Abstract
Identification of the causative pathogen is required to optimize the effective therapy in infective endocarditis (IE). The aim of this study was to assess a 16S rDNA PCR to identify bacteria from heart valve tissues and to evaluate its usefulness as a complement to blood and removed valves cultures. A total of 266 patients diagnosed with IE from January 2015 to December 2019 were evaluated. Results between 16S rDNA PCR from heart valve tissues were compared with microbiological cultures. Blood cultures were positive in 83.5% of patients diagnosed with IE, while 39.6% and 71.8% of the evaluated heart valve samples were positive by culture and 16S rDNA PCR, respectively. For 32 (12%) patients, 16S rDNA tissue PCR provided valuable information supporting the results of blood cultures in the case of bacteria characteristic from the skin microbiota. Additionally, a microorganism was identified by using 16S rDNA PCR in 36% of blood culture-negative cases. The present study reveals that molecular diagnosis using 16S rDNA tissue PCR provides complementary information for the diagnosis of IE, and it should be recommended in surgical endocarditis, especially when blood cultures are negative.
Keywords: infective endocarditis, 16S rDNA PCR, blood culture, valve heart culture, molecular diagnosis
1. Introduction
Infective endocarditis (IE) is a serious disease associated with the infection of a native or prosthetic heart valve or an implanted cardiac device [1,2]. Appropriate recognition and management of this infection remain a clinical challenge. The outcomes of affected patients are still poor, with mortality rates of up to 30% [3]. Currently, blood and heart valve tissue cultures are the gold standards for the identification of the causative pathogen, which is important for the initiation of an optimal targeted antimicrobial therapy. However, up to 10% of all IE are associated with negative blood cultures [4,5]. These are related to cases in which antibiotic therapy is initiated before sampling or with IE caused by intracellular, fastidious, slow-growing, and/or unculturable microorganisms such as Coxiella burnetii or Tropheryma whipplei [6]. Consequently, to achieve a precise diagnosis, current guidelines recommend alternative methods for pathogen detection [7].
In recent years, molecular detection methods have become an important part of IE diagnosis and represent an alternative approach, especially when the culture is negative. Amplification through polymerase chain reaction (PCR) and further sequencing of variable regions of the 16S rDNA gene is a widely accepted method for identifying bacteria [8,9]. There is growing evidence supporting the use of this approach for the detection of bacteria from removed heart valves with sensitivities around 95% [10]. Therefore, 16S rDNA valve PCR has been established as an independent, specific, and rapid method to assist pathogen detection in cases of blood and valve negative cultures and can also be useful to confirm previously detected microorganisms [11].
The aim of this study was to assess a 16S rDNA PCR method to identify bacteria from heart valve tissues in a large series of patients and to evaluate the usefulness of this technique as a complement to the culture of the valve and blood for the etiological diagnosis of IE.
2. Materials and Methods
2.1. Patients
All patients admitted to a tertiary hospital in northern Spain (Hospital Universitario Central de Asturias, Oviedo, Spain) with a diagnosis of IE from January 2015 to December 2019 were included in the study. Our institution is a 1039-bed university hospital and has a cardiovascular surgery department that provides patient care to a population of 1 million people. The diagnoses of IE were established based on clinical, microbiological, and echocardiographic findings according to the modified Duke’s criteria [12]. Patients’ clinical records were retrospectively reviewed using a standardized data collection sheet that included clinical and microbiological information such as location of IE, type of valve tissue, and blood and valve culture results.
2.2. Microbiological Cultures
Microbiology tests, including microbiological culture and bacterial identification, were carried out at the hospital Clinical Microbiology Unit. Blood cultures were performed on each patient and were inoculated in two separate sets of Standard Aerobic (SA) and Standard Anaerobic (SN) BacT/ALERT®® blood culture bottles (bioMérieux Inc., Durham, NC, USA) which were processed in a BacT/ALERT®® VIRTUO™ machine (VIRTUO™, bioMérieux Inc., Hazelwood, MO, USA). Blood cultures bottles were incubated 15 days, and, if positive, standard subcultures were performed according to IDSA guidelines [13]. Bacterial identification of growing colonies was performed by MALDI-TOF MS (Bruker Daltonik, Bremen, Germany). Heart valve samples (native and prosthetic), pacemaker catheter electrodes, and/or heart tissue as vegetations or perivalvular abscess were collected under sterile conditions in patients who underwent surgery and were processed for bacterial and fungal culture according to IDSA guidelines [13].
