Abstract
Loss of the Epstein-Barr virus (EBV) genome from Akata Burkitt lymphoma (BL) cells is coincident with a loss of malignant phenotype, despite the fact that Akata and other EBV-positive BL cells express a restricted set of EBV gene products (type I latency) that are not known to overtly affect cell growth. Here we demonstrate that reestablishment of type I latency in EBV-negative Akata cells restores tumorigenicity and that tumorigenic potential correlates with an increased resistance to apoptosis under growth-limiting conditions. The antiapoptotic effect of EBV was associated with a higher level of Bcl-2 expression and an EBV-dependent decrease in steady-state levels of c-MYC protein. Although the EBV EBNA-1 protein is expressed in all EBV-associated tumors and is reported to have oncogenic potential, enforced expression of EBNA-1 alone in EBV-negative Akata cells failed to restore tumorigenicity or EBV-dependent down-regulation of c-MYC. These data provide direct evidence that EBV contributes to the tumorigenic potential of Burkitt lymphoma and suggest a novel model whereby a restricted latency program of EBV promotes B-cell survival, and thus virus persistence within an immune host, by selectively targeting the expression of c-MYC.
Epstein-Barr virus (EBV) is a ubiquitous human herpesvirus that establishes a life-long latent infection within B lymphocytes. Latency is associated with the expression of as many as ten viral proteins, including a family of six nuclear proteins (EBNAs), three integral membrane proteins (LMPs) and the recently identified polypeptide RK-BARF0 (17, 24). Several of these, most notably the EBNA-3 proteins, elicit a strong EBV-specific cytotoxic T-lymphocyte immune response (23, 38, 43). Through differential expression of EBV proteins that are immunogenic, potentially oncogenic and those necessary for sustaining infection, an equilibrium is established between infected B cells and the host’s immune surveillance. As a consequence, a relatively stable pool of latently infected cells is maintained by the host (31, 36, 56). Thus, EBV is particularly well adapted for persistence within B lymphocytes of an immune host, and though the virus encodes the oncogenic protein LMP-1, its association with human malignancy is rare, especially considering the high incidence of EBV infection worldwide.
As a result of the balanced relationship that exists between EBV and its host, pathogenesis associated with EBV latency is generally a consequence of either an immune deficiency, leading to EBV-induced immunoblastic lymphoma, or a known or suspected secondary genetic or environmental event that promotes full oncogenic transformation of latently infected cells (43). Nevertheless, expression of LMP-1 in some tumors, most notably in EBV-positive Hodgkin’s lymphoma and nasopharyngeal carcinoma, strongly suggests that EBV indeed actively contributes to the oncogenic potential of these malignancies (5, 11, 14, 22, 41, 53, 61).
In contrast to tumors such as Hodgkin’s lymphoma and nasopharyngeal carcinoma, LMP-1 is not expressed in EBV-positive Burkitt lymphoma (BL), in which the pattern of viral gene expression, referred to as type I latency, is restricted to the proteins EBNA-1 (required for EBV DNA maintenance) and RK-BARF0 and to two small noncoding but highly expressed RNA polymerase III transcripts, EBER-1 and EBER-2 (2, 6, 17, 18, 46). Neither RK-BARF0 or the EBERs are required for EBV-mediated immortalization of B cells in vitro (44, 54). Furthermore, although EBNA-1 has been linked to development of B-cell lymphomas in some transgenic mice (59, 60), there is as yet no evidence that EBNA-1 directly contributes to tumorigenicity in EBV-associated malignancies. Overall, these findings argue against a significant contribution by EBV to tumorigenic potential in established BL. Thus, the dominant factor responsible for maintenance of tumorigenicity in BL is believed to be the deregulated expression of the c-MYC proto-oncogene (in this paper, c-MYC refers to the gene and c-myc refers to the mRNA and cDNA), which occurs as the result of characteristic chromosomal translocations that juxtapose the c-MYC gene and immunoglobulin loci (reviewed in reference 29). Indeed, programmed expression of c-MYC in the B-cell compartment of transgenic mice predisposes these animals to protracted development of lymphoma (1).
The assumption that EBV does not directly contribute to tumorigenicity in BL, however, has been challenged by the finding that a loss in tumorigenic potential is associated with spontaneous loss of the EBV genome in the Akata BL cell line, which in culture maintains the type I latency program characteristic of primary BL tumors (52). Here we report that EBV-associated tumorigenicity of Akata BL cells correlates with an increased resistance to apoptosis that is coincident with a down-regulation of c-MYC protein, but not mRNA, and increased expression of the antiapoptotic protein Bcl-2. Reinfection of EBV-negative Akata cells, but not stable expression of EBNA-1 alone, restored EBV-dependent regulation of c-MYC and tumorigenicity. These data therefore provide direct evidence that EBV can significantly contribute to tumorigenic potential in BL. The data also indicate that the type I latency program of EBV gene expression contributes to long-term survival of latently infected cells and suggest a novel model whereby EBV suppresses apoptosis by selectively down-regulating c-MYC protein expression under limiting growth conditions.
MATERIALS AND METHODS
Cell culture and EBV infection.
Cell lines were maintained in RPMI 1640 medium supplemented with 2 mM l-glutamine and 10% defined fetal bovine serum (HyClone); the exceptions were HL60 and K562, which were maintained in media containing 20% serum. Akata, SavI, KemI, and MutuI are human group I BL cell lines that maintain a type I EBV latency. IB4 is an EBV-immortalized human B-lymphoblastoid cell line (LCL) that expresses all known EBV latency-associated genes (type III latency). HL60 and K562 are human promyelocytic and erythroleukemic cell lines, respectively. Clonal EBV-positive Akata cells were derived from colonies isolated in soft agar (see below) and designated A.1, A.3, A.5, etc. The clonal EBV-negative Akata cell lines 3F2, 2C1, and 2A8 (gift of J. Sixbey) were isolated by single-cell sorting (FACStar Plus; Becton Dickinson) following treatment of parental EBV-positive Akata cells with 50 μM hydroxyurea (9). Loss of the EBV genome in these cells was confirmed by the lack of EBNA-1 expression as determined by immunoblot analysis and by the inability to amplify EBV DNA by PCR with primers specific for the large internal (BamHI-W) repeat of the EBV genome and/or the inability to detect EBV DNA by fluorescence in situ hybridization (FISH) using a cosmid DNA probe containing the EBV SalI B fragment (9).
EBV-negative Akata cells were reinfected with wild-type EBV (Akata strain) produced from parental (EBV-positive) Akata cells treated with 1% (vol/vol) anti-immunoglobulin G for 48 h to induce production of virus (55). Cells were removed by centrifugation, and the supernatant was prefiltered through a 0.8-μm-pore-size nitrocellulose membrane (Nalgene). Virus was then concentrated approximately 100-fold by positive-pressure filtration through a membrane with a molecular weight cutoff of 500. To infect, 2 × 106 cells were pelleted by centrifugation, resuspended in 1 ml of concentrated virus, and incubated at 37°C for 1 h. The cells were then washed with and resuspended in fresh growth medium (8 ml), and were reinfected approximately 5 days later when the cells required feeding. Alternatively, cells were infected once with a recombinant virus, derived from the Akata strain of EBV, that contained a neomycin resistance gene at the site of the BDLF3 (gp150) open reading frame (4). Clonal lines of reinfected Akata cells were then isolated by single-cell sorting followed by confirmation of EBNA-1 expression by immunoblotting. In some instances, Akata cells reinfected with the recombinant EBV were maintained in medium containing 400 μg of G418 (Geneticin; Gibco BRL) per ml to ensure that all cells in the culture contained the EBV genome.
Cell death assays.
Cultures were seeded at 2.5 × 105 cells per ml in standard growth medium (containing 10% serum), and cell numbers were determined daily to assess growth rate. On days 2 through 6, an aliquot of cells was removed, washed three times in growth medium that contained 0.1% serum, and then resuspended in this reduced-serum medium at 5 × 105 cells per ml. Viability was then determined daily, using trypan blue dye exclusion to monitor cell survival. For cytospin preparations, cells from 100 μl of culture were pelleted onto a glass slide in a Cyto-Tek chamber holder (Sakura), air dried, fixed in methanol for 5 min at room temperature, air dried, stained in 0.02% (wt/vol) modified Giemsa stain (Sigma) for 45 min at room temperature, rinsed several times in distilled water, air dried, and then mounted in Permount (Fisher).
