Abstract
Recent progress in identifying the catalytic subunits of histone acetyltransferase (HAT) complexes has implicated histone acetylation in the regulation of transcription. Here, we have analyzed the function of two native yeast HAT complexes, SAGA (Spt-Ada-Gcn5 Acetyltransferase) and NuA4 (nucleosome acetyltransferase of H4), in activating transcription from preassembled nucleosomal array templates in vitro. Each complex was tested for the ability to enhance transcription driven by GAL4 derivatives containing either acidic, glutamine-rich, or proline-rich activation domains. On nucleosomal array templates, the SAGA complex selectively stimulates transcription driven by the VP16 acidic activation domain in an acetyl coenzyme A-dependent manner. In contrast, the NuA4 complex facilitates transcription mediated by any of the activation domains tested if allowed to preacetylate the nucleosomal template, indicating a general stimulatory effect of histone H4 acetylation. However, when the extent of acetylation by NuA4 is limited, the complex also preferentially stimulates VP16-driven transcription. SAGA and NuA4 interact directly with the VP16 activation domain but not with a glutamine-rich or proline-rich activation domain. These data suggest that recruitment of the SAGA and NuA4 HAT complexes by the VP16 activation domain contributes to HAT-dependent activation. In addition, extensive H4/H2B acetylation by NuA4 leads to a general activation of transcription, which is independent of activator-NuA4 interactions.
Numerous biochemical and genetic studies have provided compelling evidence for a role of chromatin structures in the regulation of gene transcription (reviewed in references 20, 32 and 62). The distribution and/or structure of nucleosomes is altered at transcription control elements prior to or concurrently with transcription activation (reviewed in references 3 and 48). Several laboratories have pursued the mechanisms by which nucleosome and chromatin structure is altered in order to facilitate the interactions of sequence-specific transcription factors with DNA. These genetic and biochemical studies have revealed the participation of multiprotein complexes in the remodeling of chromatin structures. One group of these complexes are the ATP-dependent chromatin-remodeling complexes and includes the SWI/SNF, NURF, RSC, ACF, and CHRAC complexes (10, 16, 29, 54, and 58). A second group of complexes function, at least in part, by controlling histone modifications. These include histone acetyltransferase (HAT) and histone deacetylase complexes (reviewed in references 8 and 60).
The level of histone acetylation at different chromosomal loci correlates with gene activity. Increased levels of histone acetylation are often associated with transcriptional activity, whereas hypoacetylation of histones has been observed at genetically silenced regions and in heterochromatin (7, 24). Acetylation of lysine side chains within the histone N-terminal tail domains reduces the affinity of the histone tails for DNA (27). Histone deacetylase inhibitors have been shown to increase transcription from the human immunodeficiency virus type 1 (HIV-1) promoter in vivo (57) and in vitro (46), and the incorporation of acetylated histones during in vitro nucleosome assembly of 5S RNA genes is less repressive than that of nonacetylated histones to subsequent transcription (55).
Recently, several enzymes responsible for histone acetylation and deacetylation have been identified and often correspond to previously known transcriptional regulatory factors (reviewed in references 42 and 60). We have identified four native high-molecular-weight complexes from yeast extracts that contain nucleosomal HAT activities (21). One of these HAT complexes, termed NuA4 (nucleosome acetyltransferase of histone H4), is a 1.3-MDa complex of which the catalytic subunit has not been identified. NuA4 has a preference for acetylating histones H4 and H2A in the nucleosome (21). Another HAT complex, termed SAGA (Spt-Ada-Gcn5 acetyltransferase), is a 1.8-MDa complex containing several previously identified transcriptional regulators from the ADA gene products and the TBP group of Spt gene products (reviewed in reference 23). SAGA contains GCN5 as the catalytic HAT subunit and also contains Ada2, Ada3, Ada5/Spt20, Spt3, Spt8, and Spt7. This complex preferentially acetylates nucleosomal histones H3, H2B, and, to some degree, H4 (21).
The observation that several transcriptional coactivators are HATs (reviewed in references 42 and 60) suggests a role of the corresponding DNA-binding transcription activators in histone acetylation. The genetic analyses of SAGA components (i.e., the ADA genes) strongly suggest that SAGA functions, in part, through interactions with acidic transcriptional activators (reviewed in reference 23). Indeed, biochemical studies have demonstrated a direct interaction of the VP16 activation domain with ADA2 (2). Moreover, our initial functional studies demonstrate direct interactions of both the SAGA and NuA4 HAT complexes with the VP16 activation domain and that these complexes stimulated transcription in an acetyl coenzyme A (acetyl-CoA)-dependent reaction (56).
There are at least two non-mutually exclusive mechanisms by which the SAGA and NuA4 HAT complexes might stimulate transcription. (i) Overall acetylation of the nucleosomal templates by these complexes may have a general stimulatory effect on transcription. (ii) Recruitment of the SAGA or NuA4 complexes by a promoter-bound activator may lead to localized acetylation, which, in turn, stimulates transcription. The former possibility suggests that all activated (detectable) transcription should be stimulated and is consistent with observations that nucleosome templates reconstituted with acetylated histones are more permissive for transcription (36, 55). The latter possibility suggests that stimulation should be activator specific and is consistent with the observations of in vivo targeting of Gcn5-dependent acetylation to promoter-proximal regions (33) and in vitro interactions of SAGA and NuA4 with the VP16 activation domain and with GCN4 (56). To test these possibilities, we examined the function of activation domains from distinct classes (acidic, glutamine rich, and proline rich) in SAGA- and NuA4-stimulated transcription. We present evidence for a role of recruitment of the SAGA and NuA4 complexes by an acidic activation domain in transcriptional stimulation by these HAT complexes. Acidic activator interactions appear to be essential for transcriptional stimulation by SAGA and enhance transcriptional stimulation by NuA4. However, extensive acetylation of the nucleosome template by the NuA4 complex eventually leads to a general stimulation of transcription by any of the activators tested.
MATERIALS AND METHODS
Plasmid construction.
pIC-2085S/G5E4R contains the G5E4-5S array fragment (Fig. 1A) and was constructed as follows. A dinucleosome length G5E4 fragment (2,586 bp) containing adenovirus E4 gene sequences from −38 to +218 with five copies of a 17-mer GAL4-binding site was produced by PCR amplification using pG5E4T as the template (34) and oligonucleotide 5′-CCCGCTCGAGTGCATGCCTGCAGGTC for the 5′ XhoI site and oligonucleotide 5′-CCCGCTCGAGATTACAGCCCCCATAGG for the 3′ XhoI site. The XhoI-digested PCR product was subcloned into the XhoI site of pIC-2085S, which has five tandem repeats of the sea urchin 5S rRNA-encoding gene (rDNA) nucleosome-positioning sequence flanking both sides of a short linker region (49).
