Abstract
Saccharomyces cerevisiae responds to pyrimidine starvation by increasing the expression of four URA genes, encoding the enzymes of de novo pyrimidine biosynthesis, three- to eightfold. The increase in gene expression is dependent on a transcriptional activator protein, Ppr1p. Here, we investigate the mechanism by which the transcriptional activity of Ppr1p responds to the level of pyrimidine biosynthetic intermediates. We find that purified Ppr1p is unable to promote activation of transcription in an in vitro system. Transcriptional activation by Ppr1p can be observed, however, if either dihydroorotic acid (DHO) or orotic acid (OA) is included in the transcription reactions. The transcriptional activation function and the DHO/OA-responsive element of Ppr1p localize to the carboxyl-terminal 134 amino acids of the protein. Thus, Ppr1p directly senses the level of early pyrimidine biosynthetic intermediates within the cell and activates the expression of genes encoding proteins required later in the pathway. These results are discussed in terms of (i) regulation of the pyrimidine biosynthetic pathway and (ii) a novel mechanism of regulating gene expression.
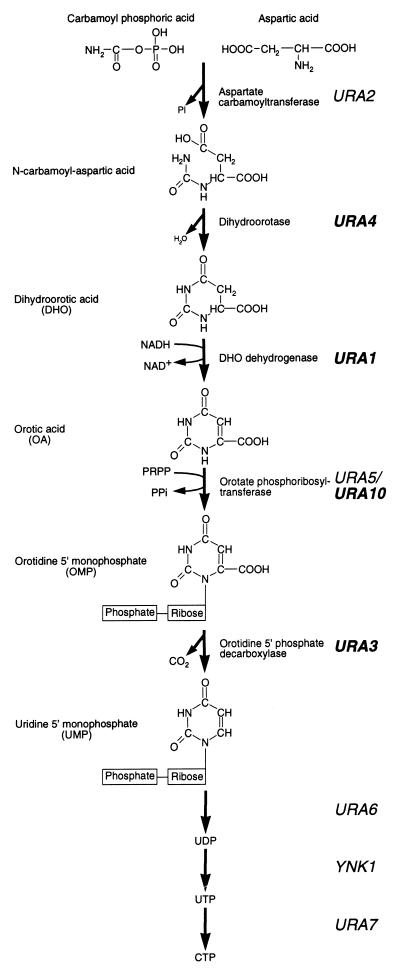
In Saccharomyces cerevisiae, the biosynthesis of pyrimidines involves the de novo synthesis of UMP from glutamine (Fig. 1). Carbamoyl phosphate, derived from glutamine, undergoes a condensation reaction with aspartic acid, resulting in the formation of N-carbamoyl aspartic acid. Both the formation and subsequent condensation of carbamoyl phosphate are performed by Ura2p. The pyrimidine ring of N-carbamoyl aspartic acid is closed by the elimination of water to form dihydroorotic acid (DHO), which is subsequently oxidized to form orotic acid (OA), and a ribose-phosphate group is then added to form orotidine 5′-monophosphate (OMP). The formation of OMP is performed by two isoenzymes, Ura5p and Ura10p (2). OMP is then decarboxylated to yield UMP, which may subsequently be processed to form other pyrimidines (3). Regulation of this pathway occurs at several levels. First, UTP down-regulates the enzymatic activity of Ura2p (1) and transcription of the URA2 gene (22). Second, under conditions of pyrimidine starvation, transcription of the URA1, URA3, URA4, and URA10 genes (the URA genes) is increased some three- to eightfold (24). This increase in transcription is dependent on a transcriptional activator, Ppr1p (16).
FIG. 1.
Pyrimidine biosynthetic pathway of S. cerevisiae. The pyrimidine ring is formed by the condensation of carbamoyl phosphoric acid and aspartic acid, followed by the elimination of a water molecule to form DHO. DHO is subsequently oxidized to OA by the product of the URA1 gene. A phosphoribose moiety, provided by 5-phosphoribose 1-pyrophosphate (PRPP), is added to OA to form OMP, which is then decarboxylated to form UMP by the product of the URA3 gene. UMP is subsequently converted to UDP and UTP before its conversion into CTP. Ppr1p is a transcriptional activator of the genes shown in bold typeface during conditions of pyrimidine starvation.
Ppr1p is a 904-amino-acid protein that bears a Zn2Cys6 binuclear cluster DNA binding motif near its amino-terminal end—amino acids 29 to 123 (8). The crystal structure of the DNA binding domain of Ppr1p complexed with its cognate DNA site has been solved (17). Like other members of this family of proteins, Ppr1p has an acidic C-terminal domain which may, as in the other family members, represent the transcriptional activation domain (25). The protein binds to defined sites (CGGN6CCG) found approximately 100 to 200 bp upstream of the translational start sites of the URA genes (10, 24). In the absence of Ppr1p, these genes are transcribed at a constitutive, basal level. A single constitutive and several noninducible mutations of the PPR1 gene have been identified (15, 16). The noninducible mutations map to the C6 zinc cluster of the DNA binding domain of Ppr1p (amino acids 43, 57, and 64) (11), while the constitutive mutation maps to amino acid 233 (ppr1-1 changes leucine 233 to serine [24a]). The noninducible mutations are therefore likely to be defective in DNA binding, but the effect of L233S invoking constitutive expression is more difficult to interpret.