Immunoblotting.
For the detection of EBNA-1, LMP-1, c-MYC, and Bcl-2 by immunoblotting, 106 cells were washed once in phosphate-buffered saline, lysed in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer (100 mM Tris-HCl [pH 6.8], 200 mM dithiothreitol [DTT], 4% SDS, protease inhibitor cocktail [Sigma], 20% glycerol, 0.2% bromophenol blue), immediately heated at 100°C for 5 min, and sonicated. Proteins were fractionated by SDS-PAGE in 10% acrylamide gels, transferred to an Immobilon P membrane (Millipore), and immunoblotted by using an enhanced chemiluminescence detection system (Amersham). Primary antibodies used for immunoblotting were mouse monoclonal antibodies to c-MYC (9E10 [13]), Bcl-2 (M0887; Dako), and LMP-1 (S12 [30]) and a polyclonal rabbit antiserum to EBNA-1 (gift of J. Hearing). Immunoreactive proteins were detected with secondary antibodies conjugated to horseradish peroxidase. All blots were subsequently stripped of antibody in 62.5 mM Tris-HCl (pH 6.8)–100 mM 2-mercaptoethanol–2% SDS (50°C for 30 min) and reprobed with a mouse monoclonal antibody to actin (N350; Amersham) as a control for protein loading. For analysis of protein expression in SCID mouse tumors, approximately equal amounts of tumor tissue were solubilized in SDS-PAGE sample buffer and processed as described above. For detection of EBNA-1 and LMP-1 in Fig. 7, 107 cells were lysed in 200 μl of 50 mM Tris (pH 8.0)–150 mM NaCl–1 mM EDTA–1% Triton X-100–protease inhibitor cocktail–0.5 mM phenylmethylsulfonyl fluoride (PMSF) and then centrifuged at 12,000 × g for 5 min to remove insoluble material. The protein concentration of the supernatant was determined by the Bradford method (Bio-Rad), and 50 μg of protein was then subjected to SDS-PAGE and immunoblotting.
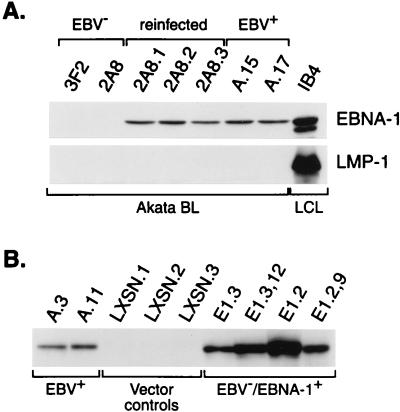
FIG. 7.
Restoration of restricted EBV latency and establishment of EBV-independent EBNA-1 expression in EBV-negative Akata cells. (A) 2A8 Akata cells reinfected with EBV (lines 2A8.1, 2A8.2, and 2A8.3) express normal levels of EBNA-1 but, like their EBV-positive counterparts A.15 and A.17, do not express detectable LMP-1. The IB4 LCL provides a positive control and reference for detection of LMP-1. (B) Stable expression of EBNA-1 in EBV-negative Akata cells. A.3 and A.11 are EBV-positive Akata cells; LXSN.1, LXSN.2, and LXSN.3 are G418-resistant EBV-negative Akata cell lines that contain the expression vector pLXSN without DNA encoding EBNA-1; E1.3,12 and E1.2,9 are clonal EBV−/EBNA-1+ Akata cells derived from the nonclonal E1.3 and E1.2 lines, respectively. All lanes contained 50 μg of total cellular protein.
FIG. 10.
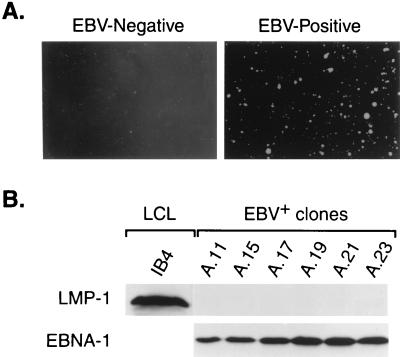
Tumors in SCID mice express EBNA-1 but not LMP-1. Representative results from immunoblot analysis of EBNA-1 and LMP-1 expression in tumors that resulted from injection of EBV-positive A.15 and reinfected 2A8 Akata cells are shown. Protein from IB4 cells was used as a positive control; tissue harvested from a site of injection of EBV-negative Akata cells served as a negative control (lane 1). Results of the analysis of all 13 tumors obtained are presented in Table 1.
Measurement of c-MYC stability and synthesis.
For cycloheximide-chase experiments, protein synthesis was inhibited in cells that were in either mid-log- or stationary-phase growth by addition of cycloheximide (Calbiochem-Novabiochem) to the cell culture at a final concentration of 25 μg/ml. An aliquot of 3 × 106 cells was harvested at 0, 15, 30, 60, 120, and 240 min after addition of cycloheximide, washed once in phosphate-buffered saline, resuspended in lysis buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 150 mM NaCl, 0.5% Nonidet P-40 [NP-40], 1 mM PMSF), and incubated for 20 min at 4°C. Lysate containing protein from the equivalent of 106 cells from each chase point was then analyzed for c-MYC expression by immunoblotting with anti-c-MYC antibody (9E10) as described above. For pulse-chase analysis, EBV-negative and -positive Akata cells (108) in log- or stationary-phase growth were washed twice in methionine- and cysteine-free RPMI 1640 medium (BioWhittaker) containing l-glutamine and 10% serum. Cells were then incubated at 37°C in this medium for 30 min, followed by a pulse-labeling period of 15 min in the same medium containing 1 mCi of Tran35S-label (ICN). Cells were then washed twice in medium containing a fivefold excess of methionine and cysteine and incubated in this medium for up to 240 min. At chase times of 0, 15, 30, 60, 120, and 240 min, 1.25 × 107 cells were removed, pelleted, resuspended in 1 ml of lysis buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 150 mM NaCl, 0.5% NP-40, 1 mM PMSF), and incubated for 20 min at 4°C. All lysates were then precleared with Protein G Plus-Agarose (Oncogene Research Products) to remove nonspecific adsorbents. Lysate containing protein from the equivalent of 107 cells was processed by immunoprecipitation with 2 μg of antibody to c-MYC (Ab-3; Oncogene Research Products) and 20 μl of Protein G Plus-Agarose. Immunoprecipitates were washed three times in SNNTE buffer (5% sucrose, 1% NP-40, 500 mM NaCl, 50 mM Tris-HCl [pH 7.5], 5 mM EDTA) and once in radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate), resuspended in 30 μl of loading buffer (100 mM Tris-HCl, 200 mM DTT, 4% SDS, 20% glycerol, 0.2% bromophenol blue), boiled for 4 min, and then subjected to SDS-PAGE in a 10% acrylamide gel. Gels were fixed in 10% acetic acid–30% methanol for 30 min, washed three times in distilled water for 10 min, soaked in fluor solution (1 M sodium salicylate, 5% glycerol) for 20 min, dried, and exposed to Kodak BioMax MR film. To calculate protein half-life, immunoprecipitated c-MYC from each chase point was quantified by phosphorimage analysis (Molecular Dynamics) and normalized for total protein, and the resulting data were subjected to linear regression analysis.
Analysis of RNA.
Total cellular RNA was isolated from 107 cells with RNAzol B as recommended by the manufacturer (Tel-Test), followed by extraction with an equal volume of phenol-chloroform and then chloroform prior to ethanol precipitation. For Northern blot analysis, 10 μg of RNA per sample was fractionated by electrophoresis in a 1.2% agarose–2.2 M formaldehyde gel (48) and blotted onto a GeneScreen Plus membrane (DuPont). RNA blots were prehybridized for 4 h at 45°C followed by hybridization at the same temperature to a 32P-labeled human c-myc cDNA probe under conditions described previously (39). Following hybridization, blots were washed at 62°C (49) and processed by autoradiography. Blots were then stripped and rehybridized to a probe specific for 28S rRNA to control for differences in RNA loading of the gel.
