Abstract
Simple Summary
Previous studies reporting large gene panels in breast cancer have mainly reported on the contribution of additional genes compared to BRCA1/2. We have shown a very large differential additional effect from non-BRCA genes dependent on a priori likelihood of BRCA1 and BRCA2 combined. We have undertaken panel testing beyond BRCA1/2 in 1398 patients with breast cancer and identified 95 (6.3%) with actionable breast cancer genes. The highest rate was found for grade 3 ER+ Her2− breast cancers. Those with the lowest likelihood of BRCA1/2 by Manchester score had a 3-fold higher rate of non-BRCA genes whereas those with predicted rates of ~80% had 5-fold higher rate of BRCA1/2. Unless those referring patients with breast cancer for extended panel testing are certain there is no loss of sensitivity for BRCA1/2 they should opt of a bespoke BRCA1/2 test first in those with high prior likelihoods of BRCA1/2 pathogenic variants.
Abstract
Whilst panel testing of an extended group of genes including BRCA1/2 is commonplace, these studies have not been subdivided by histiotype or by a priori BRCA1/2 probability. Patients with a breast cancer diagnosis undergoing extended panel testing were assessed for frequency of actionable variants in breast cancer genes other than BRCA1/2 by histiotype and Manchester score (MS) to reflect a priori BRCA1/2 likelihood. Rates were adjusted by prior testing for BRCA1/2 in an extended series. 95/1398 (6.3%) who underwent panel testing were found to be positive for actionable non-BRCA1/2 breast/ovarian cancer genes (ATM, BARD1, CDH1, CHEK2, PALB2, PTEN, RAD51C, RAD51D, TP53). As expected, PALB2, CHEK2 and ATM were predominant with 80-(5.3%). The highest rate occurred in Grade-3 ER+/HER2− breast cancers-(9.6%). Rates of non-BRCA actionable genes was fairly constant over all likelihoods of BRCA1/2 but adjusted rates were three times higher with MS < 9 (BRCA1/2 = 1.5%, other = 4.7%), but was only 1.6% compared to 79.3% with MS ≥ 40. Although rates of detection of non-BRCA actionable genes are relatively constant across BRCA1/2 likelihoods this disguises an overall adjusted low frequency in high-likelihood families which have been heavily pre-tested for BRCA1/2. Any loss of detection sensitivity for BRCA1/2 actionable variants in breast cancer panels should lead to bespoke BRCA1/2 testing being conducted first.
Keywords: breast cancer, BRCA1, BRCA2, PALB2, ATM, CHEK2, panel test
1. Introduction
In recent years there has been a massive expansion in the use of multi-gene panels to test for breast cancer predisposition. These results have been used as a type of case control study to assess genes for breast cancer associations and the increase in actionable gene identification other than BRCA1 and BRCA2 [1,2,3,4,5,6,7,8,9,10]. These studies have generally shown an almost equal rate of pathogenic variants (PVs) identified in BRCA1/2 compared to all of the additional potentially actionable genes combined. In addition, many of the additional variants have been identified in the lower penetrance moderate risk genes [11], ATM and CHEK2, that occur with a higher population prevalence. The only other actionable breast cancer gene consistently identified at a substantial rate is PALB2, which is now considered to be a high-risk gene [12]. Many studies have also included a range of organ sites, as well as including unaffected individuals [2,7,8,9].
In general, these studies have concentrated on the headline rates of pathogenic variants (PVs) identified in the additional genes and not on the differential effect of the original likelihood of a BRCA1/2 pathogenic variant. Most studies that have used a ‘control’ frequency for PVs in the relevant genes have used population databases such as gnomAD [13], which may not be ideal controls for the population tested as they may not represent the rates in certain populations especially those with founder effects. We have assessed the frequency of potentially actionable breast cancer gene variants in a large series of women with breast cancer comparing rates of detection of non-BRCA PVs with BRCA1/2 at different BRCA1/2 likelihood scores. We have also assessed the variant frequencies in different breast pathology subtypes.
2. Materials and Methods
2.1. Study Population
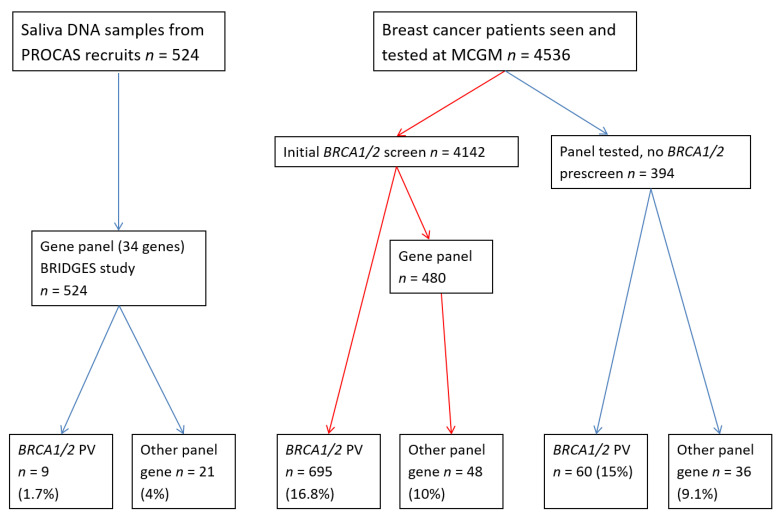
A total of 5060 women with breast cancer and 1443 control samples were included in the study, male cases were excluded. Of the patients with a breast cancer diagnosis, 4536 were referred to St Mary’s, Wythenshawe and the Christie hospitals in Manchester, consented and had blood samples taken for DNA extraction testing of breast cancer genes through the Manchester Centre for Genomic Medicine (MCGM) (Figure 1). MCGM participants agreed to return of results on genes predisposing to breast cancer. The remaining participants were recruited through the Predicting the Risk Of Cancer At Screening (PROCAS) study. This study recruited women aged 46–73 years of age from a population screening programme and included 187 breast cancers diagnosed before study entry and 337 after study entry (total n = 524). In addition, 1443 control samples without breast cancer from PROCAS [14] were tested as part of Breast Cancer Risk after Diagnostic Gene Sequencing (BRIDGES). PROCAS women only initially provided consent for return of a polygenic risk score, but also consented to further research genetic testing. We are applying for ethical approval to approach women with panel testing to assess whether they wish to receive results.
Figure 1.
Flow chart showing selection of samples for panel testing.
