Abstract
Previous work has shown that heat shock factor (HSF) plays a central role in remodeling the chromatin structure of the yeast HSP82 promoter via constitutive interactions with its high-affinity binding site, heat shock element 1 (HSE1). The HSF-HSE1 interaction is also critical for stimulating both basal (noninduced) and induced transcription. By contrast, the function of the adjacent, inducibly occupied HSE2 and -3 is unknown. In this study, we examined the consequences of mutations in HSE1, HSE2, and HSE3 on HSF binding and transactivation. We provide evidence that in vivo, HSF binds to these three sites cooperatively. This cooperativity is seen both before and after heat shock, is required for full inducibility, and can be recapitulated in vitro on both linear and supercoiled templates. Quantitative in vitro footprinting reveals that occupancy of HSE2 and -3 by Saccharomyces cerevisiae HSF (ScHSF) is enhanced ∼100-fold through cooperative interactions with the HSF-HSE1 complex. HSE1 point mutants, whose basal transcription is virtually abolished, are functionally compensated by cooperative interactions with HSE2 and -3 following heat shock, resulting in robust inducibility. Using a competition binding assay, we show that the affinity of recombinant HSF for the full-length HSP82 promoter is reduced nearly an order of magnitude by a single-point mutation within HSE1, paralleling the effect of these mutations on noninduced transcript levels. We propose that the remodeled chromatin phenotype previously shown for HSE1 point mutants (and lost in HSE1 deletion mutants) stems from the retention of productive, cooperative interactions between HSF and its target binding sites.
The ability to respond rapidly to a large constellation of environmental stresses is crucial to the survival of all organisms, from bacteria to humans (32). Almost without exception, this response is regulated at the transcriptional level. In eukaryotes, environmental stress is sensed, directly or indirectly, by a sequence-specific transcriptional activator termed heat shock factor (HSF) (31, 54). In insects and vertebrates, HSF exists in a non-DNA binding, monomeric form predominantly localized within the cytoplasm (37, 53). Upon heat shock (or other stress), HSF rapidly trimerizes via arrays of amphipathic α-helical residues in the N-terminal domain, translocates into the nucleus, and binds to its cognate promoter elements within chromatin (50). In all metazoan heat shock genes which have been examined, the promoter regions are maintained in a transcriptionally poised, nucleosome-free state by other sequence-specific regulators, which create an environment conducive for rapid, inducible HSF binding (24). Thus, a specific architecture needs to be established within the upstream regulatory regions of stress-responsive genes; HSF is in fact incapable of binding to the Drosophila hsp70 promoter in the absence of GAGA, TATA, or initiator elements (38).
In contrast, heat shock promoters of the budding yeast Saccharomyces cerevisiae appear to be maintained in a transcriptionally poised state by HSF itself. At HSC82, a 4-bp substitution within the high-affinity heat shock element (HSE) abolishes promoter-associated DNase I hypersensitivity and restriction enzyme accessibility, despite the continuous presence of a second activator, GRF2/REB1, bound adjacent to the mutated HSE (6). Similarly, deletion or substitution of the high-affinity HSE at HSP82 (creating hsp82 alleles termed ΔHSE1 and ΔHSE1·, respectively) reduces transcription ≥100-fold and results in a dramatic alteration of promoter chromatin structure: the nuclease-hypersensitive region is replaced by two stably positioned nucleosomes, one centered over the mutated HSE and the other centered over the TATA initiation site (16). However, a double-point mutation within HSE1 (termed P2), despite seriously weakening sequence-specific interactions within the HSP82 enhancer (reference 28 and this study), has no effect on promoter-associated DNase I hypersensitivity (21, 28) or on the pattern of micrococcal nuclease (MNase) cleavage (8, 12). Thus, the phenotype of the ΔHSE1 and ΔHSE1· alleles suggests an HSF-dependent mechanism for establishment of the nucleosome-free state, whereas the phenotype of hsp82-P2 argues for an HSF-independent mechanism. To clarify the role of HSF in regulating HSP82, we have used a combined mutagenesis and footprinting strategy. This approach has confirmed a central role for HSF in potentiating HSP82 promoter function and has led to the discovery that HSF binds cooperatively to the HSP82 upstream region, both in vivo and in vitro. These cooperative interactions, which are maintained even in the presence of a double-point mutation within HSE1, likely underlie the phenotypic difference between P2 and ΔHSE1.
MATERIALS AND METHODS
Cultivation conditions.
Yeast strains were cultivated at 30°C in either rich (YPDA) medium or synthetic complete medium lacking uracil (Ura− medium). Galactose shift of cells bearing a GAL1-HSF1 episomal gene was achieved by pregrowing cells in Ura− medium containing 2% raffinose and shifting them to medium containing 1.5% raffinose and 0.5% galactose.
In vitro mutagenesis and yeast strain construction.
Oligonucleotide-directed mutagenesis was performed on a 2.9-kb EcoRI fragment of HSP82 subcloned into M13mp18 as previously described (16). The in vitro-mutagenized fragment was targeted by two-step gene transplacement to the hsp82/CYH2s locus of strain SLY102. SLY102 is isogenic to SLY101, which is a haploid spore isolated from a cross between W303-1B and B-7056 (22). In all cases, successful transplacement was confirmed by Southern blot hybridization; for certain promoter mutants (G161, G162, P2, ΔHSE1, ΔHSE1·, and ΔHSE1·t [true]), the chromosomal mutation was further confirmed by genomic sequencing. Strains used in this study are listed in Table 1.
TABLE 1.
Yeast strains used
| Strain | Genotype | Source or reference |
|---|---|---|
| W303-1B | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | R. Rothstein |
| SLY101 | MATα ade2 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 cyh2r | 22 |
| SLY102 | SLY101, hsp82Δ::CYH2s | 22 |
| KEY102 | SLY101, hsp82-ΔHSE1 | 16 |
| KEY103 | SLY101, hsp82-G161 | This study |
| KEY104 | SLY101, hsp82-G162 | This study |
| KEY105 | SLY101, hsp82-ΔHSE1· | 16 |
| KEY106 | SLY101, hsp82-HSCS | This study |
| KEY107 | SLY101, hsp82-G2 | This study |
| KEY202 | KEY102, [pGAL1HSF (GAL1-HSF1 URA3 CEN)] | 16 |
| DSG101 | SLY101, hsp82-P2 | This study |
| DSG102 | SLY101, hsp82-P3 | This study |
| DSG103 | SLY101, hsp82-PG | This study |
| DSG104 | SLY101, hsp82-G5 | This study |
| DSG105 | SLY101, hsp82-ΔHSE1·t | This study |
| DSG112 | SLY101, hsp82-190 | This study |
| DSG113 | SLY101, hsp82-190/P2 | This study |
| DSG114 | SLY101, hsp82-190/G2 | This study |
| DSG115 | SLY101, hsp82-265 | This study |
| DSG116 | SLY101, hsp82-265/P2 | This study |
| DSG117 | SLY101, hsp82-265/G2 | This study |
| CSGy55 | MATα HSP82/lacZ::LEU2 leu2::hisG ho::LYS2 lys2 ura3 smo1 cyh2r-z | 43 |
| CSGy153 | CSGy55, hsp82-ΔSTRE/lacZ::LEU2 | 43 |
Northern assays.
Cultures were grown to early logarithmic phase (1 × 107 to 2 × 107 cells per ml) and then split into two aliquots. The non-heat-shocked aliquot was metabolically poisoned with 20 mM sodium azide, chilled to 0°C, and harvested. The remaining aliquot was rapidly shifted from 30 to 39°C through addition of an equivalent volume of 51°C medium. Heat shock was terminated with azide as described above, and RNA was isolated from all samples by glass bead lysis (23). RNA was resolved on 1.25% agarose–1.1 M formaldehyde gels, blotted to GeneScreen, and sequentially hybridized to HSP82- and ACT1-specific probes. HSP82 hybridization was at 45°C, using as probes primer-extended synthetic oligonucleotides (two were used with equal success: a 62-mer spanning positions +2226 to +2287 and a 100-mer spanning positions +2190 to +2289). ACT1 hybridization was conducted at 55°C, using as probe an antisense RNA corresponding to the 1.6-kb BamHI-HindIII ACT1 fragment. Following each hybridization, the blot was exposed to a PhosphorImager screen, and HSP82-specific signal, normalized to that of ACT1, quantified with ImageQuant 1.1 (Molecular Dynamics). To optimize measurement of scarce HSP82 transcripts, the background was set equal to the signal present in RNA isolated from the hsp82Δ strain SLY102.
DMS in vivo footprinting.
Cells were grown to early log phase at 30°C in rich medium, concentrated by centrifugation 100-fold to a density of ∼109 cells/ml, divided into 500-μl aliquots, then either maintained under nonstressful conditions (23°C) or subjected to a 15-min 39°C heat shock. At this point dimethyl sulfate (DMS) was added to each aliquot to a final concentration of 0.1, 0.2, 0.4, or 0.8%, and cells were incubated at either 23 or 39°C for 2 min. The reaction was quenched through the addition of an equal volume of stop buffer (1 M sorbitol, 0.1 M 2-mercaptoethanol, 20 mM sodium azide, 0.1 M EDTA, 0.1 M Tris-HCl [pH 8]), and genomic DNA was isolated and analyzed as previously described (7). HSP82-specific cleavages were revealed by amplified primer extension (AMPEX) (7) using either a lower-strand-identical primer (+26→−11) or an upper-strand-identical primer (−342→−315). These primers span HSP82 sequence from +26 to −11 or from −342 to −315, with positions numbered relative to the major transcription start site (+1) (9). Reaction products were electrophoresed on an 8% sequencing gel, detected by a PhosphorImager, and analyzed by densitometry with ImageQuant 1.1.
