Abstract
Latent Epstein-Barr virus (EBV) is maintained as a nucleosome-covered episome that can be transcriptionally activated by overexpression of the viral immediate-early protein, Zta. We show here that reactivation of latent EBV by Zta can be significantly enhanced by coexpression of the cellular coactivators CREB binding protein (CBP) and p300. A stable complex containing both Zta and CBP could be isolated from lytically stimulated, but not latently infected RAJI nuclear extracts. Zta-mediated viral reactivation and transcriptional activation were both significantly inhibited by coexpression of the E1A 12S protein but not by an N-terminal deletion mutation of E1A (E1AΔ2-36), which fails to bind CBP. Zta bound directly to two related cysteine- and histidine-rich domains of CBP, referred to as C/H1 and C/H3. These domains both interacted specifically with the transcriptional activation domain of Zta in an electrophoretic mobility shift assay. Interestingly, we found that the C/H3 domain was a potent dominant negative inhibitor of Zta transcriptional activation function. In contrast, an amino-terminal fragment containing the C/H1 domain was sufficient for coactivation of Zta transcription and viral reactivation function. Thus, CBP can stimulate the transcription of latent EBV in a histone acetyltransferase-independent manner mediated by the CBP amino-terminal C/H1-containing domain. We propose that CBP may regulate aspects of EBV latency and reactivation by integrating cellular signals mediated by competitive interactions between C/H1, C/H3, and the Zta activation domain.
Epstein-Barr virus (EBV) establishes a latent infection in human B lymphocytes that can be periodically reactivated by various cell signaling systems (reviewed in references 32 and 55). The latent virus exists as a chromatin-associated, multicopy episome with highly restricted transcription patterns (18, 59). Reactivation of latent EBV can be induced by several different chemicals, including phorbol esters (72), calcium ionophores (20), 5-azacytidine (5), and sodium butyrate (56). These reagents relieve multiple levels of transcriptional repression and promote transcriptional activation of the immediate-early genes of EBV. zta is the primary immediate-early gene of EBV that can induce viral reactivation in B-lymphocytes when sufficiently overexpressed by a heterologous promoter (13, 17, 47). Zta is a member of the b-Zip family of transcriptional activators and stimulates the transcription of multiple viral genes by binding directly to Zta response elements (ZREs) in viral promoters (21, 31, 41).
The mechanism of Zta transcriptional activation has been investigated in some detail. Zta can stimulate the formation of a preinitiation complex consisting of the general transcription factors TFIIA and TFIID (39). Zta interacts directly with several polypeptide components of TFIIA and TFIID and can alter the binding of TATA binding protein-associated factors to promoter sequences near the start site of transcription (14, 33, 42). The stable Zta-TFIID-TFIIA complex is more competent for recruiting TFIIB (15). High-level transcriptional activation in vitro by Zta requires interactions not only with TFIID, TFIIA, and TFIIB but also with a poorly defined set of transcriptional coactivators (38). All of these interactions are dependent on the amino-terminal activation domain of Zta (38, 39).
Regulation of EBV latency can occur at multiple levels, including the control of Zta transcriptional activation functions. Several transcription factors that mediate complex cellular signaling pathways bind directly to Zta and affect its ability to stimulate transcription. Zta can bind NF-κB p65, p53, and the retinoic acid receptor (RAR), all of which are mediated through the Zta dimerization domain (27, 61, 70). Transfection of p65, p53, or RAR profoundly inhibits Zta transcriptional activation function, and treatment of EBV-infected cells with RA inhibits phorbol ester-mediated reactivation (44, 62, 67), but the precise mechanism of this inhibition is not completely understood.
Multiple signaling mechanisms may be integrated by the family of coactivator proteins CREB binding protein (CBP) and p300, which interact directly with multiple activators and coactivator complexes (22, 29, 60, 64). CBP was originally found to bind to the phosphorylated form of CREB and mediate its transcription activation function (16, 37, 49). p300 was identified by virtue of its interaction with the amino-terminal domain of the adenovirus E1A oncoprotein, and this association is important for the growth-transforming activity of E1A (2, 19). p300 and CBP are highly related proteins and have many overlapping functions including the ability to bind CREB and E1A (45). CBP and p300 bind many other transcription factors, including p53, NF-κB p65, and RAR, and the association with CBP has been shown to correlate with transcription activation function (26, 30, 43, 53, 71).
CBP-p300 stimulates transcription by at least two distinct mechanisms. CBP-p300 possesses an intrinsic histone acetyltransferase activity (HAT), and this activity is important for the remodeling of chromatin which inhibits the accessibility of general transcription factors to core promoter sequences (3, 24, 50). The HAT domain of CBP functions as a transcriptional activation domain when artificially tethered to some core promoters (46), and inactivation of CBP HAT activity by mutagenesis abrogates transcription coactivation by some activators (34). Also, CBP is capable of directly acetylating nonhistone transcription factors, including p53 (25). Acetylation of p53 stimulates DNA binding activity, and this correlates with an increase in transcriptional activation function (25). CBP may also activate transcription by a HAT-independent mechanism (63). CBP mutants lacking HAT activity can stimulate transcription from some promoters (34, 63). CBP mediates interactions between transcription factors and the holo-RNA polymerase II, thus serving as a bridging factor which may stabilize the formation of a preinitiation complex (49). Recruitment of holo-RNA polymerase II by promoter-bound factors is an efficient mechanism of transcriptional activation (4, 54). Thus, CBP can modulate transcription by acetylation of histones and other factors, as well as by recruiting holo-RNA polymerase II to the promoter.
CBP-p300 can also associate with multiprotein complexes that contain additional HAT activities (60). The multiprotein P/CAF complex associates with CBP, as does p/CIP and NCoA, which can bind directly to some promoter-bound transcription factors, such as nuclear hormone receptors (12, 65, 68). The requirement for multiple HAT and coactivators that are capable of associating directly and indirectly with transcription factors, as well as with each other, raises the issues of how these multiprotein complexes contribute to transcription activation and which subsets of coactivators are required for specific activation pathways (64, 65). Some of these coactivators may have specialized functions in specific cell types and for particular genetic pathways (36). In this work, we explore the role of CBP in mediating the reactivation of latent EBV by binding directly to the Zta activation domain.
MATERIALS AND METHODS
Cells.
HeLa and D98/HR1 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Gibco BRL), glutamine, and penicillin-streptomycin (complete medium) in a 5% CO2 incubator at 37°C. D98/HR1 is a hybrid of the EBV-positive Burkitt’s lymphoma cell line P3HR1 and the human epithelial cell line D98 (23). Raji cells, derived from an EBV-positive Burkitt’s lymphoma, were maintained in RPMI medium supplemented with 10% fetal bovine serum, glutamine, and antibiotics (complete medium) in a 5% CO2 incubator at 37°C.
Plasmids.
