To reach the goal of curing currently incurable hematologic malignancies, we need to go beyond focusing on single gene mutatons and gain deeper understanding of the consequences of genetic alterations on gene-regulatory pathways. Edited by John Crispino, these 5 cutting-edge reviews from leaders in the their fields not only summarize our current understanding of key pathways that contribute to myeloid malignancies, but also discuss new therapeutic avenues related to them. They provide a springboard for further groundbreaking basic and clinical advances in hematologic malignancies.
Visual Abstract
Abstract
Protein-coding and noncoding RNAs can be decorated with a wealth of chemical modifications, and such modifications coordinately orchestrate gene expression during normal hematopoietic differentiation and development. Aberrant expression and/or dysfunction of the relevant RNA modification modulators/regulators (“writers,” “erasers,” and “readers”) drive the initiation and progression of hematopoietic malignancies; targeting these dysregulated modulators holds potent therapeutic potential for the treatment of hematopoietic malignancies. In this review, we summarize current progress in the understanding of the biological functions and underlying mechanisms of RNA modifications in normal and malignant hematopoiesis, with a focus on the N6-methyladenosine modification, as well as discuss the therapeutic potential of targeting RNA modifications for the treatment of hematopoietic malignancies, especially acute myeloid leukemia.
Introduction
Based on the central dogma, gene expression is a multilayered process that begins with the transcription of DNA into intermediate carrier messenger RNAs (mRNAs), followed by the transfer of mRNA sequence information to proteins that execute the biological functions. The modification of chromosomes without changes in DNA sequences is an important regulatory mechanism for gene expression that is termed “epigenetics.” Likewise, similar “epigenetic” modifications have been observed in RNAs, including ribosomal RNA, transfer RNA, small nuclear RNA, and mRNA, designated as “epitranscriptomics.”1 Since the discovery of the first RNA modification pseudouridine (the so-called “fifth nucleotide”) in 1957,2 >170 types of chemical modifications have been reported in protein-coding and noncoding RNA transcripts.3-7 Although many of these modifications have been identified for decades, only a few have been validated because of technical limitations, and RNA decorations were generally recognized as irreversible. Thus, related studies focusing on epitranscriptomics have largely lagged behind epigenetic studies focusing on DNA and histone modifications. The RNA epitranscriptomic landscape has been greatly stimulated in recent years, primarily as a result of the discovery of enzyme FTO (the fat mass and obesity-associated protein), which is capable of reversing N6-methyladenosine (m6A) to adenosine.8-10 Also, thanks to high-throughput next-generation sequencing and the highly sensitive mass spectrometry method for precise quantification, a set of RNA modifications has been well characterized, mapped, and investigated at the transcriptomic and global levels.11-20
Recent studies on many RNA modifications indicate that they are dynamically regulated in diverse physiological and pathological settings (eg, in normal development and malignant transformation).5,7,21 In particular, evidence is emerging that RNA modifications play important roles in normal hematopoiesis (especially myelopoiesis) and leukemogenesis.6,7,10,22-24 Among all RNA modifications, m6A represents 1 of the most prevalent and well-studied posttranscriptional modifications in mRNA, and other modifications, such as adenosine to inosine (A-to-I) editing, 5-methylcytosine (m5C), 5-hydroxymethylcytosine (hm5C), and pseudouridine, have also been reported for their functions in normal hematopoiesis and/or leukemogenesis.3,6,7,25 The correct deposition of these modifications is essential to guarantee normal hematopoiesis and prevent differentiation blockage or leukemogenesis.5-7,10,22-24 Dysregulation of RNA modification machineries has been reported in leukemia, and their abnormal expression contributes to the initiation, progression, and drug resistance of leukemia. Thus, targeting the RNA modification regulatory pathways provides an attractive avenue to develop novel therapeutic strategies for the treatment of hematopoietic malignancies. Here, we highlight recent advances in our understanding of the roles and underlying mechanisms of RNA modifications (especially the m6A modification) in regulating gene expression during normal hematopoiesis and leukemogenesis and discuss the therapeutic potential of targeting such dysregulated epitranscriptomic regulations in treating hematopoietic malignancies.
m6A modification and the modulators
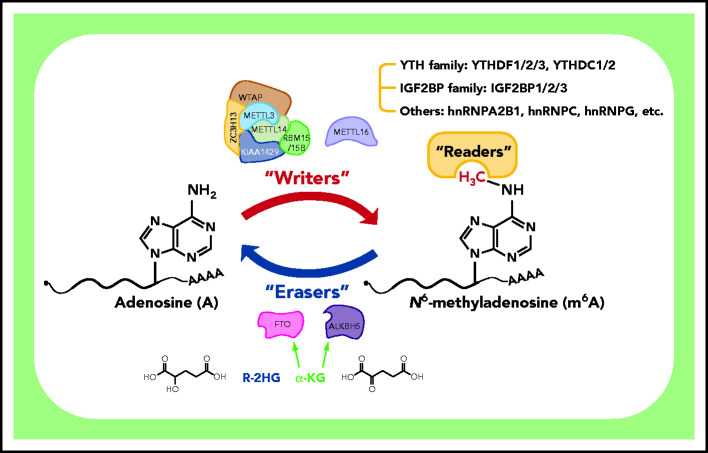
Being the most abundant internal modification of eukaryotic mRNAs, m6A mark is also present in noncoding RNAs, such as long noncoding RNAs and microRNAs.6 Based on ultrahigh-performance liquid chromatography coupled with triple-quadrupole tandem mass spectrometry analysis, the m6A/A ratios range from 0.2% to 0.5% in poly(A)+ RNAs of eukaryotes, supporting the ubiquity of m6A decorations.26,27 Transcriptome-wide mapping studies of m6A modification identified >12 000 m6A peaks in >7000 transcripts, with the m6A-modified sites distributed across entire transcripts but being especially enriched around stop codons; many m6A sites are highly conserved between humans and mice.15,21 Another study revealed a total of 9754 m6A peaks in 5578 transcripts (average, 2 peaks per transcript), including 5461 mRNAs (of 9923 mRNAs) and 117 noncoding RNAs.28 Collectively, m6A decoration is ubiquitous in mammalian cells, with ∼1 m6A peak per 2000 nucleotides or ∼2 peaks per transcript on average. The m6A content is highly variable across different tissues and cell types, indicating its dynamic nature.21 Like other epigenetic marks, such as DNA methylation and histone modifications, m6A is deposited by “writer” proteins (methyltransferases), removed by “eraser” proteins (demethylases), and recognized and bound by “reader” proteins (m6A-binding proteins) to exert its biological functions (Figure 1).6,7,29
Figure 1.
Deposition, removal, and recognition of m6A modification. The m6A modification is deposited by “writers” that function as a complex or as a single protein (METTL16). The MTC consists of 3 key components (METTL3, METTL14, and WTAP) and several auxiliary subunits (including RBM15/15B, KIAA1429, and ZC3H13). Two m6A demethylases, FTO and ALKBH5, serve as “erasers” and can reverse m6A to A in an α-ketoglutarate (α-KG)-dependent way. Notably, R-2-hydroxyglutarate (R-2HG), a metabolite produced by mutant isocitrate dehydrogenase, is reported to competitively suppress the demethylase activity of FTO as a result of its similar structure to α-KG. The broad biological functions of m6A are mediated by “readers” that are able to recognize the methyl group or m6A-induced structural changes (“the m6A switch”) to regulate gene expression of downstream targets. Currently known readers include the YTH family (YTHDF1/2/3 and YTHDC1/2), the IGF2BP family (IGF2BP1/2/3), and other proteins (eg, hnRNPA2B1, hnRNPC, and hnRNPG) that recognize m6A switches.