Purification of recombinant HSF.
All recombinant HSF proteins were purified from Escherichia coli BL21(DE3). The purification of glutathione S-transferase (GST)–S. cerevisiae HSF (ScHSF) has been previously described (6); by the criterion of Coomassie blue staining, the affinity-purified product was intact and virtually free of contaminating polypeptides (data not shown). C-terminally tagged His6-ScHSF was purified from pET3d-HSF-His-transformed bacteria (generously provided by Nick Santoro and Dennis Thiele, University of Michigan) by using nickel-charged resin (Novagen) according to the manufacturer’s protocol except that binding buffer was substituted for wash buffer in all washing steps. Following the final step, the product was relatively intact, as assessed by immunoblot assay, but not completely free of contaminating polypeptides. N-terminally tagged His6-Drosophila HSF (dHSF) was purified from pET15B/dHSF-transformed E. coli BL21 (generously provided by Paul Mason and John Lis, Cornell University) in a similar manner.
DNase I in vitro footprinting and DMS methylation protection analyses. (i) Linear templates.
hsp82 promoter fragments were generated by PCR amplification of plasmid templates (see below) using primers −294→−274 and +51→+31, the forward primer being end labelled. Binding and footprinting reactions were carried out at 23°C for 45 min, using a template concentration of ∼1 nM (10 ng/50-μl reaction) and ScHSF concentrations ranging from 1 to 100 nM. Binding reactions were conducted in HSF binding buffer, consisting of 150 mM NaCl, 1 mM CaCl2, 3 mM MgCl2, 20 mM Tris (pH 8), 0.5 mM EDTA, bovine serum albumin (100 μg/ml), 1 mM phenylmethylsulfonyl fluoride, pepstatin (2 μg/ml), leupeptin (2 μg/ml), chymostatin (0.5 μg/ml), E-64 (7.2 μg/ml), 2 mM N-ethylmaleimide, 0.01% Nonidet P-40, and 0.5 mM n-octylglucoside. Resultant protein-DNA complexes were digested with 0.1 U of DNase I for 2 min at 23°C.
(ii) Supercoiled templates.
Recombinant ScHSF, at concentrations ranging from 30 to 840 nM, was mixed with hsp82 templates (1.6 nM) at 23°C for 45 min as described above. Resultant complexes were reacted with 0.1% DMS at 23°C for 1 min. The reaction was quenched through the addition of 2-mercaptoethanol and sodium acetate to final concentrations of 0.5 and 0.75 M, respectively, and DNA was purified and subjected to AMPEX using primer +26→−11. Templates were double-stranded M13mp18 constructs bearing the hsp82 EcoRI fragment spanning positions −1300 to +1600.
Determination of dissociation constants.
Dissociation constants (Kd) were calculated from densitometric scans of DNase I digests using the equation fHSE-HSF = [HSF]f/([HSF]f + Kd), where fHSE-HSF is the fraction of template bound by HSF and [HSF]f is the concentration of free (unbound) protein for a given input concentration of ScHSF. Curve fitting was done using KaleidaGraph 3.09 (Synergy). Kd values derived for HSE2 and -3 on mutant templates are estimates based on extrapolation to the maximal protection observed on the wild-type (WT) template (see Fig. 6). A potential source of error in the Kd determinations is that the reaction between HSF and DNA was not at equilibrium before addition of the footprinting reagent. The concern is that at low protein concentrations and with a slow on rate, a 45-min incubation might have been insufficient to achieve equilibrium. To gauge how the amount of time to reach equilibrium is related to the concentrations of the two reagents and to the on- and off-rate constants, the reaction between HSF and DNA (HSF + DNA→DNA-HSF) was computer simulated (Scientist 2.01 [MicroMath]). When Kd = 1 nM and kon and koff are set to 105 mol−1 s−1 and 10−4 s−1, respectively, equilibrium between HSF and DNA is achieved within 45 min for [HSF] ≥ 10 nM (1 nM DNA), as shown by plots of the calculated fraction of template bound by HSF (fHSE-HSF versus [HSF]). These calculated plots were nearly identical to the plots determined experimentally. On the other hand, when Kd = 1 nM and the on and off rates were set to 104 mol−1 s−1 and 10−5 s−1, respectively, equilibrium was not achieved within 45 min, even for [HSF] > 10 nM. Plots derived from these simulations did not resemble those obtained experimentally. Therefore, for HSF binding to HSE1 and -2, the on and off rates are consistent with values of ∼105 mol−1 s−1 and ∼10−4 s−1, respectively. For these two cases, the apparent Kd values may overestimate the true Kd values by a factor of 2 or 3. For reactions between HSF and HSEs that had apparent Kd values greater than 10 nM, if kon ≪ 105 mol−1 s−1, then an incubation time of 45 min would have been insufficient to reach equilibrium. In such cases, the apparent Kd values may overestimate the true Kd values by a factor of 10 or more.
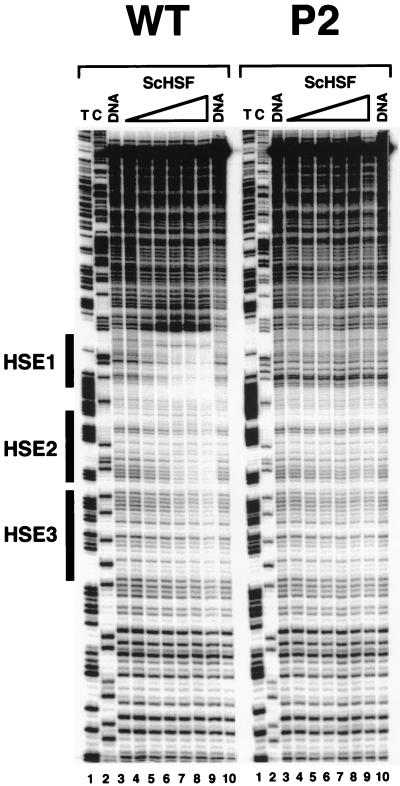
FIG. 6.
ScHSF binds cooperatively to its target HSEs on naked hsp82 templates. Linear DNA templates (1 nM), amplified by PCR and bearing the HSP82+ or hsp82-P2 upstream sequence, were titrated with increasing amounts of GST-ScHSF (0, 0.4, 1.2, 3.6, 33, and 100 nM in lanes 4 to 9, respectively). Resultant protein-DNA complexes were digested with DNase I, and the digestion products were processed, electrophoretically separated on an 8% sequencing gel, and visualized with a PhosphorImager. DNA, naked DNA digested with DNase I; T and C, dideoxy sequencing ladders.
Binding competition assays. (i) Preparation and labeling of DNA templates.
The following gel-purified hsp82 templates were used: WT (a 353-bp PCR fragment encompassing positions −295 to +45 of HSP82+ and including 13 bp of flanking sequence), G161 (363 bp, spanning −295 to +55 of hsp82-G161 with 13-bp flank), P2 (343 bp, −285 to +45), and ΔHSE1· (333 bp, −285 to +35). Templates were 32P labelled during PCR amplification to approximately the same specific activity through use of common 5′-end-labelled, gel-purified primers. For the synthetic HSE competition assay, complementary oligonucleotides bearing three or six tandem inverted arrays of the consensus HSF binding unit, nTTCT, were annealed, end labelled, and purified. Their upper-strand sequences are as follows: HSE3 (48-mer; pentameric motifs underlined), TTGCGTTGGATCCCTAATTTCTAGAACTTTCTGAGCAAGCTTTAAGCG; and HSE6 (43-mer), GGTAAGCTTCTAGAACTTTCTAGAACTTTCTAGAACCCGGGGG.
(ii) Binding reactions and gel retardation assays.
Binding reactions were carried out in 50-μl volumes at room temperature (∼23°C) for 1 h. For the promoter competition assay, ∼9 nM template (40,000 cpm) was incubated in HSF binding buffer in the presence of various concentrations of GST-ScHSF and nonspecific competitor DNA [poly(dI-dC)]. For the synthetic HSE competition assay, ∼70 pmol (100,000 cpm) each of HSE3 and HSE6 were incubated in HSF binding buffer with increasing concentrations of GST-ScHSF, His6-ScHSF, or His6-dHSF in the presence of 1.0 μg of poly(dI-dC) per ml. Reaction mixtures contained 0, 160, 320, 640, and 1,280 ng of GST-ScHSF (0, 9, 18, 36, and 71 nM, respectively); 0, 0.2, 0.4, 0.8, and 1.6 μl of His6-ScHSF; or 0, 0.5, 1.0, 2.0, and 4.0 μl of His6-dHSF (see Fig. 8A). The reactions were analyzed by electrophoresis at room temperature on 5% polyacrylamide–50 mM Tris–50 mM boric acid–0.5 mM EDTA 1-mm-thick gels run for 150 V-h. Free and bound species were detected by autoradiography, excised, and eluted in 0.5 M ammonium acetate–0.5 mM EDTA–0.1% sodium dodecyl sulfate (SDS). DNAs were directly precipitated, dried, dissolved in sequencing gel sample buffer, and electrophoresed on a 12% sequencing gel. Quantitation of radioactivity was done with a PhosphorImager as described above.
