase H and then subsequent degradation of the injected oligonucleotide (17). Often there is also reduction of the levels of the encoded protein as a result of destruction of the mRNA (17). We tested several antisense oligonucleotides against SLBP1 for the ability to destroy SLBP1 mRNA. Many of these resulted in complete cleavage of the SLBP1 mRNA (Fig. 4A, lanes 1 to 8), while a control oligonucleotide had no effect on SLBP1 mRNA (Fig. 4A, lane 13). One of these oligonucleotides, AX09 (Fig. 4A, lane 3), also showed minimal toxicity to the oocytes and was used for the subsequent studies. Extracts were prepared from oocytes 48 h after injection of antisense oligonucleotide AX09. These extracts had greatly reduced amounts of free active SLBP1 protein but the same amount of free active SLBP2 protein as control oocytes or oocytes injected with a control oligonucleotide (Fig. 4B). Thus, the antisense treatment selectively removed the SLBP1 protein.
FIG. 4.
SLBP1, but not SLBP2, is involved in histone pre-mRNA processing. (A) A series of antisense oligonucleotides (AX07 to AX12, lanes 1 to 6; AX01 to AX06, lanes 7 to 12) directed against SLBP1 mRNA were injected into Xenopus oocytes, and RNA was prepared 4 h later. The RNA was resolved by electrophoresis on a 0.8% agarose gel and probed for SLBP1 mRNA. (B) Oocytes were injected with oligonucleotide AX09 and a control oligonucleotide, and extracts were prepared 48 h later. The extracts were assayed by mobility shift assay for the formation of complexes that bind to the stem-loop. (C) Oocytes were injected with synthetic mRNAs encoding SLBP1 (lanes 2 and 3) or SLBP2 (lane 3 and 4). Forty-eight hours later, the oocytes were fractionated into nuclei and cytoplasm and assayed for the presence of complexes that bound the stem-loop. Lane 1 shows an extract from uninjected oocytes. Extracts from one oocyte were analyzed in each lane. (D) Oocytes were injected with oligonucleotide AX09 (lanes 2 to 4) or with a control oligonucleotide (lane 1). Four hours later, synthetic mRNA encoding SLBP2 (lane 3) or SLBP1 (lane 4) was injected into some of the oocytes. After 48 h, histone H2a-614 DNA was injected into the nucleus. RNA was prepared 18 h after the injection of H2a-614 DNA, and the transcripts were assayed for processed and unprocessed histone mRNA by using an S1 nuclease assay, as shown at the bottom. In a separate experiment, untreated oocytes were injected with buffer (lane 5) or mRNAs encoding frog SLBP2 (lane 6) or frog SLBP1 (lane 7) 48 h prior to injection of histone H2a-614 DNA into the nucleus. RNA was prepared 18 h after injection of the DNA and analyzed by using S1 nuclease mapping.
We then demonstrated that the levels of SLBP1 could be restored by injection of synthetic SLBP1 mRNA. In order to complement the oocytes deficient in histone pre-mRNA processing, we expressed the SLBP1 and SLBP2 proteins in the oocyte by injecting mRNAs encoding either SLBP1 or SLBP2. In each case, a large amount of active SLBP was expressed as judged by a mobility shift assay. Expression of either SLBP from injected mRNA resulted in a 20- to 30-fold increase in SLBP over the amount already present in the mature oocyte (Fig. 4C), and overexpressed SLBP1 and SLBP2 had the same subcellular distributions as endogenous SLBP1 and SLBP2 (cf. Fig. 4C and Fig. 2C and D).
To determine whether the reduction in free SLBP1 protein resulted in a reduction in the ability to process histone mRNA, we developed an S1 nuclease protection assay that allowed us to measure both processed and unprocessed mRNA from an injected mouse histone gene (Fig. 4D). This assay maps both the properly processed RNA and any transcripts which extend more than 33 nt past the 3′ end of histone mRNA. These heterogeneous longer transcripts all protect the same size fragment. The ratio of the two protected fragments is a measure of the efficiency of processing. When a control oligonucleotide was injected into the oocytes prior to injection of the mouse histone H2a-614 gene, the majority (70 to 80%) of the transcripts were processed (Fig. 4D, lane 1). When the oocytes were injected with antisense oligonucleotide AX03 prior to injection of the H2a-614 gene, reducing the amount of free SLBP1, the processing efficiency was reduced to about 35% (Fig. 4D, lane 2). Expression of SLBP1, but not SLBP2, by injection of the appropriate synthetic mRNA restored processing to >80% efficiency in vivo (Fig. 4D, lane 4).
