Abstract
The transposable elements HeT-A and TART constitute the telomeres of Drosophila chromosomes. Both are non-long terminal repeat (LTR) retrotransposons, sharing the remarkable property of transposing only to chromosome ends. In addition, strong sequence similarity of their gag proteins indicates that these coding regions share a common ancestor. These findings led to the assumption that HeT-A and TART are closely related. However, we now find that these elements produce quite different sets of transcripts. HeT-A produces only sense-strand transcripts of the full-length element, whereas TART produces both sense and antisense full-length RNAs, with antisense transcripts in more than 10-fold excess over sense RNA. In addition, features of TART sequence organization resemble those of a subclass of non-LTR elements characterized by unequal terminal repeats. Thus, the ancestral gag sequence appears to have become incorporated in two different types of elements, possibly with different functions in the telomere. HeT-A transcripts are found in both nuclear and cytoplasmic cell fractions, consistent with roles as both mRNA and transposition template. In contrast, both sense and antisense TART transcripts are almost entirely concentrated in nuclear fractions. Also, TART open reading frame 2 probes detect a cytoplasmic mRNA for reverse transcriptase (RT), with no similarity to TART sequence 5′ or 3′ of the RT coding region. This RNA could be a processed TART transcript or the product of a “free-standing” RT gene. Either origin would be novel. The distinctive transcription patterns of both HeT-A and TART are conserved in Drosophila yakuba, despite significant sequence divergence. The conservation argues that these sets of transcripts are important to the function(s) of HeT-A and TART.
Telomeres in Drosophila melanogaster are composed of multiple copies of two non-long terminal repeat (LTR) retrotransposons, HeT-A and TART, instead of the short DNA repeats generated by telomerase on the chromosome ends of most eukaryotes (13, 24). Successive transpositions of HeT-A and TART yield arrays of repeats that are larger and more irregular than the repeats produced by telomerase. Nevertheless, these transpositions are, in some sense, equivalent to the telomere-generating action of telomerase; both telomerase and the transposition of HeT-A and TART extend chromosome ends by RNA-templated additions of specific sequences.
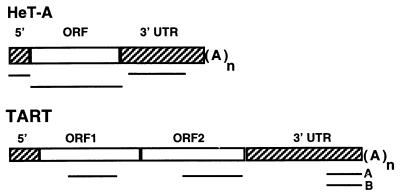
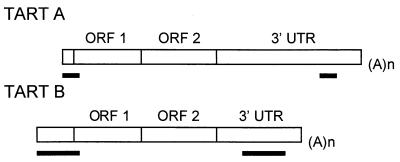
HeT-A and TART share two features that distinguish them from other known retrotransposable elements. Both transpose only to the ends of chromosomes (apparently to any chromosome end in D. melanogaster), and each contains a large segment of untranslated sequence (Fig. 1). We have recently shown that, for HeT-A, each of these distinguishing features is also conserved in related species (10), even when phylogenetic separation is great enough for the HeT-A sequences to have diverged by nearly 50%. HeT-A and TART also have some significant differences. TART encodes its own reverse transcriptase (RT); HeT-A does not. However, HeT-A seems efficient in utilizing RT from some other source because most of the documented transpositions onto broken ends have been HeT-A rather than TART (1, 2, 33). A second difference is seen in the large 3′ untranslated regions (UTRs). HeT-A elements have a distinctive pattern of A-rich segments in this region (9). Although the sequence of the HeT-A 3′ UTR diverges even faster than that of the coding region, the specific pattern of A-rich segments is conserved in other species, suggesting that it has a function (10). The 3′ UTRs of TART elements are more heterogeneous; D. melanogaster has at least two subfamilies of TART elements, A and B, whose 3′ UTRs do not cross-hybridize (33). TART 3′ ends also show much less evidence of sequence patterning than do HeT-A UTRs. However, as discussed below, TART has its own distinctive sequence features.
FIG. 1.
Diagrams of the two telomeric retrotransposons. The bars under each diagram indicate the sequences used as probes for the RNA hybridization experiments reported here. Each sequence was transcribed in vitro to yield both sense and antisense probes. TART elements can be divided into several families on the basis of their 3′ UTR sequences. The two probes marked A and B identify different subfamilies of TART and do not cross-hybridize under the conditions used here. HeT-A elements are ∼6 kb, and the TART element shown is >10 kb. (A)n, site of the poly(A) tail on RNA transposition intermediate.
What is now known about the two telomeric retrotransposons raises several questions about function and, ultimately, about evolution. Are the features shared by HeT-A and TART sufficient for either element to fulfill its telomeric function(s)? Are the differences important, so that the two elements must cooperate to form a telomere? Are the two elements derived from a single ancestor, or are they products of convergent evolution? These questions are difficult to answer directly. Every D. melanogaster stock that we have examined has multiple copies of both HeT-A and TART. We detect no pattern in the distribution of the two elements that would suggest either a functional difference or a simple way of breeding a fly with no copies of one of the elements.
To learn more about the relationship between HeT-A and TART, we have extended our studies of the transcription of these elements. We found earlier that the predominant transcript of HeT-A was, as expected for a non-LTR retrotransposon, a sense-strand copy of the entire element (6). We detected no antisense copies of this RNA. Surprisingly, we now find both sense and antisense TART RNAs. Furthermore, the antisense transcripts are more than 10-fold more abundant than sense-strand RNAs. Our analyses indicate that transcriptionally active TART elements are more variable in size and sequence than HeT-A elements; however, the distribution of variants differs among fly stocks, suggesting that these variants are functionally equivalent. The major differences in the sets of transcripts found for HeT-A and TART are not aberrations of the genetic background of a particular stock. We have surveyed several stocks and a distantly related Drosophila species and found the same general patterns.
Another unexpected finding of our studies is an RNA with similarity to the TART RT coding sequence but not to any other part of TART DNA. All the eukaryotic RTs which have been studied, with the notable exception of telomerase, have been encoded by retroelements and transcriptionally linked to the gag coding region. Thus, this new RNA is a novel RT mRNA. HeT-A is an unusual non-LTR retrotransposon in that it does not encode its own RT. It is intriguing that the RT mRNA is found in cells that are expressing HeT-A.
We have found that both subfamilies of TART for which sequence is available have one pair of remarkably conserved repeats. These repeats resemble those found in a small subgroup of non-LTR retrotransposons that have been described as having unequal, or nonidentical, terminal repeats (30). HeT-A does not have repeats of this type. This difference in sequence organization, in conjunction with the differences in transcription patterns, suggests that an ancestral gag sequence has become associated with two different subclasses of non-LTR elements to form HeT-A and TART.
MATERIALS AND METHODS
Drosophila stocks and cell lines.
Of the D. melanogaster stocks used in this work, 2057 (an isogenic y; cn bw sp stock used for the Drosophila Genome Project P1 phage library [35]) was the generous gift of E. Lozovskaya, and the stocks shown in Fig. 5 were the generous gift of A. A. Hoffmann (20). The other D. melanogaster and Drosophila yakuba stocks have been in our collection for many years. D. melanogaster cultured cells, Schneider line 2, were grown in Dulbecco’s modified Eagle’s medium with lactalbumin hydrolysate, 10% fetal bovine serum, and 10 mM nonessential amino acids. Schneider line 3 cells were grown in Schneider’s medium (Gibco BRL, Gaithersburg, Md.) plus 10% fetal bovine serum.
FIG. 5.
