Abstract
Mobile genetic elements threaten genome integrity in all organisms. MUT-2/RDE-3 is a ribonucleotidyltransferase required for transposon silencing and RNA interference (RNAi) in C. elegans1–4. When tethered to RNAs in heterologous expression systems, RDE-3 can add long stretches of alternating non-templated uridine (U) and guanosine (G) ribonucleotides to the 3’ termini of these RNAs (poly(UG) or pUG tails)5. Here we show that, in its natural context in C. elegans, RDE-3 adds pUG tails to targets of RNAi, as well as to transposon RNAs. pUG tails with more than 16 perfectly alternating 3’ U and G nucleotides convert RNA fragments into agents of gene silencing. pUG tails promote gene silencing by recruiting RNA-dependent RNA polymerases (RdRPs), which use pUG-tailed RNAs (pUG RNAs) as templates to synthesize small interfering RNAs (siRNAs). Our results show that cycles of pUG RNA–templated siRNA synthesis and siRNA-directed mRNA pUGylation underlie dsRNA-directed transgenerational epigenetic inheritance in the C. elegans germline. We speculate that this pUG RNA/siRNA silencing loop allows parents to inoculate progeny against the expression of unwanted or parasitic genetic elements
Transposable elements are mobile parasitic genetic elements present in all genomes. Transposons threaten genome integrity, and can cause disease by disrupting genes or inducing non-allelic recombination. RNA interference (RNAi) is a conserved gene silencing mechanism initiated by double-stranded RNA (dsRNA)6. Forward genetic screens to identify factors required for either transposon silencing or RNAi have been conducted in the model metazoan C. elegans1–3. These