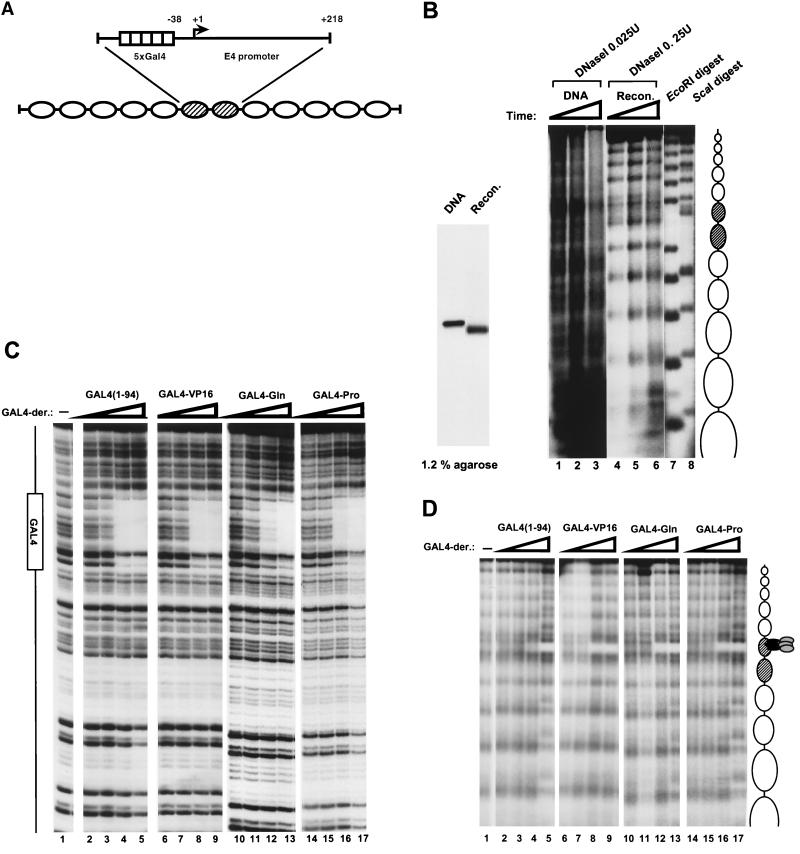
FIG. 1.
Analysis of the reconstituted nucleosome template. (A) Schematic of the G5E4-5S DNA fragment that was used to reconstituted a 12-nucleosome array by histone octamer transfer. The 2,586-bp Asp718-ClaI fragment excised from pIC208/G5E4R contains tandem repeats of the sea urchin 5S rDNA nucleosome spacing sequences. Between the repeats, there is a dinucleosome length fragment harboring five GAL4-binding sites upstream of the E4 promoter. (B) Comparison of the nucleosome-reconstituted array with the G5E4-5S DNA fragment. (Left) The end-labeled G5E4-5S DNA fragment (DNA lane) and the end-labeled nucleosome-reconstituted fragment (Recon. lane) were run on a 1.2% native agarose gel. (Right) DNase I digestion of the nucleosome-reconstituted array and the G5E4-5S DNA fragment. The end-labeled G5E4-5S DNA fragment (DNA, lanes 1 to 3) and the nucleosome-reconstituted array (Recon., lanes 4 to 6) were digested with 0.025 and 0.25 U of DNase I, respectively. The times of digestion were 0.5 min (lanes 1 and 4), 1 min (lanes 2 and 5), and 1.5 min (lanes 3 and 6). The 5S rDNA repeats were revealed by partial EcoRI digestion (lane 7) of the array, and the 5S rDNA repeats and the five GAL4-binding sites were revealed by partial ScaI digestion (lane 8) of the array. EcoRI and ScaI sites are present at the junction between the 5S repeats. A ScaI site is also present in the center of each GAL4-binding site. Nucleosome positions on the G5E4-5S DNA fragment are indicated schematically on the right of the autoradiogram. (C) DNase I footprinting of GAL4 derivatives binding to a naked DNA fragment of GUB, which contains one GAL4-binding site. Binding reaction mixtures were treated with 0.033 U of DNase I for 1 min in the presence (lanes 2 to 17) or the absence (lane 1) of GAL4 derivatives. Reaction mixtures contained twofold-increasing amounts of each GAL4 derivative. GAL4 derivatives were included at concentrations of 3.1 nM (lanes 2, 6, 10, and 14), 6.3 nM (lanes 3, 7, 11, and 15), 12.5 nM (lanes 4, 8, 12, and 16), and 25 nM (lanes 5, 9, 13, and 17) DNA-binding activity. The GAL4-binding site is shown as a box to the left of the autoradiogram. (D) DNase I digestion analysis of various GAL4 derivatives of the nucleosome-reconstituted array. Binding reaction mixtures were treated with DNase I for 0.5 min in the presence (lanes 2 to 17) or the absence (lane 1) of GAL4 derivatives. Reaction mixtures contained 0.5 nM (lanes 2, 6, 10, and 14), 2.5 nM (lanes 3, 7, 11, and 15), 12.5 nM (lane 4, 8, 12, and 16), or 62.5 nM (lanes 5, 9, 13, and 17) DNA binding activity of each GAL4 derivative. GAL4-binding sites and nucleosome positions on the G5E4-5S DNA fragment are indicated schematically on the right.
Plasmids pG3, pT7Gal 1-94/Sp1 A+B, and pP13, which direct the expression of GAL4(1-94), GAL4-Gln, and GAL4-Pro, respectively, were obtained from B. F. Pugh (51). The plasmid expressing GAL4-VP16 was obtained by S. L. Berger (4).
Plasmids for expression of glutathione S-transferase (GST)-Gln and GST-Pro in Escherichia coli were constructed as follows. A DNA fragment was synthesized by PCR using pT7Gal 1-94/Sp1 A+B (51) as the template and oligonucleotides 5′-GGGCATATGTCCGGCGGACAGGGA and 5′-CGGAAGCTTCTTACTTATCTAGAGCTCG for GST-Gln. A DNA fragment was synthesized by PCR using pP13 (51) as the template and oligonucleotides 5′-GGGCATATGGATCTTGTCTCGCTGGC and 5′-CCCGAATTCTCCCAGATACCAGGACTG for GST-Pro. After cleavage with NdeI and EcoRI, these fragments were subcloned into NdeI-EcoRI-digested pGEX-B, which was provided by M. Meiseterernst.
pHIV(D,N), which was used as an internal control plasmid template in transcription reactions for checking the recovery of the reaction, was constructed as follows. A DraIII (position −248)-NarI (position +183) restriction fragment of HIV-1 was treated sequentially with Klenow and T4 polymerase at the 5′ and 3′ overhangs, respectively, and cloned into pIC20R (49) treated with EcoRV, BamHI, and Klenow.
Purification of GAL4-derivative proteins and GST fusion proteins.
GAL4(1-94), GAL4-VP16, GAL4-Gln, and GAL4-Pro were bacterially expressed and purified as previously described (12, 34, 51), except that GAL4-VP16 was step eluted at 0.4 M NaCl from a DEAE column by T. Owen-Hughes, R. T. Utley, and J. Côté. The quality and activity of the products were checked by both sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and gel shift analysis by using the DNA with GAL4-binding sites, followed by footprinting analysis (Fig. 1). GST, GST-VP16, GST-Gln, and GST-Pro were expressed and purified by following the manufacturer’s (Pharmacia) protocol, except that the bacterial pellet suspension solution contained 10 mM β-mercaptoethanol, 2-mg/liter leupeptin, 2-mg/liter pepstatin A, 2-mg/liter benzamidine, and 2-mg/liter aprotinin for GST-VP16.
HAT purification.