It is now clear that m6A is deposited in mRNA cotranscriptionally primarily by the methyltransferase complex (MTC), which consists of 3 major subunits: METTL3 (methyltransferase-like 3), METTL14, and WTAP (Wilms tumor suppressor 1-associated protein).6,7,21,29 METTL3 is responsible for installing m6A using its catalytic subunit, and METTL14 forms a stable heterodimer with METTL3 to allosterically activate METTL3 and determine the methylation sites; WTAP directly binds to METTL3/14 and is indispensable for optimal substrate recruitment and localization of METTL3/14.26,30-36 METTL14 was also reported to be able to recognize histone H3 lysine 36 trimethylation modification and, thereby, selectively facilitate m6A deposition in mRNA cotranscriptionally.21,30 Other accessory subunits in the complex, including KIAA1429 (also named VIRMA), RBM15/RBM15B, and ZC3H13, assist in anchoring the complex to nuclear speckles and recognizing particular mRNA binding sites.26,37-42 Recent studies also show that METTL16, a METTL3 homolog, is capable of depositing m6A in U6 small nuclear RNA, MALAT1 long noncoding RNA, and MAT2A mRNA on its own.43-45 The m6A decoration can be selectively reversed to A by the demethylases FTO and ALKBH5 (AlkB homolog 5), which enable intricate and dynamic control of m6A in a wealth of physiological and pathological cellular processes.8,46 A number of m6A-binding proteins are recruited by the m6A modification and mediate the broad impact of m6A on gene expression during myriad bioprocesses, such as m6A-dependent regulation of mRNA stability, translation initiation, mRNA splicing, mRNA nuclear export, and mRNA structure.6,7,10,47-54 Currently known m6A readers primarily include 2 families: the YT521-B homology (YTH) domain family and the insulin-like growth factor 2 mRNA-binding protein (IGF2BP) family. The YTH family includes 5 members: YTHDF1/2/3 and YTHDC1/2, which are reported to play different roles in mRNA degradation (YTHDF2/3 and YTHDC2), splicing (YTHDC1), translation (YTHDF1/3 and YTHDC2), and structure modulation (YTHDC2).47,48,50,53,55 Unlike the YTH family members (ie, YTHDF2/3 and YTHDC2), which promote decay of target mRNAs, the recently identified IGF2BP reader family members (including IGF2BP1/2/3) enhance the stability of target mRNAs and facilitate their translation.54 In addition, a few other m6A readers that recognize m6A-induced RNA structural remodeling (“the m6A switch”), such as hnRNPA2B1, hnRNPC, and hnRNPG, have been reported.56-59
Normal hematopoiesis is coordinated by m6A regulation
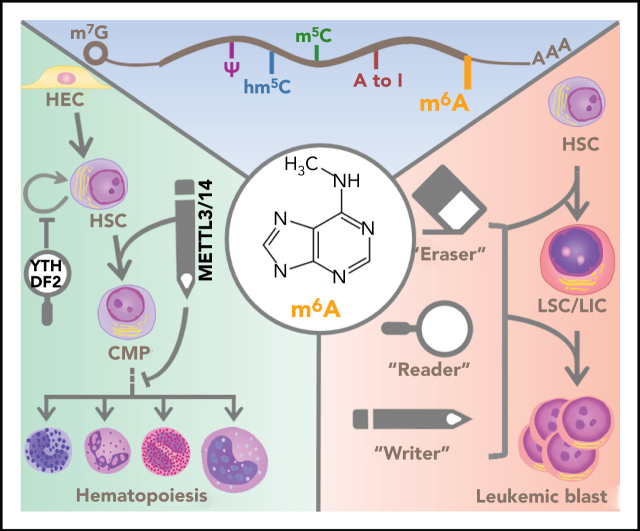
Hematopoietic stem cells (HSCs) are fundamentally characterized by their ability to generate new HSCs (termed “self-renewal”) and produce a spectrum of blood lineages and mature blood cells. During this normal hematopoiesis, the gene expression is highly orchestrated and fine-tuned at the DNA and mRNA levels.60,61 To form the blood system, hematopoiesis is organized as a cellular hierarchy, where HSCs generate a variety of multilineage progenitor cells that subsequently differentiate into precursors committed to individual lineages and, eventually, mature blood cells.61 Particularly, the multipotential HSCs give rise to the oligopotential common myeloid progenitors, which next mature into megakaryocyte-erythrocyte progenitors and granulocyte-monocyte progenitors and, ultimately, functional myeloid cells (including erythrocytes, megakaryocytes, granulocytes, monocytes, macrophages, and myeloid dendritic cells).62
In this developmental progression, m6A was found to be implicated at multiple stages as early as the emergence of hematopoietic stem and progenitor cells (HSPCs) during embryogenesis (Figure 2).63,64 HSPCs are specified from hemogenic endothelial cells through the endothelial-to-hematopoietic transition (EHT), and depletion of mettl3 in zebrafish and mouse embryos resulted in a significant decrease in m6A levels, as well as a blockage in the generation of the earliest HSPCs.63,64 In mettl3-null embryos, the reduced m6A modification in the arterial endothelial genes notch1a and rhoca resulted in a delay of YTHDF2-mediated mRNA decay and, subsequently the continuous activation of Notch signaling in arterial endothelial cells and blockage of EHT; as a result, the homozygous zebrafish mutants died 10 days postfertilization as a result of severe hematopoietic defects.64 Forced expression of mettl3 in endothelial cells, but not in fast-twitch muscle cells, rescued the HSPC defects in mettl3-null embryos, suggesting that HSPC formation is specifically regulated by m6A modifications in endothelial cells.64 Additionally, although deletion of Mettl3 in endothelial cells attenuated HSPC development and function in mouse embryos, silencing of Mettl3 at later stages in hematopoietic cells did not induce obvious changes in HSPCs, implying that the regulation of HSPCs by Mettl3 in mice primarily occurs in EHT during embryogenesis.63 In the adult murine hematopoietic system, deletion of Mettl3 blocks differentiation and markedly reduces reconstitution potential in HSCs, and causes the accumulation of impaired HSCs in bone marrow.65,66 Kharas and colleagues further revealed that Mettl3-mediated m6A methylation maintains HSC identity and permits its symmetric commitment division (in contrast to symmetric renewal or asymmetric cell division) by targeting Myc, underscoring the important role of RNA methylation in murine HSCs’ differentiation fate choice.66 In contrast, small hairpin RNA–mediated knockdown (KD) of METTL3 in human HSPCs reduces global m6A levels, promotes myeloid differentiation after 7 days, and inhibits cell growth while not inducing notable apoptosis in METTL3-depleted cells.67 Consistently, forced expression of wild-type, but not catalytic-dead, METTL3 in human HSPCs increases proliferation and inhibits myeloid differentiation, indicating the requirement of METTL3 enzymatic activity for regulating human HSPC myeloid differentiation.67
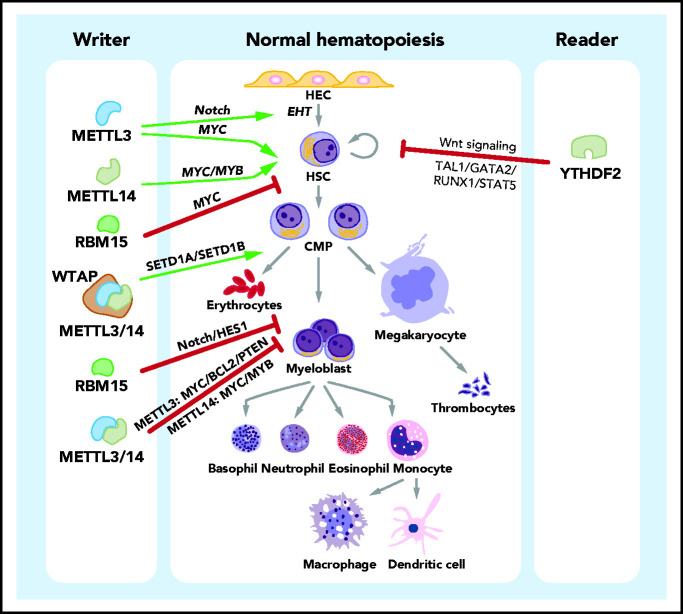
Figure 2.
Roles of m6A in normal hematopoiesis. In the process of normal hematopoiesis, m6A regulators can play promoting (green arrows) or inhibitory (red bars) roles at various stages. Specifically, METTL3 promotes the generation of HSCs, and METTL3 and METTL14 promote the self-renewal activity of HSCs. Another component of the m6A writer complex, RBM15, suppresses the transition of long-term HSCs to short-term HSCs. The 3 key subunits of the m6A writer complex, METTL3, METTL14, and WTAP, promote erythroid differentiation. In contrast, METTL3, METTL14, and RBM15 inhibit the myeloid differentiation of HSCs in normal hematopoiesis. The m6A reader YTHDF2 suppresses the self-renewal activity of HSCs. The identified downstream effectors (signaling pathways and target proteins) or mediator proteins that are critical for the function of m6A regulators are listed (in italics). CMP, common myeloid progenitor; HEC, hemogenic endothelial cell.