2.3. 16S rDNA PCR Assays
Additionally, heart samples were evaluated by a 16S rDNA-based PCR and further sequencing except for those cases in which there was an unequivocal diagnosis by culture. Bacterial DNA was extracted using MagCore (RBC Bioscience, New Taipei City, Taiwan). A PCR amplification of the 16S RNA-encoding gene and subsequent sequencing was performed as previously described [14]. Sequence results were compared with data from two different databases (Blast—Basic Local Alignment Search Tool and leBIBI—Quick Bioinformatic Phylogeny of Prokaryotes—v 1.1).
2.4. Data Analysis
Data were analyzed using SPSS software (IBM SPSS Statistics 19). Descriptive statistics were computed for all study variables. Differences between the length antimicrobial treatment groups were performed using Student’s t-test. For all comparisons, a p value of less than 0.05 was considered significant.
2.5. Ethics
Ethical approval for the study was provided by the Research Ethics Committee of the Principality of Asturias (reference CEImPA-2018-210, in 14 August 2018).
3. Results
3.1. Patient Demographics and Clinical Features
A total of 266 patients with a diagnosis of IE according to the modified Duke’s criteria were included in the study. Demographic characteristics and clinical features are displayed in Table 1. Most patients were male (n = 171, 64.3%) with a mean age of 68 years (range 6–95). Native valve endocarditis accounted for 140 (52.6%) episodes, while prosthetic valve endocarditis accounted for 115 (43.2%). All patients were treated with antibiotics before valve surgery. Valve replacement was carried out in 154 patients.
Table 1.
Clinical features of patients with infective endocarditis included in the study.
| Demographics | n (%) |
|---|---|
| Male sex | 171 (64.3) |
| Age mean ± SD (years) | 68 ± 13 |
| Medical history | |
| Native valve | 140 (52.6) |
| Prosthetic valve | 115 (43.2) |
| Intracardiac device | 18 (6.8) |
| Previous endocarditis | 20 (7.5) |
| Location | |
| Aortic | 138 (51.9) |
| Mitral | 65 (24.4) |
| Pulmonary | 2 (0.8) |
| Tricuspid | 3 (1.1) |
| Pacemaker | 15 (5.6) |
| Multiple valves | 40 (15.1) |
| Others | 3 (1.1) |
| Clinical course | |
| Surgical treatment | 154 (57.9) |
| Time of antimicrobial treatment before surgery (mean ± SD, days) | 10.3 ± 12.8 |
| Time of antimicrobial treatment after surgery (mean ± SD, days) * | 42.3 ± 19.5 |
| Time of total antimicrobial treatment (mean ± SD, days) * | 42.7 ± 22.5 |
| Deaths | 72 (27.1) |
SD, standard desviation. * Patients on suppressive treatment for more than 90 days have been removed.
3.2. Previous Antimicrobial Therapy and Its Effect on Microbiological Results
The mean duration of antimicrobial treatment before surgery was 10.3 days (range: 1–105). In order to analyze the potential effect of antimicrobial therapy on microbiological results, results from heart valves and other tissues were analyzed in relation to the length of antimicrobial treatment before surgery (Figure 1). 16S rDNA valve PCR was positive in 71.4% of patients who received antibiotics for ≥10 days before surgery, while only 24.1% of the same patients had a positive valve culture. The mean duration of preoperative treatment was similar in patients with negative and positive PCR (10.7 ± SD and 11.5 ± SD days, respectively, p = 0.8). However, the mean duration of antibiotic treatment before surgery was longer in patients with negative valve culture (11.2 ± SD and 6.7 ± SD days for those with negative and positive valve culture, respectively, p = 0.005). These data show that bacterial DNA could persist during antibiotic treatment in infected valve tissue in contrast to the valve culture, wherein the latter’s results are affected by the length of the antimicrobial therapy.
Figure 1.
Antimicrobial therapy duration before surgery and its relatedness with valve culture and 16S rDNA PCR results. Bars represent the median of days.
3.3. Microbiological Methods Results
A causative microorganism was identified in most of the patients. No microorganism was detected neither by blood culture, 16S rDNA PCR, or culture of valvular specimen for only 18 subjects. However, all these episodes were clinically classified as possible IE based on clinical and echocardiographic findings according to the modified Duke’s criteria [12].
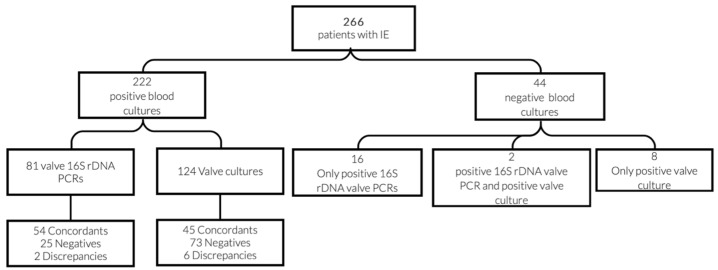
Blood cultures were performed on the 266 patients. In addition, valve culture was completed in 154. The 16S rDNA PCR was carried out in 103 out of these 154 cases. Microbiological test performances were as follows: 222 patients (222/266, 83.5%) had positive blood cultures, 74 (74/103, 71.8%) had a positive result for 16S rDNA valve PCR, and 61 (61/154, 39.6%) yielded a positive result of valve culture. Microbiological results are summarized in Figure 2.
