Abstract
cdc25A is a tyrosine phosphatase that activates G1 cyclin-dependent kinases (Cdk’s). In human keratinocytes, cdc25A expression is down-regulated after the initial drop in Cdk activity caused by cell exposure to the antimitogenic cytokine transforming growth factor β (TGF-β) or removal of serum factors. Here we show that the TGF-β-inhibitory-response element in the cdc25A promoter maps to an E2F site at nucleotides −62 to −55 from the transcription start site. This site is not required for basal transcription in keratinocytes. We provide evidence that the cell cycle arrest program activated by TGF-β in human keratinocytes includes the generation of E2F4-p130 complexes that in association with histone deacetylase HDAC1 inhibit the activity of the cdc25A promoter from this repressor E2F site. This mechanism is part of a program that places keratinocytes in the quiescent state following the initial drop in Cdk activity caused by cell exposure to TGF-β.
Mitogens and antimitogens affect cell proliferation by regulating the activity of G1 cyclin-dependent kinases (Cdk’s) that commit the cell to completing the division cycle (28, 30). G1 Cdk’s, which include the cyclin D-dependent kinases Cdk4 and Cdk6 and the cyclin E-dependent kinase Cdk2, phosphorylate the members of the retinoblastoma protein family pRb, p107, and p130, also known as “pocket proteins” (34, 39). Phosphorylation of pocket proteins regulates their interactions with transcription factors of the E2F family, i.e., E2F1 through E2F5 (3, 11, 16). In mitogenically stimulated cells, Cdk phosphorylation of pocket proteins allows E2F factors to activate the expression of components involved in DNA synthesis, such as dihydrofolate reductase, thymidine kinase, DNA polymerase α, ORC1, and Cdk components such as cyclin E, cyclin A, and cdc2 (10, 11). When the level of G1 Cdk activity in the cell is low, pocket proteins accumulate in a hypophosphorylated state that inhibits these E2F functions. However, recent evidence suggests that the binding of pocket proteins to E2F factors not only silences transcriptional activation but also generates transcriptional repressor complexes (6, 18, 37, 40). Thus, mutation of E2F sites in certain genes can lead to derepression of these genes in quiescent cells, suggesting that these sites are occupied by E2F repressor complexes during G0 (36, 47). The broader role of these repressor complexes and, in particular, their possible involvement in the action of antimitogenic cytokines such as transforming growth factor β (TGF-β) have remained unknown.
TGF-β and related family members have a wide range of biological effects, including regulation of cell proliferation (1, 28). TGF-β can promote growth in mesenchymal tissues through effects on extracellular matrix production, cell adhesion receptors, and production of autocrine mitogens. In other cell types, including epithelial, hematopoietic, and certain mesenchymal cells, TGF-β exerts antiproliferative effects through a repertoire of gene responses that varies depending on the cell type (19). TGF-β causes rapid up-regulation of the Cdk inhibitor p15Ink4B in keratinocytes, lung, thyroid, and mammary epithelial cells (7, 15, 32, 33), up-regulation of the Cdk inhibitor p21Cip1 in keratinocytes and ovarian epithelial cells (9, 12, 32), rapid down-regulation of the Cdk-activating tyrosine phosphatase cdc25A in mammary epithelial cells (19), and down-regulation of Cdk4 in lung epithelial cells emerging from serum deprivation (13); furthermore, in most of these cell types, TGF-β rapidly down-regulates c-myc expression (1, 19, 27, 31). Individually or combined, these effects cause a decrease in Cdk activity that compromises further progression through G1 and sets in motion secondary events that place the cell in a quiescent state.
One of these secondary events in human keratinocytes is cdc25A down-regulation (19). In contrast to the rapid decrease in cdc25A mRNA caused by TGF-β in mammary epithelial cells, cdc25A mRNA levels in keratinocytes decrease slowly, and this decrease follows the initial fall in Cdk activity induced by TGF-β via Cdk inhibitors (15, 32). Investigating the mechanisms underlying these events, we observed that inhibition of cdc25A promoter activity by TGF-β in human keratinocytes requires the integrity of an E2F site that binds E2F4-p130 and requires histone deacetylase for transcriptional repression.
MATERIALS AND METHODS
Cell culture, transfections, and luciferase assay.
HaCaT keratinocytes (4) and L17 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. Cells were transiently transfected by use of the DEAE-dextran transfection method as described previously (2). Luciferase activity was measured in triplicate samples after cell incubation with or without 200 pM TGF-β for 24 h, unless specified otherwise. Serum deprivation refers to cell incubation in media containing 0.2% fetal bovine serum for 24 h. In the experiments testing the effects of trichostatin A (Wako), this drug was added 24 h after transfection at the concentrations indicated below and for 24 h.
cdc25A promoter constructs.