All glutathione S-transferase (GST)-CBP fusion protein expression plasmids were constructed by PCR amplification from a full-length mouse CBP template with oligonucleotides containing appropriate restriction sites and cloned into pGEX-2T (Pharmacia). GST-C/H1 codes for amino acids (aa) 301 to 585 of CBP, encompassing the cysteine- and histidine-rich region 1 (C/H1), and was cloned as a BamHI-digested fragment into BamHI-cut pGEX2T. GST-NTAD codes for aa 227 to 460 of CBP and was cloned as a BamHI-digested fragment into BamHI-digested vector (63). GST-KIX expresses the phospho-CREB interacting region of CBP (aa 586 to 666) and was cloned as a BamHI-EcoRI-digested fragment into BamHI-EcoRI-cut pGEX2T (52). For the CBP dominant-negative-mutant experiments, oligonucleotides with inserted Asp718 and BamHI restriction sites and N-terminal HA epitope tag were PCR amplified from a full-length mouse CBP template, digested with Asp718 and BamHI, and cloned into the eukaryotic expression plasmid pRTS2, a pSG5-based vector (Stratagene) (57). The dominant negative CBP constructs CBP 1430–1915, CBP 1680–1915, CBP 1430–1680, and CBP 1194–1915 express an N-terminal HA tag with the nominal CBP amino acid sequences. Plasmid pPL609a was constructed by subcloning a BamHI fragment of mouse CBP containing a termination codon at nucleotide 3855 into pRTS2 and was used for subsequent C-terminal deletion constructs. CBP 1–1098 was created by digesting pPL609a with XbaI and religating to express aa 1 to 1098 of CBP. CBP 1–499 was constructed by digesting pPL609A with PstI and religating to express aa 1 to 499 of CBP. CBP 1–312 was constructed by digesting pPL609a with EcoRV and HindIII, blunt ended with Klenow DNA polymerase, and religated to express aa 1 to 312 of CBP. CBP 301–499 was generated by PCR with an amino-terminal HA tag and cloned with Asp718 at the 5′ end and a BamHI-BglII fusion at the 3′ end into the pRTS2 expression vector. The EBV Zta protein was expressed in transient-transfection assays from either ZtaSRα, a simian virus 40 (SV40)-based enhancer system, or Zta-pCDNA3, a cytomegalovirus-based promoter system. Plasmids pZ7E4TCAT and pZ5E4TCAT were gifts from M. Carey (9). BHLF1CAT(pDH123) and pSV2CAT have been described previously (40). 3×Dyad-TKCAT(p403.3) was a gift from J. Yates (66). The GST-CBP-containing plasmids GST(1680–1915), GST(1761–1915), and GST(1799–1915) and the eukaryotic expression vectors for CBP (1917–2441), and CBP (1430–1915) were gifts from T. Halazonetis (58). Full-length mouse RC/RSV-CBP was a gift from R. Goodman (37). The E1A wild-type and E1A Δ2–36 plasmids were gifts from Gerd Blobel (7). Expression vectors for human GCN5 and P/CAF were provided by S. Berger (8) and Y. Nakatani (68), respectively.
Transfections and CAT assays.
D98/HR1 and HeLa cells were seeded at 7 to 15% confluency in six-well plates 12 to 16 h prior to transfection. The cells were washed with Dulbecco’s phosphate-buffered saline (DPBS) and refed with DMEM 4 h prior to transfection. Plasmid effector DNA was added at 1 to 4 μg, depending upon the experiment, and the Z7E4TCAT, BHLF1CAT, 3XDyadTKCAT, and pSV2CAT target plasmids were added to 1 μg. DNA was transfected by the calcium phosphate precipitation method (11). The cells were washed with DPBS 16 to 20 h posttransfection, refed with DMEM, and harvested approximately 24 h later. Chloramphenicol acetyltransferase (CAT) assays were performed by the direct liquid scintillation assay, as previously described (10). Briefly, cell extracts were heat inactivated at 65°C to inactivate endogenous CAT activity and then pelleted to remove cellular debris. Equal amounts of extract were added along with [14C]butyryl-acetyl coenzyme A (NEN) and chloramphenicol for 2 h. Acetylated chloramphenicol was extracted with 0.5 ml of xylene and mixed with Econofluor-2 (NEN) for liquid scintillation counting. The results of CAT assays were based upon experiments performed at least in triplicate on multiple independent occasions. Expression levels of all activator proteins were monitored by Western blot analysis.
GST binding assays.
Purified GST proteins were incubated with 35S-labeled protein generated from a rabbit reticulocyte lysate-coupled transcription-translation system (Promega). Binding conditions and washings were essentially those described previously (51), except that the protein binding buffer was modified to contain 20 mM HEPES (pH 7.9), 12% glycerol, 0.5 mM EDTA, 100 mM KCl, 0.1% Nonidet P-40 (NP-40), 5 mM β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride (PMSF) (51). Bound proteins were eluted from glutathione-Sepharose 4B beads by boiling in Laemmli buffer.
Western blot analysis.
D98/HR1 and Raji cells were treated as described in the figure legends and harvested approximately 36 to 40 h after treatment. The antibodies used include EBV p52/50 anti-EA-D monoclonal antibody (Advanced Biotechnologies), anti-HA (Boehringer-Mannheim), anti-actin (Boehringer-Mannheim), and a rabbit polyclonal antibody raised against Zta. Signals were visualized by enhanced chemiluminescence (Amersham).
DNA binding assays.
Magnesium-agarose electrophoretic mobility shift assays (EMSAs) were performed as described previously (39). The probe was prepared by end labeling pZ5E4TCAT (9) at an Asp718 site with [α-32P]dATP by using Klenow polymerase and then digested with HindIII to generate the Z5E4T promoter. Zta and ΔZta (Δ2–141) were prepared as described previously (39).
Immunoprecipitation.
Raji cells were induced for lytic reactivation of EBV with 3 mM sodium butyrate and 100 ng of tetradecanoyl phorbol acetate (TPA) per ml. The cells were harvested 36 to 40 h after treatment, washed with DPBS, and resuspended in lysis buffer (20 mM NaH2PO4, 150 mM NaCl, 5 mM MgCl2, 0.1% NP-40, 1 mM PMSF, 1 mM dithiothreitol, 1 μg of pepstatin per ml, 1 μg of leupeptin per ml). They were incubated on ice for 30 min, lysed with a Dounce homogenizer, pelleted, and sonicated with a Misonix sonicator microtip on setting 5. After the debris was pelleted, extracts from lysed and sonicated cell pellets were combined and precleared with protein A-Sepharose CL4B beads. Approximately 107 cell equivalents were used for each immunoprecipitation with anti-CBP-NT and CBP-CT antibodies (Upstate Biotechnology Inc.) coupled to protein A-Sepharose CL4B beads or to control protein A-Sepharose beads and incubated for 1 h. The immunoprecipitates were washed five times with lysis buffer, and bound proteins were eluted with 0.7 M NaCl. Western blots were probed with rabbit polyclonal anti-Zta antibody.
Preparation of HeLa- and baculovirus-derived CBP.
HeLa nuclear extracts were fractionated on a P11 phosphocellulose column. The 0.3 M KCl fraction containing CBP was loaded onto a DEAE-Sephacel column, and the 0.5 M KCl fraction containing CBP was further fractionated on Q-Sepharose with a linear gradient of 100 to 600 mM KCl. The fractions containing CBP (at 200 mM KCl) were dialyzed to 10 mM potassium phosphate and loaded onto a hydoxylapatite column, which was eluted with a gradient of 10 to 500 mM potassium phosphate. The CBP eluted at 400 mM potassium phosphate and was estimated to be 500-fold enriched from nuclear extracts. Silver-staining analysis indicated that CBP was the most abundant polypeptide in this fraction. For expression of recombinant CBP (rCBP) in baculovirus, the N-terminal HA-tagged cDNA encoding mouse CBP was inserted into baculovirus expression vector pVL1392 (Pharmingen) and used to transfect Sf9 cells. Baculovirus expressing HA-CBP was amplified and used to infect Sf9 cells for 72 h. Nuclear extracts produced by Dounce homogenization were incubated with 12CA5-conjugated protein A-Sepharose (Pharmacia) overnight at 4°C. The mixture was washed three times with 20 mM HEPES–20% glycerol–400 mM NaCl–0.05% NP-40, 1 mM dithiothreitol–1 mM PMSF, and HA-CBP was eluted with 12CA5 specific peptide (1 mg/ml) in D100 buffer (20 mM HEPES, 20% glycerol, 0.2 mM EDTA, 1 mM dithiothreitol, 1 mM PMSF, 100 mM KCl).