Figure 2.
Microorganism detection by the different microbiological methods evaluated (blood culture, heart valve or vegetation culture, and/or 16S rDNA PCR).
Microbiological test results for each individual patient and results grouped by species are detailed in Supplementary Table S1 and Table 2, respectively. The main etiological agents causing IE in our series were coagulase-negative staphylococci (n = 69) followed by viridans group streptococci (n = 49), Staphylococcus aureus (n = 48) and enterococci (n = 46).
Table 2.
Microbiological results grouped by species of patients diagnosed with infective endocarditis.
| Number of Microorganisms Detected | ||||||||
|---|---|---|---|---|---|---|---|---|
| Bacterial Species | Number of Cases | Only Positive Blood Culture | Only Positive Heart Tissue Culture | Only Positive Heart Tissue 16S rDNA PCR | Positive Blood Culture | Positive Heart Tissue Culture | Positive Heart Tissue 16S rDNA PCR | Positive in All Methods |
| Staphylococcus aureus | 48 | 32 | 2 | 1 | 45 | 9 | 8 | 1 |
| Coagulase-negative staphylococci (CoNS) | 69 | 41 | 3 | 2 | 64 | 19 | 15 | 6 |
| Staphylococcus epidermidis | 56 | 33 | 1 | 1 | 54 | 16 | 13 | 6 |
| Staphylococcus lugdunensis | 5 | 2 | 0 | 1 | 4 | 1 | 2 | 0 |
| Staphylococcus auricularis | 2 | 1 | 1 | 0 | 1 | 1 | 0 | 0 |
| Staphylococcus hominis | 2 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| Staphylococcus haemolyticus | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| Staphylococcus warneri | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| Other CoNS | 2 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| Viridans group streptococci | 49 | 28 | 0 | 3 | 46 | 3 | 18 | 0 |
| Streptococcus mitis | 3 | 0 | 0 | 2 | 1 | 0 | 3 | 0 |
| Streptococcus mutans | 3 | 0 | 0 | 1 | 2 | 0 | 3 | 0 |
| Streptococcus anginosus | 2 | 1 | 0 | 0 | 2 | 1 | 0 | 0 |
| Streptococcus constellatus | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Streptococcus gallolyticus | 17 | 10 | 0 | 0 | 17 | 2 | 5 | 0 |
| Streptococcus gordonii | 6 | 4 | 0 | 0 | 6 | 0 | 2 | 0 |
| Streptococcus oralis | 6 | 3 | 0 | 0 | 6 | 0 | 3 | 0 |
| Streptococcus parasanguis | 2 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| Streptococcus salivarius | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Streptococcus sanguinis | 6 | 4 | 0 | 0 | 6 | 0 | 2 | 0 |
| Other | 2 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| beta-hemolytic streptococci | 5 | 2 | 0 | 1 | 4 | 1 | 2 | 0 |
| Streptococcus dysgalactiae | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| Streptococcus agalactiae | 4 | 2 | 0 | 1 | 3 | 0 | 2 | 0 |
| Enterobacterales and nonfermenting Gram-negative bacilli | 8 | 3 | 0 | 2 | 6 | 2 | 3 | 0 |
| Escherichia coli | 3 | 1 | 0 | 1 | 2 | 0 | 2 | 0 |
| Enterobacter cloacae | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Chryseobacterium spp. | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| Pseudomonas aeruginosa | 2 | 1 | 0 | 0 | 2 | 1 | 0 | 0 |
| Serratia marcescens | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| Enterococci | 46 | 24 | 1 | 0 | 47 | 15 | 16 | 7 |
| Enterococcus faecalis | 43 | 21 | 1 | 0 | 42 | 14 | 15 | 7 |
| Enterococcus faecium | 3 | 2 | 0 | 0 | 3 | 0 | 1 | 0 |
| Candida spp. | 2 | 1 | 0 | 0 | 2 | 1 | 0 | 0 |
| Other | 17 | 6 | 1 | 7 | 7 | 3 | 10 | 0 |
| Abiotrophia defectiva | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Aerococcus urinae | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Agregatibacter actinomycetemcomitans | 2 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| Listeria monocytogenes | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Tropheryma whipplei | 2 | 0 | 0 | 2 | 0 | 0 | 2 | 0 |
| Coxiella burnetii | 4 | 0 | 0 | 4 | 0 | 0 | 4 | 0 |
| Cutibacterium acnes | 4 | 0 | 1 | 0 | 1 | 3 | 3 | 0 |
| Bartonella henselae | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| Streptococcus pneumoniae | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Polymicrobial infection | 4 | 0 | 1 | 0 | 3 | 3 | 2 | 0 |
3.4. Concordances and Discrepancies between the Different Evaluated Methods
Concordances and discrepancies between the different evaluated microbiological methods are shown in Figure 3 and Figure 4. When both blood culture and 16S rDNA valve PCR were positive, results were concordant in all but two cases (Table 3). One of them corresponded to a case of IE by Candida albicans and Streptococcus dysgalactiae. As 16S rDNA PCR only detects bacterial DNA, the result of the valve analysis was expected to be negative. However, S. dysgalactiae was identified, and the episode was considered and treated as a polymicrobial infection. The second case corresponded to mixed endocarditis caused by S. epidermidis and Granullicatella adiacens, detected by 16S rDNA PCR and blood culture, respectively. As both germs are known etiological agents of infective endocarditis, it could not be ruled out that the infection was caused by both germs, and treatment was targeted to both pathogens.