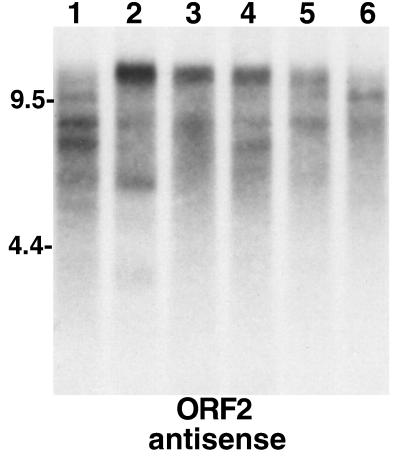
Elements yielding different-size TART transcripts can be segregated by breeding. An autoradiograph of a Northern blot of total RNA probed for TART antisense ORF2 RNA is shown. Lanes 1 to 6 contain RNA from lines selected from a mass-bred population of D. melanogaster initiated from females collected in Australia in February 1994 (20). Lines 1 to 3 were then selected on the basis of heat shock resistance, and lines 4 to 6 were parallel control lines. (Our samples, a gift of A. Hoffmann, were taken after 3 years of selection.) For our purposes the six lines illustrate the variation in TART expression patterns that can be selected from a large population of flies. Each line shows a somewhat different pattern of TART RNA. Probes for ORF1 show a similar picture. The different patterns seen clearly in the antisense strands are reflected in the sense strands (not shown), although the two strands do not give identical patterns.
Sequences for RNA probes (Fig. 1).
The following HeT-A probes were from element 23Zn-1 (GenBank accession no. U06920): 5′ UTR probe, nucleotides (nt) 982 to 1746; open reading frame (ORF) probe, nt 1746 to 4421; and 3′ UTR probe, nt 4851 to 6481. The TART ORF probes were ORF1 (accession no. U14101), nt 1801 to 3336, and ORF2 (accession no. U02279), nt 434 to 2683. The 3′ UTR sequence for TART subfamily A was amplified from genomic DNA of stock 2057, and that of subfamily B was amplified from P1 phage 13-14, using PCR primers derived from accession no. U02279 (A probe) and U14101 (B probe). The amplified sequences were cloned in Bluescript II SK (Stratagene, La Jolla, Calif.). For each probe used in this study, sense and antisense strands were transcribed from DNA fragments of identical length.
RNA extraction.
One hundred flies were homogenized in a Dounce homogenizer or 2 × 108 to 3 × 108 cultured cells were resuspended in 2 ml of buffer (100 mM NaCl, 50 mM Tris-HCl [pH 7.0], 20 mM EDTA). The solution was brought to 1% sodium dodecyl sulfate (SDS) and 50 μg of Proteinase K/ml. The homogenate was incubated at 37°C for 20 min and then extracted with phenol-chloroform and chloroform. Nucleic acids were precipitated with 3 volumes of ethanol plus 0.2 M LiCl. The pellet was resuspended in 200 μl of H2O and precipitated with an equal volume of 4 M LiCl at 4°C overnight. Finally, the pellet was resuspended in 200 μL of H2O and precipitated with 3 volumes of ethanol plus 0.3 M Na acetate.
Northern hybridization.
RNA samples (20 μg per lane) were treated with glyoxal, separated on an 0.8% agarose gel, and transferred to Hybond-N nylon membrane according to the method of Sambrook et al. (27). 32P-labeled riboprobes were transcribed in vitro from DNA fragments inserted into Bluescript II SK with T7 or T3 RNA polymerases, according to the Promega (Madison, Wis.) protocol. Hybridization was performed at 65°C in 4× SET (1× SET is 0.15 M NaCl, 0.03 M Tris-Cl [pH 7.0], and 2 mM EDTA), 5× Denhardt’s solution, 0.5% SDS, and 50 μl of salmon sperm DNA/ml (27). The filters were washed three times with 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate) plus 0.5% SDS at 65°C and then treated with 100 U of RNase T1 (Boehringer Mannheim, Indianapolis, Ind.)/ml in buffer (10 mM Tris-Cl [pH 7.5], 300 mM NaCl, 5 mM EDTA) for 1 h at 37°C, rinsed with 1× SSC–0.5% SDS, and exposed for autoradiography.
Cell fractionation.
Schneider 2 cells (2 × 108) were washed once in phosphate-buffered saline, resuspended in 8 ml of homogenization buffer (10 mM Tris HCl [pH 8], 1 mM EDTA, 10 mM NaCl, 10 mM MgCl2), and placed on ice for 10 min. The cells were disrupted in a Dounce homogenizer and centrifuged for 10 min at 800 × g. The pellet was used as the nuclear fraction. The supernatant was centrifuged again at 800 × g, and the supernatant from this spin was used as the cytoplasmic fraction. RNA was isolated from both the nuclear pellet and the second supernatant, as described above.
Nucleotide sequence accession numbers.
The sequences of the clones discussed in this study have been deposited in the GenBank database under accession no. AF072856 (A subfamily, 990-nt probe) and AF072857 (B subfamily, 950-nt probe).
RESULTS
Technical details of hybridization.
It is important to note that both HeT-A and TART are present in the genome in multiple copies with various degrees of divergence. In addition, parts of the noncoding regions of these elements are closely related to sequences in other types of repeats (5). Thus, there are many potential sources for sequences hybridizing with the probes used here, and it is important to eliminate hybridization to similar, but not identical, sequences. For this reason, all hybrids in these experiments were treated with RNase T1 before autoradiographic exposure. This treatment, adapted from RNase protection experiments, assures that the transcripts detected have strong sequence similarity to the probe, although not necessarily over their complete lengths.
HeT-A elements produce only sense-strand transcripts.
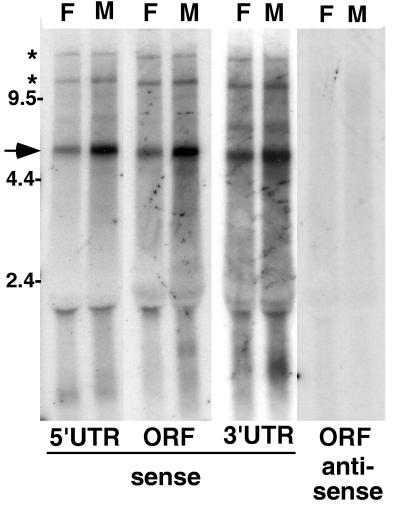
When RNA from either flies or cultured cell lines is probed with sequence from any part of HeT-A, the major species of RNA detected is a sense-strand transcript of ∼6 kb (Fig. 2). This is the size expected for a full-length transcript of the element. HeT-A elements which have been sequenced differ by multiple insertions and/or deletions (9, 25), but the net result is that the elements are approximately the same size and should be expected to migrate as the slightly broad band seen in our gels (Fig. 2; see also Fig. 7). Two minor bands of apparently larger RNAs detected by these probes are thought to be readthrough transcripts of tandem elements.
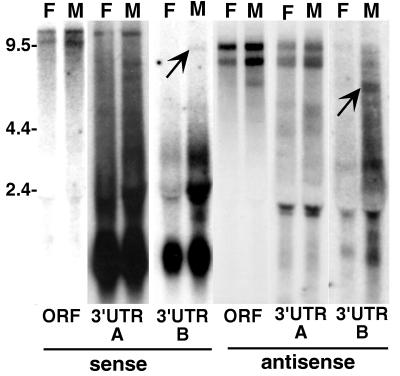
FIG. 2.