The cdc25A promoter-luciferase constructs NPGL3 and NP0.7 have been described (14). The nucleotide sequence of the human cdc25A promoter region from −460 to +129 was determined by double-strand sequencing of the BamHI/XhoI fragment of the cdc25A genomic clone in NPGL3 (14). To generate terminal deletions and to introduce mutations in the cdc25A promoter, the fragment from −460 to +129 was transferred into pGL2Basic (Promega), giving rise to NPGL2. Smaller cdc25A promoter constructs were generated from NPGL2 by PCR with Taq polymerase (Perkin-Elmer) and primers specific for cdc25A sequences and vector primers. The amplified products were cloned into the XhoI/HindIII sites of pGL2Basic. Mutations into specific transcription factor binding sites were introduced by synthesizing oligonucleotides containing the selected mutation plus convenient restriction sites for cloning and then using one of these oligonucleotides with a vector primer in a PCR. The mutated PCR product was digested and cloned into NPGL2 constructs from which the correspondent wild-type fragment had been removed. The correct sequences of truncated and mutated constructs were confirmed by complete double-strand sequencing of each promoter fragment.
Other assays.
For immunoprecipitation, cell lysates were prepared in lysis buffer containing 50 mM HEPES at pH 7.4, 150 mM NaCl, 10% glycerol, 0.1% Tween 20, and 1 mM dithiothreitol plus protease and phosphatase inhibitors. Aliquots (1 mg of total proteins) were incubated with antibodies against E2F4 (C-20; Santa Cruz Biotechnology) or against HDAC1 (Upstate Biotechnology) for 2 h at 4 C with gentle agitation. Immunocomplexes bound to protein A-Sepharose (Pharmacia) were collected by centrifugation and washed four times in lysis buffer before being resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
For Western immunoblot analysis, cell extracts (100 μg of total proteins) or immunoprecipitates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Immobilon; Millipore). Immunoblots were developed by use of chemiluminescence (ECL; Amersham). Antibodies used were against E2F1 (KH95; Santa Cruz Biotechnology), E2F4 (C-20; Santa Cruz Biotechnology), pRb (G3-245; Pharmingen), p107 (SD9; Pharmingen), and p130 (C-20; Santa Cruz Biotechnology).
Northern blot analysis was done as described previously (32). cdc25A and glyceraldehyde-3-phosphate dehydrogenase mRNAs were quantified with a phosphorimager and the results were plotted relative to the values for untreated controls.
RESULTS
cdc25A down-regulation by TGF-β independently of E-box sites.
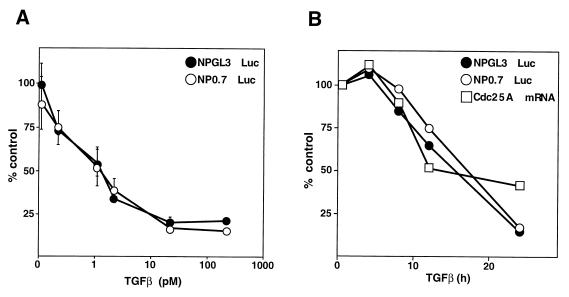
The transcription of cdc25A can be stimulated by c-myc under certain conditions (14). Since TGF-β rapidly down-regulates (half-life = 2 h) c-myc expression in primary keratinocytes (31) and in the human HaCaT keratinocyte line (data not shown), repression of cdc25A by TGF-β in these cells might result from down-regulation of c-myc. However, we previously observed in HaCaT cells that TGF-β can down-regulate a minimal cdc25A promoter region lacking discernible myc binding sites (also known as the “E box”) (19). This region of cdc25A is known as the “natural promoter” region and includes nucleotides −460 to +129 relative to the transcription start site (14) (see sequence in Fig. 2A). In order to determine whether inclusion of E-box sites might confer a heightened sensitivity of cdc25A to TGF-β, we analyzed the response of a larger cdc25A promoter segment which contains the E box (NP0.7 construct). However, the kinetics of down-regulation of the NP0.7 reporter (Fig. 1B) and its sensitivity to various TGF-β concentrations (Fig. 1A) were indistinguishable from those of the natural-promoter construct NPGL3. These results suggested that an element within nucleotides −460 to +129 mediates cdc25A down-regulation in response to TGF-β and that this element is not a typical E box.
FIG. 2.
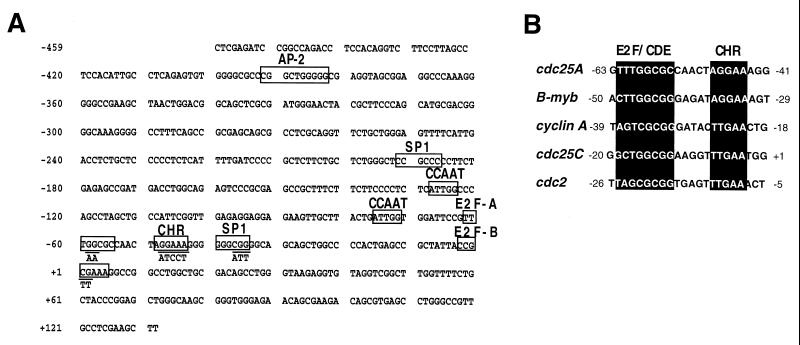
Nucleotide sequence of the human cdc25A promoter region. (A) Sequence of the human cdc25A promoter region from −460 to +129. The consensus AP-2 site, Sp1 sites, CCAAT sites, E2F-A and E2F-B sites, and CHR site are indicated. The sequences of the mutations introduced into the E2F-A and E2F-B sites, the CHR site, and the downstream Sp1 site and described in the legend to Fig. 3 are also indicated. (B) Alignment of the cdc25A promoter sequence in the region of the E2F-A site and the adjacent CHR site with the corresponding promoter sequences from B-myb, the cyclin A gene, cdc25C, and cdc2 (47).