The elegant genetic analysis of Lacroute (9) clearly demonstrated induction of the enzymes of the pyrimidine pathway by biosynthetic intermediates of the pathway itself. A yeast strain mutated in ura2, and thereby unable to synthesize DHO, will not induce the pathway in response to pyrimidine starvation. Also, a yeast strain mutated in ura1, resulting in an accumulation of DHO, induces the pathway (9). The increase in activity of the pathway enzymes in response to pyrimidine starvation is due to increased expression of the URA genes themselves (14). Altering the levels of DHO within the cell affects the induced but not the constitutive levels of URA expression (3). It has thus been speculated that DHO may act together with Ppr1p as an inducer of URA gene expression under conditions of pyrimidine starvation. To understand this phenomenon further, we have undertaken a biochemical investigation of the transcriptional properties of Ppr1p.
We begin by describing the construction of a recombinant baculovirus expressing full-length yeast Ppr1p in insect cells. We show that while purified Ppr1p interacts only weakly with DNA, strong DNA binding can be promoted by an as yet unidentified small molecule present in extracts of insect and yeast cells. Once bound to DNA, Ppr1p is transcriptionally inert in vitro. Transcriptional activity can be induced, however, by the addition of either DHO or OA to transcription reactions containing Ppr1p. We define the DHO/OA-responsive domain of Ppr1p to the carboxyl-terminal 134 amino acids of the protein, coincident with the activation domain of the protein. Thus, we envisage a model for the activation of the URA1 and URA3 genes in which DNA-bound Ppr1p directly senses the level of DHO and/or OA. Once the concentration of these molecules reaches a certain threshold, Ppr1p activates the URA genes to promote the biosynthesis of UMP.
MATERIALS AND METHODS
Media and strains.
Escherichia coli DH5α was used for all DNA manipulations, and strain XA90 was used for protein expression from the tac promoter (23). Yeast nuclear extract was prepared from S. cerevisiae BJ2168 (MATα ura3 leu2 trp1 gal2 prb1 pep4 prc1) grown in YPD medium (20). Spodoptera frugiperda Sf9 and High Five insect cells (Invitrogen) were grown in Grace’s insect medium (Invitrogen) supplemented with 10% fetal calf serum at 27°C.
Construction of recombinant baculovirus.
The coding region of PPR1 was amplified from pUC8-PPR1 (a gift of Stanley Liang) by PCR using oligonucleotides 1317 (5′-GGGGGGGATCCGATGAAGCAGAAAAAATTTAACTCC-3′) and 1318 (5′-GGGGGGAAGCTTCTAAAATATTCCACCGGATTCAGA-3′). The PCR product (∼3 kb) was cleaved with BamHI and HindIII (underlined) and cloned into the BamHI/HindIII sites of pBlueBacHisB (Invitrogen). The majority of the PPR1 coding sequence in the resulting plasmid (pRJR236) was replaced by a 2.5-kb EcoNI fragment of pUC8-PPR1. Sequencing of the remainder of the PPR1 coding sequence confirmed that no mutations had arisen (data not shown). Thus, pRJR236 contains the entire coding sequence for Ppr1p, fused to an N-terminal RGSH6 tag, under the control of the polyhedrin promoter. Plasmid pRJR236 was cotransfected with wild-type, linearized Autographa californica nuclear polyhedrosis virus DNA into S. frugiperda Sf9 insect cells, using a Bac-N-Blue transfection kit (Invitrogen) according to the manufacturer’s instructions. Recombinant viruses were identified as those yielding blue plaques on plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside. PCR analysis of the viral DNA confirmed the presence of the PPR1 gene (data not shown), and the pure, recombinant virus was used to create a high-titer viral stock, using Sf9 cells (1.25 × 108 PFU/ml).
Protein purification.
Full-length Ppr1p was purified from High Five insect cells grown in monolayer culture in 225-cm2 tissue culture flasks (Costar) infected with Ppr1p recombinant baculovirus at a multiplicity of infection of 5 and harvested 2 days postinfection. High Five cells gave increased yields of Ppr1p compared with those in Sf9 cells. Cell pellets were resuspended in buffer A (1 ml/107 infected cells; 20 mM HEPES [pH 7.8], 300 mM NaCl, 10% glycerol) containing a complete protease inhibitor cocktail (Boehringer Mannheim), sonicated, and centrifuged at 30,000 × g for 20 min at 4°C. Ppr1p was purified by nickel affinity (Ni2+-nitrilotriacetic acid [NTA]) chromatography using Pro-Bond resin (Invitrogen) equilibrated in buffer A. Following washes with buffer A and with buffer A containing 500 mM NaCl, 10 mM β-mercaptoethanol, and 30 mM imidazole, protein was eluted from the column with buffer A containing 250 mM imidazole.
The DNA binding domain of Ppr1p (residues 29 to 123) was purified as described elsewhere (17). The fusion protein Gal4p(1-93)-Ppr1p(770-904) was constructed by PCR amplification of the PPR1 gene from pUC8-PPR1, using oligonucleotides 3071 (5′-CCCGGATCCCTAAAATATTCCACC-3′) and 3072 (5′-CCCGAGCTCAACCGCATGTCAAGT-3′). The amplified DNA (414 bp) was cleaved with SacI and BamHI (underlined) and cloned into the SacI/BamHI sites of pRJR1 (23). The resulting plasmid, pRJR369, expressed Gal4p(1-93)-Ppr1p(770-904) from the tac promoter. Cells containing the plasmid were grown and induced, and the protein was purified as described elsewhere (23).
Western blotting.
Proteins were separated on 10% polyacrylamide gels containing sodium dodecyl sulfate (SDS) and then transferred to a nitrocellulose membrane by using a wet blotter (Bio-Rad). The membranes were washed in phosphate-buffered saline and blocked with phosphate-buffered saline containing 5% nonfat dried milk and 0.2% Tween 20. The RGS.His primary antibody (Qiagen) was detected with a sheep anti-mouse immunoglobulin-peroxidase conjugate (Amersham) and visualized by enhanced chemiluminescence (Amersham).
