Abstract
ras oncogene-transformed PA-1 human teratocarcinoma cells have abundant AP-2 mRNA but, paradoxically, little AP-2 transcriptional activity. We have previously shown that overexpression of AP-2 in nontumorigenic variants of PA-1 cells results in inhibition of AP-2 activity and induction of tumorigenicity similar to that caused by ras transformation of PA-1 cells. Evidence indicated the existence of a novel mechanism of inhibition of AP-2 activity involving sequestering of transcriptional coactivators. In this study, we found that PC4 is a positive coactivator of AP-2 and can restore AP-2 activity in ras-transformed PA-1 cells. Relative to vector-transfected ras cell lines, ras cell lines stably transfected with and expressing the PC4 cDNA have a diminished growth rate and exhibit a loss of anchorage-independent growth, and they are unable to induce the formation of tumors in nude mice. These data suggest that a transcriptional coactivator, like a tumor suppressor, can have a growth-suppressive effect on cells. Our experiments are the first to show that ras oncogenes and oncogenic transcription factors can induce transformation through effects on the transcription machinery rather than through specific programs of gene expression.
Signals elicited by oncogenes, growth factors, hormones, and other agents converge on various transcription factors that modulate the expression of target genes. Transcription factors play a fundamental role in genetic control of cell survival, growth, and differentiation. The overexpression of one or more of these factors can affect the cellular transcriptional profile and lead to inhibition of the activities of other transcription factors (10, 24). For example, a transcriptional interference phenomenon has been observed during the overexpression of the viral activator VP16, which inhibited its own activity and the activity of GCN4 (7). Transcriptional activation by both of these transcription factors required a common mediator present in a partially purified yeast fraction, suggesting that overexpression of GAL4-VP16 might sequester this mediator, causing inhibition of the activity of GCN4. High levels of serum response factor inhibit its own activity and the activity of GAL4-VP16 (23). The RAP74 subunit of transcription factor TFIIF relieves the transcriptional interference mediated by serum response factor in vitro (34). A transcriptional interference phenomenon has been observed during the overexpression of many transcription factors; however, the physiological relevance of this phenomenon remains unclear because most of the transcriptional interference studies have been carried out in vitro. The few in vivo studies that have been done involved artificially induced overexpression of transcription factors. The estrogen hormone receptor was shown to inhibit the transcriptional activation mediated by the progesterone and glucocorticoid receptors (21). Reciprocally, the progesterone and glucocorticoid receptors inhibit the activity of the estrogen receptor. The possible therapeutic importance of this transcriptional interplay became evident in two breast cancer cell lines that express steroid hormone receptors. Estrogen-dependent transcription was found to be blocked by the addition of agonistic ligands of the progesterone and glucocorticoid receptors. This repression was alleviated by the addition of the antiprogesterone and antiglucocorticoid ligand RU486. These studies also indicated that these steroid receptors have a common coactivator. High levels of progesterone and glucocorticoid receptors might sequester this coactivator, which is essential for estrogen-dependent transcription.
It is curious that deregulation of any one of the oncogenic transcription factors, each of which is but a small part of a growth signal transduction pathway, can oncogenically transform cells. Transcriptional interference is thought to result through effects on elements of the general transcriptional machinery which could thereby have a pleiotropic effect resulting in cellular transformation. One such protein, the transcription factor AP-2, is developmentally regulated and is associated with programmed gene expression in the neural crest cell lineage during mouse embryogenesis (22). The activity of AP-2 is regulated by at least three different signal transduction pathways. Retinoic acid (RA), a developmental morphogen, was found to transiently increase AP-2 mRNA transcription and transcriptional activity in the human teratocarcinoma cell line N Tera 2 (18). Similar effects of RA were observed in PA-1 cells (3), another human teratocarcinoma cell line (27, 28, 33). The cyclic AMP-inducible protein kinase A pathway and the phorbol ester-inducible protein kinase C pathway have been shown to increase AP-2 activity in HeLa cells (5, 12, 13). AP-2 is the major regulator of the c-erbB-2 promoter in cells that overexpress it and as such has been implicated in the causation of human mammary carcinoma (2). Recently, the ERF-1 transcription factor that is involved in the regulation of estrogen receptor gene transcription in hormonally responsive breast and endometrial carcinomas was identified as AP-2γ, a member of the AP-2 family (20).
Transcriptional interference, in which overexpression of a transcription factor results in inhibition of itself or other transcription factors, occurs physiologically in ras-transformed cells (15). We previously found that an activated ras oncogene increased the expression of AP-2 in the human teratocarcinoma cell line PA-1 and that abundant AP-2 resulted in transcription self-interference in these cells. Our studies indicated a profound physiological role for AP-2 transcriptional self-interference. PA-1 cells that overexpressed AP-2 exhibited transformed properties due to AP-2 transcriptional self-interference. Although AP-2 has been shown to be an activator of gene expression (2, 12, 31, 32), PA-1 cell lines that stably overexpress AP-2 exhibit reduced AP-2 activity. Like ras-transformed cells, the AP-2-overexpressing cell lines form colonies in soft agar (15) and are tumorigenic when injected subcutaneously into nude mice (this report). Therefore, our experiments suggest that a ras oncogene may use the mechanism of AP-2 transcriptional self-interference to transform PA-1 cells. A GAL4–AP-2 fusion protein containing the activation domain of AP-2 linked to a heterologous GAL4 DNA-binding domain retained the transcriptional self-interference activity and transformed PA-1 clone 1 cells to be tumorigenic in nude mice (this report). Thus, it appeared that the activation domain of AP-2 was sufficient for it transforming activity. Overexpression of AP-2 inhibited the activity of the GAL4–AP-2 fusion protein, and vice versa, even though they bind to two different target sequences. Overexpression of AP-2 also inhibited the activity of GAL4-VP16 (this report). These studies indicated that through its activation domain, AP-2 protein sequestered one or more coactivators needed for the function of these transcription factors. It is possible that elevation of the level of the coactivator(s) relieves AP-2 transcriptional interference and restores AP-2 activity, and such coactivators might thereby abolish ras oncogene-induced tumorigenicity. To test this hypothesis, we sought to identify AP-2-interacting proteins and then determine whether these proteins are coactivators of AP-2-dependent transcription. We report here that AP-2 physically interacts with the positive coactivator PC4, which relieves AP-2 transcriptional self-interference in transient transfection experiments using chloramphenicol acetyltransferase (CAT) reporter constructs. These results identified PC4 as one of the limiting cofactors of AP-2-mediated transcriptional activation. Stable expression of PC4 in ras-transformed PA-1 cells profoundly elevated the level of AP-2 activity in the resulting cell lines. The PC4-expressing ras cell lines had a diminished growth rate and exhibited a loss of anchorage-independent growth and tumorigenicity. Our observations demonstrate that a transcriptional coactivator can have a growth-suppressive effect on cells that is indicative of tumor suppressor properties and identify a signal transduction mechanism by which ras oncogenes, through a transcription mechanism, can induce transformation mediated by its effects on general coactivators rather than through specific gene targets.
MATERIALS AND METHODS
Cell culture and assays of growth rate and tumorigenicity.
PA-1 human teratocarcinoma cells were derived from a female ovarian germ cell tumor (33); the origin and properties of non-ras-, ras-, and AP-2B-transformed PA-1 sublines were described previously (3, 28). The cells were grown in modified Eagle’s medium with Earl’s salts (GIBCO Laboratories, Gaithersburg, Md.), supplemented with 5% fetal bovine serum (Hazelton Biologics, Lenexa, Kans.) and antibiotics, at 37°C in 5% CO2–95% air. The PC4 cDNA was cloned into the EcoRI site of the zeomycin resistance vector. PA-1 9113 cells containing an endogenous activated N-ras oncogene (4 × 105 per 100-mm-diameter culture dish) were transfected by the calcium phosphate precipitation method. After 3 weeks of selection in medium containing zeomycin, the colonies were counted and picked by using glass cloning cylinders. The cells were expanded and tested for tumorigenicity in athymic nude mice by injecting 3 × 106 cells subcutaneously (one site per mouse). For transfection of clones expressing AP-2 or GAL4–AP-2 fusion proteins, the G418-resistant cells were pooled and tested for tumorigenicity in athymic nude mice by injecting 3 × 106 cells subcutaneously. In some cases, individual colonies were picked, expanded into cells lines, and tested. Growth rates and anchorage-independent growth in 0.35% agarose were determined as previously described (3).