Preparation of yeast extracts and fractionation of the HAT complexes by Ni2+-nitrilotriacetic acid agarose and MonoQ fast protein liquid chromatography were done as previously described (21). Peak HAT fractions for NuA4 and SAGA were pooled separately and further purified. NuA4 fractions were pooled and loaded onto a histone agarose column and subjected to Superose 6 exclusion chromatography. Fractions containing SAGA were purified on a MonoS HR5/5 column (Pharmacia) and on Superose 6. For transcription studies, the pure peak fraction of either NuA4 or SAGA after Superose 6 chromatography was used. Incorporation of approximately 1,000 cpm of 3H-labeled acetyl-CoA into the G5E4-5S reconstituted nucleosome was defined as 1 U of HAT activity.
Template DNA fragment preparation.
To prepare the G5E4-5S DNA fragment for reconstitution, pIC-2085S/G5E4R was digested with Asp718 and treated with Klenow fragment in the absence (template for transcription) or the presence (template for native gel electrophoresis and DNase I digestion analysis) of [α-32P]dATP. The fragment was further digested with ClaI, SspI, and AlwNI, and the Asp718-ClaI fragment was gel purified by electroelution.
Nucleosome reconstitution.
Core histone preparation and nucleosome reconstitution on the G5E4-5S fragment were performed essentially as described previously (38). Briefly, 2 μg of HeLa core histone was mixed with 2 μg of the G5E4-5S DNA fragment in 50 mM HEPES-NaOH (pH 7.5)–2 M NaCl–1 mM EDTA–1 mM dithiothreitol (DTT)–1 mM phenylmethylsulfonyl fluoride (PMSF)–100-μg/ml bovine serum albumin (BSA) in a final volume of 10 μl. This mixture was diluted gradually with 50 mM HEPES-NaOH (pH 7.5)–2 mM EDTA, 5 mM DTT–0.5 mM PMSF at 30°C. Finally, the mixture was brought to 0.1 M NaCl by the addition of 100 μl of 10 mM Tris-HCl (pH 7.5), 2 mM EDTA, 5 mM DTT, 0.5 mM PMSF, 0.1% Nonidet P-40, 20% glycerol, and 100-μg/ml BSA. The nucleosome-reconstituted template was stored at 4°C and was stable for a month.
Binding reactions.
Binding reactions were performed in 20 μl with 10 mM HEPES-NaOH (pH 7.8)–60 mM NaCl–5 mM DTT–0.5 mM PMSF–0.25-mg/ml BSA–10 mM sodium butyrate–5% glycerol at room temperature for 10 min. Typical binding reactions contained 15 ng (10 fmol) of reconstituted DNA fragment. GAL4 derivatives were added to the reaction mixture at the final concentrations indicated in the figure legends. One unit of NuA4 or SAGA in the presence or the absence of 1.25 μM acetyl-CoA was included as indicated in the figures. Binding reactions were subjected to either native gel electrophoresis, DNase I digestion analysis, or transcription reactions.
DNase I digestion assays.
Factor-bound templates were treated with 2 μl (0.25 U for nucleosome template DNA or 0.025 U for naked template DNA) of DNase I (Boehringer Mannheim) in 50 mM MgCl2 for 0.5, 1, or 1.5 min at room temperature (Fig. 1B). Binding reaction mixtures with GAL4 derivatives (Fig. 2D) were treated with DNase I for 0.5 min, except GAL4-Gln, which was treated for 1.5 min. DNase I was terminated with stop mix (49). After ethanol precipitation, DNA was dissolved in 10 mM Tris-HCl (pH 7.5)–1 mM EDTA–50 mM NaCl and electrophoresed through 1.5% 1× Tris-borate-EDTA–agarose gel. Gels subsequently were fixed in a solution of 10% methanol–10% acetic acid and then dried.
FIG. 2.
Analysis of transcription from the naked DNA template and from the nucleosome-reconstituted array template with or without GAL4 derivatives. (A) Transcripts detected by primer extension from the E4 promoter (E4) of naked template DNA (pIC2085S/G5E4R) and that from the HIV promoter (HIV) of control plasmid pHIV(D,N) are indicated on the left. A 12.5 nM concentration of DNA-binding activity of each GAL4 derivative was included where indicated. Lane 1 is a control reaction to which no GAL4 derivatives were added. Lanes 6 to 13 show transcripts in the presence of HATs (SAGA and NuA4) and acetyl-CoA. (B) Transcripts from the G5E4-5S nucleosome array template in the presence or the absence of acetyl-CoA without or with the same amounts of various GAL4-derivative proteins as used for panel A.
DNase I footprinting of naked template DNA.
Probe GUB was generated by digestion of pGUB (1) with EcoRI, treatment with Klenow fragment in the presence of [α-32P]dATP, and then digestion with SalI. A 180-bp DNA fragment was purified by PAGE as previously described (1). Binding reactions were performed with 10 to 20 fmol of probe DNA in the presence or the absence of GAL4 derivatives at room temperature for 30 min, followed by the addition of 2 μl (0.033 U) of DNase I and incubation for 1 min at room temperature. DNase I was terminated as previously described (1). After ethanol precipitation, DNA was dissolved in formamide loading buffer, heat denatured, and resolved on 8% acrylamide–8 M urea sequencing gels (1).
Transcription reaction.
A binding reaction mixture containing 15 ng (10 fmol) of a reconstituted DNA fragment or 15 ng of pIC-2085S/G5E4R was mixed in a 50-μl (final volume) reaction mixture containing 15 mM HEPES-NaOH (pH 7.8), 6 mM MgCl2, 30 mM KCl, 2% polyvinyl alcohol, 150 ng of poly(dI-dC), HeLa cell nuclear extract (40 to 50 μg of protein prepared as previously described [17]), 10 ng of pHIV (internal control), and 0.4 mM ribonucleotides. The transcription reaction was performed at 30°C for 30 min and terminated with STOP mix (30). The resulting RNAs were detected by using 32P-labeled E4 primer (positions +110 to +86) and HIV primer (positions +81 to +50) in a mixed-primer extension assay as previously described (30), except that 50 U of Moloney murine leukemia virus reverse transcriptase (Gibco-BRL) and 400 μM deoxynucleoside triphosphates were used. The reverse-transcribed DNA products were separated on urea–8% polyacrylamide gels, visualized by autoradiography (Kodak BioMax), and quantitated following PhosphorImager scanning (Molecular Dynamics). All transcriptions were repeated at least three times.
GST pull-down assays.
Five units of NuA4 or SAGA was incubated with the indicated GST fusion proteins bound to glutathione-Sepharose 4B beads (Pharmacia) for 2 h at 4°C. The supernatant was removed, and the beads were washed five times with 80 mM bead wash buffer (80 mM NaCl, 25 mM HEPES-NaOH [pH 7.5], 50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.05% Nonidet P-40, 5 mM DTT, 0.5 mM PMSF, 10% glycerol). Equal amounts of both the supernatant and the beads were directly assayed for HAT activity on nucleosomes (see below and reference 21).
HAT assays.