screens identified an overlapping set of genes, indicating that an RNAi-related process silences transposons1–3. One gene required for both efficient transposon silencing and RNAi in C. elegans is mut-2/rde-3, which encodes a protein with homology to ribonucleotidyltransferases (rNTs)1–4. rNTs add non-templated ribonucleotides to RNAs and other substrates7,8. Recently, C. elegans MUT-2/RDE-3 (henceforth, RDE-3) was shown to add perfectly alternating U and G ribonucleotides to the 3’ termini of RNAs (termed polyUG or pUG tails) to which it was tethered either in S. cerevisiae or in X. laevis oocytes5. Taken together, these data prompted the proposal that RDE-3 may append non-templated pUG tails to the 3’ termini of RNAs during transposon silencing and/or RNAi in C. elegans5.
RDE-3 pUGylates mRNAs targeted by RNAi.
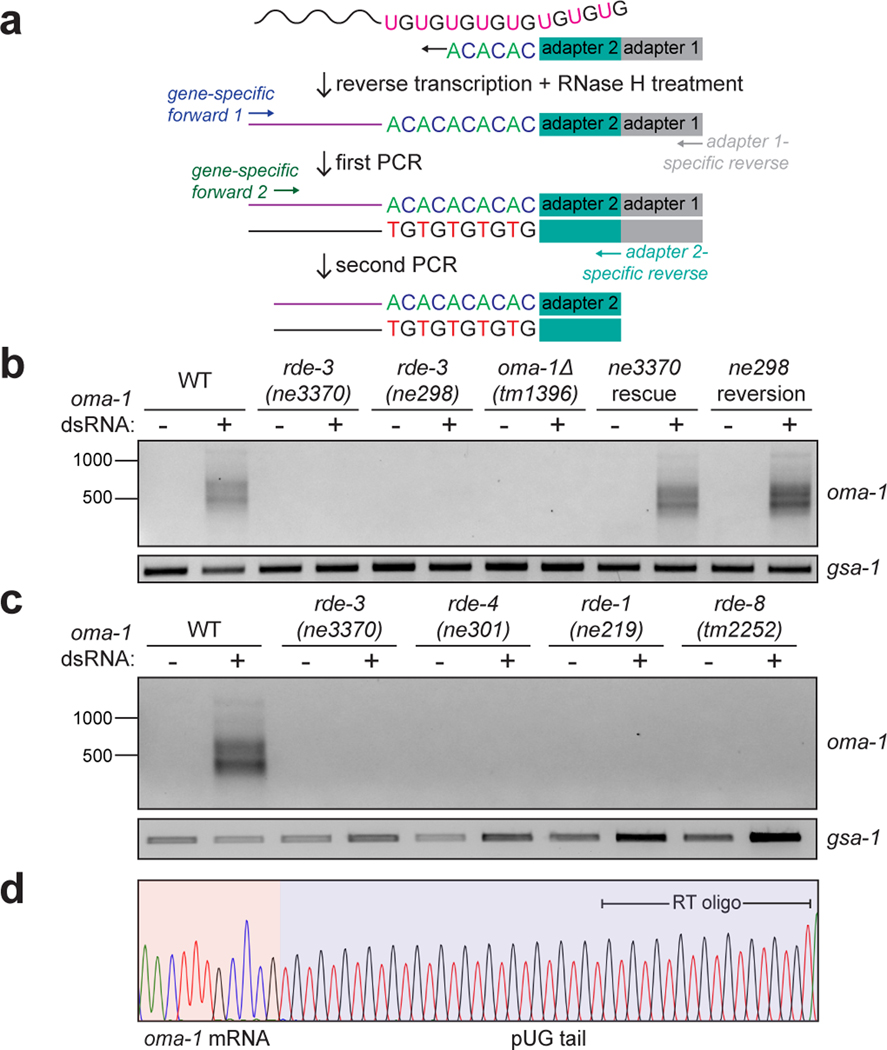
We first asked whether pUG tails are added to RNAs targeted by RNAi in C. elegans. We used an (AC)9 oligo to reverse transcribe (RT) total RNA extracted from animals exposed to dsRNA targeting the germline-expressed gene oma-19, and then performed nested PCR to try to detect oma-1 RNAs modified with 3’ pUG repeats (Fig. 1a). This approach (termed pUG PCR) detected PCR products that were dependent on oma-1 dsRNA (Fig. 1b), as well as on components of the RNAi machinery including RDE-4, which promotes dsRNA processing into siRNAs10,11; the siRNA-binding Argonaute (AGO) protein RDE-110,11; and RDE-8, which is an endonuclease that cleaves mRNAs exhibiting homology to siRNAs12 (Fig. 1c).
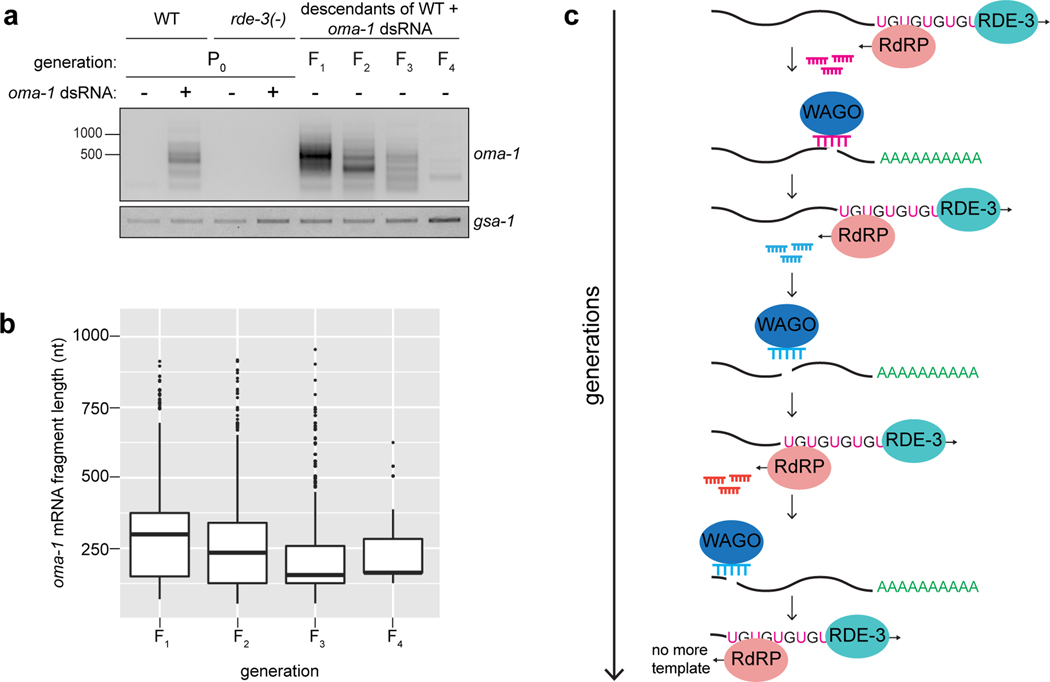
Figure 1. pUG tails are added to mRNA fragments in vivo.
