Abstract
Translation is regulated predominantly by an interplay between cis elements at the 3′ and 5′ ends of mRNAs and trans-acting proteins. Cyclosporin A (CsA), a calcineurin antagonist and blocker of interleukin-2 (IL-2) transcription in T cells, was found to inhibit translation of IL-3 mRNA in autocrine mast cell tumor lines. The mechanism involved ribosome-associated poly(A) shortening and required an intact AU-rich element in the 3′ untranslated region. FK506, another calcineurin inhibitor, shared the effect. The translational inhibition by CsA was specific to oncogenically induced lymphokines IL-3 and IL-4 but not to IL-6, c-jun, and c-myc, which are expressed in the nonmalignant precursor cells. Furthermore, no translational down-regulation of the mRNA was observed in IL-3-transfected precursor cells. These data suggest that translational silencing is associated with the tumor phenotype.
Autocrine production of cytokines has been reported to occur in a variety of experimental and clinical malignancies (18, 32, 65). In several cases, aberrant expression of the growth factor involved rearrangements of the respective genes (6, 27, 37, 56, 64). Constitutive growth factor expression due to transcript stabilization has also been described (1, 23, 27, 53).
We have been studying a murine tumor model where mastocytomas obtained from v-H-ras-transformed PB-3c cells (an interleukin-3 [IL-3]-dependent bone marrow mast cell line) are characterized by autocrine IL-3 expression (40). In the majority of the tumors (class I), IL-3 expression was due to enhanced transcript stability. In a small number of tumors (class II), however, increased IL-3 expression occurred due to the transcriptional activation of an allele via insertion of retroviral intracisternal A particle sequences into the 5′-flanking region (26, 27). Alternatively, the IL-3-dependent precursor cells could be transformed by the direct expression of IL-3 cDNA (42). These data suggested a key role for IL-3 expression in the transformation of these cells, and v-H-ras oncogene expression is thought to facilitate the activation of a signal transduction pathway leading to oncogenic IL-3 mRNA expression.
The stable mRNA in class I cells could be down-regulated and IL-3 dependence could be restored by somatic cell fusion with IL-3-dependent PB-3c cells (19, 27), arguing that transcript stability was the result of a recessive mutation and occurred by a trans-acting mechanism. In addition, IL-3 mRNA expression in class I cells was inhibited by treatment with the immunosuppressant drugs cyclosporin A (CsA), FK506, and rapamycin (2, 41). The inhibition involved destabilization of the transcripts, and the process required an intact AU-rich element (ARE) in the 3′ untranslated region (UTR) of the mRNA. These results suggested that the tumors were generated as a result of a defect in the IL-3 mRNA decay pathway operative in normal cells. The destabilization of the transcripts by the drugs restored the decay function, as did cell fusion, thus causing “tumor reversion.” With a view to identifying the possible factors that play a role in the regulation of IL-3 expression, we set out to investigate the different degradation pathways in our class I and class II tumor cells.
In normal cells, cytokine mRNAs are short-lived, with half lives in the range of 30 min to 1 h (reference 46 and references therein). In tumors, as mentioned above, decay is often slowed (45). This instability is mediated mainly by AREs ranging from 50 to 150 nucleotides, present in the 3′ UTR of most of the mRNAs for cytokines and proto-oncogenes. A pentamer motif, AUUUA, was previously suggested as the minimal destabilizing element (11, 54). Recent data indicate that the consensus nonamer UUAUUUA(U/A)(U/A), especially two juxtaposed elements, are highly active (33, 67). A mutational analysis of IL-3 has shown that a cluster of six AUUUA repeats containing the nonamer regulate decay and stabilization by Ca2+ in the PB-3c mast cells (57).
Studies on the c-fos ARE have suggested a biphasic pattern of mRNA decay (12, 46, 55). Accordingly, poly(A) shortening to 30 to 60 nucleotides is the first step of mRNA degradation (12, 55, 61). In mammalian cells, AREs of c-fos, c-myc and granulocyte-macrophage colony-stimulating factor (GM-CSF) have been implicated in accelerating cytoplasmic poly(A) shortening (9, 12, 14). Two classes of ARE have been proposed; class I ARE, present in c-fos and c-myc, directs synchronous poly(A) shortening, implying a distributive or nonprocessive nuclease cleavage of poly(A) tails, and class II ARE, present in GM-CSF and IL-3 mRNAs, mediates asynchronous deadenylation (processive ribonuclease action), resulting in the formation of fully deadenylated intermediates (12–14).
The AREs are thought to regulate mRNA decay via specific binding factors. Several such ARE binding proteins have been identified (7, 8, 36, 39, 43); however, the exact function of most of these is not clearly understood. Treatment of mast cells and T cells with calcium ionophores and/or phorbol esters (phorbol myristate acetate) has been shown to result in specific stabilization of IL-3 and GM-CSF mRNAs (62). The loss of ARE-binding activity of AU-B was found to correlate with stabilization of lymphokine mRNAs in phorbol myristate acetate-stimulated T cells, and AU-B has been shown to bind to the 3′ UTR of GM-CSF and not c-myc (7).
Studies with yeast have established the presence of non-ARE 3′ UTR instability elements that promote rapid poly(A) shortening and subsequent decay (38). Poly(A)-binding protein (PABP) has been identified as a key molecule that regulates deadenylation, since strains deficient in PABP failed to shorten poly(A) tails (35, 48). These studies have established the presence of different routes for mRNA degradation such as deadenylation-dependent or deadenylation-independent decapping with a subsequent exonucleolytic cleavage (4, 16, 47). Decapping and decay appear to initiate after poly(A) tails are shortened to less than 15 nucleotides, at which point PABP binding is minimal (49), implicating PABP as a negative regulator of decapping and subsequent exonucleolytic cleavage (4, 16).
Although factors contributing to a link between mRNA decay and translation have not been fully elucidated, ribosome-associated destabilization of short-lived mRNAs has been reported (51, 60). Recently, evidence for a physical association between mRNA decay and the translation machinery in yeast has emerged and a circular “closed loop” model has been proposed (28). Accordingly, the PABP-poly(A) tail complex interacts with the 5′ region of the mRNA via the cap-binding protein, eIF-4E, and the adapter molecule, eIF-4G. The binding affinity of PABP to the poly(A) tail could presumably be regulated by various 3′ UTR sequences and their binding factors that might function as activators or inhibitors (24, 28, 29, 50).
In the present study, we show that treatment with the immunosuppressants CsA and FK506 leads to a ribosome-associated deadenylation of the labile IL-3 mRNA in the class II tumors and that this process requires ongoing translation. However, the stable transcripts in the class I cells are most probably degraded by a non-ribosome-associated mechanism. Experiments with IL-3 transgenes suggested that this pathway operates in Jurkat T cells and that deletion of the ARE makes the transcripts resistant to CsA-mediated poly(A) tail shortening. Surprisingly, the effect was observed in the tumor cells with the oncogenically induced lymphokines IL-3 and IL-4 but not with IL-6, which is also expressed by the precursor cells. Furthermore, the IL-3 transcripts expressed by a transgene in the precursor cells were not subjected to translational silencing by CsA.
MATERIALS AND METHODS
Cell lines and tissue culture.
PB-3c is a cloned, IL-3-dependent mast cell line derived from murine DBA/2 bone marrow, and the IL-3 autocrine tumor V2D1, V4D6, and V3D7 were described previously (40). All cell lines, including Jurkat and CTL44 T cells, were cultured in Iscove modified Dulbecco medium (IMDM) as described previously (40).
Nuclear run-on assays.
