Abstract
The mdm2 gene is positively regulated by p53 through a p53-responsive DNA element in the first intron of the mdm2 gene. mdm2 binds p53, thereby abrogating the ability of p53 to activate the mdm2 gene, and thus forming an autoregulatory loop of mdm2 gene regulation. Although the mdm2 gene is thought to act as an oncogene by blocking the activity of p53, recent studies indicate that mdm2 can act independently of p53 and block the G1 cell cycle arrest mediated by members of the retinoblastoma gene family and can activate E2F1/DP1 and the cyclin A gene promoter. In addition, factors other than p53 have recently been shown to regulate the mdm2 gene. In this article, we report that thyroid hormone (T3) receptors (T3Rs), but not the closely related members of the nuclear thyroid hormone/retinoid receptor gene family (retinoic acid receptor, vitamin D receptor, peroxisome proliferation activation receptor, or retinoid X receptor), regulate mdm2 through the same intron sequences that are modulated by p53. Chicken ovalbumin upstream promoter transcription factor I, an orphan nuclear receptor which normally acts as a transcriptional repressor, also activates mdm2 through the same intron region of the mdm2 gene. Two T3R-responsive DNA elements were identified and further mapped to sequences within each of the p53 binding sites of the mdm2 intron. A 10-amino-acid sequence in the N-terminal region of T3Rα that is important for transactivation and interaction with TFIIB was also found to be important for activation of the mdm2 gene response element. T3 was found to stimulate the endogenous mdm2 gene in GH4C1 cells. These cells are known to express T3Rs, and T3 is known to stimulate replication of these cells via an effect in the G1 phase of the cell cycle. Our findings, which indicate that T3Rs can regulate the mdm2 gene independently of p53, provide an explanation for certain known effects of T3 and T3Rs on cell proliferation. In addition, these findings provide further evidence for p53-independent regulation of mdm2 which could lead to the development of tumors from cells that express low levels of p53 or that express p53 mutants defective in binding to and activating the mdm2 gene.
The mdm2 oncogene was originally identified as a gene that is amplified and overexpressed in a tumorigenic derivative of mouse 3T3 cells (3T3DM), with the amplified sequences being located on extrachromosomal double minute particles (8). Subsequent studies revealed that overexpression of the mdm2 gene was responsible for transformation of these cells (19). Amplification and overexpression of the mdm2 gene have been detected in a number of human sarcomas (41, 49), suggesting that this oncogene plays a role in human carcinogenesis. The product of the mouse mdm2 gene is a protein with a predicted molecular mass of 54 kDa, although it migrates as a ∼95-kDa protein in sodium dodecyl sulfate (SDS) gels (4).
mdm2 associates with wild-type and certain mutant p53 proteins (34, 47), and an excess of mdm2 can abrogate transcriptional activation by wild-type p53 (47). In addition, the binding of mdm2 to p53 results in a decrease in p53 levels through enhanced degradation (33, 40) by a proteosome-mediated process (40). Thus, overexpression of mdm2 serves as a negative regulator of p53 function. The mdm2 gene is regulated by wild-type p53 through an intronic sequence which contains a p53 DNA-binding region and functions as a p53 response element (69). This results in an autoregulatory loop in which mdm2 complexes with p53, thereby reducing the extent of its own expression (5, 67).
An interesting property of the mdm2 gene is that it can generate multiple transcripts which may differ in coding potential. Sequence analysis of mdm2 clones isolated from murine (19) and human (49) cDNA libraries indicates that the mdm2 gene can generate at least seven distinct mRNA species. The mdm2 splice variants differ in their ability to inhibit p53-mediated transactivation (31). Thus, the existence of multiple mdm2 proteins raises the possibility that mdm2 may elicit effects in cells independently of its known effect on p53. This is suggested by studies indicating that mdm2 can overcome a G1 cell cycle arrest mediated by members of the retinoblastoma gene family (16, 68) and that expression of mdm2 can activate E2F1/DP1 (46) and the cyclin A gene promoter (44). This is also supported by studies of mdm2 expression in mammary tissue of p53+/+ and p53−/− mice indicating that mdm2 can regulate DNA synthesis independently of its ability to inhibit p53 activity. Recent studies have also indicated that expression of mdm2 can be modulated by factors other than p53 (e.g., fibroblast growth factor [58]), suggesting that mdm2 may act and be regulated independently of p53.
The thyroid hormones influence a variety of physiological processes, including cell growth and metabolism in mammals, initiation of metamorphosis in amphibia, and development of the vertebrate nervous system (53). Most, if not all, of these actions are mediated by thyroid hormone (l-triiodothyronine; T3) nuclear receptors (T3Rs). The T3Rs are encoded by two genes (α and β) and are expressed as several isoforms (T3Rα1, T3Rβ1, and T3Rβ2) (43). The T3Rs are members of the thyroid hormone/retinoid receptor subfamily of nuclear hormone receptors, which includes the retinoic acid receptors (RARs), the retinoid X receptors (RXRs), the vitamin D receptor (VDR), the peroxisome proliferation activation receptors (PPARs), and orphan receptors such as chicken ovalbumin upstream promoter transcription factor (COUP-TF) (12, 24).
T3Rs activate transcription through DNA sequences referred to as thyroid hormone response elements (TREs). Naturally occurring TREs are organized as imperfect invert repeats and/or direct repeats (DRs) of the optimized AGGTCA half-site (7, 63). TREs have been identified in a wide variety of genes, including the long terminal repeat (LTR) of Moloney murine leukemia virus (55), the promoter and third intron of the rat growth hormone gene (54), and the promoters of the malic enzyme (13) and human α-myosin heavy-chain (62) genes, and within the NF-κB motifs of the human immunodeficiency virus type 1 (HIV-1) LTR (15). Certain TREs are also regulated by other members of the T3R/RAR subfamily (e.g., the rat growth hormone TRE) (62).
In this study, we report that T3R can stimulate the native mdm2 gene in a T3-dependent fashion. Stimulation was found to occur via a TRE(s) which we localized to the first intron of the mdm2 gene. Similar results were found with homologous sequences from the human mdm2 gene (70). In contrast with T3R, no ligand-dependent stimulation was found with RAR, RXR, VDR, or PPAR, although constitutive activation was found to occur with COUP-TFI. Two T3R-responsive DNA elements were identified in the mdm2 intron and further mapped to sequences within the putative p53 binding sites. Our findings, which indicate that T3R can regulate the mdm2 gene independently of p53, may provide an explanation for certain known effects of T3 and T3R on cell proliferation (22, 32) and in the promotion of tumor development (29).
(This study was done by J.-S.Q. and Y.Y. in partial fulfillment of the requirements for a Ph.D. degree from the Sackler Institute for Graduate Biomedical Science at the New York University School of Medicine.)
MATERIALS AND METHODS
Cell culture and transfection.
HeLa cells and GH4C1 rat pituitary cells were cultured and transfected by electroporation as previously described (1, 22). p53-null (10)1 cells (67) were transfected by calcium phosphate precipitation. The chloramphenicol acetyltransferase (CAT) reporter vectors and other plasmids used are described below. After incubation for 48 h with or without the indicated ligand(s), cells were harvested for assay of CAT activity by a thin-layer chromatography assay (1, 22). Acetylated and unreacted [14C]chloramphenicol were excised from the thin-layer plate and quantitated in a liquid scintillation counter. The amount of protein used in the assays was adjusted to keep the percent conversion of [14C]chloramphenicol below 40%, which is in the linear range. CAT activity values were normalized to represent the percentage of [14C]chloramphenicol acetylated by a specific amount of cell protein in 16 h at 37°C. All experiments were performed with duplicate or triplicate flasks, which showed variations of less than 10%, and each experiment was repeated at least three times with similar results.
Cloning and plasmid construction.