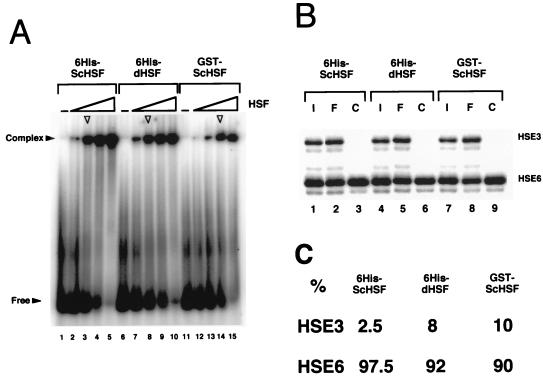
FIG. 8.
ScHSF binds cooperatively to adjacent trimeric binding sites on synthetic DNA templates. (A) EMSA illustrating the formation of HSF-HSE complexes as a function of increasing HSF concentration. The 32P-labelled DNA probe, a near-equimolar mixture of two synthetic HSEs, HSE3 (a 48-mer containing three adjacent AGAAN pentameric units) and HSE6 (a 43-mer containing six adjacent pentameric units), was titrated with increasing concentrations of His6-ScHSF, His6-dHSF, or GST-ScHSF (see Materials and Methods). The binding reactions were carried out at 23°C for 1 h; resultant complexes were separated on a 5% native polyacrylamide gel. (B) Sequencing gel analysis of input (I), free (F), and complexed (C) species, excised from the indicated lanes in panel A (arrowheads). A phosphorimage of the dried gel is shown. (C) Quantitation of results, expressed as percentage of HSE3 and HSE6 in each complex, normalized to input.
RESULTS
Dynamic interactions at activator and repressor binding sites within the HSP82 promoter.
Previous mutational and footprinting analyses of HSP82 have revealed the presence of a number of regulatory motifs within its upstream region, including a TATA box, a mitotic repressor/meiotic activator motif (URS1-ARE), and three HSEs (13, 16, 17, 21, 28, 43). These sites reside within a constitutive DNase I-hypersensitive region (45) and are occupied by sequence-specific DNA binding proteins (summarized in Fig. 1A; promoter sequence is provided in Fig. 1B). As shown in Fig. 2, DMS in vivo footprinting coupled with AMPEX demonstrates that certain protein-DNA interactions are constitutive, others are inducible, and still others are repressible (i.e., lost upon heat shock). Under noninducing conditions, binding is readily detectable at HSE1 (within both major and minor grooves), at URS1-ARE (minor groove only), and at TATA (minor groove only) (Fig. 2). Major and minor groove interactions are detected by protection/hyperreactivity of the N-7 of guanines and the N-3 of adenines, respectively (27). The AMPEX technique permits detection of both guanine and adenine modifications (6). Following heat shock, interactions at HSE1 are strengthened (e.g., G residues at −161 and −162), while those at URS1-ARE are lost (e.g., A residues at −114, −115, and −120). Moreover, prominent, heat shock-inducible interactions are evident at HSE2 and -3 and at a consensus HAP2/3/5 site located ∼3 helical turns upstream of the TATA box (Fig. 2A). Note that while protein binding to HSE2 and -3 is virtually undetectable under control conditions, in certain strain backgrounds weak constitutive interaction is seen (see Fig. 5C). Taken together, our data are consistent with the notion that activator and repressor complexes coexist at the noninduced promoter and that these complexes engage in dynamic, stress-responsive interactions.
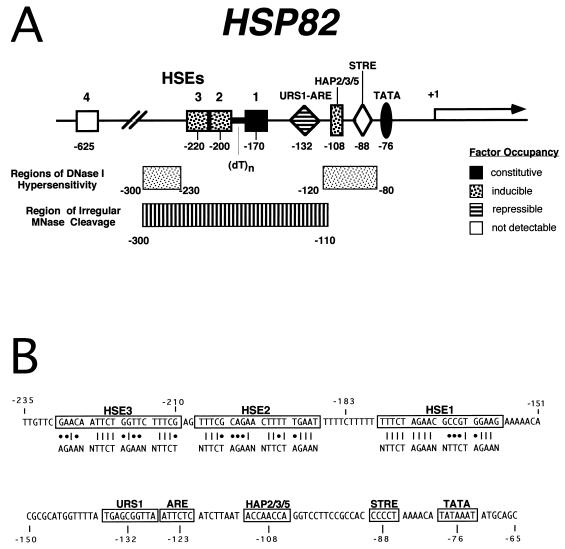
FIG. 1.
Principal regulatory motifs within the HSP82 promoter. (A) Summary of in vivo footprinting and chromatin mapping analyses. Eight cis-acting elements implicated by biochemical (references 13, 16, 17, and 28 and this study) and genetic (references 16, 28, and 43 and this study) assays, or by their match to published consensus sequences (9), are indicated, as are the approximate locations of DNase I hypersensitivity (17, 21, 28, 45) and MNase disruption (8, 16). Also indicated is the occupancy state of each promoter element. Note that HSE4 is fully dispensable for normal promoter function (Fig. 4) and is detectably occupied only under conditions of HSF overexpression (16). STRE is likewise not detectably occupied, and no function has yet been ascribed to it. (B) Sequence of the heat shock enhancer region. Matches of each HSE to the HSF consensus (consisting of a tandem inverted array of the pentameric sequence AGAAN [11]) are indicated by vertical lines; mismatches are indicated by dots. Thus, HSE1, -2, and -3 exhibit matches of 12/16, 10/16, and 9/16, respectively, to the heat shock consensus.
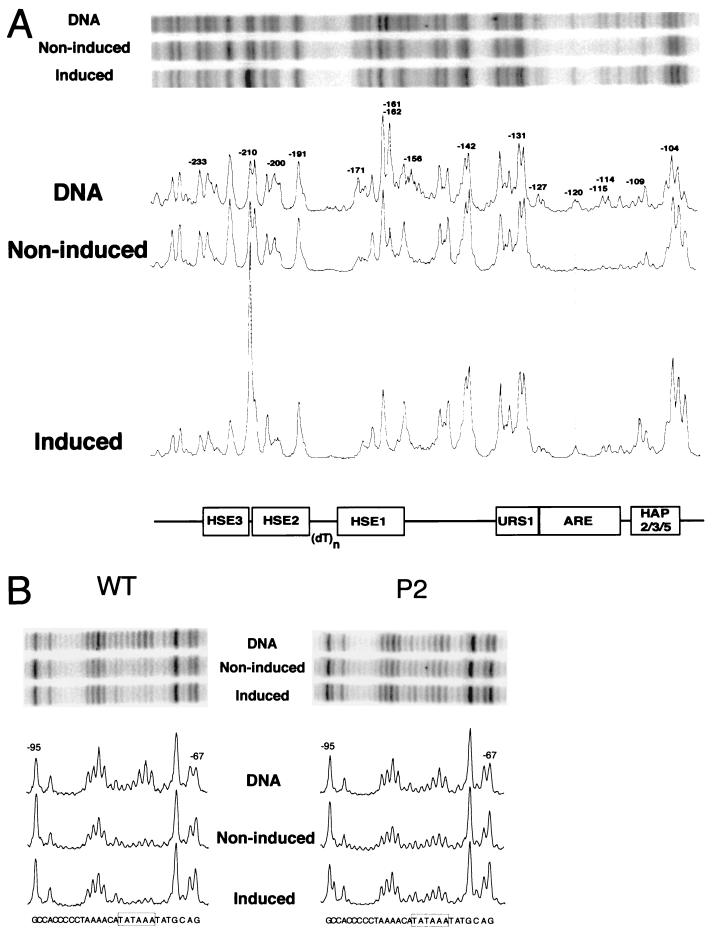
FIG. 2.
DMS in vivo footprint analysis of the hsp82 promoter region before and after a 15-min, 30-to-39°C heat shock (noninduced and induced, respectively). (A) Upstream promoter region analysis of HSP82+ (strain W303-1B). Early-log-phase cultures (50-ml equivalents) were harvested, resuspended in 0.5 ml of rich medium (maintained at either 30 or 39°C), and reacted with 0.1% DMS for 1 min. Genomic DNA was isolated and subjected to AMPEX with primer +26→−11, and the product was electrophoretically resolved on a 6% sequencing gel. Representative lanes from a single phosphorimage and corresponding densitometric scans of an upper-strand analysis are shown; their amplitudes are normalized to the −191 G peak (scans of Fig. 5 and 7 are similarly normalized). DNA, naked genomic DNA, isolated from the same strain, reacted with 0.1% DMS at 23°C for 1 min and processed as described above. (B) TATA region analysis of HSP82+ and hsp82-P2 (strains W303-1B and DSG101, respectively) conducted as described above (also using primer +26→−11). Phosphorimages of selected lanes and their corresponding scans are shown.
FIG. 5.