a, Assay to detect gene-specific pUG RNAs. Note: (AC)9 RT oligo can anneal anywhere along the pUG tail. b, oma-1 pUG PCR on total RNA isolated from animals of indicated genotypes, +/− oma-1 dsRNA (RNAi). rde-3 mutants were rescued as described in Main text and Methods. gsa-1, which has an 18nt long genomically encoded pUG repeat in its 3’UTR, is a loading control. Wild-type (WT) vs. rde-3(ne3370) and WT vs. rde-3(ne298) data is representative of >10 and 2 biologically independent experiments, respectively. c, oma-1 pUG PCR on total RNA from animals of indicated genotypes, +/− oma-1 dsRNA. Data is representative of 3 biologically independent experiments. d, Sanger sequencing chromatogram (red=T, black=G, blue=C, green=A) of an oma-1 pUG PCR product.
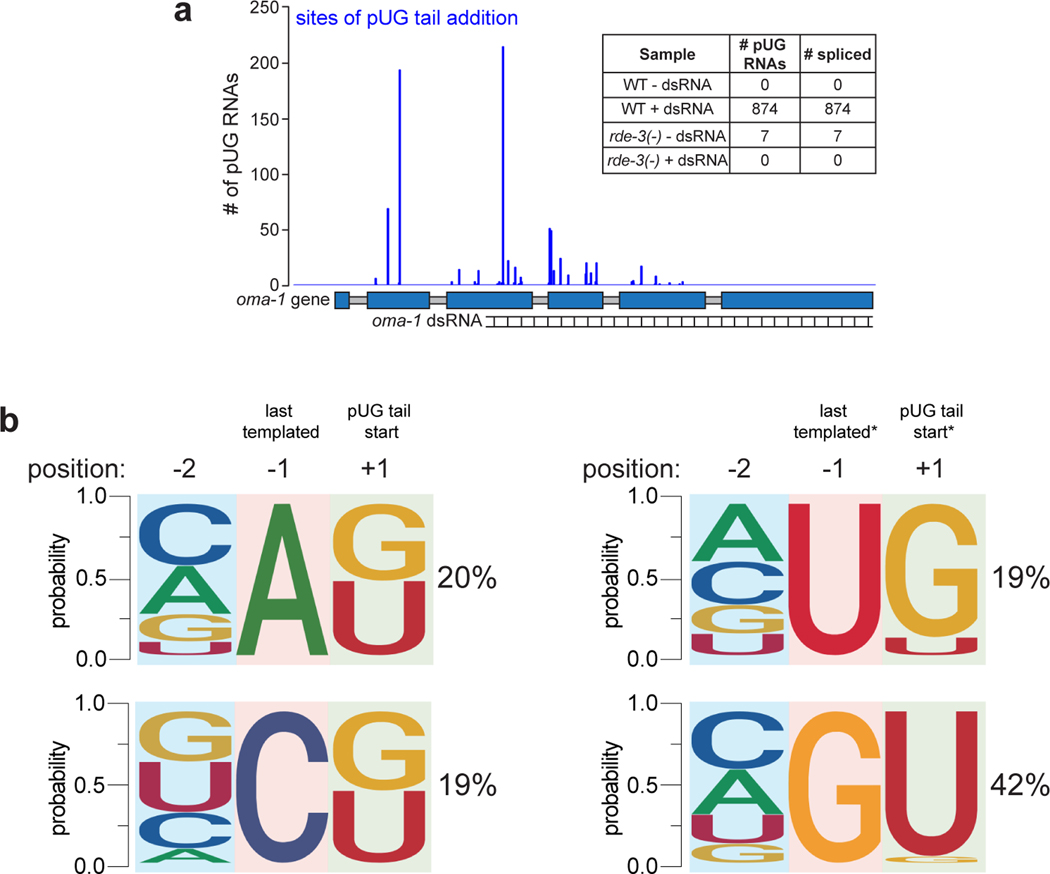
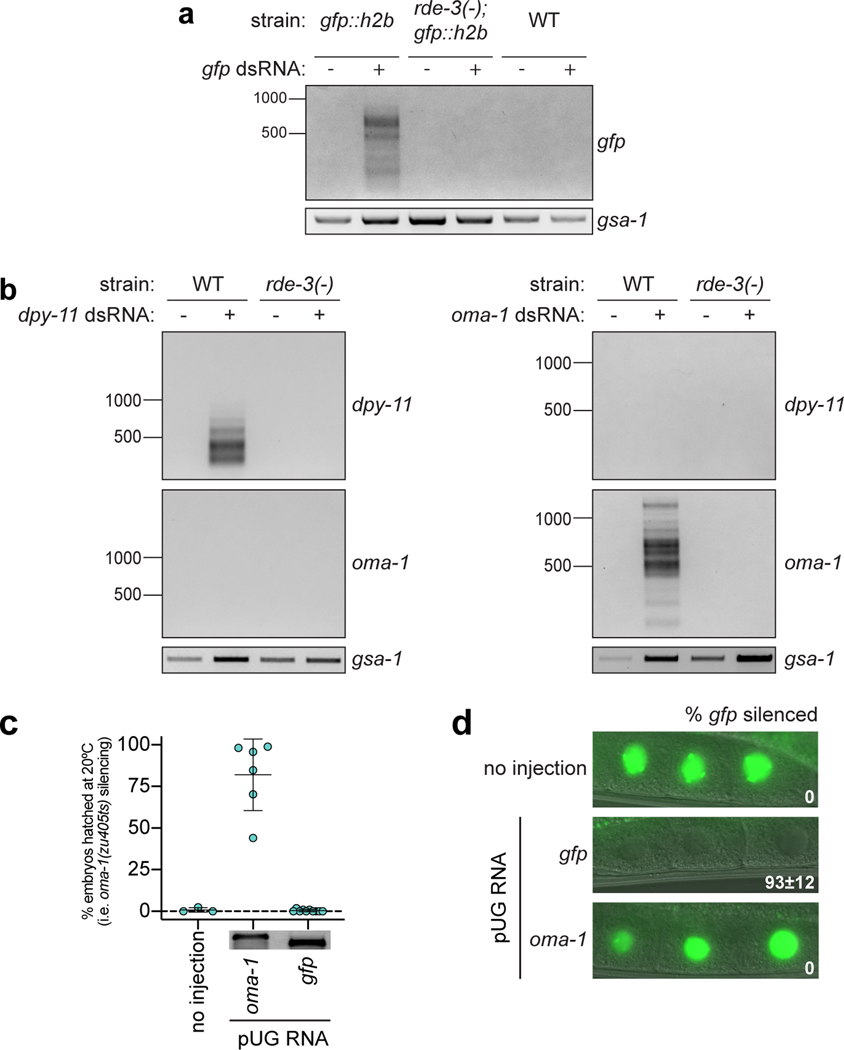
Sanger and Illumina sequencing revealed that most (>89%) pUG PCR products were derived from hybrid RNAs consisting of stretches of nearly perfectly alternating (error rate <2%, Supplementary Table 1) U and G repeats appended to the 3’ termini of sense and spliced oma-1 mRNA fragments (Fig. 1d, Extended Data Fig. 1a). pUGylation sites were non-randomly distributed along the oma-1 mRNA (Extended Data Fig. 1). Critically, most (64%) pUG tails