Preparation of nuclei, in vitro transcription, and hybridization were done as described previously (41).
Northern blot analysis.
Approximately 105 cells/ml were treated with the indicated concentrations of CsA overnight or were left untreated. For method A, the total cytoplasmic RNA was isolated by the method of Gough (21). The cells were lysed in the presence of Nonidet P-40 (NP-40) and nuclei were removed by centrifugation. RNA was recovered from the supernatant by sodium dodecyl sulfate-urea-phenol-chloroform extraction and precipitation. For method B, lysis was performed with sodium guanidinium thiocyanate, a very effective protein denaturant as well as a strong RNase inhibitor (15). By this method, nuclear as well as membrane-bound ribosome-associated RNAs were extracted. For method C, to recover the membrane-bound ribosome associated RNA, deoxycholate (DOC) was used (31) for lysis, followed by removal of nuclei by centrifugation. To inhibit the nuclear RNases that are released due to DOC treatment, 400 U of RNasin/ml was included in the lysis buffer. RNA was subsequently isolated by sodium dodecyl sulfate-urea-phenol-chloroform extraction.
Poly(A) mRNA isolation, agarose gel analysis, prehybridization, and hybridization were done as described previously (27). The IL-3 probes were generated from an SP6 vector containing IL-3 cDNA fragments; chicken β-actin, murine c-myc, c-jun, and IL-6 probes were generated by random priming (26).
Methylcellulose cloning.
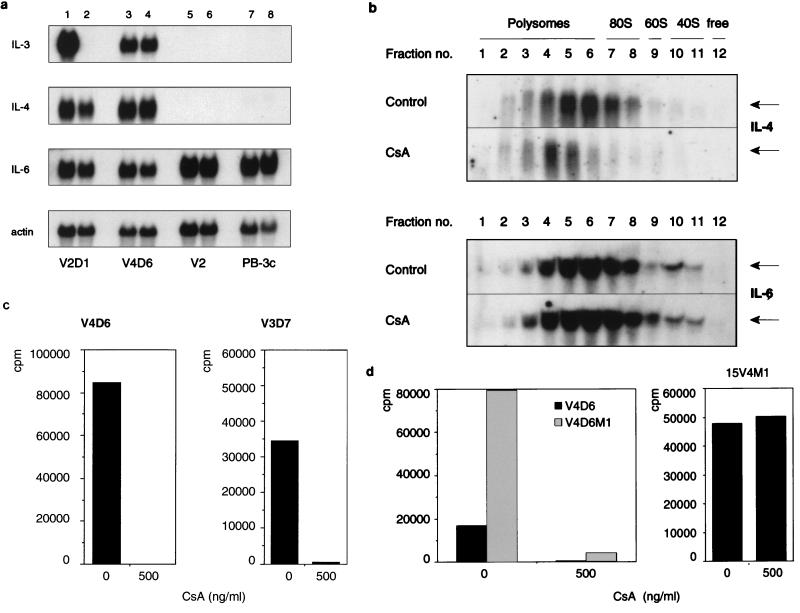
The cloning was done in 1-ml cultures as described previously (40). The cloning mixtures contained 1% methylcellulose in IMDM, 1% bovine serum albumin, 20% fetal calf serum, 300 μg of iron-saturated human transferrin per ml, and the indicated concentrations of CsA. V4D6 cells were cloned in the absence of exogenous IL-3, while the PB-3c (clone 20) precursor cells required the addition of IL-3 (41). The numbers of colonies were determined after 10 days of seeding. For V4D6, 100% corresponds to 2,600 colonies/ml, and for clone 20 cells, 100% is approximately 10,000 colonies/ml. The cloning efficiency of autocrine tumor cells in the absence of the growth factor is very much lower than that of the precursor cells, which can be cloned only in presence of the growth factor. The number of cells per clone gradually decreased with increasing concentrations of CsA with V4D6 cells, whereas no such effect was detectable with PB-3c.
Mitogenicity assay.
For preparation of the supernatants, cells were washed three times in IMDM and plated at 1 × 105/ml for V4D6 or 5 × 105/ml for V3D7, in IMDM with 10% fetal calf serum (40). Except for the dose-response experiments, 500 ng of CsA per ml was added wherever indicated at time zero. Supernatants (20%) were assayed on IL-3-dependent PB-3c cells as described previously (40). IL-4 was assayed on the IL-4-dependent murine T-cell clone CTL44 (20). For preparation of cytosol, cells were washed in IMDM and plated at 2 × 105/ml in 50 ml for V4D6, 2 × 105/ml in 150 ml for V3D7, and 2 × 105/ml in 50 ml for 15V4M1. After incubation, the cells were harvested, washed three times in phosphate-buffered saline, resuspended in 500 ml of phosphate-buffered saline, and frozen at −70°C until further use. Cytosol was prepared by sonication for 15 s with an ultrasonic disintegrater. The lysate was centrifuged at full speed in an Eppendorf centrifuge at 4°C for 10 min, and 5% supernatants were used for the assay, unless indicated otherwise.
IL-3 transgenes and electroporation.
pIL-3 M1 hph (wild-type IL-3 plasmid) and pIL-3 M1ΔAU hph (ARE-deleted IL-3 plasmid) have been described previously (41). These plasmids contain the hygromycin resistance gene (hph) and were introduced into Jurkat, V4D6, and 15V4 cells by electroporation at 300 V and 960 μF, selected for hygromycin B resistance, generating stable transfectants. The Jurkat cells used here were previously transfected with a v-H-ras oncogene expressed from the Moloney murine leukemia virus long terminal repeat (unpublished data).
Polysome analysis.
For polysome gradients, 3 × 107 control or CsA-treated cells were lysed in 300 μl of hypertonic buffer containing NP-40, DOC, and RNasin (0.26 U/ml) and centrifuged through a 5-ml sucrose gradient (17.1 to 51%) for 75 min at 50,000 rpm in an SW 55 rotor. Twelve fractions were collected, and RNA samples were prepared as described by Jeffries et al. (31).
In vitro reconstitution.
To check for the presence of RNase activity in V4D6 cells, an exogenous RNA was incubated with lysates for 0 to 2 h at 4°C (similar conditions to those for the sucrose gradient analysis), extracted, and analysed by Northern blotting. Exogenous RNA was either from V4D6 cells transfected with a plasmid carrying the full-length IL-3 gene under the control of Moloney murine leukemia virus promoter (26) or from V2D1 cells transfected with an ARE-deleted IL-3 transgene (41), which provided abundant transcripts for detection. Lysates were prepared from V4D6 cells as described for polysome analysis (31), and 15 μl was incubated with the exogenous RNA. Assays were also done in the presence of RNasin (3 U/ml) or additional polysomes from V4D6 cells. To prepare the polysomes, lysates (300 μl) from 3 × 107 V4D6 cells were layered over a cushion of 17.1% sucrose and centrifuged at 50,000 rpm for 1 h in an SW 55 rotor. The supernatant was decanted, the walls were dried, and the polysome pellet was resuspended in 300 μl of hypertonic buffer (31). A 5-μl volume from this preparation was added to the incubation mixture containing the lysate and exogenous RNA.
RNase H analysis.