Analysis of GST–AP-2-associated proteins.
The glutathione S-transferase (GST)–AP-2 fusion construct was made by cloning the 1.9-kb AP-2 cDNA into the EcoRI site of pGEX-3B (26). The GST–AP-2 fusion protein was purified from bacterial extracts as described by Smith and Concoran (26). Nuclear extracts were prepared essentially as described by Dignam et al. (6). Trichloroacetic acid-precipitable counts of nuclear extracts (8 × 106 cpm) were mixed with 20 μg of GST–AP-2 protein bound to glutathione-Sepharose beads in 1 ml of Tris-buffered saline, pH 7.4, containing 0.05% Tween 20 (TBST) and rocked for 2 h at 4°C. The mixture was washed and cleaved with blood coagulation factor Xa, as described by Smith and Cocoran (26), to release AP-2-associated proteins. The proteins were resolved on a 10% polyacrylamide gel (16). The gel was dried and exposed to Kodak X-OMAT X-ray film. When cold PA-1 nuclear extracts were used in the studies, the glutathione-Sepharose beads with bound GST–AP-2 and proteins were transferred to a Hybond-ECL nitrocellulose membrane (Amersham Corp., Arlington Heights, Ill.) and probed with a 1:4,000 dilution of antiserum raised against PC4. The signals were detected by using horseradish peroxidase-conjugated anti-rabbit antibody and enhanced chemiluminescence (ECL; Amersham Corp.) according to the manufacturer’s instructions.
Immunoprecipitation.
Four milligrams of nuclear extract in 1 ml of TBST was used in the assay. Two microliters of AP-2 (C-18), a rabbit polyclonal antibody made against a synthetic peptide corresponding to AP-2 C-terminal amino acids 420 to 437 (Santa Cruz Biotechnology, Santa Cruz, Calif.) and 2 μl of a rabbit polyclonal antibody made against a synthetic peptide corresponding to amino acids 153 to 168 of AP-2 were used for immunoprecipitation. The immunoprecipitated complex was washed four times in TBST and resolved on a sodium dodecyl sulfate 20% polyacrylamide gel. The proteins were transferred to a Hybond-ECL nitrocellulose membrane (Amersham Corp.) and probed with an antiserum raised against PC4.
An AP-2 cDNA of approximately 1.9 kb isolated from 6928 PA-1 cells (3) was cloned in the proper orientation into the EcoRI site of plasmid pSG5 (Stratagene, La Jolla, Calif.) to generate pSAP2. A 1.6-kb cDNA of AP-2B isolated from the same cell line was cloned similarly in pSG5 to generate pSAP-2B. A HindIII-SacI DNA fragment from pGAP-2/11-226 encoding fusion protein GAL4–AP-2/11-226 was blunted with the Klenow fragment of DNA polymerase and subcloned in the proper orientation into EcoRI-cut and filled-in pSG5 to generate pSGAP-2/11-226. Plasmid pSG5 contains a simian virus 40 (SV40) early promoter and β-globin intron sequences that enable efficient expression of cloned genes. The presence of the T7 promoter in pSG5 enables in vitro transcription and translation of AP-2, AP-2B, GAL4–AP-2, and PC4. In vitro synthesis of proteins was performed, using the TNT in vitro transcription-translation system (Promega Corp., Madison, Wis.) according to the manufacturer’s instructions, with 1 μg of plasmid DNA and 40 μCi of l-[35S]methionine in a 50-μl reaction volume. Proteins were mixed in 1 ml of TBST, nuclear extract (containing about 100 μg of protein) was added, and the mixture was precleaned for 4 h at 4°C with 20 μl of protein A adsorbed to agarose beads. Immunoprecipitation with 1 μl of PC4-specific antiserum or 1 μl of AP-2 N-terminus-specific antibody made against a synthetic peptide corresponding to amino acids 2 to 14 of AP-2 was carried out as described above.
Transient transfections of PA-1 cells and CAT assays.
AP-2 response element sequences from the distal basal-level element of human metallothionein gene IIA corresponding to nucleotides (nt) −188 to −161 were oligomerized, and a reporter construct, 3× AP-2REhMt-tk CAT (3× AP-2-CAT), was made by cloning three response elements adjacent to the herpes simplex virus tk gene promoter in the vector pBLCAT2 (17). Transient transfection of non-ras-transformed, differentiation-competent PA-1 9117 cells was performed by the calcium phosphate precipitation method. GAL4-VP16 expression plasmid pSGVP and GAL4 reporter plasmid G5E1bCAT were generous gifts of M. Ptashne (24). Plasmid pCH110 (Pharmacia Biotech, Piscataway, N.J.), which contains the lacZ gene under the control of an SV40 promoter, and RSV-LTRCAT (a gift of Eric Olson) were used to test the effect of PC4 on SV40 promoter- and Rous sarcoma virus (RSV) long terminal repeat (LTR)-driven transcription, respectively.
In vitro transcription.
Plasmid pcmyc-PC is a derivative of pC2AT (25a), which contains nt −44 to +4 of the human c-myc P2 promoter and a 398-bp G-free transcription cassette. Three AP-2 sites, found in the human metallothionein gene IIA basal-level promoter from nt −188 to −159, were cloned upstream of the c-myc P2 promoter to create pcmyc-AP-2. In vitro transcription reactions were performed with 50 ng of plasmid DNA and 10 μCi of [α-32P]UTP, using the HeLa Cell Extract Transcription System (Promega Corp.) essentially as described by the manufacturer. In certain experiments, AP-2 protein was removed from HeLa cell nuclear extracts by using an AP-2 C-terminus-specific antibody C-18 that had been attached to protein A-agarose beads. After incubation on ice for 15 min, the beads with the bound antibody were removed by centrifugation, and the AP-2-depleted nuclear extract was used in the assays. The transcription products were separated on a 5% polyacrylamide gel containing 7 M urea, dried, and exposed to Kodak Biomax MR film at −70°C. Plasmid p052 was used as a control plasmid in the in vitro transcription reactions. This control plasmid contains an unrelated hsp70 promoter, a 298-bp G-free transcription cassette, and a weaker, mutated form of the adenovirus major late initiator (29).
Expression plasmids.
AP-2 1-165 was constructed from pSAP-2 by deleting the C-terminal AP-2 sequences from the SmaI site at amino acid 165. AP-2/ΔI 121–165 was made by deleting the sequence in between the BamHI and SmaI sites. AP-2/ΔI 13–165 was made by deleting the sequence in between the BanI and SmaI sites. The constructs AP-2/ΔI 166–278 and AP-2/ΔI 166–398 were made by deleting the amino acids between the SmaI site and one of the two PstI sites. The reading frame of AP-2 in the construct AP-2/ΔI 166–278 was altered during the genetic manipulation and was later restored by inserting one A nucleotide at the junction sequence. The PC4 expression vector pSPC4 was constructed by inserting an EcoRI fragment of about 400 bp, isolated from pGEX-PC415, into the EcoRI site of pSG5 in the proper orientation. The GAL4 DNA-binding domain- and AP-2 activation domain-containing fusion construct pGAL4-AP2/11–226 was made by inserting amino acids 11 to 226 of AP-2 into EcoRI-cut and mung bean nuclease-blunted pSG424 (25). The nucleotide sequences and reading frames of all of these constructs were verified by double-stranded DNA sequence analysis.
RESULTS
Physical interaction of AP-2 and PC4.
We have shown that AP-2 is overexpressed in N-ras-transformed variants of the human teratocarcinoma cell line PA-1 and that abundant AP-2 protein results in transcriptional self-interference in these cells (15). In addition, non-ras-transformed cell lines forced to stably overexpress AP-2 form colonies in soft agar, just as ras-transformed cells do, and these AP-2-overexpressing cells are tumorigenic when injected into nude mice (Table 1). Therefore, our experiments suggested that a ras oncogene can use the mechanism of AP-2 transcriptional self-interference to transform PA-1 cells. Western blot analysis indicated the existence of high levels of AP-2 protein in ras-transformed cells, suggesting that mechanisms other than defective translation of AP-2 mRNA contribute to inhibition of AP-2 activity. AP-2 protein produced in ras oncogene-transformed cells could bind to AP-2 target sequences in electrophoretic mobility shift assays, indicating that the reduced AP-2 activity was not due to a defect in DNA binding.