FIG. 1.
cdc25A down-regulation by TGF-β in HaCaT keratinocytes. (A) Luciferase (Luc) reporter constructs containing the promoter region (−460 to +129) of cdc25A (NPGL3) or this region plus a myc-responsive region (NP0.7) were transiently transfected into HaCaT keratinocytes and TGF-β was added at the indicated concentrations for 24 h. Values are the averages and standard deviations of triplicate samples. The control value (100%) corresponds to cells transfected with NPGL3 and left untreated. (B) HaCaT keratinocytes were transfected with NPGL3 or NP0.7, treated with TGF-β (200 pM), and harvested at the indicated times. Samples of untransfected cells were also taken at the same times, and the amounts of cdc25A mRNA were determined by Northern blotting, quantified by phosphorimager, and plotted relative to untreated controls.
An E2F site mediating cdc25A repression in response to TGF-β.
The region of cdc25A from −460 to +129 lacks a consensus TATA box and contains potential binding sites for a variety of known transcription factors (Fig. 2A). In addition to one AP-2 site, two SP-1 sites, and two inverted CCAAT boxes, we identified two E2F sites, one (which we termed the E2F-A site) located approximately 60 bp upstream of the transcription start site and the other (termed E2F-B) spanning the transcription start site (Fig. 2A). The E2F-A site and the surrounding sequence are fully conserved in the mouse cdc25A promoter, whereas the E2F-B site is not (data not shown). The E2F-A site is followed by a 6-bp sequence corresponding to the “cell cycle gene homology region” (CHR). The CHR was previously identified in cell cycle-regulated genes, including B-myb, the cyclin A gene, cdc2, and cdc25C, as a sequence whose integrity is required for repression of these genes in G0 (46, 47). In the promoters of these genes, the CHR element is typically located 6 bp downstream from a functionally repressive E2F or CDE site. This spacing is conserved in the human and mouse cdc25A promoters (Fig. 2B; data not shown for the mouse promoter).
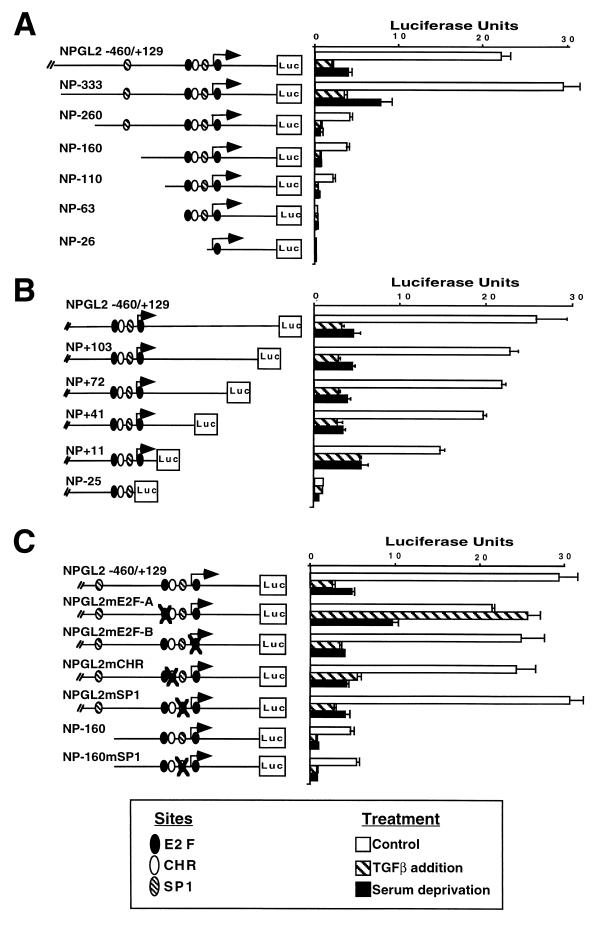
To identify DNA regions required for basal transcription and for TGF-β inhibition of the cdc25A promoter, we generated a series of terminal truncations driving the expression of a luciferase gene. The activities of these reporter constructs were tested by transient transfection into HaCaT keratinocytes in the presence or absence of TGF-β. We also tested the activities of these promoter constructs following removal of serum factors, a condition that decreases the level of cdc25A mRNA in other cell types (20). Both TGF-β addition in the presence of serum and serum deprivation inhibited cdc25A promoter activity (Fig. 3A). Analysis of constructs generated by serial deletions from the 5′ end of the cdc25A natural promoter showed that a substantial fraction of the basal activity depended on the presence of a region −333 to −260 bp from the start site. However, the shortest 5′-deletion construct with detectable basal activity (NP-110) was still inhibited by TGF-β addition or mitogen deprivation (Fig. 3A). Using 3′-deletion constructs, we found that deletion up to nucleotide +11 still allowed transcriptional activity and that this activity was inhibited by both TGF-β addition and mitogen deprivation (Fig. 3B). Further 3′ deletions prevented basal transcription.