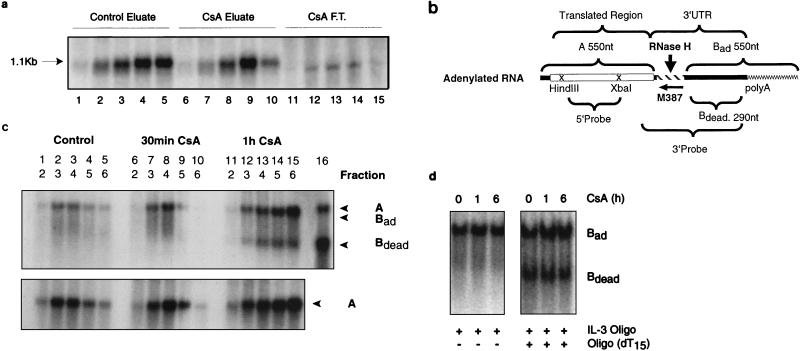
RNAs from the gradient fractions were hybridized to an IL-3-specific oligonucleotide (M387), complementary to the sequences immediately after the stop codon or in combination with oligo(dT)15 for in vitro deadenylation. The RNAs were subsequently subjected to RNase H digestion as described previously (25). This results in the generation of the following fragments (see Fig. 5b): first, fragment A (550 nucleotides [nt]), representing the 5′ coding sequence; second, a 3′ UTR fragment of 290 nt without a poly(A) tail (Bdead); and third, a fully adenylated 3′ fragment carrying approximately 250 adenylate residues (24a), yielding a fragment (Bad) of 550 nt. 3′ fragments of variable length, depending on the degree of adenylation, were also observed as deadenylation intermediates. The blots were hybridized to a 3′ probe, which detects both 5′ and 3′ fragments, or to a 5′ probe, which recognizes only the coding sequences (see Fig. 5c). As a control for the fully deadenylated IL-3 transcripts, in vitro deadenylation was performed by combining M387 and oligo(dT)15.
FIG. 5.
Polysome-associated poly(A) shortening of IL-3 mRNA. (a) Poly(A)+ mRNA from gradient fractions was selected by oligo(dT) cellulose chromatography. Prior to selection, two adjacent sucrose gradient fractions were pooled in the case of control gradients (lanes 1 to 5). With CsA-treated cells (100 ng/ml for 4 h), two adjacent fractions from four parallel gradients were used (lanes 6 to 15). The RNA in the flowthrough fraction (F.T.) from CsA-treated samples was also processed, and it migrates slightly faster than that in the eluate (lanes 11 to 15). A size control sample was included in the experiment and is marked by an arrow. (b) Schematic representation of the IL-3 cDNA and the probes used for RNase H analysis (25). A, 5′ RNase H fragment; Bad and Bdead, fully adenylated and deadenylated 3′ RNase H fragments, respectively. The numbers indicate the expected nucleotide length. M387 is an IL-3-specific oligonucleotide (not drawn to scale). The braces below indicate the two probes used. (c) The top panel shows a Northern blot probed with the 3′ probe which recognizes both 5′ fragment A and the 3′ fragments Bad and Bdead. Note that A and Bad have almost identical sizes. Bdead becomes prominent after CsA treatment. The bottom panel shows a parallel Northern blot probed with the 5′ probe recognizing only fragment A, whose size did not change following CsA treatment. Indicated below the lane numbers are the gradient fraction numbers, which correspond to the numbers in Fig. 4a. Note that the amount of RNA used for the analysis of CsA-treated material was two and three times greater for the 30-min and 1-h samples, respectively, than that used for the control. Lane 16 shows that RNA subjected to in vitro deadenylation with oligo(IL-3) in combination with oligo(dT) yielded an upper signal for A and a lower one for Bdead. (d) RNase H analysis of total RNA from V4D6 cells treated with CsA (500 ng/ml). The RNA was prepared by method A, where membrane-bound ribosomes are not recovered. In the left panel, oligo(IL-3) alone was used for hybridization. In the right panel, oligo(IL-3) and oligo(dT) were both used (in vitro deadenylation).
RESULTS
Down-regulation of the labile IL-3 mRNA in class II tumor cells by CsA was observed only when membrane-bound ribosome-associated RNA was extracted.
We have previously reported that the autocrine IL-3 expression in class I tumor cells is the result of a posttranscriptional mechanism involving transcript stabilization by a trans-acting alteration (26). The mechanism appeared to be recessive because fusion of somatic cells to the IL-3-dependent precursor PB-3c cells led to the down-regulation of IL-3 expression and reversion to IL-3 dependence (19, 27). In contrast, class II tumor cells are characterized by the presence of retroviral intracisternal A particle sequences at the 5′-flanking region of the IL-3 gene, resulting in the transcriptional activation of the gene (27). Correspondingly, transcripts were labile, as opposed to the abnormally stable IL-3 mRNA in class I cells. Figure 1a shows a nuclear run-on analysis with the enhanced IL-3 transcriptional activity in V4D6 cells compared to V2D1 cells. The v-H-ras expression was higher in V2D1 cells than in V4D6 cells. Figure 1b shows the reported (26) abnormal stability of IL-3 mRNA in the presence of actinomycin D in class I V2D1 cells (the half life is more than 3 h) compared to class II V4D6 cells (the half life is approximately 45 min). Transcripts of c-myc decayed with a similar rate (the half life is 30 min) in both cell lines, as reported previously (26). The salient features of class I and class II tumor lines are summarized in Table 1.
FIG. 1.
IL-3 mRNA expression in class I and class II cells. (a) Nuclear run-on analysis. Nuclei were prepared from V4D6 and V2D1 cells, and in vitro transcription rates were measured as described previously (27). (b) Northern blot analysis of poly(A)+ RNA in V2D1 and V4D6 cells. The cells were treated with 5 mg of actinomycin D per ml, and RNA was prepared at the indicated time points by method A. The blots were first hybridized with IL-3 and subsequently rehybridized with c-myc and β-actin probes, as described in Materials and Methods. The half-lives of IL-3 mRNA were more than 3 h and approximately 45 min in V2D1 and V4D6 cells, respectively. c-myc mRNA decayed in both tumor cells with a similar half-life of 30 min.
TABLE 1.
Features of class I and II tumor lines
| Feature | Class I tumors (prototype V2D1) | Class II tumors (prototype V4D6) |
|---|---|---|
| IL-3 gene | No rearrangement | IAP insertion |
| Nuclear IL-3 transcription | Comparable to the precursor cells | Enhanced transcription |
| IL-3 mRNA | Abnormally stable; no cis alteration | Short lived; no cis alteration |
| Somatic cell fusion with normal PB-3c | Down-regulation of IL-3; reversion to IL-3 dependence | No down-regulation of IL-3; IL-3 independence |
Previous studies showed that the autocrine proliferation of class I cells could be inhibited by treatment with the calcineurin inhibitors CsA and FK506, which acted by down-regulating the stable IL-3 mRNA. The mechanism was posttranscriptional and involved destabilization of the transcripts and required an intact 3′ UTR (41). We now tested the effect of CsA on the labile IL-3 mRNA expressed by the class II cells. Figure 2a shows the Northern blot analysis of poly(A)+ RNA from cytoplasmic lysates of V2D1 and V4D6 cells, hybridized with an IL-3-specific RNA probe. CsA did not affect the abundance of IL-3 mRNA in V4D6 cells, while the transcripts disappeared in V2D1 cells as reported previously (41).