The full-length chicken T3Rα (cT3Rα) cDNA corresponding to amino acids 1 to 408, cloned into a pEXPRESS (pEX) vector (pEX-cT3Rα), has been described previously (25). pRSVT7-cT3Rα(21–408), pEX-cT3Rα(31–408), pEX-cT3Rα(51–408), pRSV-T7-cT3Rα(21–30/51–408), and pRSV-T7-cT3Rα(13–20/51–408) have been described previously (14, 30, 57). Human T3Rβ1 (hT3Rβ1) (28) and rat T3Rβ2 (rT3Rβ2) (35) were also expressed from pEX vectors. Wild-type and mutant human p53 plasmids, pC53-SN3 and pC53(V143A) (gifts from Bert Vogelstein), respectively, utilize a pCMV-Neo-Bam vector (3) or a T7 polymerase-transcribed pBluescript vector. The 59-nucleotide murine p53 response element sequence (TGGTCAAGTTGGGACACGTCCggcgtcggctgtcggagGAGCTAAGTCCTGACATGTCT [uppercase corresponds to Seq1 and Seq2]) from the first intron of the murine mdm2 gene (67) was cloned into the HindIII site at position −88 of ΔMTV-CAT, which lacks the glucocorticoid response elements of the mouse mammary tumor virus LTR (63). This vector has been termed ΔMTV-m59-CAT. 5′-TGGTCAAGTTGGGACACGTCC-3′ (Seq1) and 5′-GAGCTAAGTCCTGACATGTCT-3′ (Seq2) from the 59-bp element were cloned upstream of nucleotide −88 in ΔMTV-CAT and have been termed ΔMTV-Seq1-CAT and ΔMTV-Seq2-CAT, respectively. The homologous 59-bp p53 response element from the human mdm2 gene (70) was also cloned at position −88 of ΔMTV-CAT (ΔMTV-h59-CAT). Cosx1CAT, which contains a 1-kb fragment from the first intron of the murine mdm2 gene subcloned upstream from the adenovirus major late TATA box-terminal deoxynucleotidyltransferase (TdT) initiation signal-CAT gene sequence (p1634CAT) (67), was a gift from Arnold J. Levine. Various deletion mutants containing different fragments of the mdm2 first-intron region cloned into the p1634CAT vector (H0.5ΔNCAT, HX0.5CAT, BN300CAT, BA200CAT, and BP100CAT) (67) were also generously provided by Arnold J. Levine. A COUP-TFI-expressing Rous sarcoma virus vector that has been described elsewhere (11) was obtained from Ming-Jer Tsai.
Gel mobility shift assays.
cT3Rα was expressed in Escherichia coli and purified to apparent homogeneity as described previously (23). Following the purification, the amount of cT3Rα was estimated with l-[125I]T3 (23). Double-stranded oligonucleotides complementary to Seq1 and Seq2, as mentioned above, were synthesized and annealed. These oligonucleotides are flanked by HindIII cohesive ends, permitting cloning and end labeling. cT3Rα was incubated with 32P-labeled Seq1 or Seq2 as described previously (23). The 30-μl incubation mixture contained 25 mM Tris (pH 7.8), 0.5 mM EDTA, 80 mM KCl, 1 mM dithiothreitol, 100 ng of aprotinin, 0.25 μg of poly(dI-dC), 0.05% (vol/vol) Triton X-100, 10% (vol/vol) glycerol, and various amounts of receptor, with or without 1 μM T3. For studies involving heterodimer binding to Seq1, the incubation mixture contained 15 fmol of reticulosyte lysate-synthesized cT3Rα and 25 fmol of baculovirus-expressed murine RXRβ (mRXRβ) as previously described (1). Samples were analyzed by electrophoresis at 4°C in nondenaturing SDS–5% polyacrylamide gels in buffer containing 10 mM Tris, 7.5 mM acetic acid, and 40 μM EDTA (pH 7.8) (23). The gels were dried and autoradiographed.
Western blot analysis of mdm2.
GH4C1 cells were cultured in hormone-depleted medium (22) for 24 h. The cells were then incubated with 500 nM T3 for 4 days, 2 days, or 1 day before the cells were harvested for assay of mdm2. Some cells were incubated without T3 for the 4-day period. At the time of harvest, the cells were about 70 to 80% confluent. Cells were then washed twice with phosphate-buffered saline, scraped from the dishes, and lysed in RIPA buffer (50 mM Tris-HCl [pH 7.5], 5 mM EDTA [pH 8.0], 0.5% Nonidet P-40, 0.5% deoxycholic acid, 0.1% SDS, 150 mM NaCl), supplemented with 1 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin per ml, 2 μg of leupeptin per ml, and 1 μg of pepstatin per ml, for 30 min on ice with frequent vortexing. Lysates were then clarified by microcentrifugation, and a small aliquot of each was removed for determination of the protein concentration. Equal quantities of lysate protein were separated in an SDS–10% polyacrylamide gel. Verification of the efficiency of transfer to nitrocellulose and the equality of loaded protein quantities were determined by staining the membrane with 0.5% Ponceau S in 10% acetic acid and by probing the blots with antiactin antibody. Blots were washed in H2O and then incubated overnight at 4°C in blocking buffer (5% nonfat dry milk in Tris-buffered saline [TBS]) and then with anti-mdm2 monoclonal antibody 2A10 (a gift from Arnold J. Levine), which recognizes the central region (amino acids 294 to 339) of hmdm2 and reacts with mdm2 proteins from a variety of species. Blots were also probed with a monoclonal antibody (Ab-1; Oncogene Science) that recognizes only the N terminus of mdm2. Blots were washed four times with TBS supplemented with Triton X-100 and then twice with unsupplemented TBS. Blots were then incubated at room temperature for 1 h with peroxidase-labeled goat anti-mouse immunoglobulin G second antibody (Kirkegaard & Perry Laboratories, Inc.) at 0.2 μg/ml, washed four times with TBS, and then visualized with an enhanced chemiluminescence detection system (E. I. Dupont de Nemours & Co.).
RESULTS
The mdm2 gene is stimulated by T3 in GH4C1 cells.
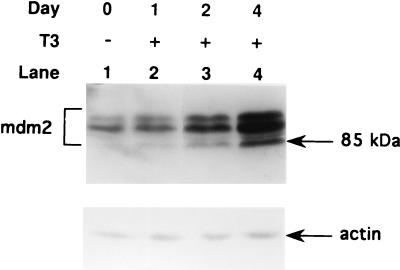
T3 is known to stimulate the growth of GH4C1 cells and related rat pituitary cell lines which express T3Rs (9, 22). This growth-stimulatory effect of T3 has been shown to result from an effect on the G1 phase of the cell cycle (32). Since these cells express functional p53 (52), and p53 is known to influence the length of G1, we assessed whether T3 might act to inhibit p53-mediated responses in GH4C1 cells. Since the expression of mdm2 is a sensitive indicator of p53 activity, we studied the effect of T3 on mdm2. Western blotting studies indicate that GH4C1 cells express three forms of mdm2 which range from 85 to 95 kDa (Fig. 1). This is consistent with previous reports indicating that the mdm2 gene encodes a number of alternatively spliced mRNAs that give rise to several isoforms of mdm2 protein in the cell (31, 50). The mdm2 isoforms, particularly the 85-kDa form, were stimulated rather than inhibited by T3, suggesting that T3R acts indirectly (i.e., via p53) or directly to activate the mdm2 gene.
FIG. 1.
The mdm2 gene is stimulated by T3 in GH4C1 cells. GH4C1 cells were cultured in hormone-depleted medium (22, 27) for 24 h. The cells were then incubated with (+) or without (−) 500 nM T3 for 4 days, 2 days, or 1 day before the cells were harvested for assay of mdm2. Day 0 cells were incubated without T3 for the entire 4-day period. Equal quantities of cell lysate protein were separated in an SDS–10% polyacrylamide gel. After transfer to nitrocellulose membranes, the samples were blotted with anti-mdm2 monoclonal antibody 2A10, which recognizes the central region of the mdm2 protein.