FIG. 3.
TGF-β down-regulates cdc25A promoter activity through an E2F site. (A) Transient-expression analysis of 5′-truncated cdc25A promoter-luciferase (Luc) constructs in proliferating HaCaT cells left untreated or treated with TGF-β, or with 0.2% serum for 24 h. Plasmids were named to indicate the 5′ truncation; e.g., NP-333 contains 333 bases upstream of the start site. All plasmids harbor a 129-bp region downstream of the start site. Symbols indicate the locations of consensus sites. Luciferase values are the averages and standard deviations of triplicate assays. (B) Same as panel A but transfections were done with 3′-truncated constructs. Plasmids were named to indicate the 3′ truncation; e.g., NP+103 contains 103 bases downstream of the start site. All plasmids, with the exception of NP-25, harbor a 460-bp region upstream of the start site. (C) Same as panel A but transfections were done with cdc25A promoter-luciferase constructs containing the regions of the cdc25A promoter from −460 to +129 or −160 to +129 with mutations in the indicated sites (crossed symbols). Mutations are shown in Fig. 2A.
Taken together, the results from the 5′- and 3′-deletion analyses indicate that DNA sequences responsible for inhibition of cdc25A promoter by TGF-β addition or mitogen deprivation reside between nucleotides −110 and +11 of the cdc25A promoter. This region includes the two E2F sites, the CHR site, and one SP1 site (Fig. 2 and 3). To determine whether any of these sites is responsible for cdc25A down-regulation, we mutated each site in the context of the natural promoter. We also mutated the SP1 site in the −160 deletion construct (which lacks the upstream SP1 site) to generate a promoter which was devoid of SP1 sites. Analysis of the resulting reporter constructs showed that the SP1 sites, the CHR site, and the E2F-B site were dispensable for basal transcription or inhibition by TGF-β addition or mitogen deprivation (Fig. 3C). Mutation of the E2F-A site did not affect the basal activity but completely prevented the inhibition by TGF-β and had a partial effect on inhibition by serum deprivation (Fig. 3C). These results suggest that (i) TGF-β action represses the cdc25A promoter through the E2F-A site, which by itself lacks any significant enhancer function, and (ii) this site only partially mediates the inhibitory effect of serum deprivation, which therefore must act through additional mechanisms.
TGF-β-induced changes in E2F4-pocket protein complexes.
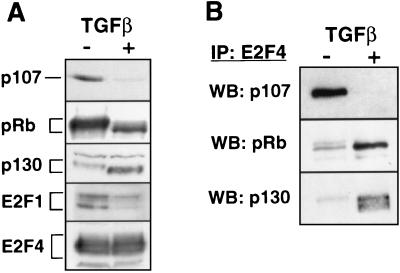
To determine whether regulation of cdc25A promoter activity by TGF-β is associated with changes in the abundance of potential E2F binding components, we measured the levels of E2F-1, E2F-4, pRb, p107, and p130 in control and TGF-β-treated HaCaT cells (Fig. 4A). As previously described for these cells and other cells that undergo G1 arrest, the results of pRb Western immunoblotting showed that TGF-β increases the mobility of pRb in a manner that corresponds to decreased phosphorylation of this protein (23) (Fig. 4A). A similar effect was observed with p130 (Fig. 4A). These effects were likely the result of the inhibition of G1 Cdk’s by TGF-β in keratinocytes (15, 32). These results also showed that TGF-β caused a decrease in the levels of pRb and p107 whereas the levels of p130 were markedly increased (Fig. 4A). TGF-β did not alter the level of E2F4 but strongly decreased the level of E2F1 in HaCaT cells (Fig. 4A). This effect might be the result of E2F1 mRNA down-regulation in TGF-β-treated HaCaT cells (24).
FIG. 4.
Effects of TGF-β on pocket protein levels and E2F4 complexes. (A) Levels of the indicated proteins were determined by Western immunoblotting of lysates from proliferating (−) and TGF-β-treated (+) HaCaT cells. (B) The level of cellular E2F4 complexes containing pocket proteins was determined by E2F4 immunoprecipitation (IP) followed by Western immunoblotting (WB) with the indicated antibodies.