2.2. Genetic Testing
Clinical pre-screening for BRCA1/2 variants was carried out on 4142 MCGM patients. Samples from 874 women were selected for Next Generation Sequencing (NGS) panels, including 480 who tested negative on BRCA1/2 pre-screening and 394 who had not undergone pre-screening. The panel included as a minimum the following genes in addition to BRCA1 and BRCA2: PALB2, CHEK2, ATM, CDH1, STK11, PTEN, TP53, RAD51C, RAD51D, BRIP1, BARD1 and NBN. Saliva DNA samples from the PROCAS study [15] were tested as part of the BRIDGES study utilising a 34 gene panel (ABRAXAS1, AKT1, ATM, BABAM2, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, EPCAM, FANCC, FANCM, GEN1, MEN1, MLH1, MRE11, MSH2, MSH6, MUTYH, NBN, NF1, PALB2, PIK3CA, PMS2, PTEN, RAD50, RAD51C, RAD51D, RECQL, RINT1, STK11, TP53, XRCC2) including the four mismatch repair (MMR) genes (MLH1, MSH2, MSH6 and PMS2) [16].
PVs were annotated according to the ACMG guidelines [17] and verified using Sanger sequencing. Additionally, the PV rate of all 5060 women with breast cancer tested for BRCA1/2 at the MCGM (1997–2021) and in PROCAS was also assessed, regardless of whether they have had (additional) panel testing. The samples clinically tested at MCGM were annotated by a clinically approved national laboratory hub. Annotation of research results from BRIDGES were initially confirmed by the BRIDGES team and confirmed by our in-house research group (EvV).
2.3. Calculated Likelihood of a BRCA1/1 Pathogenic Variant
The pathology adjusted Manchester Scoring System (MS) was used to assess likelihoods of a pathogenic variant in BRCA1/2 [18]. Briefly, each breast cancer in a direct lineage to the index case is scored based on age at diagnosis for each gene (1–6 points) as well as each non mucinous epithelial ovarian cancer (5–8 points). All breast and ovarian cancers in the index individual are scored with an adjustment for pathology. Her2+, low grade and lobular cancers receive minus scores for BRCA1 whilst triple negative and high-grade cancers receive positive scores. Pathology adjusted scores of 15–19 are equivalent to a 10% probability of a BRCA1/2 pathogenic variant with a 20–24-point score equivalent to a 20% probability [18]. The proportion of positive tests from panel testing other than BRCA1/2 was adjusted to reflect previous testing of BRCA1/2 and the proportion of positive tests for BRCA1/2 at each MS score range. Thus a 10% detection rate in BRCA1/2 negative samples where the BRCA1/2 detection was 50% translates to a 5% overall panel detection rate beyond BRCA. An actionable breast cancer gene was defined as a gene confirmed in the BRIDGES study as having a 2-fold relative risk for breast cancer [16].
3. Results
3.1. Participants
A total of 1398 people with breast cancer were included in this study. There were 874 women tested through the MCGM as part of the clinical service or research projects. Of these, 740 women had a family history of only breast cancer, with 134 having an additional personal or family history of ovarian cancer. Additionally, 524 affected women and 1443 female controls that took part in the PROCAS study were included (age range 29–75; median 58.8 years) and were tested as part of the BRIDGES study (Figure 1).
Overall, 480 women with a family history (34.3%) and MS of −1 (some score below 0 with pathology adjustment) up to 56 (median 18) had been negative for BRCA1/2 on a pre-screen.
All panels included the following genes: PALB2, CHEK2, ATM, CDH1, STK11, PTEN, TP53, RAD51C, RAD51D, BRIP1, BARD1 and NBN. All 524 population-based samples and 347 familial samples (total n = 871) were also tested for variants in mismatch repair (MMR) genes. In addition, 1443 controls without breast cancer from PROCAS aged 46–73 years were tested for the full panel, including MMR genes.
Additionally, the PV status of all 5060 women with breast cancer tested for BRCA1/2 was included in order to determine the effect of extended panel testing in women stratified by MS.
3.2. Pathogenic Variant Rate
Overall, 174/1398 (12.4%) PVs were identified in 172 women (BRCA1 (26); BRCA2 (43); ATM (31); BARD1 (1); BRIP1 (7); CDH1 (1); CHEK2 (25); NBN (3); PALB2 (24); PTEN (1); RAD51C (1); RAD51D (1); TP53 (10)) (Table 1). One woman harboured both a BRCA1 and a BRCA2 PV and one woman carried a BRCA2 PV as well as a BRIP1 PV.
Table 1.
Rates of detection of BRCA1/2 and non BRCA breast/ovarian pathogenic variants by Manchester score.
| Manchester Score | ≤8 | 9–10 | 11–12 | 13–14 | 15–19 | 20–24 | 25–29 | 30–39 | ≥40 | Total PVs Cases | PROCAS Controls PVs | OR | 95% CI | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Full extended panel cohort | ||||||||||||||
| BRCA1 | 1 | 0 | 0 | 0 | 2 | 3 | 3 | 9 | 8 | 26 | 2 | NA | NA | NA |
| BRCA2 | 5 | 1 | 1 | 4 | 11 | 4 | 9 | 7 | 1 | 43 | 6 | NA | NA | NA |
| ATM | 7 | 2 | 1 | 3 | 7 | 5 | 3 | 3 | 0 | 31 | 5 | 6.52 | 2.56–15.55 | <0.0001 |
| CHEK2 | 12 | 0 | 1 | 2 | 4 | 2 | 0 | 3 | 1 | 25 | 3 | 8.74 | 2.88–27.60 | <0.0001 |
| TP53 * | 3 | 0 | 0 | 0 | 4 | 1 | 1 | 0 | 1 | 10 | 0 | 36.02 | 6.33–392.1 | <0.0001 |
| PALB2 | 2 | 3 | 3 | 1 | 6 | 6 | 3 | 0 | 0 | 24 | 3 | 8.38 | 2.74–26.53 | <0.0001 |
| NBN | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 2 | 1.55 | 0.32–8.75 | 0.6825 |