The 3 major components of MTC (METTL3, METTL14, and WTAP) have also been reported to promote erythroid differentiation.68 By installing m6A marks into a set of target transcripts, the m6A writers promote the translation of these targets in an m6A-dependent way.68 The target genes primarily include 3 distinct groups: genes involved in the H3K4 methyltransferase network, ribosome/poly(A)+ RNA binding proteins, and genes with causal roles in erythropoiesis.68 Particularly, the MTC-mediated deposition of m6A modification stimulates the translation of H3K4me3 methyltransferases SETD1A and SETD1B, thereby facilitating the maintenance of H3K4me3 transcriptional activation marks and the binding of KLF1 (a master transcriptional regulator of erythropoiesis) to erythroid gene promoters and, eventually, driving human erythroid lineage specification.68
Durable self-renewal is another defining characteristic of HSCs, and it is required for lifelong blood production.60 Our recent study shows that METTL14 is expressed at high levels in HSPCs and is progressively downregulated during normal hematopoiesis; silencing of METTL14 partially inhibits the self-renewal activity of HSCs and induces myeloid differentiation.69 Similarly, 2 independent studies on METTL3 demonstrated that Mettl3 depletion in mouse HSCs leads to the exit of quiescence and a defect in long-term hematopoietic reconstitution capacity.66,70
RBM15 (RNA binding motif protein 15), originally known as the fusion partner of the MKL1 gene in the t(1;22)(p13;q13) translocation of acute megakaryoblastic leukemia, has recently been recognized as an auxiliary component in m6A MTC.33,42,71 Multiple previous studies have revealed the function of RBM15 in normal hematopoiesis.72-74 Rbm15 is expressed at high levels in murine HSCs but at moderate levels during hematopoietic development; conditional knockout (KO) of Rbm15 in mice leads to a block in pro/pre-B differentiation but an expansion of megakaryocytic and myeloid cells; depletion of Rbm15 causes a preference for granulocytic maturation.74 Another independent study demonstrated the high expression of Rbm15 in lineage-depleted bone marrow cells and the lower expression in differentiated macrophages and megakaryocytes; forced expression of Rbm15 attenuates myeloid differentiation.72 Mechanistically, Rbm15 activates Notch-induced HES1 transcription in hematopoietic cells to suppress myeloid differentiation.72 It has also been reported that Rbm15 KO blocks long-term HSCs to short-term HSC differentiation and causes increased, but abnormal, megakaryocyte development in mice.73 Such effects of Rbm15 on hematopoiesis are mediated, at least partially, through regulation of c-Myc levels. However, whether the loss-of-function phenotypes of RBM15 are directly attributable to alterations in m6A marks of its target RNAs has yet to be elucidated and requires further investigation.
The function of ALKBH5, a major m6A eraser, in normal hematopoiesis has also been studied. Two independent studies suggested that Alkbh5 is dispensable for normal hematopoiesis, because Alkbh5 depletion exhibited little effect on normal hematopoiesis, HSPC homeostasis, or normal HSPC self-renewal/reconstitution in physiological and hematopoietic stress conditions.75,76 The role of FTO, the other major m6A eraser, in normal hematopoiesis has yet to be investigated.
The importance of m6A readers in the regulation of myelopoiesis has been suggested by the studies of YTHDF2, an m6A reader protein that promotes degradation of its target transcripts.47 Ythdf2 deficiency in mice led to the expansion of functional HSCs under physiological conditions and hematopoietic stress, without causing abnormal lineage differentiation or hematopoietic malignancies.77,78 Ythdf2-KO murine HSPCs did not show defects in their functionality and exhibited improved repopulating and regeneration capacity as a result of the activation of Wnt signaling following deletion of Ythdf2.78 Additionally, YTHDF2 suppresses HSC self-renewal by promoting the decay of a set of critical HSC self-renewal–related transcription factor genes, including TAL1, GATA2, RUNX1, and STAT5.77 Moreover, Li et al further showed that YTHDF2 KD dramatically increased the ex vivo expansion of human umbilical cord blood HSCs, providing a promising approach to expand these cells before their transplantation.77 Despite the fact that several key findings have been made with regard to the functions of m6A in the regulation of normal hematopoiesis, more studies are needed to fully elucidate the roles of other m6A regulators during this process and to guide investigations of the molecular mechanisms underlying these developmental transitions.
The development and maintenance of myeloid malignancies are facilitated by m6A dysregulation
Abnormal or blocked differentiation, dysregulated self-renewal, and excessive proliferation of HSCs and/or myeloid progenitor cells are common features of myeloid malignancies, a spectrum of clonal hematopoietic disorders consisting of acute myeloid leukemia (AML), myelodysplastic syndrome (MDS), and myeloproliferative neoplasm (MPN). AML is 1 of the most common types of leukemia and has a very poor prognosis in general, with a 5-year survival rate of only 20% to 30%.79-82 MDS is characterized by the pathological feature of cytological dysplasia, whereas MPN is characterized by abnormal clonal myeloproliferation without dysplasia (dyserythropoiesis or dysgranulopoiesis).83-85 MDS and MPN have preleukemic features and can frequently evolve to secondary AML. Over the past 4 decades, promising and encouraging improvement has been seen in the treatment of younger AML patients, especially with the development of a number of targeted therapies in the past few years, and >50% of these patients can achieve complete remission.86,87 However, more than half of new AML cases are diagnosed in older adults (>65 years old), and progress in the treatment of older patients has been much less conspicuous than that seen in younger ones. More importantly, drug toxicity, drug resistance, and relapses are still challenging issues for current therapeutic agents.88 Therefore, there is still an urgent unmet medical need for the identification of new therapeutic targets and the development of combinatorial treatment strategies to provide durable remissions with low toxicity to AML patients, especially for the treatment of older AML patients, as well as refractory/relapsed younger AML patients. Recent studies revealing the close association between m6A dysregulation and the pathogenesis of myeloid malignancies provide novel insights into the mechanisms underlying AML pathogenesis and drug resistance that will ultimately lead to the development of improved new therapies by targeting m6A-related pathways to treat AML (Figure 3).
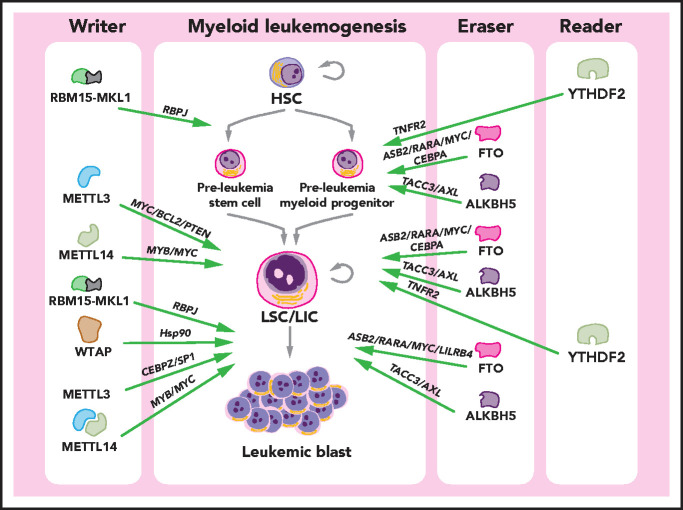
Figure 3.
Roles of m6A in myeloid leukemogenesis. In the process of myeloid leukemogenesis, multiple m6A regulators play oncogenic roles (green arrows) at different stages of leukemogenesis. METTL3 and METTL14 promote the self-renewal/proliferation of leukemia stem cells/leukemia-initiating cells (LSCs/LICs) and the proliferation/survival of leukemia cells. The RBM15-MKL1 fusion protein plays a role in acute megakaryoblastic leukemia, facilitating the transformation of megakaryocytes and inducing a differentiation bias toward the megakaryocytic lineage. WTAP promotes the proliferation of leukemia cells. FTO and ALKBH5 promote the transformation of HSCs to LSCs/LICs, maintain the self-renewal capacity of LSCs/LICs, and are required for the survival/proliferation of leukemia cells. YTHDF2 is essential for leukemia initiation and the functional integrity of LSCs/LICs. The identified downstream effectors (signaling pathways and target proteins) or mediator proteins that are critical for the function of m6A regulators are listed (in italics).