TABLE 1.
Tumorigenic properties of PA-1 cells
| Cell line | Growth in nude mice | Latent period (wks) |
|---|---|---|
| Clone 1-neo | 0/3a | >24 |
| Clone 1–AP-2 | 2/5b | 22, 26 |
| Clone 1–AP-2 | 5/6a | 16 |
| Clone 1–GAL4–AP-2 | 5/7a | 8–20 |
| Clone 1–GAL4–VP16 | 2/5b | 4, 6 |
| 9113 | 4/4 | 6 |
| 9113-zeo-PC4c | 0/2a | >32 |
Number of cell lines developed from independently picked G418r colonies that formed tumors in nude mice/total number of cell lines tested. G418r colony-derived cell lines were shown to express the transgenes by reverse transcription-PCR.
Pool of transfected G418r colonies: number of mice bearing tumors/number of mice injected with cells.
Cell lines 9113-zeo-PC4-1 and 9113-zeo-PC4-2. Note that neither of these 9113-zeo-PC4 clones expressing the PC4 cDNA could form tumors in nude mice.
Since a GAL4–AP-2 fusion protein, containing the activation domain of AP-2 linked to a heterologous GAL4 DNA-binding domain, retained the transcriptional self-interference activity (15), we tested whether transfected PA-1 clone 1 cells, which are nontumorigenic in nude mice, could be transformed by the activation domain found in the GAL4–AP-2 fusion protein. When cell lines derived from G418-resistant colonies of PA-1 clone 1 cells expressing the GAL4–AP-2 fusion protein were injected into nude mice, five of seven resulted in the development of tumors after a latent period of 8 to 20 weeks (Table 1). This tumor latency was similar to that observed for AP-2-transfected clone 1 cells, which formed tumors in five of six mice within 16 weeks of injection. Therefore, the construct consisting of the activation domain of AP-2 fused to a heterologous DNA-binding domain, just as the whole AP-2 protein, is capable of transforming clone 1 PA-1 cells.
Overexpression of AP-2 inhibits the activity of the GAL4–AP-2 fusion protein and vice versa, although they bind to two different target sequences. Overexpression of AP-2 also inhibits the activity of GAL4-VP16. Likewise, when this strong activation domain in GAL4-VP16 was transfected into clone 1 cells and a pool of G418-resistant colonies was tested in nude mice, two of five mice receiving these cells formed tumors with a latent period of 4 or 6 weeks (Table 1), which is quite fast for PA-1 cells. Therefore, when a strong activation domain like that in AP-2, GAL4–AP-2, or GAL4-VP16 is overexpressed in PA-1 clone 1 cells, cellular transformation ensues.
One possible explanation for these observations is that through its activation domain, AP-2 protein, at high levels, sequesters one or more coactivators needed for the function of these transcription factors. Elevation of the level of the coactivator(s) might relieve AP-2 transcriptional interference and restore AP-2 activity. It is also possible that the coactivators, if upregulated, suppress AP-2- and ras oncogene-induced tumorigenicity.
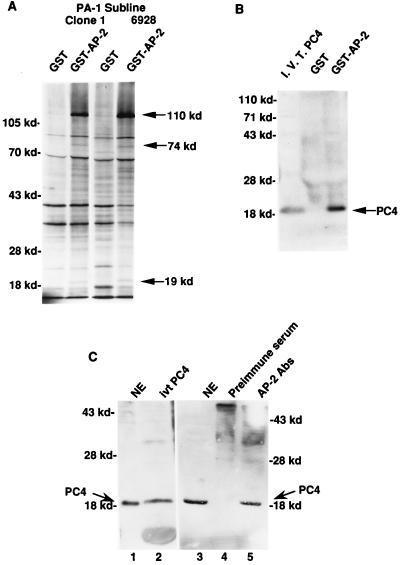
To test this hypothesis, we sought to identify AP-2-interacting proteins and then analyze whether these proteins are coactivators of AP-2-dependent transcription. Immobilized GST–AP-2 fusion protein was allowed to interact with nuclear proteins from PA-1 cells metabolically labeled with 35S. The GST–AP-2 fusion protein contains a cleavage site for blood coagulation factor Xa between the amino acid sequences of GST and AP-2. AP-2 and its associated proteins were specifically released by using blood coagulation factor Xa and resolved on a polyacrylamide gel. At least three polypeptides, of about 19, 74, and 110 kDa, were observed among the released proteins, indicating that these three polypeptides specifically associate with AP-2 (Fig. 1A). This interaction of AP-2 with the three polypeptides was observed in both PA-1 clone 1 sublines and the activated N-ras oncogene-transfected subline 6928. These three were the only proteins that reproducibly demonstrated an interaction with AP-2. The 74-kDa protein was identified as the RAP74 subunit of transcription factor TFIIF, and the 110-kDa protein was identified as the enzyme poly(ADP-ribose) polymerase (15a). Two polypeptides, both of about 60 kDa, also appeared to be interacting with AP-2; however, this interaction was not observed in every experiment. The association of the three polypeptides was not seen when GST alone was used in these assays.
FIG. 1.
Physical interaction between AP-2 and PC4. (A) Physical interaction between GST–AP-2 and proteins from PA-1 cell nuclear extracts. The GST–AP-2 fusion protein binding assays were performed as described in Materials and Methods. Nuclear extracts prepared from metabolically 35S-labeled PA-1 cells contain at least three polypeptides that specifically interact with AP-2. The polypeptides (110, 74, and 19 kDa) are marked at the right. The mobilities of the molecular markers are indicated on the left (in kilodaltons). Clone 1, a subline of PA-1; 6928, a ras oncogene-transfected clone 1 line. (B) Physical interaction between GST–AP-2 and PC4. Four-milligram quantities of unlabeled PA-1 nuclear extracts were used in these assays, and the GST–AP-2-bound proteins were electrophoresed and transferred to a nitrocellulose membrane. The membrane was probed with an antiserum raised against PC4. The mobilities of the molecular markers are indicated at the left (in kilodaltons). The 19-kDa PC4 protein is marked on the right. I.V.T. PC4, in vitro-translated PC4 protein. (C) Physical interaction of AP-2 and PC4 in PA-1 cells. Four-milligram quantities of PA-1 cell nuclear extracts were treated with AP-2-specific antibodies and analyzed for coimmunoprecipitation of PC4 as described in Materials and Methods. The molecular markers are shown on the left and on the right (in kilodaltons). The smear in lanes 4 and 5 occurred because of the immunoglobulin molecules used for immunoprecipitation. Lanes: 1, PA-1 cell nuclear extract (NE), 50 μg; 2, in vitro-translated (ivt) PC4 protein; 3, NE, 50 μg; 4, preimmune serum; 5, anti-AP-2 antibodies (Abs).
Ge and Roeder (8) identified a positive coactivator, the 19-kDa protein PC4, in the upstream stimulatory activity fraction (4) in mammalian cells. The coactivator PC4 has been shown to stimulate transcriptional activation during TFIIA-TFIID-promoter complex formation (14). Natural and recombinant PC4 proteins markedly stimulated the activity of various activation domains, including the acidic activation domain VP16, the proline-rich activation domain CTF, and the glutamine-rich activation domain SP-1 (8). The activation domain of AP-2 is rich in proline and glutamine (32). Overexpression of AP-2 inhibits the activity of VP16 (see below), suggesting that these transcriptional activators have a common cofactor. The coactivator PC4 has been shown to physically associate with immobilized GAL4-VP16 (8). The amino acid sequence of PC4 predicts a molecular mass of 14.4 kDa; however, it migrates as a 17- to 19-kDa protein during electrophoresis. Because the coactivator PC4 has a molecular mass similar to that of one of the AP-2-associated proteins (19 kDa), we tested whether PC4 could physically associate with the immobilized GST–AP-2 fusion protein. Nuclear extracts of PA-1 cells were mixed with immobilized GST–AP-2, and the bound proteins were transferred to nitrocellulose membranes. Antiserum against PC4 recognized a 19-kDa protein among the GST–AP-2-bound proteins that comigrated with in vitro-translated PC4 protein (Fig. 1B). Nuclear proteins bound by GST alone did not show association with PC4.