These changes correlated with related changes in the levels of specific E2F4 complexes, as determined by immunoprecipitation with anti-E2F4 antibodies and Western immunoblotting of the precipitates with antibodies against specific pocket proteins. Thus, control cells contained high levels of E2F4-p107 complexes and low levels of E2F4-p130 complexes, whereas the reverse was seen in TGF-β-treated cells (Fig. 4B). E2F4-pRb complexes were detected in control as well as TGF-β-treated cells, being more abundant in the latter (Fig. 4B). This might be due to a higher E2F binding affinity of hypophosphorylated pRb (16, 21). These data argue that TGF-β action specifically inhibited the formation of E2F4-p107 complexes and induced the formation of E2F4-p130 complexes.
Participation of histone deacetylase in cdc25A repression.
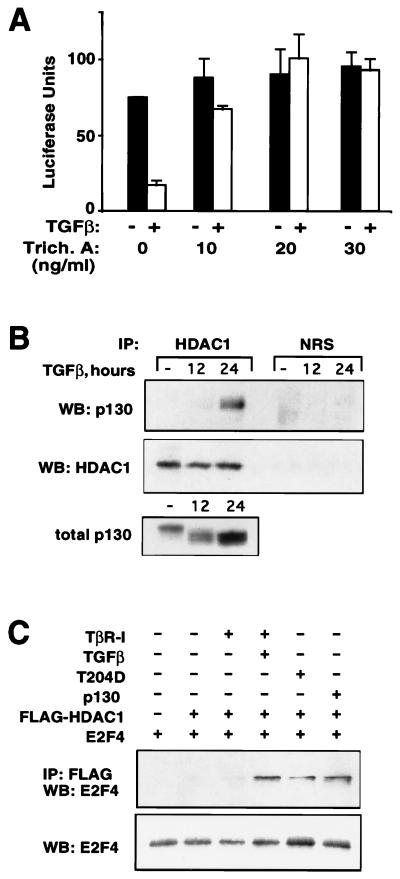
Histone deacetylation has been recently proposed as a mechanism for transcriptional repression in general (42) and for pRb-mediated repression of a subset of E2F-responsive genes in particular (5, 25, 26). The latter has been shown to result from the recruitment of the histone deacetylase HDAC1 to the target promoter through a specific interaction with pRb. To investigate whether cdc25A repression by E2F4-p130 in TGF-β-treated cells might also involve the participation of histone deacetylase, we first tested the effect of a specific histone deacetylase inhibitor, trichostatin A (44). When added simultaneously with TGF-β, trichostatin A prevented the inhibition of the cdc25A reporter (Fig. 5A). This effect was significant at trichostatin A concentrations of 10 ng/ml and above (Fig. 5A), which are well within the reported range that specifically inhibits histone deacetylase in the cell (22, 35, 44). Trichostatin A did not affect transcriptional activation of the 3TP-lux reporter by TGF-β (data not shown), indicating that it did not cause a general inhibition of TGF-β signal transduction.
FIG. 5.
Histone deacetylase participates in transcriptional repression of cdc25A promoter by TGF-β. (A) The reporter construct NPGL2 was transfected into HaCaT cells which were then incubated with or without TGF-β and the indicated concentrations of trichostatin A. Luciferase values are the averages and standard deviations of triplicate samples. (B) Extracts from proliferating or TGF-β-treated HaCaT cells were immunoprecipitated (IP) with anti-HDAC1 polyclonal antibody (HDAC1) or normal rabbit serum (NRS). Immunocomplexes were then subjected to Western immunoblotting (WB) with p130 or HDAC1 antibodies. A Western immunoblot of total cell extracts is also shown (bottom panel). (C) L17 cells were transfected with the indicated plasmids and left untreated or treated with TGF-β for 24 h. Immunoprecipitations were performed with anti-FLAG antibody followed by Western immunoblotting with anti-E2F4 antibody. The amount of E2F4 expressed for each transfection is also shown (bottom panel).
To investigate the presence of histone deacetylase in p130 complexes, we immunoprecipitated extracts from control and TGF-β-treated cells with anti-HDAC1 antibody followed by Western immunoblotting of these precipitates with anti-p130. No interaction between p130 and HDAC1 was detected in extracts from control cells. TGF-β did not increase the HDAC1 levels in the cell (Fig. 5B). However, TGF-β induced a significant association of HDAC1 with p130 (Fig. 5B). The kinetics of appearance of the p130-HDAC1 complex were parallel to those of p130 accumulation and cdc25A inhibition, with high levels being reached after 24 h of cell exposure to TGF-β (Fig. 5B; compare to Fig. 1B). An alignment of the total p130 and HDAC1-associated p130 immunoblots shown in Fig. 5B demonstrated that HDAC1 preferentially associates with the hypophosphorylated (fast-migrating) form of p130. Thus, both the decrease in p130 phosphorylation and the increase in p130 protein levels caused by TGF-β seem to contribute to the association with HDAC1.