HeT-A elements produce only sense-strand transcripts. Autoradiograph of a Northern blot of total RNA from adult D. melanogaster females (lanes F) and males (lanes M) probed with HeT-A sequences. Single-strand probes detecting sense-strand RNA were used for lanes marked “sense,” while “antisense” lanes were probed with the opposite strand. See Fig. 1 for the extent of sequence in each probe. Each probe detects a prominent sense-strand RNA of ∼6 kb (arrow). Less abundant RNAs (asterisks) of approximately twice this size and greater may represent readthrough transcripts of tandem elements. (The label at ∼2 kb is due to RNA trapped by rRNA.) Each of the probes for sense-strand transcripts also detects one or more small (<2-kb) RNAs. Some of these RNAs appear to be sex specific. No antisense transcripts are detected with any of the probes. The lanes probed for antisense ORF sequences are shown. Similar results were obtained when blots were probed for 5′ and 3′ UTR antisense sequences. The marker sizes are shown in kilobases.
FIG. 7.
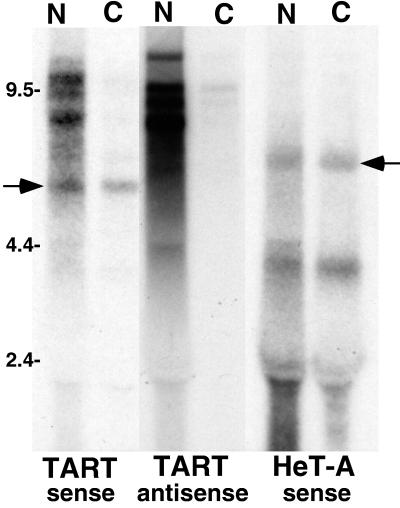
The nucleocytoplasmic distribution of TART transcripts differs from that of HeT-A transcripts. An autoradiograph of a Northern blot of RNA from cultured cells after nucleocytoplasmic fractionation is shown. N, nuclear fractions; C, cytoplasmic fractions. TART sequences were detected with ORF2 probes. HeT-A sequences were detected with a 3′ UTR probe. Full-length HeT-A RNA (right arrow) and TART RTx RNA (left arrow) are found in nuclear and cytoplasmic fractions, while the large TART transcripts (both sense and antisense) are almost entirely limited to the nuclear fraction. The 6-kb HeT-A RNA is seen with all HeT-A probes. The ∼4-kb HeT-A RNA is detected only with 3′ UTR probes and only in cultured cells; its origin is not known. The label at ∼2 kb is due to RNA trapped by rRNA.
In addition to the large sense-strand RNAs, each of these probes detect one or more much smaller (<2-kb) RNAs. These smaller RNAs are different for each probe and also show sex-specific differences in expression. We do not yet know the source of these RNAs. They may be transcribed from full-sized HeT-A elements or from truncated elements.
Although the sequence of HeT-A is transcribed into several sense-strand RNAs, no part of the sequence has been found in antisense RNA. The complete absence of hybridization seen when a Northern blot is probed for antisense RNA containing the ORF sequence (Fig. 2) is typical of the result obtained when probes from any part of the HeT-A sequence are used.
TART produces both sense and antisense transcripts of a heterogeneous array of full-length elements.
In striking contrast to HeT-A, TART produces transcripts of both strands in both flies and cultured cells. We have concentrated our study on the RNAs, both sense and antisense, that run as several distinct bands with estimated sizes between 7.5 and 12 kb (Fig. 3). The numbers, sizes, and relative intensities of these bands differ from stock to stock (compare Fig. 3 through 8). Our studies suggest that all of the RNAs in the 7.5- to 12-kb range represent full-length elements. As discussed below, TART appears to be a more heterogeneous family than HeT-A. TART elements whose 3′ UTRs differ by nearly 2 kb in length have been sequenced. The 5′ end of TART has not yet been defined. The longest published TART sequence is 10,636 nt (33), and it does not have a complete 5′ end. We do not yet have probes for the 5′ UTR, but all of the probes that we do have (ORF1, ORF2, and 3′ UTR [Fig. 3 and 4]) hybridize to the same bands in this region of the gel; thus, each species of RNA seems to have the entire sequence, even though some are shorter than 10 kb.
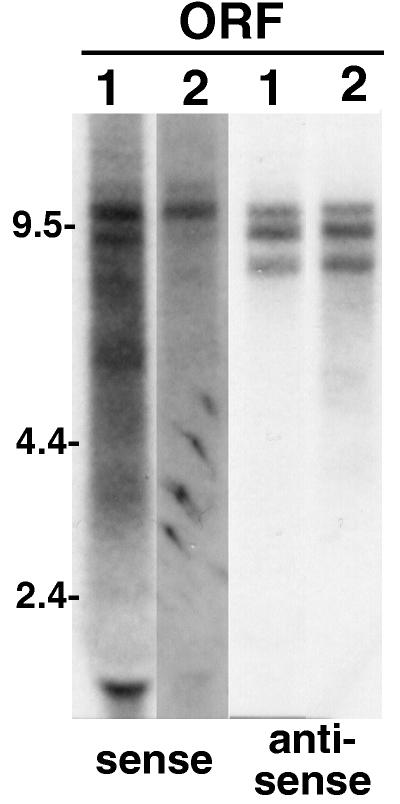
FIG. 3.
TART produces multiple full-length sequences, both sense and antisense. An autoradiograph is shown of a Northern blot of total RNA from adult Oregon R flies probed for transcripts carrying sequence of TART ORF1 (lanes 1) and ORF2 (lanes 2). Both ORFs are found in the same set of transcripts. Both sense and antisense probes detect sets of transcripts migrating between 7.5 and 12 kb; however, the sense and antisense RNAs do not exactly comigrate. The transcripts in the 7.5- to 12-kb region in Oregon R flies differ (in number and size) from those in other fly stocks and in the two cultured cell lines studied (compare Fig. 3 through 8). Note that the autoradiographic exposures shown are chosen to best display the bands produced by each probe and therefore vary from probe to probe. The detection of sense-strand RNAs requires significantly more exposure than detection of antisense transcripts. The label at ∼2 kb is due to RNA trapped by rRNA.
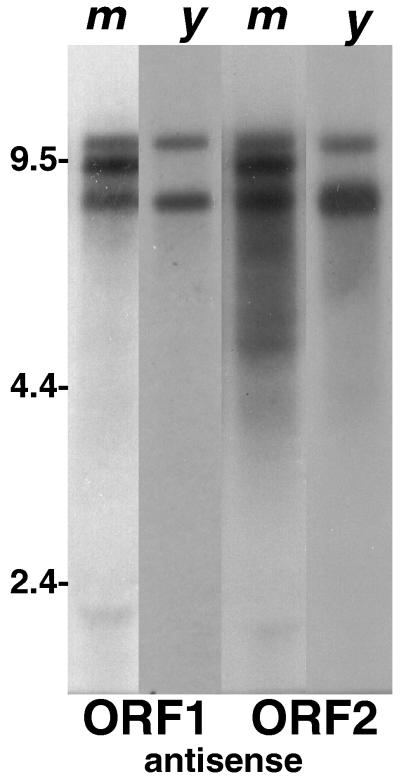
FIG. 8.
The abundant antisense transcription of TART is conserved in related Drosophila species. An autoradiograph of a Northern blot comparing TART transcripts in D. melanogaster (m) with those in D. yakuba (y) is shown. Both samples have been probed with sequences from D. melanogaster TART ORF1 and ORF2. The D. melanogaster lanes were exposed for 20 h, and the D. yakuba lanes were exposed for 44 h. Only lanes with antisense transcripts are shown. In D. yakuba, as in D. melanogaster, TART antisense transcripts are much more abundant than TART sense RNAs.