RESULTS
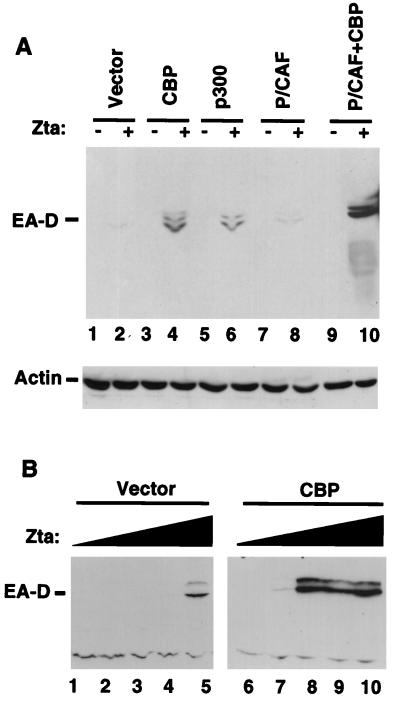
To pursue the possibility that HAT-containing coactivators participate in viral reactivation, we tested the ability of various known HATs to stimulate viral reactivation by themselves or in association with Zta overexpression. Latently infected D98/HR1 cells were transfected with CBP, p300, or P/CAF in the presence or absence of Zta and assayed for viral reactivation by Western blot analysis with antibody directed against the viral early antigen, EA-D. EA-D consists of the product of the BMRF1 open reading frame, which is transcriptionally activated by the binding of Zta to its promoter. Under these conditions, Zta by itself had a weak stimulatory effect on EA-D expression (Fig. 1A, lane 2). Transfection of CBP, p300, or P/CAF had no effect on reactivation alone (lanes 3, 5, and 7). However, cotransfection of Zta with CBP or with p300 resulted in significant expression of EA-D (lanes 4 and 6). P/CAF did not coactivate Zta-mediated expression of EA-D. However, coexpression of P/CAF with CBP further increased the levels of EA-D expression in the presence of Zta, supporting models that P/CAF functions in association with CBP.
FIG. 1.
CBP-p300 stimulates Zta reactivation of latent EBV. (A) Latently infected D98/HR1 cells were transfected with (+) or without (−) 1 μg of Zta plus 4 μg of pRTS2 vector (lanes 1 and 2) or 4 μg of various HAT-containing coactivators, CBP (lanes 3 and 4), p300 (lanes 5 and 6), P/CAF (lanes 7 and 8), or both CBP and P/CAF (lanes 9 and 10). Transfected cell extracts were analyzed for viral lytic antigen EA-D by Western blot analysis. Identical extracts were examined for actin expression by Western blot analysis (bottom). (B) D98/HR1 cells were transfected with increasing concentrations of Zta in the presence or absence of 4 μg of CBP. Zta was transfected at 0.1 μg (lanes 1 and 6), 0.5 μg (lanes 2 and 7), 1.0 μg (lane 3 and 8), 2.0 μg (lanes 4 and 9), or 4.0 μg (lanes 5 and 10). EA-D expression was assayed by Western blotting 48 h posttransfection.
The ability of CBP to enhance Zta-mediated reactivation was further characterized by titrating the amount of Zta expression plasmid required to stimulate EA-D expression (Fig. 1B). In the absence of CBP, EA-D expression was detected only when 4 μg of Zta was transfected (Fig. 1B, lane 5). However, in the presence of 4 μg of CBP, higher levels of EA-D were expressed at 1, 2, and 4 μg of Zta expression plasmid (lanes 8 to 10). Quantitation of plasmid-derived Zta in these cells is complicated by the reactivation of the endogenous virus-encoded Zta, but similar titrations and cotransfections in EBV-negative HeLa cells indicated that Zta levels increased proportionately to transfected plasmid DNA and that CBP cotransfection had no significant effect on plasmid expression levels (see Fig. 6C and 7B). Furthermore, similar observations were made whether Zta was expressed from SV40- or cytomegalovirus-based expression vectors (data not shown). These results suggest that CBP can enhance the ability of Zta to stimulate EA-D expression from cells latently infected with EBV.
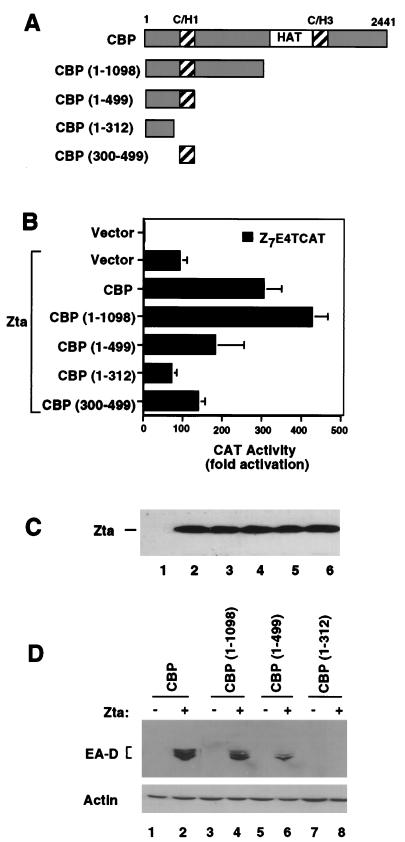
FIG. 6.
The amino-terminal C/H1 domain of CBP is required for coactivation with Zta. (A) Schematic of CBP deletion mutants used to map the amino-terminal coactivation function of CBP. (B) HeLa cells were transfected with Z7E4TCAT (1 μg) and either pRTS2 vector (1 μg) or Zta expression plasmid (1 μg). Expression plasmids for full-length CBP or the amino-terminal fragments CBP (1–1098), CBP (1–499), CBP (1–312), or CBP (300–499) were cotransfected (4 μg of each) and assayed for CAT activity 48 h posttransfection. CAT activity is reported as fold activation above that for vector alone. (C) Extracts derived from transfected HeLa cells described in panel B were assayed for levels of Zta expression by Western blot analysis. Zta is expressed in all samples except lane 1. Levels of Zta were measured when cotransfected with pRTS2 vector (lane 2), full-length CBP (lane 3), CBP (1–1098) (lane 4), CBP (1–499) (lane 5), and CBP (1–312) (lane 6). (D) D98/HR1 cells were assayed for the ability of CBP N-terminal fragments to stimulate EA-D expression. The cells were transfected with 1 μg of Zta expression plasmid (+) or pRTS2 vector (−) and cotransfected with 4 μg of either CBP, CBP (1–1098), CBP(1–499), or CBP (1–312) as indicated above the lanes. The cells were analyzed by Western blotting 48 h posttransfection for the expression of EA-D (top) or for actin (bottom).
