Abstract
HCF is a mammalian nuclear protein that undergoes proteolytic processing and is required for cell proliferation. During productive herpes simplex virus (HSV) infection, the viral transactivator VP16 associates with HCF to initiate HSV gene transcription. Here, we show that the worm Caenorhabditis elegans possesses a functional homolog of mammalian HCF that can associate with and activate the viral protein VP16. The pattern of sequence conservation, however, is uneven. Sequences required for mammalian HCF processing are not present in C. elegans HCF. Furthermore, not all elements of mammalian HCF that are required for promoting cell proliferation are conserved. Nevertheless, unexpectedly, C. elegans HCF can promote mammalian cell proliferation because a region of HCF that is conserved can promote mammalian cell proliferation better than its human counterpart. These results suggest that HCF possesses a highly conserved role in metazoan cell proliferation which is targeted by VP16 to regulate HSV infection. The precise mechanisms, however, by which HCF functions in mammals and worms appear to differ.
In infected cells, human herpes simplex virus (HSV) can cause either lytic or latent infection. During lytic infection, the viral regulatory protein VP16 (also referred to as Vmw65, α-TIF, and ICP25) forms a multiprotein-DNA complex with two cellular proteins: the POU homeodomain transcription factor Oct-1 and the cell proliferation factor HCF. VP16 first associates with HCF (also referred to as C1, VCAF, and CFF) to form a stable DNA-independent heterodimeric complex. VP16 binding to HCF facilitates its subsequent association with Oct-1 on VP16-responsive cis-regulatory TAATGARAT (R, purine) elements in the HSV immediate-early promoters, initiating a cascade of viral gene expression (for reviews, see references 22, 28, and 31).
Human HCF consists of a complex of noncovalently associated polypeptides ranging from 110 to 150 kDa (15, 16, 30, 32). These associated polypeptides are derived from a large precursor protein of over 2,000 amino acids by specific proteolytic cleavage at a series of six centrally located 26-amino acid repeats, referred to here as HCFPRO repeats (15, 30, 32). The function of the carboxy-terminal fragments is not known. Within the amino-terminal fragments, however, the amino-terminal 380 residues are sufficient to bind VP16, stabilize the VP16-induced complex with Oct-1, and activate transcription in vivo (17, 33). This region of HCF, called the HCFVIC domain, contains six sequence repeats related to “kelch” repeats found in the Drosophila egg chamber protein kelch (34). These repeats, called HCFKEL repeats, are both necessary and sufficient for efficient complex formation with VP16 (33). Among mammalian HCFs, both sets of repeats, HCFPRO and HCFKEL, are highly conserved (8, 13, 30).
In addition to its role in HSV gene expression, HCF is required for cell proliferation. A single missense mutation in the hamster cell line tsBN67, a proline-to-serine substitution at position 134 (P134S) of HCF, causes a temperature-sensitive defect in cell proliferation (8) and disrupts interaction with VP16 (33). Consistent with an involvement in cell proliferation, human HCF is expressed in proliferating cultured cells and embryonic tissues (13, 29).
HCF may have been conserved during metazoan evolution, because extracts from Spodoptera and Drosophila insect cells, but not yeast, can stabilize VP16 association with human Oct-1 (14, 31). We show here that the nematode Caenorhabditis elegans expresses a functional homolog of human HCF. Although C. elegans HCF lacks some sequence elements important for human HCF function, the C. elegans HCF homolog can support mammalian cell proliferation, suggesting that HCF plays a highly conserved role in metazoan cell proliferation.
MATERIALS AND METHODS
Extract preparation and electrophoretic mobility retardation analysis.
Wild-type Bristol N2 strain C. elegans worms were grown as previously described (4). All worms were grown at 20°C. Worms were freed of bacteria (18) and collected, and worm pellets were suspended in 10 mM HEPES (pH 7.6) containing 10 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 0.5 mM EGTA, 10 mM benzamidine, 1 mM Na metabisulfite, 2 μg of leupeptin/ml, and 2 μg of aprotinin/ml. The worm suspension was passed through a French press at 1,500 psi. The homogenate was then centrifuged at 85,000 × g for 30 min at 4°C. The supernatant solution was taken as total cell extract. About 2 μg of cell extract (about 1 to 3 μg/μl) was used for electrophoretic mobility retardation analysis. Human HCF was purified by immunoprecipitation with amino-terminal αHCFN18 antipeptide antisera (8) and elution with the HCFN18 peptide. The Oct-1 POU domain and VP16 lacking its transcriptional activation domain (VP16ΔC) were prepared for VP16-induced complex formation as glutathione S-transferase (GST) fusion proteins. The proteins were expressed and purified from Escherichia coli as previously described (30, 33); the Oct-1 POU domain, but not VP16ΔC, was separated from the GST moiety by treatment with thrombin. Electrophoretic mobility retardation analysis was performed with the (OCTA+)TAATGARAT probe as previously described (30).