Figure 3.
Concordance between blood and heart valve culture and/or 16S rDNA valve PCR results.
Figure 4.
Distribution of microorganisms with concordant results between blood and valve culture in comparison with 16S rDNA valve PCR.
Table 3.
Discrepancies between the different evaluated methods for the diagnosis of infective endocarditis.
| Blood Cultures | Heart Valve or Vegetation Cultures | 16S rDNA Heart Valves PCR |
| Discrepancies between blood cultures and 16S rDNA heart valve PCR | ||
| Candida albicans | Candida albicans | Streptococcus dysgalactiae |
| Granulicatella adiacens | Negative | Staphylococcus epidermidis |
| Discrepancies between heart valve or vegetation cultures and the others | ||
| Streptococcus oralis | Cutibacterium acnes | Streptococcus oralis |
| Staphylococcus warneri | Staphylococcus warneri and Cutibacterium acnes | Negative |
| Streptococcus dysgalactiae | Streptococcus dysgalactiae and Staphylococcus aureus | Not performed |
| Agregatibacter actinomycetemcomitans | Klebsiella pneumoniae | Not performed |
| Enterococcus faecalis | Staphylococcus cohnii | Enterococcus faecalis |
| Staphylococcus aureus | Cutibacterium acnes | Negative |
| Streptococcus mutans | Staphylococcus haemolyticus | Streptococcus mutans |
Regarding valve cultures, several discrepancies were found (Table 3). These discrepancies were mostly considered as contaminations of valve cultures, as microorganisms detected by the other evaluated methods were more plausible.
3.5. Diagnostic Benefit from Valve PCR
16S rDNA valve PCR contributed with clinically relevant information, allowing an etiological diagnosis in 16 cases of IE with valve and blood culture-negative results (Table 4). Moreover, this was the only diagnostic method that detected species such as C. burnetii and T. whipplei. Additionally, in 16 other cases, 16S rDNA PCR supported the results of blood and/or valve cultures in case of IE caused by coagulase-negative staphylococci or other bacteria from the skin microbiota, such as Cutibacterium acnes (Table 4).
Table 4.
Diagnostic benefit of 16S rDNA PCR for patients with infective endocarditis.
| Blood Cultures | Heart Valve or Vegetation Cultures | 16S rDNA Heart Valves PCR | Number of Cases |
|---|---|---|---|
| Patients with only positive valve PCR | |||
| Negative | Negative | Coxiella burnetii | 4 |
| Negative | Negative | Tropheryma whipplei | 2 |
| Negative | Negative | Streptococcus mitis | 2 |
| Negative | Negative | Staphylococcus aureus | 1 |
| Negative | Negative | Staphylococcus epidermidis | 1 |
| Negative | Negative | Escherichia coli | 1 |
| Negative | Negative | Staphylococcus lugdunensis | 1 |
| Negative | Negative | Streptococcus agalactiae | 1 |
| Negative | Negative | Bartonella henselae | 1 |
| Negative | Negative | Streptococcus mutans | 1 |
| Negative | Negative | Chyseobacterium spp. | 1 |
| Patients with coagulase-negative staphylococci and skin commensals in blood cultures that are confirmed by concordant valve PCR | |||
| Staphylococcus epidermidis | Staphylococcus epidermidis | Staphylococcus epidermidis | 6 |
| Staphylococcus epidermidis | Negative | Staphylococcus epidermidis | 6 |
| Staphylococcus lugdunensis | Negative | Staphylococcus lugdunensis | 1 |
| Negative | Cutibacterium acnes | Cutibacterium acnes | 2 |
| Cutibacterium acnes | Negative | Cutibacterium acnes | 1 |
4. Discussion
The present study evaluates the use of 16S rDNA heart tissue PCR combined with culture-based methods for the identification of the etiological microorganism in IE. Our results confirm that blood culture-negative endocarditis (BCNE) benefits by adding microbiological molecular assays [7]. In addition, the present work also confirms previous evidence that 16S rDNA heart valve PCR testing is particularly useful in comparison to tissue culture in patients undergoing surgery [15] and to corroborate microorganisms that can sometimes be considered contaminants.