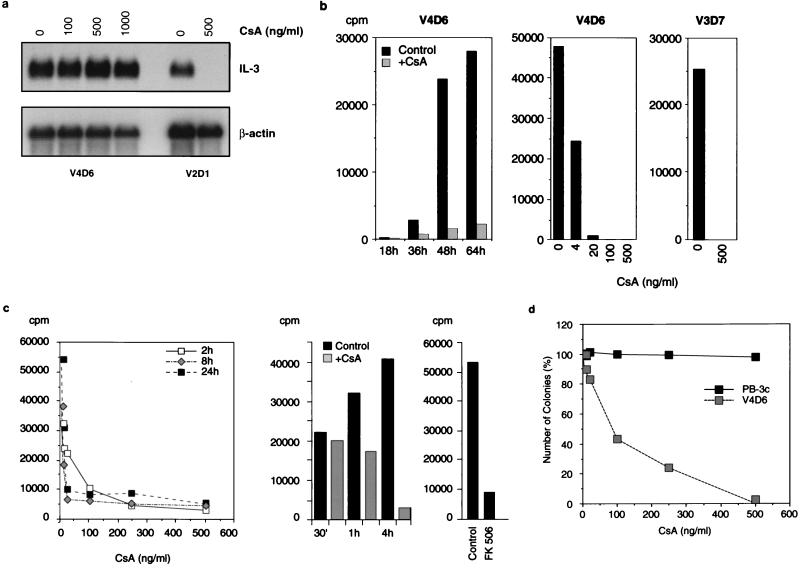
FIG. 2.
CsA and FK506 inhibit translation of IL-3 and in vitro growth of class II cells. (a) Poly(A)+ mRNA (RNA prepared by method A) from control and CsA-treated (for 16 h) class II V4D6 and class I V2D1 cells was hybridized with an IL-3 5′ probe (see Fig. 5b) and rehybridized with a β-actin probe. Drug concentrations are indicated above the lanes. (b) The left panel shows the time course. The mitogenic activity of a 20% supernatant of CsA-treated (100 ng/ml; grey bars) or untreated (black bars) V4D6 cells, assayed on IL-3-dependent PB-3c cells, is shown. CsA does not affect the mitogenic activity of exogenous IL-3 on PB-3c cells (41), excluding carryover of drug as an explanation. The middle panel shows the dose response. The cells were grown for 72 h at the indicated CsA concentrations, and 20% supernatants were tested on PB-3c. The right panel shows the result when 20% of the 10×-concentrated supernatant from V3D7 was tested. (c) The left and middle panels show the dose-response and time course, respectively, of CsA-treated and untreated cytosol (5%) from V4D6 cells assayed on PB-3c, as described for panel b. The right panel shows the result when cytosol (5%) from V4D6 cells was treated with 20 ng of FK506 per ml for 8 h. (d) Effect of CsA on colony formation. A total of 2 × 104 V4D6 or IL-3-dependent PB-3c cells per ml were plated in the absence or presence, respectively, of the indicated concentrations of CsA. The number of colonies is expressed as a percentage. Methylcellulose preparation and cloning were performed as described previously (40).
Surprisingly, however, CsA inhibited the release of IL-3 in the class II tumor lines when assayed for mitogenicity on PB-3c cells (Fig. 2b). Half-maximal inhibition was observed at 4 ng/ml, which is very similar to the reported concentration for mRNA destabilization in class I cells (41). To distinguish between inhibition of secretion and translation, cytosolic extracts from CsA-treated cells were tested for mitogenic activity. The inhibition was detectable as early as 1 h (Fig. 2c, middle panel) and the inhibitory concentration was again approximately 4 ng/ml (Fig. 2c, left panel). Similar results were obtained with V3D7, another class II tumor line (Fig. 2b, right panel, and data not shown). FK506 was also inhibitory (Fig. 2c, right panel).
We next examined the effect of CsA on colony formation by V4D6 cells in methylcellulose and observed a dose-dependent inhibition by CsA (Fig. 2d). However, as described above, the precursor cells requiring IL-3 for growth (PB-3c) were not affected by the drug, indicating that the effect is tumor specific.
To approach the apparent paradox that CsA affects IL-3 production but not cytoplasmic mRNA, we reinvestigated IL-3 mRNA levels by using different RNA preparation methods. For the preparation of RNA from hematopoietic cells which contain high concentrations of RNases, method A has routinely been used (Fig. 2a). It involves lysis of the cells with a mild detergent (NP-40) and subsequent removal of the undamaged nuclei by centrifugation (21). This ensures that the RNases present in the nuclei do not degrade the RNA during lysis. However, the membrane-bound ribosomes are not efficiently released and are thereby removed along with the nuclei. We now tested two additional methods which use very strong detergents that allow efficient extraction of the membrane-bound ribosome-associated RNA. In method B (15), the nuclear RNA also is extracted, while in method C, the nuclei are removed prior to RNA extraction. Figure 3 (upper panel, A) shows that in V4D6 cells, IL-3 transcripts were not down-regulated by CsA when NP-40 alone was used for the lysis. However, CsA-treated cells showed a reduced IL-3 signal when method B (Fig. 3, top, panel B) and method C (Fig. 3, top, panel C) were used to prepare RNA. In contrast to V4D6 cells, the CsA-induced inhibition in V2D1 cells was evident irrespective of the method by which RNA was isolated (Fig. 3, bottom). Taken together the results obtained from different RNA preparation methods as well as the data from secreted and intracellular IL-3 activity suggested that CsA inhibited IL-3 expression at the translational level.
FIG. 3.
Membrane-bound ribosome-associated RNA extraction reveals the effect of CsA. (Top) V4D6 cells. (Bottom) V2D1 cells. (A) RNA was extracted after lysis with NP-40 alone. (B) RNA was prepared by extraction with sodium guanidinium thiocyanate. (C) Membrane-bound ribosome-associated RNA was recovered by the addition of DOC.
IL-3 mRNA in class II cells is specifically degraded on the polysomes and requires ongoing translation.
To study the mechanism of translational inhibition by CsA, we examined polysome association of IL-3 transcripts by sucrose gradient analysis. In untreated V4D6 cells, IL-3 mRNA gave a broad signal over the gradient, including the ribosomal and polysomal fractions (Fig. 4a, upper panel). Following CsA or FK506 treatment, two effects were evident: (i) the total IL-3 signal intensity was reduced, and (ii) a lower band became more prominent, particularly in the polysomal fractions with a concomitant shift of the upper band toward the ribosomal fractions in drug-treated cells. A 2-h treatment was sufficient to induce these effects, and incubation for up to 24 h did not change the pattern further (data not shown).
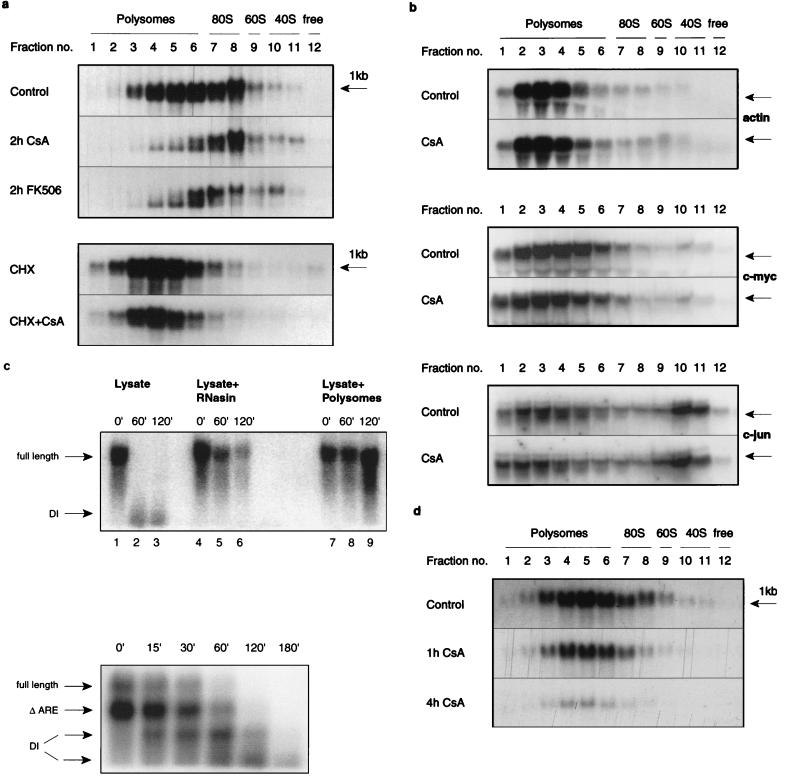
FIG. 4.