Generation of EBV−/EBNA-1+ cell lines.
To generate EBV-negative Akata cells that stably expressed EBNA-1, the EBNA-1 open reading frame was inserted into the BamHI restriction site of the retroviral expression vector pLXSN (33) to yield pLE1SN, in which expression of EBNA-1 is under control of the retroviral 5′ long terminal repeat. Ten micrograms of pLE1SN or pLXSN (to generate control cell lines) was used to transfect 8 × 106 EBV-negative Akata cells by electroporation as described previously (50). At 48 h posttransfection, cells were placed in 24-well tissue culture plates (104 cells per well) in medium containing 200 μg of G418 per ml. To generate clonal EBV−/EBNA-1+ cell lines, single cells were isolated from pools of G418-resistant EBNA-1-positive cells by cell sorting (FACStar Plus; Becton Dickinson) into 96-well plates containing conditioned medium. Following expansion of these cells under selection with G418, clones that expressed EBNA-1 were identified by immunoblotting.
Growth in soft agar and tumorigenicity assays.
For the analysis of growth potential in soft agar, 104 cells were suspended in 3 ml of 0.33% agarose (SeaPlaque; FMC Corporation) in standard growth medium (without phenol red) and plated onto a 3-ml layer of solidified 0.66% agarose in the same medium in a 6-cm-diameter tissue culture dish. Once the top layer of agarose had solidified, the cells were incubated at 37°C in a humidified atmosphere of 5% CO2 for 4 weeks and fed weekly with 1 ml of liquid medium containing 0.33% agarose. Each cell line tested was plated in triplicate.
Tumorigenicity of Akata BL cells was assessed by the ability to induce tumors in SCID mice. Male SCID mice (C.B-17/lcr//SJ scid/scid), obtained from the colony maintained by the St. Jude Children’s Research Hospital Animal Resource Center, were injected subcutaneously in each hind flank with 2 × 107 cells in phosphate-buffered saline (200 μl). Each mouse received an injection in the right flank of either EBV-positive Akata, reinfected EBV-negative Akata, or EBV−/EBNA-1+ Akata cells and an injection in the left flank of the appropriate EBV-negative Akata cells as a control. Animals were monitored for 8 weeks for tumor growth and then sacrificed, and their tumors were excised for analysis of EBV protein expression when approximately 1 cm in diameter (5 to 7 weeks postinjection).
RESULTS
Enhanced growth potential of BL is associated with type I EBV latency.
EBV-negative Akata BL cells are nontumorigenic relative to their EBV-positive counterparts when assayed for growth in soft agar and tumor induction in athymic nude mice (52). Akata BL cells may therefore provide an ideal system to address the contribution of EBV to tumorigenicity in BL. However, the low plating efficiency of EBV-positive Akata cells in soft agar (1 to 3% [52]) raised the possibility that enhanced growth potential associated with EBV infection in Akata BL cells was not a stable phenotype. We therefore initially addressed whether EBV-positive Akata cells and several clones of EBV-negative Akata cells had consistent and measurably different growth properties. The EBV-negative clones analyzed either had lost the EBV genome spontaneously (two clones) or were generated by treatment with hydroxyurea (three clones), which hastens the loss of EBV episomes in these cells (9). Under the low dose of hydroxyurea used (50 μM), which is approximately 40-fold lower than that used therapeutically, no toxic effects were observed, nor did we observe any increase in genomic instability as a result of treatment with hydroxyurea (9). Loss of the EBV episome was confirmed by failure to detect EBV DNA by PCR and/or fluorescence in situ hybridization (FISH) (data not shown). As illustrated by representative data presented in Fig. 1A, EBV-positive Akata cells consistently exhibited a greater growth potential in soft agar relative to EBV-negative cells. Although an EBV-negative cell line would occasionally exhibit a plating efficiency similar to those of EBV-positive cells (1 to 3%), the resulting colonies from these cells were always markedly smaller than those derived from EBV-positive cells (see below). Thus, our findings were consistent with previous observations suggesting that EBV contributes to the growth potential of Akata BL cells (52).
FIG. 1.
Enhanced growth potential of EBV-positive Akata cells is independent of LMP-1. (A) Clonal EBV-negative and parental EBV-positive Akata cells were plated in 0.33% agarose at a density of 104 cells per 6-cm-diameter dish. Colonies were photographed 4 weeks after plating. (B) Individual colonies derived from the EBV-positive parental Akata cells were transferred from agarose to liquid culture, briefly expanded, and analyzed for expression of EBNA-1 and LMP-1 by immunoblotting. Each lane contained protein from 106 cells. IB4 is an EBV-immortalized LCL used as a control for detection of LMP-1 expression.
Because of the low plating efficiency of EBV-positive cells in soft agar and the potential of some BL cells to drift to a type III latency (full expression of latency-associated genes) and become LMP-1 positive (18, 45), the enhanced growth potential of EBV-positive Akata cells may have been due to the induction of LMP-1 expression under the restrictive growth conditions of this assay. We therefore isolated a number of colonies from soft agar and evaluated EBV gene expression in these cells. Whereas all six Akata BL clones examined expressed EBNA-1, none expressed detectable levels of LMP-1 (Fig. 1B), and at no time after culture in liquid growth medium have these EBV-positive Akata cell clones exhibited intercellular adhesion that is characteristic of type III latency. Thus, the greater growth potential of EBV-positive than of EBV-negative Akata BL cells is associated with a type I latency program and is independent of LMP-1. Finally, when these clonal EBV-positive Akata cell lines were reassayed for growth in soft agar (see below), their plating efficiencies were identical to that of the parental EBV-positive Akata line. Therefore, clonal lines derived in this manner do not represent a subpopulation of cells that have growth potentials greater than that of the general pool of parental EBV-positive cells.
Type I EBV latency in BL confers resistance to apoptosis under growth-limiting conditions.
Although EBV-positive Akata cells demonstrated a greater growth potential than their EBV-negative counterparts in soft agar, we did not observe a significant difference in growth properties during standard culture in liquid medium, as EBV-positive and -negative Akata cells exhibited identical growth rates and reached similar saturation densities (Fig. 2A). However, EBV-negative cells rapidly lost viability after 5 to 6 days in culture without feeding, whereas EBV-positive cells often remained viable for an additional 2 to 3 days (data not shown). This finding suggested that the mechanism(s) responsible for the increased growth potential associated with EBV in Akata BL cells was manifest only under growth-limiting conditions, such as would occur in soft agar or in vivo.
FIG. 2.
The type I EBV latency program promotes BL cell survival following serum deprivation. (A) Clonal EBV-positive (shaded symbols) and EBV-negative (open symbols) Akata cells were seeded at 2.5 × 105 cells per ml in standard growth medium containing 10% serum, and cell concentration was monitored daily for 6 days to assess growth rate. (B) Cells from the log (day 2) and stationary (day 5) phases of growth were washed and transferred to growth medium containing 0.1% serum, in triplicate, at a density of 5 × 105 cells per ml. Cell viability was determined daily by trypan blue dye exclusion. Results are representative of seven independent experiments.
Activation of c-MYC in BL augments the endogenous apoptotic program (35). We therefore reasoned that under growth-limiting conditions, EBV might enhance cell survival by suppressing c-MYC-induced apoptosis. To address this issue, we shifted cells from the logarithmic and stationary phases of the growth cycle into medium containing 0.1% serum and assessed cell viability over a 4-day period. When cells from log-phase growth were shifted to low serum, EBV-positive and -negative Akata cells died at the same rate (Fig. 2B, left panel). However, when EBV-positive Akata BL cells from stationary-phase growth were shifted to low serum, following an initial 10 to 20% loss in viability, there was a pronounced delay in the rate of cell death. By contrast, EBV-negative BL stationary-phase cells died at rates comparable to rates for these cells at log phase (Fig. 2B, right panel). Furthermore, when taken from stationary-phase cultures, EBV-positive but not EBV-negative Akata BL cells continued to proliferate at a low rate for the first 3 days in 0.1% serum. This was not observed when these cells were taken from log-phase growth.