We also injected mRNAs encoding either SLBP1 or SLBP2 into oocytes that had not been injected with antisense oligonucleotides. After 48 h, to allow expression of SLBP, the mouse histone H2a-614 gene was injected into the oocyte nucleus, and RNA was prepared from the oocytes 16 h later. This batch of oocytes processed about 80% of the histone transcripts, as did oocytes injected with the SLBP2 mRNA (Fig. 4D, lanes 5 and 6). Oocytes injected with the SLBP1 mRNA processed >95% of the histone transcripts (Fig. 4D, lane 7). Thus, both of these experiments demonstrate that SLBP1, but not SLBP2, functions in histone pre-mRNA processing in vivo.
Differential expression of SLBP1 and SLBP2 during development.
We used Northern blots, mobility shift assays, and Western blots to measure the expression of SLBP mRNAs and protein during oogenesis and embryogenesis. Northern blots were done to compare the levels of histone H3 mRNA, SLBP1 mRNA, and SLBP2 mRNA. SLBP1 is encoded by an mRNA which is 6 to 7 kb long (the largest cDNA clone we obtained was 4.5 kb), SLBP2 is encoded by a 1.3-kb mRNA, and histone H3 is encoded by a family of mRNAs about 500 nt in length. Therefore, we were able to resolve these three mRNA species by hybridizing a single blot with all three probes. The specific activity of the probe for histone mRNA was deliberately made 20 times lower than the two SLBP probes, which were of similar specific activities, to allow the abundant histone mRNA to be readily detected on the same blot. Some histone mRNA was already present in stage I oocytes, and it reached a constant level by stage III (Fig. 5A). There was no further increase in histone mRNA until the MBT (Fig. 5A), when transcription is activated, as previously reported (42). SLBP1 mRNA and SLBP2 mRNA are expressed in stage I oocytes and reach constant levels by stage III of oogenesis. SLBP1 mRNA remains present at the same level in early embryogenesis, and the amount of SLBP1 mRNA increases after the MBT. In contrast, SLBP2 mRNA is destroyed by the MBT (Fig. 5A, stages 7 and 9) and is not expressed later in embryogenesis or in somatic cells (not shown).
To measure the relative amounts of the SLBP1 and SLBP2 proteins, extracts were prepared from the six stages of oogenesis and several embryonic stages and assayed by mobility shift assay (Fig. 5B) and Western blotting (Fig. 5C). The mobility shift assay measures soluble, active SLBP. It may not detect SLBPs that are tightly complexed with RNA (16) and will not detect any SLBPs that exist in modified forms which do not bind RNA with high affinity. There were higher levels of SLBP2 binding activity than of SLBP1 binding activity in stage I oocytes, and these high ratios persisted through stage III of oogenesis (Fig. 5B). Later in oogenesis (stages IV to VI), the amount of SLBP1 activity was higher than the amount of SLBP2 activity, due largely to an increase in the amount of active SLBP1, although there was also a decrease in the amount of active SLBP2 (Fig. 5B). In eggs and early embryos, there was very little SLBP2 activity and large amounts of SLBP1 activity (Fig. 5B). Note that in these assays, which are done in probe excess, we measure the actual relative amounts of free active SLBP1 and SLBP2 protein, since they bind the stem-loop with similar affinities (Fig. 2D).
Qualitatively similar results were obtained when total SLBP1 and SLBP2 protein were measured by Western blotting. We assayed total protein from equal numbers of oocytes from different stages. SLBP2 was readily detectable in stage I oocytes and increased through stages III and IV. There was a subsequent slight decline in SLBP2 levels as oocytes matured to stage VI (Fig. 5C, bottom). The levels of SLBP2 protein dropped dramatically in eggs compared to stage VI oocytes, indicating that most of the SLBP2 is degraded in the transition from oocytes to eggs. SLBP2 was also degraded when oocytes were matured in vitro by treatment with progesterone (Fig. 5C, lane P). In contrast, the levels of SLBP1 were very low in stage I oocytes and gradually increased throughout oogenesis (Fig. 5C, top). There was a further twofold increase in SLBP1 when the oocytes were matured to eggs (Fig. 5C). The levels of SLBP1 continued to increase during early embryogenesis (not shown).