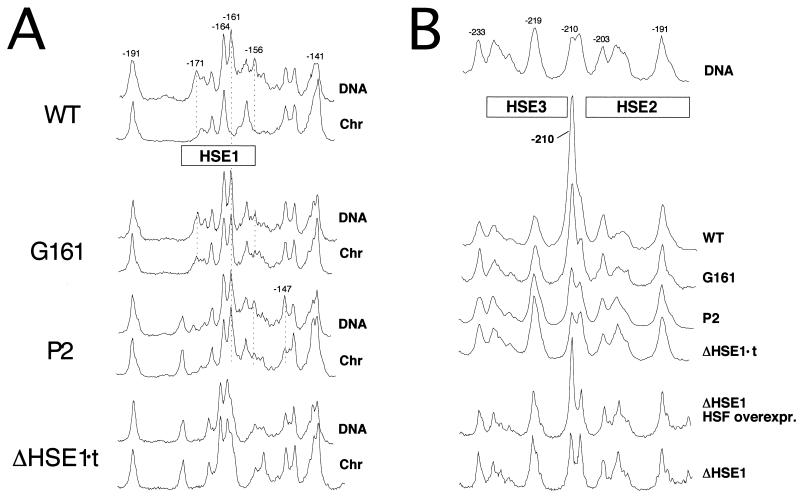
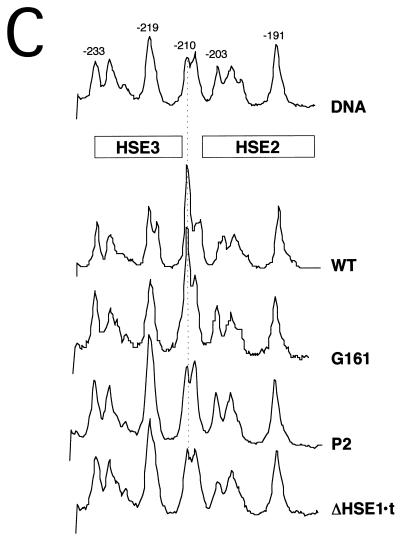
HSF binds cooperatively to its target HSEs at HSP82 in vivo. (A) In vivo methylation protection analysis of the HSE1 region from induced cells. Densitometric scans are shown. Strains harboring the indicated hsp82 alleles were reacted with DMS and subjected to AMPEX analysis using primer +26→−11 as described in the legend to Fig. 2. Chr, DNA isolated from 15-min-heat-shocked cells treated with DMS during the last 2 min of heat shock; DNA, methylation profile of naked genomic DNA; WT, strain W303-1B. (B) In vivo footprint analysis of the HSE2-HSE3 region from induced cells. Cells were heat-shocked and reacted with DMS, and DNA was isolated and analyzed as for panel A. ΔHSE1 HSF overexpr., strain KEY202 subjected to a 3.5-h galactose shift prior to heat shock. Note that the bottom two tracings are derived from a separate gel. DNA, isolated from WT and analyzed as in panel A. (C) In vivo footprint analysis of noninduced cultures. Cells from cultures maintained at 30°C were subjected to DMS treatment and DNA isolated and analyzed as described above. WT, strain SLY101.
Induced HSP82 expression is resilient to the effects of point mutations within HSE1.
Previous work has shown that the P2 double-point mutation, when targeted to the native chromosomal locus of HSP82, virtually abolishes noninduced expression while diminishing induced expression severalfold (21, 28). Accompanying this expression phenotype are loss of all detectable interactions over HSE1, both before and after heat shock (see Fig. 5). Paralleling the loss of HSF-HSE1 interactions at hsp82-P2 is a weakening of the TATA binding protein (TBP) footprint (Fig. 2B). This observation is consistent with previous findings (16) indicating a critical role for HSF in facilitating TBP binding to the promoter.
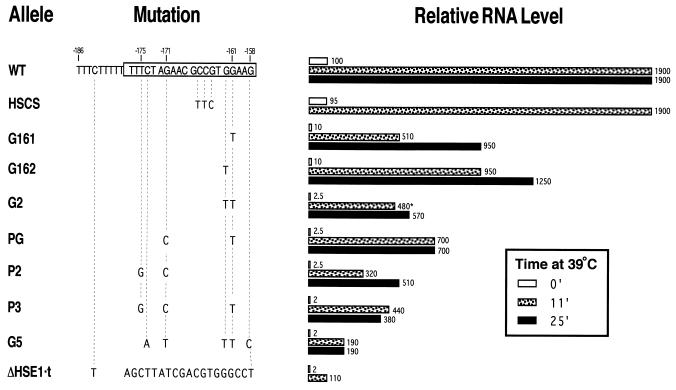
To determine whether P2 was unusual in its impact on expression, we constructed isogenic hsp82 strains bearing different nucleotide substitutions within HSE1. The consensus HSE consists of a tandem inverted array of AGAAN pentameric units (Fig. 1B) (11). As the first two AGAAN modules of HSE1 were mutated in P2 (at positions −171 and −175 [Fig. 3]), it was of interest to know whether other point mutations of HSE1, in particular those involving the fourth conserved module (GGAAG [Fig. 1B]), would similarly affect expression. As shown in Fig. 3, single nucleotide substitutions at positions −161 and −162, corresponding to upper-strand guanines that are strongly protected from DMS methylation in vivo (Fig. 2A), reduce basal expression 1 order of magnitude but have relatively little effect on induced expression levels. The G162 mutant demonstrates that despite being less conserved than A (11), G at position 1 (GGAAG) is as pivotal to function as the highly conserved G at position 2 (GGAAG; cf. allele G161). When the two point transversions are combined, creating an allele termed G2, basal transcription is nearly abolished. Induced expression, on the other hand, is reduced only three- to fourfold. This phenotype, which is also seen with other combinations of two- or three-point substitutions (alleles PG and P3), resembles that of P2. A slightly more severe phenotype accompanies a five-point mutation of HSE1 in which the three consensus AGAAN modules plus a fourth overlapping one (−159 to −156) are mutated (allele G5). As a control, we introduced a triple-point mutation within the degenerate third module (improving the match of HSE1 to the AGAAN consensus to 15/16); this had no phenotype (allele HSCS). Thus, HSE1 functions as a “gapped” HSE (36, 39); within this context, the severity of the expression phenotype parallels the severity of the lesion. Moreover, the difference in induced RNA levels between the point mutants and a 20-bp substitution of HSE1 (termed ΔHSE1·t), suggests that HSE1 retains function in the presence of at least three, and perhaps as many as five, point substitutions (Fig. 3). Notably, promoter-associated DNase I hypersensitivity and irregular MNase cleavage ladders are still evident within the point-mutated alleles (12, 21, 28), implying that the nucleosome-disrupted state characteristic of the WT promoter is largely preserved in these mutants.
FIG. 3.
Expression phenotypes of HSE1 point mutants. Isogenic hsp82 strains were constructed, and RNA was isolated from cells grown under nonstressful conditions (30°C; non-heat shocked [0′]) or cells subjected to a 30→39°C thermal upshift for either 11 or 25 min (11′ or 25′). HSP82 transcript levels were assayed by Northern hybridization and quantitated by normalization to ACT1 as described previously (16). Values are means (±15%) and are based on a minimum of three independent experiments. ∗, 15-min time point.
HSE2 and -3 can functionally compensate for mutations within HSE1.
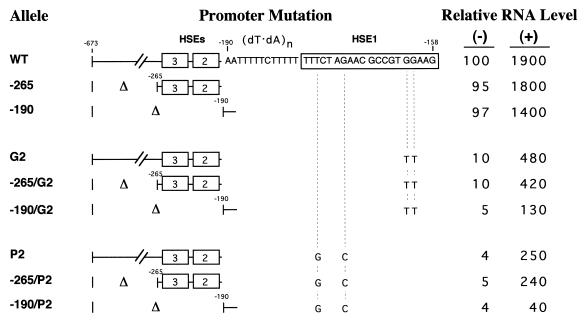
The relatively mild heat shock phenotype of HSE1 point mutants could be a consequence of compensating, stress-inducible binding of HSF to the lower-affinity, upstream HSEs (Fig. 2A). To test this idea, we constructed strains bearing 5′ deletions within the chromosomal copy of hsp82. The endpoint for these deletions was a ClaI site at position −673, located within the promoter of the adjacent, divergently transcribed YAR1 gene (26). As shown in Fig. 4, deletion of the sequence between −265 and −673 is without phenotype in WT, G2, and P2 contexts (alleles −265, −265/G2, and −265/P2, respectively). Therefore, it is unlikely that HSP82 regulatory elements exist upstream of position −265. In contrast, when the region encompassing HSE2 and -3 is also deleted (−190 to −265), a 25% reduction in induced transcript levels is seen in the otherwise WT promoter (allele −190) in response to either acute or chronic heat stress (Fig. 4 and data not shown). Thus, HSE2 and -3, despite serving as binding sites for HSF, appear to play only a minor role in HSP82 regulation in the context of a WT HSE1. Strikingly, when the −190 upstream deletion is combined with double-point HSE1 mutations, a synergistic effect is seen. Induced expression of hsp82-190/G2 is nearly fourfold lower than that of hsp82-G2, while a sixfold reduction is seen when the P2 mutation is combined with the same upstream deletion. This analysis argues that while HSE2 and -3 enhance heat shock-induced transcription of the WT allele 30 to 40%, they boost hsp82-G2 expression nearly 300% and hsp82-P2 expression nearly 500%. Minimal functional compensation is seen under noninducing conditions, consistent with the low level of occupancy of these elements (Fig. 2A; see also Fig. 5C). We conclude that under stressful conditions, HSE2 and -3 can functionally compensate for mutations within HSE1. Note that while HSE2 and -3 preserve robust inducibility, the absolute magnitude of activated expression in HSE1 point mutants is reduced (Fig. 3).
FIG. 4.
HSE2 and -3 compensate for point mutations within HSE1. Expression phenotypes of hsp82 promoter mutants bearing deletions between −265 and −673 or between −190 and −673 in WT, P2, or G2 contexts are shown. Isogenic S. cerevisiae strains were constructed, RNA was isolated from non-heat-shocked (−) and 15-min-heat-shocked (+) cells, and HSP82 transcript levels were assayed as described in the legend to Fig. 3. Values represent means (±15%) of three independent experiments.
HSF binds cooperatively to its target HSEs in vivo.