A total of 154 (57.9%) patients in this series required surgical treatment of IE. Although preoperative blood cultures detected the etiological agent in almost two-thirds of the cases, 16S rDNA valve PCR analysis and/or valve tissue culture contributed to the microbiologic diagnosis in 9.8% (26/266) of patients. The test performance was 39.6% (61/154) for valve culture and 71.8% (74/103) for 16S rDNA valve PCR. Other studies have reported performances of 46% and 92% for the same methods, respectively [16], in line with our results. Due to the high performance of molecular methods in comparison to culture, some studies have questioned the need for routine culture of heart valves when 16S rDNA PCR analysis is available [17], and current evidence suggests that 16S rDNA PCR should be preferentially conducted after a blood test to identify the causative pathogen. However, applying only molecular methods to valves cannot be recommended, as the etiological agent is not recovered and therefore, the performance of antimicrobial susceptibility testing to guide the instauration of an optimal targeted antimicrobial therapy is not possible. Therefore, culture-based diagnostic techniques are an essential part of IE management.
Our series has a proportion of BCNE of 16.5% (44/266). BCNE rates are highly variable in the literature, ranging from 2.5 to 31% [18]. This variability is conditioned by several factors, such as antimicrobial treatments prior to culture and/or the incidence of non-cultivable or fastidious microorganisms [19]. Molecular analysis of heart valves was found to be the only method to establish the etiology of BCNE, giving consistent results in 16 out of 44 patients (36%). These results agree with recent articles where 16S rDNA valve PCR was positive in 41% of surgical blood culture-negative endocarditis [20].
Most of the patients analyzed in the present work were under antibiotic therapy at the time of heart valve excision. Although this is the usual clinical practice, only a few studies have documented the average length of antibiotic treatment given prior to surgery. The mean duration of previous antimicrobial treatment in our series was similar in patients with negative and positive 16S rDNA valve PCR, respectively. However, we found a significant difference in the duration of antibiotic treatment before surgery between patients with negative and positive valve cultures. Previous works reported that the sensitivity of 16S rDNA valve PCR is independent of the length of antibiotic treatment before surgery, in contrast to culture sensitivity [21,22]. This can be explained by the persistence of DNA in spite of antibiotic treatment that reduces microorganism viability and consequently impairs their growth in microbiological cultures [23].
A potentially cultivable microorganism was detected by 16S rDNA in seven cases of BCNE from our series. In four of them, Streptococcus spp. was identified, and three were positive for Staphylococcus spp. Similar studies reported other potentially cultivable microorganisms such as enterococci and nutritionally variant streptococci in BCNE [7]. Additionally, 16S rDNA valve PCR significantly increases the ability to detect intracellular organisms such as Coxiella spp., Tropheryma spp., and Bartonella spp. [24]. In the present work, we reported four, two, and one cases of C. burnetti, T. whipplei, and B. henselae IE, respectively, all detected by 16S rDNA valve PCR. Even though the epidemiology of these pathogens has regional differences [15], underdiagnosis has been demonstrated in southern European countries after the incorporation of new molecular tools [25]. In a series of 1383 patients in France, C. burnetii accounted for at least 5% of all IE [26]. According to several authors, the diagnosis is often delayed for several months [27], and the infection is involved in 37–45% of BCNE [28,29]. Additionally, in a recent multicenter study carried out in Spain, 28.2% of BCNE reported were caused by Coxiella burnetii and 10.3% by Tropheryma whipplei [30]. These are all emerging pathogens, perhaps not because of the increase in cases but because of the improvement in diagnostic techniques that allow their detection.
Furthermore, the performance of 16S rDNA PCR in our study was useful to interpret blood culture results, for instance, to confirm infections by bacteria from the skin microbiota, a benefit that has already been highlighted in other works [20].