FIG. 4.
TART A- and B-subfamily elements yield full-length transcripts of several sizes. An autoradiograph of a Northern blot of total RNA from adult 2057 flies probed with TART sequences is shown. In this stock ORF probes detect two bands of sense-strand RNA migrating above the 9.5-kb marker. (Both ORF1 and ORF2 probes give this result; ORF2 is shown.) The A-subfamily 3′ UTR probe identifies the top band as TART A RNA, while the B-subfamily 3′ UTR probe labels a band corunning with the lower band but seen only in RNA from males (indicated by the arrow in the B 3′ UTR lane). Because females in this stock lack full-length B elements, we conclude that the band just above 9.5 kb hybridizing with ORF probes in females represents a third subfamily of TART found both in males and females. This third-subfamily RNA comigrates with the B transcript in males. Both 3′ UTR probes detect very abundant short sense RNAs (<2 kb). Probes for antisense ORF sequences detect two transcripts whose gel mobilities differ slightly from that of the large sense RNAs. The larger antisense transcript may comigrate with the smaller sense RNA. Both of the large antisense RNAs hybridize with A-subfamily 3′ UTR probes. The B-subfamily 3′ UTR detects a smaller and less abundant antisense RNA seen only in males with either ORF or B 3′ UTR probes (indicated by the arrow in the B 3′ UTR lane). The label at ∼2 kb is due to RNA trapped by rRNA.
Although both sense and antisense transcripts appear to contain the complete TART sequence, they do not exactly comigrate in the gel. This suggests that they are transcribed from different copies of TART. The failure to comigrate may be partly explained by their biased base compositions. Sense-strand transcripts are twice as A rich as the antisense strands, and this difference in base composition might affect gel mobility. However, if this were the entire explanation we would expect that in each stock the two sets of RNAs would have the same numbers and relative intensities of hybridizing bands, with one set somewhat offset from the other in the gel. This is not what is observed.
Like HeT-A, TART probes also hybridize to a heterogeneous set of small (<2-kb) RNAs. TART probes detect unusually large amounts of small sense-strand transcripts complementary to the A and B 3′ UTR sequences but not to other parts of the element (Fig. 4). As with the small HeT-A RNAs, it is not clear whether these small RNAs are transcribed from full-length or partial elements.
TART elements form a heterogeneous family and yield a complex set of transcripts.
Both TART (33) and HeT-A (9) can be divided into subfamilies on the basis of their 3′ UTR sequences. The intrafamily divergence of TART subfamilies is greater than that of HeT-A subfamilies. The 3′ UTRs of different TART subfamilies are unable to cross-hybridize under the conditions used in this work, while different HeT-A subfamily 3′ UTRs do cross-hybridize. TART 3′ UTRs also have greater length differences than HeT-A elements. The small number of nearly full-length elements which have been sequenced show TART 3′ UTRs of 5.1 and 3.3 kb, while HeT-A 3′ UTRs are 2.6 to 2.3 kb. These limited data are supported by the sizes of the apparent full-length transcripts of these elements. The TART sense transcripts migrate on gels as though they were 7.5 to 12 kb, while HeT-A transcripts run in a single broad band of ∼6.0 kb.
The transcription of TART subfamilies is complex and differs among fly stocks. The 3′-most sequences of A and B elements do not cross-hybridize and can be used as probes to identify transcripts of the subfamilies. Each probe hybridizes to a subset of the presumed full-length transcripts. Other transcripts hybridize with neither probe and presumably indicate the presence of additional subfamilies. A typical example of these studies is shown in Fig. 4, taken from our analysis of stock 2057. In this stock, the A probe hybridizes with the larger sense transcript, consistent with the longer 3′ UTR in the published TART A sequence. Also consistent with the size of the published TART B 3′ UTR, the B probe hybridizes with a smaller sense-strand transcript found only in males. The limitation to males is understandable because full-sized B elements are found only in DNA from males (data not shown) and must be on the Y chromosome. We assume that 2057 has transcripts of a third TART subfamily because TART ORF probes detect a sense RNA in females which is the same size as the B subfamily in males (running with the 9.5-kb marker). This RNA does not hybridize with either A or B probes in females and must comigrate with the B-subfamily transcripts in males. The relation between TART subfamily and transcript size is not always simple. Both of the major antisense transcripts in 2057 hybridize with the A probe. The B probe binds a smaller antisense transcript, again seen only in males. In Oregon R, sense and antisense B probes hybridize to RNAs of at least two different sizes (data not shown).
The sharply defined bands in the 7.5 to 12 kb region suggest that TART elements have a limited number of variant forms. To see whether this limitation was due to the small breeding populations in the laboratory, we examined TART elements in a series of lines (Fig. 5) derived from a wild-caught population, collected in Australia in 1994 and therefore geographically distant from any other stocks we have studied (20). We examined these lines after 3 years of segregation (see the legend to Fig. 5). The TART bands in these new lines show transcripts of many different sizes. The sources of these transcripts appear to be in the process of being segregated in the different lines. We conclude that there are many variants of TART in wild populations. In the same populations, HeT-A transcripts migrate in a single broad band, as do those of laboratory stocks.
In spite of their differences, the TART subfamilies may be functionally interchangeable. In stock 2057 we detect full-length B-subfamily elements only on the Y chromosome, and these elements make only a minor contribution to the transcripts in the male. In contrast, in the Oregon R stock, B-subfamily elements are abundant in both males and females and contribute strongly to the pool of transcripts in both sexes. No transcripts of A elements are detected in Oregon R flies (data not shown).
TART antisense transcripts are more than 10 times as abundant as sense transcripts.
Blots probed for sense-strand RNAs require significantly longer autoradiographic exposures than do blots probed for antisense RNA. To obtain a more quantitative estimate of the relative amounts of the two sets of transcripts, we have used equal amounts of each probe and varied autoradiographic exposures until they yielded bands of nearly equal densities for sense and antisense transcripts. These comparisons show clearly that there is a marked excess of antisense RNAs. Figure 6 shows a typical example of these comparisons. The exposure for the lanes of sense-strand RNAs is six times that for lanes of antisense RNAs. One more factor must be taken into account in estimating the relative amounts of transcripts of the two strands. The sense strands are twice as A rich as the antisense strands. The probes used for these experiments are labeled with [32P]UTP by in vitro transcription. Therefore, the specific activity of the probe depends on the specific activity of the nucleotide mix and the sequence transcribed. We have used the same nucleotide mix for both sense and antisense probes. The fragment transcribed to give the probe for the sense strand has approximately twice (1.94 times) as many A residues as does its complement, and therefore it will be labeled 1.94 times as heavily as probes for the other strand. Taken together, the probe strength and the exposure time used for Fig. 6 lead to the conclusion that antisense strands are approximately 12 times as abundant as sense strands.
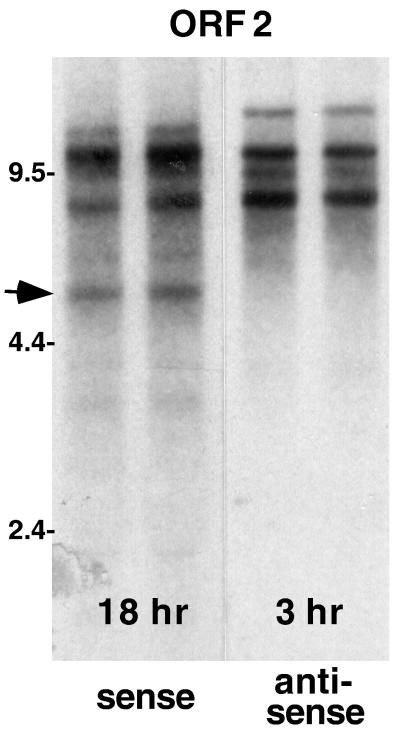
FIG. 6.