The m6A modification was first linked with myeloid leukemia in the study reporting the oncogenic role of FTO in AML.89 Analysis of a microarray dataset of >100 human AML patient samples revealed that FTO is aberrantly upregulated in t(11q23)/MLL-rearranged AML, t(15;17)/PML-RARA AML, and normal karyotype AML with FLT3-ITD and/or NPM1 mutations. Furthermore, these leukemia fusion genes and/or mutant oncogenes, including MLL-AF9 (MA9), PML-RARA, FLT3ITD, and NPM1 mutation, promote expression of FTO. Overexpression of wild-type, but not catalytic-dead, FTO significantly promotes leukemia cell growth/proliferation in vitro and accelerates MLL-AF9–induced leukemogenesis in vivo, whereas FTO depletion suppresses leukemia cell growth/proliferation and delays leukemogenesis.89 Importantly, FTO exerts its biological function as an m6A demethylase: it removes m6A modifications from its targets, including ASB2 and RARA, to downregulate their expression (which indicates that these transcripts may be recognized by m6A readers that promote stability of their target mRNAs [eg, IGF2BPs]). This signaling pathway is also responsible for the all-trans retinoic acid–induced differentiation of acute promyelocytic leukemia cells where FTO is significantly downregulated upon all-trans retinoic acid treatment.89
There have been some arguments that FTO also recognizes and demethylates N6,2-O-dimethyladenosine (m6Am), with the same chemical structure in the base moiety as m6A and a higher reaction efficacy than m6A.90,91 However, unlike m6A, m6Am primarily locates immediately downstream of the 5′ cap, and its presence only accounts for 1/20th to 1/30th of that of m6A in AML cells.27 A recent comprehensive evaluation of FTO enzymatic activity and function demonstrates that the transcript levels of mRNAs possessing m6A are affected by FTO-mediated demethylation to a much greater extent than are the ones with m6Am.92 In addition, all of the reported functionally important target transcripts of FTO, including ASB2, RARA, MYC, and CEBPA, contain m6A modifications but not m6Am.27,89 Thus, all of the available evidence suggests that the oncogenic function of FTO in AML is attributed to its m6A (rather than m6Am) demethylase activity.
The m6A demethylase activity of FTO requires the presence of its cofactor α-ketoglutarate (α-KG), and this activity can be competitively inhibited by the metabolite R-2-hydroxyglutarate (R-2HG) as a result of its similar structure to α-KG (Figure 1). R-2HG is produced at high levels by mutant isocitrate dehydrogenase 1/2, which can be found in 10% to 20% of AML patients.93-96 Although R-2HG was previously considered an oncometabolite,97-100 surprisingly, we found that it actually exhibits a broad antitumor activity in leukemia and glioma by targeting the FTO/m6A/MYC/CEBPA axis.27 At physiologically relevant levels, R-2HG significantly inhibited cell proliferation and viability in a time- and dose-dependent manner in the vast majority of the 27 leukemia cell lines that we tested, and it significantly delayed leukemogenesis in xenotransplantation mouse models. R-2HG also synergizes with first-line chemotherapy drugs, including hypomethylating agents (eg, 5-azacytidine and decitabine), to inhibit AML cell survival/proliferation in vitro and in vivo.27 These findings highlight the therapeutic potential of targeting FTO in the treatment of AML.
Indeed, by developing selective FTO inhibitors (FB23 and FB23-2), we provided proof-of-concept evidence that pharmacological inhibition of FTO could significantly inhibit AML growth in vitro and in vivo.101 In a subsequent study, we identified 2 more effective FTO inhibitors (ie, CS1 and CS2) with 50% inhibitory concentration values in the low nanomolar range that are 10 times more effective than previously developed FTO inhibitors.102 CS1 and CS2 showed potent antileukemia efficacy in a series of preclinical animal models, including patient-derived xenotransplantation models in which primary AML cells collected from relapsed AML patients were used as donor cells; treatment with CS1 or CS2 alone often resulted in doubled overall survival in treated mice compared with control mice.102 Furthermore, we found that FTO is particularly highly expressed in leukemia stem cells/leukemia-initiating cells (LSCs/LICs), and CS1/CS2 treatment significantly attenuates the self-renewal capacity of LSCs/LICs.102 Moreover, pharmacological inhibition of FTO by CS1 or CS2 also downregulates the expression of immune checkpoint genes, especially LILRB4, which is expressed at levels that are 40- to 50-fold higher than PD-L1 and PD-L2 in AML, sensitizing AML cells to T-cell cytotoxicity and HMAs by overcoming immune evasion.102 Overall, these results demonstrate the critical role of FTO as a major m6A eraser in AML development/maintenance, LSC/LIC self-renewal, and immune evasion and suggest that pharmacological inhibition of FTO with effective inhibitors, alone or in combination with other therapeutics (eg, chemotherapy, targeted therapy, and immunotherapy), represents an effective novel therapeutic strategy to treat AML. In addition, we found that FTO plays a critical role in various types of solid tumors, and FTO inhibitors show potent antitumor efficacy in treating multiple types of solid tumors, highlighting the broad therapeutic potential of targeting FTO in treating various types of cancers.102 Interestingly, CS1 (NSC337766, also named bisantrene) and CS2 (NSC368390, also named brequinar) have been introduced into clinical trials as an anthracene compound and an inhibitor of dihydroorotate dehydrogenase, respectively; most patients tolerate both drugs well.103-115 Thus, CS1 and CS2 could be readily applied in the clinic, alone or in combination with other therapeutic agents, to treat FTO-overexpressing cancers, such as AML, in the near future.
Two recent independent studies found that the other m6A eraser, ALKBH5, is also essential for AML development/maintenance and LSC/LIC self-renewal, while being dispensable for normal hematopoiesis and normal HSPC self-renewal.75,76 Similar to FTO, ALKBH5 is overexpressed in AML patient samples, especially in LSCs/LICs, compared with healthy controls. Notably, the increased expression of ALKBH5 is correlated with a poor prognosis in AML patients.75 The expression of ALKBH5 is regulated by KDM4C, a histone demethylase that increases the chromatin accessibility of the ALKBH5 locus via removal of repressive H3K9me3 marks and, thereby, enhances binding of transcriptional factors and RNA polymerase II.76 Although ALKBH5 and FTO play oncogenic roles in AML as m6A demethylases, they appear to target largely distinct transcripts in many shared pathways.75 Two functionally important targets of ALKBH5 have been identified: TACC3 and AXL. ALKBH5 regulates the mRNA stability of TACC3 and AXL in an m6A-dependent manner, and both targets can mediate, at least in part, the overall function of ALKBH5 in AML.75,76 Thus, these findings highlight the therapeutic potential of targeting ALKBH5 in the treatment of AML.75,76,116 It is very important to develop potent small molecule inhibitors targeting ALKBH5 as effective and safe novel therapeutics to cure AML by selectively eliminating LSCs/LICs while sparing normal hematopoiesis.
In addition to m6A erasers, multiple components from the writer complex have been implicated in myeloid malignancies, including METTL3, METTL14, WTAP, and RBM15.67,69,71,117-120 Results from 2 independent studies support the oncogenic role of METTL3 in AML.67,118 Human AML cells express higher levels of METTL3 than do healthy HSPCs.67 Ablation of METTL3 in human and mouse AML cells inhibits cell growth/proliferation, induces cell cycle arrest and apoptosis, and reverses the myeloid differentiation block, consistent with the proliferation-promoting and differentiation-inhibitory effects of METTL3 on normal HSPCs.67,118 Furthermore, loss of METTL3 delays in vivo development of AML in a xenotransplantation mouse model.67,118 METTL3 has been shown to exert an oncogenic role by promoting the translation of its targets, such as MYC, BCL2, and PTEN.67 In addition, guided by CEBPZ, METTL3 can be recruited to the transcriptional start sites of its target genes SP1 and SP2, and it deposits m6A modifications on the target transcripts to increase their expression, which, in turn, leads to enhanced activity of the downstream MYC pathways.118
Similarly, METTL14, another key component of the writer complex, is detected at higher levels in AML cells than in normal mononuclear cells, and exerts an oncogenic role in AML.69 METTL14 KD in vitro leads to inhibition of cell growth and induction of differentiation and apoptosis, whereas disruption of METTL14 in vivo significantly delays AML onset and prolongs survival in allotransplantation and xenotransplantation mouse models.69 METTL14 positively regulates the expression of MYB and MYC via an m6A-dependent mechanism and, thereby, enhances the self-renewal/proliferation of LSCs/LICs to facilitate AML initiation and maintenance.69 Notably, although KD of METTL14 showed minimal effect on the survival/proliferation of human normal HSPCs,69 KD of METTL3 significantly inhibited growth/proliferation of human normal HSPCs.63 Such a discrepancy might be attributed to METTL3’s additional function in mRNA translation in cytosol that is independent from METTL14, whereas METTL14 exerts its function solely through its interaction with METTL3 for m6A deposition in the nucleus.26,31,34-36,121-123 Thus, targeting METTL3 alone may cause evident side effects because it is also required for the maintenance and proliferation of human normal HSPCs. These data provide a strong rationale for the development of pharmacological approaches to inhibit the METTL3 and METTL14 interaction and, thereby, suppress their m6A deposition activity as novel therapeutic strategies for AML treatment.