Coimmunoprecipitation studies were carried out to test whether the interaction of the AP-2 and the PC4 proteins occurred in cells. Nuclear extracts of PA-1 cells were immunoprecipitated with AP-2-specific antibodies. Two anti-AP-2 antibodies, one specific for the C terminus and the other specific for the middle region of the protein, were used to ensure the precipitation of AP-2. The presence of PC4 in the AP-2-immunoprecipitated complex was determined by separating the complex by SDS-polyacrylamide gel electrophoresis, performing Western blot analysis, and probing the blot with a PC4-specific antiserum. As shown in Fig. 1C, the PC4 antiserum recognized a prominent single band in the nuclear extracts of PA-1 cells (lane 1) that comigrated with the in vitro-synthesized PC4 protein (lane 2). The complex immunoprecipitated from the PA-1 cell nuclear extract with AP-2-specific antibodies also contained a band recognized by the PC4 antiserum (lane 5), and this band comigrated with the PC4 band of nuclear extracts (lane 3). When preimmune serum was used in the experiment, the immunoprecipitated complex did not contain PC4 (lane 4). These experiments demonstrated that AP-2 and PC4 proteins are physically associated in PA-1 cells.
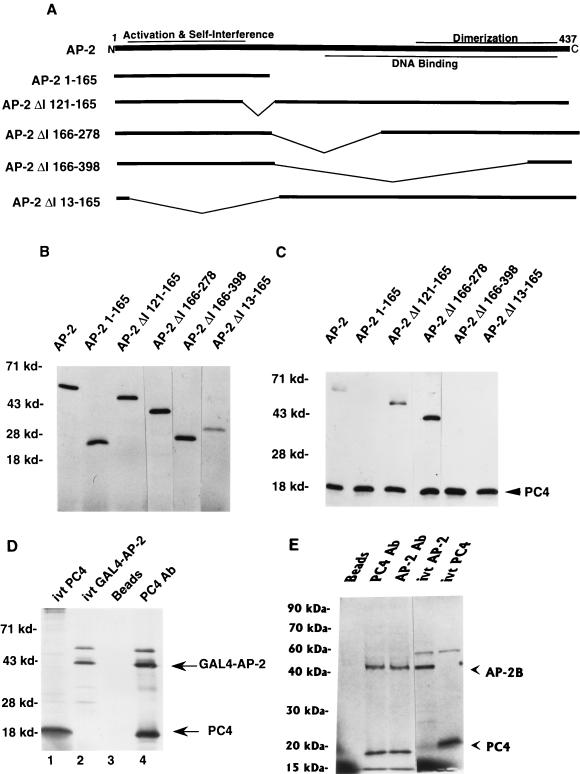
PC4 interaction requires the N-terminal region of AP-2.
The self-interference function of AP-2 resides in the N-terminal region of the protein, between amino acids 11 and 121 (Fig. 2) (15). The activation domain of AP-2 is found between amino acids 50 and 121 (15, 32). The C-terminal two-thirds of the AP-2 molecule, from amino acid 203, is necessary for sequence-specific DNA binding. The dimerization domain of AP-2, situated between amino acids 278 and 409, is an integral part of the DNA-binding domain. Coimmunoprecipitation of AP-2 and PC4 provided a means of determining the region of the AP-2 molecule that interacts with PC4. PC4 and various deletion mutants of AP-2 were translated in vitro with [35S]methionine labeling (Fig. 2B) and mixed with PA-1 cell nuclear extracts that were depleted of endogenous AP-2. Addition of PA-1 cell nuclear extracts enhanced their coimmunoprecipitation. Antiserum against PC4 was used for coimmunoprecipitation. As shown in Fig. 2C, the immunoprecipitated complex contained in vitro-translated AP-2 protein along with PC4. Coimmunoprecipitation with in vitro-translated PC4 was observed with two other AP-2 proteins, from which were deleted the internal amino acids from positions 121 to 165 (AP-2/ΔI 121–165) and from positions 166 to 278 (AP-2/ΔI 166–278). These deletion constructs of AP-2 contained the N-terminal activation and self-interference regions and the C-terminal dimerization region. The DNA-binding motif that is present in the former construct was destroyed in the latter. Internal deletion beyond amino acid 278, which destroys the dimerization motif of AP-2, as in the mutant AP-2/ΔI 166–398, eliminated the interaction of AP-2 with PC4. Interestingly, the AP-2 polypeptide containing N-terminal amino acid 1 to 165 did not coimmunoprecipitate with PC4. An AP-2 polypeptide in which these N-terminal activation and self-interference domains were deleted (encoded by the construct AP-2/ΔI 13–165) also did not coimmunoprecipitate with PC4. These results indicated that both N-terminal amino acid 11 to 121 and C-terminal amino acids from position 279 on were necessary for interaction with PC4. These regions contain the activation/self-interference and dimerization functions of AP-2, respectively. These results suggested either that the N terminus of AP-2 interacted with PC4 as a dimer or that a conformation of the AP-2 protein consisting of both the N and C termini was necessary for the interaction with PC4.
FIG. 2.
Activation domain of AP-2 interacts with PC4. AP-2, GAL4–AP-2, AP-2B, and PC4 proteins were synthesized by using a TNT T7 polymerase kit as described in Materials and Methods. (A) Deletion mutants of AP-2 used for coimmunoprecipitation with PC4. Functional regions on the AP-2 protein that have been characterized are shown on the full-length AP-2 molecule. (B) In vitro-translated AP-2 proteins. AP-2 proteins were synthesized in vitro and separated on an SDS–10% polyacrylamide gel. The panel with the wild-type AP-2 protein contains one-fifth of the amount of input protein that was used for coimmunoprecipitation. The mobilities of molecular markers are indicated at the left (in kilodaltons). (C) Coimmunoprecipitation of PC4 and various deletion mutants of AP-2. Coimmunoprecipitation studies were carried out with PC4 antiserum as described in Materials and Methods. The immunoprecipitated proteins were separated on an SDS–15% polyacrylamide gel. The molecular markers are shown on the left, and the PC4 protein is indicated on the right. (D and E) Physical interaction of PC4 and the GAL4–AP-2 fusion protein (D) or AP-2B (E). The experiments were carried out as described in Materials and Methods. For in vitro-translated (ivt) PC4, ivt GAL4–AP-2, and ivt–AP-2; one-fifth of the amounts of their respective proteins that were used in immunoprecipitation assays were employed. The molecular markers are shown at the left, and the GAL4–AP-2, AP-2B, and PC4 proteins are indicated on the right.
To test these possibilities, we fused the N-terminal domain of AP-2 from amino acids 11 to 226 to the heterologous DNA-binding domain GAL4. The GAL4–AP-2 fusion protein was synthesized in vitro and mixed with in vitro-synthesized PC4 protein. Antiserum raised against PC4 coimmunoprecipitated GAL4–AP-2, indicating that the PC4 and GAL4–AP-2 proteins interacted with each other (Fig. 2D). PC4 did not coimmunoprecipitate the DNA-binding domain of GAL4 protein alone (data not shown), indicating that PC4 interacted with the N-terminal region of AP-2. These results confirmed that the activation domain in the N-terminal region of AP-2 was needed for the interaction of PC4.
We reported earlier the identification of an alternatively spliced form of AP-2, AP-2B, that has the same N-terminal region of AP-2 containing the activation domain but, due to alternate splicing, different C-terminal amino acids, which eliminates the dimerization and DNA-binding domains of AP-2 (3). AP-2B retains the tumorigenic properties of AP-2 in that both can transform clone 1 PA-1 cells. We tested whether the in vitro-synthesized PC4 and AP-2B proteins could interact. A PC4-specific antiserum coimmunoprecipitated AP-2B with PC4 (Fig. 2E), and an N-terminus-specific AP-2 antibody coimmunoprecipitated PC4 with AP-2B. These results confirm that the N-terminal 293 amino acids that are conserved in AP-2 and AP-2B are necessary for the interaction with PC4. These data strengthen the observation that the GAL4–AP-2 fusion protein N-terminal amino acids 11 to 226 are sufficient for interaction with PC4 in the presence of a heterologous DNA-binding domain (see above). It is intriguing that the protein containing amino acids 1 to 165 of AP-2 did not exhibit interaction with PC4 as shown above. Perhaps the region containing the dimerization and DNA-binding domains of AP-2 or GAL4 or the C-terminal amino acids of AP-2B are necessary for AP-2 to be in a conformation that makes the activation domain accessible for efficient interaction with PC4. Moreover, the efficient interaction of PC4 with GAL4–AP-2 or AP-2B did not require the presence of PA-1 cell nuclear extract, indicating that their interaction was direct and did not require additional factors. As we mentioned above, the interaction of PC4 and AP-2 occurred in the absence of the nuclear extracts; however, the interaction was more efficient in the presence of PA-1 cell nuclear extract. The proteins present in the nuclear extract may have modified AP-2 and induced additional conformational changes in AP-2 that are necessary for efficient interaction with PC4.