The first intron of the mdm2 gene contains a ligand-dependent TRE.
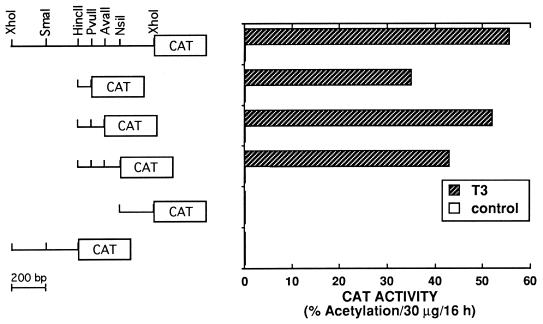
The first intron of the mdm2 gene contains a p53 binding site which, when placed upstream of a minimal promoter, can stimulate a reporter gene in a p53-dependent fashion (38, 67). To determine whether T3R regulates mdm2 without indirectly affecting p53, T3-dependent stimulation of Cosx1CAT was analyzed in HeLa cells, which express very little if any p53 (56). Cosx1CAT contains a 1-kb DNA fragment from the first intron of the mdm2 gene cloned just upstream of the adenovirus major late TATA box and TdT initiator sequence which is linked to the CAT reporter gene (67). HeLa cells were transfected with Cosx1CAT with or without vectors expressing cT3Rα. T3 stimulated expression of Cosx1CAT about 15-fold (Fig. 2). Expression of the control minimal-promoter CAT reporter gene, which lacks the 1-kb mdm2 intronic region (p1634CAT) (not shown), was not altered by T3. This suggests that T3R can activate the mdm2 gene independently of p53 and that the first intron of the mdm2 gene contains a response element(s) for T3R in addition to a response element(s) for p53.
FIG. 2.
The first intron of the mdm2 gene contains a T3-dependent response element. HeLa cells were transfected by electroporation with 5 μg of Cosx1CAT or the indicated deletion mutants of Cosx1CAT with 4 μg of a vector expressing cT3Rα. Cosx1CAT contains a 1-kb XhoI-XhoI fragment from the first intron of the murine mdm2 gene cloned upstream from the adenovirus major late TATA box-TdT initiation signal-CAT gene sequence (67). Following transfection, cells were incubated for 48 h without or with 500 nM T3 prior to determination of CAT activity.
The mdm2 TRE is embedded within the p53-responsive sequence.
The TRE within the mdm2 gene was mapped to an 85-bp HincII-PvuII sequence in the first intron of mdm2 (Fig. 2). T3-dependent stimulation by cT3Rα was found only with constructs containing this 85-bp sequence, which has been reported to also contain the p53 response element of the gene (67). To further map the TRE, a 59-bp putative p53 response sequence (Fig. 3) from the 85-bp HincII-PvuII region of the murine mdm2 gene was cloned at nucleotide −88 of ΔMTV-CAT to create ΔMTV-m59-CAT. This 59-bp sequence can be divided into two regions (Seq1 and Seq2) (Fig. 3), which were individually cloned at position −88 of ΔMTV-CAT, resulting in ΔMTV-Seq1-CAT and ΔMTV-Seq2-CAT, respectively. Underlined in Seq1 and Seq2 of Fig. 3 are hexanucleotide sequences which show a high degree of homology to those found in known TREs (6, 10, 15).
FIG. 3.
Nucleotide sequences of Seq1 and Seq2 of the 59-bp intronic p53 response element of the mdm2 gene. Underlined are hexanucleotide sequences showing a high degree of homology to known native TREs, suggesting that the TRE in Seq1 is organized as a DR+4 TRE. The sequence underlined in Seq2 has on its complementary strand AGGACT. This is identical to sequences found in a TRE from the human thyroid-stimulating hormone α gene (10) and from the TRE sequences at positions −88 to −83 and −102 to −97 of the NF-κB binding sites of the HIV-1 LTR (15).
T3 and cT3Rα strongly stimulated ΔMTV-m59-CAT and ΔMTV-Seq1-CAT in HeLa cells but only weakly activated ΔMTV-Seq2-CAT (Table 1). These results suggest that Seq1 contains a strong TRE while Seq2 contains a weak TRE. Stimulation of Seq1 by T3 was similar to that found with ΔMTV-TREp-CAT, which contains an optimized TRE (TREp; AGGTCA TGACCT) (Table 1). The ΔMTV-m59-CAT reporter gene was also strongly stimulated by wild-type p53. p53 or cT3Rα activated ΔMTV-Seq1-CAT to the same extent as ΔMTV-m59-CAT, while ΔMTV-Seq2-CAT was much less efficiently activated by p53 or cT3Rα (Table 1). None of these plasmids showed significant CAT expression without expression of p53 or cT3Rα, which is consistent with the notion that HeLa cells contain very low levels of p53 (56) and T3R (23). Our findings are consistent with the prior prediction that the 59-bp sequence of the mdm2 gene’s first intron contains two putative p53 binding sites (Seq1 and Seq2) (38, 67). However, our results indicate that these two sites can function independently and that the 5′ site (Seq1) functions more efficiently as a p53 response element than the 3′ site (Seq2). The similar relative extents of activation of ΔMTV-m59-CAT, ΔMTV-Seq1-CAT, and ΔMTV-Seq2-CAT by p53 and T3R suggest that these factors interact with similar sequences contained within Seq1 or Seq2.
TABLE 1.
A T3R-responsive sequence is embedded within the p53 response element of the mdm2 genea
| Transfection | CAT activityb
|
|
|---|---|---|
| Basal | T3 | |
| ΔMTV-CAT | 0.3 | 0.4 |
| +cT3Rα | 0.2 | 0.3 |
| +p53 | 0.2 | NDc |
| ΔMTV-TREp-CAT | 0.25 | 0.35 |
| +cT3Rα | 0.15 | 48.4 |
| +p53 | 0.17 | ND |
| ΔMTV-m59-CAT | 0.19 | 0.24 |
| +cT3Rα | 0.3 | 46.7 |
| +p53 | 54.6 | ND |
| ΔMTV-Seq1-CAT | 0.22 | 0.37 |
| +cT3Rα | 0.2 | 37.5 |
| +p53 | 52.0 | ND |
| ΔMTV-Seq2-CAT | 0.3 | 0.36 |
| +cT3Rα | 0.2 | 3.0 |
| +p53 | 6.5 | ND |
HeLa cells were transfected by electroporation with 5 μg of the indicated CAT reporter genes along with 4 μg of cT3Rα or p53 expression vector. Cells were incubated with and without 500 nM T3 and harvested for determination of CAT activity 48 h later.
Percentage of [14C]chloramphenicol acetylated per 20 μg of cell protein in 16 h.
ND, not determined.
To assess whether the human mdm2 p53 response element is also T3R responsive, we cloned the homologous 59-bp fragment from the human gene into ΔMTV-CAT and examined activation of the resulting constructs, ΔMTV-m59-CAT and ΔMTV-h59-CAT, by the T3R isoforms cT3Rα, hT3Rβ1, and rT3Rβ2 as well as by p53 (Table 2). ΔMTV-m59-CAT and ΔMTV-h59-CAT were similarly activated by cT3Rα, hT3Rβ1, and rT3Rβ2, while the response to p53 was slightly greater for ΔMTV-h59-CAT. This slight increase in response of human mdm2 to p53 was a consistent finding. Although the T3Rs can stimulate the 59-bp mdm2 sequence in HeLa cells, these cells may express low levels of p53. To determine whether T3R can stimulate the 59-bp mdm2 sequence independently of an action of p53, we studied the effect of T3Rs on stimulation of ΔMTV-m59-CAT in (10)1 cells which express no p53 (67) (Table 3). ΔMTV-m59-CAT was stimulated by p53 as well as by cT3Rα or hT3Rβ1 in the presence of T3. In the absence of T3, cT3Rα or hT3Rβ1 inhibited stimulation by p53, supporting the notion that p53 and the T3Rs may bind to the same or overlapping sequences. Similar studies with p53-null SAOS-2 cells also documented T3-dependent activation of ΔMTV-m59-CAT (data not shown).