Two other components of the writer complex, WTAP and RBM15, have also been recognized as oncogenic proteins in AML, either on its own (WTAP) or as part of a fusion protein (RBM15-MKL1).71,117,119,120 However, these findings were made before their identification as part of the m6A installing machinery,33,38 and it remains to be elucidated whether their effects in AML are attributed to their regulation of m6A modifications. The newly discovered m6A writer, METTL16, has also been shown to be essential for leukemia cell growth in 2 CRISPR-Cas9 screens,43,45,118 yet further studies are needed to better understand its role in hematopoietic malignancies.
Intriguingly, both m6A writers (METTL3 and METTL14) and erasers (FTO and ALKBH5) are oncogenic in AML. One possible explanation for this seemingly paradoxical phenomenon is that the writers and erasers may regulate different sets of target genes, which leads to somewhat similar downstream biological effects.10 The MTC (containing METTL3, METTL14, WTAP, and RBM15) is responsible for the installation of m6A into thousands of transcripts in a given cell type, whereas FTO and ALKBH5 only target a relatively small portion of all m6A-modified transcripts.8,46,92 Thus, MTC disruption is presumed to exert more dramatic effects than FTO or ALKBH5 disruption, because many more transcripts will be affected in the former situation; this is in agreement with the aforementioned observations that murine normal hematopoiesis is significantly affected by depletion of METTL3 or METTL14 but not ALKBH5. Moreover, the fact that MYC mRNA stability is promoted by METTL14 (via regulation of m6A on the 3'-terminal exon of MYC) and FTO (via regulation of m6A on the 5'-terminal and internal exons of MYC) provides an additional explanation: the same target transcript may be recognized by different readers on different regions, which, in turn, results in distinct RNA fates.27,69 Collectively, 1 inspiration from current evidence is that perturbation of RNA m6A methylation homeostasis by dysregulation of writers or erasers could induce imbalanced gene expression and facilitate the development and maintenance of myeloid malignancies.
The association between the m6A reader, YTHDF2, and myeloid malignancies has also been established recently.124 Overexpression of YTHDF2 is observed in a broad spectrum of AML subtypes, and YTHDF2 protects the functional integrity of LSCs/LICs to allow AML initiation and propagation in humans and mice. It is noteworthy that YTHDF2 deficiency, like ALKBH5 depletion, selectively compromises the survival and engraftment capacity of human AML cells, whereas normal HSC activity is enhanced by loss of YTHDF2, indicating that YTHDF2 appears to be a feasible novel target for AML therapeutics.77,78,124 Nonetheless, further studies are required for the development and assessment of specific small molecule inhibitors against YTHDF2.
Other RNA modifications in normal and malignant hematopoiesis
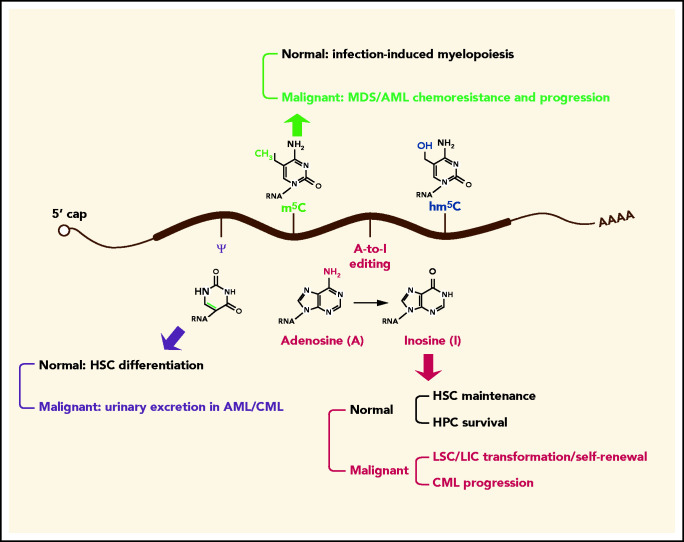
To date, >170 chemical modifications have been described in coding and noncoding RNAs,3,4,7 although the majority of such modifications have not been validated, and the prevalence of many modifications in RNAs could be very low. In comparison with m6A, our understanding of the biological functions of other RNA modifications in normal and malignant hematopoiesis remains very limited, but it is beginning to consolidate with the development of high-throughput sequencing technologies that enable transcriptome-wide mapping of specific RNA modifications (Figure 4).11-14,16-20 Here, we discuss the roles of several other well-validated RNA modifications (including A-to-I editing, m5C, hm5C, and pseudouridine) in normal and malignant hematopoiesis.
Figure 4.
Roles of other RNA modifications in normal and malignant hematopoiesis. In addition to m6A, other RNA modifications, such as A-to-I editing, m5C, and pseudouridine (ψ), are implicated in normal and malignant hematopoiesis. Specifically, ψ synthase is essential for efficient HSC differentiation, and ψ is highly excreted in urine in AML and chronic myeloid leukemia (CML). Increased levels of m5C and its writers confer chemoresistance to MDS/AML cells, m5C writers are positively associated with MDS/AML progression, and m5C eraser TET2 promotes infection-induced myelopoiesis. The writer for A-to-I editing is required for maintenance of HSCs and survival of hematopoietic progenitor cell (HPCs), whereas the same writer promotes transformation of myeloid progenitors into LSCs/LICs and enhances LSC/LIC self-renewal activity. The level of A-to-I editing is positively correlated with CML progression.
Site-selective editing of adenosine to inosine (“A-to-I editing”) is mediated by the ADAR (adenosine deaminases acting on RNA) family, which includes ADAR1 (ADAR), ADAR2 (ADARB1), and ADAR3 (ADARB2).125 ADAR1 is required for the maintenance of HSCs and the survival of differentiating hematopoietic progenitor cells.126,127 Within the myeloid lineage, myeloid progenitors express ADAR1 at low levels, whereas mature granulocytes exhibit much higher ADAR1 expression, which correlates with a global increase in A-to-I editing during myeloid leukemia cell maturation.128,129 However, erythroid- and myeloid-restricted depletion of Adar1 in mouse models suggests that ADAR1 is dispensable for normal myelopoiesis, although it is essential for normal erythropoiesis.128,130 In chronic myeloid leukemia (CML), a subcategory of MPN, the level of A-to-I editing is increased in parallel with disease progression.131 ADAR1 activation promotes malignant reprogramming of myeloid progenitors into LSCs/LICs, enhances LSC/LIC self-renewal, and promotes CML progenitor propagation through hyperediting of cell cycle regulatory and tumor suppressor RNAs.131-133 Because m6A and A-to-I editing both occurs on adenosines, there is a possibility that m6A writers and ADARs compete for the same adenosines, with the fate of the modified transcripts determined by the intricate regulation of both modifications. Indeed, Yang and colleagues found a negative correlation between m6A and A-to-I editing using a genome-wide analysis of A-to-I editing abundance in m6A+ and m6A− transcripts.134 Suppression of m6A writers (METTL3/METTL14) and m6A erasers (FTO) resulted in an increase and decrease in global A-to-I editing ratios, respectively, without altering ADAR expression. The m6A-modified transcripts have lower binding affinities to ADAR1 for further A-to-I editing, which accounts, at least in part, for the negative modulation of A-to-I editing by m6A.134
The methylation of carbon 5 in cytosine (m5C) in RNA is catalyzed by the NOL1/NOP2/SUN domain (NSUN) family of methyltransferases or DNMT2 and m5C is subject to oxidation by TET proteins to generate hm5C and other oxidative metabolites.135-137 The m5C marks and its methyltransferases NSUNs/DMNT2 are markedly increased in hypomethylating agent–resistant MDS/AML cells.138 The mechanistic study revealed that NSUNs/DNMT2 directly interact with hnRNPK to recruit RNA-polymerase II/CTD9 and erythroid/myeloid lineage–determining transcription factors GATA1 and SPI1/PU.1 to nascent RNAs to form a drug-responsive active chromatin structures to confer chemoresistance to MDS/AML cells.138 Consistently, the expression of m5C methyltransferases and hnRNPK is positively correlated with MDS/AML progression.138 By contrast, the m5C eraser TET2 is reported to promote infection-induced myelopoiesis by oxidizing the m5C modifications in SOCS3 mRNAs, which subsequently allows the binding of ADAR1 to unmethylated SOCS3 mRNA to repress SOCS3 expression in an RNA editing–independent manner.139
Pseudouridylation of ribosomal RNA, first discovered in 1957, is catalyzed by dyskerin and guided by the H/ACA box small nucleolar RNAs.2,140 During the past 6 decades, some clues have been provided by previous studies about the involvement of pseudouridine in normal and malignant myelopoiesis. The urinary excretion of pseudouridine was elevated in >80% of cases of untreated AML and CML.141 Additionally, the pseudouridine synthase activity of dyskerin is required for efficient differentiation of HSCs.140 However, the clinical implications and the underlying mechanisms for dysregulation of pseudouridine in myeloid malignancies have yet to be elucidated.