Effect of CsA and FK506 on the polysome association of IL-3 mRNA. (a) In the top panel, cells were treated where indicated for 2 h with CsA (100 ng/ml) or FK506 (20 ng/ml), and lysates from control and treated cells were subjected to sucrose density gradient centrifugation (31) and Northern blot analysis. In the bottom panel, cells were pretreated with cycloheximide (5 μg/ml) for 30 min prior to the addition of CsA for a further 2 h. As a control, cells were also treated with cycloheximide alone for 2.5 h. (b) Northern blots from sucrose density gradients were first hybridized with labelled c-jun and subsequently rehybridized with c-myc and β-actin probes. (c) Lysates of V4D6 cells were prepared as described for polysome preparation. In the top panel, the lysate was incubated at 4°C for the indicated times with exogenous total RNA from V4D6 cells transfected with full-length IL-3 transgene (lanes 1 to 3), with added RNasin (3 U/ml) (lanes 4 to 6) or with exogenous polysome preparation (lanes 7 to 9). The bottom panel shows the time course of the decay. Here, the exogenous RNA was from V2D1 cells transfected with an ARE-lacking transgene. These cells express the endogenous full-length IL-3 transcripts as well as the ARE-lacking transcripts from the transgene (see the arrows). DI, decay intermediate. (d) RNA from polysome gradients of V2D1 was analyzed as described for V4D6 cells.
When V4D6 cells were pretreated for 30 min with cycloheximide, the effects of CsA were abolished (Fig. 4a, lower panel), indicating a possible role for translational elongation. Cycloheximide stabilizes unstable mRNAs in yeast (3), and an indirect role for it in V4D6 cells cannot be ruled out. In contrast to IL-3 mRNA, CsA did not affect the polysome-associated β-actin, c-myc, or c-jun transcripts (Fig. 4b), indicating that the drug effect was specific.
Note that no signal representing the free, non-ribosome-associated IL-3 mRNA could be detected, even in the untreated gradients. To address this question, in vitro reconstitution experiments were performed. Lysates were depleted from polysomes by centrifugation and incubated with exogenous total RNA isolated from V4D6 cells expressing an IL-3 transgene which provided abundant IL-3 transcripts. The Northern blot data showed that the free IL-3 mRNA was degraded rapidly following lysis-induced decompartmentalization and that addition of exogenous polysomes or excess of RNasin prevented this process (Fig. 4c, top panel). Figure 4c, bottom panel, shows the time course of the in vitro decay process with exogenous RNA containing full-length and ARE-deleted IL-3 transcripts. Decay of both transcripts could be observed at 4°C as early as 15 min, and distinct decay intermediates could be detected. These data provided an explanation for the lack of IL-3 transcript detection in the free fractions of the sucrose gradients, because there is a time lapse of about 2 h between lysis and collection of the fractions and RNA extraction.
In contrast to the data shown with V4D6 cells, the polysome profile of class I V2D1 showed no ribosome-associated shortening of the IL-3 mRNA (Fig. 4d), but an overall decrease in signal intensity was observed following CsA treatment. This is consistent with the observed reduction in cytoplasmic mRNA (Fig. 2a and Fig. 3, bottom panel). Furthermore, different methods of RNA preparation did not alter the effect of CsA in V2D1 (Fig. 3, bottom panel). It thus appears that IL-3 mRNA degradation in V2D1 cells is cytoplasmic.
IL-3 mRNA in V4D6 cells undergoes a polysome-associated poly(A) shortening.
We next determined if the short fragment observed in the polysome gradients from V4D6 cells represented deadenylated transcripts. RNA from gradient fractions was subjected to oligo(dT)-cellulose chromatography and analyzed by Northern blotting. Poly(A) selection eliminated the qualitative difference between control and CsA-treated samples, and now the lower band was not detectable in the eluates (Fig. 5a, lanes 1 to 10). However, the lower band could now be detected in the flowthrough fraction (lanes 11 to 15). This suggested that the transcripts which formed the lower band lacked a poly(A) tail. To confirm this, we performed an RNase H analysis, schematically described in Fig. 5b. Hybridization with an IL-3-specific oligonucleotide (M387) followed by RNase H digestion generates adenylated and deadenylated 3′ fragments (Bad, Bdead), as well as an invariant 5′ fragment termed A (25). Northern blots were hybridized with the 3′ probe recognizing both the 5′ fragment A and the 3′ fragments Bad and Bdead. The pattern from control cells (Fig. 5c, top, lanes 1 to 5) showed a single band of about 550 nt resulting from an overlap of the 5′-terminal A fragment and the fully adenylated 3′ fragment (Bad) carrying about 250 adenylate residues. After 30 min of CsA treatment (lanes 6 to 10), a smear indicative of partial deadenylation was observed, and after 1 h (lanes 11 to 15), there was clear evidence of extensive deadenylation, as indicated by the appearance of a lower band, Bdead, of the expected size (290 nt). A probe detecting only the 5′ fragment A gave a band which did not change in size with CsA treatment (Fig. 5c, bottom), consistent with the interpretation that CsA promotes deadenylation but does not affect the 5′ portion of the transcript. No signal corresponding to fully deadenylated IL-3 was observed in V4D6 cells when cytoplasmic mRNA from CsA-treated cells prepared by method A (which does not include membrane-bound ribosome associated RNA) was subjected to RNase H analysis (Fig. 5d, left).
ARE-lacking transcripts do not undergo poly(A) shortening.
To facilitate the analysis of transcripts from an ARE-deleted transgene and to test if the effect of CsA could also be observed in another cellular background, we transfected murine IL-3 transgenes (41) into human Jurkat T cells. The full-length transcripts displayed a half-life similar to that of V4D6 cells, whereas mRNA lacking the ARE was stable for several hours (data not shown). The abundance of cytoplasmic mRNA from both full-length and ARE-deleted constructs was not altered by CsA treatment (Fig. 6a). As with V4D6 cells, gradient analysis of Jurkat cells indicated a polysome-associated degradation of the full-length transcripts (Fig. 6b, top). These transcripts displayed a characteristic pattern representing the adenylated (upper band) and the deadenylated (lower band) species in untreated cells, indicating the rapid physiological degradation. In CsA-treated cells, a decrease in the polysome-association of the adenylated mRNA and a shift of this species to the 80S ribosomal subunit can be observed. Deletion of the ARE, however, prevented CsA from exerting an effect on IL-3 mRNA associated with the ribosomes (Fig. 6b, bottom).
FIG. 6.