TABLE 2.
Stimulation of the murine and human mdm2 p53 response sequence by different isoforms of T3Ra
| Transfection | CAT activityb
|
|
|---|---|---|
| Basal | T3 | |
| ΔMTV-m59-CAT | 2.9 | 4.2 |
| +p53 | 76.9 | 72.0 |
| +cT3Rα | 2.1 | 42.7 |
| +hT3Rβ1 | 0.9 | 43 |
| +rT3Rβ2 | 1.1 | 45.3 |
| ΔMTV-h59-CAT | 3.4 | 1.8 |
| +p53 | 145 | 155 |
| +cT3Rα | 2.3 | 73 |
| +hT3Rβ1 | 3.2 | 47.8 |
| +rT3Rβ2 | 2.8 | 47 |
HeLa cells were transfected by electroporation with 5 μg of ΔMTV-m59-CAT or ΔMTV-h59-CAT along with 1 μg of cT3Rα, hT3Rβ1, rT3Rβ2, or p53 expression vector. Cells were incubated with and without 500 nM T3 and harvested for determination of CAT activity 48 h later.
Percentage of [14C]chloramphenicol acetylated per 20 μg of cell protein in 16 h.
TABLE 3.
Ligand-dependent stimulation of the murine mdm2 p53 response sequence by T3R in p53-null cellsa
| Transfection | CAT activityb
|
|
|---|---|---|
| Basal | T3 | |
| ΔMTV-m59-CAT | 6.6 | 6.3 |
| +p53 | 35.9 | 39.7 |
| +cT3Rα | 5.1 | 20.1 |
| +cT3Rα + p53 | 10.2 | 39.4 |
| +hT3Rβ1 | 6.9 | 31 |
| +hT3Rβ1 + p53 | 16.7 | 48.8 |
p53-null (10)1 cells (67) were transfected by calcium phosphate coprecipitation in 9-cm2 wells with (per well) 1.25 μg of ΔMTV-m59-CAT alone or with 0.25 μg of cT3Rα, 0.25 μg of hT3Rβ1, or 0.25 μg of p53 expression vector as indicated. Cells were incubated with and without 500 nM T3 and harvested for determination of CAT activity 48 h later. Cells which were transfected with only p53 also received 0.2 μg of control expression vector which lack the cT3Rα or hT3Rβ1 cDNAs.
Percentage of [14C]chloramphenicol acetylated per 20 μg of cell protein in 16 h.
To assess whether the mdm2-responsive sequence is activated by endogenously expressed p53 or T3R, GH4C1 cells were transfected with ΔMTV-m59-CAT, ΔMTV-TREp-CAT, or ΔMTV-CAT (Table 4). As found in HeLa cells (Tables 1 and 2), very low levels of basal expression were evident for ΔMTV-TREp-CAT and ΔMTV-CAT. However, in contrast with the results for HeLa cells, high levels of basal activity was found with ΔMTV-m59-CAT (Table 4), which is consistent with previous studies indicating that GH4C1 cells contain functional p53 (52). Addition of T3 resulted in further activation of ΔMTV-m59-CAT by endogenous T3R and a similar stimulation of ΔMTV-TREp-CAT but no stimulation of ΔMTV-CAT (Table 4). These results, along with the findings for HeLa cells and (10)1 cells (Tables 1 to 3), support the notion that endogenous T3Rs in GH4C1 cells (26, 27) can activate the mdm2 gene through the same intronic 59-bp p53-responsive sequence.
TABLE 4.
Transcriptional activation of ΔMTV-m59-CAT by endogenous p53 and T3Rs in GH4C1 cellsa
| Transfection | CAT activityb
|
|
|---|---|---|
| Basal | T3 | |
| ΔMTV-CAT | 0.3 | 0.3 |
| ΔMTV-TREp-CAT | 0.35 | 52 |
| ΔMTV-m59-CAT | 15.2 | 42.5 |
GH4C1 cells were transfected by electroporation with 5 μg of the CAT reporter genes as indicated. Cells were incubated with and without 500 nM T3 and harvested for determination of CAT activity 48 h later.
Percentage of [14C]chloramphenicol acetylated per 20 μg of cell protein in 16 h.
T3R binds to Seq1 and Seq2 of the p53-responsive element.
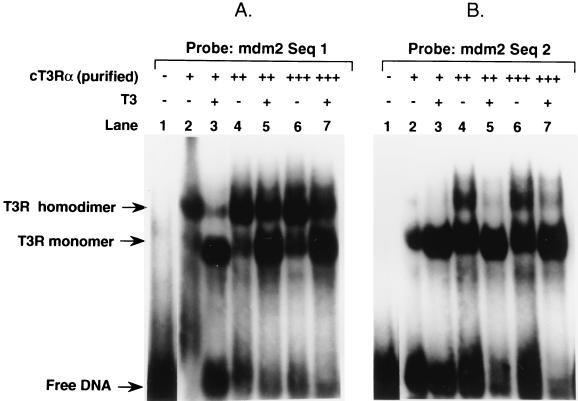
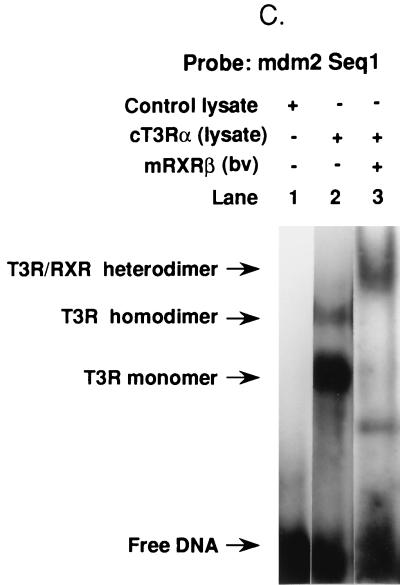
TREs of native genes are organized as DRs and/or inverted repeats of hexanucleotide half-sites separated by variously sized nucleotide gaps (6, 48, 64). The half-site sequences of native genes show significant diversity, indicating that the T3Rs are capable of activating a wide variety of structurally divergent TREs. To study the binding of T3R to the 59-bp sequence of the murine element, gel mobility shift assays were performed with 32P-labeled Seq1 or Seq2 as the probe. cT3Rα bound to both Seq1 and Seq2 as monomers and homodimers, but it exhibited a higher degree of affinity for Seq1 (Fig. 4A and B). T3 increased the binding of monomers but decreased the binding of homodimers to both sequences (Fig. 4A and B). This effect of T3 is commonly seen with DR elements, suggesting that the Seq1 and Seq2 TREs have such an organization. T3Rs bind preferentially to DNA as heterodimers with the RXRs in vitro, and we and others have provided evidence that the T3R-RXR heterodimer is the functional form of the receptor on most native response elements in vivo (1, 51). The ability of cT3Rα and mRXRβ to bind as a heterodimer to Seq1 and Seq2 was studied. cT3Rα and mRXRβ bound as an abundant heterodimer to Seq1 (Fig. 4C), while the extent of heterodimer binding to Seq2 was very low (not illustrated), which paralleled the functional activity of T3-dependent stimulation of these sequences (Table 1).
FIG. 4.
Binding of T3R to Seq1 and Seq2 of the mdm2 p53 response sequence. Gel mobility shift assays were used to study the binding of purified cT3Rα, at 15 (+), 30 (++), or 45 (+++) fmol, to 32P-labeled Seq1 (A) or Seq2 (B) in the absence (−) or presence (+) of 1 μM T3. Shifted complexes are indicated by arrows. (C) Binding of 15 fmol of cT3Rα plus 25 fmol of mRXRβ. For the experiment in panel C, the cT3Rα used was synthesized in vitro, using rabbit reticulocyte lysate (2 μl) (1), while mRXRβ was synthesized by using a baculovirus (bv) expression vector (45). Two microliters of unprogrammed reticulocyte lysate was used as a control. The migration positions of cT3Rα monomers, homodimers, and cT3Rα-mRXRβ heterodimers have been previously established (1, 23).