C. elegans hcf-1 cDNA isolation and in vitro protein expression.
The amino acid sequence of the human HCFVIC domain (residues 1 to 380) was used to search the C. elegans Sanger Center Network database (26) with TBLASTN (2). Significant sequence similarity to a predicted gene present on the overlapping cosmids C33H5 and C46A5 was found. A cDNA copy of this gene was prepared by PCR with oligonucleotides containing C. elegans sequence corresponding to predicted initiation (ATGGACGAAGATGTCGGTTTAG) and termination (TTACTGATGATCGAAACGAGCTC) codons (underlined) and a mixed-stage C. elegans cDNA library (a kind gift of R. Barstead, Oklahoma Medical Research Foundation). A 2.4-kb DNA fragment was amplified and isolated for sequence analysis and subcloning. Sequences encoding the full-length (782 residues) and the amino-terminal 395 amino acids of C. elegans HCF were cloned into the in vitro transcription and translation expression plasmid pNCITE as previously described (33). In vitro transcription and translation reactions were performed with the TNT system (Promega, Inc.). The anti-C. elegans HCF antiserum αCeHCFN16 was raised in rabbits against a 17-amino-acid peptide containing the 16 amino-terminal residues of C. elegans HCF (MDEDVGLEATNYSRGDC) (underlined) coupled to keyhole limpet hemocyanin via the carboxy-terminal cysteine residue as previously described (10).
Synchronization of C. elegans, extract preparation, and Northern hybridization analysis.
Viable Bristol N2 strain C. elegans embryos were prepared by treating a mixed population of worms with alkaline hypochlorite (11). The embryos were synchronized by letting the worms hatch in the absence of nutrients at 20°C overnight. The hatched worms were subsequently transferred to NGM plates with E. coli OP50 as a food source. L1-stage worms were harvested 6 h after feeding; the harvesting times (hours) after feeding for worms at other stages were as follows: L2, 20; L3, 29; L4, 40 h. Young adult worms were harvested 52 h after feeding (18). The stage of worm development was verified by microscopic examination. Protein extracts were prepared and electrophoretic mobility retardation analyses were performed as described above. RNA extracts were prepared from cleaned worms by freezing the pelleted worms in liquid nitrogen and grinding them into a powder in a mortar for total RNA preparation as previously described (12). Twenty-five micrograms of total RNA was used to perform Northern hybridization analysis as previously described (23) with 32P-labeled, random-primed DNA fragments corresponding to full-length C. elegans hcf-1 as the probe.
Mammalian-cell HCF expression vectors and rescue of the tsBN67 temperature-sensitive defect.
The human HCF expression constructs pCGNHCFFL, pCGNHCFN1011, and pCGNHCFN380 have been described previously (30, 33). The C. elegans HCF expression constructs pCGNCeHCFFL and pCGNCeHCFN395 contain coding sequences corresponding to full-length HCF and the amino-terminal 395 residues of CeHCF, respectively, in the cytomegalovirus promoter and hemagglutinin epitope-tagged expression construct pCGN (30). Rescue of the tsBN67 temperature-sensitive defect was performed as described previously (8). tsBN67 cells were seeded at 2 × 105 cells/100-mm-diameter dish and cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum at permissive temperature (33.5°C) for 20 h prior to transfection. Expression constructs for human or C. elegans HCF (2 μg/dish) were then cotransfected by calcium-phosphate coprecipitation with the plasmid pSV2neo (0.5 μg/dish) into tsBN67 cells (5). Following transfection, the cells were incubated at 33.5°C for 20 h, washed with serum-free medium, and incubated at 33.5°C for an additional 24 h before selection for 2 weeks at the nonpermissive temperature of 39.5°C in the presence of G418 (0.8 mg/ml). Colonies were identified by fixation and staining with crystal violet.
Nucleotide sequence accession number.
The GenBank accession number for the C. elegans HCF sequence is AF072907.
RESULTS
VP16-induced complex-forming activity in C. elegans extracts.
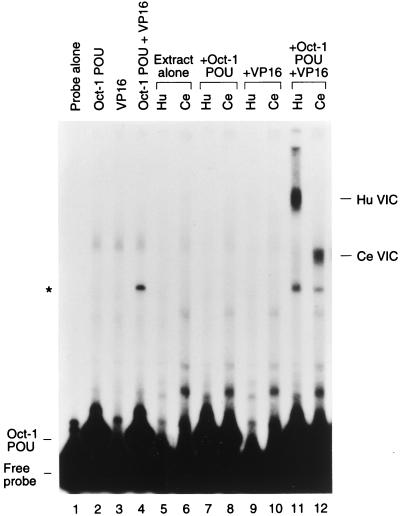
Analyses of insect cells have revealed an activity capable of stabilizing VP16 association with human Oct-1, suggesting the existence of an HCF homolog in insects (14, 31). To determine whether such an activity is generally present in metazoans, we asked whether extracts from the nematode C. elegans can also stabilize VP16 association with human Oct-1, as shown in Fig. 1. We prepared cell extract from mixed-stage worms and compared the VP16-induced complex formation of this extract with that of purified human HCF by using an electrophoretic mobility retardation assay with an HSV TAATGARAT site. The TAATGARAT site, an (OCTA+)TAATGARAT site, contained the octamer sequence ATGCTAAT, which serves as a binding site for Oct-1. For this assay, we used VP16 lacking its transcriptional-activation domain and the human Oct-1 DNA-binding POU domain expressed and purified from E. coli. In the absence of human HCF or C. elegans extract, the Oct-1 POU domain, but not VP16, bound DNA on its own (Fig. 1, cf. lanes 1 to 3). Together, the Oct-1 POU domain and VP16 formed a low level of HCF-independent VP16-induced complex (Fig. 1, lane 4). Addition of purified human HCF (lane 5) did not result in any new complexes, whereas addition of the C. elegans extract alone resulted in a low level of a novel complex (lane 6), for which we do not know the identity. Addition of human HCF or C. elegans extract to either the Oct-1 POU domain or VP16 resulted in superimposition of the individual patterns of complex formation (Fig. 1, cf. lanes 2 and 3 and lanes 5 to 10). However, addition of either the human HCF preparation or the C. elegans extract together with VP16 to the Oct-1 POU domain induced novel VP16-induced complexes (cf. lanes 11 and 12 with lanes 7 and 8). These results suggest that C. elegans worms contain an activity that can replace human HCF in stabilizing the VP16-induced complex.