To test whether a complex between E2F4 and HDAC1 was generated upon activation of TGF-β signaling and whether this effect could be obtained by overexpression of p130, we used the L17 cell line, which lacks TGF-β receptor type I (TβRI). Transfection of FLAG-tagged HDAC1 and E2F4 in L17 cells was not sufficient to generate an association between these proteins, indicating a requirement for additional factors. An efficient binding between HDAC1 and E2F4 was detectable only when wild-type TβRI was transfected in the presence of TGF-β or when TGF-β signaling was activated by transfecting constitutively active TβRI (T204D) (41). The association between HDAC1 and E2F4 was also generated by the introduction of p130. These data show that E2F4 can associate with HDAC1 upon TGF-β treatment and suggest that p130 is capable of mediating this interaction.
DISCUSSION
Rapid repression of cdc25A by TGF-β causes the inactivation of Cdk4 and Cdk6 in MCF-10A human mammary epithelial cells in the absence of any effect on Cdk inhibitors (19). cdc25A down-regulation is also observed in keratinocytes, in which the primary TGF-β response includes the induction of Cdk inhibitors such as p15Ink4B and p21CIP1 (15, 32). cdc25A down-regulation in keratinocytes is a delayed effect and may represent the implementation of a program of mitotic quiescence during the TGF-β antiproliferative response. In the present work, we have studied the inhibition of cdc25A in the context of this set of events, and we have identified E2F-mediated repression of transcription as the specific mechanism underlying this gene response in HaCaT keratinocytes.
The natural promoter of cdc25A, nucleotides −460 to +129 (14), was sufficient to mediate repression by TGF-β in HaCaT keratinocytes. Our promoter deletion and transcription factor site mutation analysis suggests that the delayed repression of cdc25A expression induced by TGF-β in HaCaT cells is mediated through an E2F site. Interestingly, this site participated only partially in the repression of cdc25A caused by deprivation of serum mitogens, indicating that E2F-mediated repression is uniquely effective when triggered by TGF-β. Our data also show that the E2F-A site in the context of the cdc25A promoter did not contribute to basal transcriptional activity; thus, it functioned as a pure repressor site. The E2F-A site is contiguous to a CHR sequence that in other genes appears to mediate repression in concert with upstream E2F sites (46, 47). In the TGF-β response, however, the integrity of the CHR site was not required for the repressive effect on the cdc25A promoter, at least when tested under our experimental conditions.
Although E2F was initially proposed to activate transcription, E2F sites in several promoters were recently shown to repress transcription (47). In vivo footprinting of the promoter of E2F-responsive genes, such as B-myb, the cyclin A gene, and cdc2, demonstrated occupancy of the E2F site exclusively in G0/G1, when the expression of these genes was repressed (17, 36, 45). Loss of E2F site binding occurred when cells were released from growth arrest and coincided with transcriptional activation of these genes. Based on our analysis of the cdc25A promoter in the TGF-β antiproliferative response, we propose that E2F-mediated repression of this promoter by TGF-β might be sustained by a similar mechanism in vivo.
Several lines of evidence suggest that, among E2F family members, E2F4 is a major participant in the repressor complex which binds to E2F sites during quiescence (29, 38). Whereas E2F1, -2, -3, and -5 together comprise less than 30% of the endogenous E2F species and their expression is strongly suppressed during the G0/G1 phase of the cell cycle, E2F4 accounts for the majority of E2F complexes, particularly during quiescence (29, 38). Our results with HaCaT keratinocytes treated with TGF-β showed that transcriptional repression by E2F is associated with a marked change in the composition of E2F4 complexes, switching from E2F4-p107 in proliferating cells to E2F4-p130 in TGF-β-treated cells. This switch coincides with, and is likely to be caused by, the dephosphorylation and accumulation of p130 and the down-regulation of p107 caused by TGF-β.
How do E2F4-p130 complexes mediate transcriptional repression? An answer is suggested by our results with the specific inhibitor of histone deacetylase, trichostatin A. Trichostatin A prevents transcriptional repression of the cdc25A promoter by TGF-β, suggesting a requirement for histone deacetylase activity. Recruitment of histone deacetylases by DNA binding proteins has been proposed as a mechanism for transcriptional repression (43). Furthermore, recent reports have shown an association of HDAC1 with pRb in transcriptional repressor complexes (5, 25, 26). Here we show that p130 interacts with HDAC1 in response to TGF-β. No association between p130 and HDAC1 was detectable in proliferating cells. The interaction between endogenous HDAC1 and p130 was observed in cells treated with TGF-β for 24 h, although some association was already detectable after 12 h. The kinetics suggests that this process requires first the dephosphorylation and then the accumulation of sufficient levels of p130 protein for effective binding with HDAC1. Therefore, one function of the E2F4-p130 complexes abundant in quiescent cells (8, 37) may be the recruitment of histone deacetylase to the promoters of E2F-repressible genes such as cdc25A, thus allowing transcriptional silencing. Our data showing an association between E2F4 and HDAC1 in TGF-β-treated cells together with the ability of p130 to promote formation of this complex suggests a mechanism in which E2F4 gains the ability to repress transcription by associating with p130 and possibly with pRb in TGF-β-treated cells. This leads to the formation of E2F4-p130 complexes that are able to recruit histone deacetylase, thus repressing transcription. We propose that the initial decline in Cdk activity by TGF-β in HaCaT keratinocytes triggers a program of orderly withdrawal from the cell cycle, and repression of cdc25A transcription by E2F and histone deacetylase through the mechanism described here is an event representative of this program.