TART elements produce many more antisense- than sense-strand transcripts. An autoradiograph of a Northern blot of total RNA (replicate preparations) from the D. melanogaster Schneider 2 cell line probed with sequence from TART ORF2 is shown. Autoradiographic exposures were chosen so that probes for sense- and antisense-strand RNA produced bands of approximately equal intensity. The exposure used for sense-strand transcripts (18 h) is approximately six times that for antisense RNA (3 h). Because of the strong strand bias in A residues, the probe used to detect sense-strand RNA has incorporated nearly twice as much 32P as the complementary probe, indicating that antisense transcripts are more than 10-fold more abundant than sense RNA. ORF1 and 3′ UTR probes from TART produce similar blots, with the exception that only the ORF2 probe (containing RT sequences) detects a sense-strand transcript of 5.5 kb, which we call RTx (arrow).
TART probes detect an unusual RT mRNA.
The RT of retroviruses and retrotransposons is typically encoded by their full-length sense-strand transcripts. The enzyme is translated as part of a polyprotein linked to the product of the gag gene either by a frameshift or by readthrough of a leaky stop codon (14). Full-length TART transcripts partially fit the general pattern. They contain both ORF1, the gag gene, and ORF2, which has the RT coding sequence; however, this ORF2 appears to require an internal initiation for translation, unlike many retrotransposons (33). It was surprising, therefore, to find a sense-strand RNA that hybridized with TART ORF2 probes (Fig. 6) but not with TART sequence either 5′ or 3′ of the RT sequence (data not shown). Thus, this RNA appears to be an mRNA for RT, and we refer to it as RTx RNA. Because it hybridizes with TART at very high stringency, it might be a processed product. However, if so, it must have undergone removal of both 5′ and 3′ sequences, something that has not been reported for other retrotransposons. RTx RNA might also be the product of a “free-standing” RT gene, perhaps related to the RT of TART in much the same way that viral oncogenes are related to their cellular counterparts. No antisense copy of RTx RNA has been detected.
RTx RNA is present in both lines of cultured Drosophila cells which we maintain (Schneider 2 and Schneider 3). We have not yet detected RTx RNA in intact flies; however, all of our studies thus far have used RNA from whole flies. If RTx RNA is restricted to a subset of tissues or developmental periods, it might well have been missed in analyses of bulk RNA. The presence of RTx RNA in the two immortal cell lines studied is intriguing. The two lines had separate origins (29), and they also have different patterns of full-size TART RNA (data not shown). In spite of these differences both lines now express RTx RNA.
The intracellular distribution of TART transcripts differs from that of HeT-A transcripts.
Full-length sense-strand transcripts of typical retrotransposons serve as mRNA for the products of the gag and pol genes. The same transcripts can also serve as templates for the reverse transcription required for transposition to a new chromosomal site. For non-LTR retrotransposons, such as HeT-A and TART, this reverse transcription occurs in the nucleus, primed by chromosomal DNA at the site of integration (19). Therefore, the dual roles of full-length transcripts suggest that these transcripts will be found in both the nucleus and the cytoplasm. There is no reason to expect the intracellular distribution of HeT-A transcripts to be different from that of TART transcripts, yet the distributions of these transcripts differ markedly (Fig. 7). When we assay RNA from cell fractions, the sets of presumed full-length TART transcripts, both sense and antisense, are found almost entirely in the nuclear fraction. Therefore, these TART RNAs must be either inside the nucleus or tightly associated with it. In contrast, a significant fraction of full-length HeT-A RNA (which is sense strand only) is present in the cytoplasmic fraction. RTx RNA is also detected in the cytoplasmic fraction, consistent with the supposition that it is an mRNA.
Transcription patterns of the telomeric transposons are conserved across species.
We have begun our studies of telomeric elements in other species with those of D. yakuba, thought to be separated from D. melanogaster by 5 to 15 million years (15). Previously, we cloned HeT-A elements from D. yakuba (HeT-Ayak) and found that, although the overall sequence identity between HeT-Ayak and HeT-Amel is only 55%, the D. yakuba HeT-A elements show all the unusual features of HeT-Amel: they transpose only to telomeres, where they form long arrays; they have the unusual, long 3′ UTR; and they lack RT coding sequences (9). Our studies now show that, like HeT-Amel, HeT-Ayak produces only sense-strand RNA and its full-length transcripts are relatively homogeneous in size, migrating as a single rather broad band in our gels (data not shown).
TART elements have not yet been cloned from D. yakuba; however preliminary evidence suggests that D. yakuba has its own TART elements. Southern blots of D. yakuba DNA probed with TART coding sequences from D. melanogaster show multiple faint bands of hybridizing restriction fragments (data not shown). This result would be expected if D. yakuba has a family of TART elements whose sequence has diverged from the D. melanogaster TART sequences, much as the HeT-A elements have diverged. Because D. melanogaster TART probes detect TART sequences in D. yakuba DNA, we have used these D. melanogaster probes to study D. yakuba TART transcripts. These probes detect multiple transcripts migrating approximately with full-length TART transcripts from D. melanogaster (Fig. 8). ORF1 and ORF2 probes hybridize to all of the bands in this set, as expected from our studies of D. melanogaster. Significantly, TART antisense transcripts are much more abundant than sense transcripts in D. yakuba, as they are in D. melanogaster. Northern blots of D. yakuba RNA require much longer autoradiographic exposures than blots of D. melanogaster RNA, presumably reflecting the lower level of hybridization with the D. melanogaster probe. Although we can obtain reasonable pictures of the blots showing antisense transcripts, the lower levels of sense-strand RNA yield pictures that are very difficult to reproduce and therefore are not shown here.
Comparisons of the HeT-A and TART gag coding sequences give evidence of a common ancestor.
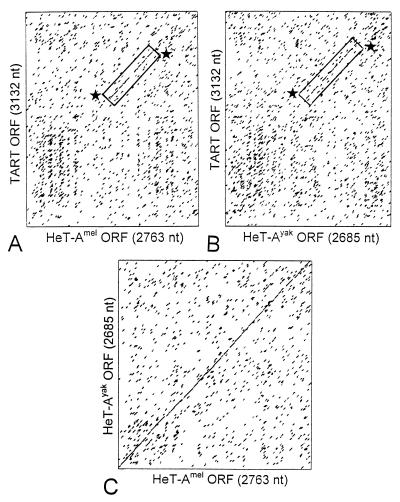
The coding region of HeT-A appears to be closely related to ORF1 of TART. Both sequences are clearly related to the ORF1 sequences of several insect non-LTR elements, but HeT-A and TART are most closely related to each other (25). Dot matrix analyses show that the 3′ half of the D. melanogaster TARTmel ORF1 has a high level of nucleotide identity with the equivalent part of the coding regions of HeT-A elements from both D. melanogaster and D. yakuba, even though the coding regions of the two HeT-A elements are only ∼65% identical to each other at the nucleotide level (Fig. 9).
FIG. 9.