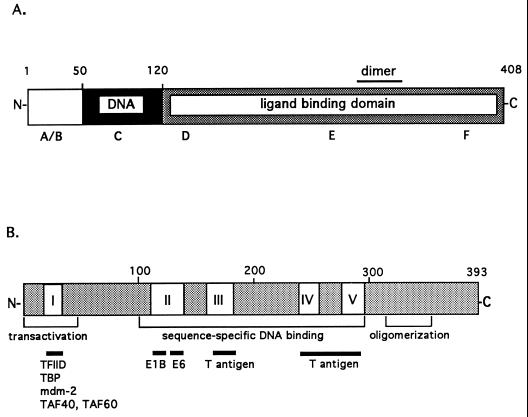
A 10-amino-acid sequence in the A/B region of T3Rα, which is important for interaction with TFIIB, is also required for activation of mdm2.
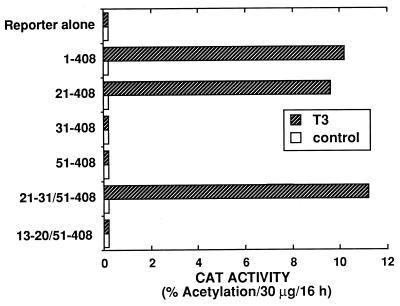
The domain structures of cT3Rα and p53 are illustrated in Fig. 5. The N-terminal A/B region is the least-conserved region among members of the thyroid/retinoid receptor subfamily and the T3R isoforms. The highly conserved 68-amino-acid C domain is organized into two zinc finger DNA-binding structures. High-affinity ligand binding requires the D, E, and F domains, a region which also contains the τi and heptad repeats thought to be involved in protein-protein interactions (1, 24, 51). Amino acids 21 to 30 (DGKRKRKSSQ), in the A/B region, contain a cluster of five basic amino acids important for transcriptional activation of native TREs and for interaction with the general transcription factor TFIIB (30). This 10-amino-acid sequence also appears to be important for activation via the mdm2 intronic TRE (Fig. 6). Several N-terminal deletion mutants of cT3Rα were compared for their ability to transactivate Cosx1CAT. Transfection studies indicate that cT3Rα(21–408), but not cT3Rα(31–408), activated Cosx1CAT similarly to wild-type cT3Rα (Fig. 6). These results indicate that amino acids 21 to 31 are required for transcriptional stimulation by the mdm2 intronic TRE and that TFIIB likely plays a role in this activation. This was further confirmed by functional studies indicating that cT3Rα(21–30/51–408), but not cT3Rα(13–20/51–408), can stimulate Cosx1CAT (Fig. 6).
FIG. 5.
Domain structures of cT3Rα (A) and p53 (B). (A) cT3Rα, like other members of the nuclear receptor family, can be divided into six distinct regions (A to F) (18). The N-terminal A/B region is the least-conserved region among the nuclear hormone receptors. The highly conserved 68-amino-acid C domain (amino acids 51 to 119) is organized into two zinc finger DNA-binding structures. D, E, and F comprise the ligand-binding domain (amino acids 120 to 408) (37). (B) p53 can be divided into three regions. The N terminus (amino acids 1 to 43) contains a strong transcription regulatory region (20, 61, 69) and interacts with a number of proteins, including mdm2 (39). The central region (amino acids 100 to 300) contains the sequence-specific DNA-binding domain (69) and four regions (II to V) that are evolutionarily conserved within all vertebrate species (59). The C-terminal region contains an oligomerization domain that dictates the formation of stable p53 tetramers (amino acids 340 to 393) (60) and a nonspecific nucleic acid-binding domain (amino acids 330 to 393) (66).
FIG. 6.
A 10-amino-acid sequence in the N-terminal A/B domain of cT3Rα is required for activation of the mdm2 TRE. HeLa cells were transfected by electroporation with 5 μg of Cosx1CAT alone or with 4 μg of vector expressing wild-type cT3Rα or the indicated N-terminal deletion mutants of cT3Rα. Following transfection, cells were incubated for 48 h without or with 500 nM T3 prior to determination of CAT activity.
The mdm2 p53/T3R response element is also activated by COUP-TFI but not by RAR, VDR, RXR, or PPAR.
COUP-TFI was initially characterized by its binding to the COUP element (positions −90 to −70) (2, 65). Analysis of COUP-TFI binding sites revealed that COUP-TF could bind to many A/GGGTCA repeats with different spacings and orientations (12). COUP-TFI is known to repress hormonal induction of many target genes by VDR, T3R, and RAR (11). Transfer of the putative ligand binding domain of COUP-TFI to the GAL4 DNA binding domain suggested that it possesses an active silencing function within its C-terminal domain (11).
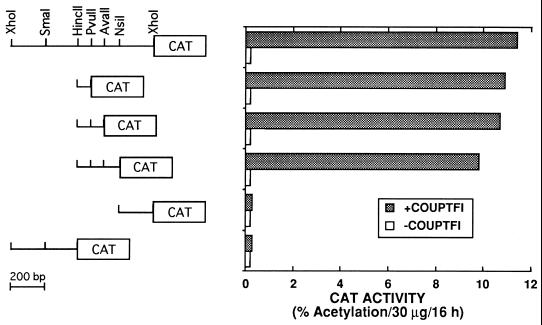
To determine whether COUP-TFI had an effect on the transcriptional regulation of Cosx1CAT, HeLa cells were transfected with Cosx1CAT with or without a vector expressing COUP-TFI. Surprisingly, CAT activity was stimulated rather than repressed by COUP-TFI (Fig. 7). COUP-TFI stimulation was localized to the same 85-bp p53/T3R response region (Fig. 7), indicating that COUP-TFI, which normally represses many promoters, can also function as a transcriptional activator on specific response sequences. In contrast with T3R and COUP-TFI, hRARα, hVDR, hRXRα, and hPPARγ did not stimulate Cosx1CAT without or with their cognate ligands (data not shown).
FIG. 7.
COUP-TFI activates the mdm2 p53 response sequence. HeLa cells were transfected by electroporation with 4 μg of the indicated reporters that contain different fragments of the first intron of the mdm2 gene and 4 μg of a pRSV control vector (−COUPTFI) or pRSV-COUP-TFI (+COUPTFI). Following transfection, cells were incubated in hormone-depleted medium for 48 h prior to determination of CAT activity.
DISCUSSION
Previous studies have shown that p53 stimulates the expression of the mdm2 gene through a p53 response sequence in the first intron of the mdm2 gene (38, 67). The segment conferring p53 responsiveness includes two elements which display significant homology to a consensus p53 binding site (17). These two elements have been termed Seq1 and Seq2 (Fig. 3). We showed that the 59-bp p53 DNA-binding site contains two TREs (Table 1) and that the major TRE in the 59-bp region is contained within Seq1. T3R binds as a monomer or homodimer with a higher affinity for Seq1 than for Seq2 (Fig. 4A and B), or it binds to Seq1 as a heterodimer with RXR. This difference in affinity for T3R parallels the difference in the abilities of Seq1 and Seq2 to function as TREs (Table 1). The strong TRE found in Seq1 resembles a DR with a 4-bp gap (DR+4), which is characteristic of many native TREs (Fig. 3). Inspection of Seq1 indicates that it contains the sequence TGGTCAagttGGGACA, which resembles an idealized TRE half-site (AGGTCA) organized as an imperfect DR with a 4-bp gap (DR+4). Examination of Seq2 indicated that it contains the sequence AGTCCT. The half-site on the complementary strand (AGGACT) is identical to half-sites found in a TRE from the human thyroid-stimulating hormone α gene (10) and in the TRE sequences at positions −88 to −83 and −102 to −97 of NF-κB binding sites of the HIV-1 LTR (15).