FIG. 1.
Extract from the nematode C. elegans can stabilize a VP16-induced complex. Partially purified human HCF and cell extract from mixed-stage C. elegans were assayed for VP16-induced complex formation by using an electrophoretic mobility retardation assay with bacterially expressed Oct-1 POU domain (Oct-1 POU), GST-VP16ΔC fusion protein (VP16), and labeled HSV (OCTA+)TAATGARAT probes. Results obtained with probe alone (lane 1) and with probes with the Oct-1 POU domain (lane 2), GST-VP16 (lane 3), and the Oct-1 POU domain and GST-VP16 (lane 4) are shown. Purified human HCF or 2 μg of C. elegans cell extract was assayed with probe alone (lanes 5 and 6) and with probes with the Oct-1 POU domain (lanes 7 and 8), GST-VP16 (lanes 9 and 10), and the Oct-1 POU domain and GST-VP16 (lanes 11 and 12). The positions of the free probe, the Oct-1 POU domain-DNA complex, and the VP16-induced complexes from human (Hu VIC) and C. elegans (Ce VIC) extracts are indicated. The asterisk indicates the position of an HCF-independent VP16-induced complex.
The mobility of the C. elegans VP16-induced complex (Ce VIC) is between that of the HCF-independent VP16-induced complex and that of the human HCF VP16-induced complex (Hu VIC) (Fig. 1), suggesting that a C. elegans protein of a different size from that of human HCF, probably smaller, is incorporated into the C. elegans VP16-induced complex. A similar analysis of insect extracts suggested that insect HCFs are also smaller than human HCF (31); direct comparison of VP16-induced complexes stabilized by insect and C. elegans extracts suggests that the C. elegans HCF-like protein is smaller than its insect counterparts (28a).
Functional C. elegans HCF homolog.
The surprising finding that invertebrate cell extracts can stabilize association of a viral protein with a human transcription factor (i.e., VP16 with Oct-1) (this study and references 14 and 31), prompted us to search the C. elegans genome sequence for human HCFVIC domain-related sequences. This search, however, was hampered by the general similarity of the HCFVIC domain to kelch repeat-containing proteins. For example, in our first attempt to identify a C. elegans HCF homolog, we identified sequence similarity to the predicted C. elegans gene F33C8.1 (Q19981), a kelch repeat-containing gene. The product of this gene, however, failed to stabilize the VP16-induced complex (data not shown).
Subsequent determination of the C. elegans genome sequence revealed a sequence on C. elegans chromosome IV which was related to both the amino- and carboxy-terminal regions of human HCF. This sequence similarity has also been noted by others (13, 17). The predicted coding sequence for this gene presented in the database and reproduced in these published studies (13, 17) does not, however, reveal similarity to the entire region of human HCF that is required for stabilization of the VP16-induced complex (i.e., the HCFVIC domain) (33). Further examination of the C. elegans genomic sequence neighboring this gene revealed possible exons that would provide similarity to the entire human HCFVIC domain. Encouraged by this observation, we designed primers that we predicted would match the translational initiation and stop codons of the C. elegans HCF gene and used them for PCR with a C. elegans cDNA library. A cDNA thus isolated revealed sequence similarity to the human HCF gene across the entire HCFVIC domain-coding sequences. Further PCR analysis showed that the transcript from this gene contains a trans-spliced SL1 leader sequence positioned five nucleotides upstream of the predicted initiation codon (data not shown).
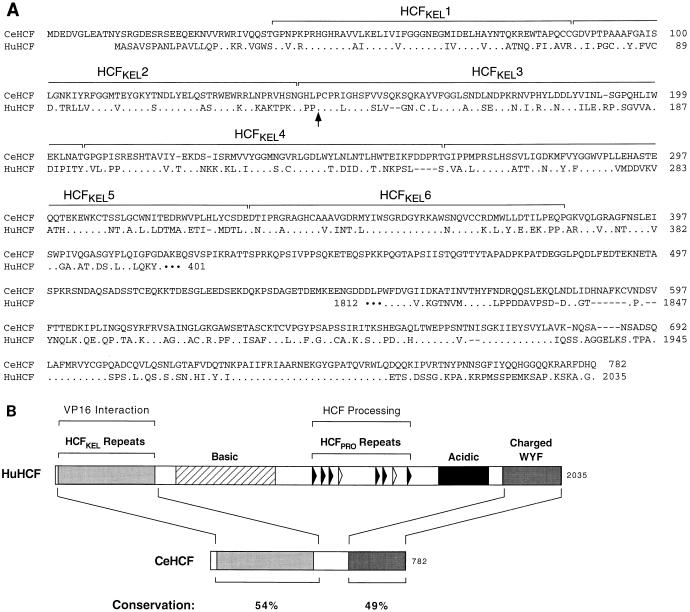
Figure 2 shows a comparison of the amino-acid sequence of this C. elegans HCF-related protein with that of human HCF. In contrast to human HCF, which is approximately 2,000 amino acids long, the predicted C. elegans protein contains only 782 amino acids, in which the amino- and carboxy-terminal regions of human HCF are highly conserved. As illustrated in Fig. 2B, the amino-terminal HCFVIC domain of human HCF is 54% identical to the corresponding region of the C. elegans HCF-related protein and the carboxy-terminal 230 amino acids of human HCF are 49% identical to the carboxy-terminal region of the C. elegans protein (see also Fig. 2A). The remaining central 136 amino acids of the C. elegans protein share less similarity to human HCF, displaying only little, if any, similarity to the basic and acidic regions in human HCF. Significantly, as noted previously (13, 17), this C. elegans protein contains no evident similarity to the human HCFPRO repeats required for HCF processing in human cells (32). This comparison suggests that, if this protein is the functional C. elegans homolog of human HCF, C. elegans HCF either is not processed or is processed by a different mechanism.