Mobility shift assay.
The PPR1 probe used was a double-stranded oligonucleotide (5-TCTTCGGTAATCTCCGAAGC-3′), representing the high-affinity Ppr1p binding site from the URA3 promoter (24). Reactions mixes (20 μl) contained 20 mM HEPES-KOH (pH 7.5), 150 mM NaCl, 10% glycerol, 5 mM MgCl2, 1 mM ZnSO4, 690 μg of sonicated salmon sperm DNA per ml, 10 pM 32P-labelled probe DNA, and protein at the indicated concentrations. Reaction mixes were incubated for 30 min at room temperature and were then subjected to electrophoresis through a prerun 5% polyacrylamide gel containing 0.5× Tris-borate-EDTA and 1% glycerol for 90 min at 150 V. Gels were dried and then analyzed by autoradiography.
In vitro transcription.
Yeast nuclear extract was prepared as described elsewhere (20, 21). Transcription reaction mixes (25 μl) contained 10 mM HEPES (pH 7.5), 10 mM MgSO4, 5 mM EGTA, 10% glycerol, 2.5 mM dithiothreitol, 100 mM potassium glutamate, 10 mM magnesium acetate, 2% polyvinyl alcohol, 8 mM phosphoenolpyruvate, 0.31 nM either pG5E4 (28) or pPPR17E4 (a gift of Josh Brickman), 4 nM pGEM3Z (Promega), and 3 μl of yeast nuclear extract (60 mg/ml). Reaction mixes were supplemented with pyrimidine biosynthetic intermediates (Sigma) at the concentrations indicated and were then incubated with Gal4p derivatives or Ppr1p for 10 min at 25°C. Nucleoside triphosphates were added to a final concentration of 1 mM, and the reactions were allowed to proceed for an additional 45 min at 25°C. Primer extension analysis of the RNA produced during these reactions was performed with an oligonucleotide complementary to the E4 coding sequence (5′-GCGGCAGCCTAACAGTCAGCCTTACCAGTA-3′) (12, 13). The extension products were separated on a 10% polyacrylamide gel containing 1× Tris-borate-EDTA and 5 M urea and then analyzed by autoradiography.
RESULTS
Purification of Ppr1p.
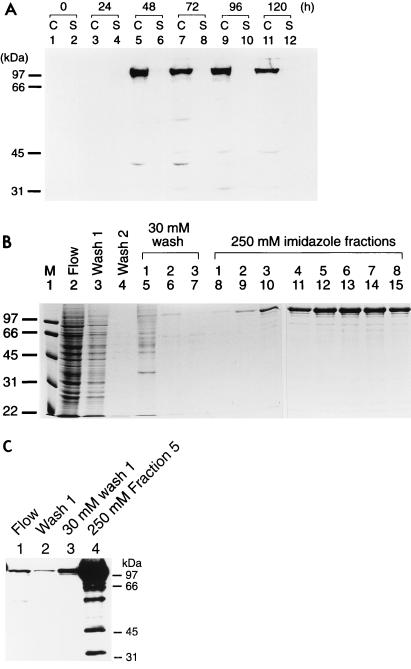
A recombinant baculovirus expressing full-length PPR1 from the polyhedrin promoter was constructed and used to infect High Five insect cells. Protein expression was monitored by Western blotting (Fig. 2A). While Ppr1p could not be detected in uninfected insect cells (Fig. 2A, lanes 1 and 2) or in cells infected with a wild-type virus (data not shown), at 2 days postinfection, the Ppr1p recombinant virus gave rise to a band of approximately 106 kDa, corresponding to the expected size of the full-length protein. The Ppr1p protein accumulated within the cells 24 to 48 h postinfection (Fig. 2A, lanes 5 and 7). At later times, the amount of full-length Ppr1p within the cells diminished, and a number of degradation products could be observed (Fig. 2A, lane 11). Based on these data, cells were harvested 2 days postinfection, before the onset of cell lysis. Ppr1p was purified from infected cells by nickel affinity chromatography (Fig. 2B). Buffer containing 30 mM imidazole (Fig. 2B, lanes 5 to 7) eluted a significant number of contaminants from the purification column, while Ppr1p was eluted from the column with buffer containing 250 mM imidazole (Fig. 2B, lanes 8 to 15). Ppr1p produced in this way is estimated to be >95% pure, with 2.25 mg of protein being obtained from 108 insect cells in adherent cell culture. Western blot analysis of the purified protein (Fig. 2C) shows that a number of degradation products of Ppr1p are present in the purified material (Fig. 2C, lane 4). However, a comparison of these lower-molecular-weight bands with the Coomassie-stained gel (Fig. 2B, lane 12) suggests that they are of low abundance.
FIG. 2.
Overproduction and purification of Ppr1p. (A) Time course of Ppr1p expression in baculovirus-infected insect cells. Cultures of High Five insect cells (50% confluent) were infected at time zero with recombinant baculovirus containing the PPR1 gene under the control of the polyhedrin promoter. Samples of the cells (lanes C) or the culture supernatant (lanes S) were taken at the times indicated and analyzed by Western blotting. (B) Purification of Ppr1p. Insect cells infected with recombinant PPR1 baculovirus were harvested 2 days postinfection. Cell extracts were prepared, and soluble protein was applied to a Ni2+-NTA agarose column. The column flowthrough is shown in lane 2. The column was washed with loading buffer (lanes 3 and 4) and then with buffer containing 30 mM imidazole (lane 5 to 7). Ppr1p was eluted with buffer containing 250 mM imidazole (lane 8 to 15). Samples of each fraction were run on an SDS-polyacrylamide gel that was stained with Coomassie brilliant blue. Sizes of molecular weight standards (M; in kilodaltons) are indicated. (C) Western blot analysis of the purification of Ppr1p. Samples from lanes 2, 3, 5, and 12 in panel B were separated by SDS-polyacrylamide gel electrophoresis and subjected to Western blotting. Sizes of molecular weight standards are indicated.