Although the results of the Western blot and mobility shift assays were qualitatively similar, there were significant quantitative differences. Western blotting measures all of the SLBP protein, while the mobility shift assay measures the amount of free active protein. First, there is a large increase in the SLBP1 activity measured by the mobility shift assay at stage IV of oogenesis. In contrast, there is a gradual increase in SLBP1 protein from stages II to V of oogenesis, as analyzed by Western blotting. These results are consistent with some SLBP1 being released from histone mRNA or becoming activated between stage III and stage IV of oogenesis. Second, there is not a dramatic decrease in the amount of free SLBP2 binding activity as oocytes are matured to eggs, as measured by the mobility shift assay, while there is a dramatic drop as measured by Western blotting. This result suggests that much of the SLBP2 protein in stage VI oocytes is either bound to histone mRNA or is in an inactive form. These results are consistent with the accumulation of free SLBP1 late in oogenesis, while most of the SLBP2 is bound to stored histone mRNA. At oocyte maturation, the degradation of SLBP2 may then allow SLBP1 to associate with the histone mRNA.
Different SLBPs are associated with histone mRNA during different developmental stages.
To determine which SLBPs were associated with histone mRNAs at each stage of oogenesis, we used antibodies to the two SLBPs to precipitate complexes containing the SLBPs from oocyte and egg extracts. RNA was prepared from the antibody precipitates and the histone H2a mRNA detected by S1 nuclease mapping of the bound and unbound mRNAs. A portion of the unbound fraction was analyzed by Western blotting to demonstrate that the SLBPs had indeed been removed from the extracts (not shown). In each experiment we performed the precipitations with each extract with the three antibodies (SLBP1, SLBP2, and mouse SLBP) in parallel. The absolute amounts of the histone mRNA precipitated varied some from experiment to experiment, but the relative amounts precipitated by anti-SLBP1 and anti-SLBP2 were similar for extracts from the same stage in different experiments. When stage IV or VI oocytes were analyzed, very little histone H2a mRNA was precipitated by anti-SLBP1. In stage II oocytes, about twice as much histone mRNA was precipitated by anti-SLBP2 as by anti-SLBP1 (Fig. 6, lanes 1 and 5). More than 70% of the histone H2a mRNA was precipitated by anti-SLBP2 in stage IV oocytes (Fig. 6, lanes 6), and less than 10% was precipitated by anti-SLBP1. In stage VI oocytes, three times as much histone mRNA was precipitated by the SLBP2 antibody as the SLBP1 antibody, even though most of the SLBP present in stage VI oocytes is SLBP1. Thus, throughout oogenesis, the bulk of the histone mRNA is associated with SLBP2. Since in both stage IV and stage VI oocytes the most abundant free SLBP is SLBP1 (Fig. 5B), while most of the histone mRNA isolated by antibody precipitation is bound to SLBP2, it is unlikely that there has been significant rearrangement of the SLBPs during the isolation and precipitation procedures.
FIG. 6.
Determination of SLBPs bound to histone mRNA during oogenesis. Extracts from stage II (lanes 1, 5, and 9), stage IV (lanes 2, 6, and 10), and stage VI (lanes 3, 7, 11, 13, 15, and 17) oocytes, oocytes matured in vitro with progesterone (lanes 4, 8, and 12), or eggs (lanes 14, 16, and 18) were incubated with antibodies to either frog SLBP1 (lanes 1 to 4, 13, and 14), frog SLBP2 (lanes 5 to 8, 15, and 16), or mouse SLBP (lanes 9 to 12, 17, and 18). The RNAs bound to the antibodies were isolated and assayed by S1 nuclease mapping for frog histone H2a mRNA (top). RNA was also prepared from the supernatant after incubation with antibody and protein A-agarose and assayed for frog histone H2a mRNA by S1 nuclease mapping (bottom). The protected fragments were resolved by gel electrophoresis and detected by autoradiography. The results are representative of three independent experiments. A diagram of the S1 nuclease assay is shown. The mRNA from this H2a gene protects a 224-nt fragment (H2a), and the mRNAs from other H2a genes protect a 180-nt fragment (H2ax) which maps to the stop codon.