FIG. 2.
C. elegans HCF displays uneven sequence similarity to human HCF. (A) Amino acid sequence alignment of C. elegans HCF with corresponding regions of human HCF. Dots represent positions of identity between human HCF and C. elegans HCF sequences. The limits of the six HCFKEL repeats (33) are indicated in brackets. The conserved proline residue (P145 in C. elegans HCF and P134 in human HCF) that is mutated in tsBN67 HCF (8) is indicated by the arrow. (B) Schematic structure comparison of human and C. elegans HCFs. The schematic representation of human HCF is as shown previously (30). Conservation represents the percentages of identical residues between C. elegans and human HCF in the regions indicated. Charged/WYF, region enriched in charged and large hydrophobic residues.
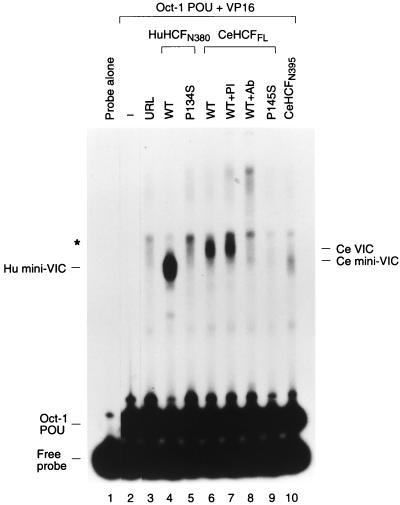
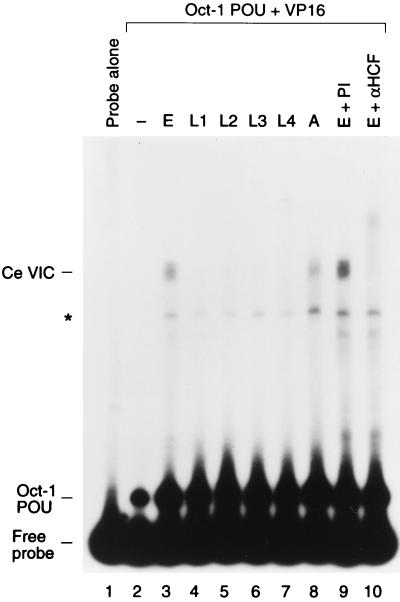
To determine whether the C. elegans HCF-related protein is functionally related to human HCF, we assayed the ability of this protein to stabilize the VP16-induced complex by using an electrophoretic mobility retardation assay as shown in Fig. 3. To compare the activities of the human and C. elegans proteins, we synthesized the HCFVIC domain of human HCF and the full-length HCF protein and putative HCFVIC domain of C. elegans by in vitro translation. The in vitro translation extract used for this experiment generated a low level of contaminating complex in the electrophoretic mobility retardation assay (Fig. 3, cf. lane 3 with lanes 1 and 2). Consistent with previous results (17, 33), the wild-type human HCFVIC domain (lane 4) but not the mutant human HCFVIC domain carrying the tsBN67 P134S mutation (Fig. 3, lane 5) directed formation of a prominent VP16-induced complex, referred to as Hu mini-VIC. The full-length C. elegans protein synthesized in vitro also directed VP16-induced complex formation (Fig. 3, lane 6); this complex comigrates with the VP16-induced complex generated by C. elegans extract (data not shown). Sensitivity of the C. elegans VP16-induced complex to antisera directed against the amino terminus of the predicted C. elegans protein sequence (Fig. 3, lane 8) but not to the corresponding preimmune antisera (lane 7) indicates that, as with the human protein, the C. elegans protein was incorporated into the VP16-induced complex. These results suggest that this C. elegans HCF-related protein is a functional homolog of human HCF. Following the convention of the C. elegans research community, we designate this protein HCF-1 and its cognate gene hcf-1. Here, however, we refer to HCF-1 as C. elegans HCF or CeHCF.
FIG. 3.
In vitro-translated CeHCF promotes VP16-induced complex formation. Human and C. elegans HCFs were synthesized in vitro and assayed for HCF activities by using an electrophoretic mobility retardation assay. Lane 1 contains probe alone, and lanes 2 to 10 contain probes with the Oct-1 POU domain and GST-VP16 proteins. Samples contained in addition unprogrammed reticulocyte lysate (lane 3) and reticulocyte lysates programmed with templates for wild type (lane 4) or P134S (lane 5) human HCFN380, full-length wild type (lanes 6 to 8), P145S (lane 9) CeHCF and CeHCFN395 (lane 10). Preimmune serum (lane 7) and anti-CeHCF antiserum (lane 8) were added to the binding reactions. The positions of free probe, the Oct-1 POU domain complex, and the VP16-induced complex with human HCFN380 (Hu mini-VIC) and full-length (Ce VIC) and amino-terminal (Ce mini-VIC) CeHCFs are indicated. The asterisk indicates a nonspecific complex generated by the reticulocyte lysate.