Conclusions
RNA modifications are essential players in cell fate decision during hematopoietic development, and our knowledge of the roles of RNA modifications in various biological processes is growing rapidly as a result of improved detection methods, especially next-generation sequencing to map RNA marks at the transcriptome level. Recent discoveries have positioned these modifications (primarily m6A) as key regulatory mechanisms in normal and malignant hematopoiesis. Aberrant expression of m6A modulators (writers, erasers, and readers) has been described and investigated for most aspects of leukemogenesis; their pharmacological inhibition, especially the m6A eraser FTO, has achieved promising antileukemic effects and represents novel therapeutic strategies for AML. Targeting dysregulated m6A machinery with effective inhibitors, alone or in combination with other therapeutics, represents an attractive novel approach to treat leukemia patients, especially refractory/relapsed patients who are resistant to available therapies.
Most recent m6A modification studies in hematopoietic malignancies have focused on AML. The functions and underlying molecular mechanisms of m6A modification and the associated machinery in other types of hematopoietic malignancies, such as acute lymphoblastic leukemia, chronical lymphocytic leukemia, CML, MDS, and MPN, have yet to be investigated. It would be interesting to determine whether a given m6A modulator plays distinct roles and regulates largely different sets of targets across different types of hematopoietic malignancies.
Unlike m6A decoration, the investigation of the roles of many other modifications, such as N4-acetylcytidine, N1-methyladenosine, m5C, and hm5C, is still restricted by the sensitivity and precision of current detection techniques. Their impact on normal hematopoiesis and leukemogenesis, their abundance in leukemia patients in contrast to healthy controls, and their biological functions in determining RNA fate remain to be investigated. Therefore, it is imperative to develop more advanced detection methods and tools for the identification and quantification of genuine RNA modifications with higher sensitivity, precision, and resolution. From a clinical perspective, such knowledge is essential to reveal their biological functions during the different steps of hematopoiesis and leukemogenesis and is critical for the identification and characterization of novel potent biomarkers and druggable targets for the treatment of hematopoietic malignancies.
Acknowledgments
This work was supported, in part, by National Institutes of Health grants R01 CA243386, R01 CA214965, R01 CA236399, R01 CA211614 (National Cancer Institute), and R01 DK124116 (National Institute of Diabetes and Digestive and Kidney Disease) (all to J.C.), The Margaret Early Medical Research Trust (R.S.), and a Held Foundation Fellowship (Y.Q.). J.C. is a Leukemia and Lymphoma Society Scholar. The authors apologize to colleagues whose work could not be cited because of space constraints.
Authorship
Contribution: Y.Q., R.S., and J.C. wrote the manuscript and created the figures.
Conflict-of-interest disclosure: J.C. is a scientific founder of Genovel Biotech Corp and holds equity interest in the company. The remaining authors declare no competing financial interests.
Correspondence: Jianjun Chen, City of Hope Comprehensive Cancer Center, 1218 Fifth Ave, Monrovia, CA 91016; e-mail: jianchen@coh.org.
REFERENCES
- 1.Frye M, Jaffrey SR, Pan T, Rechavi G, Suzuki T.. RNA modifications: what have we learned and where are we headed? Nat Rev Genet. 2016;17(6):365-372. [DOI] [PubMed] [Google Scholar]
- 2.Davis FF, Allen FW.. Ribonucleic acids from yeast which contain a fifth nucleotide. J Biol Chem. 1957;227(2):907-915. [PubMed] [Google Scholar]
- 3.Boccaletto P, Machnicka MA, Purta E, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46(D1):D303-D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xuan J-J, Sun W-J, Lin P-H, et al. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 2018;46(D1):D327-D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frye M, Harada BT, Behm M, He C.. RNA modifications modulate gene expression during development. Science. 2018;361(6409):1346-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang H, Weng H, Chen J.. m6A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37(3):270-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang H, Weng H, Deng X, Chen J.. RNA modifications in cancer: functions, mechanisms, and therapeutic implications. Annu Rev Cancer Biol. 2020;4(1):221-240. [Google Scholar]
- 8.Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO [published correction appears in Nat Chem Biol. 2012;8(12):1008]. Nat Chem Biol. 2011;7(12):885-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng X, Su R, Stanford S, Chen J.. Critical enzymatic functions of FTO in obesity and cancer. Front Endocrinol (Lausanne). 2018;9:396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng X, Su R, Weng H, Huang H, Li Z, Chen J.. RNA N6-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28(5):507-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530(7591):441-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arango D, Sturgill D, Alhusaini N, et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell. 2018;175(7):1872-1886.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV.. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515(7525):143-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delatte B, Wang F, Ngoc LV, et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016;351(6270):282-285. [DOI] [PubMed] [Google Scholar]
- 15.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201-206. [DOI] [PubMed] [Google Scholar]
- 16.Schwartz S, Bernstein DA, Mumbach MR, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159(1):148-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki T, Ueda H, Okada S, Sakurai M.. Transcriptome-wide identification of adenosine-to-inosine editing using the ICE-seq method. Nat Protoc. 2015;10(5):715-732. [DOI] [PubMed] [Google Scholar]
- 18.Hussain S, Sajini AA, Blanco S, et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4(2):255-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khoddami V, Cairns BR.. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol. 2013;31(5):458-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Squires JE, Patel HR, Nousch M, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40(11):5023-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang H, Weng H, Chen J.. The biogenesis and precise control of RNA m6A methylation. Trends Genet. 2020;36(1):44-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin GH, Park CY.. Meddling with METTLs in normal and leukemia stem cells. Cell Stem Cell. 2018;22(2):139-141. [DOI] [PubMed] [Google Scholar]
- 23.Vu LP, Cheng Y, Kharas MG.. The biology of m6A RNA methylation in normal and malignant hematopoiesis. Cancer Discov. 2019;9(1):25-33. [DOI] [PubMed] [Google Scholar]
- 24.Weng H, Huang H, Chen J.. RNA N6-methyladenosine modification in normal and malignant hematopoiesis. Adv Exp Med Biol. 2019;1143:75-93. [DOI] [PubMed] [Google Scholar]
- 25.Cantara WA, Crain PF, Rozenski J, et al. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39(Database issue):D195-D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su R, Dong L, Li C, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell. 2018;172(1-2):90-105.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Batista PJ, Molinie B, Wang J, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15(6):707-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roundtree IA, Evans ME, Pan T, He C.. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang H, Weng H, Zhou K, et al. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature. 2019;567(7748):414-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM.. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3(11):1233-1247. [PMC free article] [PubMed] [Google Scholar]
- 32.Geula S, Moshitch-Moshkovitz S, Dominissini D, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347(6225):1002-1006. [DOI] [PubMed] [Google Scholar]