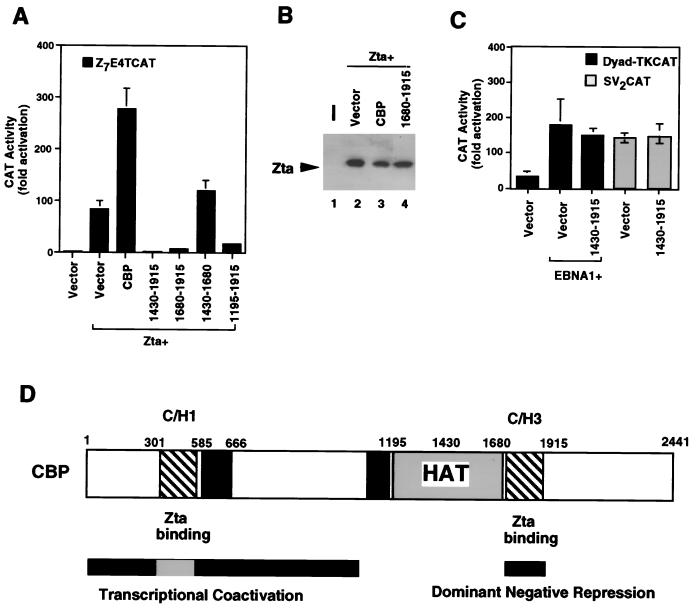
FIG. 7.
Dominant negative inhibition of Zta transcription activation by the CBP C/H3 domain. HeLa cells were cotransfected with the Z7E4TCAT reporter and either vector or Zta expression vector (as indicated). Zta-transfected cells were cotransfected with 4 μg of either vector or expression plasmids for CBP, CBP (1430–1915), CBP (1680–1915), CBP (1430–1680), or CBP (1195–1915). CAT activity is presented as fold activation above that for vector alone. (B) Expression levels of Zta in cotransfected cell extracts were measured by Western blotting. Zta levels are shown with pRTS2 vector (lane 2), CBP (lane 3), or CBP (1680–1915) (lane 4). (C) The CBP C/H3 domain did not inhibit EBNA1 or pSV2CAT activity. HeLa cells were cotransfected with the EBNA1 and the 3×Dyad-TKCAT reporter in the presence of 4 μg of pRTS2 vector or CBP (1430–1915) expression plasmid. Similarly, pSV2CAT was cotransfected with 4 μg of pRTS2 vector or CBP (1430–1915). CAT activity is presented as units above that for vector alone. (D) Summary schematic of CBP domains which bind Zta and either coactivate or function as a dominant negative repressor in cotransfection assays.
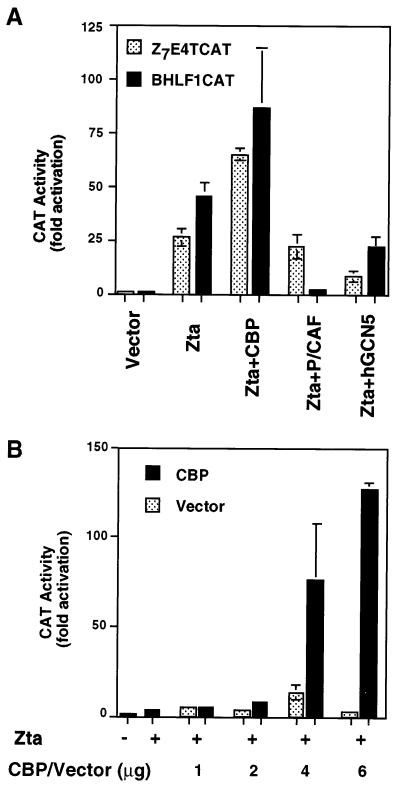
CBP was next examined for its ability to costimulate the transcriptional activation function of Zta from a reporter plasmid in an EBV-negative cell line. HeLa cells were cotransfected with Zta and either CBP, P/CAF, or hGCN5 for the ability to stimulate transcription from Zta-responsive reporter plasmids, Z7E4TCAT or BHLF1CAT. BHLF1 is an EBV-derived promoter that contains four ZREs interspersed with several cellular activator binding sites. Z7E4T is a synthetic promoter with seven ZREs upstream of the adenovirus E4 TATA and initiator element, and it is strictly dependent on Zta for its activation. We found that CBP stimulated Zta activation of Zta-responsive promoters whereas P/CAF and hGCN5 were largely inhibitory in this assay (Fig. 2A). While CBP typically amplified Zta transcription activation 2.5- to 5-fold, we were able to identify optimal conditions where CBP could stimulate transcription levels as high as 12-fold above those of Zta alone (Fig. 2B). These results suggest that CBP can be made limiting for transcriptional activation by Zta and that cellular conditions may contribute to the relative availability of CBP.
FIG. 2.
CBP coactivates Zta transcription activation in EBV-negative cells. (A) HeLa cells were cotransfected with 1 μg of the reporter plasmid Z7E4TCAT (shaded) or BHLF1CAT (black) and 1 μg of Zta or pRTS2 vector control. Coactivator expression vectors (1 μg) CBP, P/CAF, or hGCN5 were cotransfected as indicated. CAT activity is reported as fold activation above that due to vector. (B) Titration of CBP or pRTS2 vector reveals the optimal conditions for coactivation. Zta (1 μg) was cotransfected with 1, 2, 4, or 6 μg of either CBP (black) or pRTS2 vector (shaded) as indicated.
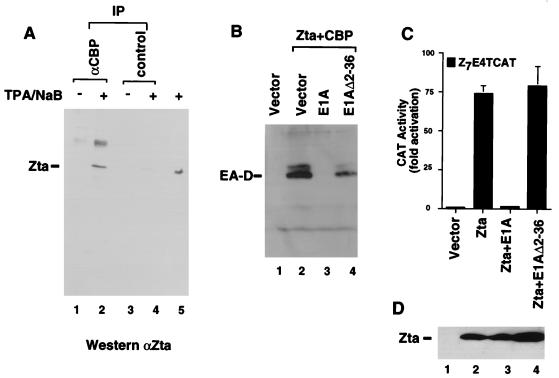
The ability of CBP to coactivate transcription by an activator typically correlates with the formation of a stable complex between CBP and the activator. To determine if Zta and CBP form a stable complex in vivo, extracts were derived from latently infected Raji cells or from Raji cells treated with TPA and sodium butyrate to stimulate EBV reactivation. The extracts were subjected to immunoprecipitation with polyclonal antibody directed against CBP or with control protein A-Sepharose beads. Immunoprecipitated complexes were then eluted with 0.7 M NaCl, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and immunoblotted with a polyclonal antibody directed against Zta. We found that Zta could be detected in CBP-specific immunoprecipitates derived from treated extracts (Fig. 3A, lane 2) but not from untreated extracts (lane 1) or from control immunoprecipitates (lanes 3 and 4). This suggests that Zta forms a stable complex with CBP in latently infected B cells that had been reactivated by TPA and sodium butyrate treatment. The relative amount of Zta that could be coimmunoprecipitated with CBP was a small fraction of the input Zta (less than 5%), suggesting that only a fraction of Zta is complexed with CBP or that a Zta-CBP complex is only partly stable throughout the immunoprecipitation reaction.
FIG. 3.
Physical and functional association between CBP and Zta in vivo. (A) Nuclear extracts derived from untreated (−) or TPA- plus-sodium butyrate (NaB)-treated (+) Raji cells were subjected to immunoprecipitation (IP) with two antibodies specific for CBP (αCBP) or with protein A-Sepharose beads (control). Immunoprecipitates were eluted with 0.7 M NaCl and analyzed by Western blotting with anti-Zta-specific serum (αZta). The induced cell input (5%) is shown in lane 5. The 33-kDa Zta protein is indicated. (B) E1A inhibits Zta-plus-CBP coactivation of latent EBV. D98/HR1 cells were transfected with vector (lane 1) or with Zta (1 μg) plus CBP (4 μg) (lanes 2 to 4). The specificity of CBP was analyzed by the cotransfection of 4 μg of either the E1A 12S expression vector (lane 3) or the N-terminal deletion mutant of E1A (Δ2–36) (lane 4). EA-D expression was detected by Western blotting. (C) E1A inhibits Zta transcriptional activation in HeLa cells. The Z7E4TCAT reporter plasmid (1 μg) was cotransfected with 1 μg of either pRTS2 (Vector) or with Zta alone, Zta plus E1A 12S, or Zta plus E1A (Δ2–36). CAT activity is reported as fold activation above vector alone. (D) Extracts derived from transfected cells in panel C were examined by Western blotting for expression levels of Zta.