Consistent with the observed functional relationship between the C. elegans and human HCF proteins, the proline that is changed to a serine in the temperature-sensitive tsBN67 mutation is conserved in the C. elegans HCF protein (Fig. 2A). To determine whether a mutation in C. elegans HCF which is analogous to the hamster tsBN67 mutation also disrupts VP16-induced complex formation, we engineered the proline-to-serine mutation into the corresponding position (residue 145) of full-length C. elegans HCF (P145S). As in human and hamster HCF (8, 33), this tsBN67 proline-to-serine point mutation disrupts VP16-induced complex formation (Fig. 3, cf. lanes 9 and 6).
We also assayed the ability of the region of C. elegans HCF corresponding to the human HCFVIC domain (CeHCF residues 1 to 395 [CeHCFN395]) to stabilize the VP16-induced complex. Curiously, although CeHCFN395 stabilized the VP16-induced complex (Fig. 3, lane 10), it did so less efficiently than either the human HCFVIC domain or the full-length CeHCF (Fig. 3, cf. lanes 4, 6, and 10). Thus, the C. elegans HCF region that contributes to stabilization of the VP16-induced complex may extend beyond the HCFKEL-repeat region.
Developmental regulation of HCF expression in C. elegans.
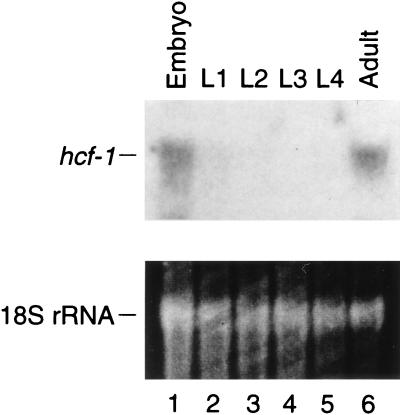
Southern hybridization analysis using the cloned C. elegans hcf-1 cDNA as probe demonstrated that there is a single copy of the hcf-1 gene in the C. elegans genome (data not shown). To reveal the expression of hcf-1 during development, worms were synchronized by embryo isolation and subsequent collection of worms at the embryonic, L1 to L4 larval, and gravid (embryo-containing adults) developmental stages. Total RNA was isolated and probed for hcf-1 mRNA expression by Northern hybridization analysis as shown in Fig. 4. This analysis revealed a single hcf-1 mRNA of 2.6 kb expressed primarily in embryos and adults (Fig. 4, cf. lanes 1 and 6 with the larval stages in lanes 2 to 5). The embryonic expression parallels the embryonic expression of human HCF mRNA (13, 29). The adult expression may represent either adult cell expression (i.e., somatic or germline) or expression in the embryos present in gravid adults; we have not distinguished between these possibilities.
FIG. 4.
The expression of the hcf-1 gene is developmentally regulated. C. elegans worms were synchronized as described in Materials and Methods. Total RNAs were isolated from embryos; L1, L2, L3, and L4 larvae; and adults. RNAs were probed by Northern hybridization analysis using labeled hcf-1 cDNA as a probe. The levels of total RNA in each lane were assayed by staining the 18S rRNA with ethidium bromide as shown at the bottom.
We also assayed the abilities of extracts from C. elegans at different stages of development to stabilize the VP16-induced complex. We prepared cell extracts from worms at different developmental stages and assayed their abilities to support VP16-induced complex formation by using an electrophoretic mobility retardation assay as shown in Fig. 5. As with the hcf-1 mRNA, CeHCF activity was observed in extracts from embryos and adult worms (Fig. 5, lanes 3 and 8) but not in extracts from larvae (lanes 4 to 7). The authenticity of the observed complex with the embryo extract was confirmed by use of the antisera directed against the amino terminus of the predicted C. elegans protein sequence (αHCF) and preimmune antisera (Fig. 5, lanes 9 and 10), which shows that the predicted amino-terminal sequence presented in Fig. 2 is correct. Thus, during C. elegans development, VP16-induced complex-forming activity correlates with hcf-1 gene expression.
FIG. 5.
CeHCF activity correlates with hcf-1 gene expression during C. elegans development. Total-cell extracts from worms at different developmental stages were assayed for VP16-induced complex formation by using an electrophoretic mobility retardation assay. Lane 1 contains probe alone, and lanes 2 to 10 contain probes with the Oct-1 POU domain and GST-VP16 proteins. Samples contained in addition extract from embryos (lanes 3, 9, and 10); L1, L2, L3, or L4 larvae (lanes 4 to 7); and adults (lane 8). Preimmune serum (lane 9) and anti-CeHCF antiserum (lane 10) were added to the binding reaction. The positions of free probe, the Oct-1 POU domain complex, and the VP16-induced complex (Ce VIC) are indicated. The asterisk indicates a weak HCF-independent VP16-induced complex.
Rescue of the tsBN67 cell proliferation defect by CeHCF.
The studies described above demonstrate that the ability of HCF to stabilize the VP16-induced complex has been conserved during metazoan evolution (Fig. 1 and 3). Therefore, we asked whether the ability of HCF to promote cell proliferation has also been conserved. A priori, examination of the C. elegans HCF sequence would suggest that the C. elegans HCF protein cannot overcome the hamster tsBN67 cell-proliferation defect because it lacks sequences corresponding to the basic region of human HCF (Fig. 2); in human HCF, the basic region sequences are required to rescue the tsBN67 cell proliferation defect (33). Nevertheless, to test the ability of C. elegans HCF to promote mammalian cell proliferation, we transfected tsBN67 cells with mammalian CeHCF expression vectors and assayed the transfected cells for colony formation at a nonpermissive temperature as shown in Fig. 6. Analysis of protein expression in a separate short-term assay showed that all of the proteins assayed are faithfully expressed (data not shown).