To investigate the effect of HSE1 point mutations on in vivo protein-DNA interactions, we subjected selected strains to DMS genomic footprinting as described above. Under noninducing conditions, the G161 single-base substitution abolishes detectable binding (data not shown). Under inducing conditions, slight protection is seen at HSE1 (Fig. 5A), consistent with weak or fractional occupancy. To determine whether occupancy of HSE2 and -3 is facilitated by cooperative interactions with the protein complex bound at HSE1, we assayed DMS-induced hyperreactivity of the −210 G residue. If the HSEs are bound cooperatively, then mutations that weaken the stability of the HSE1-HSF complex will also impair occupancy of HSE2 and -3. In striking support of this idea, the G→T point mutation at −161 substantially diminishes the DMS hyperreactivity of the −210 G residue (Fig. 5B, allele G161). Moreover, the P2 double-point mutation, which diminishes the HSF-HSE1 interaction even further (Fig. 5A), has a more marked effect on the occupancy of HSE2-HSE3 (Fig. 5B). The ΔHSE1·t mutation, a 20-bp substitution of HSE1, eliminates all detectable interactions at HSE2 and -3, even following heat shock. Notably, 10- to 30-fold overexpression of HSF restores DMS hyperreactivity to the −210 G of hsp82-ΔHSE1, concomitant with an ∼20-fold increase in induced transcript levels (16). Taken together, these data are consistent with the notion that HSF does in fact bind to HSE2 and -3 in vivo, that its binding is cooperative, and that inducible HSF occupancy of the low-affinity HSEs is facilitated by the presence of the constitutive HSF-HSE1 complex.
Is there cooperative binding under noninducing conditions? As mentioned above, in certain genetic backgrounds (e.g., strain SLY101), weak constitutive binding to HSE2 and -3 is evident (Fig. 5C). Constitutive binding to HSE3 is also detectable in this background using an in vivo dam methyltransferase assay (35). Such low-level binding is not apparent in all strains (e.g., W303 [Fig. 2A] and YPH102 [13]). However, as the hsp82 mutants used in this study are isogenic to SLY101 (Table 1), this allowed us to test the effects of introducing single- and multiple-point mutations within HSE1 on constitutive HSF-HSE2/3 interactions. As clearly shown in Fig. 5, these interactions are also sensitive to HSE1 lesions (compare P2 or ΔHSE1·t with WT in Fig. 5C), although less so than under inducing conditions (compare G161 with WT in Fig. 5B and C). We conclude that HSF binds cooperatively to its target HSEs under both noninducing and inducing conditions.
HSF binds cooperatively to its target HSEs in vitro.
Cooperative binding to HSE2 and -3 could stem from a number of unrelated mechanisms. First, it could reflect direct protein-protein interactions, whereby HSF bound at HSE1 facilitates the binding of additional HSF trimers to HSE2 and -3 (i.e., classic cooperativity). Second, binding of HSF to its low-affinity sites could be dependent on an altered DNA topology generated by the nearby HSF-HSE1 complex. Third, the HSF bound to HSE1 might antagonize nucleosomal assembly of the enhancer region, rendering HSE2 and -3 more accessible to HSF within chromatin, thereby permitting binding to the weak sites. To help distinguish between these possibilities, we assayed binding of affinity-purified recombinant HSF (GST-ScHSF) to linear DNA templates bearing the WT, P2, and ΔHSE1·t promoter regions (Fig. 6 and data not shown). Reactions were conducted in a buffer containing Nonidet P-40 and n-octylglucoside, conditions which result in high-affinity binding of HSF to DNA (46), and binding was assessed by DNase I footprinting. Mimicking its behavior in vivo, ScHSF binds to the WT but not the mutated HSE1 sequence on a linear template (Fig. 6, lanes 4 to 9). Moreover, HSF binding to the HSE2-HSE3 region is markedly reduced on the P2 template (Fig. 6) and is eliminated altogether on a ΔHSE1·t template (data not shown; see Fig. 7). The in vitro DNase I cleavage profiles of WT and P2 templates bound to GST-ScHSF are virtually identical to DNase I genomic footprints obtained from the corresponding strains (8, 17, 28).
FIG. 7.
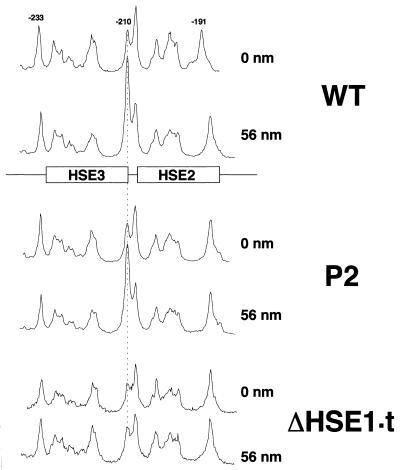
DMS in vitro methylation analysis of supercoiled templates bearing WT, P2, or ΔHSE1·t sequences. Plasmid templates (1.6 nM) were reacted with either 0 or 56 nM GST-ScHSF as indicated and then methylated with DMS. DNA was then purified and subjected to AMPEX analysis. Densitometric scans of the HSE2-HSE3 region of the sequencing gel are shown.
As determined by quantitative densitometry (not shown), GST-ScHSF binds with high affinity to its target sites on the WT template (apparent Kds of 1.1, 3.6, and 26 nM for HSE1, -2, and -3, respectively). By comparison, the apparent dissociation constants for HSE2 and -3 increase to 32 and 260 nM on the P2 template and at least another order of magnitude on the ΔHSE1·t template (no interaction detectable at the highest concentration of ScHSF used [100 nM] [data not shown]). Thus, cooperative interactions with HSE1 enhance the apparent affinity of HSF for HSE2 and -3 by as much as 2 orders of magnitude (but see Materials and Methods). In the presence of a double-point HSE1 mutation, binding to the weak sites still takes place, albeit at a 10-fold-reduced level. Entirely consistent results are seen when supercoiled templates are substituted for linear ones and binding is assayed by DMS methylation protection: HSF binds HSE2 and -3 on WT and P2 templates but not on the ΔHSE1·t template (Fig. 7). In particular, the signature DMS-hyperreactive site between HSE2 and -3 is evident on the P2 template but not on the ΔHSE1·t template, arguing that productive HSF-HSE1 interactions are possible at P2 and that these are sufficiently stable to seed HSF binding to the weak sites.
A caveat to the foregoing experiments is that they were conducted with a GST fusion protein. Therefore, it is possible that cooperative interactions are artificially enhanced by the presence of the 26-kDa GST moiety, which can form homodimers (48). This could give rise to the presence of nonphysiological HSF multimers (e.g., dimers and hexamers). To rule this out, we repeated the DNase I protection assay using His6-ScHSF (attempts to excise the GST domain from ScHSF by thrombin cleavage [15] proved unsuccessful). As was the case with GST-ScHSF, His6-ScHSF bound cooperatively to the hsp82 promoter templates, both linear and supercoiled (data not shown).
To provide independent confirmation of ScHSF cooperativity, we performed a binding competition assay. We reacted two forms of recombinant ScHSF, GST-ScHSF and His6-ScHSF, or recombinant His6-dHSF, with synthetic templates containing either three (HSE3) or six (HSE6) 5-bp units (AGAAN) in a tandem inverted array in an electrophoretic mobility shift assay (EMSA). These templates are capable of binding one or two trimers, respectively (51). Reactions were conducted with increasing concentrations of protein at 23°C for 1 h in HSF buffer, and then resultant HSF-DNA complexes were separated from the free DNA species by native gel electrophoresis (Fig. 8A). The complexed and free DNA species were purified from the indicated reactions (Fig. 8B, lanes 3, 8, and 14) and resolved on a sequencing gel alongside the input DNA. Quantitation of the two species indicates that recombinant ScHSF binds HSE6 with 10- to 40-fold-higher affinity than HSE3 (Fig. 8C), consistent with cooperative binding. Indeed, the marked preference of ScHSF for the HSE6 template is virtually identical to that exhibited by dHSF (Fig. 8B, lane 6; summarized in Fig. 8C). We conclude that ScHSF, like dHSF, binds DNA with a high degree of cooperativity. Taken together, our data argue that HSF cooperatively binds the HSP82 upstream region, both in vivo and in vitro, through classic, protein-protein interactions. A mechanism involving an altered DNA topology, while not formally ruled out, is rendered unlikely by the fact that the cooperativity seen is irrespective of template conformation or nucleotide sequence (Fig. 6 to 8).
Overall affinity of ScHSF for the HSP82 promoter is dramatically reduced by a single-nucleotide substitution within HSE1.