DNA binding activity of purified Ppr1p.
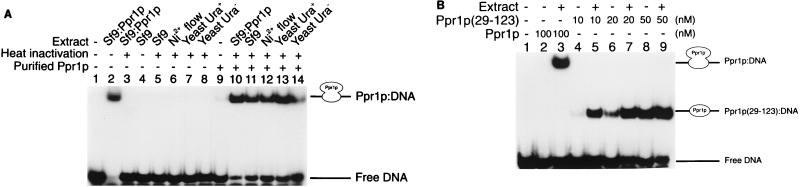
The DNA binding activity of full-length Ppr1p was initially analyzed in crude extracts of insect cells infected with a baculovirus producing Ppr1p. Using electrophoretic mobility shift assays, we were unable to detect binding of proteins to DNA bearing a Ppr1p binding site from uninfected Sf9 or High Five cells (Fig. 3A, lane 4, and data not shown) or from Sf9 or High Five cells infected with wild-type baculovirus (data not shown). However, extracts made from insect cells infected with recombinant Ppr1p-producing baculovirus resulted in the formation of a specific DNA-protein complex (Fig. 3A, lane 2).
FIG. 3.
DNA binding properties of purified Ppr1p. (A) DNA binding by full-length Ppr1p. Electrophoretic mobility shift assays were performed as described in Materials and Methods. Reactions contained 32P-labelled DNA comprising a single Ppr1p binding site and, where indicated, 100 nM purified Ppr1p. The reactions were supplemented with cell extracts from Sf9 insect cells infected with a baculovirus producing Ppr1p (lanes 2, 3, and 10), Sf9 insect cells (lanes 4, 5, and 11), the flowthrough of a Ni2+-NTA column purification of Ppr1p from Sf9 insect cells infected with a baculovirus producing Ppr1p (lanes 6 and 12), or yeast cells (strain JPY5) grown in either medium containing uracil (lanes 7 and 13) or medium lacking uracil (lanes 8 and 14). Where indicated, the supplemented extract was placed in a boiling water bath for 5 min and centrifuged. The supernatant was then added to the binding reactions. Positions of the free DNA and the Ppr1p-DNA complex are indicated. (B) DNA binding properties of Ppr1p(29-123). Electrophoretic mobility shift assays were performed as described above, and Ppr1p or Ppr1p(29-123) was added at the concentrations indicated. Where indicated, a heat-treated extract of yeast cells (strain JPY5) was added to the reactions. Positions of the free DNA and the protein-DNA complexes are indicated.
While detection of a Ppr1p-DNA complex in cell extracts was straightforward, detection of DNA binding by purified Ppr1p proved to be much more difficult. The purified protein (Fig. 3A, lane 9) formed a weak complex with DNA, even though there was considerably more Ppr1p in these reactions than in the samples prepared with the crude cell extracts. Efficient DNA binding by Ppr1p was recovered by supplementing the binding reactions with various cell extracts. For example, wild-type Sf9 cells did not contain a Ppr1p binding activity (Fig. 3A, lanes 4 and 5). However, the Sf9 extract, either the untreated material or the supernatant resulting from boiling and subsequent centrifugation, promoted high-affinity DNA binding of purified Ppr1p (Fig. 3A; compare lanes 9 to 11). The ability to promote purified Ppr1p DNA binding was not limited to insect cell extracts. Heat-inactivated yeast cell extracts grown in either uracil-rich or uracil-lacking medium, which exhibit no Ppr1p DNA binding activity in their own right, also promoted the formation of a Ppr1p-DNA complex (Fig. 3A, lanes 13 and 14). Yeast nuclear extracts prepared for in vitro transcription reactions (see below) also supported high-level Ppr1p DNA binding activity (data not shown).
The ability of these extracts to promote high-affinity DNA binding of Ppr1p does not appear to be the function of a protein. The extracts could be heat treated (Fig. 3A) or treated with proteinase K (data not shown), and their ability to aid high-affinity DNA binding of Ppr1p was not diminished. Dialysis of the extracts, however, significantly impaired their ability to promote high-affinity Ppr1p DNA binding. We therefore speculate that a small molecule may be involved. The DNA binding-promoting factor was also resistant to high levels of EDTA (up to 0.1 M [final concentration]). We tested the ability of metabolic intermediates of the pyrimidine biosynthetic pathway (those shown in Fig. 1, including DHO and OA) and several intermediates of purine biosynthesis to promote DNA binding of Ppr1p but observed no effects (data not shown). Addition of further zinc or magnesium ions to the binding reactions, known to be required for DNA binding of Ppr1p (17), did not promote high-affinity binding of full-length Ppr1p (data not shown).
The effect of extracts on the DNA binding activity was not limited to the full-length protein. Figure 3B shows the results of a heat-treated yeast cell extract on the DNA binding activity of full-length Ppr1p and on the isolated DNA binding domain, amino acids 29 to 123 (10). At low concentrations of Ppr1p(29-123), the extract increased the amount of the protein-DNA complex approximately threefold (compare Fig. 3B, lanes 4 and 5). At higher concentrations of Ppr1p(29-123), the effect of the extract was less obvious (Fig. 3B, lanes 8 and 9). Thus, we believe that the extracts provide a small molecule involved in the stabilization of the Ppr1p-DNA complex.