| CDH1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | n/a | ||
| RAD51C | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | n/a | ||
| RAD51D | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | n/a | ||
| BRIP1 | 1 | 0 | 0 | 2 | 1 | 3 | 0 | 0 | 0 | 7 | 2 | 3.63 | 0.82–17.31 | 0.1037 |
| BARD1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1.03 | 0.05–19.62 | >0.9999 |
| PTEN | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | n/a | ||
| Total PVs | 34 | 7 | 6 | 13 | 36 | 24 | 21 | 22 | 11 | 174 | 24 | |||
| Total tested | 525 | 78 | 91 | 106 | 255 | 149 | 95 | 72 | 27 | 1398 | 1443 | |||
| % PVs | 6.5% | 9.0% | 6.6% | 12.3% | 14.1% | 16.1% | 22.1% | 30.6% | 40.7% | 12.4% | 1.7% | |||
| Total non-BRCA PVs | 28 | 6 | 5 | 9 | 23 | 17 | 9 | 6 | 2 | 105 | ||||
| % non-BRCA PVs | 5.3% | 7.7% | 5.5% | 8.5% | 9.0% | 11.4% | 9.5% | 8.3% | 7.4% | 7.5% | ||||
| % adjusted for all BRCA1/2 tested | 5.3% | 7.4% | 5.3% | 8.0% | 8.1% | 8.8% | 6.4% | 3.6% | 1.5% | 6.4% | ||||
| No pre-screen | ||||||||||||||
| BRCA1 | 1 | 0 | 0 | 0 | 2 | 3 | 3 | 9 | 8 | 26 | 2 | 21.00 | 5.41–90.02 | <0.0001 |
| BRCA2 | 5 | 1 | 1 | 4 | 11 | 4 | 9 | 7 | 1 | 43 | 6 | 11.77 | 5.05–25.65 | <0.0001 |
| ATM | 6 | 2 | 0 | 3 | 3 | 3 | 1 | 1 | 0 | 19 | 5 | 6.08 | 2.43–14.96 | <0.0001 |
| CHEK2 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 13 | 3 | 6.89 | 2.04–22.80 | 0.001 |
| TP53 * | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 10.91 | 1.27–158.2 | 0.0647 |
| PALB2 | 2 | 3 | 0 | 0 | 3 | 3 | 2 | 0 | 0 | 13 | 3 | 6.89 | 2.04–22.80 | 0.001 |
| NBN | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 1.57 | 0.25–10.06 | 0.6448 |
| CDH1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | n/a | ||
| RAD51C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | n/a | ||
| RAD51D | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | n/a | ||
| BRIP1 | 1 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 5 | 2 | 3.95 | 0.85–19.9 | 0.1172 |
| BARD1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1.57 | 0.08–29.9 | >0.9999 |
| PTEN | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | n/a | ||
| Total PVs | 32 | 7 | 1 | 10 | 19 | 15 | 15 | 18 | 9 | 126 | 24 | |||
| Total tested | 487 | 66 | 63 | 53 | 108 | 59 | 37 | 33 | 12 | 918 | 1443 | |||
| % PVs | 6.6% | 10.6% | 1.6% | 18.9% | 17.6% | 25.4% | 40.5% | 54.5% | 75.0% | 13.7% | 1.7% | |||
| Total non-BRCA PVs | 26 | 6 | 0 | 6 | 6 | 8 | 3 | 2 | 0 | 57 | ||||
| % non-BRCA PVs | 5.3% | 9.1% | 0.0% | 11.3% | 5.6% | 13.6% | 8.1% | 6.1% | 0.0% | 6.2% | ||||
| % BRCA1/2 PVs | 1.2% | 1.5% | 1.6% | 7.5% | 12.0% | 11.9% | 32.4% | 48.5% | 75.0% | 7.5% | ||||
| % adjusted for all BRCA1/2 tested | 5.3% | 8.8% | 0.0% | 10.6% | 5.0% | 10.4% | 5.5% | 2.6% | 0.0% | 5.3% | ||||
| Negative pre-screen | ||||||||||||||
| ATM | 1 | 0 | 1 | 0 | 4 | 2 | 2 | 2 | 0 | 12 | 5 | 7.4 | 2.7–19.0 | <0.0001 |
| CHEK2 | 0 | 0 | 1 | 2 | 4 | 2 | 0 | 2 | 1 | 12 | 3 | 12.3 | 3.5–41.0 | <0.0001 |
| TP53 * | 1 | 0 | 0 | 0 | 4 | 1 | 1 | 0 | 1 | 8 | 0 | 84.7 | 13.2–941.5 | <0.0001 |
| PALB2 | 0 | 0 | 3 | 1 | 3 | 3 | 1 | 0 | 0 | 11 | 3 | 11.3 | 3.5–37.9 | <0.0001 |
| NBN | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 2 | 1.50 | 0.1–12.9 | 0.5777 |
| CDH1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | n/a | ||
| RAD51C | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | n/a | ||
| RAD51D | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | n/a | ||
| BRIP1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 2 | 3.02 | 0.5–19.3 | 0.261 |
| BARD1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | n/a | ||
| PTEN | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | n/a | ||
| Total PVs | 2 | 0 | 5 | 3 | 17 | 9 | 6 | 4 | 2 | 48 | 16 | |||
| Total tested | 38 | 12 | 28 | 53 | 147 | 90 | 58 | 39 | 15 | 480 | 1443 | |||
| % PVs | 5.3% | 0.0% | 17.9% | 5.7% | 11.6% | 10.0% | 10.3% | 10.3% | 13.3% | 10.0% | 1.1% | |||
| % adjusted for all BRCA1/2 tested | 5.2% | 0.0% | 17.1% | 5.3% | 10.3% | 7.7% | 7.0% | 4.5% | 2.8% | 8.5% | ||||
| All samples tested for BRCA1/2 | ||||||||||||||
| BRCA1 | 4 | 5 | 5 | 16 | 49 | 87 | 60 | 94 | 67 | 387 | ||||
| BRCA2 | 9 | 7 | 15 | 22 | 82 | 86 | 66 | 65 | 25 | 377 | ||||
| Total tested | 878 | 326 | 468 | 619 | 1240 | 745 | 387 | 281 | 116 | 5060 | ||||
| % BRCA1/2 PVs | 1.5% | 3.7% | 4.3% | 6.1% | 10.6% | 23.2% | 32.6% | 56.6% | 79.3% | 15.1% | ||||
* Population frequency estimate for local population taken as 1 in 5000; NA-Not assessable as 480 pre-screened for BRCA1/2.
Of the 874 women tested via MCGM, 60 BRCA1/2 PVs were identified in 59 women (BRCA1 (25); BRCA2 (35) (one harboured both a BRCA1 and a BRCA2 PV), however 480 samples had already screened negative for BRCA1/2. This thus represents a detection rate of 60/394 (15%). In the remaining 815 women, 80 PVs were identified in non-BRCA1/2 genes (ATM (25); BARD1 (1); BRIP1 (5); CDH1 (1); CHEK2 (15); NBN (2); PALB2 (20); RAD51C (1); RAD51D (1); TP53 (9)).
Of the 524 women recruited through PROCAS, 34 PVs in 33 women were identified (BRCA2 (8); BRCA1 (1); ATM (6); BRIP1 (2); CHEK2 (10); NBN (1); PALB2 (4); PTEN (1); TP53 (1); one woman harboured both a BRCA2 and a BRIP1 PV).