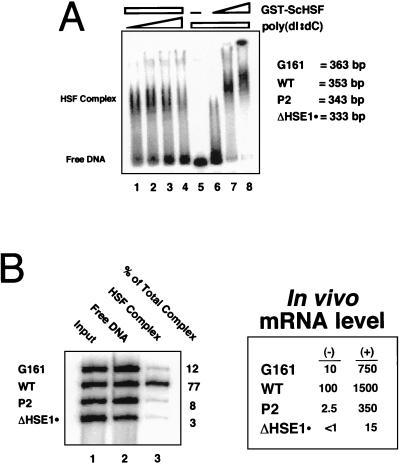
As recombinant HSF binds to naked DNA templates in a manner that closely parallels its binding in vivo, we tested the overall relative affinity of the HSP82 promoter and its mutated derivatives for ScHSF in a binding competition assay. If the expression phenotypes of HSE1 mutants are principally a consequence of reduced affinity of HSF for the hsp82 promoter, then one would predict a correlation between overall affinity of HSF for each hsp82 promoter region and its respective expression. To test this, we incubated equimolar concentrations of end-labelled DNA fragments corresponding to the promoter regions of the WT, G161, P2, and ΔHSE1· alleles (each comprising ∼300 bp of regulatory sequence and differing in length by 10-bp increments) with 50 nM GST-ScHSF in the presence of increasing concentration of nonspecific competitor DNA. Following a 1-h incubation at 23°C, resultant HSF-DNA complexes were separated from the free DNA species by native gel electrophoresis (Fig. 9A, lanes 1 to 4). The complexed and free DNA species from the sample in lane 2 were purified and resolved on a sequencing gel alongside the input DNA (Fig. 9B). Relative binding constants for the three mutant hsp82 derivatives were then quantified relative to WT by using the equation KWT/Kn = (CWT/DWT)/(Cn/Dn), where C and D represent intensities of the complex and free DNA, respectively (25). Such quantitation indicates that the G161 point substitution reduces the intrinsic affinity of the hsp82 promoter for ScHSF more than sixfold (77 versus 12). This dramatic reduction in overall affinity is seen despite the presence of unmutated HSE2 and -3. Even greater reductions in affinity are seen for the more extensively mutated derivatives, 15-fold for the P2 (2-bp) mutation and >25-fold for the ΔHSE1· (32-bp) substitution. These results are entirely consistent with the apparent Kds determined above. As illustrated in Fig. 9B, a comparison of the relative expression levels of the corresponding alleles reveals a remarkable correlation between overall HSF affinity and noninduced transcription. This finding suggests that non-heat shock expression phenotypes are dictated largely by the intrinsic affinity of HSF for HSE1 in vivo. The preference of recombinant ScHSF for the WT template determined by this assay is a minimum estimate since dissociation of HSF from tight binding sites can require >100 h (51).
FIG. 9.
Overall affinity of ScHSF for the HSP82 promoter is dramatically reduced by a single-point mutation within HSE1. (A) Formation of HSF-HSE complexes as resolved by EMSA. The 32P-labelled DNA probe, an equimolar mixture of four promoter templates, WT, G161, P2, and ΔHSE1·, was reacted either with a constant level of GST-ScHSF (50 nM) in the presence of increasing concentrations of poly(dI-dC) (0.5, 1, 2, and 4 μg per 50-μl reaction; lanes 1 to 4) or with increasing concentrations of GST-ScHSF (0, 25, 100, and 200 nM; lanes 5 to 8) in the presence of a constant level of competitor DNA (1 μg per 50-μl reaction). Binding reactions were carried out as for Fig. 8; HSF complexes were separated from free DNA by electrophoresis on a native 2% agarose gel. (B) Sequencing gel analysis of the input probe and of free and complexed DNA isolated from lane 2 of panel A. The percentage of each species in the HSF complex is provided on the right. The corresponding expression level of each hsp82 allele is shown in the inset. (−), non-heat shocked; (+), heat shocked for 15 min.
DISCUSSION
The principal findings of this work are as follows: (i) constitutive, stress-inducible, and stress-repressible protein-DNA interactions take place at the HSP82 promoter; (ii) HSF binds cooperatively to its target HSEs within the HSP82 promoter, both in vivo and in vitro; (iii) binding of HSF to HSE2 and -3 functionally compensates for mutations within HSE1; and (iv) noninduced HSP82 transcript levels directly correlate with the intrinsic affinity of ScHSF for the gene’s promoter, whereas induced transcript levels correlate with the extent of HSF interaction at HSE2 and -3.
Dynamic protein-DNA interactions at the HSP82 promoter.
High-resolution DMS in vivo footprinting reveal that certain sequences within the HSP82 promoter are constitutively occupied, others are inducibly occupied, and still others are occupied before but not after heat shock. The principal enhancer and core promoter elements, HSE1 and TATA, are strongly occupied under noninducing conditions, as is the URS1-ARE repressor element. Following heat shock, HSE2 and -3 and a consensus HAP2/3/5 site are also occupied, and the interactions at HSE1 and TATA are strengthened. In contrast, interactions at URS1-ARE are lost. That the protections and enhancements within HSE1, -2, and -3 reflect HSF binding is supported by in vitro footprinting assays and in vivo overexpression assays (this study and reference 13). A schematic model of protein-DNA interactions at the HSP82 promoter, both before and after heat shock, is presented in Fig. 10.
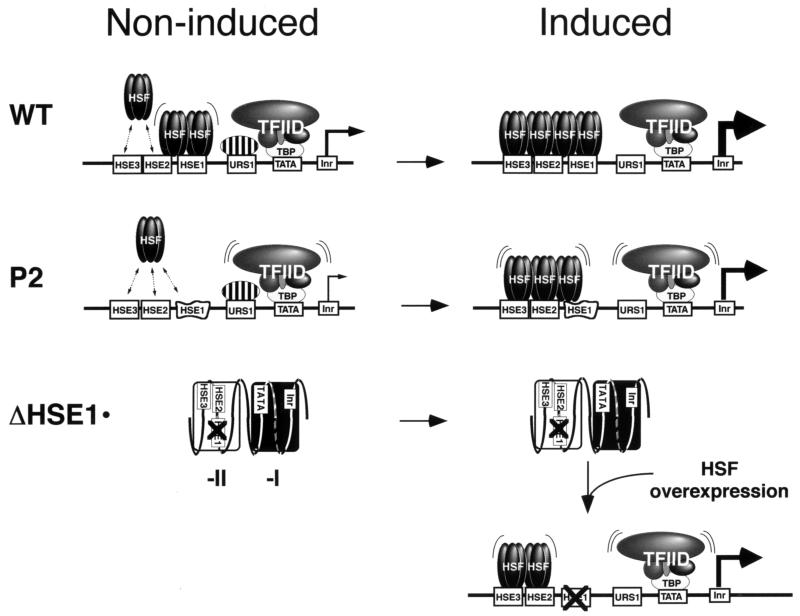
FIG. 10.
Three classes of hsp82 promoter architecture, as epitomized by the WT, P2, and ΔHSE1· alleles. Models are based on biochemical and genetic data reported here and elsewhere (13, 16, 17, 21, 28, 43). Fractional occupancy of promoter elements is indicated by dotted arrows for transient protein-DNA interactions, parallel curved lines for weak interactions, and single curved lines for moderately strong interactions; all other interactions are stable. The relative rate of transcription of each allele is symbolized by the thickness of the arrow. Identity of the protein complex (striped oval) exhibiting stress-repressible binding to URS1-ARE is unknown (but see text). The distorted box represents a 2-bp substitution of HSE1, while the crossed-out box represents a 32-bp substitution of HSE1 and flanking sequence. Not shown is the presence of open RNA polymerase II complexes thought to exist on the induced WT allele upstream of the initiator site (Inr) and paused elongation complexes downstream of it (13). Whereas the WT and P2 promoters have nuclease-hypersensitive, nucleosome-free (or disrupted) structures, the ΔHSE1· promoter is characterized by the presence of two stably positioned nucleosomes, one translationally positioned and the other rotationally phased (open and filled rectangles, respectively). cis-acting elements are depicted on the dinucleosome at their approximate mapped locations (8). The structure of the ΔHSE1·t promoter, based on genomic footprinting assays (8), appears to be intermediate between that of P2 and ΔHSE1·.
Our data thus confirm earlier reports that a high level of HSE binding activity exists in S. cerevisiae prior to heat shock (3, 17, 19, 28, 41, 42). They are also fully consistent with more recent findings (13) that HSE occupancy, at least at HSP82, markedly increases following heat shock. Extrapolation from in vitro footprinting assays suggest that the endogenous DNA binding activity of ScHSF increases approximately 1 order of magnitude upon heat shock, in agreement with earlier estimates (13). This compares to the 2- to 3-order-of-magnitude increase in DNA binding activity estimated for Drosophila and mammalian HSFs, which undergo a monomer-to-trimer transition prior to being targeted to the nucleus (reviewed in references 24 and 50). Interestingly, heat shock-inducible binding has not been observed at the HSEs of two other yeast heat shock promoters, HSC82 (6, 7) and HSP26 (3). While HSC82 is only slightly (2- to 3-fold) inducible, HSP26 is strongly (>50-fold) induced by heat shock. Thus, the basis for this difference is unclear.
Stress-repressible interactions at URS1-ARE represent, to our knowledge, the first example of such in vivo binding by a transcriptional regulator. URS1 is a negative regulatory element of meiotically induced genes (reviewed in reference 29). It is thought to mediate cis repression by recruitment of the Ume6-Sin3-Rpd3 histone deacetylase complex (HDAC) (20), which specifically binds to the URS1 motif (2) and deacetylates histones H3 and H4 within an ∼360-bp region of the target promoter (34). Our experiments indicate that a minor-groove binding activity, centered at the bipartite URS1-ARE element (−127 to −112), is lost upon heat shock. This may reflect the binding and heat shock-specific release of the Ume6 complex. This stress-repressible interaction, although subtle, has been seen in three different genetic backgrounds and in a number of hsp82 promoter mutants (8). Importantly, it is not seen in those alleles that have undergone the dinucleosomal transition (ΔHSE1 and ΔHSE1·), nor is it seen at an hsp82 allele bearing an 10-bp substitution of the URS1 sequence and exhibiting a twofold increase in noninduced transcription (data not shown). The loss of the URS1-ARE footprint at hsp82-ΔHSE1 may indicate that the putative HDAC is incapable of binding to the surface of a sequence-positioned nucleosome. Further investigation is necessary to identify the factor binding to this site, and to test these and other intriguing possibilities.