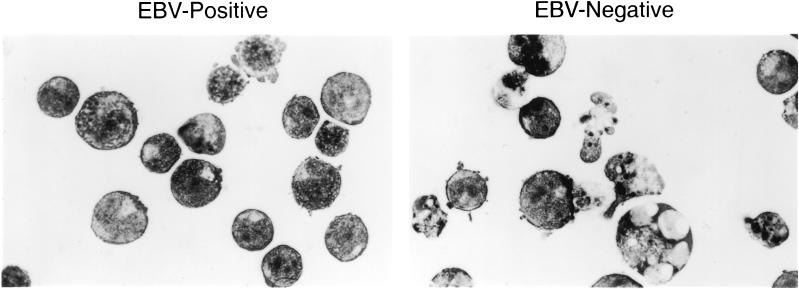
Previous studies have demonstrated that BL cells die by apoptosis in response to the withdrawal of survival factors (serum deprivation) (21). To confirm that the death of Akata BL cells in our assays was due to apoptosis, we examined cytospin preparations of stationary-phase EBV-negative and -positive cells that had been maintained in 0.1% serum for 3 days. As demonstrated in Fig. 3, EBV-negative cells displayed changes typical of apoptosis, including condensation of chromatin into micronuclei, vacuolation, and cell debris. By contrast, the majority of EBV-positive cells appeared relatively healthy. Thus, as predicted, the greatest difference in sensitivity to apoptosis of EBV-positive versus -negative Akata cells occurred under growth-limiting conditions.
FIG. 3.
Serum deprivation induces morphological changes in BL cells that are characteristic of apoptosis. EBV-positive (A.3) and -negative (2C1) Akata cells were maintained in medium containing 0.1% serum for 3 days after reaching the stationary phase of the growth cycle. Cytospin preparations shown here indicate that the EBV-negative cells displayed changes typical of apoptotic cells, including condensation of chromatin into micronuclei and vacuolation. By contrast, EBV-positive cells appeared relatively healthy.
Suppression of apoptosis by EBV is associated with down-regulation of c-MYC expression.
To determine if altered expression of c-MYC might account for EBV-mediated suppression of apoptosis, we examined c-MYC protein levels in EBV-positive and EBV-negative Akata cells at specific intervals of the cell growth cycle. Additionally, we examined the expression of several anti- and proapoptotic proteins of the Bcl-2 family. During log-phase growth (Fig. 4, days 1 to 3), there were no detectable differences in the levels of c-MYC protein between EBV-positive and EBV-negative cells. However, consistent with their fate when deprived of survival factors, c-MYC was dramatically down-regulated in EBV-positive cells as they entered the stationary phase of growth. By contrast, EBV-negative cells expressed c-MYC at levels comparable to those observed during log-phase growth. Both EBV-positive and EBV-negative Akata cells showed increased expression of the antiapoptotic protein Bcl-2 as they approached the stationary phase of growth, and EBV-positive Akata BL cells generally had higher steady-state levels of Bcl-2 protein than the EBV-negative Akata cells (Fig. 4). We did not observe any EBV- or growth phase-associated differences in the expression of the antiapoptotic proteins Mcl-1 and Bcl-XL or the proapoptotic protein Bax (data not shown).
FIG. 4.
EBV- and growth phase-associated expression of c-MYC and Bcl-2. Clonal EBV-positive (A.3 and A.15) and EBV-negative (3F2, 2C1, and 2A8) Akata BL cells were seeded (2.5 × 105 per ml) in complete growth medium, and 106 cells were harvested daily from the experiment presented in Fig. 2A for the first 5 days in culture to monitor expression of c-MYC and Bcl-2 by immunoblot analysis. Detection of actin served as a loading control. Results are representative of three independent experiments.
Because growth phase-associated regulation of c-MYC levels clearly correlated with EBV infection in Akata BL cells, we examined whether this phenomenon was also characteristic of other BL cell lines that maintain type I latency. Although we lacked paired EBV-positive and -negative group I BL lines as for the Akata BL, the same growth phase-associated down-regulation of c-MYC was evident in the three other EBV-positive BL lines examined (Fig. 5). Thus, this regulation of c-MYC is a common property of BL cells that maintain type I EBV latency.
FIG. 5.
c-MYC is a common target for down-regulation in group I BL cell lines. The group I BL cell lines KemI, SavI, and MutuI were seeded in complete growth medium (2.5 × 105 per ml), and cell samples were harvested on days 1 through 5 to monitor expression of c-MYC as for Fig. 4.
To determine the mechanism whereby c-MYC expression was down-regulated in EBV-positive cells, we initially examined effects of growth phase on c-myc mRNA levels. Unlike c-MYC protein in EBV-positive cells, c-myc transcripts did not diminish in stationary-phase cells (Fig. 6A), suggesting that the effect on c-MYC expression was translational or posttranslational in nature. To address potential effects on c-MYC protein half-life, (t1/2), we performed cycloheximide-chase experiments with EBV-negative and -positive Akata cells from both log- and stationary-phase growth. As demonstrated in Fig. 6B (upper two panels), the t1/2 of c-MYC was equivalent in EBV-negative and -positive Akata cells and did not differ with respect to the growth phase of the cells. However, as expected, the steady-state levels of c-MYC were significantly decreased in EBV-positive cells in stationary phase relative to EBV-negative cells or either cell type in log phase.
FIG. 6.
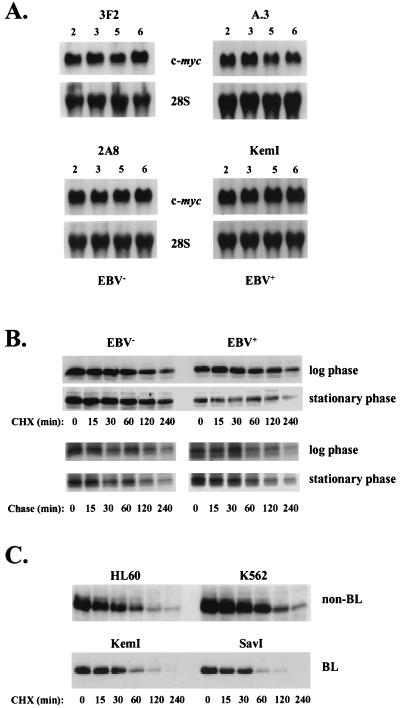
EBV down-regulates c-MYC expression through a posttranscriptional mechanism. (A) Northern blot analysis of c-myc mRNA and 28S rRNA (loading control) levels in EBV-negative (3F2 and 2A8) and EBV-positive (A.3 and KemI) BL cells at 2, 3, 5, and 6 days after seeding in complete growth medium at 2.5 × 105 cells per ml. Each lane contained 10 μg of total cellular RNA. (B) Cycloheximide (CHX)- and pulse-chase analysis of c-MYC t1/2 in EBV-negative (3F2) and EBV-positive (A.3) Akata BL cells during the log and stationary phases of the cell growth cycle. At chase times of 0, 15, 30, 60, 120, and 240 min, c-MYC levels were analyzed by immunoblotting (CHX-chases) or by immunoprecipitation and SDS-PAGE of pulse-labeled protein. Following quantification by phosphorimage analysis of immunoprecipitated c-MYC at each chase point, the data were subjected to linear regression analysis to determine the t1/2 of c-MYC (see text). (C) Half-lives of c-MYC during log-phase growth in the human promyelocytic and erythroleukemia cell lines HL60 and K562, respectively, and two additional group I BL cell lines (KemI and SavI), were analyzed by CHX-chase experiments as in panel B.