HAT assays for the G5E4-5S nucleosome array template were performed as follows. A 20-μl volume of a binding reaction mixture was incubated with 1 U of NuA4 or SAGA and 3H-labeled acetyl-CoA (0.125 μCi) in HAT buffer (50 mM Tris-HCl [pH 8.0], 50 mM KCl, 5% glycerol, 0.1 mM EDTA, 1 mM DTT, 1 mM PMSF, 10 mM sodium butyrate) at 30°C. The reaction mixture was spotted onto a P81 membrane filter (Whatman). The membrane was washed three times in 50 mM NaHCO3-Na2CO3 pH 9.2) and briefly rinsed, and radioactivity was counted in a liquid scintillation counter (Beckman). For fluorography as shown in Fig. 3B, a binding reaction mixture containing 150 ng of nucleosome template DNA was incubated at 30°C for 30 min and subjected to SDS–15% PAGE. Gels were Coomassie stained and prepared for fluorography by using the manufacturer’s protocol (Enhance; Du Pont NEN). For the experiment whose results are shown in Fig. 6, gel filtration columns (MicroSpin S-300 HR Column; Pharmacia Biotech) were used by following the manufacturer’s protocol.
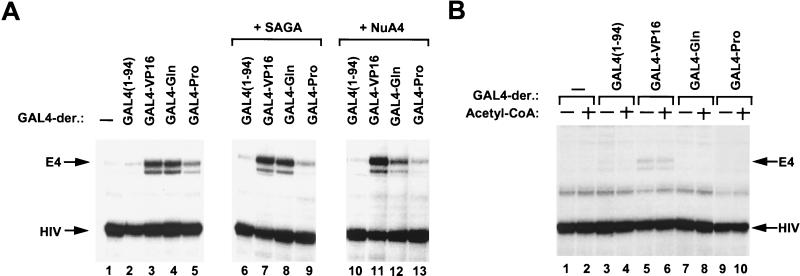
FIG. 3.
Effects of HATs on GAL4 derivative-mediated transcription of the nucleosome array templates. (A) Diagram of the transcription protocol. Subsequent to the factor-binding reaction, 1 U of NuA4 or SAGA, in the presence or the absence of acetyl-CoA, was included and incubated at 30°C for 30 min. After the incubation, transcription reactions were started by adding HeLa cell nuclear extract and nucleotides. The same amounts of various GAL4-derivative proteins were used as for Fig. 2A. NTPs, nucleoside triphosphates; RT, room temperature. (B) HAT assays of the G5E4-5S nucleosome array template. Nucleosome templates were incubated in a binding reaction mixture with NuA4 or SAGA in the presence (lane 2) or the absence (lane 1) of 3H-labeled acetyl-CoA. After incubation, SDS-sample buffer and 3 μg of core histone were added, and the mixture was loaded onto a gel. Lanes 3 and 4 show Coomassie staining of the same gel, indicating the migratory positions of the four core histones. (C) Transcripts from the G5E4-5S array template, which were incubated with SAGA, in the presence or the absence of acetyl-CoA, without (−) or with various GAL4 derivative proteins. (D) Transcripts from the G5E4-5S array template which were incubated with NuA4 in the presence or the absence of acetyl-CoA without (−) or with various GAL4-derivative proteins.
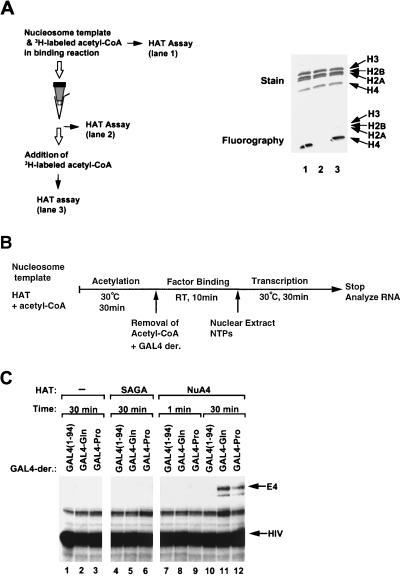
FIG. 6.
Extensive acetylation by NuA4 in the absence of activators facilitates transcription driven by GAL4-Gln and GAL4-Pro. (A) Diagram (left) and SDS-gel (right) showing the prevention of NuA4 HAT activity through the removal of acetyl-CoA by gel filtration. HAT activity was monitored in the input reaction mixture containing the G5E4 nucleosomal template, 3H-labeled acetyl-CoA, and NuA4 (lane 1), after gel filtration to remove the acetyl-CoA (lane 2), and after gel filtration followed by the readdition of 3H-labeled acetyl-CoA (lane 3). (B) Diagram of the transcription protocol. Subsequent to acetylation with 1 U of SAGA (30 min of incubation) or NuA4 (1 or 30 min of incubation), gel filtration was performed to remove the acetyl-CoA. Recovered samples were subjected to a factor-binding reaction with GAL4(1-94), GAL4-Gln, and GAL4-Pro, followed by transcription reactions. RT, room temperature; NTPs, nucleoside triphosphates. (C) G5E4 array templates were incubated with SAGA (lanes 4 to 6) or NuA4 (lanes 7 to 12) for the indicated time periods and assayed for transcription as described in panel B.
RESULTS
Nucleosomes repress activation domain function.
To directly test the function of the native yeast HAT complexes in transcription activation, we developed an in vitro transcription system using a nucleosome array template reconstituted from defined components in a purified system (i.e., DNA and histones). The DNA fragment used for nucleosome reconstitution was a 2,586-bp DNA fragment (G5E4-5S) that contains a dinucleosome length sequence with five GAL4-binding sites and the adenovirus E4 promoter (34) flanked on either side by five repeats of a nucleosome-positioning sequence from the sea urchin 5S rDNA (47). This fragment is illustrated schematically in Fig. 1A. The fragment was reconstituted into an array of nucleosomes by using purified histone octamers (38).
Reconstitution of the G5E4-5S DNA fragment into an array of nucleosomes was assayed by mobility shift and nuclease digestion. The nucleosome-assembled G5E4-5S fragment and the G5E4-5S fragment as naked DNA were analyzed by native agarose gel electrophoresis (Fig. 1B, left panel). As previously observed (37), the nucleosome-reconstituted array fragment (Recon. lane) was found to have faster mobility than the same fragment as histone-free DNA (DNA lane). Importantly, the reconstituted array migrated as a discrete band, suggesting that most molecules contain similar numbers of nucleosomes. The histone-free DNA fragment and the array fragment were also subjected to DNase I digestion, followed by agarose gel electrophoresis (Fig. 1B, right panel). A nucleosome length repeating pattern of cleavage and protection was observed on the reconstituted array fragment (lanes 4 to 6) that was not apparent upon digestion of the naked DNA fragment (lanes 1 to 3). This repeating pattern corresponded nicely to the repeating 5S DNA sequences (indicated by the partial EcoRI digest), indicating the reconstitution of a single nucleosome core per repeat. Importantly, nucleosome protection from DNase I cleavage extended over the region of the GAL4-binding sites and the E4 promoter (shaded ovals), which could be localized by partial ScaI digestion (lane 8). Thus, the repeating array of spaced nucleosomes established by the 5S nucleosome positioning sequences continued through the dinucleosome length promoter insert.
To test the function of different transcription activation domains in HAT complex-driven transcription, we used GAL4 derivatives representing three functionally distinct classes of activation domains. GAL4-VP16 is a potent acidic activation domain from the virion of herpes simplex virus (45, 53) fused with a GAL4 DNA-binding domain. GAL4-Gln is the glutamine-rich activation domain of human Sp1 fused with GAL4(1-94) (51). GAL4-Pro is the proline-rich activation domain of human CTF/NFI fused with GAL4(1-94) (51). GAL4(1-94) consists of the amino-terminal 94 amino acids of the yeast GAL4 protein containing the DNA-binding and dimerization domains but lacking an activation domain.