were longer (range=19–75nt) than the (AC)9 oligo used for RT (Fig. 1d, Supplementary Table 1), indicating that the detected pUG RNAs were not the result of priming from genomically encoded UG-rich sequences. RDE-3 was required for addition of pUG repeats to mRNA fragments (termed pUGylation): rde-3 mutants, including rde-3(ne3370) animals, which harbor a deletion that removes residues required for catalysis within the rNT domain of RDE-3 (henceforth rde-3(−))5, failed to produce oma-1 pUG RNAs in response to oma-1 dsRNA (Fig. 1b). pUGylation defects in rde-3 mutants were rescued by introducing a wild-type copy of rde-3 into rde-3(−) animals or by CRISPR/Cas9-mediated reversion of a missense allele (ne298) of rde-3 to wild-type (Fig. 1b). Furthermore, RNA pUGylation was a general response to RNAi: animals exposed to dsRNA targeting a germline-expressed gfp::h2b transgene or the hypodermis-expressed dpy-11 gene13 produced RDE-3–dependent gfp and dpy-11 pUG RNAs, respectively (Extended Data Fig. 2a,b). Finally, pUGylation was sequence-specific, since dpy-11 dsRNA did not induce oma-1 pUG RNA biogenesis and vice versa (Extended Data Fig. 2b). Together, these data indicate that RDE-3 adds pUG tails to mRNAs targeted for silencing by RNAi.
pUG RNAs drive gene silencing.