TART ORF1 has significant sequence similarity with the HeT-A coding region. (A and B) Dot matrix comparisons of the nucleotide sequence of TART ORF1 from D. melanogaster with sequences of the entire coding regions of HeT-A elements from D. melanogaster (A) and D. yakuba (B). (C) The two HeT-A elements have 65% sequence identity distributed evenly over the coding region. HeT-Amel has 38% identity with TART, and HeT-Ayak has 41% identity with TART. For both HeT-A elements, the regions of identity with TART are strongest in the C-terminal end of the coding region (boxed and marked by stars).
When comparisons are based on amino acid sequences, TARTmel ORF1 has similarities throughout the HeT-A coding region (25) and is as similar to the D. yakuba HeT-A as to the D. melanogaster HeT-A. HeT-A elements of D. melanogaster and D. yakuba show 64% amino acid sequence similarity (identities plus conservative replacements). A 707-amino-acid stretch of the TARTmel ORF1 product shows 50% amino acid sequence similarity with HeT-Amel and 52% similarity with HeT-Ayak. Furthermore, two-thirds of the similar amino acids in each comparison between TART and HeT-A are positions that are also conserved when the two HeT-A products are compared. (We note that HeT-A elements within D. melanogaster can vary both in sequence and in length [3, 25]. Therefore, these percentages vary with the particular element used in each comparison. The numbers given are representative.) Thus, the sequences that are conserved in the HeT-A ORF show a strong tendency to be conserved in TART ORF1, a situation that suggests that they are evolving from a common ancestor.
HeT-A and TART belong to different subfamilies of non-LTR retrotransposons.
Our sequence analyses reveal that TART displays several features which it does not share with Het-A; instead, these features characterize a small subgroup of non-LTR transposons, elements with unusual terminal repeats (see Fig. 10). A striking feature of non-LTR retrotransposons is their tendency to be truncated at the 5′ end. This tendency led to the suggestion that these elements are reverse transcribed into their site of transposition, with variable failure of reverse transcription to complete the copy of the 5′ end (31). This basic mechanism of transposition has been elegantly confirmed for the R2Bm element (19). HeT-A and TART are also frequently truncated at the 5′ end, although the extent to which this is due to failure of reverse transcription as opposed to erosion of the chromosome end is unclear. The complete 5′ end of HeT-A has been defined by the analysis of several elements that had identical junctions with an upstream element, a 5′ UTR, and a complete coding region (7).
FIG. 10.
Diagrams of TART A and TART B showing locations of perfect repeat sequences (indicated by the black bars under the elements). Both elements are truncated at the 5′ end. The TART B diagram is based on data from reference 33;0. (A)n, site of the poly(A) tail on RNA transposition intermediate.
No junctions between apparently complete TART elements and other sequences have been found. The longest available TART sequence is that of a B element extending 962 bp 5′ of the start of ORF1. Sheen and Levis (33) noted that this element had a perfectly identical repeat of 1,046 bp (Fig. 10). The 5′ copy of this repeat extended from the start of the sequence to bp 84 of ORF1. The 3′ copy of this repeat was in the 3′ UTR and ended 560 bp from the 3′ end of the element. We have recently cloned a junction between two TART A elements. In this junction, the poly(A) tail of the distal element is joined to the proximal element 33 bp before the start of ORF1 (26a). This sequence showed that the A subfamily also has two perfect repeats, which differ in sequence from the B repeats but have similar positions in the element. The 5′ repeat of the A element extends from the junction 117 bp into ORF1, and the 3′ repeat ends 374 bp from the poly(A) end of the element. Because of the 5′ truncation of the cloned element, the full size of the repeat is unknown. In view of the absolute sequence conservation between repeats within each element, the divergence between elements is striking, the more so because most of the sequence that we have to compare is within ORF1. The 84 bp of B repeat that lie in ORF1 have only 71% identity with the first 84 bp of the A repeat lying in ORF1. The amino acid sequence encoded by these nucleotides has only 70% identity.
The complete sequence identity of the repeats within TART elements contrasts sharply with the divergence between TART elements. Similar conserved repeats have been reported for other non-LTR retrotransposons, most notably DER from Dictyostelium discoideum (30) and TOC1 from Chlamydomonas reinhardtii (11). In these elements a 5′ repeat is completely identical with a 3′ repeat that, like the TART repeats, is some distance from the 3′ end of the element. These repeats have been called unequal terminal repeats because the terminal 3′ sequences have no match at the 5′ end; the region of identity is in the subterminal sequence. The identity of the repeats suggests that they are replicated from the same template, as are retroviral LTRs (12). Because of this resemblance, Day et al. (11) have proposed that these elements use a variation of the retroviral integration mechanism.
DISCUSSION
The Drosophila telomeric transposons present intriguing evolutionary questions.
Drosophila telomeres are a striking exception to the almost-universal eukaryotic telomere, which is maintained by the enzyme telomerase. Drosophila must have shared ancestors with organisms that now have telomerase. Therefore, it is interesting to speculate on the evolution of the Drosophila retrotransposon-type telomere. We have suggested that telomere retrotransposons have evolved from components of telomerase (24, 26). This hypothesis has recently received support from evidence that the catalytic subunit of telomerase resembles the RTs of non-LTR retrotransposons (18, 21, 22). This finding is particularly intriguing, since both HeT-A and TART are non-LTR retrotransposons. Nevertheless, there is no compelling evidence to eliminate other possible explanations for the Drosophila telomere, such as the “domestication” of existing, parasitic retrotransposons to replace telomerase in Drosophila or the evolution of telomerase from retrotransposons, with Drosophila demonstrating the ancestral mechanism.
Regardless of the explanation for the evolutionary origin of Drosophila telomeres, the question of the relationship between HeT-A and TART is important. Are these two elements simply variants, fulfilling a function that is so simple that it places few constraints on the form of the element? For example, if the only role of telomere sequences is to protect genes by providing extra DNA on the end of the chromosome, any sequence that can be added to the chromosome should be adequate. If so, the differences between HeT-A and TART would be entirely irrelevant, reflecting only chance differences. On the other hand, these differences might be evidence that these two elements have evolved to fulfill different roles in the cell. For example, both sequences might be required to form an effective telomere. One obvious possibility is that TART provides the RT for HeT-A, which does not encode the enzyme. This raises the question of why HeT-A exists at all, if TART owns the RT. The question is even more puzzling because HeT-A is significantly more abundant than TART in the genome. If TART supplies the RT, it would seem that HeT-A must also have something to contribute. An alternative explanation for the two elements is that one or both of them, or an encoded protein, might have a second task in the cell, a task not necessarily related to forming the telomere. This possibility is raised by studies of Saccharomyces cerevisiae, where multiple copies of genes adaptive for certain environments are associated with chromosome ends, suggesting that the association with telomeres facilitates their amplification (23). The functional importance of the yeast genes is easy to see because we understand the gene products. The roles of the transposon products are more cryptic, but they too might be exploiting the ease with which gene amplification can occur at telomeres.
HeT-A and TART belong to different subgroups of non-LTR retrotransposons.
The studies reported here offer a new insight into the relationship between HeT-A and TART. TART displays several features which it does not share with HeT-A but which are found in a different subgroup of non-LTR retrotransposons. This evidence that the two elements belong to different subgroups suggests that the evolutions of HeT-A and TART converged by acquiring related gag proteins, which may be responsible for their telomere localization.