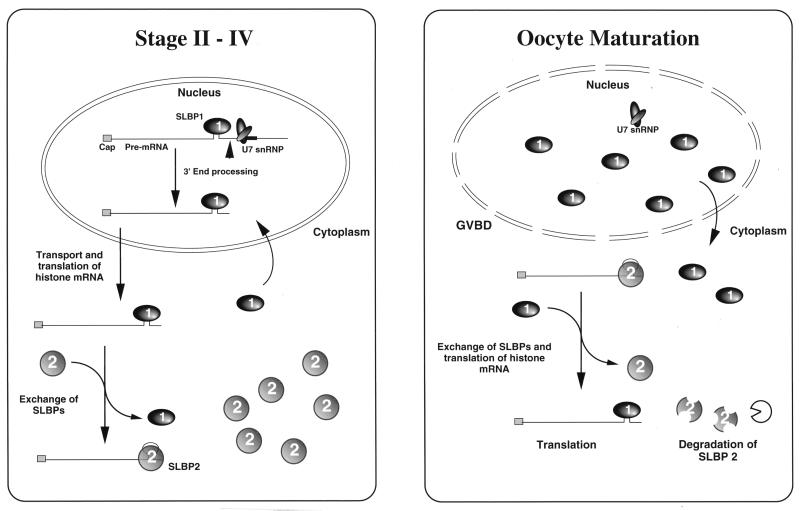
When eggs were analyzed by an identical procedure using the same two antibodies, the results were strikingly different. More histone mRNA was precipitated by the anti-SLBP1 antibody than by the anti-SLBP2 antibody (Fig. 6, lanes 14 and 16). Essentially identical results were obtained when extracts from oocytes matured by treatment with progesterone in vitro were analyzed (Fig. 6, lanes 4 and 8). During this period, there is a dramatic decrease in the total amount of SLBP2 protein measured by Western blotting (Fig. 5C). Thus, during the transition from an oocyte to an egg, much of the histone mRNA loses its SLBP2 protein and then binds to SLBP1 (Fig. 7), and the SLBP2 protein is degraded. This transition occurs at the time of activation of histone mRNA translation (51), suggesting that this is one of the molecular events involved in translational regulation.
FIG. 7.
Model for histone mRNA metabolism during oogenesis in Xenopus.
DISCUSSION
Early in oogenesis, the frog stores most of the maternal mRNAs which will be utilized in early embryogenesis. Storage of mRNAs occurs concomitantly with the biosynthesis of the mRNA, and some of the essential events, including association with the general translational repressor FRGY2, require that the RNA be synthesized in the nucleus (6, 26, 29). The stored inactive mRNAs generally have short poly(A) tails (47). Other mRNAs that are translated in oocytes have longer poly(A) tails and are presumably not complexed with FRGY2 or other translational repressors. At oocyte maturation, a translational program is activated (48) which controls development until the MBT, when zygotic transcription is activated (31). This program involves default deadenylation of the majority of mRNAs (12) and selective cytoplasmic adenylation of many RNAs that are then translated (5, 15). In addition, there are general and sequence-specific RNA binding proteins that are involved in translational repression, which must also be removed from mRNAs at this time (15). Similar programs of translational control are present in many early embryos (15, 44, 48).
A fraction of the frog oocyte histone mRNA binds to oligo(dT) cellulose (20, 34) due to a small number of adenosines added to the end of the stem-loop (4). These terminal adenosines are removed at oocyte maturation (4). However, only a small fraction of the stored histone mRNAs with stem-loops in stage VI oocytes are polyadenylated, as judged by binding to oligo(dT) cellulose (35b). This modification does not affect the relative affinity of the two different SLBPs (45a), and hence this modification cannot be essential for the global activation of histone mRNA translation.
In organisms with very rapid cleavage stages, there is an exponentially increasing demand for histone proteins to assemble chromatin as development proceeds. Many different strategies have evolved for dealing with this problem. Some organisms (e.g., sea urchins) have a set of histone genes that are present in multiple copies and are expressed only during the rapid cleavage stages (27). These zygotic genes supply histone mRNAs during the cleavage stages and then are not used again. In Drosophila, there is stored histone mRNA utilized initially as well as synthesis of histone mRNA from a set of repeated genes (2, 3).