To determine whether EBV-dependent regulation of c-MYC levels in Akata cells was occurring at the level of translation, as well as to more accurately measure the t1/2 of c-MYC, pulse-chase analyses were performed. As demonstrated in Fig. 6B (lower panels), the rate of c-MYC synthesis did not differ between EBV-negative and -positive cells during log phase, consistent with the equivalent steady-state levels of c-MYC observed in these cells during exponential growth (Fig. 4). Surprisingly, when the rate of c-MYC synthesis was examined in cells within stationary phase, we did not observe a lower level of c-MYC synthesis in EBV-positive versus EBV-negative Akata cells, as one would expect if EBV-dependent repression of c-MYC expression was translational. However, when we analyzed total c-MYC levels by immunoblotting of these stationary-phase cell lysates, it was evident that in response to being placed in fresh medium (containing 10% serum) during the initial washing, preincubation, and pulse-labeling steps of the procedure, EBV-positive cells had begun to synthesize c-MYC, and this occurred at a rate higher than in EBV-negative cells (data not shown). This finding is consistent with our observations that EBV-positive Akata cells reenter the growth cycle much faster than the EBV-negative cells once they have reached stationary phase. Thus, although the EBV-associated decrease in c-MYC levels during stationary phase is likely due to an inhibition of translation (as neither c-myc mRNA levels or protein half-life is reduced), rapid de novo synthesis of c-MYC in EBV-positive relative to EBV-negative Akata cells during the pulse-chase procedure prevents direct measurement of translation rates.
The cycloheximide- and pulse-chase experiments indicated that the t1/2 of c-MYC in Akata BL cells is on the order of 120 min (Fig. 6B), severalfold greater than previously reported c-MYC t1/2 values of 20 to 30 min (20). To our knowledge the t1/2 of c-MYC in BL cells has not been reported; to address if the relatively long t1/2 of c-MYC in Akata cells is representative of other BL cells, we performed cycloheximide-chase analyses of c-MYC stability in KemI and SavI group I BL cells, as well as in two non-BL lines, HL60 and K562. As shown in Fig. 6C, the t1/2 of c-MYC in each of these cell lines is in agreement with previously reported values of 20 to 30 min. Thus, although the t1/2 of c-MYC in Akata BL cells is longer than that of c-MYC in the two other BL cell lines examined, this difference in c-MYC half-life does not account for the shared down-regulation of steady-state levels of c-MYC observed as Akata and other group I BL cell lines approach stationary phase (compare Fig. 4 to Fig. 5).
EBV infection, but not EBNA-1, restores resistance to apoptosis and tumorigenicity to EBV-negative BL.
It was possible that the parallel loss of the EBV genome and tumorigenic potential in some Akata cells was merely coincidental or that a propensity to lose the EBV genome was an effect, rather than the cause, of the spontaneous loss of tumorigenicity. To directly test the contribution of EBV to tumorigenicity, we reinfected EBV-negative Akata cells and assessed the consequences of a restored EBV latency on tumorigenic potential. Moreover, since it had been suggested that EBNA-1 alone might directly contribute to the oncogenic potential of EBV (59, 60), we also established several Akata cell lines that constitutively expressed EBNA-1 in an EBV-negative background.
Attempts to reestablish type I latency in clonal EBV-negative Akata cells revealed that infection was not equally sustained in all Akata clones. Although the EBV-negative cells were infectable as indicated by the expression of EBNA-1 protein and detection of EBV DNA by PCR during the first 2 weeks postinfection, over a period of several weeks to months the number of infected cells in the culture decreased. One EBV-negative Akata BL clone, 2A8, maintained a relatively stable infection and was therefore chosen for analysis. EBV-positive clones were isolated from the pool of reinfected 2A8 cells, and three clonal lines (2A8.1, 2A8.2, and 2A8.3) were evaluated. All three lines expressed EBNA-1 at levels comparable to those for wild-type EBV-positive cells and, as expected for type I latency, failed to express LMP-1 (Fig. 7A). Similarly, several EBV-negative Akata lines stably transfected with an EBNA-1 expression construct expressed moderate to high levels of EBNA-1 (Fig. 7B).
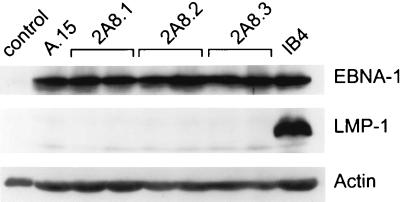
To determine whether EBV reinfection or EBNA-1 alone could restore the ability of EBV-negative Akata cells to down-regulate c-MYC protein and survive under growth-limiting conditions, cells were analyzed throughout the growth cycle as described above. To ensure that all cells harbored EBV, the reinfected 2A8 cells, which contained a recombinant EBV carrying a neomycin resistance gene, were placed under selection with G418. Reinfected cells did indeed down-regulate c-MYC in a manner similar to that observed in wild-type EBV-positive cells (compare the left panel of Fig. 8A to Fig. 4). By contrast, enforced overexpression of EBNA-1 alone in EBV-negative Akata cells was not sufficient to mediate down-regulation of c-MYC (Fig. 8A, right panel). Interestingly, reinfection with EBV did not further enhance Bcl-2 expression as cells approached stationary phase, nor did reinfected cells express notably higher levels of Bcl-2 relative to the EBV-negative 2A8 cells (data not shown). Nonetheless, as expected from their ability to down-regulate c-MYC, the reinfected 2A8 cells were more resistant to apoptosis than were the EBV-negative 2A8 cells (Fig. 8B, left panel). By contrast, cells engineered to express EBNA-1 in an EBV-negative Akata cell background were not protected from apoptosis following serum deprivation (Fig. 8B, right panel).
FIG. 8.
EBV infection, but not EBNA-1 alone, regulates c-MYC levels and sensitivity to apoptosis. (A) Immunoblot analysis of c-MYC throughout the cell growth cycle in reinfected and EBV−/EBNA-1+ Akata cells. Cells were seeded as described in Fig. 2 and harvested at the indicated day postseeding. (B) EBV infection, but not EBNA-1 alone, protects Akata cells from apoptosis during stationary-phase growth. The indicated cells were deprived of serum, and percent viability was determined at the indicated intervals by trypan blue dye exclusion. Results are representative of two independent experiments.
To determine whether reinfection with EBV conferred enhanced growth potential in assays more indicative of tumorigenicity, we analyzed the EBV-negative and reinfected Akata cells for growth in soft agar and for tumor induction in SCID mice (representative results are presented in Fig. 9). Although EBV-negative 2A8 cells exhibited a limited ability to produce colonies in soft agar, reinfection of 2A8 cells clearly enhanced growth potential, as indicated by the formation of larger colonies. Most importantly, although the reinfected 2A8 cells produced colonies somewhat smaller than those of the EBV-positive A.15 control cells, reinfected 2A8 cells always produced tumors when injected into SCID mice, whereas the parental 2A8 line and another EBV-negative line analyzed (3F2) never induced tumors (Fig. 9B and Table 1). Tumors resulting from reinfected 2A8 cells developed approximately 1 week later than those derived from the EBV-positive A.15 cells, which correlated with the augmented rate of growth of A.15 cells in soft agar relative to reinfected 2A8 cells (Fig. 9A). It should be noted that the reinfected cells had, on average, a lower EBV genome copy number relative to A.15 as determined by FISH, which may be in part responsible for their lower rate of growth in these assays. When analyzed by immunoblotting for EBV gene expression, each tumor expressed EBNA-1 but not LMP-1 (Fig. 10 and Table 1), characteristic of the cells prior to injection.
FIG. 9.
EBV infection restores tumorigenic potential to EBV-negative Akata cells. (A) Growth of EBV-positive (A.15), EBV-negative (2A8), and reinfected 2A8 (2A8.2) Akata cells in soft agar. Shown are low- and high-magnification micrographs of cell colonies resulting from growth in 0.33% agarose. (B) Tumorigenicity of Akata BL cells assayed by tumor induction in SCID mice (representative results). Each mouse received an injection of EBV-negative cells (3F2 or 2A8) in the left hind flank and EBV-positive (A.15) or reinfected 2A8 (2A8.2) cells in the right flank (2 × 107 cells per injection). The mouse that received the 3F2 and A.15 cells was photographed 5 weeks postinjection; the mouse that received the 2A8 and 2A8.2 cells was photographed 7 weeks postinjection. The results from all SCID mouse assays are presented in Table 1.
TABLE 1.