- 33.Patil DP, Chen CK, Pickering BF, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537(7620):369-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Feng J, Xue Y, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex [published correction appears in Nature. 2017;542(7640):260]. Nature. 2016;534 (7608):575-578. [DOI] [PubMed] [Google Scholar]
- 35.Śledź P, Jinek M.. Structural insights into the molecular mechanism of the m6A writer complex. eLife. 2016;5:e18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang P, Doxtader KA, Nam Y.. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell. 2016;63(2):306-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haussmann IU, Bodi Z, Sanchez-Moran E, et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540(7632):301-304. [DOI] [PubMed] [Google Scholar]
- 38.Ping X-L, Sun B-F, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24(2):177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz S, Mumbach MR, Jovanovic M, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep. 2014;8(1):284-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wen J, Lv R, Ma H, et al. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol Cell. 2018;69(6):1028-1038.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yue Y, Liu J, Cui X, et al. VIRMA mediates preferential m6A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knuckles P, Lence T, Haussmann IU, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32(5-6):415-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pendleton KE, Chen B, Liu K, et al. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169(5):824-835.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown JA, Kinzig CG, DeGregorio SJ, Steitz JA.. Methyltransferase-like protein 16 binds the 3'-terminal triple helix of MALAT1 long noncoding RNA. Proc Natl Acad Sci USA. 2016;113(49):14013-14018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warda AS, Kretschmer J, Hackert P, et al. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18(11):2004-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014; 505(7481):117-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao W, Adhikari S, Dahal U, et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing [published correction appears in Mol Cell. 2016;61(6):925]. Mol Cell. 2016;61(4):507-519. [DOI] [PubMed] [Google Scholar]
- 49.Roundtree IA, Luo GZ, Zhang Z, et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife. 2017;6:e31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Zhao BS, Roundtree IA, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du H, Zhao Y, He J, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7(1):12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li A, Chen YS, Ping XL, et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27(3):444-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi H, Wang X, Lu Z, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27(3):315-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang H, Weng H, Sun W, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. [published corrections appear in Nat Cell Biol. 2018;20(9):1098; Nat Cell Biol. 2020;22(10):1288]. Nat Cell Biol. 2018;20(3):285-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsu PJ, Zhu Y, Ma H, et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27(9):1115-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF.. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162(6):1299-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T.. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518(7540):560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou KI, Shi H, Lyu R, et al. Regulation of co-transcriptional pre-mRNA splicing by m6A through the low-complexity protein hnRNPG. Mol Cell. 2019;76(1):70-81.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T.. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45(10):6051-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Doulatov S, Notta F, Laurenti E, Dick JE.. Hematopoiesis: a human perspective. Cell Stem Cell. 2012;10(2):120-136. [DOI] [PubMed] [Google Scholar]
- 61.Orkin SH, Zon LI.. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schultze JL, Mass E, Schlitzer A.. Emerging principles in myelopoiesis at homeostasis and during infection and inflammation. Immunity. 2019;50(2):288-301. [DOI] [PubMed] [Google Scholar]
- 63.Lv J, Zhang Y, Gao S, et al. Endothelial-specific m6A modulates mouse hematopoietic stem and progenitor cell development via Notch signaling. Cell Res. 2018;28(2):249-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang C, Chen Y, Sun B, et al. m6A modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549(7671):273-276. [DOI] [PubMed] [Google Scholar]
- 65.Lee H, Bao S, Qian Y, et al. Stage-specific requirement for Mettl3-dependent m6A mRNA methylation during haematopoietic stem cell differentiation. Nat Cell Biol. 2019;21(6):700-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng Y, Luo H, Izzo F, et al. m6A RNA methylation maintains hematopoietic stem cell identity and symmetric commitment. Cell Rep. 2019;28(7):1703-1716.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vu LP, Pickering BF, Cheng Y, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23(11):1369-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuppers DA, Arora S, Lim Y, et al. N6-methyladenosine mRNA marking promotes selective translation of regulons required for human erythropoiesis. Nat Commun. 2019;10(1):4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weng H, Huang H, Wu H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell. 2018;22(2):191-205.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yao QJ, Sang L, Lin M, et al. Mettl3-Mettl14 methyltransferase complex regulates the quiescence of adult hematopoietic stem cells. Cell Res. 2018;28(9):952-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ma Z, Morris SW, Valentine V, et al. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat Genet. 2001;28(3):220-221. [DOI] [PubMed] [Google Scholar]
- 72.Ma X, Renda MJ, Wang L, et al. Rbm15 modulates Notch-induced transcriptional activation and affects myeloid differentiation. Mol Cell Biol. 2007;27(8):3056-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Niu C, Zhang J, Breslin P, Onciu M, Ma Z, Morris SW.. c-Myc is a target of RNA-binding motif protein 15 in the regulation of adult hematopoietic stem cell and megakaryocyte development. Blood. 2009;114(10):2087-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raffel GD, Mercher T, Shigematsu H, et al. Ott1(Rbm15) has pleiotropic roles in hematopoietic development. Proc Natl Acad Sci USA. 2007;104(14):6001-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shen C, Sheng Y, Zhu AC, et al. RNA demethylase ALKBH5 selectively promotes tumorigenesis and cancer stem cell self-renewal in acute myeloid leukemia. Cell Stem Cell. 2020;27(1):64-80.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang J, Li Y, Wang P, et al. Leukemogenic chromatin alterations promote AML leukemia stem cells via a KDM4C-ALKBH5-AXL signaling axis. Cell Stem Cell. 2020;27(1):81-97.e8. [DOI] [PubMed] [Google Scholar]
- 77.Li Z, Qian P, Shao W, et al. Suppression of m6A reader Ythdf2 promotes hematopoietic stem cell expansion [published correction appears in Cell Res. 2018;28(10):1042]. Cell Res. 2018;28(9):904-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang H, Zuo H, Liu J, et al. Loss of YTHDF2-mediated m6A-dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res. 2018;28(10):1035-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Döhner H, Estey EH, Amadori S, et al. ; European LeukemiaNet. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115 (3):453-474. [DOI] [PubMed] [Google Scholar]
- 80.Döhner H, Weisdorf DJ, Bloomfield CD.. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136-1152. [DOI] [PubMed] [Google Scholar]
- 81.Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2020. CA Cancer J Clin. 2020;70 (1):7-30. [DOI] [PubMed] [Google Scholar]
- 82.Howlader N, Noone A, Krapcho M, et al. SEER Cancer Statistics Review (CSR) 1975-2017. Available at: https://seer.cancer.gov/csr/1975_2017/. Accessed 5 October 2020.