ARE-lacking IL-3 mRNA is resistant to ribosome-associated poly(A) shortening. (a) The middle panel shows a Northern blot analysis of total RNA from Jurkat cells transfected with full-length and ARE-deleted IL-3 transgenes, hybridized with an IL-3 probe. The full-length transgene expressed much less RNA than did the ARE-deleted transgene (data not shown), and as a consequence, the signal from the full-length IL-3 transcripts in lanes 1 and 2 is weak. The top panel shows a longer exposure of the full-length IL-3. The bottom panel shows the same blot rehybridized with β-actin probe. Lanes: 1 and 3, untreated cells; 2 and 4, cells treated with 1 mg of CsA per ml for 8 h; 1 and 2, cells transfected with full-length IL-3 construct; 3 and 4, cells transfected with ARE-deleted construct. (b) RNA from polysome gradients of transfected Jurkat cells was analyzed for the IL-3 signal. The top panel shows cells transfected with the full-length construct. The bottom panel shows cells transfected with the ARE-deleted construct. The cells were treated with 500 ng of CsA per ml for 8 h. The bottom panel (control and CsA-treated, ARE-deleted IL-3) was exposed approximately 10-fold less than the top panel (control and CsA-treated, full-length IL-3).
Translational silencing of ARE-containing transcripts requires a tumor phenotype.
In addition to IL-3, IL-4 is expressed by both class I (V2D1) and class II tumors (V4D6, V3D7) but not by v-H-ras-transfected premalignant precursor (V2) or by the normal nonmalignant precursor (PB-3c) cells (Fig. 7a). IL-4 synergizes with IL-3 in enhancing the in vitro growth of PB-3c mast cells (data not shown). Furthermore, in PB-3c cells, IL-4 is regulated similarly to IL-3 in that both are induced by ionomycin via a posttranscriptional mechanism. In contrast to IL-3 and IL-4, IL-6 is also expressed by PB-3c cells (Fig. 7a). A comparison between Northern blot analysis of cytoplasmic RNA (RNA made with the mild detergent NP-40) and polysomal RNA shows that IL-4 mRNA is subjected to translational inhibition by CsA (Fig. 7b, top). A pathway involving ribosome-associated poly(A) shortening for IL-4 mRNA requires further investigation. IL-4 measurements with a dependent T-cell clone showed that CsA inhibited the production of biologically active IL-4 in two class II tumors, V4D6 and V3D7 (Fig. 7c), further confirming the translational down-regulation by CsA. However, CsA treatment did not affect the polysome association pattern of IL-6 (Fig. 7b, bottom).
FIG. 7.
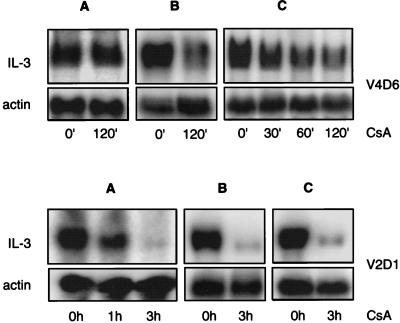
Effect of cellular background on translational inhibition by CsA. (a) Northern blot analysis of poly(A)+ RNA (RNA was extracted by method A) from PB-3c, V2, V4D6, and V2D1 cells. Lanes: 1, 3, 5, and 7, RNA from untreated cells; 2, 4, 6, and 8, RNA from cells treated with 500 ng of CsA per ml for 12 h. The blots were first hybridized with IL-6 and then rehybridized with IL-4 and IL-3 probes as described previously (41). (b) RNA from V4D6 sucrose gradient fractions was first hybridized with IL-6 (bottom) and subsequently rehybridized with IL-4 probes (top). CsA treatment (500 ng/ml) was done for 4 h. (c) IL-4 biological assay. The supernatants were those used in Fig. 2b. The assay was performed as described for IL-3, except that the IL-4-dependent T-cell clone CTL44 was used as the indicator cells. (d) IL-3 biological assay. Cytosolic extracts were prepared and assayed as described in Fig. 2b. V4D6M1 cells (2 × 105/ml in 25 ml) were treated with 500 ng of CsA per ml for 4 h, and the lysate was assayed at a concentration of 2%. 15V4M1 cells (2 × 105 cells per ml in 50 ml) were treated for 24 h, and the lysate was assayed at 15%.
To test for the requirement of tumor cellular background for CsA inhibition, full-length IL-3 transgene (pIL-3M1 hph) was introduced into V4D6 as well as the ras-expressing PB-3c clone, 15V4. Figure 7d, left, shows that V4D6 cells expressing the transgene (V4D6M1) produce much larger amounts of biologically active IL-3 than do V4D6 cells and that CsA substantially inhibits this production. However, the IL-3-transfected precursor, 15V4M1, continued to produce biologically active IL-3 in the presence of the drug (Fig. 7d, right). Taken together, these data suggest that translational silencing of IL-3 requires tumor progression.
DISCUSSION
CsA accelerates ribosome-associated poly(A) shortening.
IL-3 mRNA in V4D6 cells has a short half-life but is efficiently translated, producing substantial amounts of biologically active protein that is used for autocrine growth (26, 27, 40). Here we show that treatment with CsA or FK506 abolishes the production of IL-3 via a mechanism acting at the translational level. The drug appears to accelerate a cotranslational IL-3 mRNA decay process operating physiologically in V4D6 cells. The major role of CsA and FK506 is in the enhancement of deadenylation in a ribosome-associated manner. This may oppose poly(A)-dependent reinitiation events (28, 50) and be the prelude for further degradation (see below). Our data are reminiscent of the results obtained from in vitro analysis of c-myc mRNA (9, 10). Here, the initial step in the degradation was poly(A) shortening with a transient accumulation of poly(A)-deficient mRNA. Reconstitution experiments revealed the presence of an activity in the cytosolic fraction that destabilized polysome-associated c-myc mRNA in vitro. This destabilizing activity (AUF1) was subsequently purified and cloned (8, 17, 66). Several studies have suggested that the rate of poly(A) shortening is the key determinant of the half-life of mRNA (5, 9, 34, 55, 61).
IL-3 mRNAs in class I and class II cells decay via different routes.
We have previously shown that the oncogenically stabilized IL-3 mRNA in V2D1 cells (class I tumor) could be destabilized by CsA and FK506 (41). The data presented in Fig. 2a confirm this result and show in addition that the bulk of the labile transcripts in V4D6 class II cells were not sensitive to CsA. However, this mRNA underwent a ribosome-associated deadenylation (Fig. 4a). In contrast, gradient analysis of V2D1 cells showed that upon treatment with CsA, there is a substantial reduction in the IL-3 signal associated with ribosomal monosomes and polysomes, but no qualitative difference is observed (Fig. 4d). Thus, the pattern shown mirrors the reduction in the cytoplasmic mRNA levels following drug treatment shown in Fig. 2a. Importantly, the shortened fragments observed in V4D6 gradients were absent in V2D1 and no redistribution of signal over the gradient was observed, which argues for the absence of ribosome-associated decay in V2D1 cells. Furthermore, in contrast to V4D6, CsA-induced inhibition was observed in V2D1, even when membrane-bound ribosome-associated RNA was not extracted (Fig. 3, bottom). These data strongly suggest different CsA-induced IL-3 mRNA decay routes for class I and class II tumor cells.
ARE-lacking transcripts are resistant to CsA-induced poly(A) shortening.
To avoid interference from endogenous transcripts, we have introduced murine IL-3 transgenes into Jurkat cells, a human T-cell line. As expected, IL-3 mRNA from ARE deletion constructs was expressed at much higher levels than that from the full-length construct. CsA did not inhibit the expression of cytoplasmic IL-3 mRNA from either of the transgenes (Fig. 6a). However, the ribosome-associated degradation of the full-length transcripts also occurred in Jurkat cells, indicating that the findings made with V4D6 are not restricted to mast cell tumor lines. Importantly, the ARE-lacking transcripts were resistant to poly(A) shortening mediated by CsA (Fig. 6b).
In vitro growth inhibition of V4D6 cells by CsA.