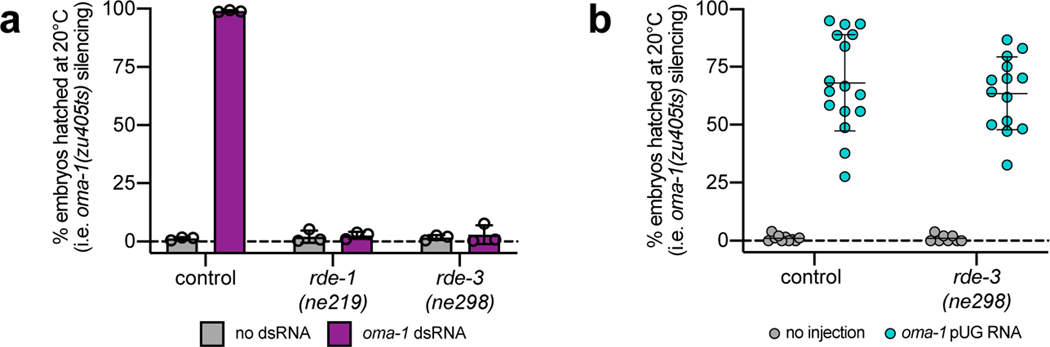
pUG tails could either mark mRNA fragments for degradation or convert mRNA fragments into active mediators of gene silencing. To differentiate these possibilities, we asked whether in vitro transcribed pUG RNAs possess gene silencing activity. Indeed, injection of a gfp pUG RNA (i.e. 18 3’-terminal pUG repeats appended to the first 369nt of the gfp mRNA) into animals expressing a germline-expressed gfp::h2b transgene was sufficient to silence gfp::h2b expression (Fig. 2a). The same gfp mRNA fragment without a 3’ tail or with 18 3’-terminal pAU, pGC or pAC repeats lacked gene silencing activity (Fig. 2a). Note: to control for potential dsRNA contamination in our in vitro transcription reactions, all RNAs were injected into rde-1(ne219) mutants, which cannot respond to dsRNA (Fig. 2a, b)3. The ability of a pUG tail to confer gene silencing activity on an mRNA fragment was both general and sequence-specific. oma-1(zu405ts) animals lay arrested embryos at 20°C unless oma-1(zu405ts) is silenced14. An in vitro transcribed 541nt long oma-1 mRNA fragment with 18 3’ pUG repeats (hereafter, oma-1 pUG RNA)—but not 18 3’ pAU, pGC or pAC repeats—was capable of silencing oma-1(zu405ts) (Fig. 2b). Additionally, an oma-1 pUG RNA injection did not silence gfp::h2b and vice versa (Extended Data Fig. 2c,d). Finally, while RDE-3 was required for efficient oma-1 RNAi (Extended Data Fig. 3a), this requirement could be bypassed by an oma-1 pUG RNA injection (Extended Data Fig. 3b), establishing that RDE-3-mediated pUGylation is necessary for RNAi. We conclude that pUG tails convert otherwise inert mRNA fragments into agents of gene silencing.
Figure 2. pUG tails convert inert RNA fragments into agents of gene silencing.