Since there is no transcription in frog embryos until the MBT (approximately 4,000 cells), the frog embryo must rely solely on stored histone protein and increased histone mRNA translation to supply the histones necessary to assemble chromatin prior to the MBT. Histone synthesis is translationally regulated in early frog embryos. During oogenesis, there is synthesis of histones in stages II and III of oogenesis and then a decrease to a very low level in oocyte stages IV to VI (51). There is a 17-fold increase in the rate of histone protein synthesis at oocyte maturation (1, 51). Adamson and Woodland (1) showed that the increase in histone mRNA translation is greatly reduced in enucleated oocytes, suggesting that some component in the nucleus is necessary for activation of translation of histone mRNA, as it is for regulation of polyadenylation of other oocyte mRNAs (11). There is a further increase in the translation rate during early development, prior to the increase in histone mRNA levels after the MBT, resulting in an ultimate 50-fold increase in the rate of histone protein synthesis (51). The same histone mRNAs are expressed in oogenesis, during early development, and in somatic cells (33). Since histone mRNAs are not polyadenylated, they must be translationally regulated by a mechanism distinct from the regulation of polyadenylated mRNAs. The different SLBPs are likely candidates to play a key role in this translational regulation, and SLBP2 may be a protein that has evolved as a specific translational repressor.
The 3′ end of histone mRNA is essential for association of the mRNA with polyribosomes (39) and translation of histone mRNA (13) in mammalian cells. Since the mammalian homologue of the SLBP1 is a component of the histone mRNP (16), frog SLBP1 is probably associated with the translationally active histone mRNA. There is synthesis of histone protein during stage II of oogenesis and a lower rate of synthesis in mature oocytes (51, 52). During stage II of oogenesis, the oocyte synthesizes about 75% of the histone protein necessary for early cleavage over a period of at least 1 to 2 weeks (9). The other 25% of the histone protein is synthesized from stored maternal RNA in a few hours. Only a small proportion of the total histone mRNA would be required for production of the histone protein in stage II oocytes. It is possible that a fraction of the histone mRNA synthesized in stage II remains associated with SLBP1 and is translated while the remainder of the histone mRNA associates with SLBP2 and is stored.
In support of this possibility, we have found a larger proportion of histone mRNA bound to SLBP1 in stage II oocytes than in stage IV oocytes (Fig. 6). Early in oogenesis, when histone mRNA is synthesized, the histone mRNA must be processed by SLBP1 and subsequently associate with SLBP2 (Fig. 7). A fraction of the histone mRNAs may remain associated with SLBP1 and be translated in stage II, while the rest is transferred to SLBP2 and is stored. Alternatively, all of the histone mRNAs may be translated for some time prior to association with SLBP2, resulting in storage. There is probably not sufficient SLBP1 to associate with all of the histone mRNA in stage II, implying that SLBP1 may function essentially catalytically rather than stoichiometrically during this stage. By stage IV of oogenesis, accumulation of histone mRNA has ceased and the great majority of the histone mRNA is bound to SLBP2. Once bound to SLBP2, the histone mRNA remains bound to SLBP2 and is translationally inactive throughout the remainder of oogenesis, even while the oocyte accumulates large amounts of SLBP1 protein. At oocyte maturation, there is a loss of SLBP2 from much of the histone mRNA, the histone mRNA is then bound by SLBP1, and SLBP2 is degraded (Fig. 7). One of the changes which occurs to activate translation of the histone mRNA at oocyte maturation likely involves removal of SLBP2, followed by association of SLBP1 with the stored histone mRNA. During embryogenesis, there is a further increase in the amount of histone mRNA translation, presumably as the rest of the SLBP2 is removed from the histone mRNA and is replaced by SLBP1.
ACKNOWLEDGMENTS
This work was supported by grants GM29832 and GM27789 from the NIH to W.F.M. T.C.I. was supported by a postdoctoral fellowship from the Andrew W. Mellon Foundation.
We thank Sally Kornbluth for some of the frog eggs used in these experiments and Mike Whitfield for advice on the mRNA immunoprecipitation experiments and for critical comments on the manuscript.
Z.-F.W. and T.C.I. contributed equally to this work.