EBV, but not EBNA-1, restores tumorigenicity to EBV-negative Akata cells
| Akata cell line(s)a | EBV/EBNA-1 status | No. of mice that developed tumors | No. of tumor in which LMP-1 was detectedb |
|---|---|---|---|
| A.15 | +/+ | 3/3 | 0/3 |
| 3F2(3) and 2A8(9) | −/− | 0/12 | NAc |
| Reinfected | +/+ | 10/10 | 0/10 |
| LXSN | −/− | 0/12 | NA |
| EBNA-1 | −/+ | 0/8 | NA |
Numbers in parentheses indicate number of mice injected with each cell line; three clonal lines of EBV-reinfected 2A8 cells were each injected in triplicate, and one mouse received a pool of reinfected 3F2 cells; each of three clones of pLXSN-containing 3F2 cells (vector controls) was injected in quadruplicate; two pools of 3F2 cells expressing EBNA-1 were injected in triplicate, and two cloned 3F2 lines expressing EBNA-1 were each injected into individual mice.
Expression of LMP-1 in tumor cells, relative to an LCL-positive control, was determined by immunoblot analysis; all tumors were EBNA-1 positive.
NA, not applicable.
An additional reinfected EBV-negative Akata cell clone, 3F2, which did not readily maintain EBV upon reinfection in vitro, was also injected into a SCID mouse several weeks after reinfection, at a time when EBNA-1 expression was no longer detectable by immunoblotting. These cells, like the reinfected 2A8 cells, induced tumor formation, and analysis of the tumor cells indicated that they were indeed EBNA-1 positive and LMP-1 negative (Table 1). Interestingly, analysis of the EBV genome status in these cells by FISH revealed a lack of episomal EBV DNA and integration of the viral genome into the host genome at two sites. Therefore, there is clearly a strong in vivo selection for EBV in establishment of tumorigenic BL. By contrast, EBV-negative Akata cells that stably expressed EBNA-1 did not exhibit greater growth potential in soft agar (data not shown) and were never capable of inducing tumors in SCID mice (Table 1). Thus, establishment of a type I latency in EBV-negative Akata BL cells, but not EBNA-1 alone, was clearly sufficient to restore tumorigenic potential, a finding which correlates with EBV’s ability to inhibit c-MYC expression under growth-limiting conditions.
DISCUSSION
We have demonstrated that EBV can directly contribute to the tumorigenic potential of BL in the context of a type I latency program. With the possible exception of EBNA-1 (see below), EBV genes expressed during type I latency have not been directly linked to either B-cell immortalization or oncogenic transformation associated with EBV infection (24). In fact, the EBER and RK-BARF0 genes are dispensable for EBV-induced immortalization of B lymphocytes in vitro (44, 54). B-cell immortalization by EBV, however, occurs in the context of a type III latency (expression of all 12 EBV latency-associated genes), and thus one cannot exclude a possible role for EBNA-1, RK-BARF0, or the EBERs in growth-related functions of the type I latency program. The reservoir of EBV in healthy infected individuals consists of B lymphocytes that, like BL cells, preclude the expression of known growth-promoting EBV proteins (7, 37, 42, 57). Thus, one or more of the EBV genes expressed during the type I latency program may perform a critical role in modulating B-cell growth or survival in vivo.
The concept that the type I latency program of EBV promotes BL cell growth and survival is strongly supported by data presented here demonstrating that (i) EBV-positive Akata BL cells are more resistant to apoptosis than are EBV-negative Akata cells under growth-limiting conditions; (ii) under conditions that favor increased sensitivity to apoptotic stimuli, there is an EBV-dependent reduction in c-MYC, an oncoprotein known to augment the apoptotic program in BL (35) and other cell lineages (3, 12, 40); (iii) EBV-positive Akata BL cells generally, but not always, express higher levels of the antiapoptotic protein Bcl-2 than do their EBV-negative counterparts throughout all stages of the cell growth cycle; and (iv) tumorigenicity in Akata BL cells is strictly dependent on a latent EBV infection characteristic of BL tumors. Based on these observations, we propose that EBV contributes to tumorigenicity in BL by inhibiting c-MYC-induced apoptosis through at least two mechanisms: a modest up-regulation of Bcl-2 expression and, most importantly, a concomitant decrease in c-MYC expression, apparently at the level of translation, under growth-limiting conditions.
Although earlier studies failed to detect Bcl-2 expression in BL cells that maintain type I latency (21, 34), our results clearly indicate that Bcl-2 is expressed in Akata and other group I BL cell lines (Fig. 4 and data not shown). However, the level of Bcl-2 expression needed to effectively inhibit apoptosis in group I BL cells equals or exceeds the much higher levels of Bcl-2 observed in LCLs and BL cells that maintain type III latency (34). Because EBV-negative Akata cells express only slightly lower levels of Bcl-2 than their EBV-positive counterparts, Bcl-2 levels characteristic of EBV-positive BL cells that maintain type I latency seem unlikely to be sufficient alone to fully restore tumorigenicity to EBV-negative Akata BL cells.
The striking finding that EBV targets the down-regulation of c-MYC protein under growth-limiting conditions in BL suggests that this is the principal mechanism by which EBV promotes cell survival, particularly given that c-MYC is the primary mediator of apoptosis in BL (35) and that Akata BL cells are null for p53 (15). To determine if down-regulation of c-MYC is indeed responsible for the increased survival of BL cells, we have attempted to repress expression of c-MYC in EBV-negative Akata cells during stationary phase, using antisense c-myc oligonucleotides that have been successfully used in BL (35). Unfortunately, we were unable to demonstrate a repression of c-MYC by such means and therefore have been unable to directly test whether repression of c-MYC is sufficient to promote group I BL cell survival. Interestingly, our observations regarding BL cells contrast those for EBV-transformed lymphoblastoid cells, in which expression of c-MYC protein is sustained under growth-restrictive conditions (8), most likely through an EBV-dependent stabilization of c-myc mRNA that occurs during type III latency (26, 27). Although these opposing effects of EBV on c-MYC expression likely result from the dominant influence of the respective EBV latency program maintained by a cell, we cannot exclude the possibility that the EBV-dependent decrease in c-MYC protein observed here is unique to BL. Presumably, the documented ability of cells that maintain type III latency to substantially up-regulate the antiapoptotic proteins Bcl-2 and A20 would protect such cells having sustained c-MYC expression (16, 19, 21, 28, 47).
Although expression of the EBV EBNA-1 protein may be responsible for the protracted development of B-cell lymphoma in some lines of transgenic mice (59, 60), our observations indicate that neither EBV-dependent tumorigenicity nor regulation of c-MYC and Bcl-2 expression in BL can be attributed to EBNA-1 alone. Other EBV gene products that are consistently expressed in BL are the small RNAs EBER-1 and EBER-2 and the more recently identified RK-BARF0 protein (2, 17). Whereas the function of RK-BARF0 is unknown, at least two functions have been ascribed to the EBER RNAs: first, EBERs bind to the double-stranded RNA-dependent protein kinase PKR and disrupt the ability of PKR, a protein with potential tumor suppressor activity, to inhibit translation (10, 25, 32, 51); second, EBER RNAs bind to the ribosomal protein L22 and sequester L22 to the nucleus (58). Given that the EBERs are capable of targeting two proteins associated with the translational process and our data suggesting that down-regulation of c-MYC expression is most likely mediated at the level of translation, the possibility that the EBERs contribute to cell survival as defined here is particularly attractive.
Although our observations provide important insights into the long-standing debate over the role of EBV in BL, equally important are the implications of these findings with respect to the maintenance of EBV latency in the healthy EBV-immune host. Because the EBV-dependent down-regulation of c-MYC in BL appears to occur through a posttranscriptional mechanism, this type of regulation may also be operational within normal latently infected B cells in vivo. Thus, EBV may contribute to its persistence by promoting the survival of proliferating latently infected B cells. Although a major reservoir of EBV in the peripheral blood appears to be resting B cells that lack EBNA-1 mRNA but which express LMP-2A transcripts (37), cells that express EBNA-1 mRNA in the absence of transcripts that encode the other EBNA proteins and LMP-1 are detectable as well (7, 57). This finding suggests that there is a subpopulation of infected B cells in vivo that maintains a pattern of EBV gene expression similar or identical to that observed in BL. Such B cells may be actively dividing in response to physiological signals and essential to maintain a critical pool of latently infected B cells that is necessary to sustain long-term infection (reviewed in reference 56). The potential of EBV to usurp physiological pathways of cell proliferation, coupled with its ability to limit cell death under growth-restrictive conditions as shown here, would enable EBV to circumvent the need for virus-induced proliferation and the associated expression of the LMP-1 oncoprotein and other EBV proteins, such as the EBNA-3 family, that are capable of evoking a strong cellular immune response.