To ensure that any transcription effect of the HAT complexes was not due to increased GAL4 derivative binding due to histone acetylation (59), we used concentrations of each GAL4 derivative that led to saturation of the nucleosome array template. The amounts of the GAL4 derivatives were first equalized based on DNA-binding activity (Fig. 1C), and then the GAL4 derivatives were compared for relative affinity on nucleosome arrays (Fig. 1D). All of the GAL4 derivatives were found to possess similar affinities for the nucleosome array template (Fig. 1D). Thus, the presence or absence of these activation domains did not appear to influence the relative affinity of each derivative for nucleosomal DNA. This is in agreement with previous studies demonstrating that nucleosome binding by GAL4 derivatives is activation domain independent (52, 65). For the transcription experiments in the following sections, we used the amount of each GAL4 derivative at which the footprint in the nucleosome array appeared complete (Fig. 1D, lanes 4, 8, 12, and 16). This represented 12.5 nM DNA-binding activity of each GAL4 derivative.
To determine if each GAL4 derivative was functional in transcription, we first tested each in transcription assays from DNA templates. As shown in Fig. 2A, each of these proteins activated transcription in vitro from naked DNA. Inclusion of GAL4-VP16 resulted in sevenfold stimulation of transcription (lane 3) relative to basal transcription (lane 1). GAL4-Gln also brought about sevenfold stimulation of transcription (lane 4). The activity of GAL4-Pro was slightly less, resulting in fourfold stimulation (lane 5). Thus, each of the GAL4 derivatives was functional as a transcription activator in vitro from naked DNA.
Next, we examined whether these proteins might have the ability to activate transcription from the preassembled nucleosome array template (Fig. 2B). Following the reconstitution of nucleosomes, basal transcription was suppressed to almost undetectable levels (lanes 1 and 2), in agreement with previous findings (63). In contrast to the observations with naked DNA, transcription from the nucleosome array template was not substantially enhanced by the inclusion of either GAL4-Gln or GAL4-Pro in the presence or the absence of acetyl-CoA (lanes 7 to 10). Inclusion of GAL4-VP16 resulted in weak transcriptional enhancement (approximately twofold; lane 5). However, this stimulation was not effected by acetyl-CoA (lane 6), indicating that it was not mediated by endogenous HAT activity (see below). The failure of these activation domains to significantly effect transcription on the preassembled nucleosome array template illustrates that the dramatic repression mediated by nucleosomes is not readily reversed by transcription activators alone.
NuA4 and SAGA HAT complexes differentially stimulate transcription by distinct activation domains.
The fact that the inclusion of acetyl-CoA in the transcription reactions shown in Fig. 2B did not result in transcription stimulation illustrates that the transcription system did not contain significant endogenous acetyltransferase activities that affected the assay. This allowed us to use this system to test the abilities of these different activation domains to exploit the HAT activities of the NuA4 and SAGA complexes. Figure 3A shows the experimental scheme we used. After a 10-min incubation to allow the binding of the GAL4 derivatives (factor binding), the nucleosome template was incubated with either NuA4 or SAGA in the presence or the absence of acetyl-CoA for 30 min (acetylation reaction). During the 30-min incubation, the NuA4 and SAGA complexes acetylated the 5S-G5E4 nucleosome array with the distinct histone preferences detected previously (21). While NuA4 primarily acetylated histone H4, the SAGA complex primarily acetylated H3 (Fig. 3B). Following the acetylation reaction, the abilities of the modified templates to function in transcription driven by the various GAL4 derivatives were assayed.
As shown in Fig. 3C, acetyl-CoA-dependent transcriptional stimulation by SAGA was specific for VP16-driven transcription. While SAGA enhanced transcription driven by GAL4-VP16 (lanes 5 and 6), it did not stimulate transcription in the presence of GAL4-Gln (lanes 7 and 8) or GAL4-Pro (lanes 9 and 10). Moreover, basal transcription (lanes 1 and 2) and transcription in the presence of GAL4(1-94) (lanes 3 and 4) was not affected by SAGA. Thus, transcriptional stimulation from the nucleosome array template by the SAGA complex was found to be specific for an acidic activation domain. By contrast, NuA4 (Fig. 3D) stimulated transcription driven by either GAL4-VP16 (lanes 5 and 6), GAL4-Gln (lanes 7 and 8), or GAL4-Pro (lanes 9 and 10). As with SAGA, we did not detect stimulation of basal transcription (lanes 1 and 2) or transcription in the presence of GAL4(1-94) (lanes 3 and 4) by NuA4. It is important to note that in all instances, the transcriptional stimulation by SAGA and NuA4 was acetyl-CoA dependent, indicating that the acetyltransferase activity of SAGA and NuA4 was required for the observed stimulation.
To test the possibility that the primary role in transcription activation by SAGA and NuA4 might result from acetylation of nonhistone protein (e.g., general transcription factors), we also tested transcription by using DNA templates in the presence of these HATs (Fig. 2A, lanes 6 to 13). SAGA and NuA4 had little effect on transcription from the DNA templates. While this observation does not exclude the possibility that nonhistone proteins are acetylated, it does illustrate that the stimulation observed in our assay is histone dependent.
NuA4 and SAGA preferentially interact with an acidic activation domain.
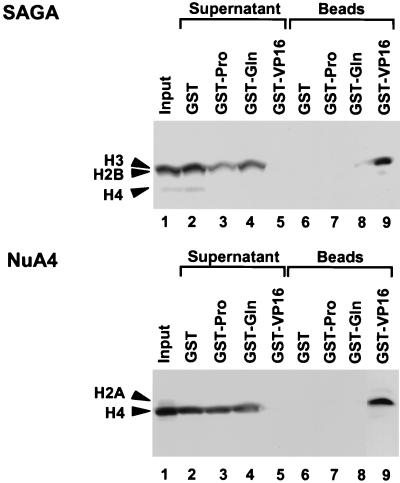
We have found that the SAGA and NuA4 HAT complexes interact directly with the VP16 activation domain (56). The different responses of SAGA and NuA4 in the transcription assays containing GAL4-Gln or GAL4-Pro may indicate that NuA4 also interacts with both of the Gln- and Pro-rich activation domains. To examine this possibility, we performed GST pull-down assays with GST fusion proteins bound to glutathione-Sepharose (Fig. 4). After incubation of GST-activation domain-beads with each HAT, the supernatant and beads were separated and analyzed for the presence of the HAT activity of NuA4 and SAGA. The HAT activity of SAGA was depleted from the supernatant (top panel, lane 5) and bound to the beads (lane 9) when the GST fusion protein contained the VP16 activation domain. By contrast, when beads were bound by GST, GST-Gln, or GST-Pro, the SAGA activity remained in the supernatant (lanes 2 to 4) and was not found bound to the beads (lanes 6 to 8). Thus, while the SAGA complex efficiently interacted with the acidic VP16 activation domain, it failed to interact with either a Gln-rich or a Pro-rich activation domain.
FIG. 4.
GST pull-down assays with various GST activation domains and HATs NuA4 and SAGA. These HAT activities were incubated with GST, GST-VP16, GST-Gln, or GST-Pro, which were bound with glutathione-Sepharose beads. The supernatants and the Sepharose beads were assayed for HAT activities on nucleosome substrates and then subjected to SDS–15% PAGE analysis and fluorography.