Activation by wild-type p53 parallels the findings with T3R (i.e., the 59-bp fragment containing Seq1 and Seq2 is about as active as that containing only Seq1, and both are much more active than that containing only Seq2) (Table 1). This suggests that T3R and p53 may contact similar sequences in the 59-bp mdm2 sequence. This is further supported by the finding that expression of unliganded T3R blocks p53-mediated stimulation of ΔMTV-m59-CAT (Table 3). All isoforms of T3R activate the mdm2 response sequence as well as the homologous sequence from the human mdm2 gene (Table 2). Hadzic et al. (30) showed that amino acids 21 to 30 (DGKRKRKSSQ) in the A/B region of cT3Rα contain a cluster of five basic amino acids that are important for transcriptional activation of native TREs and for interaction with the general transcription factor TFIIB. Similarly, this 10-amino-acid region was also found to be important for transcriptional activation of Cosx1CAT. The related receptors hRARα, hRXRα, hPPARγ, and hVDR do not transcriptionally activate these sequences, which is consistent with the apparent DR+4 organization of the TRE. In contrast, the orphan receptor COUP-TF, which commonly represses the activity of TREs, activates the mdm2 response sequence in Cosx1CAT (Fig. 7).
The mdm2 protooncogene is amplified in a variety of tumors and approximately 30% of sarcomas (41, 49). Overexpression of mdm2 inhibits the prolongation or blockade of G1 progression mediated by p53 as well as the ability of p53 to suppress transformation of cells in culture (21). Although the action of mdm2 as an oncogene could occur solely by inhibiting the function of p53 (4, 67), it could also function through p53-independent pathways. This is supported by recent studies indicating that mdm2 can overcome the G1 cell cycle arrest mediated by members of the retinoblastoma gene family (16, 68) and that expression of mdm2 can activate E2F1/DP1 (46) and the cyclin A gene promoter (44). The mdm2 gene encodes a number of alternatively spliced mRNAs that give rise to multiple protein forms (31, 50). Some isoforms lack the N-terminal epitopes required for the mdm2-p53 interaction (31, 50), which further implies a function for mdm2 in addition to its ability to block p53-mediated responses.
Landers et al. (42) found that high levels of mdm2 proteins are present in two choriocarcinoma cell lines that also overexpress wild-type p53. In this study, we found that GH4C1 cells, which appear to express functional p53 (Table 4), express high levels of mdm2 (Fig. 1). GH4C1 cells express T3Rα1, T3Rβ1, and T3Rβ2 (36). Several forms of mdm2, particularly the ∼85-kDa form, were found to be stimulated by endogenous T3Rs when GH4C1 cells were treated with T3 (Fig. 1). This ∼85-kDa form of mdm2 was not detected with monoclonal antibody Ab-1 (Oncogene Science), which recognizes the N terminus of mdm2 (data not shown), suggesting that the ∼85-kDa form of mdm2 does not possess the N-terminal amino acid sequence required for interaction with p53. This suggests that the ∼85-kDa form of mdm2 may mediate functions of mdm2 independently of its ability to block p53-mediated functions. Our results do not document that the regulation of mdm2 by T3R is through a direct pathway, because it takes at least 24 h to detect the stimulation (Fig. 1). However, one possible mechanism to account for the lag period in a direct activation pathway is that the mdm2 stimulated by T3 binds with endogenous p53, which in turn acts to decrease p53-mediated mdm2 stimulation (4, 67). This reduction in mdm2 stimulation by p53 offsets the initial extent of stimulation by T3R, resulting in an apparent lag in the time until stimulation by T3 is detected. This is consistent with the finding that endogenous levels of mdm2 appear to be sufficient to regulate p53 levels and that increased expression of mdm2 can reduce the level of p53 (40).
Our results also indicate that p53 independently transactivates Seq1 and Seq2 (Table 1), consistent with the prediction that both Seq1 and Seq2 resemble a p53 consensus sequence (4, 67). Interestingly, activation by Seq1 is similar to that found for the entire 59-bp response sequence, suggesting that Seq1 predominantly contributes to the response of the mdm2 element to p53. In addition, p53 and T3R appear to compete for the same or overlapping DNA sequences (Table 3). These results are consistent with a model in which T3R regulates the mdm2 gene, particularly in cells that lack or contain low levels of p53 or contain p53 mutants deficient in DNA binding. Mutations in the DNA binding region of p53 are common in human cancer, and the wild-type p53 allele is often concomitantly deleted. Our findings suggest that under these conditions, T3R or related factors such as COUP-TF may act to regulate the mdm2 gene. Such stimulation would also occur in cells containing levels of endogenous p53 which only partially compete with T3R or COUP-TF for the mdm2 gene-responsive sequence. This notion is supported by the finding that endogenous T3R can further activate ΔMTV-m59-CAT in the presence of endogenous levels of p53 in GH4C1 cells (Table 4). The regulation of mdm2 by T3R and COUP-TF, as well as other, as-yet-undefined transcription factors, may provide a mechanism for the growth-promoting effects of the T3Rs or other factors on certain cell types as well as for the mdm2-mediated development of tumors in cells containing p53 mutants or low levels of p53.
ACKNOWLEDGMENTS
We thank A. J. Levine for Cosx1CAT, the Cosx1CAT deletion mutants, the p1634CAT vector control, monoclonal antibody 2A10 against human and rodent mdm2, and p53-null (10)1 cells. We also thank B. Vogelstein for vectors expressing wild-type human p53.
This research was supported by NIH grant DK16636 to H.H.S. During this work, V.D.-Y. was an Aaron Diamond Foundation fellow (grant HRI817-5332F), and this work was supported in part by The Aaron Diamond Foundation. Oligonucleotide synthesis was provided by the NYUMC General Clinical Research Center (NIH NCRR grant M01RR00096). H.H.S. and V.D.-Y. are members of the NYUMC Cancer Center (funded by grant CA16087). Sequence analysis and database searches were through the NYUMC Research Computing Resource, which received support from the National Science Foundation (grant DIR-8908095).