ACKNOWLEDGMENTS
We thank J. Hearing for EBNA-1 antiserum, C. Sample for S12 antibody, J. Sixbey for the 3F2, 2C1, and 2A8 cell lines, J. Downing for HL60 and K562 cells, D. Henson, E. White, and C. Yang for excellent technical assistance, and G. Zambetti and C. Sample for advice and critical reading of the manuscript. E. Kieff and B. Tomkinson each provided an EBV-negative Akata cell line used in preliminary studies.
This work was supported by Public Health Service grants DE-11116 (to L.M.H.-F.), CA-76379 and DK44158 (to J.L.C.), and CA-73544 and CA-56639 (to J.T.S.), Cancer Center (CORE) grant CA-21765, and the American Lebanese Syrian Associated Charities. I.K.R. was supported by PHS grant T32-AI-07372.
ADDENDUM IN PROOF
Similar data indicating that resistance to apoptosis and tumorigenic potential in Akata BL cells is dependent on EBV was recently reported by K. Komano et al. (J. Virol. 72:9150–9156, 1998).
REFERENCES
- 1.Adams J M, Harris A W, Pinkert C A, Corcoran L M, Alexander W S, Cory S, Palmiter R D, Brinster R L. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 2.Arrand J R, Rymo L. Characterization of the major Epstein-Barr virus-specific RNA in Burkitt lymphoma-derived cells. J Virol. 1982;41:376–389. doi: 10.1128/jvi.41.2.376-389.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Askew D S, Ashmun R A, Simmons B C, Cleveland J L. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell-cycle arrest and accelerates apoptosis. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- 4.Borza C M, Hutt-Fletcher L M. Epstein-Barr virus recombinant lacking expression of glycoprotein gp150 infects B cells normally but is enhanced for infection of the epithelial cells. J Virol. 1998;72:7577–7582. doi: 10.1128/jvi.72.9.7577-7582.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks L, Yao Q Y, Rickinson A B, Young L S. Epstein-Barr virus latent gene transcription in nasopharyngeal carcinoma cells: coexpression of EBNA1, LMP1, and LMP2 transcripts. J Virol. 1992;66:2689–2697. doi: 10.1128/jvi.66.5.2689-2697.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks L A, Lear A L, Young L S, Rickinson A B. Transcripts from the Epstein-Barr virus BamHI A fragment are detectable in all three forms of virus latency. J Virol. 1993;67:3182–3190. doi: 10.1128/jvi.67.6.3182-3190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen F, Zou J Z, di Renzo L, Winberg G, Hu L F, Klein E, Klein G, Ernberg I. A subpopulation of normal B cells latently infected with Epstein-Barr virus resembles Burkitt lymphoma cells in expressing EBNA-1 but not EBNA-2 or LMP-1. J Virol. 1995;69:3752–3758. doi: 10.1128/jvi.69.6.3752-3758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherney B W, Bhatia K, Tosato G. A role for deregulated c-Myc expression in apoptosis of Epstein-Barr virus-immortalized B cells. Proc Natl Acad Sci USA. 1994;91:12967–12971. doi: 10.1073/pnas.91.26.12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chodosh J, Holder V P, Gan Y, Belgaumi A, Sample J, Sixbey J W. Eradication of latent Epstein-Barr virus by hydroxyurea alters the growth-transformed cell phenotype. J Infect Dis. 1998;177:1194–1201. doi: 10.1086/515290. [DOI] [PubMed] [Google Scholar]
- 10.Clarke P A, Schwemmle M, Schickinger J, Hilse K, Clemens M J. Binding of Epstein-Barr virus small RNA EBER-1 to the double-stranded RNA-activated protein kinase DAI. Nucleic Acids Res. 1991;19:243–248. doi: 10.1093/nar/19.2.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deacon E M, Pallesen G, Niedobitek G, Crocker J, Brooks L, Rickinson A B, Young L S. Epstein-Barr virus and Hodgkin’s disease: transcriptional analysis of virus latency in the malignant cells. J Exp Med. 1993;177:339–349. doi: 10.1084/jem.177.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evan G, Wyllie C S, Gilbert C S, Land H, Brooks M, Littlewood T, Waters C, Hancock D. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 13.Evan G I, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fahraeus R, Li-Fu H, Ernberg I, Finke J, Rowe M, Klein G, Falk K, Nilsson E, Yadaf M, Busson P, Tursz T, Kallin B. Expression of the Epstein-Barr virus genome in nasopharyngeal carcinoma. Int J Cancer. 1988;42:329–338. doi: 10.1002/ijc.2910420305. [DOI] [PubMed] [Google Scholar]
- 15.Farrell P J, Allan G J, Shanathan F, Vousden K H, Crook T. p53 is frequently mutated in Burkitt’s lymphoma cell lines. EMBO J. 1991;10:2879–2887. doi: 10.1002/j.1460-2075.1991.tb07837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fries K L, Miller W E, Raab-Traub N. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through induction of the A20 gene. J Virol. 1996;70:8653–8659. doi: 10.1128/jvi.70.12.8653-8659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fries K L, Sculley T B, Webster-Cyriaque J, Rajadurai P, Sadler R H, Raab-Traub N. Identification of a novel protein encoded by the BamHI A region of the Epstein-Barr virus. J Virol. 1997;71:2765–2771. doi: 10.1128/jvi.71.4.2765-2771.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregory C D, Rowe M, Rickinson A B. Different Epstein-Barr virus-B cell interactions in phenotypically distinct clones of a Burkitt’s lymphoma cell line. J Gen Virol. 1990;71:1481–1495. doi: 10.1099/0022-1317-71-7-1481. [DOI] [PubMed] [Google Scholar]
- 19.Gregory C D, Dive C, Henderson S, Smith C A, Williams G T, Gordon J, Rickinson A B. Activation of Epstein-Barr virus latent genes protects human B cells from death by apoptosis. Nature. 1991;349:612–614. doi: 10.1038/349612a0. [DOI] [PubMed] [Google Scholar]
- 20.Hann S R, Eisenman R N. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol. 1984;4:2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson S, Rowe M, Gregory C, Croom-Carter D, Wang F, Longnecker R, Kieff E, Rickinson A. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell. 1991;65:1107–1115. doi: 10.1016/0092-8674(91)90007-l. [DOI] [PubMed] [Google Scholar]
- 22.Herbst H, Dallenbach F, Hummel M, Niedobitek G, Pileri S, Muller-Lantzsch N, Stein H. Epstein-Barr virus latent membrane protein expression in Hodgkin and Reed-Sternberg cells. Proc Natl Acad Sci USA. 1991;88:4766–4770. doi: 10.1073/pnas.88.11.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khanna R, Burrows S R, Kurilla M G, Jacob C A, Misko I S, Sculley T B, Kieff E, Moss D J. Localization of Epstein-Barr virus cytotoxic T cell epitopes using recombinant vaccinia: implications for vaccine development. J Exp Med. 1992;176:160–176. doi: 10.1084/jem.176.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, et al., editors. Fields virology. 3rd ed. Vol. 2. New York, N.Y: Raven Press; 1996. pp. 2343–2396. [Google Scholar]
- 25.Koromilas A E, Roy S, Barber G N, Katze M G, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 26.Lacy J, Summers W P, Watson M, Glazer P M, Summers W C. Amplification and deregulation of MYC following Epstein-Barr virus infection of a human B-cell line. Proc Natl Acad Sci USA. 1987;84:5838–5842. doi: 10.1073/pnas.84.16.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lacy J, Summers W P, Summers W C. Post-transcriptional mechanisms of deregulation of MYC following conversion of a human B cell line by Epstein-Barr virus. EMBO J. 1989;8:1973–1980. doi: 10.1002/j.1460-2075.1989.tb03603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laherty C D, Hu H M, Opipari A W, Wang F, Dixit V M. The Epstein-Barr virus LMP1 gene product induces A20 zinc finger protein expression by activating NF-κB. J Biol Chem. 1992;267:24157–24160. [PubMed] [Google Scholar]