A total of 95 of 1398 (6.3%) women who underwent panel testing were found to be positive for actionable non-BRCA1/2 breast/ovarian cancer genes (ATM, BARD1, CDH1, CHEK2, PALB2, PTEN, RAD51C, RAD51D, TP53). As expected, PALB2, CHEK2 and ATM were the most frequent with 80 (5.3%) PVs accounting for 84.2% non BRCA1/2 actionable results. Of the other high-risk genes there were 10 TP53, one CDH1 and one PTEN PVs, but none in STK11. All the expected breast cancer associated genes (ATM, CHEK2, PALB2, TP53) had significantly increased odds ratios of above 2-fold compared to the 1443 PROCAS control samples (Table 1). Other PVs were identified in genes that have a less clear association with breast cancer such as: BRIP1, NBN, RAD50 and RECQL, but none of these showed significant associations.
3.3. Detection Rate by Manchester Score
The detection rates of PVs in non-BRCA1/2 genes in the full extended panel cohort did not vary substantially by MS (ranging between 5.3–11.4%). However, when controlling for those with prior BRCA1/2 testing, the added value of the extended panel was actually lower in those with a higher MS (i.e., those individuals with a higher probability of a BRCA1/2 pathogenic variant). Only 2.97% of those with MS ≥ 30 tested positive for a non-BRCA1/2 PV when allowing for the higher likelihood of a BRCA1/2 positive result of 63.2% (251/397) in the group of all individuals tested for BRCA PVs.
There was no significant difference between rates of the most commonly detected non-BRCA PVs in those with MS ≥ 15 and <15 (ATM PVs (3.0% and 1.6% p = 0.098), CHEK2 PVs (1.7% and 1.9% p = 0.84) and PALB2 PVs (2.5% and 1.1% p = 0.0605)). Although the combined total of all PVs in all three genes with MS ≥ 15 was statistically significantly higher at 7.2% (43/598) vs. 4.6% (37/800)-p = 0.05 this did not take into account pre-screening for BRCA1/2.
Of all women tested for BRCA1/2 PVs (n = 5060), 764 (15.10%) harboured a PV in BRCA1/BRCA2 (Table 1). In this group, 92/116 (79.3%) women with breast cancer and a MS of ≥40 tested positive for a BRCA1/2 PV (BRCA1 (n = 67) or BRCA2 (n = 25)). In the study cohort, only an additional two non-BRCA PVs were identified in the group of women with a MS ≥ 40 (2/15 (13.3%) and a negative pre-screen). The adjusted proportion of PVs in non-BRCA genes of 75/1204 (6.2%) for MS ≤ 10 was significantly higher than for those with scores ≥30 (12/405-3.0%-p = 0.01).
Of the 918 women in the study cohort without pre-screening, 68 (7.4%) tested positive for PVs in BRCA1 (n = 25), BRCA2 (n = 42) or both (n = 1). There was an increase in BRCA1/2 PV detection rate with increasing MS, similar to the 5060 women tested only for BRCA1/2 PVs.
Of the 871 women tested for variants in the MMR genes, very few PVs were identified. There were only two cases with an MSH6 PV (Supplementary Table S1). No PVs in MLH1 or MSH2 were identified. Similarly, in the control group there was only one MSH6 and one MSH2 PV identified.
Only 9/524 (1.7%) population-based PROCAS samples tested positive for a BRCA1/2 PV (MS: 0–33 median = 3, BRCA1 (n = 1), BRCA2 (n = 8)). Only 28/524 samples met the MS ≥ 15 score (indicating a 10% threshold was met) and only 2/28 (7.1%) had a BRCA1/2 PV (BRCA1 = 1, BRCA2 = 1). However, two women in this group had a PALB2 PV. In those who did not meet the 10% threshold (MS < 15), there were 7/496 (1.4%) BRCA1/2 PVs detected. In familial samples, 37/124 (29.8%) with MS ≥ 20, 11/91 (12.1%) with MS 15–19 and 5/173 (2.9%) with MS < 15 testing positive for BRCA1/2.
A total of 134 women with breast cancer had either a personal (n = 25) or family history (n = 109) of ovarian cancer. Detection rates were similar to those with breast cancer only with 8/134 (6%) having a PV beyond BRCA1/2 (PALB2 = 2) and 2/25 (8%) with breast and ovarian double primaries (Supplementary Tables S2 and S3).
3.4. Detection Rate by Tumour Pathology
Breast cancer cases with tumour pathology reports, including human epidermal growth factor receptor 2 (HER2) status are presented in Table 2. The lowest overall detection rate (6.1%). for any PV, including BRCA1/2, was in grade 1 estrogen receptor (ER)+/HER2− breast cancers. The highest detection rates were in grade 3 ER+/HER2− (18.7%) and triple negative cases (17.5%).
Table 2.
Rate of PVs by pathology and Manchester score.
| All Patients Tested for BRCA1/2 PVs (n = 5060) | Patients Included for Panel Testing (n = 1398) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| BRCA1 | % | BRCA2 | % | Total | Total PVs BRCA1/2 | Panel Positive | % | Total Panel Tested | Non BRCA | Genes | |
| Lobular total | 4 | 1.2% | 19 | 5.6% | 341 | 6.74% | 11 | 8.6% | 128 | 7.8% | ATM(4), BRCA2(1), CDH1(1), CHEK2 (1), TP53(1), PALB2(3) |
| Lobular MSS ≥ 20 | 2 | 2.7% | 8 | 10.7% | 75 | 13.3% | 5 | 11.9% | 42 | 11.9% | ATM(2), TP53 (1), PALB2(2) |
| Lobular MSS < 20 | 2 | 0.75% | 11 | 4.1% | 266 | 4.9% | 6 | 6.98% | 86 | 5.8% | ATM(2), BRCA2(1), CDH1(1), CHEK2(1), PALB2(1) |
| Grade 1 total | 3 | 0.8% | 12 | 3.3% | 363 | 4.1% | 8 | 6.1% | 132 | 4.55% | ATM(2), BRCA2(2), BRIP1(2), CHEK2(1), TP53(1) |
| Grade 1 MSS ≥ 20 | 3 | 4.8% | 3 | 4.7% | 63 | 9.5% | 1 | 6.7% | 15 | 6.7% | BRIP1(1) |
| Grade 1 MSS < 20 | 0 | 0.0% | 9 | 3.0% | 300 | 3.0% | 7 | 5.98% | 117 | 4.3% | ATM(2), BRCA2(2), BRIP1(1), CHEK2(1), TP53(1) |
| IDC Grade 2 ER+ total | 10 | 1.3% | 70 | 9.2% | 763 | 10.5% | 23 | 8.98% | 256 | 4.3% | ATM(6), BRCA2(13), BRIP1(1), CHEK2(2), PALB2(1), RAD51C(1) |
| IDC Grade 2 ER+ MSS ≥ 20 | 8 | 4.3% | 38 | 20.5% | 185 | 24.8% | 13 | 25.5% | 51 | 7.8% | ATM(2), BRCA2(9), PALB2(1), RAD51C(1) |
| IDC Grade 2 ER+ MSS < 20 | 2 | 0.35% | 32 | 5.5% | 578 | 5.9% | 10 | 4.9% | 205 | 3.4% | ATM(4), BRCA2(3), BRIP1(1), CHEK2(2) |