To further demonstrate a role of CBP in mediating Zta transcriptional activation and viral reactivation, we tested whether the adenovirus E1A 12S protein could specifically inhibit Zta activity (Fig. 3B). E1A 12S binds directly to CBP-p300 and inhibits its function in transcription coactivation for multiple activators. To determine if E1A inhibited viral reactivation by Zta, we cotransfected D98/HR1 cells with the expression plasmids for Zta, CBP, and E1A and assayed them for EA-D expression. As shown above, cotransfection of Zta and CBP stimulates EA-D expression from latent EBV in D98/HR1 cells (Fig. 3B, lane 2). Cotransfection of E1A 12S with Zta and CBP completely eliminated EA-D expression (lane 3). An amino-terminal deletion mutant of E1A (Δ2–36), which is defective for binding CBP, had only a minor inhibitory effect on reactivation by Zta plus CBP (lane 4). Thus, E1A 12S inhibits Zta- and CBP-mediated reactivation of latent EBV, and this inhibition is dependent on the ability of E1A to bind CBP.
To further explore the role of CBP in mediating Zta transcriptional activation function, we tested the effect of E1A on the ability of Zta to activate a reporter plasmid in EBV-negative HeLa cells. In these experiments, Zta stimulated Z7E4TCAT almost 70-fold (Fig. 3C, lane 2). Cotransfection of E1A 12S protein resulted in a substantial reduction of Zta to less than threefold activation (lane 3). Cotransfection of the E1A (Δ2–36) deletion mutant had no effect on Zta transcription activation function, suggesting that the inhibition by E1A was largely CBP dependent (lane 4). The expression levels of Zta protein were examined by Western blotting and found not to vary significantly by cotransfection of E1A or E1A (Δ2–36) (Fig. 3D). Thus, E1A does not reduce Zta protein abundance and therefore most probably inhibits Zta transcription activation function.
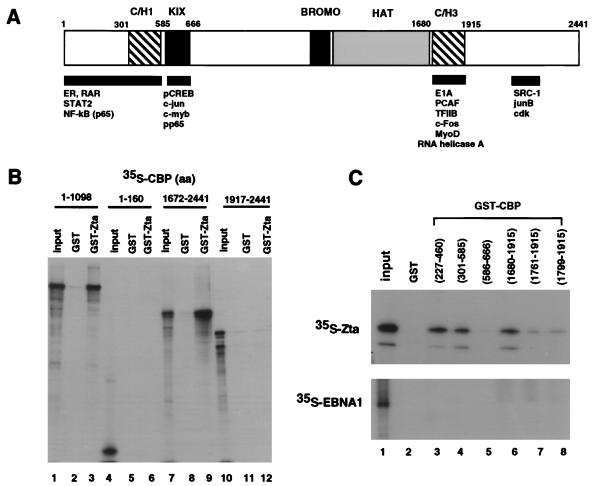
We next tested the ability of Zta to bind directly to CBP in vitro by using the GST binding assay. We found that GST-Zta was capable of interacting with two independent domains of CBP with relatively similar affinity (Fig. 4B). Zta bound to an amino-terminal fragment of CBP (aa 1 to 1098) but did not bind to a smaller amino-terminal fragment (aa 1 to 160), which has been reported to interact with nuclear hormone receptors. Similarly, Zta bound to a C-terminal fragment (aa 1672 to 2441) but not to a smaller fragment (aa 1917 to 2441), which contains the SRC-1 binding domain (Fig. 4B). Further mapping of the interaction by using previously characterized domains of CBP fused to GST revealed that Zta bound to a small N-terminal fragment and a small C-terminal fragment (Fig. 4C, top). Zta bound to residues 227 to 460, referred to as the CBP N-terminal activation domain (NTAD). It also bound to partially overlapping residues 301 to 585, which contains cysteine- and histidine-rich region 1 (C/H1). Zta did not bind to residues 585 to 666, the phospho-CREB binding domain (KIX). Zta also bound to the C-terminal C/H3, encompassing aa 1680 to 1915, with some residual binding to aa 1761 to 1915 and aa 1799 to 1915, which partially truncate the C/H3 domain. These GST-CBP fusion proteins did not interact with EBNA1, an EBV-encoded replication protein, suggesting that interaction with Zta is relatively specific (Fig. 4C, bottom). These results indicate that Zta can bind to two independent but related regions of CBP and that the homologous C/H1 and C/H3 domains are the likely interaction sites.
FIG. 4.
Zta interacts with two separate domains of CBP in vitro. (A) Schematic of CBP depicting sequence motifs and some previously characterized interaction domains. (B) GST-Zta was tested for the ability to bind to in vitro-translated fragments of CBP. CBP fragments 1 to 1098 (lanes 1 to 3), 1 to 160 (lanes 4 to 6), 1672 to 2441 (lanes 7 to 9), or 1917 to 2441 (lanes 10 to 12) were labeled with [35S]methionine and tested for the ability to bind GST or GST-Zta, as indicated. Input represents 10% of the total [35S]CBP fragment used in the binding reactions. (C) GST-CBP fragments bind to in vitro-translated Zta but not EBNA1 in vitro. GST-CBP fragments spanning aa 227 to 460 (lane 3), 301 to 585 (lane 4), 585 to 666 (lane 5), 1680 to 1915 (lane 6), 1761 to 1915 (lane 7), or 1799 to 1915 (lane 8) were tested for binding 35S-labeled Zta (top panel) or EBNA1 (bottom panel). Input represents 10% of the total 35S-labeled protein used in the initial binding reaction.
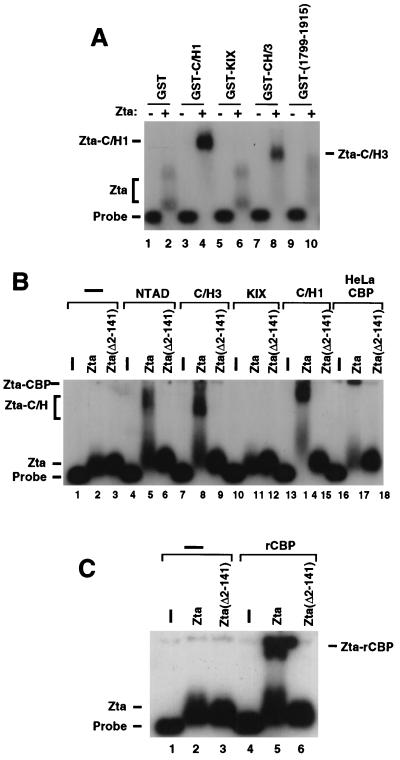
To determine if Zta can bind to CBP in the context of promoter DNA, we tested the ability of the CBP fragments to alter the mobility of Zta in EMSA. Previously, we had found that large multiprotein complexes were best resolved in magnesium-agarose gels. Incubation of Zta with a DNA probe containing five ZREs produced a small shift in this gel system (Fig. 5A, lane 2). The addition of the C/H1 domain produced a stable complex with different mobility relative to Zta alone (lane 4). Similarly, the C/H3 domain also formed a complex with Zta (lane 8), while the KIX domain (aa 586 to 666) (lane 6) and the C/H3 deletion (aa 1799 to 1915) (lane 8) did not produce stable shifts with Zta in these assays. These results clearly show that CBP subdomains can interact with Zta when bound to promoter DNA.