Despite the advantages of valvular 16S rDNA PCR, it should be noted that this test was negative in 25 samples of patients with a positive blood culture. This was unexpected; however, a potential explanation is that PCR result is restricted by the site of the excised tissue, and infection is not distributed homogeneously in the valve [31].
Metagenomic sequencing methods are currently being developing to provide information on the etiological diagnosis and its antibiotic susceptibility profile in patients with IE [32], potentially offering a powerful method to deal with negative blood cultures, especially in patients who are not undergoing surgical treatment, and to deal with polymicrobial infections [31,33]. However, Santibáñez et al. recently reported that the use of a metagenomic approach in the study of IE in resected heart valves provided no additional significant information compared to conventional 16S rDNA PCR, except for polymicrobial IE [34].
The present study has limitations, such as its retrospective nature or the fact that 16S rDNA PCR was not performed on all valve samples. However, it is a very large series compared to other previous works published in the literature, and it serves to support the usefulness of this approach for the diagnosis and management of EI. These additive results contribute to establishing the advantages and limitations of emerging techniques.
5. Conclusions
Our results illustrate the role of 16S rDNA heart tissue PCR in the microbiological diagnosis of infective endocarditis. This technique may provide additional information which can guide antibiotic therapy, especially in patients with negative blood cultures. Furthermore, this study supports the use of this method to confirm the results of patients with positive blood cultures suspected as contaminants, such as those cases caused by bacteria from the skin microbiota.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/diagnostics11081372/s1, Table S1: Microbiological results grouped by species of patients diagnosed with infective endocarditis.
Author Contributions
Conceptualization, R.R.-G. and J.F.; Data curation, M.Á.R.-E., J.F.-S., A.M., E.G.-C., C.M., and G.M.A.; Formal analysis, R.R.-G.; Investigation, R.R.-G. and J.F.; Methodology, R.R.-G., J.F.-S., and J.F.; Supervision, M.T. and J.F.; Writing—original draft, R.R.-G. and J.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of Hospital Universitario Central de Asturias, Oviedo, Spain (protocol code n 210/18, in 14 August 2018).
Informed Consent Statement
Patient consent was waived as this is a retrospective study with a large number of patients, which makes it difficult to collect informed consent. In addition, the database has been totally anonymized by the researchers who have guaranteed the confidentiality of the personal data.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Habib G., Erba P.A., Iung B., Donal E., Cosyns B., Laroche C., Popescu B.A., Prendergast B., Tornos P., Sadeghpour A., et al. Clinical presentation, aetiology and outcome of infective endocarditis. Results of the ESC-EORP EURO-ENDO (European infective endocarditis) registry: A prospective cohort study. Eur. Heart J. 2019;40:3222–3232. doi: 10.1093/eurheartj/ehz620. [DOI] [PubMed] [Google Scholar]
- 2.Cahill T.J., Prendergast B.D. Infective endocarditis. Lancet. 2016;387:882–893. doi: 10.1016/S0140-6736(15)00067-7. [DOI] [PubMed] [Google Scholar]
- 3.Pettersson G.B., Hussain S.T. Current AATS guidelines on surgical treatment of infective endocarditis. Ann. Cardiothorac. Surg. 2019;8:630–644. doi: 10.21037/acs.2019.10.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Werner M., Andersson R., Olaison L., Hogevik H.D. Swedish Society of Infectious Diseases Quality Assurance Study Group for Endocarditis. A 10-year survey of blood culture negative endocarditis in Sweden: Aminoglycoside therapy is important for survival. Scand. J. Infect. Dis. 2008;40:279–285. doi: 10.1080/00365540701642112. [DOI] [PubMed] [Google Scholar]
- 5.Lamas C.C., Eykyn S.J. Blood culture negative endocarditis: Analysis of 63 cases presenting over 25 years. Heart. 2003;89:258–262. doi: 10.1136/heart.89.3.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fournier P., Thuny F., Richet H., Lepidi H., Casalta J., Arzouni J., Maurin M., Célard M., Mainardi J.-L., Caus T., et al. Comprehensive Diagnostic Strategy for Blood Culture–Negative Endocarditis: A Prospective Study of 819 New Cases. Clin. Infect. Dis. 2010;51:131–140. doi: 10.1086/653675. [DOI] [PubMed] [Google Scholar]