- 83.Tefferi A, Vardiman JW.Myelodysplastic syndromes. N Engl J Med. 2009;361(19):1872-1885. [DOI] [PubMed] [Google Scholar]
- 84.Grinfeld J, Nangalia J, Baxter EJ, et al. Classification and personalized prognosis in myeloproliferative neoplasms. N Engl J Med. 2018;379(15):1416-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Swerdlow S, Campo E, Harris NL, et al. , eds. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Vol. 2. Revised 4th ed. Lyon, France: International Agency for Research on Cancer; 2017. [Google Scholar]
- 86.Blum WG, Mims AS.. Treating acute myeloid leukemia in the modern era: a primer. Cancer. 2020;126(21):4668-4677. [DOI] [PubMed] [Google Scholar]
- 87.Kadia TM, Ravandi F, O’Brien S, Cortes J, Kantarjian HM.. Progress in acute myeloid leukemia. Clin Lymphoma Myeloma Leuk. 2015;15(3):139-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Konig H, Zeidner JF.. Acute myeloid leukemia: changing treatment paradigms and novel agents in development. Curr Cancer Drug Targets. 2020;20(7):471-472. [DOI] [PubMed] [Google Scholar]
- 89.Li Z, Weng H, Su R, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell. 2017;31(1):127-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mauer J, Luo X, Blanjoie A, et al. Reversible methylation of m6Am in the 5' cap controls mRNA stability. Nature. 2017;541(7637):371-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mauer J, Sindelar M, Despic V, et al. FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat Chem Biol. 2019;15(4):340-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wei J, Liu F, Lu Z, et al. Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell. 2018;71(6):973-985.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate [published correction appears in Nature. 2010;465(7300):966]. Nature. 2009;462 (7274):739-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Okoye-Okafor UC, Bartholdy B, Cartier J, et al. New IDH1 mutant inhibitors for treatment of acute myeloid leukemia. Nat Chem Biol. 2015;11(11):878-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Losman JA, Looper RE, Koivunen P, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339(6127):1621-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huang Y, Su R, Sheng Y, et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. 2019;35(4):677-691.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Su R, Dong L, Li Y, et al. Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell. 2020;38 (1):79-96.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yap HY, Yap BS, Blumenschein GR, Barnes BC, Schell FC, Bodey GP.. Bisantrene, an active new drug in the treatment of metastatic breast cancer. Cancer Res. 1983;43(3):1402-1404. [PubMed] [Google Scholar]
- 104.Rothman J.The rediscovery of bisantrene: a review of the literature. Int J Cancer Res Ther. 2017;2(2):1-10. [Google Scholar]
- 105.Peters GJ, Kraal I, Pinedo HM.. In vitro and in vivo studies on the combination of brequinar sodium (DUP-785; NSC 368390) with 5-fluorouracil; effects of uridine. Br J Cancer. 1992;65(2):229-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pratt CB, Sinkule JA, Etcubanas E, et al. Phase I clinical and pharmacokinetic study of bisantrene in refractory pediatric solid tumors. Invest New Drugs. 1986;4(2):149-153. [DOI] [PubMed] [Google Scholar]
- 107.Coltman CA Jr, Osborne CK.. Bisantrene, biological and clinical effects. Cancer Treat Rev. 1984;11(4):285-288. [DOI] [PubMed] [Google Scholar]
- 108.Yap BS, Yap HY, Blumenschein GR, Bedikian AY, Pocelinko R, Bodey GP.. Phase I clinical evaluation of 9,10-anthracenedicarboxyaldehyde[bis(4,5-dihydro-1H-imidazol-2-yl)hydrazone]dihydrochloride (bisantrene). Cancer Treat Rep. 1982;66(7):1517-1520. [PubMed] [Google Scholar]
- 109.Spiegel RJ, Blum RH, Levin M, et al. Phase I clinical trial of 9,10-anthracene dicarboxaldehyde (bisantrene) administered in a five-day schedule. Cancer Res. 1982;42(1):354-358. [PubMed] [Google Scholar]
- 110.Cowan JD, Gehan E, Rivkin SE, Jones SE.. Phase II trial of bisantrene in patients with advanced sarcoma: a Southwest Oncology Group Study. Cancer Treat Rep. 1986;70(5):685-686. [PubMed] [Google Scholar]
- 111.Osborne CK, Von Hoff DD, Cowan JD, Sandbach J.. Bisantrene, an active drug in patients with advanced breast cancer. Cancer Treat Rep. 1984;68(2):357-360. [PubMed] [Google Scholar]
- 112.de Forni M, Chabot GG, Armand JP, et al. Phase I and pharmacokinetic study of brequinar (DUP 785; NSC 368390) in cancer patients. Eur J Cancer. 1993;29A(7):983-988. [DOI] [PubMed] [Google Scholar]
- 113.Noe DA, Rowinsky EK, Shen HS, et al. Phase I and pharmacokinetic study of brequinar sodium (NSC 368390). Cancer Res. 1990;50(15):4595-4599. [PubMed] [Google Scholar]
- 114.Schwartsmann G, Dodion P, Vermorken JB, et al. Phase I study of brequinar sodium (NSC 368390) in patients with solid malignancies. Cancer Chemother Pharmacol. 1990;25(5):345-351. [DOI] [PubMed] [Google Scholar]
- 115.Dodion PF, Wagener T, Stoter G, et al. Phase II trial with brequinar (DUP-785, NSC 368390) in patients with metastatic colorectal cancer: a study of the Early Clinical Trials Group of the EORTC. Ann Oncol. 1990;1(1):79-80. [DOI] [PubMed] [Google Scholar]
- 116.Cheng Y, Luo H, Kharas MG.. Rubbing out leukemia stem cells by erasing the eraser. Cell Stem Cell. 2020;27(1):3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bansal H, Yihua Q, Iyer SP, et al. WTAP is a novel oncogenic protein in acute myeloid leukemia [published correction appears in Leukemia. 2014;28(12):2427]. Leukemia. 2014;28(5):1171-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Barbieri I, Tzelepis K, Pandolfini L, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature. 2017;552(7683):126-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mercher T, Coniat MB, Monni R, et al. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc Natl Acad Sci USA. 2001;98(10):5776-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mercher T, Raffel GD, Moore SA, et al. The OTT-MAL fusion oncogene activates RBPJ-mediated transcription and induces acute megakaryoblastic leukemia in a knockin mouse model. J Clin Invest. 2009;119(4):852-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC.. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16(2):191-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Choe J, Lin S, Zhang W, et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561(7724):556-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lin S, Choe J, Du P, Triboulet R, Gregory RI.. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62(3):335-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Paris J, Morgan M, Campos J, et al. Targeting the RNA m6A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell. 2019;25(1):137-148.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Levanon EY, Eisenberg E, Yelin R, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22(8):1001-1005. [DOI] [PubMed] [Google Scholar]
- 126.Hartner JC, Walkley CR, Lu J, Orkin SH.. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling [published correction appears in Cell Res. 2018;28(10):1042]. Nat Immunol. 2009;10(1):109-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.XuFeng R, Boyer MJ, Shen H, et al. ADAR1 is required for hematopoietic progenitor cell survival via RNA editing. Proc Natl Acad Sci U S A. 2009;106(42):17763-17768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liddicoat BJ, Hartner JC, Piskol R, et al. Adenosine-to-inosine RNA editing by ADAR1 is essential for normal murine erythropoiesis. Exp Hematol. 2016;44(10):947-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rossetti C, Picardi E, Ye M, et al. RNA editing signature during myeloid leukemia cell differentiation. Leukemia. 2017;31(12):2824-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang Q, Khillan J, Gadue P, Nishikura K.. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290(5497):1765-1768. [DOI] [PubMed] [Google Scholar]
- 131.Jiang Q, Crews LA, Barrett CL, et al. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc Natl Acad Sci USA. 2013;110(3):1041-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zipeto MA, Court AC, Sadarangani A, et al. ADAR1 activation drives leukemia stem cell self-renewal by impairing Let-7 biogenesis. Cell Stem Cell. 2016;19(2):177-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jiang Q, Isquith J, Zipeto MA, et al. Hyper-editing of cell-cycle regulatory and tumor suppressor RNA promotes malignant progenitor propagation. Cancer Cell. 2019;35 (1):81-94.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Xiang J-F, Yang Q, Liu C-X, Wu M, Chen L-L, Yang L.. N6-methyladenosines modulate A-to-I RNA editing. Mol Cell. 2018;69(1):126-135.e6. [DOI] [PubMed] [Google Scholar]
- 135.Trixl L, Lusser A.. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip Rev RNA. 2019;10(1):e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bohnsack KE, Höbartner C, Bohnsack MT.. Eukaryotic 5-methylcytosine (m5C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes (Basel). 2019;10(2): 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Huber SM, van Delft P, Mendil L, et al. Formation and abundance of 5-hydroxymethylcytosine in RNA. ChemBioChem. 2015;16(5):752-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cheng JX, Chen L, Li Y, et al. RNA cytosine methylation and methyltransferases mediate chromatin organization and 5-azacytidine response and resistance in leukaemia [published correction appears in Nat Commun. 2018;9(1):2286]. Nat Commun. 2018;9(1):1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shen Q, Zhang Q, Shi Y, et al. Tet2 promotes pathogen infection-induced myelopoiesis through mRNA oxidation. Nature. 2018;554(7690):123-127. [DOI] [PubMed] [Google Scholar]
- 140.Bellodi C, McMahon M, Contreras A, et al. H/ACA small RNA dysfunctions in disease reveal key roles for noncoding RNA modifications in hematopoietic stem cell differentiation. Cell Rep. 2013;3(5): 1493-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nielsen HR, Killmann S-A.. Urinary excretion of β-aminoisobutyrate and pseudouridine in acute and chronic myeloid leukemia. J Natl Cancer Inst. 1983;71(5):887-891. [PubMed] [Google Scholar]