FIG. 5.
CBP binds Zta complexed to promoter DNA. (A) The Z5E4T promoter probe was bound by Zta (even lanes) in the presence of purified GST (lanes 1 and 2), GST-C/H1 (lanes 3 and 4), GST-KIX (lanes 5 and 6), GST-C/H3 (lanes 7 and 8), or GST-(1799–1915) (lanes 9 and 10). Bound complexes were resolved in magnesium-agarose gels. (B) The Zta activation domain is required for CBP binding. Full-length Zta (lanes 2, 5, 8, 11, 14, and 17) and an amino-terminal deletion, Zta (Δ2–141) (lanes 3, 6, 9, 12, 15, and 18), were compared for their ability to bind to CBP fragments in magnesium-agarose EMSA, as in panel A. GST-NTAD, GST-C/H1, GST-KIX, GST-C/H3, and partially purified HeLa-derived CBP (CBP) are indicated above their respective lanes. (C) HA-tagged rCBP expressed and immunoaffinity purified from baculovirus-infected Sf9 cells was tested for its ability to interact with Zta (lane 5) or Zta (Δ2–141) (lane 6) in magnesium-agarose EMSA.
The interactions of the CBP subdomains were tested for their dependence on the Zta activation domain. The magnesium-agarose EMSA was used to compare the ability of CBP subdomains to bind to either full-length Zta or an amino-terminal deletion mutant of Zta (Δ2–141), which completely eliminates the Zta activation domain (Fig. 5B). We found that the interactions between Zta and the subdomains, NTAD (aa 227 to 460), C/H1 (aa 310 to 585), and C/H3 (aa 1680 to 1915), were absolutely dependent on the Zta activation domain (Fig. 5B, lanes 5, 8, and 14). Again, we did not observe any interaction between Zta and the KIX domain, which interacts specifically with phosphorylated CREB and NF-κB. Importantly, we found that a partially purified fraction of human CBP also interacted specifically with Zta in an activation domain-specific manner (lane 17). To further verify that full-length CBP could interact with Zta, immunopurified HA-tagged CBP was generated from a baculovirus vector and assayed for an association with Zta in EMSA (Fig. 5C). rCBP had no effect on the DNA probe mobility by itself (Fig. 5C, lane 4). rCBP formed a complex with full-length Zta (lane 5) that was largely retained in the loading well, similar to that observed for the HeLa-derived CBP (Fig. 5B, lane 17). In contrast, rCBP had no effect on Zta(Δ2–141) (Fig. 5C, lane 6), further indicating that the activation domain of Zta is required for the stable interaction of Zta with full-length CBP.
We next compared the ability of the various subdomains of CBP to affect Zta function in transcriptional activation assays. Interestingly, we found that amino-terminal fragments of CBP which retain the ability to interact with Zta were capable of stimulating Zta transcriptional activation in CAT assays and in viral reactivation assays (Fig. 6). In transfection assays with HeLa cells, full-length CBP cotransfection produced an additional ca. threefold stimulation of Z7E4TCAT over activation by Zta alone (Fig. 6B). The N-terminal CBP fragment (aa 1 to 1098) produced ∼4.5-fold stimulation of Zta activation on this promoter. A fragment expressing aa 1 to 499 also activated transcription, although not as efficiently as full-length CBP, while a smaller fragment (aa 1 to 312), which lacks the C/H1 domain, eliminated all of the coactivation function. The C/H1 domain by itself produced a small but reproducible stimulation of Zta activation, suggesting that some of the Zta coactivation function resides in this small domain of CBP. Levels of Zta protein were not affected by cotransfection of CBP, indicating that CBP influenced Zta transcription activation function (Fig. 6C). A similar pattern of activation by the N-terminal fragments of CBP was found for reactivation of the virus, as measured by the expression of EA-D. Full-length CBP and CBP (1–1098) stimulated EA-D expression to near equal levels (Fig. 6D, lanes 2 and 4). CBP (1–499) stimulated EA-D expression, but the expression was reduced relative to that for full-length CBP (lane 6). In contrast, CBP (1–312) had no effect on EA-D expression (lane 8). Cotransfection of CBP with Zta had no effect on cellular actin levels, indicating that coactivation was relatively specific for Zta-responsive genes (Fig. 6D, bottom). Again, these results suggest that interaction of Zta with the amino-terminal half of CBP containing the C/H1 domain is important for transcriptional activation and reactivation of latent viral genes.
We next examined the effect of the C/H3 domain on the ability of Zta to activate transcription. In contrast to the C/H1 domain, we found that overexpression of the C/H3 domain resulted in a strong repression of Zta transcriptional activation function (Fig. 7). Again, full-length CBP produced about a 3.5-fold stimulation of Zta activation from the Z7E4TCAT reporter (Fig. 7A). Cotransfection of the C/H3-containing fragments from aa 1430 to 1915 or 1680 to 1915 resulted in a strong inhibition of Zta activation function. Cotransfection of a region outside of the C/H3 domain, aa 1430 to 1680, had no significant effect on Zta transcription activation. A fragment containing a functional HAT domain in association with the C/H3 domain (aa 1195 to 1915) was also inhibitory for Zta, although somewhat less so than were the small C/H3-containing fragments. Again, the Zta expression levels were shown not to be significantly affected by cotransfection of the CBP or C/H3 domain (Fig. 7B). The inhibitory activity of the C/H3 domain was found to be relatively specific for Zta, since it did not inhibit EBNA1 transactivation or the SV40 enhancer expression in pSV2CAT (Fig. 7C). These results indicate that the C/H3 domain, in isolation from the intact CBP, can function as a potent inhibitor of Zta transcriptional activation function.
DISCUSSION
The cellular coactivators CBP and p300 have been implicated in the transcriptional function of multiple transcription factors. In this work, we show that CBP enhances Zta transcriptional activation and reactivation of lytic EBV gene expression (Fig. 1 and 2). Zta and CBP formed a stable complex that could be immunoprecipitated from EBV-positive B lymphocytes chemically treated to stimulate viral reactivation (Fig. 3A). Zta bound directly to two distinct regions of CBP in vitro, referred to as C/H1 and C/H3 (Fig. 4 and 5). These domains have significant sequence similarity to each other and to the yeast coactivator, ADA2 (6). The interaction of C/H1 and C/H3 with Zta was dependent upon the amino-terminal transcriptional activation domain of Zta, and this interaction could be measured when Zta was bound to functional ZREs in promoter DNA (Fig. 5B). Additionally, full-length CBP partially purified from HeLa cells or as a recombinant protein expressed in baculovirus was capable of interacting with Zta in an activation domain-dependent manner. Furthermore, the transcriptional activation properties of Zta were significantly reduced by coexpression of the adenovirus E1A 12S protein, which binds the C/H3 domain of CBP (Fig. 3B), or by coexpression of the CBP C/H3 domain itself, which binds avidly to the Zta activation domain (Fig. 7). Taken together, these results strongly support a role for CBP in mediating transcriptional activation functions of Zta.
HAT-independent coactivation by CBP.