HSF cooperatively binds to the HSP82 promoter both in vivo and in vitro.
A major conclusion of this work is that binding of HSF to HSE2 and -3 is cooperative with its binding to the high-affinity HSE1 site, both in vivo and in vitro. Cooperative binding in vivo is revealed by in situ mutations in HSE1 which not only impair HSF binding to the mutated element but also cause a pronounced reduction in occupancy of the upstream HSEs. Paralleling the diminished binding in vivo, the apparent affinity of the upstream HSEs for recombinant HSF in vitro decreases by an order of magnitude as a consequence of a double-point HSE1 mutation (P2), and by at least two orders of magnitude as a consequence of a full substitution (ΔHSE1·t). That GST-ScHSF faithfully recapitulates normal HSF function is suggested by the fact that yeast cells expressing the GST fusion protein as the sole source of HSF are viable, show no temperature sensitivity, and transactivate HSP82 normally (8). Whether cooperativity exists between sites 2 and 3 is unknown since comparable experiments have not been performed with single HSE2 or HSE3 mutants.
Similar to what we have described here, HSF binds cooperatively to two closely spaced HSEs upstream of the Drosophila hsp70 gene, both in vitro (1, 47) and in vivo (1, 4). Such cooperativity is sensitive to the relative helical orientation of the two HSEs (HSEI and -II) and is abrogated by insertions of >18 bp (1). Therefore, propinquity of the two HSEs (exhibiting a center-to-center distance of 24 bp) is a principal determinant of cooperative binding at hsp70 (10). Whether a similar requirement applies at HSP82, where the center-to-center distance between HSE1 and -2 is 30 bp, is unknown. Unlike HSP82, hsp70 is strongly dependent on the upstream degenerate HSE for activated transcription. Deletion of HSEII causes a 90% reduction in transcription (1, 47); in contrast, deletion of HSE2 and -3 has no effect on non-induced HSP82 transcription and results in only a 25% reduction in heat shock-induced transcription. Thus, cooperative binding of HSF to HSE2 and -3 contributes only modestly to HSP82 transcript levels. Interestingly, cooperative interactions between HSE1 and HSE2 and -3 are pivotal to the activation of the divergently transcribed YAR1 gene in the context of the −673 to −265 chromosomal deletion (hsp82-265). Northern analysis of a strain bearing this allele reveals that YAR1 RNA levels, rather than being heat shock-repressed as found for WT (26), are strongly induced, and with kinetics that parallel those of HSP82. However, mutagenesis of either HSE2/3 (hsp82-190) or HSE1 (hsp82-265/P2) obviates this stress induction (35).
We have considered a potential role for the consensus stress response element (STRE), CCCCT, located 10 bp upstream of the TATA box and target of the Msn2p and Msn4p stress-responsive activators, in mediating hsp82 transcriptional induction. Such a sequence has been shown to mediate thermal and HOG pathway signals in the promoters of DNA damage-responsive genes (reviewed in references 30 and 33). However, as assayed by DMS footprinting, the sequence is not detectably occupied in either control or induced cells (data not shown); further, a 5-bp substitution of the STRE is virtually without phenotype in response to either thermal or osmotic shock (44). We conclude that HSF is the principal, if not sole, regulator of HSP82 stress responsiveness.
Overall affinity of HSF for the HSP82 promoter correlates with noninduced transcription levels.
A novel finding of this study is the striking correlation between the intrinsic affinity of HSF for the HSP82 upstream region (defined as spanning −300 to +50), and non-heat shock transcript levels. This implies that the expression level of HSP82 in noninduced cells is dictated largely, if not exclusively, by the overall affinity of HSF for its target HSEs. A similar relationship between in vivo binding affinity and transactivation has been shown for GAL4 in its regulation of the GAL1 and GAL10 genes (14). This straightforward relationship is not likely to apply following heat shock, however, even though an approximate linear correlation exists between induced expression levels and the strength of the HSF-HSE2/3 interaction (as assayed hyperreactivity of −210 G). Increased occupancy of the heat shock enhancer reflects an increase in the intracellular concentration of competent HSF. However, HSE2 and -3 together make only a minor contribution to the induced expression phenotype, as the constitutively bound HSF-HSE1 complex itself drives over 70% of induced HSP82 transcription (Fig. 4). Thus, a regulated step subsequent to DNA binding—such as stress-induced phosphorylation and/or a conformational change in HSF (18, 40, 42)—is more likely responsible for the 15- to 20-fold increase in transcription of HSP82 following heat shock than is the recruitment of HSF trimers to HSE2 and -3.
Cooperative DNA binding of HSF to the point mutants explains retention of the remodeled chromatin phenotype.
One of the motivations for this study was to elucidate the molecular basis for the disparate expression and structural phenotypes of the ΔHSE1 and P2 alleles. As discussed above (see the introduction), a paradoxical finding has been that HSE1 point mutants such as P2, despite showing no evidence of protein-DNA interaction at the mutated HSE, nonetheless retain the 5′ DNase I-hypersensitive site in chromatin (21, 28) and a disrupted MNase cleavage profile over the promoter (8, 12), virtually indistinguishable from WT. In contrast, hsp82 mutants in which HSE1 and flanking nucleotides have been either fully deleted or substituted undergo a dramatic remodeling in which a stable dinucleosomal structure replaces the accessible, nuclease-hypersensitive structure characteristic of the WT promoter (16). This paradox can now be understood in light of the present results. In P2 and other HSE1 point mutants (single, double, and triple), as in WT, the three HSEs are cooperatively bound by HSF, both before and after heat shock. In the alleles in which HSE1 has been fully deleted or substituted, no such cooperativity is possible, since HSE2 and -3 have <1% of the affinity for HSF as they do in their native context and <10% of the affinity for HSF as they do in the P2 context. Clearly, in the case of the HSE1 point mutants, HSF binding is fractional, and at least under noninducing conditions, a single trimer may rapidly exchange between the three weak sites. Models of three hypothetical classes of hsp82 promoter structure, epitomized by the WT, P2, and ΔHSE1· alleles, are depicted in Fig. 10.
It is of interest that as for ScHSF, GAL4 binding to a weak site is associated with remodeling of the underlying nucleosome in absence of stable GAL4-UASG interactions (52). Likewise, a single TRE binding site for the thyroid hormone receptor-retinoid X receptor (TR-RXR) heterodimer is equally effective in chromatin disruption as four clustered TREs, yet only the latter efficiently transactivates a linked promoter (49). For activators such as ScHSF, GAL4, and TR-RXR, chromatin remodeling is perhaps a more fundamental activity than transcriptional activation itself.
ACKNOWLEDGMENTS
We thank Stephan Witt for assistance in deriving affinity constants and for conducting computer simulations of template-ligand interactions; Karen English, Chris Adams, and Tuba Diken for technical assistance; Bill Garrard and Christina Bourgeois Venturi for critical reading of the manuscript; and John Lis, Paul Mason, Nick Santoro, Chris Szent-Gyorgyi, and Dennis Thiele for gifts of strains and reagents.
This work was supported by grants to D.S.G. from the National Institute of General Medical Sciences (GM45842), the American Cancer Society, Inc. (NP-945), and the Center for Excellence in Cancer Research at LSUMC—Shreveport.
REFERENCES
- 1.Amin J, Fernandez M, Ananthan J, Lis J T, Voellmy R. Cooperative binding of heat shock transcription factor to the Hsp70 promoter in vivo and in vitro. J Biol Chem. 1994;269:4804–4811. [PubMed] [Google Scholar]
- 2.Anderson S F, Steber C M, Esposito R E, Coleman J E. UME6, a negative regulator of meiosis in Saccharomyces cerevisiae, contains a C-terminal Zn2Cys6 binuclear cluster that binds the URS1 DNA sequence in a zinc-dependent manner. Protein Sci. 1995;4:1832–1843. doi: 10.1002/pro.5560040918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J, Pederson D S. A distal heat shock element promotes the rapid response to heat shock of the HSP26 gene in the yeast Saccharomyces cerevisiae. J Biol Chem. 1993;268:7442–7448. [PubMed] [Google Scholar]
- 4.Cohen R S, Meselson M. Periodic interactions of heat shock transcriptional elements. Nature. 1988;332:856–858. doi: 10.1038/332856a0. [DOI] [PubMed] [Google Scholar]
- 5.Diken, T., and D. S. Gross. 1994. Unpublished results.
- 6.Erkine A M, Adams C C, Diken T, Gross D S. Heat shock factor gains access to the yeast HSC82 promoter independently of other sequence-specific factors and antagonizes nucleosomal repression of basal and induced transcription. Mol Cell Biol. 1996;16:7004–7017. doi: 10.1128/mcb.16.12.7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erkine A M, Adams C C, Gao M, Gross D S. Multiple protein-DNA interactions over the yeast HSC82 heat shock gene promoter. Nucleic Acids Res. 1995;23:1822–1829. doi: 10.1093/nar/23.10.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erkine, A. M., and D. S. Gross. 1998. Unpublished results.
- 9.Erkine A M, Szent-Gyorgyi C, Simmons S F, Gross D S. The upstream sequences of the HSP82 and HSC82 genes of Saccharomyces cerevisiae: regulatory elements and nucleosome positioning motifs. Yeast. 1995;11:573–580. doi: 10.1002/yea.320110607. [DOI] [PubMed] [Google Scholar]
- 10.Fernandes M, Xiao H, Lis J T. Binding of heat shock factor to and transcriptional activation of heat shock genes in Drosophila. Nucleic Acids Res. 1995;23:4799–4804. doi: 10.1093/nar/23.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandes M, Xiao H, Lis J T. Fine structure analyses of the Drosophila and Saccharomyces heat shock factor-heat shock element interactions. Nucleic Acids Res. 1994;22:167–173. doi: 10.1093/nar/22.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, M., S. Lee, and D. S. Gross. 1993. Unpublished results.