Transcriptional activity of purified Ppr1p.
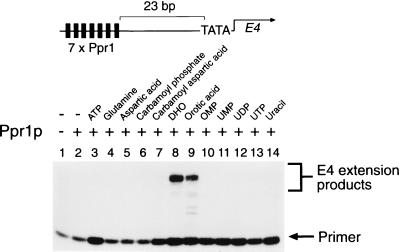
The transcriptional activity of Ppr1p was investigated in a yeast nuclear extract-based in vitro transcription system (28). Plasmid DNA bearing seven consensus Ppr1p binding sites (10) upstream of the E4 gene, shown at the top in Fig. 4, was incubated with purified Ppr1p and yeast nuclear extract. Transcripts from the E4 gene were then analyzed by primer extension. In the absence of other added factors, Ppr1p was found to be unable to promote transcription of the E4 gene (Fig. 4; compare lanes 1 and 2).
FIG. 4.
Transcriptional activity of purified Ppr1p. In vitro transcription reactions contained 50 nM purified Ppr1p where indicated and were supplemented with the compounds indicated, each at a final concentration of 1 mM, except for aspartic acid (1.6 mM) and carbamoyl aspartic acid (0.8 mM). Transcription products from the template shown at the top were analyzed by primer extension. Positions of the primer and the E4 extension products are indicated.
To investigate the effect of pyrimidine biosynthetic intermediates on the transcriptional activity of Ppr1p, in vitro transcription reactions were performed in the presence of the intermediates. Figure 4 shows that either DHO or OA allows transcriptional activation in the presence of Ppr1p (Fig. 4, lanes 8 and 9). Other intermediates of the pyrimidine biosynthetic pathway had no effect on the ability of Ppr1p to activate transcription (Fig. 4, lanes 3 to 7 and 10 to 14).
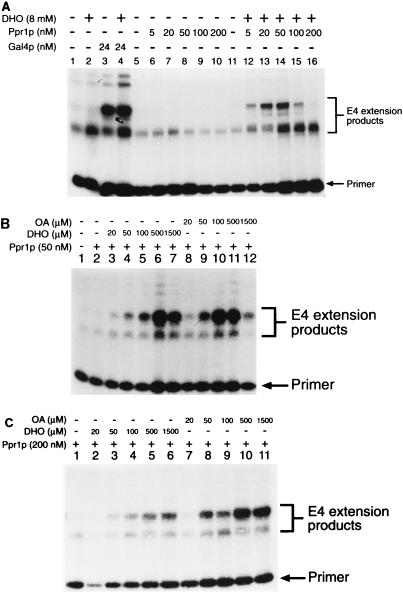
The ability of DHO and OA to promote transcriptional activation is specific to Ppr1p. DHO had no effect on the basal level of transcription in vitro (Fig. 5A, lanes 1 and 2) or on the levels of activated transcription derived from a Gal4p-based activator (Fig. 5A, lanes 3 and 4). Titration of Ppr1p into the transcription reactions in the absence of DHO (Fig. 5A, lanes 6 to 10) showed that even at high concentrations, Ppr1p alone did not activate transcription. At high concentrations, Ppr1p actually seemed to repress the basal level of transcriptional activity. In the presence of DHO, Ppr1p activated transcription (Fig. 5A, lanes 12 to 16). Under the conditions used in this experiment, activation increased as the level of Ppr1p increased. However, above a concentration of 50 nM Ppr1p (Fig. 5A, lane 14), the level of activation decreased. To investigate this phenomenon further, we set up transcription reactions containing fixed concentrations of Ppr1p and then titrated in either DHO or OA (Fig. 5B and C). At relatively low concentrations of Ppr1p (50 nM [Fig. 5B]), DHO and OA were both able to promote transcriptional activation. OA was approximately twofold more efficient than DHO at activating Ppr1p (Fig. 5B; compare lanes 5 and 10). At high concentrations of either inducer molecule, however, transcriptional activity was inhibited (Fig. 5B, lanes 7 and 12). Higher concentrations of Ppr1p (200 nM [Fig. 5C]) overcome the effect of transcriptional inhibition by high concentrations of either DHO or OA (Fig. 5C, lanes 6 and 11). OA remained more efficient at promoting transcription mediated by Ppr1p (Fig. 5C; compare lanes 3 and 8) at this higher concentration of protein. The decrease in transcriptional activity at high concentrations of DHO and OA cannot be attributed to loss of Ppr1p DNA binding. Results of electrophoretic mobility shift assays indicated that DNA binding by Ppr1p in the presence of cell extracts was unaffected by either DHO or OA present at concentrations of up to 10 mM (data not shown).
FIG. 5.
Concentration effects of Ppr1p, DHO, and OA. (A) In vitro transcription reactions contained pG5E4 (lanes 1 to 4) or pPPR7E4 (lanes 5 to 16) as template DNA. Reactions contained Gal4p (amino acids 1 to 93 fused to 768 to 881) or Ppr1p at the levels indicated. DHO was added to the reactions as indicated. (B and C) In vitro transcription reactions were performed with pPPR7E4 as the template DNA. Reactions contained Ppr1p and either DHO or OA at the concentrations indicated. Positions of the primer and the E4 extension products in each case are indicated.
Localization of the DHO/OA-responsive domain of Ppr1p.