- 29.Magrath I. The pathogenesis of Burkitt’s lymphoma. Adv Cancer Res. 1990;55:133–270. doi: 10.1016/s0065-230x(08)60470-4. [DOI] [PubMed] [Google Scholar]
- 30.Mann K P, Staunton D, Thorley-Lawson D A. Epstein-Barr virus-encoded protein found in plasma membranes of transformed cells. J Virol. 1985;55:710–720. doi: 10.1128/jvi.55.3.710-720.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masucci M, Ernberg I. Epstein-Barr virus: adaptation to a life within the immune system. Trends Microbiol. 1994;2:125–130. doi: 10.1016/0966-842x(94)90599-1. [DOI] [PubMed] [Google Scholar]
- 32.Meurs E F, Galabru J, Barber G N, Katze M G, Hovanessian A G. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1993;90:232–236. doi: 10.1073/pnas.90.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller A D, Rosman G. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 34.Milner A E, Johnson G D, Gregory C D. Prevention of programmed cell death in Burkitt lymphoma cell lines by bcl-2-dependent and -independent mechanisms. Int J Cancer. 1992;52:636–644. doi: 10.1002/ijc.2910520424. [DOI] [PubMed] [Google Scholar]
- 35.Milner A E, Grand R J A, Waters C M, Gregory C D. Apoptosis in Burkitt lymphoma cells is driven by c-myc. Oncogene. 1993;8:3385–3391. [PubMed] [Google Scholar]
- 36.Miyashita E M, Yang B, Lam K M C, Crawford D H, Thorley-Lawson D A. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell. 1995;80:593–601. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- 37.Miyashita E M, Yang B, Babcock G J, Thorley-Lawson D A. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J Virol. 1997;71:4882–4891. doi: 10.1128/jvi.71.7.4882-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murray R, Kurilla M, Brooks J, Thomas W, Rowe M, Kieff E, Rickinson A. Identification of target antigens for the human cytotoxic T cell response to Epstein-Barr virus (EBV): implications for the immune control of EBV positive malignancies. J Exp Med. 1992;176:157–168. doi: 10.1084/jem.176.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nonkwelo C, Henson E B D, Sample J. Characterization of the Epstein-Barr virus Fp promoter. Virology. 1995;206:183–195. doi: 10.1016/s0042-6822(95)80033-6. [DOI] [PubMed] [Google Scholar]
- 40.Packham G, Cleveland J L. c-Myc and apoptosis. Biochim Biophys Acta. 1995;1242:11–28. doi: 10.1016/0304-419x(94)00015-t. [DOI] [PubMed] [Google Scholar]
- 41.Pallesen G, Hamilton-Dutoit S J, Rowe M, Young L S. Expression of Epstein-Barr virus latent gene products in tumour cells of Hodgkin’s disease. Lancet. 1991;337:320–322. doi: 10.1016/0140-6736(91)90943-j. [DOI] [PubMed] [Google Scholar]
- 42.Qu L, Rowe D T. Epstein-Barr virus latent gene expression in uncultured peripheral blood lymphocytes. J Virol. 1992;66:3715–3724. doi: 10.1128/jvi.66.6.3715-3724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, et al., editors. Fields virology. 3rd ed. Vol. 2. New York, N.Y: Raven Press; 1996. pp. 2397–2446. [Google Scholar]
- 44.Robertson E S, Tomkinson B, Kieff E. An Epstein-Barr virus with a 58-kilobase-pair deletion that includes BARF0 transforms B lymphocytes in vitro. J Virol. 1994;68:1449–1458. doi: 10.1128/jvi.68.3.1449-1458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rooney C M, Gregory C D, Rowe M, Finerty S, Edwards C, Rupani H, Rickinson A B. Endemic Burkitt’s lymphoma: phenotypic analysis of tumor biopsy cells and of derived tumor cell lines. J Natl Cancer Inst. 1986;77:681–687. doi: 10.1093/jnci/77.3.681. [DOI] [PubMed] [Google Scholar]
- 46.Rowe D T, Rowe M, Evan G I, Wallace L E, Farrell P J, Rickinson A B. Restricted expression of EBV latent genes and T-lymphocyte-detected membrane antigen in Burkitt’s lymphoma cells. EMBO J. 1986;5:2599–2607. doi: 10.1002/j.1460-2075.1986.tb04540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rowe M, Peng-Pilon M, Huen D S, Hardy R, Croom-Carter D, Lundgren E, Rickinson A B. Upregulation of bcl-2 by the Epstein-Barr virus latent membrane protein LMP1: a B-cell-specific response that is delayed relative to NFκB activation and to induction of cell surface markers. J Virol. 1994;68:5602–5612. doi: 10.1128/jvi.68.9.5602-5612.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 49.Sample J, Kieff E. Transcription of the Epstein-Barr virus genome during latency in growth-transformed lymphocytes. J Virol. 1990;64:1667–1674. doi: 10.1128/jvi.64.4.1667-1674.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sample J, Henson E B D, Sample C. The Epstein-Barr virus nuclear protein 1 promoter active in type I latency is autoregulated. J Virol. 1992;66:4654–4661. doi: 10.1128/jvi.66.8.4654-4661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharp T V, Schwemmle M, Jeffrey I, Laing K, Mellor H, Proud C G, Hilse K, Clemens M J. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 1993;21:4483–4490. doi: 10.1093/nar/21.19.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimizu N, Tanabe-Tochikura A, Kuroiwa Y, Takada K. Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV-positive Burkitt’s lymphoma (BL) line Akata: malignant phenotypes of BL cells are dependent on EBV. J Virol. 1994;68:6069–6073. doi: 10.1128/jvi.68.9.6069-6073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith P, Griffin B. Differential expression of Epstein-Barr viral transcripts for two proteins (TP1 and LMP) in lymphocyte and epithelial cells. Nucleic Acids Res. 1991;19:2435–2440. doi: 10.1093/nar/19.9.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swaminathan S, Tomkinson B, Kieff E. Recombinant Epstein-Barr virus with small RNA (EBERS) genes deleted transforms lymphocytes and replicates in vitro. Proc Natl Acad Sci USA. 1991;88:1546–1550. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takada K, Ono Y. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J Virol. 1989;63:445–449. doi: 10.1128/jvi.63.1.445-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thorley-Lawson D A, Miyashita E M, Khan G. Epstein-Barr virus and the B cell: that’s all it takes. Trends Microbiol. 1996;4:204–208. doi: 10.1016/s0966-842x(96)90020-7. [DOI] [PubMed] [Google Scholar]
- 57.Tierney R J, Steven N, Young L S, Rickinson A B. Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection and in the carrier state. J Virol. 1994;68:7374–7385. doi: 10.1128/jvi.68.11.7374-7385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toczyski D P, Matera A G, Ward D C, Steitz J A. The Epstein-Barr virus (EBV) small RNA EBER1 binds and relocalizes ribosomal protein L22 in EBV-infected human B lymphocytes. Proc Natl Acad Sci USA. 1994;91:3463–3467. doi: 10.1073/pnas.91.8.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson J B, Levine A J. The oncogenic potential of Epstein-Barr virus nuclear antigen 1 in transgenic mice. Curr Top Microbiol Immunol. 1992;182:325–384. doi: 10.1007/978-3-642-77633-5_48. [DOI] [PubMed] [Google Scholar]
- 60.Wilson J B, Bell J L, Levine A J. Expression of Epstein-Barr virus nuclear antigen-1 induces B cell neoplasia in transgenic mice. EMBO J. 1996;15:3117–3126. [PMC free article] [PubMed] [Google Scholar]
- 61.Young L S, Dawson C W, Clark D, Rupani H, Busson P, Tursz T, Johnson A, Rickinson A B. Epstein-Barr virus gene expression in nasopharyngeal carcinoma. J Gen Virol. 1988;69:1051–1065. doi: 10.1099/0022-1317-69-5-1051. [DOI] [PubMed] [Google Scholar]