REFERENCES
- 1.Au-Fliegner M, Helmer E, Casanova J, Raaka B M, Samuels H H. The conserved ninth C-terminal heptad in thyroid hormone and retinoic acid receptors mediates diverse responses by affecting heterodimer but not homodimer formation. Mol Cell Biol. 1993;13:5725–5737. doi: 10.1128/mcb.13.9.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagchi M K, Tsai S Y, Tsai M-J, O’Malley B W. Purification and characterization of chicken ovalbumin gene upstream promoter transcription factor from homologous oviduct cells. Mol Cell Biol. 1987;7:4151–4158. doi: 10.1128/mcb.7.12.4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker S J, Markowitz S, Fearon E R, Willson J K U, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 4.Barak Y T, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barettino D, Bugge T H, Bartunek P, Ruiz M V, Sonntag-Buck V, Beug H, Zenke M, Stunnenberg H G. Unliganded T3R, but not its oncogenic variant, v-erbA, suppresses RAR-dependent transactivation by titrating out RXR. EMBO J. 1993;12:1343–1354. doi: 10.1002/j.1460-2075.1993.tb05779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brent G A, Harney J W, Chen Y, Warne R L, Moore D D, Larsen P R. Mutations of the rat growth hormone promoter which increase and decrease response to thyroid hormone define a consensus thyroid hormone response element. Mol Endocrinol. 1989;3:1996–2004. doi: 10.1210/mend-3-12-1996. [DOI] [PubMed] [Google Scholar]
- 7.Brent G A, Larsen P R, Harney J W, Koenig R J, Moore D D. Functional characterization of the rat growth hormone promoter elements required for induction by thyroid hormone with and without a co-transfected beta type thyroid hormone receptor. J Biol Chem. 1989;264:178–182. [PubMed] [Google Scholar]
- 8.Cahilly-Snyder L, Yang-Feng T, Francke U, George D L. Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line. Somat Cell Mol Genet. 1987;13:235–244. doi: 10.1007/BF01535205. [DOI] [PubMed] [Google Scholar]
- 9.Casanova J, Horowitz Z D, Copp R P, McIntyre W R, Pascual A, Samuels H H. Photoaffinity labeling of thyroid hormone nuclear receptors: influence of n-butyrate and analysis of the half-lives of the 57,000 and 47,000 molecular weight receptor forms. J Biol Chem. 1984;259:12084–12091. [PubMed] [Google Scholar]
- 10.Chatterjee V K K, Lee J-K, Rentoumis A, Jameson J L. Negative regulation of the thyroid stimulating hormone alpha gene by thyroid hormone: receptor interaction adjacent to the TATA box. Proc Natl Acad Sci USA. 1989;86:9114–9118. doi: 10.1073/pnas.86.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooney A J, Leng X, Tsai S Y, O’Malley B W, Tsai M-J. Multiple mechanisms of chicken ovalbumin upstream promoter transcription factor-dependent repression of transactivation by the vitamin D, thyroid hormone, and retinoic acid receptors. J Biol Chem. 1993;268:4152–4160. [PubMed] [Google Scholar]
- 12.Cooney A J, Tsai S Y, O’Malley B W, Tsai M-J. Chicken ovalbumin upstream promoter transcription factor (COUP-TF) dimers bind to different GGTCA response elements, allowing COUP-TF to repress hormonal induction of the vitamin D3, thyroid hormone, and retinoic acid receptors. Mol Cell Biol. 1992;12:4153–4163. doi: 10.1128/mcb.12.9.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deavergne B, Petty K J, Nikodem V M. Functional characterization and receptor binding studies of the malic enzyme thyroid hormone response element. J Biol Chem. 1991;266:1006–1013. [PubMed] [Google Scholar]
- 14.Desai-Yajnik V, Hadzic E, Modlinger P, Malhotra S, Gechlik G, Samuels H H. Interactions of thyroid hormone receptor with the human immunodeficiency virus type 1 (HIV-1) long terminal repeat and the HIV-1 Tat transactivator. J Virol. 1995;69:5103–5112. doi: 10.1128/jvi.69.8.5103-5112.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desai-Yajnik V, Samuels H H. The NF-κB and Sp1 DNA motifs of the human immunodeficiency virus type 1 long terminal repeat function as novel thyroid hormone response elements. Mol Cell Biol. 1993;13:5057–5069. doi: 10.1128/mcb.13.8.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubs-Poterszman M C, Tocque B, Wasylyk B. MDM2 transformation in the absence of p53 and abrogation of the p107 G1 cell-cycle arrest. Oncogene. 1995;11:2445–2449. [PubMed] [Google Scholar]
- 17.El-Deiry W S, Kern S E, Pietenpol J A, Kinzler K W, Vogelstein B. Human genomic DNA sequences define a consensus binding site for p53. Nat Genet. 1992;1:44–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 18.Evans R M. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farkharzadeh S S, Trusko S P, George D L. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J. 1991;10:1565–1569. doi: 10.1002/j.1460-2075.1991.tb07676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fields S, Jang S K. The p53 proto-oncogene can act as a suppressor of transformation. Science. 1990;249:1046–1049. [Google Scholar]
- 21.Finlay C A. The mdm-2 oncogene can overcome wild-type p53 suppression of transformed cell growth. Mol Cell Biol. 1993;13:301–306. doi: 10.1128/mcb.13.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flug F, Copp R P, Casanova J, Horowitz Z D, Janocko L, Plotnick M, Samuels H. cis-acting elements of the rat growth hormone gene which mediate basal and regulated expression by thyroid hormone. J Biol Chem. 1987;262:6373–6382. [PubMed] [Google Scholar]
- 23.Forman B M, Casanova J, Raaka B M, Ghysdael J, Samuels H H. Half-site spacing and orientation determines whether thyroid hormone and retinoic acid receptors and related factors bind to DNA response elements as monomers, homodimers, or heterodimers. Mol Endocrinol. 1992;6:429–442. doi: 10.1210/mend.6.3.1316541. [DOI] [PubMed] [Google Scholar]
- 24.Forman B M, Samuels H H. Interactions among a subfamily of nuclear hormone receptors: the regulatory zipper model. Mol Endocrinol. 1990;4:1293–1301. doi: 10.1210/mend-4-9-1293. [DOI] [PubMed] [Google Scholar]
- 25.Forman B M, Samuels H H. pEXPRESS: a family of expression vectors containing a single transcription unit active in prokaryotes, eukaryotes and in vitro. Gene. 1991;105:9–15. doi: 10.1016/0378-1119(91)90507-8. [DOI] [PubMed] [Google Scholar]
- 26.Forman B M, Yang C-R, Au M, Casanova J, Ghysdael J, Samuels H H. A domain containing leucine zipper like motifs mediates novel in vivo interactions between the thyroid hormone and retinoic acid receptors. Mol Endocrinol. 1989;3:1610–1626. doi: 10.1210/mend-3-10-1610. [DOI] [PubMed] [Google Scholar]
- 27.Forman B M, Yang C-R, Stanley F, Casanova J, Samuels H H. c-erbA protooncogenes mediate thyroid hormone-dependent and -independent regulation of the rat growth hormone and prolactin genes. Mol Endocrinol. 1988;2:902–911. doi: 10.1210/mend-2-10-902. [DOI] [PubMed] [Google Scholar]
- 28.Geffner M E, Su F, Ross N, Hershman J M, Dop C V, Menke J B, Hao E-H, Stanzak R K, Eaton T, Samuels H H, Usala S J. An arginine to histidine mutation in codon 311 of the c-erbAβ gene results in a mutant thyroid hormone receptor which does not mediate a dominant negative phenotype. J Clin Invest. 1993;91:538–546. doi: 10.1172/JCI116233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guernsey D L, Fisher P B. Thyroid hormone and neoplastic transformation. Crit Rev Oncog. 1990;1:389–408. [PubMed] [Google Scholar]
- 30.Hadzic E, Desai-Yajnik V, Helmer E, Guo S, Wu S, Koudinova N, Casanova J, Raaka B M, Samuels H H. A 10-amino-acid sequence in the N-terminal A/B domain of thyroid hormone receptor α is essential for transcriptional activation and interaction with the general transcription factor TFIIB. Mol Cell Biol. 1995;15:4507–4517. doi: 10.1128/mcb.15.8.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haines D S, Landers J E, Engle L J, George D L. Physical and functional interaction between wild-type p53 and mdm2 proteins. Mol Cell Biol. 1994;14:1171–1178. doi: 10.1128/mcb.14.2.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halperin Y, Surks M I, Shapiro L E. l-Triiodothyronine (T3) regulates cellular growth rate, growth hormone production, and levels of nuclear T3 receptors via distinct dose-response ranges in cultured GC cells. Endocrinology. 1990;126:2321–2326. doi: 10.1210/endo-126-5-2321. [DOI] [PubMed] [Google Scholar]