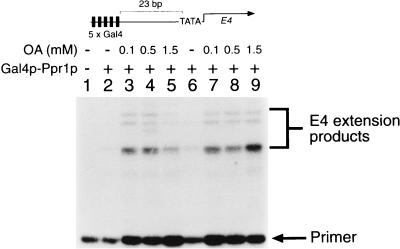
To define the activation domain of Prp1p, we constructed a number of chimeric proteins in which the carboxyl-terminal end of Ppr1p was fused to the DNA binding domain of Gal4p. The transcriptional properties of one of these chimeric proteins, Gal4p(1-93)–Ppr1p(770-904), measured from a template containing Gal4p binding sites is shown in Fig. 6. The chimera had no transcriptional activity on its own (Fig. 6, lane 2), but transcriptional activity of the protein was observed in the presence of OA (Fig. 6, lane 3 to 5 and 7 to 9). As we observed for full-length Ppr1p, the inhibition of transcriptional activity of the Gal4p-Ppr1p fusion at high levels of OA (Fig. 6, lane 5) was overcome by increasing the protein concentration (Fig. 6, lane 9). These data provide strong evidence that both the transcriptional activation domain of Ppr1p and the DHO-OA interaction region are localized within the C-terminal 134 amino acids of the protein.
FIG. 6.
The C-terminal region of Ppr1p is responsive to OA-induced stimulation of transcriptional activity. Transcription reactions contained plasmid pG5E4 and Gal4p(1-93)–Ppr1p(770-904) at either 250 nM (lanes 2 to 5) or 450 nM (lanes 6 to 9). The reactions were supplemented with OA at the concentrations indicated. Positions of the primer and the E4 extension products are indicated.
DISCUSSION
The regulation of de novo pyrimidine biosynthesis in yeast is complex and occurs at several levels. The enzymatic activities of the proteins of the pathway are subject to endpoint inhibition, e.g., the inhibition of Ura2p activity by UTP (1, 22). High levels of UTP also down-regulate URA2 gene expression (22). Elegant genetic and biochemical experiments (9) have led to the suggestion that an accumulation of certain early intermediates of the pyrimidine biosynthetic pathway (DHO) increase the activity of enzymes further down the pathway. This increased activity has been attributed to increased expression of the genes encoding those enzymes (14). Here we show that DHO and OA convert a transcription factor, Ppr1p, from a transcriptionally inert to an active state. Thus, the regulation of the pyrimidine biosynthetic pathway may be considered as occurring in the following manner.
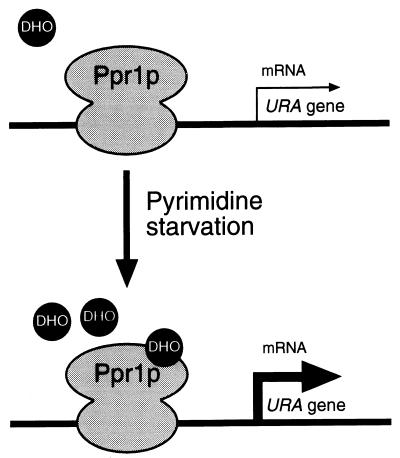
In conditions of pyrimidine abundance, the URA genes (Fig. 1) are transcribed at a basal constitutive level. The enzymatic activity of Ura2p, and the expression of the URA2 gene itself, is down-regulated by UTP (1, 7, 22). As the levels of pyrimidines become limiting, the negative effects on Ura2p are alleviated, leading to a buildup of N-carbamoyl aspartic acid, presumably within the nucleus since Ura2p is exclusively a nuclear enzyme (19). N-Carbamoyl aspartic acid will then be converted to DHO by the action of Ura4p. The role of DHO is twofold. First, it serves as a substrate for Ura1p-mediated conversion to OA in the cytoplasm (18). Second, DHO will activate Ppr1p bound constitutively upstream of the URA genes (24) (Fig. 7). Activation of Ppr1p will lead to increased production of the enzymes of the pyrimidine biosynthetic pathway and will promote the synthesis of UMP and eventually UTP. High levels of DHO and OA may have the effect of inhibiting the transcriptional activity of Ppr1p and consequently down-regulate the pathway.
FIG. 7.
Model for the activation of Ppr1p. DNA-bound Ppr1p senses the level of DHO and/or OA within the nucleus. At low DHO levels, the URA genes are transcribed at a constitutive level. Once the concentration of DHO rises above a critical level, Ppr1p promotes the transcription of its target genes (URA1, URA3, URA4, and URA10). DHO interacts with the carboxyl-terminal region of Ppr1p to release the activation domain and allow it to interact with the RNA polymerase II transcriptional machinery. Activating the URA genes will promote the conversion of DHO to OA and eventually the biosynthesis of UTP and CTP. UTP will down-regulate the enzymatic activity of Ura2p, thereby modulating the induction of DHO formation.
We find that both DHO and OA are capable of converting Ppr1p from a transcriptionally inactive state to an active one. It is possible that this effect is mediated though a component of the nuclear extract used in the in vitro transcription assays rather than a direct effect on Ppr1p itself. We believe that a direct effect is more likely since the nuclear extract is produced from yeast cells grown in rich (uracil-containing) medium, under which conditions Ppr1p should not be active. Further biochemical experiments will be required to determine if Ppr1p directly binds DHO/OA. OA is more efficient than DHO at the conversion of Ppr1p into a transcriptional activator. This finding raises the question as to the nature of the physiological inducer of Ppr1p. Ura2p is known to be an exclusively nuclear enzyme in S. cerevisiae (19). The cellular location of Ura4p has not been identified, but Ura1p (which converts DHO to OA) is found in the cytoplasm (18). Therefore, assuming that neither DHO or OA is nuclear excluded, either should be available for interaction with Ppr1p.