REFERENCES
- 1.Adamson E D, Woodland H R. Changes in the rate of histone synthesis during oocyte maturation and very early development of Xenopus laevis. Dev Biol. 1977;57:136–149. doi: 10.1016/0012-1606(77)90360-8. [DOI] [PubMed] [Google Scholar]
- 2.Anderson K V, Lengyel J A. Changing rates of histone mRNA synthesis and turnover in Drosophila embryos. Cell. 1980;21:717–727. doi: 10.1016/0092-8674(80)90435-3. [DOI] [PubMed] [Google Scholar]
- 3.Anderson K V, Lengyel J A. Histone gene expression in Drosophila development: multiple levels of gene regulation. In: Stein G, Marzluff W F, editors. Histone genes: structure, organization and regulation. New York, N.Y: John Wiley and Sons; 1984. pp. 135–162. [Google Scholar]
- 4.Ballantine J E, Woodland H R. Polyadenylation of histone mRNA in Xenopus oocytes and embryos. FEBS Lett. 1985;180:224–228. doi: 10.1016/0014-5793(85)81075-9. [DOI] [PubMed] [Google Scholar]
- 5.Barkoff A, Ballantyne S, Wickens M. Meiotic maturation in Xenopus requires polyadenylation of multiple mRNAs. EMBO J. 1998;17:3168–3175. doi: 10.1093/emboj/17.11.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouvet P, Wolffe A P. A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell. 1994;77:931–942. doi: 10.1016/0092-8674(94)90141-4. [DOI] [PubMed] [Google Scholar]
- 7.Craig A W, Haghighat A, Yu A T, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 8.Dominski Z, Sumerel J, Hanson R J, Marzluff W F. The polyribosomal protein bound to the 3′ end of histone mRNA can function in histone pre-mRNA processing. RNA. 1995;1:915–923. [PMC free article] [PubMed] [Google Scholar]
- 9.Dumont J N. Oogenesis in Xenopus laevis (Daudin). 1. Stage of oocyte development in laboratory maintained animals. J Morphol. 1972;136:153–180. doi: 10.1002/jmor.1051360203. [DOI] [PubMed] [Google Scholar]
- 10.Evans J P, Kay B K. Biochemical fractionation of oocytes. Methods Cell Biol. 1991;36:133–148. doi: 10.1016/s0091-679x(08)60275-7. [DOI] [PubMed] [Google Scholar]
- 11.Fox C A, Sheets M D, Wickens M P. Poly(A) addition during maturation of frog oocytes: distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes Dev. 1989;3:2151–2162. doi: 10.1101/gad.3.12b.2151. [DOI] [PubMed] [Google Scholar]
- 12.Fox C A, Wickens M. Poly(A) removal during oocyte maturation: a default reaction selectively prevented by specific sequences in the 3′ UTR of certain maternal mRNAs. Genes Dev. 1990;4:2287–2298. doi: 10.1101/gad.4.12b.2287. [DOI] [PubMed] [Google Scholar]
- 13.Gallie D R, Lewis N J, Marzluff W F. The histone 3′-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res. 1996;24:1954–1962. doi: 10.1093/nar/24.10.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gebauer F, Xu W, Cooper G M, Richter J D. Translational control by cytoplasmic polyadenylation of c-mos mRNA is necessary for oocyte maturation in the mouse. EMBO J. 1994;13:5712–5720. doi: 10.1002/j.1460-2075.1994.tb06909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hake L E, Richter J D. Translational regulation of maternal mRNA. Biochim Biophys Acta. 1997;1332:M31–M38. doi: 10.1016/s0304-419x(96)00039-x. [DOI] [PubMed] [Google Scholar]
- 16.Hanson R J, Sun J-H, Willis D G, Marzluff W F. Efficient extraction and partial purification of the polyribosomal-associated stem-loop binding protein bound to the 3′ end of histone mRNA. Biochemistry. 1996;35:2146–2156. doi: 10.1021/bi9521856. [DOI] [PubMed] [Google Scholar]
- 17.Heasman J, Crawford A, Goldstone K, Garner-Hamrick P, Gumbiner B, McCrea P, Kintner C, Noro C Y, Wylie C. Overexpression of cadherins and underexpression of beta-catenin inhibit dorsal mesoderm induction in early Xenopus embryos. Cell. 1994;79:791–803. doi: 10.1016/0092-8674(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 18.Jacobson A. PolyA metabolism and translation: the closed loop model. In: Hershey J W, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 451–480. [Google Scholar]