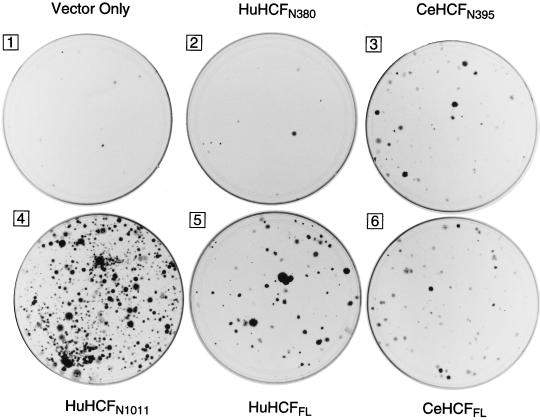
FIG. 6.
CeHCF rescues the hamster tsBN67 cell proliferation defect. Hamster tsBN67 cells were transfected with the human pCGNHCF or C. elegans pCGNCeHCF HCF expression construct as described in Materials and Methods. Plate 1, pCGN vector alone; plate 2, pCGNHCFN380; plate 3, pCGNCeHCFN395; plate 4, pCGNHCFN1011; plate 5, pCGNHCFFL; plate 6, pCGNCeHCFFL. Following the transfection protocol, the cells were incubated at 39.5°C in the presence of G418 for 2 weeks. The colonies were then fixed and stained with crystal violet.
Full-length human HCF rescued the tsBN67 defect in this assay (Fig. 6, cf. plates 5 and 1) as described previously (8, 33). Also consistent with the results of previous studies (33), the amino-terminal half of human HCF (HuHCFN1011) rescued the temperature-sensitive cell proliferation defect better than full-length HCF (Fig. 6, cf. plates 4 and 5), whereas just the HCFVIC domain (HuHCFN380) lacking the basic region failed to rescue the defect (cf. plates 1 and 2). To our surprise, full-length C. elegans HCF complemented the tsBN67 defect even though it lacks a conserved basic region (cf. plates 1 and 6). Thus, a role for HCF in cell proliferation has been conserved during metazoan evolution, but interestingly, this role is not dependent on conservation of a basic region.
Further, to our surprise and in stark contrast to the results obtained with the human protein, the region of C. elegans HCF corresponding to the human HCFVIC domain (CeHCFN395) complemented the tsBN67 defect nearly as well as the full-length C. elegans HCF protein (Fig. 6, cf. plates 3 and 6) and better than its human HCFVIC domain counterpart (cf. plates 2 and 3). This result explains why C. elegans HCF can rescue the tsBN67 defect even though it lacks an evident basic region. It also suggests that, while the cellular function of HCF has been conserved during metazoan evolution, the relative roles of different regions of HCF in promoting cell proliferation have changed: in human HCF, the HCFVIC domain cooperates with the neighboring basic region to promote cell proliferation, whereas in C. elegans, the region corresponding to the human HCFVIC domain can promote cell proliferation on its own.
DISCUSSION
Through identification and characterization of a functional homolog of mammalian HCF in C. elegans we have shown that selected sequences and functions of HCF have been conserved in metazoans.
Uneven HCF sequence conservation during metazoan evolution.
The complete sequence of C. elegans HCF reveals a striking pattern of sequence conservation: the amino- and carboxy-terminal regions of HCF are highly conserved, but as noted previously (13, 17), the central regions, including the HCFPRO repeats required for human HCF processing (32), are poorly conserved (Fig. 2). When the sequence of human HCF was first identified, there was no known sequence relationship to other proteins, but it was noted that both the amino-terminal and carboxy-terminal regions are enriched in charged and large hydrophobic residues (Charged/WYF [30]). We now know that the amino-terminal Charged/WYF region is the HCFVIC domain (17, 33) and is also involved in cell proliferation (8). Although implicated in stabilization of a VP16-induced complex with full-length VP16 (17), the cellular function of the carboxy-terminal Charged/WYF region remains unknown. Its high level of conservation in C. elegans suggests that, like its amino-terminal counterpart, it also plays an important role in HCF function, perhaps also in cell proliferation.
The lack of conservation of the central region of mammalian HCF in a functional homolog of this protein in C. elegans has important implications for our understanding of the evolution of HCF. For example, the lack of HCFPRO repeats in C. elegans HCF suggests that in C. elegans HCF processing is not essential for the function of the conserved amino- and carboxy-terminal regions of HCF. A priori, we imagined two potential structures for a functional homolog(s) of HCF in distantly related species: (i) the HCFPRO repeats and processing would be conserved, or (ii) the HCFPRO repeats would be lost and the amino- and carboxy-terminal regions would be encoded by separate genes. Instead, in C. elegans HCF, the HCFPRO repeats have been lost but the amino- and carboxy-terminal regions remain part of a single gene. These results make the purpose of HCF processing a continuing enigma. We suspect that mammalian HCF processing and controlled association of the resulting fragments provide mammalian HCF greater versatility in its cellular function such as in the control of cell proliferation.