The features that TART shares with the subgroup of non-LTR retrotransposons are not limited to the 5′ and subterminal 3′ perfect repeats. In addition, the 5′ repeat of TART elements extends some distance into ORF1, another unusual characteristic of this group (30). Our preliminary studies indicate that the placement of TART promoters differs from that of HeT-A promoters and instead may resemble that of the promoters described for the imperfect repeat element, DER (30). In earlier work we showed that the promoter of HeT-A is in an unusual position (8): it is at the 3′ end of the element and serves to promote transcription of its downstream neighbor. We have begun a similar study on TART promoter activity (13a) and have found that the region of TART equivalent to the HeT-A promoter, the 3′-most 900 bp, of both TART A and TART B has little, if any, sense-strand promoter activity. However, this region of both A and B does have good promoter activity for antisense RNA and therefore resembles the antisense promoter of DER. DER has been shown to have a promoter for sense-strand transcription in the 5′ UTR, just upstream of the 5′ repeat region (30). If the similarity between TART and DER is complete, the TART sense-strand promoter should be at the 5′ end. We are attempting to identify this 5′ end to test this possibility.
Antisense transcripts have been reported for very few non-LTR retrotransposons.
From their structure, it would appear that a sense transcript of non-LTR retrotransposons should contain all of the information necessary both for translation of element-specific proteins and for templating of the DNA copy when the element transposes. HeT-A conforms to this expectation. We have studied transcripts in both males and females of several stocks of D. melanogaster, in two immortal cell lines, and in one stock of D. yakuba. In every case we find only sense-strand transcripts of HeT-A. In contrast, parallel studies of TART show, in every case, a large excess of antisense transcripts. Since these studies detect steady-state RNA, the excess could be due to more active promoters for the antisense RNA, to a slower turnover of the RNA, or both.
It is not obvious why TART produces antisense RNA when HeT-A does not. One possible use for antisense RNA could be to allow an element to encode additional proteins; however, for several reasons, it seems unlikely that TART antisense RNA provides extra coding capacity. First, the antisense strand has only very small ORFs, no larger than those on the antisense strand of HeT-A, which is not transcribed. Second, most, if not all, TART RNA remained with the nucleus in our cell fractionation experiments. Thus, it is not likely to be serving as a template for protein synthesis; however, we cannot yet rule out the possibility that TART RNA is on polysomes that preferentially fractionate with the nucleus.
Several reports of antisense RNAs for retrotransposons have suggested that these RNAs were accidental products of readthrough transcription. Mammalian LINE-1 elements transpose to many chromosomal sites and thus can come under the control of irrelevant promoters. These irrelevant promoters probably produce many of the heterogeneous transcripts detected by LINE-1 hybridization, while transcripts capable of transposition are produced by a small subset of the elements (28, 34). Although irrelevant promoters could produce the TART antisense transcripts, this explanation seems very unlikely. TART elements are intermingled with HeT-A elements in long head-to-tail arrays on telomeres. The only promoters for which we have any evidence are the TART antisense promoters mentioned earlier, which would yield bona fide TART RNA, and HeT-A promoters, which in principle could drive TART transcription but should yield only sense transcripts.
Other retrotransposons which produce defined sets of antisense transcripts (16, 30) suggest additional roles for TART antisense RNA. The Drosophila retrotransposon, micropia, produces an antisense copy of its RT and RNase H coding sequence in the testis but not in other tissues. It has been suggested that the testis-specific transcript blocks transposition of micropia in the male germ line (16). This seems an unlikely model for TART antisense RNA. If antisense RNA acts as an inhibitor, it should block expression from TART in all tissues, because antisense RNA appears to be present wherever TART sense RNA is found. Antisense depression of transcription might explain why TART is much less abundant than HeT-A in the genome.
Another example of a role for antisense RNA is offered by the Dictyostelium element, DRE, mentioned earlier. The DRE sense strand is not a complete copy of the element; it lacks the 3′-most sequence. This 3′-most information is found on an antisense RNA, which is also an incomplete copy of the element but lacks sequence from the 5′ end of the element. It has been proposed that, by pairing of the overlapping regions, the two strands form a structure in which both strands can be extended by RT to produce a mosaic RNA-DNA hybrid with the complete DRE sequence. This hybrid could then insert into the new chromosomal site (30). A similar role in transposition is an intriguing possibility for the TART antisense RNA. However, the invariant orientation of TART elements on chromosome ends is more easily explained by a transposition mechanism thought to be more typical of non-LTR retrotransposons. The mechanism has been demonstrated for the R2Bm retrotransposon: RNA is reverse transcribed at the site of integration, primed on the 3′ OH of the chromosomal DNA (19). The polar orientation seen for TART is consistent with its reverse transcription being primed off the chromosome end. Thus, any model for integration of a double-stranded TART requires a mechanism for maintaining orientation on the chromosome. Clearly, many questions about the abundant antisense TART transcripts remain.
A novel RNA encoding RT.
Our Northern blots show a sense-strand RNA that hybridizes at very high stringency with the probe for TART ORF2 (the RT coding sequence) but does not hybridize with TART sequences either 5′ or 3′ of the RT gene. Because this RNA appears to encode RT, we have named it RTx RNA. Our data are consistent with three possible explanations for the origin of RTx RNA: (i) it could be transcribed from TART with posttranscriptional processing or altered transcriptional initiation and termination; (ii) it could be the transcript of a new retrotransposon; or (iii) it could be the mRNA product of a cellular gene, as opposed to a retroelement gene. (We refer to this as the “free-standing gene” alternative.)
Each of the possible origins for RTx is interesting. Alternative transcripts of non-LTR retrotransposons are rare. The Neurospora non-LTR retrotransposon, Tad, produces an alternative transcript that would encode RT activity (32), although it does not require the 3′ processing needed to derive RTx from TART. If RTx represents a new retrotransposon, that retrotransposon must be a telomere-specific element, like HeT-A and TART, because the TART ORF2 probe hybridized in situ only to telomeres (reference 17 and our unpublished observations). None of the analyses of cloned telomere DNA in our laboratory or others have identified an additional telomere transposon.
The third alternative, mRNA from a free-standing gene, is consistent with our preferred hypothesis for the evolutionary origin of HeT-A (see above). This hypothesis suggests that the Drosophila gene for the catalytic subunit of telomerase is still present in the genome and that it produces the RT for HeT-A (24, 26). Thus, Drosophila may have a cellular gene capable of providing the RT for the non-LTR element, HeT-A. If RTx RNA proves to be mRNA from a cellular gene, it will identify the first cellular RT gene. It may also provide insight into the evolution of retrotransposon-type telomeres.
It is interesting that we have detected RTx RNA in the two immortal cell lines that we work with. These two lines have separate origins and express different sets of TART RNAs. Therefore, the expression of RTx RNA must have occurred independently in the two lines. Bodnar et al. (4) have expressed the catalytic subunit of human telomerase in normal human cells and have shown that the life span is extended (at this time, apparently indefinitely) while the cells still maintain the normal karyotype. Thus it is possible that the expression of RTx RNA is associated with the immortality of the Drosophila cell lines.
ACKNOWLEDGMENTS
We thank E. Lozovskaya and A. A. Hoffmann for Drosophila stocks, D. Nurminsky for assistance in screening the P1 phage library, and Ky Lowenhaupt for comments on the manuscript.
This work was supported by grants from the National Institutes of Health (GM 50315 and GM57006).