How might interaction with DHO or OA convert Ppr1p into a transcriptional activator? The DHO-OA interaction region of Ppr1p colocalizes with the transcriptional activation domain since Ppr1p(770-904) fused to a heterologous DNA binding domain activates transcription in a DHO/OA-dependent fashion. This finding confirms that the predominantly acidic carboxyl-terminal end of Ppr1p (8) is indeed the activation domain and also suggests that activation and response to DHO and/or OA are closely linked. It is tempting to speculate that in the absence of DHO or OA, the activation domain of Ppr1p is constrained in such a way that it is not visible to the transcriptional machinery. Upon binding of either DHO or OA, Ppr1p undergoes a conformational change to release the activation domain, thereby promoting transcription. Parallels can be drawn between our results with Ppr1p and those obtained for Leu3p, the activator of the branched-chain amino acid metabolic genes (5, 6). Leu3p is also activated by a small-molecule intermediate of the pathway it controls, α-isopropylmalate (26). The precise effect of α-isopropylmalate on Leu3p is unclear, but it appears to disrupt an intramolecular interaction between the activation domain (the C-terminal end of Leu3p) and an internal region of the protein (4, 27). The release of the activation domain presumably allows transcriptional activation to occur. The colocalization of the activation domain and DHO/OA-responsive region that we observe for Ppr1p may suggest that if this type of intramolecular repression occurs with Ppr1p, then it does so over a more limited range.
In vitro, we observe that high concentrations of either DHO or OA inhibit the transcriptional activity of Ppr1p. This inhibition can be relieved by higher concentrations of Ppr1p. High levels of DHO or OA have no effect on other activators (Fig. 5A), and so it appears that this effect is specific for Ppr1p. We have no evidence that this inhibition is physiologically significant, but it is possible that Ppr1p contains two binding sites for DHO and OA. Binding to the first site induces Ppr1p activity, while binding at both sites—presumably requiring a higher concentration of DHO and OA—inhibits Ppr1p activity. This may provide another mechanism for controlling the flux through the pyrimidine biosynthetic pathway. The intracellular levels of DHO, OA, and Ppr1p within yeast have not yet been experimentally determined.
It has previously been shown that DNA binding by Ppr1p is independent of DHO (24). We also observe constitutive Ppr1p DNA binding in vitro, but high-affinity binding of the full-length protein is dependent on a component found in a number of cell extracts (Fig. 3). The identity of this component is unclear, although it does not appear to be a protein or a pyrimidine or purine biosynthetic intermediate. It should also be noted that other C6 zinc cluster proteins purified from baculovirus-infected insect cells are able to bind to their cognate DNA sites without the requirement for added cell extracts (3a). The effects of the extracts on the binding of full-length Ppr1p are large. In the absence of the extract, high concentrations of Ppr1p are required to observe even modest levels of DNA binding. The extracts also aid DNA binding of the isolated Ppr1p DNA binding domain, although the effect is observed only at low concentrations of Ppr1p(29-123). We therefore suggest that the effect of the compound in the extract is to stabilize the Ppr1p-DNA interaction. The relevance of this effect in vivo is not clear at present, and its potential ramifications for the activation of URA gene expression await identification of the factor from the extracts.
ACKNOWLEDGMENTS
We thank Josh Brickman, Stanley Liang, and Ronen Marmorstein for the gifts of plasmids; Judith Stanway, Ian Taylor, and Rob Hockney, Zeneca Pharmaceuticals, for help and assistance in making proteins in insect cells; and Adam Platt, Cristina Merlotti, and Ronen Marmorstein for carefully reading the manuscript.
This work was supported by grants from the Biotechnology and Biological Sciences Research Council to R.J.R.
REFERENCES
- 1.Antonelli R, Estevez L, Denis-Duphil M. Carbamyl-phosphate synthetase domain of the yeast multifunctional protein Ura2 is necessary for aspartate transcarbamylase inhibition by UTP. FEBS Lett. 1998;422:170–174. doi: 10.1016/s0014-5793(98)00010-6. [DOI] [PubMed] [Google Scholar]
- 2.de Montigny J, Kern L, Hubert J C, Lacroute F. Cloning and sequencing of URA10, a second gene encoding orotate phosphoribosyl transferase in Saccharomyces cerevisiae. Curr Genet. 1990;17:105–111. doi: 10.1007/BF00312853. [DOI] [PubMed] [Google Scholar]
- 3.Denis-Duphil M. Pyrimidine biosynthesis in Saccharomyces cerevisiae: the ura2 cluster gene, its multifunctional enzyme product, and other structural or regulatory genes involved in de novo UMP synthesis. Biochem Cell Biol. 1989;67:612–631. doi: 10.1139/o89-094. [DOI] [PubMed] [Google Scholar]
- 3a.Flynn, P. J., and R. J. Reece. Unpublished observations.