Although transcription driven by GAL4-Gln or GAL4-Pro was enhanced by the NuA4 complex following a preacetylation step, NuA4 did not interact with these activation domains in GST pull-down assays. GST-VP16 efficiently depleted the supernatant of NuA4 activity (Fig. 4, lower panel, lane 5), which was then recovered on the GST-VP16 glutathione-Sepharose beads (lane 9). By contrast, GST, GST-Gln, and GST-Pro failed to deplete the supernatant of NuA4 HAT activity (lanes 2 to 4).
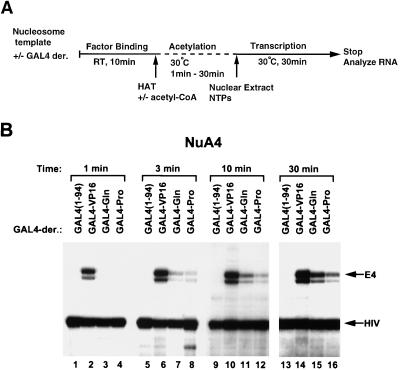
NuA4 stimulates transcription only by the VP16 activation domain when template acetylation is limiting.
The lack of ability of the Gln-rich or a Pro-rich activation domain to interact directly with NuA4 led us to suspect that stimulation of GAL4-Gln- and GAL4-Pro-driven transcription by NuA4 might be a consequence of overall acetylation of the nucleosome array template independent of interactions with the transcription activation domains. To address this possibility, we performed a time course analysis of histone acetylation by the NuA4 complex prior to the measurement of transcription activation (Fig. 5A). As shown in Fig. 5B, following a 30-min acetylation reaction, inclusion of NuA4 led to strong activation with each GAL4 derivative (Fig. 5B, lanes 13 to 16). However, when the acetylation step was shortened, transcription from the GAL4-VP16-bound nucleosome array was preferentially stimulated. Indeed, when the acetylation step was limited to only 1 min, transcription stimulation by NuA4 appeared to be as selective for VP16-driven transcription (lanes 1 to 4) as that observed for the SAGA complex (Fig. 3C). These data are consistent with the notion that NuA4 can be recruited by an acidic activation domain, which then enhances the acetylation of the factor-bound nucleosome array template. However, these data also suggest that extensive acetylation by NuA4 leads to permissive stimulation of transcription by activators that do not interact directly with NuA4.
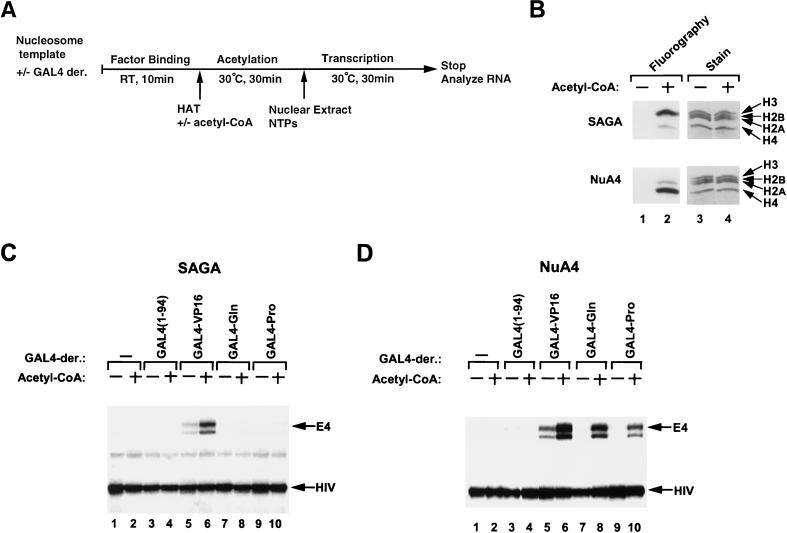
FIG. 5.
Effects of HATs on GAL4 derivative-mediated transcription of the nucleosome-reconstituted templates with limited incubation time. (A) Diagram of the transcription protocol. Subsequent to the factor-binding reaction, 1 U of NuA4, in the presence or the absence of acetyl-CoA, was included and incubated at 30°C. Incubation times were 1, 3, 10, and 30 min. The same amounts of various GAL4-derivative proteins were used as for Fig. 2A. RT, room temperature; NTPs, nucleoside triphosphates. (B) Transcripts from the G5E4 array template which were incubated with NuA4 in the presence of acetyl-CoA without (−) or with various GAL4-derivative proteins.
Extensive nontargeted acetylation of the nucleosomal template by NuA4 stimulates transcription by different activation domains.
To further confirm that extensive acetylation by NuA4 stimulates transcription mediated by activators not directly interacting with the HAT complex, we examined transcription from templates that were acetylated in the absence of GAL4-Gln and GAL4-Pro. Following acetylation of nucleosomal templates, acetyl-CoA was removed from the reactions by gel filtration. This was performed to prevent further acetylation by NuA4 upon the addition of activators to the transcription reaction mixtures (Fig. 6A). After removal of acetyl-CoA, factor binding and transcription were performed (Fig. 6B). As shown in Fig. 6C, 30 min of preacetylation with NuA4 (lanes 7 to 9), but not 1 min (lanes 10 to 12), enhanced transcription activation by GAL4-Gln and GAL4-Pro. In contrast, preacetylation with SAGA did not activate transcription mediated by GAL4-Gln or GAL4-Pro (lanes 5 and 6), consistent with the results of Fig. 3C, demonstrating that SAGA does not permit transcriptional stimulation by these activators. Thus, the increase in transcription by GAL4-Gln and GAL4-Pro corresponds to the overall acetylation of the template by NuA4 and does not require HAT activity subsequent to activator binding.
DISCUSSION
Several studies have shown that the assembly of template DNA into nucleosomes increases the degree of transcriptional regulation conferred by upstream activators in vitro (reviewed in references 39 and 40). While these studies demonstrate a role of acidic activation domains in overcoming nucleosome-mediated repression, they have neither revealed the important target of the activation domains (discussed in reference 64) nor distinguished this function from that of other activation domains. In this study, we have employed an in vitro transcription system using a well-defined nucleosome array template assembled with purified components. Thus, formation of the nucleosome template does not include the use of nucleosome assembly factors which may later contribute to nucleosome disruption (13, 61). Another important feature of this work relative to several previous studies (for example, see references 41 and 64) is that we used a preassembled nucleosome array template. Preassembly of the nucleosome array template allows separation of the effects of transcription factors during chromatin remodeling from effects occurring instead during nucleosome assembly, which can make a significant difference in the degree of transcriptional stimulation (e.g., see references 31 and 46). By using this approach, we have illustrated that preassembled nucleosomes can substantially repress the function of activation domains in vitro and that HAT complexes function to relieve this repression. However, the ability of a HAT complex to alleviate nucleosome repression is related to its ability to interact with activation domains of transcription activators. The data in this report illustrate that transcriptional stimulation by the SAGA complex is dependent on interaction with an acidic activation domain. Transcriptional stimulation by NuA4 is enhanced by interactions with an acidic activation domain, allowing for stimulation under conditions of limited histone acetylation. Thus, these data implicate HAT complexes as important targets for activation domain activity on chromatin templates.