| IDC Grade 3 ER+ total | 43 | 8.4% | 74 | 14.4% | 513 | 22.8% | 28 | 18.7% | 150 | 10.0% | ATM(4), BRCA1(6), BRCA1&BRCA2(1), BRCA2(6), CHEK2(3), PALB2(5), RAD51D(1), TP53(2) |
| IDC Grade 3 ER+ MSS ≥ 20 | 34 | 17.3% | 54 | 27.4% | 197 | 44.7% | 17 | 38.6% | 44 | 15.9% | ATM(2), BRCA1(6), BRCA1&BRCA2(1), BRCA2(3),CHEK2(1), PALB2(2), RAD51D(1), TP53(1) |
| IDC Grade 3 ER+ MSS < 20 | 9 | 2.85% | 20 | 6.33% | 316 | 9.2% | 11 | 10.4% | 106 | 7.55% | ATM(2), BRCA2(3), CHEK2(2), PALB2(3), TP53(1) |
| TNT total | 199 | 20.35% | 61 | 6.24% | 978 | 26.6% | 31 | 17.5% | 177 | 7.3% | ATM(1), BRCA1(14), BRCA2(4), BARD1(1), BRIP1(1), CHEK2(2), PALB2(7), RECQL(1) |
| TNT MSS ≥ 20 | 158 | 33.40% | 46 | 9.73% | 473 | 43.1% | 19 | 24.4% | 78 | 6.4% | BRCA1(12), BRCA2(2), BRIP1(1), CHEK2(1), PALB2(3) |
| TNT MSS < 20 | 41 | 8.12% | 15 | 2.97% | 505 | 11.1% | 12 | 12.1% | 99 | 8.1% | ATM(1), BRCA1(2), BRCA2(2), BARD1(1), CHEK2(1), PALB2(4), RECQL(1) |
| HER2+ total | 5 | 1.72% | 13 | 4.5% | 290 | 6.2% | 10 | 9.8% | 102 | 7.8% | ATM(3), BRCA2(2), CHEK2(1), PALB2(1), TP53(3) |
| HER2+ MSS ≥ 20 | 2 | 4.88% | 3 | 7.3% | 41 | 12.2% | 1 | 8.3% | 12 | 8.3% | PALB2(1) |
| HER2+ MSS < 20 | 3 | 1.20% | 10 | 4.2% | 249 | 5.2% | 9 | 10.0% | 90 | 7.8% | ATM(3), BRCA2(2), CHEK2(1), TP53(3) |
| DCIS total | 8 | 2.80% | 22 | 7.7% | 286 | 10.5% | 18 | 15.25% | 118 | 9.3% | ATM(4), BRCA2(7), BRIP1(1), CHEK2(3), PALB2(2), TP53(1) |
| DCIS MSS ≥ 20 | 4 | 7.55% | 11 | 20.75% | 53 | 28.3% | 5 | 33.3% | 15 | 13.3% | ATM(1), BRCA2(3), TP53(1) |
| DCIS MSS < 20 | 4 | 1.72% | 11 | 4.7% | 233 | 6.4% | 13 | 12.6% | 103 | 8.7% | ATM(3), BRCA2(4), BRIP1(1), CHEK2(3), PALB2(2) |
| NOS total | 115 | 7.54% | 106 | 6.95% | 1526 | 14.5% | 41 | 12.2% | 335 | 8.4% | ATM(7), BRCA1(5), BRCA2(8), BRIP1(1), CHEK2(12), PALB2(5), PTEN(1), TP53(2) |
| NOS MSS ≥ 20 | 97 | 21.95% | 79 | 17.9% | 442 | 39.8% | 16 | 18.6% | 86 | 10.5% | ATM(4), BRCA1(4), BRCA2(3), BRIP1(1), CHEK2(4) |
| NOS MSS < 20 | 18 | 1.66% | 27 | 2.5% | 1084 | 4.15% | 25 | 10.0% | 249 | 7.6% | ATM(3), BRCA1(1), BRCA2(5), CHEK2(8), PALB2(5), PTEN(1), TP53(2) |
| Total overall | 387 | 7.65% | 377 | 7.45% | 5060 | 15.1% | 170 | 12.2% | 1398 | 7.3% | ATM(31), BRCA1(25), BRCA2(42),BRCA1&BRCA2(1), BARD1(1), BRIP1(6), CDH1(1), CHEK2(25), PALB2(24), PTEN(1), TP53(10), RAD51C (1), RAD51D(1), RECQL(1) |
| MSS ≥ 20 | 308 | 20.14% | 242 | 15.8% | 1529 | 35.9% | 77 | 22.45% | 343 | 9.9% | ATM (11), BRCA1(22), BRCA2(20), BRCA1&BRCA2(1), BRIP1(3), CHEK2(6), PALB2(9), TP53(3), RAD51C (1), RAD51D(1) |
| MSS < 20 | 79 | 2.24% | 135 | 3.8% | 3531 | 6.1% | 93 | 8.8% | 1055 | 6.4% | ATM (20), BRCA1(3), BRCA2(22), BARD1(1), BRIP1(3), CDH1(1), CHEK2(19), PALB2(15), PTEN(1), TP53(7), RECQL(1) |
4. Discussion
The present study has confirmed some utility for breast cancer gene panels extended beyond BRCA1/2 in women with breast cancer. Nonetheless, the increased diagnostic yield is dominated by just three additional genes (PALB2, ATM and CHEK2) accounting for over 80% of non-BRCA1/2 variants found. Additionally, although BRCA1/2 pre-testing increases the rate of pathogenic variant detection in these genes, with higher familial scores represented by the MS, the added benefit from a panel is lower when adjusting for a higher likelihood of BRCA1 and BRCA2 PVs. For instance, an index case with a MS ≥ 40 would have an ~80% chance of a BRCA1/2 PV, but only a 1–2% chance of a PV in another actionable gene. In contrast, with a MS ≤ 8 there is only around a 1% chance of a BRCA1/2 PV, but a 5% chance of finding another PV (mainly in ATM or CHEK2). Previous studies have not addressed this large differential effect. For those with a very high likelihood of a BRCA1/2 PV by an accepted method (MS, BOADICEA/CANRISK, BRCAPRO), any loss in sensitivity of identification of a BRCA1/2 PV would mitigate any additional benefit from an extended panel vs. a targeted BRCA1/2 approach. Some loss of sensitivity is accepted in most panel tests but even a drop by 2% for a family with a MS ≥ 40 would be sufficient to warrant bespoke BRCA1/2 testing as the first test. Although most NGS panels claim to identify large single or multiple exon copy number variations (CNVs), the proven sensitivity of these tests is less clear. In Manchester, 20% of non-Jewish BRCA1 PVs are CNVs that in the past have been detected by multiple ligation-dependent probe amplification (MLPA). Given that any additional gene identified is likely to have lower penetrance than BRCA1 or BRCA2 e.g., in a large family with multiple breast/ovarian cancer cases (MS ≥ 40), it is unlikely that this will be the entire explanation for the pattern observed. For instance we observed a RAD51C PV in a large breast/ovary family which probably only fully explains the ovarian cancer predisposition [19], and for the ATM and CHEK2 variants in six families with MS of 30–39 and one CHEK2 in a MS of >40, these are unlikely to provide a full explanation for the pattern of disease in these families (Table 1) [11]. We have previously shown that a pathogenic RAD51D variant in a family with three women with ovarian cancer and four with breast cancer(MS = 55) fully segregated with disease and that BRCA1/2 testing, including RNA analysis was negative [19]. Given that only a two-fold increased likelihood of breast cancer is associated with RAD51D PVs, it is still unlikely that RAD51D fully explains the breast cancer risk in this family, although polygenic risk could add somewhat to the effects of RAD51D. Although results from BRIDGES [16] and other studies clearly differentiate PALB2 as a high-risk gene and CHEK2 as moderate-risk, this is not reflected in the similar ORs for CHEK2 and PALB2 of around eight-fold, but these risk estimates have large confidence intervals due to the small number of controls with identified variants and are based on testing many familial families with pre-screens of BRCA1/2.