- 19.Kuge H, Richter J D. Cytoplasmic 3′ poly(A) addition induces 5′ cap ribose methylation: implications for translational control of maternal mRNA. EMBO J. 1995;14:6301–6310. doi: 10.1002/j.1460-2075.1995.tb00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levenson R G, Marcu K B. On the existence of polyadenylated histone mRNA in Xenopus oocytes. Cell. 1976;9:311–322. doi: 10.1016/0092-8674(76)90121-5. [DOI] [PubMed] [Google Scholar]
- 21.Li J-M, Parsons R A, Marzluff W F. Transcription of the sea urchin U6 gene in vitro requires a TATA-like box, a proximal sequence element, and sea urchin USF, which binds an essential E box. Mol Cell Biol. 1994;14:2191–2200. doi: 10.1128/mcb.14.3.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin F, Schaller A, Eglite S, Schümperli D, Müller B. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997;16:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marzluff W F. Histone 3′ ends: essential and regulatory functions. Gene Expr. 1992;2:93–97. [PMC free article] [PubMed] [Google Scholar]
- 24.Marzluff W F, Whitfield M L, Dominski Z, Wang Z-F. Identification of the protein that interacts with the 3′ end of histone mRNA. In: Richter J D, editor. mRNA formation and function. San Diego, Calif: Academic Press; 1997. pp. 163–193. [Google Scholar]
- 25.Matsumoto K, Meric F, Wolffe A P. Translational repression dependent on the interaction of the Xenopus Y-box protein FRGY2 with mRNA—role of the cold shock domain, tail domain, and selective RNA sequence recognition. J Biol Chem. 1996;271:22706–22712. doi: 10.1074/jbc.271.37.22706. [DOI] [PubMed] [Google Scholar]
- 26.Matsumoto K, Wassarman K M, Wolffe A P. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 1998;17:2107–2121. doi: 10.1093/emboj/17.7.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maxson R, Mohun T J, Cohn R, Kedes L. Expression and organization of histone genes. Annu Rev Genet. 1983;17:239–277. doi: 10.1146/annurev.ge.17.120183.001323. [DOI] [PubMed] [Google Scholar]
- 28.Meric F, Matsumoto K, Wolffe A P. Regulated unmasking of in vivo synthesized maternal mRNA at oocyte maturation—a role for the chaperone nucleoplasmin. J Biol Chem. 1997;272:12840–12846. doi: 10.1074/jbc.272.19.12840. [DOI] [PubMed] [Google Scholar]
- 29.Meric F, Searfoss A M, Wormington M, Wolffe A P. Masking and unmasking maternal mRNA—the role of polyadenylation, transcription, splicing, and nuclear history. J Biol Chem. 1996;271:30804–30810. doi: 10.1074/jbc.271.48.30804. [DOI] [PubMed] [Google Scholar]
- 30.Munroe D, Jacobson A. Tales of poly(A): a review. Gene. 1990;91:151–158. doi: 10.1016/0378-1119(90)90082-3. [DOI] [PubMed] [Google Scholar]
- 31.Newport J, Kirschner M A. A major developmental transition in early Xenopus embryos. II. Control of the onset of transcription. Cell. 1982;3:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- 32.Perry M, Thomsen G H, Roeder R G. Genomic organization and nucleotide sequence of two distinct histone clusters from Xenopus laevis. J Mol Biol. 1985;185:479–499. doi: 10.1016/0022-2836(85)90065-8. [DOI] [PubMed] [Google Scholar]
- 33.Perry M, Thomsen G H, Roeder R G. Major transitions in histone gene expression do not occur during development in Xenopus laevis. Dev Biol. 1986;116:532–538. doi: 10.1016/0012-1606(86)90154-5. [DOI] [PubMed] [Google Scholar]
- 34.Ruderman J V, Pardue M L. A portion of all major classes of histone messenger RNA in amphibian oocytes is polyadenylated. J Biol Chem. 1978;253:2018–2025. [PubMed] [Google Scholar]
- 35.Sallés F J, Lieberfarb M E, Wreden C, Gergen J P, Strickland S. Coordinate initiation of Drosophila development by regulated polyadenylation of maternal messenger RNAs. Science. 1994;266:1996–1999. doi: 10.1126/science.7801127. [DOI] [PubMed] [Google Scholar]
- 35a.Sanchez, R., and Z. Dominski. Unpublished data.