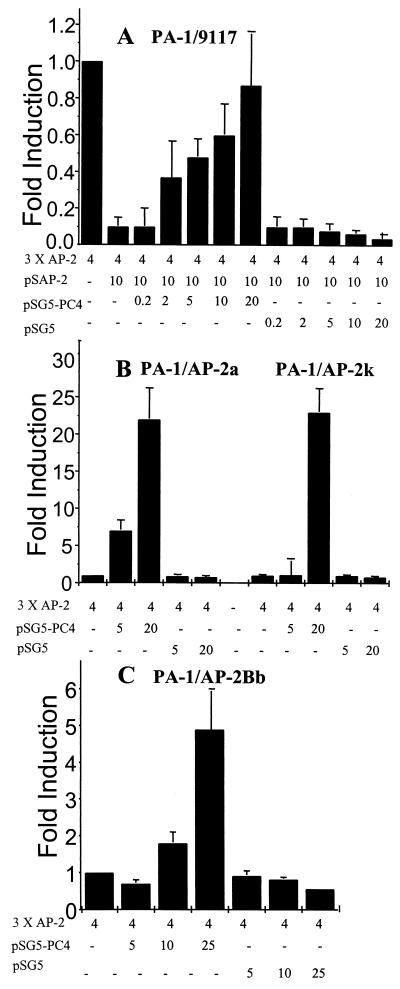
PC4 relieves AP-2 transcriptional self-interference.
Transcriptional interference phenomena have been observed with many transcription factors (10, 24, 34). Earlier studies had indicated that sequestration of a common coactivator is the cause of transcriptional interference (7, 24). If PC4 were the coactivator that was sequestered by AP-2, then excess amounts of PC4 would relieve AP-2 transcriptional self-interference. This possibility was tested as follows. A sufficient amount of AP-2 expression plasmid was transfected into non-ras-transformed 9117 PA-1 cells to induce inhibition of endogenous AP-2 activity. A PC4 expression plasmid under the control of an SV40 promoter was cotransfected into these cells to test for relief of self-interference by measuring the AP-2 transactivation activity, using AP-2 target sequences linked to a CAT reporter. Using the amount of transfected AP-2 expression plasmid (10 μg) that causes a 90% inhibition of the endogenous AP-2 activity (Fig. 3A), cotransfection of the PC4 expression plasmid restored AP-2 transactivation activity in a dose-dependent manner. The AP-2 transactivation activity was maximally relieved with 20 μg of the PC4 expression plasmid, and this level of activity was comparable to the endogenous level of AP-2 activity. PC4 did not significantly alter CAT gene expression from the parental reporter plasmid pBLCAT2, which does not have AP-2 binding sites (data not shown). Transfection of the parental expression vector pSG5, in which PC4 was cloned, did not restore AP-2 activity (Fig. 3A). PC4 did not affect expression from the SV40 promoter, which controls the AP-2 gene in plasmid pSAP-2 (data not shown).
FIG. 3.
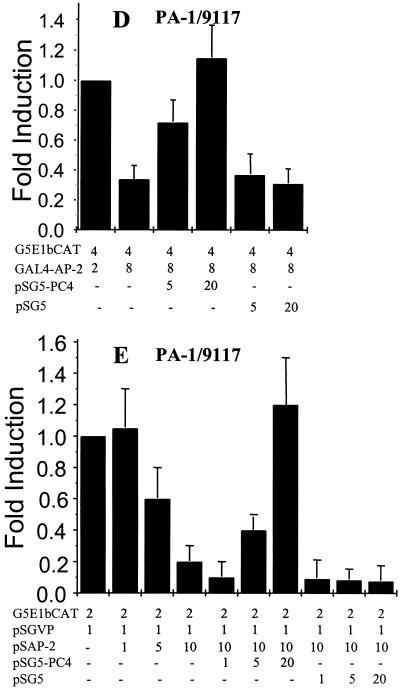
PC4 relieves AP-2 transcriptional self-interference in PA-1 cells. Transient transfections and CAT assays were performed as described in Materials and Methods to determine the effect of PC4 on AP-2 activity. The amounts of the PC4, AP-2, GAL4–AP-2, and GAL4-VP16 expression plasmids and the reporter plasmids for AP-2 (3× AP-2–CAT) and GAL4 (G5E1bCAT) transfected in each assay are shown at the bottom. The fold activity shown on each panel was calculated by measuring the percent conversion of acetylated forms of [14C]chloramphenicol, assuming the endogenous activity to be 1. (A) Transfection of an expression plasmid of PC4 relieves AP-2 transcriptional self-interference in the 9117 PA-1 subline. (B) PC4 relieves AP-2 transcriptional self-interference in PA-1 cells stably overexpressing AP-2, PA-1/AP-2a, or PA-1/AP-2k. (C) PC4 relieves AP-2 transcriptional interference in PA-1/AP-2Bb cells, PA-1 cells stably overexpressing AP-2B (see reference 3). (D) PC4 relieves GAL4–AP-2 transcriptional self-interference. Note that PA-1 cells do not have endogenous GAL4 activity, and hence the GAL4 activity determined at low-level transfection of the GAL–AP-2 expression plasmid (1 μg) was taken as 1. (E) PC4 relieves AP-2 transcriptional cross-interference and restores VP16 activity.
To demonstrate that the effect of PC4 on transcription rates is specific, plasmid pCH110, which contains a lacZ gene encoding beta-galactosidase under the control of an SV40 promoter, was cotransfected with the PC4 expression plasmid. Beta-galactosidase enzyme activity was analyzed in the presence and in the absence of PC4, and no significant alteration was apparent (data not shown). In addition, in cotransfection experiments with an RSV LTR-driven CAT gene (4 μg), PC4 (20 μg) increased the CAT activity by about 20%. Therefore, PC4 does not appear to alter transcription nonspecifically, and the restoration of AP-2 activity observed in the experiments described above was due to the relief of AP-2 transcriptional self-interference.
These observations indicate that PC4 is a positive coactivator of AP-2-mediated transcriptional activation and that PC4 is capable of relieving AP-2 transcriptional self-interference. In the absence of exogenously added AP-2 (i.e., an AP-2 expression plasmid), PC4 doubled the endogenous AP-2 activity in the same PA-1 subline, 9117 (data not shown). This doubling of AP-2 activity is significant considering that AP-2 was in a functional excess relative to PC4. Indirect evidence suggested that AP-2 was in a functional excess; small amounts of exogenous AP-2 caused inhibition rather than induction of its activity (15). A dramatic effect on endogenous AP-2 activity was seen when PC4 was transfected into AP-2-overexpressing cell lines. PA-1/AP-2a and PA-1/AP-2k are PA-1 cell line derivatives of a nontumorigenic variant, clone 1, that stably overexpress AP-2 and have tumorigenic properties (15) (Table 1). These cell lines exhibit AP-2 transcriptional self-interference and have low levels of AP-2 transactivation activity (Fig. 3B). The PC4 expression plasmid restored AP-2 transactivation activity in both AP-2-transformed cell lines. When 20 μg of the PC4 expression plasmid was cotransfected, AP-2 transactivation activity in both cell lines increased more than 25-fold relative to cells that were not cotransfected with PC4. pSG5, the parental expression vector of PC4, did not induce AP-2 activity but rather inhibited the activity slightly.