The other region of human HCF of known function but not conserved in C. elegans HCF is the basic region between the HCFVIC domain and the HCFPRO repeats (Fig. 2). In human HCF, the basic region, together with the HCFVIC domain, is required to rescue the tsBN67 cell proliferation defect (33). Because of the lack of a corresponding basic region in C. elegans HCF, we were surprised to find that C. elegans HCF can rescue the tsBN67 defect. C. elegans HCF rescues the tsBN67 phenotype because its HCFVIC-like domain is more potent than its human counterpart in rescuing the tsBN67 phenotype (Fig. 6, panels 2 and 3). Thus, in certain respects, the distantly related C. elegans HCF protein functions better than the human protein in mammalian cells. Perhaps, in human HCF, the HCFVIC domain and the basic region cooperate to promote cell proliferation, whereas in C. elegans HCF, the HCFVIC-like region can drive cells through the cell cycle independently of a basic region. Curiously, however, although the C. elegans HCFVIC-like region is more potent for rescue of the tsBN67 cell proliferation defect, it is less effective for stabilization of the VP16-induced complex (Fig. 3). We do not know the reason for this difference.
The smaller size of C. elegans HCF compared to that of human HCF demonstrates that HCFs can differ considerably in size. This flexibility may explain the nature of the C1 and C2 VP16-induced complexes described by Kristie and colleagues (14) using VP16 purified from Spodoptera cells after baculovirus expression. We suggest that the faster-migrating complex called C1 in that study contained Spodoptera HCF (or in some cases possibly amino-terminal fragments of human HCF) and the C2 complex contained full-length human HCF.
The viral protein VP16 targets a cellular protein that is highly conserved in metazoans.
VP16 is a viral protein of the human pathogen HSV. Thus, the ability of VP16 to associate with HCF from animals as distantly related to humans as C. elegans probably results because VP16 binds to a surface of HCF that is used in a cellular function conserved during metazoan evolution. We hypothesize that this conserved cellular function of HCF is its association with the basic leucine zipper protein, LZIP. LZIP, also known as Luman (19), binds HCF as VP16 does, and LZIP and VP16 share a tetrapeptide motif (E/DHxY) that directs association with HCF (7, 20). Like HCF, LZIP has been conserved during metazoan evolution. In Drosophila melanogaster, the LZIP homolog is BBF-2/dCREB-A (1, 24), which also possesses an E/DHxY motif that directs association with HCF (7, 20). Thus, in its association with HCF, VP16 probably mimics how LZIP binds HCF. Together with the studies described here, these observations suggest that C. elegans probably also possesses a functional LZIP homolog, although its identity has yet to be determined.
In contrast to the high level of conservation of the HCF surface that directs association with VP16, the surface of Oct-1 that directs association with VP16 has not been highly conserved. Indeed, owing to differences on the VP16-interaction surface of the mouse Oct-1 homeodomain, VP16 fails to associate effectively with mouse Oct-1 (6, 27). Thus, apparently the VP16-interaction surface of Oct-1 is less important for Oct-1 cellular function than the VP16-interaction surface of HCF is for HCF cellular function. Consistent with this hypothesis, VP16 is not known to mimic a cellular factor in its interaction with Oct-1. Indeed, just the opposite, a cellular factor that is known to interact with the Oct-1 homeodomain, the B-cell Oct-1 coregulator OCA-B (9, 21, 25), interacts with a different surface of the Oct-1 homeodomain than does VP16 (3).
VP16 targets a conserved cell proliferation function to control HSV infection.
One of the curiosities of HSV infection is that the viral transactivator VP16 requires association with two cellular proteins to activate viral gene expression. We have hypothesized that the requirement for productive association with cellular proteins serves as a checkpoint to gauge whether the state of the infected cell is appropriate for productive lytic infection (30). The results described here suggest that, in its association with HCF, VP16 targets a protein with an ancient role in cell proliferation. This association may serve as a mechanism to link HSV infection to the cell cycle status of the infected cell.
ACKNOWLEDGMENTS
We thank Richard Freiman, Loren Peña, and Angus Wilson for their involvement in the early phase of these studies; Serge Lichsteiner and Robert Tjian for a C. elegans extract; Deborah Aufiero for DNA sequencing; Robert Barstead for a C. elegans cDNA library; Georgia Binns for synthesis of the CeHCFN16 peptide; James Duffy for graphic arts; and Michele Cleary, Nouria Hernandez, and William Tansey for critical readings of the manuscript.
M.O.H. is a Rita Allen Foundation Scholar. Y.L. was supported in part by PHS training grant CA09176. These studies were supported by PHS grant GM54598.