In latently infected B lymphocytes, EBV gene expression is packaged in chromatin, which is likely to contribute to the repression of lytic gene expression (18, 59). Viral reactivation can be stimulated by addition of sodium butyrate, a pleiotropic agent which is a potent inhibitor of histone deacetylases, suggesting that chromatin and histone acetylation may regulate EBV reactivation (35, 56). Thus, it seems likely that HATs should promote viral reactivation. Consistent with this hypothesis, we have found that the CBP-p300 family of HAT-containing proteins costimulates viral reactivation by Zta. Ironically, we also found that the HAT domain of CBP was dispensible for costimulation of Zta transcriptional activation and viral reactivation (Fig. 6). This suggests that CBP stimulates Zta transcription activation by a HAT-independent mechanism. CBP functions as a bridging factor for CREB, by mediating an association between CREB and holo-RNA polymerase II (49). The interaction of CBP with RNA polymerase II is further mediated by RNA helicase A, which binds to the C/H3 domain of CBP (49). However, we found that the C/H3 domain of CBP was also dispensable for coactivation of Zta. A similar observation that the amino-terminal region of CBP-p300 was sufficient for transcription coactivation with CREB or NF-κB has also been reported (28, 63). Thus, it is possible that CBP costimulates Zta transcription by a mechanism distinct from histone acetylation or C/H3-dependent recruitment of holo-RNA polymerase II.
Mechanism of CBP coactivation.
CBP binding to CREB is essential but not sufficient for transcriptional activation. CREB activation requires the function of a second activation domain that binds to the TAF135 component of TFIID (49). Transcriptional activation by CREB has been proposed to function by the dual recruitment of TFIID and holo-RNA polymerase II through interactions with TAF135 and CBP, respectively. Like CREB, Zta also targets TFIID by stimulating the recruitment of a TFIID-TFIIA complex onto promoter DNA (39). Zta induces a conformational change in the TAFs of the TFIID complex, which results in an increase binding to promoter sequences near and downstream of the transcriptional initiation site. At present, we have no evidence to suggest that CBP further stimulates the association of Zta with TFIIA or TFIID. However, CBP interacts stably with TBP in coimmunoprecipitation reactions and in vitro binding reactions (1, 63, 69). Thus, it is possible that CBP further modifies the association of Zta with TBP, TFIIA, and TAFs. Zta was also shown to require preincubation with several additional activities to produce high-level transcription activation in vitro (38). These additional activities include TFIIB and a crude coactivator fraction containing USA components. While the bulk of cellular CBP is excluded from these fractions, it is possible that a fraction of CBP is present in the crude coactivator fraction or as a substoichiometric component in our HeLa-derived holo-TFIID (data not shown). Future biochemical characterization of CBP in transcription reactions may help to determine if CBP has these additional functions in preinitiation complex assembly.
Functional similarity between C/H1 and C/H3.
CBP binds to multiple transcriptional factors through several distinct domains. We have found that Zta binds to the C/H1- and C/H3-containing domains of CBP with near equal affinity. The C/H1 and C/H3 domains of CBP have significant sequence similarity to each other and are likely to share some biological functions. C/H3 binds to many factors characterized to interact with CBP, including p53, E1A, P/CAF, and RNA helicase A (26, 43, 45, 48, 68). While the C/H1 and C/H3 domains bind Zta with equal affinity, our transfection data indicate that they have distinct functional properties. We found that overexpression of the C/H3 domain inhibited Zta transcriptional activation function (Fig. 7). Presumably, the association of C/H3 with Zta prevents the functional association of Zta with the appropriate full-length target of CBP. Alternatively, overexpression of C/H3 may inhibit other CBP-associated components, like P/CAF, from forming a stable complex essential for mediating Zta transcriptional functions (28). In contrast, overexpression of CBP amino-terminal fragments containing the C/H1 domain stimulated Zta transcription activation. At least two activation domains of CBP have been mapped by fusion of CBP fragments to a heterologous DNA binding domain in transient-transfection assays (63). The NTAD overlaps the C/H1 interaction domain and is included within our mapping of a CBP transcriptional coactivation function for Zta. The C-terminal activation domain has no apparent effect on Zta function but may be important for relieving the dominant negative activity of C/H3 (data not shown).
CBP as a signal integrator.
The interaction of CBP with a multitude of diverse transcriptional activators has led to the proposal that CBP integrates transcription for multiple signaling pathways (60, 64). Antagonistic interactions between several different classes of activators, such as nuclear receptors and AP-1, are relieved by overexpression of CBP (30). Thus, CBP may be rate limiting for these interactions in vivo, and competition for CBP by these various activators may regulate the transcription output from various signal inputs. In this respect, it is interesting that RA represses EBV lytic reactivation and that RAR, NF-κB, and p53, which all bind CBP, inhibit Zta transcriptional activation. It is tempting to speculate that inhibition of Zta by these different activators may be regulated, in part, by competition for CBP. Future studies are required to determine if signals regulating the reactivation of EBV are integrated by competition for limiting concentrations of CBP.
ACKNOWLEDGMENTS
We thank S. Berger, G. Blobel, R. Goodman, T. Halazonetis, S. Kenney, and Y. Nakatani for generously providing plasmids and reagents.
This work was supported by grants from the NIH (GM54687-02) and the Leukemia Society of America. D.Z. was supported by NIH Training Program in Cancer Research (CA09171).
REFERENCES
- 1.Abraham S E, Lobo S, Yaciuk P, Wang H-G H, Moran E. p300, and p300-associated proteins, are components of TATA-binding protein (TBP) complexes. Oncogene. 1993;8:1639–1647. [PubMed] [Google Scholar]
- 2.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 3.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 4.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinbagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Sasson S A, Klein G. Activation of the Epstein-Barr virus genome by 5-aza-cytidine in latently infected human lymphoid lines. Int J Cancer. 1981;28:131–135. doi: 10.1002/ijc.2910280204. [DOI] [PubMed] [Google Scholar]
- 6.Berger S L, Pina B, Silverman N, Marcus G A, Agapite J, Regier J L, Triezenberg S J, Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992;70:252–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 7.Blobel G A, Nakajima T, Eckner R, Montminy M, Orkin S H. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Candau R, Moore P A, Wang L, Barlev N, Ying C Y, Rosen C A, Berger S L. Identification of human proteins functionally conserved with the yeast putative adapter ADA2 and GCN5. Mol Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carey M, Kolman J, Katz D A, Gradoville L, Barberis L, Miller G. Transcriptional synergy by the Epstein-Barr virus transactivator ZEBRA. J Virol. 1992;66:4803–4813. doi: 10.1128/jvi.66.8.4803-4813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cassinoti P, Weitz M. Increasing the sensitivity of a common CAT assay. BioTechniques. 1994;17:36–39. [PubMed] [Google Scholar]
- 11.Chen C, Okayama H. High-efficiency transfection of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, Lin R J, Schlitz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 13.Chevallier G A, Manet E, Chavrier P, Mosnier C, Daillie J, Sergeant A. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 1986;5:3243–3249. doi: 10.1002/j.1460-2075.1986.tb04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chi T, Carey M. Assembly of the isomerized TFIIA-TFIID-TATA ternary complex is necessary and sufficient for gene activation. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- 15.Chi T, Lieberman P, Ellwood K, Carey M. A general mechanism for transcriptional synergy by eukaryotic activators. Nature. 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- 16.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 17.Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyson P J, Farrell P J. Chromatin structure of Epstein-Barr virus. J Gen Virol. 1985;66:1931–1940. doi: 10.1099/0022-1317-66-9-1931. [DOI] [PubMed] [Google Scholar]