Our results shown in Fig. 2d indicate that the growth of V4D6 cells in vitro was inhibited by CsA in a dose-dependent manner. It is noteworthy that this effect is tumor specific and does not occur in normal precursor cells. With CsA-treated class I V2D1 cells, we have previously shown that the addition of exogenous IL-3 led to almost complete restoration of growth (41). However, exogenous IL-3 did not restore the growth of V4D6 cells (data not shown). This indicates that the inhibitory mechanism of the drug on growth is more complex and cannot be explained solely by the loss of the autocrine regulator IL-3. The drug may inhibit other elements vital for growth. In fact, translation of IL-4, a lymphokine for which these cells have receptors, is also impaired (Fig. 7). That only tumor cells, not normal IL-3-dependent cells, are growth inhibited by CsA indicates that this drug antagonizes tumor-specific growth-promoting mechanisms.
Translational inhibition is associated with tumor progression.
The CsA-induced translational inhibition of IL-3 and IL-4, the two lymphokines associated with the tumor phenotype, indicates that this process involves some cellular factors that are altered during tumor progression. It is interesting that CsA affects IL-3 and IL-4, the only two lymphokines which stimulate the growth of these cells in vitro. It is tempting to speculate that the same factors that are involved in the upregulation of the lymphokines in the tumors serve as the target for the action of CsA. Both CsA and FK506 are calcineurin antagonists, indicating a possible involvement of calcineurin phosphatase activity and hence a role for phosphorylation-dephosphorylation in the regulation of mRNA turnover linked to translation. Rapamycin, another immunosuppressive drug and inhibitor of the FRAP/RAFT kinase pathway, antagonizes initiation events of translation acting through the binding protein of eIF-4E or through inhibition of the kinase p70S6K (22, 30, 44). Recent reports show that eIF-4G mediates poly(A) tail-stimulated translation and that PABP associates with eIF-4E via eIF-4G (58, 59). Any break introduced in this “closed loop” could make the mRNA vulnerable to degradation. It is noteworthy that the poly(A) tail and PABP play apparent roles in the translation as well as the decay of short-lived mRNAs (4, 28, 59, 63). Although our data demonstrate that IL-3 mRNA degradation in class II cells requires a physical association with the translational machinery, an additional inhibition of initiation by CsA cannot be ruled out.
In conclusion, our experiments have identified a novel target mechanism of two clinically important immunosuppressive drugs at the level of translation. Selective inhibition of oncogenic regulators by drug-induced stimulation of mRNA deadenylation and subsequent degradation represents a potentially important strategy for opposing neoplastic growth.
ACKNOWLEDGMENTS
We thank George Thomas, Harold Jeffries, and Witold Filipowicz for advice; Adrian Wyss, Lyndall Brennan, and Corina Gyssler-Frey for technical assistance; members of the laboratory for their comments on the manuscript; and Nicole Vehlinger for secretarial assistance.
REFERENCES
- 1.Algate P A, McCubrey J A. Autocrine transformation of hemopoietic cells resulting from cytokine message stabilization after intracisternal A particle transposition. Oncogene. 1993;8:1221–1232. [PubMed] [Google Scholar]
- 2.Banholzer R, Nair A P K, Hirsch H H, Ming X-F, Moroni C. Rapamycin destabilizes IL-3 mRNA in autocrine tumor cells by a mechanism requiring an intact 3′-untranslated region. Mol Cell Biol. 1997;17:3254–3260. doi: 10.1128/mcb.17.6.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beelman C A, Parker R. Differential effects of translational inhibitors in cis and in trans on the decay of the unstable yeast MF2 mRNA. J Biol Chem. 1994;269:9687–9692. [PubMed] [Google Scholar]
- 4.Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein P, Peltz S W, Ross J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol Cell Biol. 1989;9:659–670. doi: 10.1128/mcb.9.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blankenstein T, Qin Z, Li W, Diamantstein T. DNA rearrangement and constitutive expression of the interleukin 6 gene in a mouse plasmocytoma. J Exp Med. 1990;171:965–970. doi: 10.1084/jem.171.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohjanen P R, Petryniak B, June C H, Thompson C B, Lindsten T. An inducible cytoplasmic factor (AU-B) binds selectively to AUUUA multimers in the 3′ untranslated region of lymphokine mRNA. Mol Cell Biol. 1991;11:3288–3295. doi: 10.1128/mcb.11.6.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brewer G. An A+U-rich element RNA-binding factor regulates c-myc stability in vitro. Mol Cell Biol. 1991;11:2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brewer G, Ross J. Poly(A) shortening and degradation of the 3′ A+U-rich sequences of human c-myc mRNA in a cell-free system. Mol Cell Biol. 1988;8:1697–1708. doi: 10.1128/mcb.8.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brewer G, Ross J. Regulation of c-myc mRNA stability in vitro by a labile destabilizer with an essential nucleic acid component. Mol Cell Biol. 1989;9:1996–2006. doi: 10.1128/mcb.9.5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C-Y A, Shyu A-B. Selective degradation of early-response-gene mRNAs: functional analyses of sequence features of the AU-rich elements. Mol Cell Biol. 1994;14:8471–8482. doi: 10.1128/mcb.14.12.8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C-Y A, Shyu A-B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 14.Chen C-Y A, Xu N, Shyu A-B. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol Cell Biol. 1995;15:5777–5788. doi: 10.1128/mcb.15.10.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 16.Decker C J, Parker R. Mechanisms of mRNA degradation in eukaryotes. Trends Biochem Sci. 1994;19:336–340. doi: 10.1016/0968-0004(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 17.DeMaria C T, Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J Biol Chem. 1996;27:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 18.Demetri G D, Ernst T J, Pratt II E S, Zenzie B W, Rheinwald J G, Griffin J D. Expression of ras oncogenes in cultured human cells alters the transcriptional and posttranscriptional regulation of cytokine genes. J Clin Investig. 1990;86:1261–1269. doi: 10.1172/JCI114833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diamantis I D, Nair A P K, Hirsch H H, Moroni C. Tumor suppression involves down-regulation of interleukin 3 expression in hybrids between autocrine mastocytoma and interleukin-3-dependent parental mast cells. Proc Natl Acad Sci USA. 1989;86:9299–9302. doi: 10.1073/pnas.86.23.9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Favre N, Erb P. Use of the CTL44 cell line, a derivative of CTL/L cells, to identify and quantify mouse interleukin-4 by bioassay. J Immunol Methods. 1993;164:213–220. doi: 10.1016/0022-1759(93)90314-w. [DOI] [PubMed] [Google Scholar]
- 21.Gough N M. Rapid quantitative preparation of cytoplasmic RNA from small numbers of cells. Anal Biochem. 1988;173:93–95. doi: 10.1016/0003-2697(88)90164-9. [DOI] [PubMed] [Google Scholar]
- 22.Haghighat A, Mader S, Pause A, Sonnenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henics T, Sanfridson A, JoNell Hamilton B, Nagy E, Rigby W F C. Enhanced stability of interleukin-2 mRNA in MLA 144 cells. J Biol Chem. 1994;269:5377–5383. [PubMed] [Google Scholar]
- 24.Hentze M W. eIF4G: a multipurpose ribosome adapter. Science. 1997;275:500–501. doi: 10.1126/science.275.5299.500. [DOI] [PubMed] [Google Scholar]
- 24a.Hirsch, H. H. Unpublished data.