- 33.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 34.Hinds P W, Finlay C A, Quartin R S, Baker S J, Fearon E R, Vogelstein B, Levine A J. Mutant p53 cDNAs from human colorectal carcinomas can cooperate with ras in transformation of primary rat cells: a comparison of the “hot spot” mutant phenotypes. Cell Growth Differ. 1990;1:571–580. [PubMed] [Google Scholar]
- 35.Hodin R, Lazar M A, Wintman B I, Darling D S, Koenig R J, Larsen P R, Moore D D, Chin W W. Identification of a thyroid hormone receptor that is pituitary-specific. Science. 1989;244:76–79. doi: 10.1126/science.2539642. [DOI] [PubMed] [Google Scholar]
- 36.Horowitz Z D, Sahnoun H, Pascual A, Casanova J, Samuels H H. Analysis of photoaffinity label derivatives to probe thyroid hormone receptor in human fibroblasts, GH1 cells and soluble receptor preparations. J Biol Chem. 1988;263:6636–6642. [PubMed] [Google Scholar]
- 37.Horowitz Z D, Yang C-R, Forman B M, Casanova J, Samuels H H. Characterization of the domain structure of chick c-erbA by deletion mutation: in vitro translation and cell transfection studies. Mol Endocrinol. 1989;3:148–156. doi: 10.1210/mend-3-1-148. [DOI] [PubMed] [Google Scholar]
- 38.Juven T, Barak Y, Zauberman A, George D L, Oren M. Wild-type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene. 1993;8:3411–3416. [PubMed] [Google Scholar]
- 39.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 40.Kubbutat M H, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 41.Landanyl M, Cha C, Lewis R, Jhanwar S C, Hacos A G, Healy J H. MDM2 gene amplification in metastatic osteosarcoma. Cancer Res. 1993;53:16–18. [PubMed] [Google Scholar]
- 42.Landers J E, Haines D S, Strauss III J F, George D L. Enhanced translation: a novel mechanism of mdm2 oncogene overexpression identified in human tumor cells. Oncogene. 1994;9:2745–2750. [PubMed] [Google Scholar]
- 43.Lazar M A. Thyroid hormone receptors: multiple forms, multiple possibilities. Endocr Rev. 1993;14:184–193. doi: 10.1210/edrv-14-2-184. [DOI] [PubMed] [Google Scholar]
- 44.Leveillard T, Wasylyk B. The MDM2 C-terminal region binds to TAFII250 and is required for MDM2 regulation of the cyclin A promoter. J Biol Chem. 1997;272:30651–30661. doi: 10.1074/jbc.272.49.30651. [DOI] [PubMed] [Google Scholar]
- 45.Marks M S, Levi B-Z, Segars J H, Driggers P H, Hirschfeld S, Nagata T, Appella E, Ozato K. H-2RIIBP expressed from a baculovirus vector binds to multiple hormone response elements. Mol Endocrinol. 1992;6:219–230. doi: 10.1210/mend.6.2.1569965. [DOI] [PubMed] [Google Scholar]
- 46.Martin K, Trouche D, Hagemeler C, Sorensen T S, Thangue N B L, Kouzarides T. Stimulation of E2F1/DP1 transcriptional activity by mdm2 oncoprotein. Nature. 1995;375:691–694. doi: 10.1038/375691a0. [DOI] [PubMed] [Google Scholar]
- 47.Momand J, Zambetti G P, Olson D C, George D, Levine A J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 48.Naar A M, Boutin J-M, Lipkin S M, Yu V C, Holloway J M, Glass C K, Rosenfeld M G. The orientation and spacing of core DNA-binding motifs dictate selective transcriptional responses to three nuclear receptors. Cell. 1991;65:1267–1279. doi: 10.1016/0092-8674(91)90021-p. [DOI] [PubMed] [Google Scholar]
- 49.Oliner J D, Kinzler K W, Meltzer P S, George D, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 50.Olson D, Marechal V, Momand J, Chen J, Romocki C, Levine A J. Identification and characterization of multiple mdm-2 proteins and mdm-2–p53 protein complexes. Oncogene. 1993;8:2353–2360. [PubMed] [Google Scholar]
- 51.Qi J-S, Desai-Yajnik V, Greene M E, Raaka B M, Samuels H H. The ligand-binding domains of the thyroid hormone/retinoid receptor gene subfamily function in vivo to mediate heterodimerization, gene silencing, and transactivation. Mol Cell Biol. 1995;15:1817–1825. doi: 10.1128/mcb.15.3.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qi J-S, Desai-Yajnik V, Yuan Y, Samuels H H. Constitutive activation of gene expression by thyroid hormone receptor results from reversal of p53-mediated repression. Mol Cell Biol. 1997;17:7195–7207. doi: 10.1128/mcb.17.12.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Samuels H H, Forman B M, Horowitz Z D, Ye Z-S. Regulation of gene expression by thyroid hormone. J Clin Invest. 1988;81:957–967. doi: 10.1172/JCI113449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sap J, deMagistris L, Stunnenberg H, Vennstrom B. A major thyroid hormone response element in the third intron of the rat growth hormone gene. EMBO J. 1990;9:887–896. doi: 10.1002/j.1460-2075.1990.tb08186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sap J, Munoz A, Schmitt J, Stunnenberg H, Vennstrom B. Repression of transcription mediated at a thyroid hormone response element by the v-erb-A oncogene product. Nature. 1989;340:242–244. doi: 10.1038/340242a0. [DOI] [PubMed] [Google Scholar]
- 56.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;61:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 57.Selmi S, Samuels H H. Thyroid hormone receptor/c-erbA and v-erbA: a single amino acid difference in the C-terminal region influences dominant negative activity and receptor dimer formation. J Biol Chem. 1991;266:11589–11593. [PubMed] [Google Scholar]
- 58.Shaulian E, Resnitzky D, Shifman O, Blandino G, Amsterdam A, Yayon A, Oren M. Induction of Mdm2 and enhancement of cell survival by bFGF. Oncogene. 1997;15:2717–2725. doi: 10.1038/sj.onc.1201453. [DOI] [PubMed] [Google Scholar]
- 59.Soussi T, deFromentel C C, May P. Structural aspects of the p53 protein in relation to gene evolution. Oncogene. 1990;5:945–952. [PubMed] [Google Scholar]
- 60.Sturzbecher H W, Brain R, Addison C, Rudge K, Remm M, Grimaldi M, Keenan E, Jenkins J R. A C-terminal α-helix plus basic region motif is the major structural determinant of p53 tetramerization. Oncogene. 1992;7:1513–1523. [PubMed] [Google Scholar]
- 61.Subler M A, Martin D W, Deb S. Overlapping domains on the p53 protein regulate its transcriptional activation and repression functions. Oncogene. 1994;9:1351–1359. [PubMed] [Google Scholar]
- 62.Umesono K, Evans R M. Determinants of target gene specificity for steroid/thyroid hormone receptors. Cell. 1989;57:1139–1146. doi: 10.1016/0092-8674(89)90051-2. [DOI] [PubMed] [Google Scholar]
- 63.Umesono K, Giguere V, Glass C K, Rosenfeld M G, Evans R M. Retinoic acid and thyroid hormone induce gene expression through a common responsive element. Nature. 1988;336:262–265. doi: 10.1038/336262a0. [DOI] [PubMed] [Google Scholar]
- 64.Umesono K, Murakami K K, Thompson C C, Evans R M. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L-H, Tsai S Y, Cook R G, Beattle W G, Tsai M-J, O’Malley B W. COUP transcription factor is a member of the steroid receptor superfamily. Nature. 1989;340:163–166. doi: 10.1038/340163a0. [DOI] [PubMed] [Google Scholar]
- 66.Wang Y, Reed M, Wang P, Stenger J E, Mayr G, Anderson M E, Schwedes J F, Tegtmeyer P. p53 domains: identification and characterization of two autonomous DNA-binding regions. Genes Dev. 1993;7:2575–2586. doi: 10.1101/gad.7.12b.2575. [DOI] [PubMed] [Google Scholar]
- 67.Wu X, Bayle J H, Olson D, Levine A J. The p53–mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 68.Xiao Z X, Chen J, Levine A J, Modjtahedi N, Xing J, Sellers W R, Livingston D M. Interaction between the retinoblastoma protein and oncoprotein mdm2. Nature. 1995;375:694–697. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 69.Zambetti G P, Bargonetti J, Walker K, Prives C, Levine A J. Wild-type p53 mediates positive regulation of gene expression through a specific DNA sequence element. Genes Dev. 1992;6:1143–1152. doi: 10.1101/gad.6.7.1143. [DOI] [PubMed] [Google Scholar]
- 70.Zauberman A, Flusberg D, Haupt V, Barak Y, Oren M. A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res. 1995;23:2584–2592. doi: 10.1093/nar/23.14.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]