- 7.Fournier P.-E., Gouriet F., Casalta J.-P., Lepidi H., Chaudet H., Thuny F., Collart F., Habib G., Raoult D. Blood culture-negative endocarditis: Improving the diagnostic yield using new diagnostic tools. Medicine. 2017;96:e8392. doi: 10.1097/MD.0000000000008392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fredricks D.N., Relman D. Application of Polymerase Chain Reaction to the Diagnosis of Infectious Diseases. Clin. Infect. Dis. 1999;29:475–486. doi: 10.1086/598618. [DOI] [PubMed] [Google Scholar]
- 9.Church D.L., Cerutti L., Gürtler A., Griener T., Zelazny A., Emler S. Performance and Application of 16S rRNA Gene Cycle Sequencing for Routine Identification of Bacteria in the Clinical Microbiology Laboratory. Clin. Microbiol. Rev. 2020;33 doi: 10.1128/CMR.00053-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marín M., Muñoz P., Sánchez M., del Rosal M., Alcalá L., Rodríguez-Créixems M., Bouza E. Molecular Diagnosis of Infective Endocarditis by Real-Time Broad-Range Polymerase Chain Reaction (PCR) and Sequencing Directly From Heart Valve Tissue. Medicine. 2007;86:195–202. doi: 10.1097/MD.0b013e31811f44ec. [DOI] [PubMed] [Google Scholar]
- 11.Vollmer T., Piper C., Horstkotte D., Körfer R., Kleesiek K., Dreier J. 23S rDNA real-time polymerase chain reaction of heart valves: A decisive tool in the diagnosis of infective endocarditis. Eur. Heart J. 2010;31:1105–1113. doi: 10.1093/eurheartj/ehp600. [DOI] [PubMed] [Google Scholar]
- 12.Li J.S., Sexton D.J., Mick N., Nettles R., Fowler V.G., Jr., Ryan T., Bashore T., Corey G.R. Proposed Modifications to the Duke Criteria for the Diagnosis of Infective Endocarditis. Clin. Infect. Dis. 2000;30:633–638. doi: 10.1086/313753. [DOI] [PubMed] [Google Scholar]
- 13. [(accessed on 28 June 2021)]; Available online: https://www.idsociety.org/practice-guideline/laboratory-diagnosis-of-infectious-diseases.
- 14.Xu J., Millar B., Moore J., Murphy K., Webb H., Fox A., Cafferkey M., Crowe M. Employment of broad-range 16S rRNA PCR to detect aetiological agents of infection from clinical specimens in patients with acute meningitis—Rapid separation of 16S rRNA PCR amplicons without the need for cloning. J. Appl. Microbiol. 2003;94:197–206. doi: 10.1046/j.1365-2672.2003.01839.x. [DOI] [PubMed] [Google Scholar]
- 15.Vondracek M., Sartipy U., Aufwerber E., Julander I., Lindblom D., Westling K. 16S rDNA sequencing of valve tissue improves microbiological diagnosis in surgically treated patients with infective endocarditis. J. Infect. 2011;62:472–478. doi: 10.1016/j.jinf.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 16.Miller R.J.H., Chow B., Pillai D., Church D. Development and evaluation of a novel fast broad-range 16S ribosomal DNA PCR and sequencing assay for diagnosis of bacterial infective endocarditis: Multi-year experience in a large Canadian healthcare zone and a literature review. BMC Infect. Dis. 2016;16:1–10. doi: 10.1186/s12879-016-1476-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muñoz P., Bouza E., Marín M., Alcalá L., Créixems M.R., Valerio M., Pinto A., on behalf of the Group for the Management of Infective Endocarditis of the Gregorio Marañón Hospital Heart Valves Should Not Be Routinely Cultured. J. Clin. Microbiol. 2008;46:2897–2901. doi: 10.1128/JCM.02173-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faraji R., Behjati-Ardakani M., Moshtaghioun S.M., Kalantar S.M., Namayandeh S.M., Soltani M., Emami M., Zandi H., Firoozabadi A.D., Kazeminasab M., et al. The diagnosis of microorganism involved in infective endocarditis (IE) by polymerase chain reaction (PCR) and real-time PCR: A systematic review. Kaohsiung J. Med. Sci. 2018;34:71–78. doi: 10.1016/j.kjms.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godfrey R., Curtis S., Schilling W., James P.R. Blood culture negative endocarditis in the modern era of 16S rRNA sequencing. Clin. Med. 2020;20:412–416. doi: 10.7861/clinmed.2019-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armstrong C., Kuhn T.C., Dufner M., Ehlermann P., Zimmermann S., Lichtenstern C., Soethoff J., Katus H.A., Leuschner F., Heininger A. The diagnostic benefit of 16S rDNA PCR examination of infective endocarditis heart valves: A cohort study of 146 surgical cases confirmed by histopathology. Clin. Res. Cardiol. 2020;110:332–342. doi: 10.1007/s00392-020-01678-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voldstedlund M., Pedersen L.N., Baandrup U., Klaaborg K.E., Fuursted K. Broad-range PCR and sequencing in routine diagnosis of infective endocarditis. APMIS. 2008;116:190–198. doi: 10.1111/j.1600-0463.2008.00942.x. [DOI] [PubMed] [Google Scholar]