a-c, To control for potential dsRNA contamination in in vitro transcription reactions, RNAs were injected into rde-1(ne219) mutants, which cannot respond to dsRNA3. a, Fluorescence micrographs showing −1 to −3 oocytes of adult progeny of rde-1(ne219); gfp::h2b animals injected in the germline with in vitro transcribed RNAs consisting of the first 369nt of gfp mRNA with the indicated 3’ terminal repeats. Mean % progeny with gfp::h2b silenced is indicated ± standard deviation (s.d.). # of injected animals (n) = 3 (no injection); 9 [gfp dsRNA, (AU)18]; 10 [no tail, (GC)18, (AC)18]; and 16 [(UG)18]. b-c, oma-1(zu405ts) animals lay arrested embryos at 20°C unless oma-1(zu405ts) is silenced14. Adult rde-1(ne219); oma-1(zu405ts) animals were injected in the germline with in vitro transcribed RNAs consisting of the first 541nt of oma-1 mRNA with b, indicated 3’ terminal repeats or c, varying 3’ pUG tail lengths; different 3’ UG repeat sequences; or with (UG)18 on the 3’ end, 5’ end or in the middle of the oma-1 mRNA. For all oma-1 pUG RNA injection data, each point represents % hatched embryos laid by 5 progeny derived from one injected animal at 20°C (see Methods). Error bars: s.d. of the mean. Insets: injected RNAs run on 2% agarose gel to assess RNA integrity. For b, n=6 (no injection); 10 (oma-1 dsRNA, no tail); 12 [(UG)18]; 9 [(GC)18]; and 8 [(AU)18, (AC)18]. For c, n=9 [no injection #1, (U18G18), (UUGG)9]; 10 [(UG)0, 1, 8, 14, scrambled UGs, 3’ (UG)18, internal (UG)18]; 8 [(UG)5, 5’ (UG)18]; 12 [(UG)18 #1]; 5 [(UG)18 #2]; 11 [(UG)40]; 6 (no injection #2) and 3 (no injection #3).
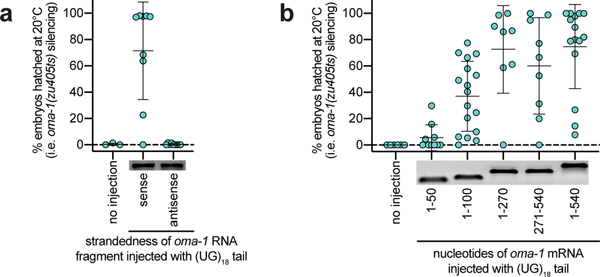
We used our pUG RNA injection assay to define the features of pUG RNAs required for biological activity. We injected animals with the same oma-1 mRNA fragment harboring varying numbers of 3’ UG repeats and found that oma-1 pUG RNAs with 14, 18, or 40—but not 1, 5, or 8—UG repeats were capable of triggering oma-1 gene silencing (Fig. 2c). We also found that while perfectly alternating 3’ U and G repeats conferred silencing activity on an mRNA fragment, 3’ tails with scrambled UG sequence or other combinations of Us and Gs did not (Fig. 2c). Moreover, while an oma-1 mRNA fragment with a 3’ pUG tail triggered oma-1(zu405ts) silencing, oma-1 mRNA fragments with 5’ or internal UG repeats did not (Fig. 2c). Finally, the oma-1 segment of an oma-1 pUG RNA had to possess the sense coding sequence (Extended Data Fig. 4a) and be >50nt in length for pUG RNA functionality (Extended Data Fig. 4b). Together, these data show that a pUG RNA must consist of >8 3’ UG repeats appended to >50nt of sense RNA in order to trigger gene silencing.
RDE-3 pUGylates germline-expressed RNAs.
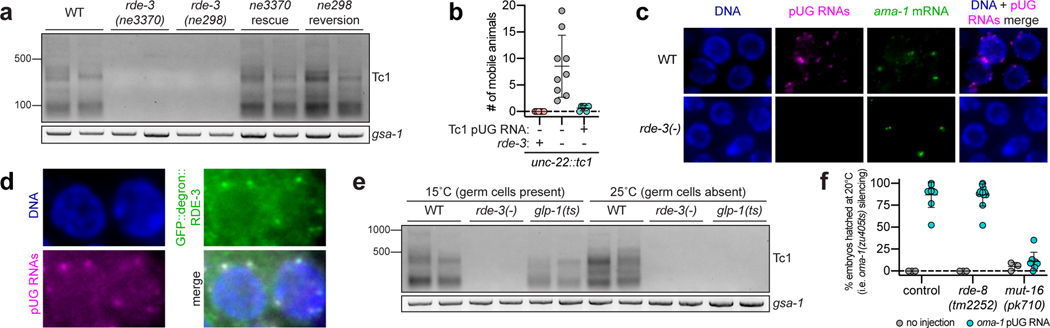
We next asked whether endogenous mRNAs are pUGylated in C. elegans. Tc1 is the most common DNA transposon in the C. elegans genome15. In the absence of RDE-3, Tc1 transposase RNA is upregulated and Tc1 mobilizes1, suggesting that Tc1 RNA might be pUGylated in wild-type animals. Indeed, using a Tc1-specific pUG PCR assay (Fig. 1a), we observed RDE-3–dependent pUG tails appended to Tc1 RNA fragments (Fig. 3a, Supplementary Table 1). In addition, Tc1 mobilization caused by rde-3 mutation was suppressed by injection of a Tc1 pUG RNA (Fig. 3b). We conclude that RDE-3–based pUGylation silences the Tc1 transposon in C. elegans.