Case control analysis is arguably the most informative method to identify breast cancer gene associations as it also provides confirmation of the level of any increased risk. Many recent large-scale studies have published regarding this, including recent evidence that variants in the MMR genes MSH6 and PMS2 may be associated with an increased risk of breast cancer [2,20]. Although the first of these studies was a true case control study, there has been concern that one of the variant calls in MSH6 (c.2945delC; p.(Pro982Leufs)) that was very frequent in cases, may be spurious due to a sequencing artifact [21]. This study also used gnomAD controls [13] rather than a matched, local population control series. The second study was not a true case control study and assessed risk in carriers identified on panel testing without adjusting for the proportion of referred cases with breast cancer [20]. The very high population frequencies of variants in MSH6 and PMS2 without typical Lynch syndrome associated cancers may ironically have falsely elevated the odds ratio [22]. Furthermore, no evidence for any association between these variants in MMR genes and breast cancer has been seen in larger case control studies [1], segregation analysis [23], nor in prospective analysis of PV carriers [24], with the possible exception of MSH6 in BRIDGES, which was borderline significant at two-fold risk [16]. As such, patients with MMR PVs, with the possible exception of MSH6, should not be advised that they are at increased risk of breast cancer.
The lowest detection rate overall both for BRCA1/2 and additional panel genes was for grade 1 cancers with 4.1% and 4.5% respectively. The highest detection rate was for grade 3 ER+ HER2− breast cancers with a non BRCA1/2 panel detection rate of 10%. There has been very little in the literature on the effect of pathology on panel detection rates in non-BRCA1/2 genes although we have recently reported an association with PALB2 and grade 3 [25]. A Brazilian study recently also confirmed a higher detection rate in high grade tumours [26]. Although this was a small study (n = 224; 61 PVs-19 BRCA1/2) and the association was likely driven primarily by BRCA1/2. The BRIDGES study did not assess grade, but there was a clear association between RAD51C, RAD51D, PALB2 and BARD1, with ER− breast cancer and ATM and CHEK2 with ER+ breast cancer with no association of ATM with ER− [16].
The present study does have some limitations. The size of the control population was probably not large enough to generate very reliable odds ratio estimates. We had to use our previous population estimate for TP53 [27], but had no such estimate for other rare genes such as CDH1. The study was not large enough to statistically assert that BRIP1 is not an actionable breast cancer gene [28]. Nor was it large enough to address the variant detection rates for the ovarian susceptibility genes RAD51C and RAD51D that have previously been excluded as breast cancer susceptibility genes [29,30], but which were more recently confirmed as breast cancer genes just reaching two-fold relative risk [16]. There is also some previous evidence that variants in RAD51D may predispose specifically to triple negative breast cancer [16]. The BRIDGES study also only called definitive PVs using NGS and did not call CNVs. As already stated, this could have reduced PV detection sensitivity in the UK population to below 80% for BRCA1. The odds ratios for BRCA1 could therefore be exaggerated by the fact that in-house testing of over 4536 cases in Table 1 included MLPA, since CNVs were not reported in the control samples tested in the BRIDGES study. Despite this, our study also has a number of strengths. This is a large study with well documented family history, including first and second-degree relatives. A high proportion of the study patients had full pathology available, including HER2 status, which was not generally available before 2000. We have also used matching local controls known not to have breast cancer.
5. Conclusions
Although the trend towards panel testing beyond BRCA1/2 is likely to be irreversible, there are some lessons to be learnt from the current study. The majority of the increased diagnostic yield results from just three genes PALB2, CHEK2 and ATM. Only PALB2 can be considered a strong enough risk factor on its own and the actionability of ATM and CHEK2 pathogenic variants needs to be taken in the context of other risk factors, including a polygenic risk score, ideally through a validated model such as BOADICEA/CanRisk [31]. Women with certain tumours, and in particular grade 1 invasive breast cancers, should be informed that there is a very low likelihood of any meaningful result from panel testing. Finally, the low adjusted diagnostic uplift in those with very high a priori likelihood of BRCA1/2 means that clinicians should ensure that panels used have high sensitivity for BRCA1/2 PVs present in their population or opt for bespoke BRCA1/2 testing in the first instance.
Acknowledgments
D.G.E., E.M.v.V., E.F.H., E.R.W., M.J.S., W.G.N., and S.J.H. are supported by the all Manchester NIHR Biomedical Research Centre (IS-BRC-1215-20007) and some aspects of the panel testing were also supported by this grant. The BRIDGES study was funded through a number of sources. The sequencing and analysis for this project was funded by the European Union’s Horizon 2020 Research and Innovation Programme (BRIDGES: grant number 634935) and the Wellcome Trust [grant No.: v203477/Z/16/Z]. BCAC co-ordination was additionally funded by the European Union’s Horizon 2020 Research and Innovation Programme (BRIDGES: grant number 634935, BCAST: grant number 633784) and by Cancer Research UK (C1287/A16563). Prevent Breast Cancer (GA19-002) funded panel testing of lobular breast cancers.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13164154/s1, Table S1: Results from MSH6 testing of 871 women with breast cancer (524 from PROCAS) and 1443 controls; Table S2: Panel testing results for women with a personal (n = 25) or family (n = 109) history of ovarian cancer; Table S3: Panel testing results for women with a personal (n = 25) history of breast and ovarian cancer.