- 4.Friden P, Reynolds C, Schimmel P. A large internal deletion converts yeast LEU3 to a constitutive transcriptional activator. Mol Cell Biol. 1989;9:4056–4060. doi: 10.1128/mcb.9.9.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friden P, Schimmel P. LEU3 of Saccharomyces cerevisiae activates multiple genes for branched-chain amino acid biosynthesis by binding to a common decanucleotide core sequence. Mol Cell Biol. 1988;8:2690–2697. doi: 10.1128/mcb.8.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friden P, Schimmel P. LEU3 of Saccharomyces cerevisiae encodes a factor for control of RNA levels of a group of leucine-specific genes. Mol Cell Biol. 1987;7:2708–2717. doi: 10.1128/mcb.7.8.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaquet L, Serre V, Lollier M, Penverne B, Herve G, Souciet J L, Potier S. Allosteric regulation of carbamoylphosphate synthetase-aspartate transcarbamylase multifunctional protein of Saccharomyces cerevisiae: selection, mapping and identification of missense mutations define three regions involved in feedback inhibition by UTP. J Mol Biol. 1995;248:639–652. doi: 10.1006/jmbi.1995.0248. [DOI] [PubMed] [Google Scholar]
- 8.Kammerer B, Guyonvarch A, Hubert J C. Yeast regulatory gene PPR1. I. Nucleotide sequence, restriction map and codon usage. J Mol Biol. 1984;180:239–250. doi: 10.1016/s0022-2836(84)80002-9. [DOI] [PubMed] [Google Scholar]
- 9.Lacroute F. Regulation of pyrimidine biosynthesis in Saccharomyces cerevisiae. J Bacteriol. 1968;95:824–842. doi: 10.1128/jb.95.3.824-832.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang S, Marmorstein R, Harrison S C, Ptashne M. DNA sequence preferences of GAL4 and PPR1: how a subset of Zn2Cys6 binuclear cluster proteins recognizes DNA. Mol Cell Biol. 1996;16:3773–3780. doi: 10.1128/mcb.16.7.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liljelund P, Losson R, Kammerer B, Lacroute F. Yeast regulatory gene PPR1. II. Chromosomal localization, meiotic map, suppressibility, dominance/recessivity and dosage effect. J Mol Biol. 1984;180:251–265. doi: 10.1016/s0022-2836(84)80003-0. [DOI] [PubMed] [Google Scholar]
- 12.Lillie J W, Green M, Green M R. An adenovirus E1a protein region required for transformation and transcriptional repression. Cell. 1986;46:1043–1051. doi: 10.1016/0092-8674(86)90704-x. [DOI] [PubMed] [Google Scholar]
- 13.Lin Y-S, Carey M F, Ptashne M, Green M R. GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell. 1988;54:659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- 14.Loison G, Jund R, Nguyen-Juilleret M, Lacroute F. Evidence for transcriptional regulation of dihydroorotic acid dehydrogenase in Saccharomyces cerevisiae. Curr Genet. 1981;3:119–123. doi: 10.1007/BF00365715. [DOI] [PubMed] [Google Scholar]
- 15.Loison G, Losson R, Lacroute F. Constitutive mutants for orotidine 5 phosphate decarboxylase and dihydroorotic acid dehydrogenase in Saccharomyces cerevisiae. Curr Genet. 1980;2:39–44. doi: 10.1007/BF00445692. [DOI] [PubMed] [Google Scholar]
- 16.Losson R, Lacroute F. Cloning of a eukaryotic regulatory gene. Mol Gen Genet. 1981;184:394–399. doi: 10.1007/BF00352511. [DOI] [PubMed] [Google Scholar]
- 17.Marmorstein R, Harrison S C. Crystal structure of a PPR1-DNA complex: DNA recognition by proteins containing a Zn2Cys6 binuclear cluster. Genes Dev. 1994;8:2504–2512. doi: 10.1101/gad.8.20.2504. [DOI] [PubMed] [Google Scholar]
- 18.Nagy M, Lacroute F, Thomas D. Divergent evolution of pyrimidine biosynthesis between anaerobic and aerobic yeasts. Proc Natl Acad Sci USA. 1992;89:8966–8970. doi: 10.1073/pnas.89.19.8966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagy M, Laporte J, Penverne B, Herve G. Nuclear localization of aspartate transcarbamoylase in Saccharomyces cerevisiae. J Cell Biol. 1982;92:790–794. doi: 10.1083/jcb.92.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohashi Y, Brickman J M, Furman E, Middleton B, Carey M. Modulating the potency of an activator in a yeast in vitro transcription system. Mol Cell Biol. 1994;13:2731–2739. doi: 10.1128/mcb.14.4.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponticelli A S, Struhl K. Analysis of Saccharomyces cerevisiae his3 transcription in vitro: biochemical support for multiple mechanisms of transcription. Mol Cell Biol. 1990;10:2832–2839. doi: 10.1128/mcb.10.6.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Potier S, Lacroute F, Hubert J C, Souciet J L. Studies on transcription of the yeast URA2 gene. FEMS Microbiol Lett. 1990;60:215–219. doi: 10.1016/0378-1097(90)90374-y. [DOI] [PubMed] [Google Scholar]
- 23.Reece R J, Rickles R J, Ptashne M. Overproduction and single-step purification of GAL4 fusion proteins from Escherichia coli. Gene. 1993;126:105–107. doi: 10.1016/0378-1119(93)90596-u. [DOI] [PubMed] [Google Scholar]
- 24.Roy A, Exinger F, Losson R. cis- and trans-acting regulatory elements of the yeast URA3 promoter. Mol Cell Biol. 1990;10:5257–5270. doi: 10.1128/mcb.10.10.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Roy, A., and R. Losson. Personal communication.
- 25.Schjerling P, Holmberg S. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 1996;24:4599–4607. doi: 10.1093/nar/24.23.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sze J-Y, Woontner M, Jaehning J A, Kohlhaw G B. In vitro transcriptional activation by a metabolic derivative: activation of Leu3 depends on α-isopropylmalate. Science. 1992;258:1143–1145. doi: 10.1126/science.1439822. [DOI] [PubMed] [Google Scholar]
- 27.Wang D, Hu Y, Zheng F, Zhou K, Kohlhaw G B. Evidence that intramolecular interactions are involved in masking the activation domain of the transcriptional activator Leu3p. J Biol Chem. 1997;272:19383–19392. doi: 10.1074/jbc.272.31.19383. [DOI] [PubMed] [Google Scholar]
- 28.Wu Y, Reece R J, Ptashne M. Quantitation of putative activator-target affinities predicts transcriptional activating potentials. EMBO J. 1996;15:3951–3963. [PMC free article] [PubMed] [Google Scholar]