- 25.Hirsch H H, Backenstoss V, Moroni C. Impaired interleukin-3 mRNA decay in autocrine mast cell tumors after transient calcium ionophore stimulation. Growth Factors. 1996;13:99–110. doi: 10.3109/08977199609034570. [DOI] [PubMed] [Google Scholar]
- 26.Hirsch H H, Nair A P K, Backenstoss V, Moroni C. Interleukin-3 mRNA stabilization by a trans-acting mechanism in autocrine tumors lacking interleukin-3 gene rearrangements. J Biol Chem. 1995;270:20629–20635. doi: 10.1074/jbc.270.35.20629. [DOI] [PubMed] [Google Scholar]
- 27.Hirsch H H, Nair A P K, Moroni C. Suppressible and nonsuppressible autocrine mast cell tumors are distinguished by insertion of an endogenous retroviral element (IAP) into the interleukin 3 gene. J Exp Med. 1993;178:403–411. doi: 10.1084/jem.178.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobson A. Poly(A) metabolism and translation: the closed-loop model. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 451–480. [Google Scholar]
- 29.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 30.Jefferies H B J, Fumagalli S, Dennis P B, Reinhard C, Pearson R, Thomas G. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jefferies H B J, Reinhard C, Kozma S C, Thomas C. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc Natl Acad Sci USA. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawano M, Hirano T, Matsuda T, Taga T, Horii Y, Iwato K, Asaoku H. Autocrine generation and requirement of BSF 2/IL 6 for human multiple myelomas. Nature. 1988;332:83–85. doi: 10.1038/332083a0. [DOI] [PubMed] [Google Scholar]
- 33.Lagnado C A, Brown C Y, Goodall G J. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A) Mol Cell Biol. 1994;14:7984–7995. doi: 10.1128/mcb.14.12.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laird-Offringa I A, de Wit C L, Elfferich P, van der Eb A J. Poly(A) tail shortening is the translation-dependent step in c-myc mRNA degradation. Mol Cell Biol. 1990;10:6132–6140. doi: 10.1128/mcb.10.12.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lowell J E, Rudner D Z, Sachs A B. 3′-UTR-dependent deadenylation by the yeast poly(A) nuclease. Genes Dev. 1992;6:2088–2099. doi: 10.1101/gad.6.11.2088. [DOI] [PubMed] [Google Scholar]
- 36.Malter J S. Identification of an AUUUA-specific messenger RNA binding protein. Science. 1989;246:664–666. doi: 10.1126/science.2814487. [DOI] [PubMed] [Google Scholar]
- 37.Meeker T, Hardy D, Willman C, Hogan T, Abrams J. Activation of interleukin-3 gene by chromosome translocation in acute lymphocytic leukemia with eosinophiilia. Blood. 1990;76:285–289. [PubMed] [Google Scholar]
- 38.Muhlrad D, Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 39.Myer V E, Fan X C, Steitz J A. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nair A P K, Diamantis I D, Conscience J-F, Kindler V, Hofer P, Moroni C. A v-H-ras-dependent hemopoietic tumor model involving progression from a clonal stage of transformation competence to autocrine interleukin 3 production. Mol Cell Biol. 1989;9:1183–1190. doi: 10.1128/mcb.9.3.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nair A P K, Hahn S, Banholzer R, Hirsch H H, Moroni C. Cyclosporin A inhibits growth of autocrine tumour cell lines by destabilizing interleukin-3 mRNA. Nature. 1994;369:239–242. doi: 10.1038/369239a0. [DOI] [PubMed] [Google Scholar]
- 42.Nair A P K, Hirsch H H, Moroni C. Mast cells sensitive to v-H-ras transformation are hyperinducible for interleukin-3 expression and have lost tumor-suppressor activity. Oncogene. 1992;7:1963–1972. [PubMed] [Google Scholar]
- 43.Nakagawa J, Waldner H P, Meyer-Monard S, Hofsteenge J, Jenö P, Moroni C. AUH, a gene encoding an AU-specific RNA binding protein with intrinsic enoyl-CoA hydratase activity. Proc Natl Acad Sci USA. 1995;92:2051–2055. doi: 10.1073/pnas.92.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pause A, Belsham G J, Gingras A-C, Donzé O, Lin T-A, Lawrence J C, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 45.Ross H J, Sato N, Ueyama Y, Koeffler H P. Cytokine messenger RNA stability is enhanced in tumor cells. Blood. 1991;77:1787–1795. [PubMed] [Google Scholar]
- 46.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sachs A B. Messenger RNA degradation in eukaryotes. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- 48.Sachs A B, Davis R W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 49.Sachs A B, Davis R W, Kornberg R D. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 51.Schiavi S C, Wellington C L, Shyu A-B, Chen C-Y A, Greenberg M E, Belasco J G. Multiple elements in the c-fos protein-coding region facilitate mRNA deadenylation and decay by a mechanism coupled to translation. J Biol Chem. 1994;269:3441–3448. [PubMed] [Google Scholar]
- 52.Schreiber S L, Crabtree G R. The mechanism of action of cyclosporin A and FK506. Immunol Today. 1992;13(4):136–142. doi: 10.1016/0167-5699(92)90111-J. [DOI] [PubMed] [Google Scholar]
- 53.Schuler G D, Cole M D. GM-CSF and oncogene mRNA stabilities are independently regulated in trans in a mouse monocytic tumor. Cell. 1988;55:1115–1122. doi: 10.1016/0092-8674(88)90256-5. [DOI] [PubMed] [Google Scholar]
- 54.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 55.Shyu A-B, Belasco J G, Greenberg M E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 56.Stocking C, Löliger C, Kawai M, Suciu S, Gough N, Ostertag W. Identification of genes involved in growth autonomy of hematopoietic cells by analysis of factor independent mutants. Cell. 1988;53:869–879. doi: 10.1016/s0092-8674(88)90329-7. [DOI] [PubMed] [Google Scholar]
- 57.Stoecklin G, Hahn S, Moroni C. Functional hierarchy of AUUUA motifs in mediating rapid interleukin-3 mRNA decay. J Biol Chem. 1994;269:28591–28597. [PubMed] [Google Scholar]
- 58.Tarun Jr S Z, Wells S E, Deardorff J A, Sachs A B. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tarun S Z, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 60.Veyrune J L, Carillo S, Vié A, Blanchard J M. c-fos mRNA instability determinants present within both the coding and the 3′ non coding region link the degradation of this mRNA to its translation. Oncogene. 1995;11:2127–2134. [PubMed] [Google Scholar]
- 61.Wilson T, Treismann R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature. 1988;336:396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- 62.Wodnar-Filipowicz A, Moroni C. Regulation of interleukin 3 mRNA expression in mast cells occurs at the posttranscriptional level and is mediated by calcium ions. Proc Natl Acad Sci USA. 1990;87:777–781. doi: 10.1073/pnas.87.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wormington M, Searfoss A M, Hurney C A. Overexpression of poly(A) binding protein prevents maturation-specific deadenylation and translational inactivation in Xenopus oocytes. EMBO J. 1997;15:900–909. [PMC free article] [PubMed] [Google Scholar]
- 64.Ymer S, Tucker W Q J, Sanderson C J, Hapel A, Campbell H D, Young I. Constitutive synthesis of interleukin-3 by leukemia cell line WEHI-3B is due to retroviral insertion near the gene. Nature. 1985;317:255–258. doi: 10.1038/317255a0. [DOI] [PubMed] [Google Scholar]
- 65.Young D C, Wagner K, Griffin J D. Constitutive expression of the granulocyte-macrophage colony-stimulating factor gene in acute myeloblastic leukemia. J Clin Investig. 1987;79:100–106. doi: 10.1172/JCI112769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang W, Wagner B J, Ehrenman K, Schaefer A W, DeMaria C T, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zubiaga A M, Belasco J G, Greenberg M E. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]