Author Contributions
Conceptualization, D.G.E.; methodology, D.G.E., E.M.v.V.; validation, D.G.E., E.M.v.V.; formal analysis, D.G.E., E.M.v.V.; investigation, D.G.E.; resources, D.G.E.; data curation, D.G.E.; writing, all original draft preparation, D.G.E., E.M.v.V.; writing—review and editing, all authors; funding acquisition, D.G.E. All authors have read and agreed to the published version of the manuscript.
Funding
D.G.E., E.M.v.V., E.F.H., E.R.W., M.J.S., W.G.N., and S.J.H. are supported by the all Manchester NIHR Biomedical Research Centre (IS-BRC-1215-20007) and some aspects of the panel testing were also supported by this grant. The BRIDGES study was funded through a number of sources. The sequencing and analysis for this project was funded by the European Union’s Horizon 2020 Research and Innovation Programme (BRIDGES: grant number 634935) and the Wellcome Trust (grant No.: v203477/Z/16/Z). BCAC co-ordination was additionally funded by the European Union’s Horizon 2020 Research and Innovation Programme (BRIDGES: grant number 634935, BCAST: grant number 633784) and by Cancer Research UK (C1287/A16563). Prevent Breast Cancer (GA19-002) funded panel testing of lobular breast cancers.
Institutional Review Board Statement
Clinical or research consent was given for extended testing of breast cancer associated genes (approval from the North Manchester Research Ethics Committee, reference 09/H1008/81(PROCAS) and 08/H1006/77).
Informed Consent Statement
Clinical or research consent was given for extended testing of breast cancer associated genes. No individual identifiable information is in the manuscript.
Data Availability Statement
Data is available on request.
Conflicts of Interest
D.G.E. has consultancies from AstraZeneca, Springworks and Recusrion not directly relevant to the manuscript. All other authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Couch F.J., Shimelis H., Hu C., Hart S.N., Polley E.C., Na J., Hallberg E., Moore R., Thomas A., Lilyquist J., et al. Associations Between Cancer Predisposition Testing Panel Genes and Breast Cancer. JAMA Oncol. 2017;3:1190–1196. doi: 10.1001/jamaoncol.2017.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu H.M., Li S., Black M.H., Lee S., Hoiness R., Wu S., Mu W., Huether R., Chen J., Sridhar S., et al. Association of Breast and Ovarian Cancers with Predisposition Genes Identified by Large-Scale Sequencing. JAMA Oncol. 2019;5:51–57. doi: 10.1001/jamaoncol.2018.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimelis H., LaDuca H., Hu C., Hart S.N., Na J., Thomas A., Akinhanmi M., Moore R.M., Brauch H., Cox A., et al. Triple-Negative Breast Cancer Risk Genes Identified by Multigene Hereditary Cancer Panel Testing. J. Natl. Cancer Inst. 2018;110:855–862. doi: 10.1093/jnci/djy106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hauke J., Horvath J., Gross E., Gehrig A., Honisch E., Hackmann K., Schmidt G., Arnold N., Faust U., Sutter C., et al. Gene panel testing of 5589 BRCA1/2-negative index patients with breast cancer in a routine diagnostic setting: Results of the German Consortium for Hereditary Breast and Ovarian Cancer. Cancer Med. 2018;7:1349–1358. doi: 10.1002/cam4.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tung N., Lin N.U., Kidd J., Allen B.A., Singh N., Wenstrup R.J., Hartman A.R., Winer E.P., Garber J.E. Frequency of Germline Mutations in 25 Cancer Susceptibility Genes in a Sequential Series of Patients with Breast Cancer. J. Clin. Oncol. 2016;34:1460–1468. doi: 10.1200/JCO.2015.65.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buys S.S., Sandbach J.F., Gammon A., Patel G., Kidd J., Brown K.L., Sharma L., Saam J., Lancaster J., Daly M.B. A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer. 2017;123:1721–1730. doi: 10.1002/cncr.30498. [DOI] [PubMed] [Google Scholar]
- 7.Li J., Meeks H., Feng B.J., Healey S., Thorne H., Makunin I., Ellis J., kConFab Investigators. Campbell I., Southey M., et al. Targeted massively parallel sequencing of a panel of putative breast cancer susceptibility genes in a large cohort of multiple-case breast and ovarian cancer families. J. Med. Genet. 2016;53:34–42. doi: 10.1136/jmedgenet-2015-103452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desmond A., Kurian A.W., Gabree M., Mills M.A., Anderson M.J., Kobayashi Y., Horick N., Yang S., Shannon K.M., Tung N., et al. Clinical Actionability of Multigene Panel Testing for Hereditary Breast and Ovarian Cancer Risk Assessment. JAMA Oncol. 2015;1:943–951. doi: 10.1001/jamaoncol.2015.2690. [DOI] [PubMed] [Google Scholar]
- 9.Kapoor N.S., Curcio L.D., Blakemore C.A., Bremner A.K., McFarland R.E., West J.G., Banks K.C. Multigene Panel Testing Detects Equal Rates of Pathogenic BRCA1/2 Mutations and has a Higher Diagnostic Yield Compared to Limited BRCA1/2 Analysis Alone in Patients at Risk for Hereditary Breast Cancer. Ann. Surg. Oncol. 2015;22:3282–3288. doi: 10.1245/s10434-015-4754-2. [DOI] [PubMed] [Google Scholar]
- 10.Fanale D., Incorvaia L., Filorizzo C., Bono M., Fiorino A., Calo V., Brando C., Corsini L.R., Barraco N., Badalamenti G., et al. Detection of Germline Mutations in a Cohort of 139 Patients with Bilateral Breast Cancer by Multi-Gene Panel Testing: Impact of Pathogenic Variants in Other Genes beyond BRCA1/2. Cancers. 2020;12:2415. doi: 10.3390/cancers12092415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Easton D.F., Pharoah P.D., Antoniou A.C., Tischkowitz M., Tavtigian S.V., Nathanson K.L., Devilee P., Meindl A., Couch F.J., Southey M., et al. Gene-panel sequencing and the prediction of breast-cancer risk. N. Engl. J. Med. 2015;372:2243–2257. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antoniou A.C., Casadei S., Heikkinen T., Barrowdale D., Pylkas K., Roberts J., Lee A., Subramanian D., De Leeneer K., Fostira F., et al. Breast-cancer risk in families with mutations in PALB2. N. Engl. J. Med. 2014;371:497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alfoldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans D.G., Brentnall A., Byers H., Harkness E., Stavrinos P., Howell A., FH-risk study Group. Newman W.G., Cuzick J. The impact of a panel of 18 SNPs on breast cancer risk in women attending a UK familial screening clinic: A case-control study. J. Med. Genet. 2017;54:111–113. doi: 10.1136/jmedgenet-2016-104125. [DOI] [PubMed] [Google Scholar]