Previous studies have implicated targets of acidic, glutamine-rich, or proline-rich activation domains. The acidic activation domain of VP16 is capable of binding to TBP and TFIIB (28, 35, 50). The glutamine-rich activation domain of Sp1 binds directly and specifically to the C-terminal domain of TBP (19) and Drosophila TAF110 (26). The combination of TAF250, TAF150, TAF110, and TBP can support robust Sp1 activation (14). Other cofactors and histone H1 are suggested to be involved in transcription mediated by the Pro-rich activation domain of CTF/NFI (18). The data presented in this report indicate a distinct function of the VP16 activation domain that is not shared with Gln-rich or Pro-rich activation domains. We found that this acidic activation domain has the ability to interact directly with the NuA4 and SAGA HAT complexes, a property not found in Gln-rich or Pro-rich activation domains. The specificity of this interaction can account for the preferential stimulation of VP16-driven transcription by the SAGA and NuA4 HAT complexes. Therefore, one of the crucial roles of acidic activation domains in yeast may be to recruit HAT complexes to promoters in chromatin. The acetylation of nucleosomes resulting from specific interaction between a regulatory factor and the HATs may facilitate the binding of additional upstream activators and general transcription factors to their corresponding recognition elements. Factors containing activation domains which cannot recruit HATs may benefit from HAT recruitment provided by other activators. On naturally occurring promoters involving multiple different regulatory factors, one or more factors may work as a key regulator at early steps important for alleviating the repressive effects of chromatin (9). Such factors may function largely by HAT recruitment.
It is interesting that the NuA4 and SAGA complexes display distinct functions in transcription activation. Preacetylation of the nucleosome templates by NuA4 permitted transcription activation by any of the activation domains tested here. In contrast, even after a preacetylation step, the SAGA complex maintained its specificity for VP16-mediated activation. These observations suggest that acetylation of histone H4 (i.e., by NuA4) is more permissive to transcription than acetylation of H3 (i.e., by SAGA). This is consistent with earlier studies illustrating that H4 acetylation appears to play a greater role in stimulating factor binding to nucleosomes (59). In addition, the selectivity of SAGA for VP16-driven transcription indicates that its function in transcription activation requires other features of the complex in addition to its HAT activity. This is consistent with the fact that the SAGA complex contains regulatory proteins from the ADA, SPT, and TAF groups of gene products (21, 22), many of which are phenotypically more important than GCN5 (44). Furthermore, the activation domain selectivity of SAGA in transcription stimulation seen here is consistent with previous studies done with yeast. These studies have shown that while GCN4, VP16 (5), ADR1 (15), TFE3 (6), NFκB p65 (6), human foamy virus protein, Bel-1 (6), the τ1 domain of the glucocorticoid receptor (25), and the activation subdomain of the p53 tumor suppressor protein (11) require the ADA2 gene product to function, the transactivation domains of GAL4 (43) and HAP4 (5, 43) do not. Thus, there appear to be SAGA-dependent and SAGA-independent classes of transcription activators in vitro and in vivo. In addition to histone acetylation, SAGA is likely to play a role in preinitiation complex formation and/or transcriptional elongation. These functions will become clearer as the roles of additional SAGA components are identified (22).
ACKNOWLEDGMENTS
We are grateful to Jacques Côté, Rhea T. Utley, Thomas Owen-Hughes, Ahmed Hanssan, and Michael Meisterernst for providing materials. We thank Sam John, Patrick A. Grant, Thomas Owen-Hughes, Rhea T. Utley, and Jacques Côté for many helpful discussions and critical reading of the manuscript. We also thank Kiyoshi Kawakami and Marissa Vignali for encouragement of this work.
This work was supported by a grant from National Institute of General Medical Sciences. D.J.S. was supported by a postdoctoral fellowship from the Cancer Research Institute. A.E. is a recipient of a postdoctoral fellowship from the Austrian Science Foundation. J.L.W. is an HHMI Associate Investigator.
REFERENCES
- 1.Adams C C, Workman J L. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol Cell Biol. 1995;15:1405–1421. doi: 10.1128/mcb.15.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlev N A, Candau R, Wang L, Darpino P, Silverman N, Berger S L. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. doi: 10.1074/jbc.270.33.19337. [DOI] [PubMed] [Google Scholar]
- 3.Becker P B. The establishment of active promoters in chromatin. BioEssays. 1994;16:541–547. doi: 10.1002/bies.950160807. [DOI] [PubMed] [Google Scholar]
- 4.Berger S L, Cress W D, Cress A, Triezenberg S J, Guarente L. Selective inhibition of activated but not basal transcription by the acidic activation domain of VP16: evidence for transcriptional adaptors. Cell. 1990;61:1199–1208. doi: 10.1016/0092-8674(90)90684-7. [DOI] [PubMed] [Google Scholar]
- 5.Berger S L, Piña B, Silverman N, Marcus G A, Agapite J, Regier J L, Triezenberg S J, Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 6.Blair W S, Bogerd H, Cullen B R. Genetic analysis indicates that the human foamy virus Bel-1 protein contains a transcription activation domain of the acidic class. J Virol. 1994;68:3803–3808. doi: 10.1128/jvi.68.6.3803-3808.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 8.Brownell J E, Allis C D. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 9.Bunker C A, Kingston R E. Activation domain-mediated enhancement of activator binding to chromatin in mammalian cells. Proc Natl Acad Sci USA. 1996;93:10820–10825. doi: 10.1073/pnas.93.20.10820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 11.Candau R, Scolnick D M, Darpino P, Ying C Y, Halazonetis T D, Berger S L. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene. 1997;15:807–816. doi: 10.1038/sj.onc.1201244. [DOI] [PubMed] [Google Scholar]
- 12.Chasman D I, Leatherwood J, Carey M, Ptashne M, Kornberg R D. Activation of yeast polymerase II transcription by herpesvirus VP16 and GAL4 derivatives in vitro. Mol Cell Biol. 1989;9:4746–4749. doi: 10.1128/mcb.9.11.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H, Li B, Workman J L. A histone-binding protein, nucleoplasmin, stimulates transcription factor binding to nucleosomes and factor-induced nucleosome disassembly. EMBO J. 1994;13:380–390. doi: 10.1002/j.1460-2075.1994.tb06272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J-L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 15.Chiang Y-C, Komarnitsky P, Chase D, Denis C L. ADR1 activation domains contact the histone acetyltransferase GCN5 and the core transcriptional factor TFIIB. J Biol Chem. 1996;271:32359–32365. doi: 10.1074/jbc.271.50.32359. [DOI] [PubMed] [Google Scholar]
- 16.Côté J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 17.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dusserre Y, Mermod N. Purified cofactors and histone H1 mediate transcriptional regulation by CTF/NF-I. Mol Cell Biol. 1992;12:5228–5237. doi: 10.1128/mcb.12.11.5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emili A, Greenblatt J, Ingles C J. Species-specific interaction of the glutamine-rich activation domains of Sp1 with TATA box-binding protein. Mol Cell Biol. 1994;14:1582–1593. doi: 10.1128/mcb.14.3.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992;355:219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- 21.Grant P A, Duggan L, Côté J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 22.Grant P A, Schieltz D, Pray-Grant M G, Steger D J, Reese J C, Yates III J R, Workman J L. A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 23.Hampsey M. A SAGA of histone acetylation and gene expression. Trends Genet. 1997;13:427–429. doi: 10.1016/s0168-9525(97)01292-4. [DOI] [PubMed] [Google Scholar]