ACKNOWLEDGMENTS
We thank Charles Zhang for expert technical assistance and Stacy Blain for critically reading the manuscript.
A.I. was the recipient of a fellowship from the Clinical Scholars Training Program of the Memorial Sloan-Kettering Cancer Center. J.M. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Alexandrow M G, Moses H L. Transforming growth factor beta and cell cycle regulation. Cancer Res. 1995;55:1452–1457. [PubMed] [Google Scholar]
- 2.Attisano L, Cárcamo J, Ventura F, Weis F M B, Massagué J, Wrana J L. Identification of human activin and TGF-β type I receptors that form heteromeric kinase complexes with type II receptors. Cell. 1993;75:671–680. doi: 10.1016/0092-8674(93)90488-c. [DOI] [PubMed] [Google Scholar]
- 3.Beijersbergen R, Kerkhoven R M, Zhu L, Carlee L, Voorhoeve P M, Bernards R. E2F-4, a new member of the E2F gene family, has oncogenic activity and associates with p107 in vivo. Genes Dev. 1994;8:2680–2690. doi: 10.1101/gad.8.22.2680. [DOI] [PubMed] [Google Scholar]
- 4.Boukamp P, Petrussevska R T, Breitkreutz D, Hornung J, Markham A, Fusenig N E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 6.Cao L, Faha B, Dembski M, Tsai L-H, Harlow E, Dyson N. Independent binding of the retinoblastoma protein and p107 to the transcription factor E2F. Nature. 1992;355:176–179. doi: 10.1038/355176a0. [DOI] [PubMed] [Google Scholar]
- 7.Carneiro C, Alvarez C V, Zalvide J, Vidal A, Dominguez F. TGF-beta1 actions on FRTL-5 cells provide a model for the physiological regulation of thyroid growth. Oncogene. 1998;16:1455–1465. doi: 10.1038/sj.onc.1201662. [DOI] [PubMed] [Google Scholar]
- 8.Corbeil H B, Branton P E. Characterization of an E2F-p130 complex formed during growth arrest. Oncogene. 1997;15:657–668. doi: 10.1038/sj.onc.1201224. [DOI] [PubMed] [Google Scholar]
- 9.Datto M B, Li Y, Panus J F, Howe D J, Xiong Y, Wang X-F. Transforming growth factor β induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc Natl Acad Sci USA. 1995;92:5545–5549. doi: 10.1073/pnas.92.12.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 12.Elbendary A, Berchuck A, Davis P, Havrilesky L, Bast J R C, Iglehart J D, Marks J R. Transforming growth factor b1 can induce CIP1/WAF1 expression independent of the p53 pathway in ovarian cancer cells. Cell Growth Differ. 1994;5:1301–1307. [PubMed] [Google Scholar]
- 13.Ewen M E, Sluss H K, Whitehouse L L, Livingston D M. TGFβ inhibition of Cdk4 synthesis is linked to cell cycle arrest. Cell. 1993;74:1009–1020. doi: 10.1016/0092-8674(93)90723-4. [DOI] [PubMed] [Google Scholar]
- 14.Galaktionov K, Chen X, Beach D. Cdc25 cell-cycle phosphatase as a target of c-myc. Nature. 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- 15.Hannon G J, Beach D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 16.Helin K, Lees J A, Vidal M, Dyson N, Harlow E, Fattaey A. A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell. 1992;70:337–350. doi: 10.1016/0092-8674(92)90107-n. [DOI] [PubMed] [Google Scholar]
- 17.Huet X, Rech J, Plet A, Vié A, Blanchard J M. Cyclin A expression is under negative transcriptional control during the cell cycle. Mol Cell Biol. 1996;16:3789–3798. doi: 10.1128/mcb.16.7.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurford R K, Cobrinik D, Lee M-H, Dyson N. pRB and p107/130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 19.Iavarone A, Massagué J. Repression of the Cdk activator Cdc25A and cell-cycle arrest by cytokine TGFbeta in cells lacking the Cdk inhibitor p15. Nature. 1997;387:417–422. doi: 10.1038/387417a0. [DOI] [PubMed] [Google Scholar]
- 20.Jinno S, Suto K, Nagata A, Igarashi M, Kanaoka Y, Nojima H, Okayama H. Cdc25A is a novel phosphatase functioning early in the cell cycle. EMBO J. 1994;13:1549–1556. doi: 10.1002/j.1460-2075.1994.tb06417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaelin W G, Jr, Krek W, Sellers W R, DeCaprio J A, Ajchenbaum F, Fuchs C S, Chittenden T, Li Y, Farnham P J, Blanar M A, Livingston D M, Flemington E K. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 22.Laherty C D, Yang W-M, Sun J-M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 23.Laiho M, DeCaprio J A, Ludlow J W, Livingston D M, Massagué J. Growth inhibition by TGF-β1 linked to suppression of retinoblastoma protein phosphorylation. Cell. 1990;62:175–185. doi: 10.1016/0092-8674(90)90251-9. [DOI] [PubMed] [Google Scholar]