- 13.Giardina C, Lis J T. Dynamic protein-DNA architecture of a yeast heat shock promoter. Mol Cell Biol. 1995;15:2737–2744. doi: 10.1128/mcb.15.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giniger E, Ptashne M. Cooperative DNA binding of the yeast transcriptional activator GAL4. Proc Natl Acad Sci USA. 1988;85:382–386. doi: 10.1073/pnas.85.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodson M L, Sarge K D. Heat-inducible DNA binding of purified heat shock transcription factor 1. J Biol Chem. 1995;270:2447–2450. doi: 10.1074/jbc.270.6.2447. [DOI] [PubMed] [Google Scholar]
- 16.Gross D S, Adams C C, Lee S, Stentz B. A critical role for heat shock transcription factor in establishing a nucleosome-free region over the TATA-initiation site of the yeast HSP82 heat shock gene. EMBO J. 1993;12:3931–3945. doi: 10.1002/j.1460-2075.1993.tb06071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross D S, English K E, Collins K W, Lee S. Genomic footprinting of the yeast HSP82 promoter reveals marked distortion of the DNA helix and constitutive occupancy of heat shock and TATA elements. J Mol Biol. 1990;216:611–631. doi: 10.1016/0022-2836(90)90387-2. [DOI] [PubMed] [Google Scholar]
- 18.Hoj A, Jakobsen B K. A short element required for turning off heat shock transcription factor: evidence that phosphorylation enhances deactivation. EMBO J. 1994;13:2617–2624. doi: 10.1002/j.1460-2075.1994.tb06552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakobsen B K, Pelham H R B. Constitutive binding of yeast heat shock factor to DNA in vivo. Mol Cell Biol. 1988;8:5040–5042. doi: 10.1128/mcb.8.11.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 21.Lee M S, Garrard W T. Uncoupling gene activity from chromatin structure: promoter mutations can inactivate transcription of the yeast HSP82 gene without eliminating nucleosome-free regions. Proc Natl Acad Sci USA. 1992;89:9166–9170. doi: 10.1073/pnas.89.19.9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S, Gross D S. Conditional silencing: The HMRE mating-type silencer exerts a rapidly reversible position effect on the yeast HSP82 heat shock gene. Mol Cell Biol. 1993;13:727–738. doi: 10.1128/mcb.13.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindquist S. Regulation of protein synthesis during heat shock. Nature. 1981;293:311–314. doi: 10.1038/293311a0. [DOI] [PubMed] [Google Scholar]
- 24.Lis J T, Wu C. Protein traffic on the heat shock promoter: parking, stalling, and trucking along. Cell. 1993;74:1–4. doi: 10.1016/0092-8674(93)90286-y. [DOI] [PubMed] [Google Scholar]
- 25.Liu-Johnson H-N, Gartenberg M, Crothers D. The DNA binding domain and bending angle of E. coli CAP protein. Cell. 1986;47:995–1005. doi: 10.1016/0092-8674(86)90814-7. [DOI] [PubMed] [Google Scholar]
- 26.Lycan D E, Stafford K A, Bollinger W, Breeden L L. A new Saccharomyces cerevisiae ankyrin repeat-encoding gene required for a normal rate of cell proliferation. Gene. 1996;171:33–40. doi: 10.1016/0378-1119(96)00023-6. [DOI] [PubMed] [Google Scholar]
- 27.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 28.McDaniel D, Caplan A J, Lee M S, Adams C C, Fishel B R, Gross D S, Garrard W T. Basal-level expression of the yeast HSP82 gene requires a heat shock regulatory element. Mol Cell Biol. 1989;9:4789–4798. doi: 10.1128/mcb.9.11.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell A P. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morano K A, Liu P C C, Thiele D J. Protein chaperones and the heat shock response in Saccharomyces cerevisiae. Curr Opin Microbiol. 1998;1:197–203. doi: 10.1016/s1369-5274(98)80011-8. [DOI] [PubMed] [Google Scholar]
- 31.Morimoto R I. Cells in stress: transcriptional activation of heat shock genes. Science. 1993;259:1409–1410. doi: 10.1126/science.8451637. [DOI] [PubMed] [Google Scholar]
- 32.Morimoto R I, Tissières A, Georgopoulos C. The stress response, function of the proteins, and perspectives. In: Morimoto R, Tissières A, Georgopoulos C, editors. Stress proteins in biology and medicine. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. pp. 1–36. [Google Scholar]
- 33.Ruis H, Schuller C. Stress signaling in yeast. Bioessays. 1995;17:959–965. doi: 10.1002/bies.950171109. [DOI] [PubMed] [Google Scholar]
- 34.Rundlett S E, Carmen A A, Suka N, Turner B M, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 35.Sekinger, E. A., and D. S. Gross. 1998. Unpublished results.
- 36.Sewell A K, Yokoya F, Yu W, Miyagawa T, Murayama T, Winge D R. Mutated yeast heat shock transcription factor exhibits elevated basal transcriptional activation and confers metal resistance. J Biol Chem. 1995;270:25079–25086. doi: 10.1074/jbc.270.42.25079. [DOI] [PubMed] [Google Scholar]
- 37.Sheldon L A, Kingston R E. Hydrophobic coiled-coil domains regulate the subcellular localization of human heat shock factor 2. Genes Dev. 1993;7:1549–1558. doi: 10.1101/gad.7.8.1549. [DOI] [PubMed] [Google Scholar]
- 38.Shopland L S, Hirayoshi K, Fernandes M, Lis J T. HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes Dev. 1995;9:2756–2769. doi: 10.1101/gad.9.22.2756. [DOI] [PubMed] [Google Scholar]
- 39.Silar P, Butler G, Thiele D J. Heat shock transcription factor activates transcription of the yeast metallothionein gene. Mol Cell Biol. 1991;11:1232–1238. doi: 10.1128/mcb.11.3.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sorger P K. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell. 1990;62:793–805. doi: 10.1016/0092-8674(90)90123-v. [DOI] [PubMed] [Google Scholar]
- 41.Sorger P K, Lewis M J, Pelham H R B. Heat shock factor is regulated differently in yeast and HeLa cells. Nature. 1987;329:81–84. doi: 10.1038/329081a0. [DOI] [PubMed] [Google Scholar]
- 42.Sorger P K, Pelham H R B. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988;54:855–864. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- 43.Szent-Gyorgyi C. A bipartite operator interacts with a heat shock element to mediate early meiotic induction of Saccharomyces cerevisiae HSP82. Mol Cell Biol. 1995;15:6754–6769. doi: 10.1128/mcb.15.12.6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szent-Gyorgyi, C., G. Alba, and D. S. Gross. 1998. Unpublished results.
- 45.Szent-Gyorgyi C, Finkelstein D B, Garrard W T. Sharp boundaries demarcate the chromatin structure of a yeast heat-shock gene. J Mol Biol. 1987;193:71–80. doi: 10.1016/0022-2836(87)90628-0. [DOI] [PubMed] [Google Scholar]
- 46.Taylor I C, Workman J L, Schuetz T J, Kingston R E. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 1991;5:1285–1298. doi: 10.1101/gad.5.7.1285. [DOI] [PubMed] [Google Scholar]
- 47.Topol J, Ruden D M, Parker C S. Sequences required for in vitro transcriptional activation of a Drosophila hsp70 gene. Cell. 1985;42:527–537. doi: 10.1016/0092-8674(85)90110-2. [DOI] [PubMed] [Google Scholar]
- 48.Walker J, Crowley P, Moreman A D, Barrett J. Biochemical properties of cloned glutathione S-transferases from Schistosoma mansoni and Schistosoma japonicum. Mol Biochem Parasitol. 1993;61:255–264. doi: 10.1016/0166-6851(93)90071-5. [DOI] [PubMed] [Google Scholar]
- 49.Wong J, Shi Y-B, Wolffe A. Determinants of chromatin disruption and transcriptional regulation instigated by the thyroid hormone receptor: hormone-regulated chromatin disruption is not sufficient for transcriptional activation. EMBO J. 1997;16:3158–3171. doi: 10.1093/emboj/16.11.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 51.Xiao H, Perisic O, Lis J T. Cooperative binding of Drosophila heat shock factor to arrays of a conserved 5 bp unit. Cell. 1991;64:585–593. doi: 10.1016/0092-8674(91)90242-q. [DOI] [PubMed] [Google Scholar]
- 52.Xu M, Simpson R T, Kladde M P. Gal4p-mediated chromatin remodeling depends on binding site position in nucleosomes but does not require DNA replication. Mol Cell Biol. 1998;18:1201–1212. doi: 10.1128/mcb.18.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zandi E, Tran T T, Chamberlain W, Parker C S. Nuclear entry, oligomerization, and DNA binding of the Drosophila heat shock transcription factor are regulated by a unique nuclear localization sequence. Genes Dev. 1997;11:1299–1314. doi: 10.1101/gad.11.10.1299. [DOI] [PubMed] [Google Scholar]
- 54.Zhong M, Orosz A, Wu C. Direct sensing of heat and oxidation by Drosophila heat shock transcription factor. Mol Cell. 1998;2:101–108. doi: 10.1016/s1097-2765(00)80118-5. [DOI] [PubMed] [Google Scholar]