The PA-1/AP-2Bb cell line stably overexpresses AP-2B and has low levels of AP-2 activity with tumorigenic properties in nude mice, similar to AP-2-expressing cell lines. If PC4 interacts with the N-terminal regions of AP-2 and AP-2B, then PC4 should restore AP-2 activity in the PA-1/AP-2Bb cell line. When 25 μg of the PC4 expression plasmid was cotransfected, AP-2 transactivation activity increased more than fivefold in this cell line (Fig. 3C). The PC4 expression plasmid was also cotransfected with the GAL4–AP-2 fusion constructs GAL4–AP-2/11–121 and GAL4–AP-2/11–226, and the transactivation activity was measured using a 5× GAL4-CAT reporter construct. PC4 significantly restored GAL4 transactivation activity. This is consistent with our observation that the N-terminal region of AP-2 interacted with PC4 when fused to the GAL4 DNA-binding domain (Fig. 2D). Figure 3D shows the relief of self-interference for one of the fusion constructs, GAL4–AP-2/11–121. The GAL4–AP-2 transcriptional self-interference resulted in an approximately threefold reduction in CAT activity, and cotransfection of PC4 relieved this inhibition. These results also suggest that the N-terminal region of AP-2 is the main interaction domain for PC4. The assay for direct binding of PC4 and AP-2 (Fig. 2C) indicated that the region between amino acids 11 and 121 is necessary but not sufficient for this interaction. This is consistent with the assay results showing that this region in GAL4–AP-2/11–121 inhibits AP-2 activity and that this inhibition can be relieved by PC4. The fact that the region in the C-terminus is necessary for the interaction of the in vitro-synthesized proteins indicates that there exist inherent differences in these assays. One involves the use of transcription to measure the minimal region that can interact with PC4, and the other relies on immunoprecipitation to identify regions necessary for a stable interaction.
PC4 has been shown to be a coactivator of VP16 activity (8). Overexpression of AP-2 can cross-interfere with the activator VP16 and inhibit its activity (Fig. 3E). If PC4 were the coactivator that was sequestered by AP-2, then high levels of PC4 should reduce their cross-interference and restore VP16 activity. We cotransfected the PC4 expression plasmid with cross-interfering amounts of AP-2. VP16 activity was measured using a GAL4-VP16 fusion plasmid and GAL4 reporter sequences. As shown in Fig. 3E, PC4 significantly restored GAL4-VP16 transactivation activity. These experiments confirmed that sequestration of the coactivator PC4 occurred in these cells and that limiting amounts of PC4 could result in AP-2-induced tumorigenicity of PA-1 cells.
PC4 relieves AP-2 transcriptional self-interference in vitro.
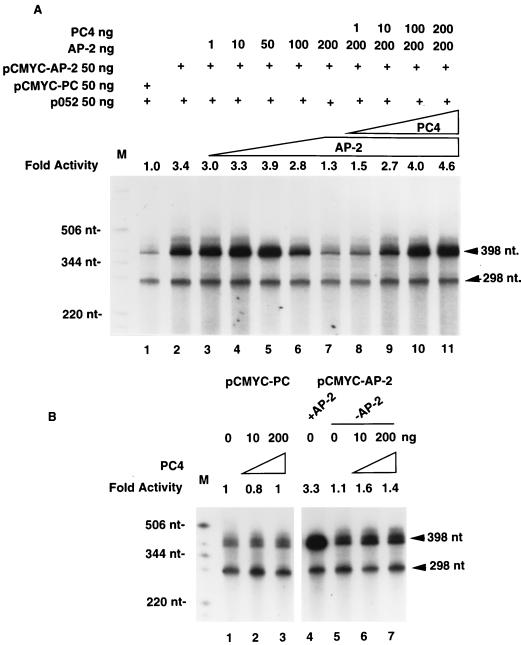
Transient transfection of the PC4 expression plasmid relieved AP-2 transcriptional self-interference in PA-1 cells. We performed AP-2 in vitro transcription experiments to test whether PC4 could restore AP-2 transcriptional self-interference in vitro as well. The effect of purified PC4 protein on AP-2-mediated transcription was examined using HeLa cell nuclear extracts. Two AP-2-binding sites were cloned upstream of a c-myc minimal promoter linked to a 398-bp DNA sequence that lacks G residues. The absence of G residues enabled the use of RNase T1 to degrade nonspecific transcripts encoded by the vector sequence (25a). As an internal control in these in vitro transcription experiments, we used a plasmid, p052, which contains an unrelated hsp70 heat shock promoter and a 298-bp DNA sequence with no G residues. Figure 4A shows the transcription products of the two plasmids.
FIG. 4.
PC4 relieves AP-2 transcriptional self-interference in vitro. In vitro transcription reactions using HeLa cell nuclear extracts were performed as described in Materials and Methods. The template plasmids used in each assay are indicated at the top. The amount of recombinant AP-2 protein or recombinant PC4 protein added to each in vitro transcription reaction is shown. A 398-nt transcription product from pcmyc-PC or pcmyc-AP-2 and a 298-nt transcription product from the control plasmid, p052, are shown on the right. End-labeled nucleotide markers (M) are marked on the left. The fold activity was calculated by scanning the autoradiographic image of the 398-nt transcripts with a DU70 spectrophotometer (Beckman Instruments Inc., Fullerton, Calif.), assuming the transcriptional activity of the parental plasmid pcmyc-PC to be 1. The values shown on each lane are adjusted for the 298-nt transcript of the internal control. (A) PC4 restores AP-2 transcription. (B) PC4 does not affect the expression of plasmid pcmyc-AP-2 in the absence of AP-2 protein and the expression of the parental plasmid pcmyc-PC. +AP-2, HeLa cell nuclear extract containing endogenous levels of AP-2 was used in the in vitro transcription assay; −AP-2, AP-2-depleted HeLa cell nuclear extract was used in the assays.
The presence of AP-2 target sequences in the plasmid pcmyc-AP-2 enhanced transcription more than threefold compared to that with the parental plasmid, pcmyc-PC (Fig. 4A, compare lanes 1 and 2), indicating the existence of endogenous AP-2 activity in HeLa cell nuclear extracts. Small quantities of AP-2 protein purified from bacteria enhanced the transcription to a level slightly higher than that with the pcmyc-AP-2 plasmid. The addition of more than 50 ng of AP-2 protein inhibited transcription from the pcmyc-AP-2 plasmid (lanes 6 and 7). Maximal inhibition was observed at 200 ng of AP-2 protein, and the transcription level was 30% higher than that of the parent plasmid, pcmyc-PC. These results indicate that AP-2 transcriptional self-interference occurs in vitro as well.
When recombinant PC4 protein was added with 200 ng of AP-2 protein, transcription from pcmyc-AP-2 was restored in a dose-dependent manner (Fig. 4A, lanes 8 to 11). The transcriptional activity of pcmyc-AP-2 was 4.6-fold higher than that of the parental plasmid, pcmyc-PC, when 200 ng of recombinant PC4 protein was used in the assay. Transcription from the control plasmid, p052, was not significantly altered in these experiments. PC4 protein did not affect transcription from the pcmyc-PC plasmid (Fig. 4B, lanes 1 to 3), indicating that the AP-2 sites in pcmyc-AP-2 are necessary for PC4-mediated restoration of transcription. AP-2 protein was depleted from the HeLa cell nuclear extracts with an AP-2 antibody, and these extracts were used in transcription assays. Transcription from the pcmyc-AP-2 plasmid was reduced significantly (compare lanes 4 and 5), as expected. This confirms that AP-2 protein is needed for activated transcription from pcmyc-AP-2. PC4 protein was not capable of enhancing the activity from pcmyc-AP-2 in the absence of AP-2 protein (lanes 6 and 7). These experiments indicate that the AP-2 protein is necessary for the PC4-mediated restoration of transcription from pcmyc-AP-2. The in vitro transcription experiments show that AP-2 transcriptional self-interference can be relieved by recombinant PC4 protein and confirm that PC4 is a positive coactivator of AP-2-mediated transcription.
PC4 suppresses ras oncogene-induced transformation.