REFERENCES
- 1.Abel T, Bhatt R, Maniatis T. A drosophila CREB/ATF transcriptional activator binds to both fat body- and liver-specific regulatory elements. Genes Dev. 1992;6:466–480. doi: 10.1101/gad.6.3.466. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Babb R, Cleary M A, Herr W. OCA-B is a functional analog of VP16 but targets a separate surface of the Oct-1 POU domain. Mol Cell Biol. 1997;17:7295–7305. doi: 10.1128/mcb.17.12.7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C A, Okayama H. Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. BioTechniques. 1988;6:632–638. [PubMed] [Google Scholar]
- 6.Cleary M A, Stern S, Tanaka M, Herr W. Differential positive control by Oct-1 and Oct-2: activation of a transcriptionally silent motif through Oct-1 and VP16 corecruitment. Genes Dev. 1993;7:72–83. doi: 10.1101/gad.7.1.72. [DOI] [PubMed] [Google Scholar]
- 7.Freiman R N, Herr W. Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev. 1997;11:3122–3127. doi: 10.1101/gad.11.23.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goto H, Motomura S, Wilson A C, Freiman R N, Nakabeppu Y, Fukushima K, Fujishiman M, Herr W, Nishimoto T. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- 9.Gstaiger M, Knoepfel L, Georgiev O, Schaffner W, Hovens C M. A B-cell coactivator of octamer-binding transcription factors. Nature. 1995;373:360–362. doi: 10.1038/373360a0. [DOI] [PubMed] [Google Scholar]
- 10.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 11.Johnson K, Hirsh D. Patterns of proteins synthesized during development of Caenorhabditis elegans. Dev Biol. 1979;70:241–248. doi: 10.1016/0012-1606(79)90020-4. [DOI] [PubMed] [Google Scholar]
- 12.Krause M. Techniques for analyzing transcription and translation. Methods Cell Biol. 1995;48:513–529. doi: 10.1016/s0091-679x(08)61401-6. [DOI] [PubMed] [Google Scholar]
- 13.Kristie T M. The mouse homologue of the human transcription factor C1 (host cell factor) J Biol Chem. 1997;272:26749–26755. doi: 10.1074/jbc.272.42.26749. [DOI] [PubMed] [Google Scholar]
- 14.Kristie T M, LeBowitz J H, Sharp P A. The octamer-binding proteins form multi-protein-DNA complexes with the HSV αTIF regulatory protein. EMBO J. 1989;8:4229–4238. doi: 10.1002/j.1460-2075.1989.tb08608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kristie T M, Pomerantz J L, Twomey T C, Parent S A, Sharp P A. The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J Biol Chem. 1995;270:4387–4394. doi: 10.1074/jbc.270.9.4387. [DOI] [PubMed] [Google Scholar]
- 16.Kristie T M, Sharp P A. Purification of the cellular C1 factor required for the stable recognition of the Oct-1 homeodomain by the herpes simplex virus α-trans-induction factor (VP16) J Biol Chem. 1993;268:6525–6534. [PubMed] [Google Scholar]
- 17.LaBoissiere S, Walker S, O’Hare P. Concerted activity of host cell factor subregions in promoting stable VP16 complex assembly and preventing interference by the acidic activation domain. Mol Cell Biol. 1997;17:7108–7118. doi: 10.1128/mcb.17.12.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis J A, Fleming J T. Basic culture methods. Methods Cell Biol. 1995;48:3–29. [PubMed] [Google Scholar]
- 19.Lu R, Yang P, O’Hare P, Misra V. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol Cell Biol. 1997;17:5117–5126. doi: 10.1128/mcb.17.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu R, Yang P, Padmakumar S, Misra V. The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, Luman, in its interaction with HCF. Mol Cell Biol. 1998;72:6291–6297. doi: 10.1128/jvi.72.8.6291-6297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo Y, Roeder R G. Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol Cell Biol. 1995;15:4115–4124. doi: 10.1128/mcb.15.8.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Hare P. The virion transactivator of herpes simplex virus. Semin Virol. 1993;4:145–155. [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Smolik S M, Rose R E, Goodman R H. A cyclic AMP-responsive element-binding transcriptional activator in Drosophila melanogaster, dCREB-A, is a member of the leucine zipper family. Mol Cell Biol. 1992;12:4123–4131. doi: 10.1128/mcb.12.9.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strubin M, Newell J W, Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell. 1995;80:497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- 26.Sulston J, Du Z, Thomas Z K, Wilson R, Hilier L, Staden R, Halloran N, Green P, Thierry-Mieg J, Qiu L, Dear S, Coulson A, Craxton M, Durbin R, Berks M, Metzstein M, Hawkins T, Ainscough R, Waterston R. The C. elegans genome sequencing project: a beginning. Nature. 1992;356:37–41. doi: 10.1038/356037a0. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki N, Peter W, Ciesiolka T, Gruss P, Schöler H R. Mouse Oct-1 contains a composite homeodomain of human Oct-1 and Oct-2. Nucleic Acids Res. 1993;21:245–252. doi: 10.1093/nar/21.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson C C, McKnight S L. Anatomy of an enhancer. Trends Genet. 1992;8:232–236. [Google Scholar]
- 28a.Wilson, A., and W. Herr. Unpublished results.
- 29.Wilson A C, Parrish J E, Massa H F, Nelson D L, Trask B J, Herr W. The gene encoding the VP16-accessory protein HCF (HCFC1) resides in human Xq28 and is highly expressed in fetal tissues and the adult kidney. Genomics. 1995;25:462–468. doi: 10.1016/0888-7543(95)80046-o. [DOI] [PubMed] [Google Scholar]
- 30.Wilson A C, LaMarco K, Peterson M G, Herr W. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell. 1993;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- 31.Wilson A C, Cleary M A, Lai J-S, LaMarco K, Peterson M G, Herr W. Combinatorial control of transcription: the herpes simplex virus VP16-induced complex. Cold Spring Harbor Symp Quant Biol. 1993;58:167–178. doi: 10.1101/sqb.1993.058.01.021. [DOI] [PubMed] [Google Scholar]
- 32.Wilson A C, Peterson M G, Herr W. The HCF repeat is an unusual proteolytic cleavage signal. Genes Dev. 1995;9:2445–2458. doi: 10.1101/gad.9.20.2445. [DOI] [PubMed] [Google Scholar]
- 33.Wilson A C, Freiman R N, Goto H, Nishimoto T, Herr W. VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol Cell Biol. 1997;17:6139–6146. doi: 10.1128/mcb.17.10.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue F, Cooley L. Kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell. 1993;72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]