- 22.Rovery C., Greub G., Lepidi H., Casalta J.-P., Habib G., Collart F., Raoult D. PCR Detection of Bacteria on Cardiac Valves of Patients with Treated Bacterial Endocarditis. J. Clin. Microbiol. 2005;43:163–167. doi: 10.1128/JCM.43.1.163-167.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barnes P., Crook D. Culture negative endocarditis. J. Infect. 1997;35:209–213. doi: 10.1016/S0163-4453(97)92662-1. [DOI] [PubMed] [Google Scholar]
- 24.Dolmans R.A.V., Boel C.H.E., Lacle M.M., Kusters J.G. Clinical Manifestations, Treatment, and Diagnosis of Tropheryma whipplei Infections. Clin. Microbiol. Rev. 2017;30:529–555. doi: 10.1128/CMR.00033-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karageorgou I., Kogerakis N., Labropoulou S., Hatzianastasiou S., Mentis A., Stavridis G., Angelakis E. Q Fever Endocarditis and a New Genotype of Coxiella burnetii, Greece. Emerg. Infect. Dis. 2020;26:2527–2529. doi: 10.3201/eid2610.191616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raoult D., Tissot-Dupont H., Foucault C., Gouvernet J., Fournier P.E., Bernit E., Stein A., Nesri M., Harle J.R., Weiller P.J. Q Fever 1985-1998: Clinical and Epidemiologic Features of 1,383 Infections. Medicine. 2000;79:109–123. doi: 10.1097/00005792-200003000-00005. [DOI] [PubMed] [Google Scholar]
- 27.Houpikian P., Raoult D. Blood culture-negative endocarditis in a reference center: Etiologic diagnosis of 348 cases. Medicine. 2005;84:162–173. doi: 10.1097/01.md.0000165658.82869.17. [DOI] [PubMed] [Google Scholar]
- 28.Subedi S., Jennings Z., Chen S.-A. Laboratory Approach to the Diagnosis of Culture-Negative Infective Endocarditis. Heart Lung Circ. 2017;26:763–771. doi: 10.1016/j.hlc.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 29.Jang Y.-R., Song J.S., Jin C.E., Ryu B.-H., Park S.Y., Lee S.-O., Choi S.-H., Kim Y.S., Woo J.H., Song J.-K., et al. Molecular detection of Coxiella burnetii in heart valve tissue from patients with culture-negative infective endocarditis. Medicine. 2018;97:e11881. doi: 10.1097/MD.0000000000011881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suardi L.R., de Alarcón A., García M.V., Ciezar A.P., Tenorio C.H., Martinez-Marcos F.J., Concejo-Martínez E., Lima J.D.L.T., García D.V., Márquez R.L., et al. Blood culture-negative infective endocarditis: A worse outcome? Results from a large multicentre retrospective Spanish cohort study. Infect. Dis. 2021:1–9. doi: 10.1080/23744235.2021.1925342. [DOI] [PubMed] [Google Scholar]
- 31.Oberbach A., Schlichting N., Feder S., Lehmann S., Kullnick Y., Buschmann T., Blumert C., Horn F., Neuhaus J., Neujahr R., et al. New insights into valve-related intramural and intracellular bacterial diversity in infective endocarditis. PLoS ONE. 2017;12:e0175569. doi: 10.1371/journal.pone.0175569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan W.-S., Au C.-H., Leung H.C.-M., Ho D.N., Li D., Chan T.-L., Lam T.-W., Ma E., Tang B.S.-F. Potential utility of metagenomic sequencing for improving etiologic diagnosis of infective endocarditis. Futur. Cardiol. 2019;15:411–424. doi: 10.2217/fca-2018-0088. [DOI] [PubMed] [Google Scholar]
- 33.Lieberman J.A., Li C.N., Lamb G.S., Kane D.A., Stewart M.K., Mamedov R.A., Cookson B.T., Salipante S.J. Case Report: Comparison of Plasma Metagenomics to Bacterial PCR in a Case of Prosthetic Valve Endocarditis. Front. Pediatr. 2021;8:575674. doi: 10.3389/fped.2020.575674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santibáñez P., Portillo A., Santibáñez S., García-Álvarez L., de Toro M., Oteo J.A. What does 16S rRNA gene-targeted next generation sequencing contribute to the study of infective endocarditis in valve tissue? MedRxiv. 2021 doi: 10.1101/2021.01.23.21250364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the article.