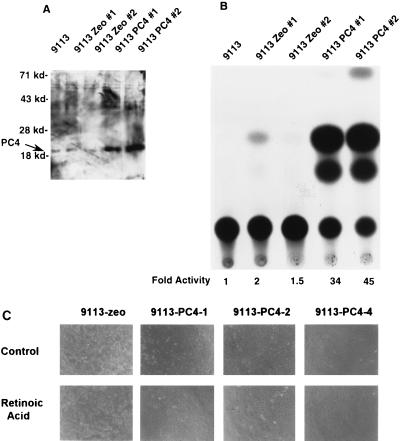
We next tested whether PC4 could restore AP-2 activity in ras-transformed cells that express high levels of AP-2 but have low AP-2 activity. ras-transformed 9113 PA-1 cells were stably transfected with the pZeoSV-PC4 expression vector, and zeomycin-resistant colonies were isolated. The transfection efficiency with plasmid PC4 (23 colonies on three plates) was not significantly different from that obtained with the pZeoSV vector containing no PC4 cDNA insert (31 colonies on three plates). All of the vector control-transfected colonies survived and could be established into cell lines. In stark contrast to the vector controls, only three PC4 colonies were picked and survived establishment in culture, resulting in cell lines 9113-zeo-PC4-1, -2, and -4. Those three PC4-transfected 9113 colonies that grew did so very slowly (Table 2). The cell lines expressed PC4 (Fig. 5A) and were found to form significantly fewer anchorage-independent colonies—on average, 100-fold fewer colonies than were observed for the vector-transfected cells (Table 2). One PC4-expressing 9113 cell line, 9113-zeo-PC4-4, which exhibited the most pronounced morphological change, flattening, grew for five passages but then rapidly reverted to the ras-transformed phenotype. RA-induced differentiation is inhibited by ras transformation (27). During this initial period, we found that 9113-zeo-PC4-4 could differentiate in medium containing RA and that its growth was inhibited 90% by this treatment, compared to the 90% resistance to treatment determined for 9113-zeo control cells (data not shown). The PC4-expressing ras cell lines 9113-zeo-PC4-1 and -2 were highly growth suppressed and failed to form tumors in nude mice (Table 1).
TABLE 2.
Growth properties of PC4-transfected 9113 ras-Transformed PA-1 cellsa
| Cell line | Cell countb | % Anchorage-independent growthc |
|---|---|---|
| 9113-zeo-1 | 12 × 106 | 8.5 |
| 9113-zeo-2 | 15 × 106 | 11 |
| 9113-zeo-PC4-1 | 1.5 × 106 | 0.1 |
| 9113-zeo-PC4-2 | 1.5 × 106 | 0.06 |
| 9113-zeo-PC4-4 | 0.9 × 106 | 0.13 |
Comparison of the growth properties of PA-1 9113 ras-transformed cells stably transfected with the vector control (zeo-1 or zeo-2) or the PC4 expression vector (PC4-1, PC4-2, or PC4-4).
Averages of counts from duplicate dishes. Cells (105) were seeded in duplicate in 60-mm-diameter culture dishes, trypsinized after 8 days of growth, and counted with the aid of a hemocytometer.
Cells (104) were seeded in 0.35% agarose in complete medium (modified Eagle’s medium with 10% fetal bovine serum) and overlaid on a bed of 0.7% agarose in complete medium. The numbers of colonies were determined after 14 days.
FIG. 5.
The ras oncogene-transformed PA-1 cell lines constitutively expressing PC4 have a nontumorigenic phenotype and are sensitive to RA-induced differentiation. PC4 cDNA was cloned into an SV40-driven pZeoSV vector that carries a gene encoding resistance to zeomycin for selection in human cells. 9113, ras-transformed PA-1 cells; 9113 Zeo #1 and #2, control transfections of 9113 cells with pZeoSV vector; 9113 PC4 #1 and #2, PC4-transfected 9113 cells. (A) PC4-transfected ras-transformed PA-1 cells have high levels of PC4. Nuclear extracts were prepared from the cells, subjected to Western blotting, and probed with a PC4-specific antiserum. The mobilities of molecular markers are shown on the left. (B) PC4-transfected ras-transformed PA-1 cells have a high endogenous level of AP-2 activity. Four micrograms of 3× AP-2–CAT was transfected into the cells, and the CAT activity was determined. The low-level endogenous AP-2 activity of 9113 cells was set to 1 to calculate the fold inductions, which are shown at the bottom. (C) The ras-transformed PA-1 cells constitutively expressing PC4 grow slowly, with a flattened morphology, and are sensitive to RA.
If PC4 is a limiting cofactor critical to the transcriptional mechanism of self-interference by which AP-2 and ras transform cells, then restoration of high levels of AP-2 transcriptional activity should accompany the loss of anchorage-independent growth and tumorigenicity observed in PC4-expressing 9113 cells. When AP-2–CAT reporter assays were performed on the PC4-expressing ras-transformed 9113 PA-1 cells, we found that PC4 restored AP-2 activity to a high level (Fig. 5B). Western blot analysis indicated that the PC4-expressing cell lines had the same level of AP-2 as the parental cell lines and pZeoSV vector-transfected cell lines, indicating that PC4 is not exerting its effects by altering the level of AP-2 protein (data not shown). The PC4-transfected 9113 colonies had a differentiated morphology which was further enhanced in the presence of 10−5 M RA (Fig. 5C), a further indication of a reversion of ras transformation. In summary, based on the poor efficiency of obtaining colonies of PC4-expressing 9113 cells, the slow growth and differentiated morphology of such cells, and their lack of tumorigenicity, we concluded that the growth-enhancing and tumorigenic effects of the ras oncogene were abrogated by PC4.
DISCUSSION
Our previous studies indicated that the ras oncogene induces high levels of AP-2 mRNA. Overexpression of AP-2 causes transcriptional self-interference, and this process leads to tumorigenicity in the human teratocarcinoma cell line PA-1. We identified three proteins (of 19, 74, and 110 kDa) that specifically interacted with the GST–AP-2 fusion protein and characterized their role in AP-2 transcriptional activation. The 74-kDa protein was identified as the RAP74 subunit of transcription factor TFIIF, and the 110-kDa protein was identified as the enzyme poly(ADP-ribose) polymerase (15a). The role of these other proteins in AP-2 transcriptional activation is currently under investigation. AP-2 and VP16 have a common coactivator, PC4. In this report we have shown that this coactivator specifically binds AP-2 and positively regulates AP-2 transcriptional activity. Ge et al. (8, 9) found that PC4 interacts specifically with the activation domains of a number of activators, including VP16, CTF, and SP-1. Their recent model suggests that several PC4 molecules are involved in stabilizing the interaction among multiple proteins of the preinitiation complex via several pairwise interactions (19). Our data indicate that PC4 is titrated away during AP-2 overexpression. Presumably both free and target DNA-bound AP-2 molecules compete for interaction with PC4. When the PC4 level is elevated, this protein becomes readily available for the target DNA-bound AP-2 molecules, thus restoring normal AP-2 transcriptional activity. While coactivator relief of transcriptional interference in vitro has been reported (24), the present work is the first example of a study in which a transcriptional cofactor was able to relieve transcriptional interference in vivo and was associated with a physiological process, reversal of tumorigenic transformation.
We have identified PC4 as a target protein, involved in AP-2 self-interference, that can reverse ras transformation. This work demonstrates that an oncogenic transcription factor, AP-2, transforms cells through its effects on a general transcriptional coactivator rather than by regulating the subset of cellular genes normally controlled by the transcription factor. Alteration of the level of this coactivator, PC4, results in reversion of the oncogenic signal. Aside from its implications as to the mechanism of transformation by ras and oncogenic transcription factors, these findings may provide a more general mechanism to exploit in reversing cellular transformation by manipulating transcriptional cofactors. Since PC4 can cause reversion of the transformed phenotype of ras-transformed cells, our data provide a new perspective of this signal transduction pathway. The cascade of phosphorylation events leading to changes in the levels of AP-2 mRNA and protein results in a transcriptional imbalance of this key coactivator, PC4. This balance of the coactivator PC4 is so critical to the mechanism of ras transformation that manipulating the level of PC4 can revert ras-transformed cells strictly by a transcriptional mechanism. The growth-inhibiting activity of PC4 can be mediated through AP-2 or any other transcription factor that requires PC4 as a coactivator.
Since PC4 maps to human chromosome 5p13, a location frequently associated with loss of heterozygosity in lung and bladder tumors (1, 30), which often contain mutations in ras protooncogenes, PC4 may play a role as a tumor suppressor in the natural occurrence of these cancers. A potential application for this approach may be in breast cancer where Her2/neu-overexpressing cells also express high levels of AP-2 and as part of a possible regulatory loop, AP-2 regulates the Her2/neu promoter in those breast cancer cells (2, 11).