- 24.Li J M, Hu P P, Shen X, Yu Y, Wang X-F. E2F4-RB and E2F4-p107 complexes suppress gene expression by transforming growth factor beta through E2F binding sites. Proc Natl Acad Sci USA. 1997;94:4948–4953. doi: 10.1073/pnas.94.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 26.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 27.Malliri A, Yeudall W A, Nikolic M, Crouch D H, Parkinson E K, Ozanne B. Sensitivity to transforming growth factor beta 1-induced growth arrest is common in human squamous cell carcinoma cell lines: c-MYC down-regulation and p21waf1 induction are important early events. Cell Growth Differ. 1996;7:1291–1304. [PubMed] [Google Scholar]
- 28.Massagué J. The transforming growth factor-β family. Annu Rev Cell Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 29.Moberg K, Starz M A, Lees J A. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moses H L, Yang E Y, Pietenpol J. TGF-β stimulation and inhibition of cell proliferation: new mechanistic insights. Cell. 1990;63:245–247. doi: 10.1016/0092-8674(90)90155-8. [DOI] [PubMed] [Google Scholar]
- 31.Pietenpol J A, Stein R W, Moran E, Yacuik P, Schlegel R, Lyons R M, Pittelkow R M, Münger K, Howley P M, Moses H L. TGF-β1 inhibition of c-myc transcription and growth in keratinocytes is abrogated by viral transforming protein with pRB binding domains. Cell. 1990;61:777–785. doi: 10.1016/0092-8674(90)90188-k. [DOI] [PubMed] [Google Scholar]
- 32.Reynisdóttir I, Polyak K, Iavarone A, Massagué J. Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-β. Genes Dev. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 33.Sandhu C, Garbe J, Bhattacharya N, Daksis J, Pan C-H, Yaswen P, Koh J, Slingerland J M, Stampfer M R. Transforming growth factor β stabilizes p15INK4B protein, increases p15INK4B-cdk4 complexes, and inhibits cyclin D1-cdk4 association in human mammary epithelial cells. Mol Cell Biol. 1997;17:2458–2467. doi: 10.1128/mcb.17.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherr C J. G1 phase progression: cycling on cue. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 35.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 36.Tommasi S, Pfeifer G P. In vivo structure of the human cdc2 promoter: release of a p130–E2F-4 complex from sequences immediately upstream of the transcription initiation site coincides with induction of cdc2 expression. Mol Cell Biol. 1995;15:6901–6913. doi: 10.1128/mcb.15.12.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vairo G, Livingston D M, Ginsberg D. Functional interaction between E2F-4 and p130: evidence for distinct mechanisms underlying growth suppression by different retinoblastoma protein family members. Genes Dev. 1995;9:869–881. doi: 10.1101/gad.9.7.869. [DOI] [PubMed] [Google Scholar]
- 38.Verona R, Moberg K, Estes S, Starz M, Vernon J P, Lees J A. E2F activity is regulated by cell cycle-dependent changes in subcellular localization. Mol Cell Biol. 1997;17:7268–7282. doi: 10.1128/mcb.17.12.7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 40.Weintraub S J, Prater C A, Dean D C. Retinoblastoma protein switches the E2F site from positive to negative element. Nature. 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- 41.Wieser R, Wrana J L, Massagué J. GS domain mutations that constitutively activate TβRI, the downstream signaling component in the TGFβ receptor complex. EMBO J. 1995;14:2199–2208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolffe A P. Histone deacetylase: a regulator of transcription. Science. 1996;272:371–372. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- 43.Wolffe A P. Sinful repression. Nature. 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida M, Horinouchi S, Beppu T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 45.Zwicker J, Liu N, Engeland K, Lucibello F C, Muller R. Cell cycle regulation of E2F site occupation in vivo. Science. 1996;271:1595–1597. doi: 10.1126/science.271.5255.1595. [DOI] [PubMed] [Google Scholar]
- 46.Zwicker J, Lucibello F C, Wolfraim L A, Gross C, Truss M, Engeland K, Muller R. Cell cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J. 1995;14:4514–4522. doi: 10.1002/j.1460-2075.1995.tb00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zwicker J, Muller R. Cell-cycle regulation of gene expression by transcriptional repression. Trends Genet. 1997;13:3–6. doi: 10.1016/s0168-9525(96)30112-1. [DOI] [PubMed] [Google Scholar]