- 35b.Sanchez, R. Unpublished data.
- 36.SenGupta D J, Zhang B L, Kraemer B, Prochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheets M D, Wu M, Wickens M. Polyadenylation of c-mos mRNA as a control point in Xenopus meiotic maturation. Nature. 1995;374:511–516. doi: 10.1038/374511a0. [DOI] [PubMed] [Google Scholar]
- 38.Stebbins-Boaz B, Hake L E, Richter J D. CPEB controls the cytoplasmic polyadenylation of cyclin, Cdk2 and c-mos mRNAs and is necessary for oocyte maturation in Xenopus. EMBO J. 1996;15:2582–2592. [PMC free article] [PubMed] [Google Scholar]
- 39.Sun J-H, Pilch D R, Marzluff W F. The histone mRNA 3′ end is required for localization of histone mRNA to polyribosomes. Nucleic Acids Res. 1992;20:6057–6066. doi: 10.1093/nar/20.22.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tarun S Z, Jr, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 41.Tarun S Z, Jr, Wells S E, Deardorff J A, Sachs A B. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Dongen W M A M, Moorman A F M, Destree O H J. Histone gene expression in early development of Xenopus laevis. Differentiation. 1983;24:226–233. doi: 10.1111/j.1432-0436.1983.tb01324.x. [DOI] [PubMed] [Google Scholar]
- 43.Vassalli J-D, Huarte J, Belin D, Gubler P, Vassalli A, O’Connell M L, Parton L A, Rickles R J, Strickland S. Regulated polyadenylation controls mRNA translation during meiotic maturation of mouse oocytes. Genes Dev. 1989;3:2163–2171. doi: 10.1101/gad.3.12b.2163. [DOI] [PubMed] [Google Scholar]
- 44.Verrotti A C, Thompson S R, Wreden C, Strickland S, Wickens M. Evolutionary conservation of sequence elements controlling cytoplasmic polyadenylylation. Proc Natl Acad Sci USA. 1996;93:9027–9032. doi: 10.1073/pnas.93.17.9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker J, Dale M, Standart N. Unmasking mRNA in clam oocytes: role of phosphorylation of a 3′ UTR masking element-binding protein at fertilization. Dev Biol. 1996;173:292–305. doi: 10.1006/dbio.1996.0024. [DOI] [PubMed] [Google Scholar]
- 45a.Wang, Z.-F. Unpublished data.
- 46.Wang Z-F, Whitfield M L, Ingledue T I, Dominski Z, Marzluff W F. The protein which binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- 46a.Whitfield, M. L., and W. F. Marzluff. Unpublished data.
- 47.Wickens M. In the beginning is the end: regulation of poly(A) addition and removal during early development. Trends Biochem Sci. 1990;15:320–324. doi: 10.1016/0968-0004(90)90022-4. [DOI] [PubMed] [Google Scholar]
- 48.Wickens M P, Kimble J, Strickland S. Translational control of developmental decisions. In: Hershey J W, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 411–450. [Google Scholar]
- 49.Williams A S, Ingledue T C, Kay B K, Marzluff W F. Changes in the stem-loop at the 3′ terminus of histone mRNA affects its nucleocytoplasmic transport and cytoplasmic regulation. Nucleic Acids Res. 1994;22:4660–4666. doi: 10.1093/nar/22.22.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams A S, Marzluff W F. The sequence of the stem and flanking sequences at the 3′ end of histone mRNA are critical determinants for the binding of the stem-loop binding protein. Nucleic Acids Res. 1995;23:654–662. doi: 10.1093/nar/23.4.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woodland H R. Histone synthesis during the development of Xenopus. FEBS Lett. 1980;121:1–7. doi: 10.1016/0014-5793(80)81252-x. [DOI] [PubMed] [Google Scholar]
- 52.Woodland H R, Adamson E D. The synthesis and storage of histones during oogenesis of Xenopus laevis. Dev Biol. 1977;57:118–135. doi: 10.1016/0012-1606(77)90359-1. [DOI] [PubMed] [Google Scholar]