REFERENCES
- 1.Biessmann H, Mason J M, Ferry K, d’Hulst M, Valgeirsdottir K, Traverse K L, Pardue M-L. Addition of telomere-associated HeT DNA sequences “heals” broken chromosome ends in Drosophila. Cell. 1990;61:663–673. doi: 10.1016/0092-8674(90)90478-w. [DOI] [PubMed] [Google Scholar]
- 2.Biessmann H, Champion L E, O’Hair M, Ikenaga K, Kasravi B, Mason J M. Frequent transpositions of Drosophila melanogaster HeT-A elements to receding chromosome ends. EMBO J. 1992;11:4459–4469. doi: 10.1002/j.1460-2075.1992.tb05547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biessmann H, Kasravi B, Bui T, Fujiwara G, Champion L E, Mason J M. Comparison of two active Het-A retroposons of Drosophila melanogaster. Chromosoma. 1994;103:90–98. doi: 10.1007/BF00352317. [DOI] [PubMed] [Google Scholar]
- 4.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C-P, Morin G B, Harley C B, Shay J W, Lichsteiner S, Wright W E. Extension of life span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 5.Danilevskaya O N, Lofsky A, Kurenova E V, Pardue M L. The Y chromosome of Drosophila melanogaster contains a distinctive subclass of HeT-A-related repeats. Genetics. 1993;134:531–543. doi: 10.1093/genetics/134.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danilevskaya O N, Slot F, Pavlova M, Pardue M L. Structure of the Drosophila HeT-A transposon: a retrotransposon-like element forming telomeres. Chromosoma. 1994;103:215–224. doi: 10.1007/BF00368015. [DOI] [PubMed] [Google Scholar]
- 7.Danilevskaya O N, Slot F, Traverse K L, Hogan N C, Pardue M L. The Drosophila telomere transposon HeT-A produces a transcript with tightly bound protein. Proc Natl Acad Sci USA. 1994;91:6679–6682. doi: 10.1073/pnas.91.14.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danilevskaya O N, Arkhipova I R, Traverse K L, Pardue M L. Promoting in tandem: the promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell. 1997;86:647–655. doi: 10.1016/s0092-8674(00)81907-8. [DOI] [PubMed] [Google Scholar]
- 9.Danilevskaya O N, Lowenhaupt K, Pardue M L. Conserved subfamilies of the Drosophila HeT-A telomere-specific retrotransposon. Genetics. 1998;148:233–242. doi: 10.1093/genetics/148.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danilevskaya O N, Tan C, Wong C J, Alibhai M, Pardue M L. Unusual features of the Drosophila melanogaster telomere transposable element HeT-A are conserved in D. yakuba telomere elements. Proc Natl Acad Sci USA. 1998;95:3770–3775. doi: 10.1073/pnas.95.7.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day A, Schirmer-Rahire M, Kuchka M R, Mayfield S P, Rochaix J-D. A transposon with an unusual arrangement of long terminal repeats in the green alga Chlamydomonas reinhardtii. EMBO J. 1988;7:1967–1972. doi: 10.1002/j.1460-2075.1988.tb03029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilboa E, Mitra S W, Goff S, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 13.Greider C W. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 13a.Haynes, K., O. N. Danilevskaya, and M. L. Pardue. Unpublished data.
- 14.Jacks T. Translational suppression in gene expression in retroviruses and retrotransposons. Curr Top Microbiol Immunol. 1990;157:93–124. doi: 10.1007/978-3-642-75218-6_4. [DOI] [PubMed] [Google Scholar]
- 15.Lachaise D, Cariou M-L, David J R, Lemeunier F, Tsacas L, Ashburner M. Historical biogeography of the Drosophila melanogaster species subgroup. Evol Biol. 1988;22:159–225. [Google Scholar]
- 16.Lankenau S, Corces V G, Lankenau D-H. The Drosophila micropia retrotransposon encodes a testis-specific antisense RNA complementary to reverse transcriptase. Mol Cell Biol. 1994;14:1764–1775. doi: 10.1128/mcb.14.3.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levis R W, Ganesan R, Houtchens K, Tolar L A, Sheen F-M. Transposons in place of telomere repeats at a Drosophila telomere. Cell. 1993;75:1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- 18.Ligner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 19.Luan D D, Korman M H, Jakubczak J L, Eickbush T H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 20.McColl G, Hoffmann A A, McKechnie S W. Response of two heat shock genes to selection for knock-down heat resistance in D. melanogaster. Genetics. 1996;143:1615–1627. doi: 10.1093/genetics/143.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyerson M, Counter C M, Eaton E N, Ellison L W, Steiner P, Caddle S D, Ziaugra L, Beijersbergen R L, Davidoff M J, Liu Q, Bacchetti S, Haber D A, Weinberg R A. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Ligner J, Harley C B, Cech T R. Telomerase catalytic subunit homologues from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 23.Ness F, Aigle M. RTM1: a member of a new family of telomeric repeated genes in yeast. Genetics. 1995;140:945–956. doi: 10.1093/genetics/140.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pardue M-L, Danilevskaya O N, Lowenhaupt K, Slot F, Traverse K L. Drosophila telomeres: new views on chromosome evolution. Trends Genet. 1996;12:48–52. doi: 10.1016/0168-9525(96)81399-0. [DOI] [PubMed] [Google Scholar]
- 25.Pardue M-L, Danilevskaya O N, Lowenhaupt K, Wong J, Erby K. The “gag” coding region of the Drosophila telomeric retrotransposon, HeT-A, has an internal frame shift and a length polymorphic region. J Mol Evol. 1996;43:572–583. doi: 10.1007/BF02202105. [DOI] [PubMed] [Google Scholar]
- 26.Pardue M-L, Danilevskaya O N, Traverse K L, Lowenhaupt K. Evolutionary links between telomeres and transposable elements. Genetica. 1997;100:73–84. [PubMed] [Google Scholar]
- 26a.Rodriguez, J., O. N. Danilevskaya, and M. L. Pardue. Unpublished data.
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 28.Schichman S A, Severynse D M, Edgell M H, Hutchison C A., III Strand-specific LINES-1 transcription in mouse F9 cells originates from the youngest phylogenic subgroup of LINE-1 elements. J Mol Biol. 1992;224:559–574. doi: 10.1016/0022-2836(92)90544-t. [DOI] [PubMed] [Google Scholar]
- 29.Schneider I. Cell lines derived from late embryonic stages of D. melanogaster. J Embryol Exp Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- 30.Schumann G, Zündorf I, Hoffmann J, Marschalek R, Dingermann T. Internally located and oppositely oriented polymerase II promoters direct convergent transcription of a LINE-like retroelement, the Dictyostelium repetitive element from Dictyostelium discoideum. Mol Cell Biol. 1994;14:3074–3084. doi: 10.1128/mcb.14.5.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarz-Sommer Z, Leclercq L, Gobel E, Saedler H. Cin4, an insert altering the structure of the AI gene in Zea mays, exhibits properties of non-viral retrotransposons. EMBO J. 1987;6:3873–3880. doi: 10.1002/j.1460-2075.1987.tb02727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sewell E, Kinsey J A. Tad, a Neurospora LINE-like retrotransposon, exhibits a complex pattern of transcription. Mol Gen Genet. 1996;252:137–145. [PubMed] [Google Scholar]
- 33.Sheen F-M, Levis R W. Transposition of the LINE-like retrotransposon, TART, to Drosophila chromosome termini. Proc Natl Acad Sci USA. 1994;91:12510–12514. doi: 10.1073/pnas.91.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skowronski J, Fanning T G, Singer M F. Unit-length LINE-1 transcripts in human teratocarcinoma cells. Mol Cell Biol. 1988;8:1385–1397. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smoller D A, Petrov D, Hartl D L. Characterization of bacteriophage P1 library containing inserts of Drosophila DNA of 75–100 kilobase pairs. Chromosoma. 1991;100:487–494. doi: 10.1007/BF00352199. [DOI] [PubMed] [Google Scholar]