- 15.van Veen E.M., Brentnall A.R., Byers H., Harkness E.F., Astley S.M., Sampson S., Howell A., Newman W.G., Cuzick J., Evans D.G.R. Use of Single-Nucleotide Polymorphisms and Mammographic Density Plus Classic Risk Factors for Breast Cancer Risk Prediction. JAMA Oncol. 2018;4:476–482. doi: 10.1001/jamaoncol.2017.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breast Cancer Susceptibility Collaboration. Dorling L., Carvalho S., Allen J., Gonzalez-Neira A., Luccarini C., Wahlstrom C., Pooley K.A., Parsons M.T., Fortuno C., et al. Breast Cancer Risk Genes–Association Analysis in More than 113,000 Women. N. Engl. J. Med. 2021;384:428–439. doi: 10.1056/NEJMoa1913948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans D.G., Harkness E.F., Plaskocinska I., Wallace A.J., Clancy T., Woodward E.R., Howell T.A., Tischkowitz M., Lalloo F. Pathology update to the Manchester Scoring System based on testing in over 4000 families. J. Med. Genet. 2017;54:674–681. doi: 10.1136/jmedgenet-2017-104584. [DOI] [PubMed] [Google Scholar]
- 19.Byers H., Wallis Y., van Veen E.M., Lalloo F., Reay K., Smith P., Wallace A.J., Bowers N., Newman W.G., Evans D.G. Sensitivity of BRCA1/2 testing in high-risk breast/ovarian/male breast cancer families: Little contribution of comprehensive RNA/NGS panel testing. Eur. J. Hum. Genet. 2016;24:1591–1597. doi: 10.1038/ejhg.2016.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts M.E., Jackson S.A., Susswein L.R., Zeinomar N., Ma X., Marshall M.L., Stettner A.R., Milewski B., Xu Z., Solomon B.D., et al. MSH6 and PMS2 germ-line pathogenic variants implicated in Lynch syndrome are associated with breast cancer. Genet. Med. 2018;20:1167–1174. doi: 10.1038/gim.2017.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanco A.M., Topper S., Nussbaum R.L. Association Between Invasive Lobular Breast Cancer and Mutations in the Mismatch Repair Gene MSH6. JAMA Oncol. 2019;5:120–121. doi: 10.1001/jamaoncol.2018.6911. [DOI] [PubMed] [Google Scholar]
- 22.Evans D.G., Woodward E.R., Lalloo F., Moller P., Sampson J., Burn J., Moeslein G., Capella G. Are women with pathogenic variants in PMS2 and MSH6 really at high lifetime risk of breast cancer? Genet. Med. 2019;21:1878–1879. doi: 10.1038/s41436-018-0401-1. [DOI] [PubMed] [Google Scholar]
- 23.Ten Broeke S.W., van der Klift H.M., Tops C.M.J., Aretz S., Bernstein I., Buchanan D.D., de la Chapelle A., Capella G., Clendenning M., Engel C., et al. Cancer Risks for PMS2-Associated Lynch Syndrome. J. Clin. Oncol. 2018;36:2961–2968. doi: 10.1200/JCO.2018.78.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moller P., Seppala T., Bernstein I., Holinski-Feder E., Sala P., Evans D.G., Lindblom A., Macrae F., Blanco I., Sijmons R., et al. Cancer incidence and survival in Lynch syndrome patients receiving colonoscopic and gynaecological surveillance: First report from the prospective Lynch syndrome database. Gut. 2017;66:464–472. doi: 10.1136/gutjnl-2015-309675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodward E.R., van Veen E.M., Forde C., Harkness E.F., Byers H.J., Ellingford J.M., Burghel G.J., Schlech H., Bowers N.L., Wallace A.J., et al. Clinical utility of testing for PALB2 and CHEK2 c.1100delC in breast and ovarian cancer. Genet. Med. 2021 doi: 10.1038/s41436-021-01234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandoval R.L., Leite A.C.R., Barbalho D.M., Assad D.X., Barroso R., Polidorio N., Dos Anjos C.H., de Miranda A.D., Ferreira A., Fernandes G.D.S., et al. Germline molecular data in hereditary breast cancer in Brazil: Lessons from a large single-center analysis. PLoS ONE. 2021;16:e0247363. doi: 10.1371/journal.pone.0247363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lalloo F., Varley J., Ellis D., Moran A., O’Dair L., Pharoah P., Evans D.G., Early Onset Breast Cancer Study Group Prediction of pathogenic mutations in patients with early-onset breast cancer by family history. Lancet. 2003;361:1101–1102. doi: 10.1016/S0140-6736(03)12856-5. [DOI] [PubMed] [Google Scholar]
- 28.Easton D.F., Lesueur F., Decker B., Michailidou K., Li J., Allen J., Luccarini C., Pooley K.A., Shah M., Bolla M.K., et al. No evidence that protein truncating variants in BRIP1 are associated with breast cancer risk: Implications for gene panel testing. J. Med. Genet. 2016;53:298–309. doi: 10.1136/jmedgenet-2015-103529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loveday C., Turnbull C., Ruark E., Xicola R.M., Ramsay E., Hughes D., Warren-Perry M., Snape K., Breast Cancer Susceptibility Collaboration. Eccles D., et al. Germline RAD51C mutations confer susceptibility to ovarian cancer. Nat. Genet. 2012;44:475–476. doi: 10.1038/ng.2224. author reply 476. [DOI] [PubMed] [Google Scholar]
- 30.Loveday C., Turnbull C., Ramsay E., Hughes D., Ruark E., Frankum J.R., Bowden G., Kalmyrzaev B., Warren-Perry M., Snape K., et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat. Genet. 2011;43:879–882. doi: 10.1038/ng.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee A., Mavaddat N., Wilcox A.N., Cunningham A.P., Carver T., Hartley S., de Villiers C.B., Izquierdo A., Simard J., Schmidt M.K., et al. BOADICEA: A comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet. Med. 2019;21:1708–1718. doi: 10.1038/s41436-018-0406-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is available on request.