- 24.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henriksson A, Almlöf T, Ford J, McEwan I J, Gustafsson J-Å, Wright A P H. Role of the Ada adaptor complex in gene activation by the glucocorticoid receptor. Mol Cell Biol. 1997;17:3065–3073. doi: 10.1128/mcb.17.6.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoey T, Weinzierl R O J, Gill G, Chen J-L, Dynlacht B D, Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 27.Hong L, Schroth G P, Matthews H R, Yau P, Bradbury E M. Studies of the DNA binding properties of histone H4 amino terminus. J Biol Chem. 1993;268:305–314. [PubMed] [Google Scholar]
- 28.Ingles C J, Shales M, Cress W D, Trienzenberg S J, Greenblatt J. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature. 1991;351:588–590. doi: 10.1038/351588a0. [DOI] [PubMed] [Google Scholar]
- 29.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 30.Jones K A, Yamamoto K R, Tjian R. Two distinct transcription factors bind to the HSV thymidine kinase promoter in vitro. Cell. 1985;42:559–572. doi: 10.1016/0092-8674(85)90113-8. [DOI] [PubMed] [Google Scholar]
- 31.Kamakaka R T, Bulger M, Kadonaga J T. Potentiation of RNA polymerase II transcription by GAL4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev. 1993;7:1779–1795. doi: 10.1101/gad.7.9.1779. [DOI] [PubMed] [Google Scholar]
- 32.Kornberg R D, Lorch Y. Chromatin structure and transcription. Annu Rev Cell Biol. 1992;8:563–587. doi: 10.1146/annurev.cb.08.110192.003023. [DOI] [PubMed] [Google Scholar]
- 33.Kuo M-H, Zhou J, Jambeck P, Churchill M E A, Allis C D. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Y-S, Carey M F, Ptashne M, Green M R. GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell. 1988;54:659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- 35.Lin Y-S, Ha I, Maldonado E, Reinberg D, Green M R. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991;353:569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- 36.Nightingale K P, Wellinger R E, Sogo J M, Becker P B. Histone acetylation facilitates RNA polymerase II transcription of the Drosophila hsp26 gene in chromatin. EMBO J. 1998;17:2865–2876. doi: 10.1093/emboj/17.10.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owen-Hughes T, Workman J L. Remodeling the chromatin structure of a nucleosome array by transcription factor-targeted trans-displacement of histones. EMBO J. 1996;15:4702–4712. [PMC free article] [PubMed] [Google Scholar]
- 38.Owen-Hughes, T., R. T. Utley, D. J. Steger, J. M. West, S. John, J. Côté, K. M. Havas, and J. L. Workman. Analysis of nucleosome disruption by ATP-driven chromatin remodeling complexes. In P. B. Becker (ed.), Chromatin protocols, in press. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 39.Owen-Hughes T A, Workman J L. Experimental analysis of chromatin function in transcription control. Crit Rev Eukaryot Gene Expr. 1994;4:403–441. [PubMed] [Google Scholar]
- 40.Paranjape S M, Kamakaka R T, Kadonaga J T. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- 41.Paranjape S M, Krumm A, Kadonaga J T. HMG17 is a chromatin-specific transcriptional coactivator that increases the efficiency of transcription initiation. Genes Dev. 1995;9:1978–1991. doi: 10.1101/gad.9.16.1978. [DOI] [PubMed] [Google Scholar]
- 42.Pennisi E. Opening the way to gene activity. Science. 1997;275:155–157. doi: 10.1126/science.275.5297.155. [DOI] [PubMed] [Google Scholar]
- 43.Piña B, Berger S, Marcus G A, Silverman N, Agapite J, Guarente L. ADA3: a gene, identified by resistance to GAL4-VP16, with properties similar to and different from those of ADA2. Mol Cell Biol. 1993;13:5981–5989. doi: 10.1128/mcb.13.10.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts S M, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/Mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sadowski I, Ma J, Triezenburg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 46.Sheridan P L, Mayall T P, Verdin E, Jones K A. Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes Dev. 1997;11:3327–3340. doi: 10.1101/gad.11.24.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simpson R T, Thoma F, Brubaker J M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- 48.Steger D J, Workman J L. Remodeling chromatin structures for transcription: what happens to the histones? BioEssays. 1996;18:875–884. doi: 10.1002/bies.950181106. [DOI] [PubMed] [Google Scholar]
- 49.Steger D J, Workman J L. Stable co-occupancy of transcription factors and histones at the HIV-1 enhancer. EMBO J. 1997;16:2463–2472. doi: 10.1093/emboj/16.9.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stringer K F, Ingles C J, Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 51.Tanese N, Pugh B F, Tjian R. Coactivators for a proline-rich activator purified from the multisubunit human TFIID complex. Genes Dev. 1991;5:2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- 52.Taylor I C A, Workman J L, Schuetz T J, Kingston R E. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 1991;5:1285–1298. doi: 10.1101/gad.5.7.1285. [DOI] [PubMed] [Google Scholar]
- 53.Triezenberg S J, LaMarco K L, McKnight S L. Evidence of DNA: protein interactions that mediate HSV-1 immediate early gene activation by VP16. Genes Dev. 1988;2:730–742. doi: 10.1101/gad.2.6.730. [DOI] [PubMed] [Google Scholar]
- 54.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 55.Ura K, Kurumizaka H, Dimitrov S, Almouzni G, Wolffe A P. Histone acetylation: influence on transcription, nucleosome mobility and positioning, and linker histone-dependent transcriptional repression. EMBO J. 1997;16:2096–2107. doi: 10.1093/emboj/16.8.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Utley R T, Ikeda K, Grant P A, Côté J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 57.Van Lint C, Emiliani S, Ott M, Verdin E. Transcriptional activation and chromatin remodeling of the HIV promoter in response to histone acetylation. EMBO J. 1996;15:1112–1120. [PMC free article] [PubMed] [Google Scholar]
- 58.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 59.Vettese-Dadey M, Grant P A, Hebbes T R, Crane-Robinson C, Allis C D, Workman J L. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 60.Wade P A, Wolffe A P. Chromatin: histone acetyltransferases in control. Curr Biol. 1997;7:R82–R84. doi: 10.1016/s0960-9822(06)00042-x. [DOI] [PubMed] [Google Scholar]
- 61.Walter P P, Owen-Hughes T A, Côté J, Workman J L. Stimulation of transcription factor binding and histone displacement by nucleosome assembly protein 1 and nucleoplasmin requires disruption of the histone octamer. Mol Cell Biol. 1995;15:6178–6187. doi: 10.1128/mcb.15.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 63.Workman J L, Roeder R G. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987;51:613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- 64.Workman J L, Taylor I C A, Kingston R E. Activation domains of stably bound GAL4 derivatives alleviate repression of promoters by nucleosomes. Cell. 1991;64:533–541. doi: 10.1016/0092-8674(91)90237-s. [DOI] [PubMed] [Google Scholar]
- 65.Workman J L, Kingston R E. Nucleosome core displacement in vitro via a metastable transcription factor-nucleosome complex. Science. 1992;258:1780–1784. doi: 10.1126/science.1465613. [DOI] [PubMed] [Google Scholar]