- 19.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300 kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 20.Faggioni A, Zompetta C, Grimaldi S, Barile G, Frati L, Lazdins J. Calcium modulation activates Epstein-Barr virus genome in latently infected cells. Science. 1986;232:1554–1556. doi: 10.1126/science.3012779. [DOI] [PubMed] [Google Scholar]
- 21.Farrell P J, Rowe D T, Rooney C M, Kouzarides T. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 1989;8:127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giles R H, Peters D J M, Bruening M H. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 23.Glaser R, Nonoyama M. Host cell regulation of induction of Epstein-Barr virus. J Virol. 1974;14:174–176. doi: 10.1128/jvi.14.1.174-176.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 25.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 26.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 27.Gutsch D E, Holley-Guthrie E A, Zhang Q, Stein B, Blanar M A, Baldwin A S, Kenney S C. The bZIP transactivator of Epstein-Barr virus, BZLF1, functionally and physically interacts with the p65 subunit of NF-κB. Mol Cell Biol. 1994;14:1939–1948. doi: 10.1128/mcb.14.3.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hottinger M O, Felzien L K, Nabel G J. Modulation of cytokine-induced HIV gene expression by competitive binding of transcription factors to the coactivator p300. EMBO J. 1998;17:3124–3134. doi: 10.1093/emboj/17.11.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janknecht R, Hunter T. Transcription. A growing coactivator network. Nature. 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- 30.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 31.Kenney S, Kamine J, Holley-Guthrie E, Lin J-C, Mar E-C, Pagano J. The Epstein-Barr virus (EBV) BZLF1 immediate-early gene product differentially affects latent versus productive EBV promoters. J Virol. 1988;63:1729–1736. doi: 10.1128/jvi.63.4.1729-1736.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2343–2396. [Google Scholar]
- 33.Kobayashi N, Boyer T G, Berk A J. A class of activation domains interacts directly with TFIIA and stimulates TFIIA-TFIID-promoter complex assembly. Mol Cell Biol. 1995;15:6465–6473. doi: 10.1128/mcb.15.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 35.Kruh J. Effects of sodium butyrate, a new pharmacological agent, on cells in culture. Mol Cell Biochem. 1982;42:65–82. doi: 10.1007/BF00222695. [DOI] [PubMed] [Google Scholar]
- 36.Kurokawa R, Kalafus D, Ogliastro M H, Kioussi C, Xu L, Torchia J, Rosenfeld M G, Glass C K. Differential use of CREB binding protein-coactivator complexes. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 37.Kwok R J, Lundblad J, Chrivia J, Richards J, Bachinger H, Brennan R, Roberts S, Green M, Goodman R. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 38.Lieberman P. Identification of functional targets of the Zta transcriptional activator by formation of stable preinitiation complex intermediates. Mol Cell Biol. 1994;14:8365–8375. doi: 10.1128/mcb.14.12.8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieberman P M, Berk A J. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA-promoter DNA complex formation. Genes Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 40.Lieberman P M, Hardwick J M, Hayward S D. Responsiveness of the Epstein-Barr virus NotI repeat promoter to the Z transactivator is mediated in a cell-type-specific manner by two independent signal regions. J Virol. 1989;63:3040–3050. doi: 10.1128/jvi.63.7.3040-3050.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lieberman P M, Hardwick J M, Sample J, Hayward G S, Hayward S D. The zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J Virol. 1990;64:1143–1155. doi: 10.1128/jvi.64.3.1143-1155.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lieberman P M, Ozer J, Gursel D B. Requirement for TFIIA-TFIID recruitment by an activator depends on promoter structure and template competition. Mol Cell Biol. 1997;17:6624–6632. doi: 10.1128/mcb.17.11.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 44.Lin J C, Smith M C, Pagano J S. Induction of replication of Epstein-Barr virus DNA by 12-O-tetradecanoyl-phorbol-13-acetate. II. Inhibition by retinoic acid and 9-(2-hydroxyethoxymethyl) guanine. Virology. 1981;111:294–298. doi: 10.1016/0042-6822(81)90675-9. [DOI] [PubMed] [Google Scholar]
- 45.Lundblad J R, Kwok R P, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 46.Martinez-Balbas M A, Bannister A J, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller G. The switch between latency and replication of Epstein-Barr virus. J Infect Dis. 1990;161:833–844. doi: 10.1093/infdis/161.5.833. [DOI] [PubMed] [Google Scholar]
- 48.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 49.Nakajima T, Uchida C, Anderson S F, Parvin J D, Montminy M. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 50.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 51.Ozer J, Moore P A, Bolden A H, Lee A, Rosen C A, Lieberman P M. Molecular cloning of the small (gamma) subunit of human TFIIA reveals functions critical for activated transcription. Genes Dev. 1994;8:2324–2335. doi: 10.1101/gad.8.19.2324. [DOI] [PubMed] [Google Scholar]
- 52.Parker D, Ferreri K, Nakajima T, LaMorte V J, Evans R, Koerber S C, Hoeger C, Montminy M R. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol Cell Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 54.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 55.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 56.Saemundsen A K, Kallin B, Klein G. Effect of n-butyrate on cellular and viral DNA synthesis in cells latently infected with Epstein-Barr virus. Virology. 1980;107:557–561. doi: 10.1016/0042-6822(80)90326-8. [DOI] [PubMed] [Google Scholar]
- 57.Sarisky R T, Gao Z, Lieberman P M, Fixman E D, Hayward G S, Hayward S D. Evidence for a replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J Virol. 1996;70:8340–8347. doi: 10.1128/jvi.70.12.8340-8347.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scolnick D M, Chehab N H, Stavridi E S, Lien M C, Caruso L, Moran E, Berger S L, Halazonetis T D. CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 1997;57:3693–3696. [PubMed] [Google Scholar]
- 59.Shaw J E, Levinger L F, Carter C W. Nucleosomal structure of Epstein-Barr virus DNA in transformed cell lines. J Virol. 1979;29:657–665. doi: 10.1128/jvi.29.2.657-665.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shikama N, Lyon J, La Thangue N B. The p300/CBP family: integration signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 61.Sista N D, Pagano J S, Liao W, Kenney S. Retinoic acid is a negative regulator of the Epstein-Barr virus protein (BZLF1) that mediates disruption of latent infection. Proc Natl Acad Sci USA. 1993;90:3894–3898. doi: 10.1073/pnas.90.9.3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sundar S K, Levine P H, Ablashi D V, Menezes J. Retinoic acid and steroids inhibit Epstein-Barr virus-induced nuclear antigen, DNA synthesis and lymphocyte transformation. Anticancer Res. 1984;4:415–418. [PubMed] [Google Scholar]
- 63.Swope D L, Mueller C L, Chrivia J C. CREB-binding protein activates transcription through multiple domains. J Biol Chem. 1996;271:28138–28145. doi: 10.1074/jbc.271.45.28138. [DOI] [PubMed] [Google Scholar]
- 64.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 65.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 66.Wysokenski D A, Yates J L. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J Virol. 1989;63:2657–2666. doi: 10.1128/jvi.63.6.2657-2666.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamamoto N, Bister K, zur Hausen H. Retinoic acid inhibition of Epstein-Barr virus induction. Nature. 1979;278:553–554. doi: 10.1038/278553a0. [DOI] [PubMed] [Google Scholar]
- 68.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 69.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Q, Gutsch D, Kenney S. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol Cell Biol. 1994;14:1929–1938. doi: 10.1128/mcb.14.3.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhong H, Voll R E, Ghosh S. Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with coactivator CBP/p300. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- 72.zur Hausen H, O’Neill F J, Freese U K, Hecher E. Persisting oncogenic herpesvirus induced by tumor promoter TPA. Nature. 1978;272:373–375. doi: 10.1038/272373a0. [DOI] [PubMed] [Google Scholar]