ACKNOWLEDGMENTS
We thank Robert Roeder and Hui Ge for their generous gifts of plasmid construct pGEX-PC4 and the rabbit antiserum raised against PC4. We are grateful to Michael Van Dyke for helpful discussions. We acknowledge Sun Yim and Yihong Yu for technical assistance in cell culture.
This work was supported by National Cancer Institute grant CA53475 to M.A.T. and by NIH core center grant 16672 and National Cancer Institute grant CA67036 to P.K.
REFERENCES
- 1.Bohm M, Kirch H, Otto T, Rubben H, Wieland I. Deletion analysis at the DEL-27, APC and MTS1 loci in bladder cancer: LOH at the DEL-27 locus on 5p13-12 is a prognostic marker of tumor progression. Int J Cancer. 1997;74:291–295. doi: 10.1002/(sici)1097-0215(19970620)74:3<291::aid-ijc10>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 2.Bosher J M, Williams T, Hurst H C. The developmentally regulated transcription factor AP-2 is involved in c-erbB-2 overexpression in human mammary carcinoma. Proc Natl Acad Sci USA. 1995;92:744–747. doi: 10.1073/pnas.92.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buettner R, Kannan P, Imhof A, Bauer R, Yim S O, Glockshuber R, Van Dyke M W, Tainsky M A. An alternatively spliced mRNA from the AP-2 gene encodes a negative regulator of transcriptional activation by AP-2. Mol Cell Biol. 1993;13:4174–4185. doi: 10.1128/mcb.13.7.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang C M, Ge H, Wang Z, Hoffmann A, Roeder R G. Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerases II and III. EMBO J. 1993;12:2749–2762. doi: 10.1002/j.1460-2075.1993.tb05936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiu R, Imagawa M, Imbra R J, Bockoven J R, Karin M. Multiple cis- and trans-acting elements mediate the transcriptional response to phorbol esters. Nature. 1987;329:648–651. doi: 10.1038/329648a0. [DOI] [PubMed] [Google Scholar]
- 6.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flanagan P M, Kelleher R J D, Sayre M H, Tschochner H, Kornberg R D A. Mediator required for activation of RNA polymerase II transcription in vitro. Nature. 1983;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- 8.Ge H, Roeder R G. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994;78:513–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 9.Ge H, Zhao Y, Chait B T, Roeder R G. Phosphorylation negatively regulates the function of coactivator PC4. Proc Natl Acad Sci USA. 1994;91:12691–12695. doi: 10.1073/pnas.91.26.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 11.Hollywood D P, Hurst H C. A novel transcription factor, OB2-1, is required for overexpression of the proto-oncogene c-erbB-2 in mammary tumour lines. EMBO J. 1993;12:2369–2375. doi: 10.1002/j.1460-2075.1993.tb05891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hyman S E, Comb M, Pearlberg J, Goodman H M. An AP-2 element acts synergistically with the cyclic AMP- and phorbol ester-inducible enhancer of the human proenkephalin gene. Mol Cell Biol. 1989;9:321–324. doi: 10.1128/mcb.9.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imagawa M, Chiu R, Karin M. Transcription factor AP-2 mediates induction by two different signal-transduction pathways: protein kinase C and cAMP. Cell. 1987;51:251–260. doi: 10.1016/0092-8674(87)90152-8. [DOI] [PubMed] [Google Scholar]
- 14.Kaiser K, Stelzer G, Meisterernst M. The coactivator p15 (PC4) initiates transcriptional activation during TFIIA-TFIID-promoter complex formation. EMBO J. 1995;14:3520–3527. doi: 10.1002/j.1460-2075.1995.tb07358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kannan P, Buettner R, Chiao P J, Yim S O, Sarkiss M, Tainsky M A. N-ras oncogene causes AP-2 transcriptional self-interference, which leads to transformation. Genes Dev. 1994;8:1258–1269. doi: 10.1101/gad.8.11.1258. [DOI] [PubMed] [Google Scholar]
- 15a.Kannan, P., and M. A. Tainsky. Unpublished data.
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Luckow B, Schutz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987;15:5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luscher B, Mitchell P J, Williams T, Tjian R. Regulation of transcription factor AP-2 by the morphogen retinoic acid and by second messengers. Genes Dev. 1989;3:1507–1517. doi: 10.1101/gad.3.10.1507. [DOI] [PubMed] [Google Scholar]
- 19.Malik S, Guermah M, Roeder R G. A dynamic model for PC4 coactivator function in RNA polymerase II transcription. Proc Natl Acad Sci USA. 1998;95:2192–2197. doi: 10.1073/pnas.95.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McPherson L A, Baichwal V R, Weigel R J. Identification of ERF-1 as a member of the AP2 transcription factor family. Proc Natl Acad Sci USA. 1997;94:4342–4347. doi: 10.1073/pnas.94.9.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer M E, Gronemeyer H, Turcotte B, Bocquel M T, Tasset D, Chambon P. Steroid hormone receptors compete for factors that mediate their enhancer function. Cell. 1989;57:433–442. doi: 10.1016/0092-8674(89)90918-5. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell P J, Timmons P M, Hebert J M, Rigby P W, Tjian R. Transcription factor AP-2 is expressed in neural crest cell lineages during mouse embryogenesis. Genes Dev. 1991;5:105–119. doi: 10.1101/gad.5.1.105. [DOI] [PubMed] [Google Scholar]
- 23.Prywes R, Zhu H. In vitro squelching of activated transcription by serum response factor: evidence for a common coactivator used by multiple transcriptional activators. Nucleic Acids Res. 1992;20:513–520. doi: 10.1093/nar/20.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- 25.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 25a.Sawardogo M, Roeder R G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985;43:165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- 26.Smith D B, Concoran M L. Unit 16-7. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidmen J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Green Publishing Associates and Wiley Interscience; 1991. pp. 1–8. [Google Scholar]
- 27.Tainsky M A, Krizman D B, Chiao P J, Yim S O, Giovanella B C. PA-1, a human cell model for multistage carcinogenesis: oncogenes and other factors. Anticancer Res. 1988;8:899–913. [PubMed] [Google Scholar]
- 28.Tainsky M A, Cooper C S, Giovanella B C, Vande Woude G F. An activated rasN gene: detected in late but not early passage human PA1 teratocarcinoma cells. Science. 1984;225:643–645. doi: 10.1126/science.6740333. [DOI] [PubMed] [Google Scholar]
- 29.Wang J C, Van Dyke M W. Sp1, USF, and GAL4 activate transcription independently of TFIID-downstream promoter interactions. Biochim Biophys Acta. 1994;1218:308–314. doi: 10.1016/0167-4781(94)90182-1. [DOI] [PubMed] [Google Scholar]
- 30.Wieland I, Bohm M, Arden K C, Ammermuller T, Bogatz S, Viars C S, Rajewsky M F. Allelic deletion mapping on chromosome 5 in human carcinomas. Oncogene. 1996;12:97–102. [PubMed] [Google Scholar]
- 31.Williams T, Admon A, Luscher B, Tjian R. Cloning and expression of AP-2, a cell-type-specific transcription factor that activates inducible enhancer elements. Genes Dev. 1988;2:1557–1569. doi: 10.1101/gad.2.12a.1557. [DOI] [PubMed] [Google Scholar]
- 32.Williams T, Tjian R. Analysis of the DNA-binding and activation properties of the human transcription factor AP-2. Genes Dev. 1991;5:670–682. doi: 10.1101/gad.5.4.670. [DOI] [PubMed] [Google Scholar]
- 33.Zeuthen J, Norgaard J D R, Avner P, Fellows M, Wartiovaara J, Vaheri A, Rosen J, Giovanella B C. Characterization of a human ovarian teratocarcinoma-derived cell line. Int J Cancer. 1980;25:19–32. doi: 10.1002/ijc.2910250104. [DOI] [PubMed] [Google Scholar]
- 34.Zhu H, Joliot V, Prywes R. Role of transcription factor TFIIF in serum response factor-activated transcription. J Biol Chem. 1994;269:3489–3497. [PubMed] [Google Scholar]