Figure 3. Endogenous pUG RNAs exist and localize to germline Mutator foci.
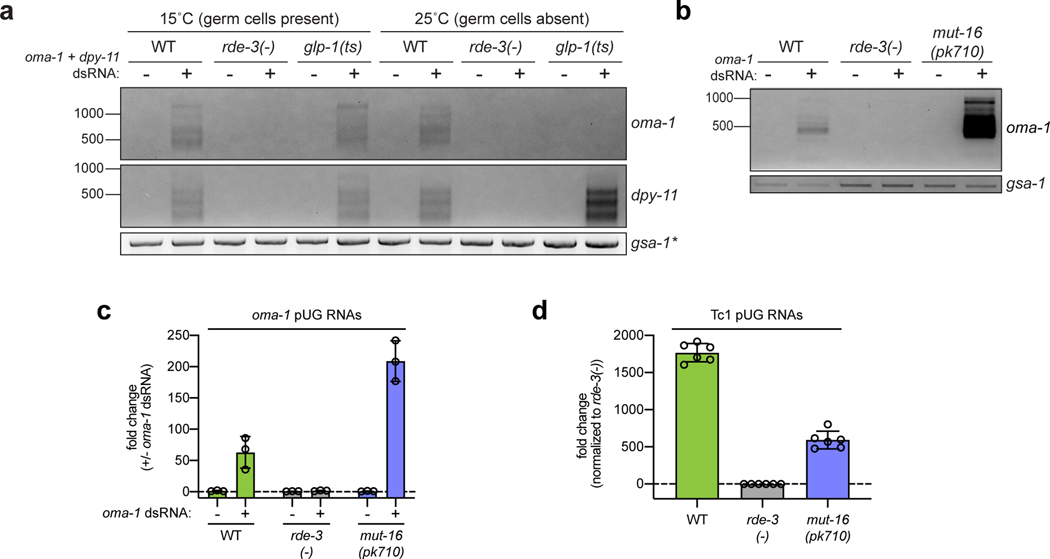
a, Tc1 pUG PCR (Fig. 1a) on total RNA from two replicates of indicated genotypes (rescue/reversion as in Fig. 1b). WT vs. rde-3(ne3370) and WT vs. rde-3(ne298) is representative of >5 and 2 biologically independent experiments, respectively. b, 18 and 22 rde-3(−); unc-22::tc1 animals were injected with Tc1 pUG RNA + co-injection marker or co-injection marker alone, respectively. Each data point (n) represents # of mobile progeny (indicating Tc1 mobilized from unc-22) laid by 25 randomly pooled co-injection marker–expressing progeny derived from injected animals (see Methods). n=9 for co-injection marker only, 6 for Tc1 pUG RNA + co-injection marker, 6 for noninjected rde-3(+); unc-22::tc1. Error bars: s.d. of the mean. c-d, Fluorescence micrographs of adult pachytene stage germ cell nuclei. DNA stained with 4’,6-diamidino-2-phenylindole (DAPI, blue). Data is representative of 3 biologically independent experiments. c, RNA FISH to detect pUG RNAs on germlines dissected from WT or rde-3(−) animals using (AC)9 DNA oligo conjugated to Alexa 647 (magenta). Positive control: RNA FISH to detect ama-1 mRNA (green). d, pUG RNA FISH (magenta) combined with immunofluorescence to detect GFP::degron::RDE-3 (green). e, Tc1 pUG PCR on total RNA isolated from replicates of glp-1(q224/ts) animals grown at 15°C (germ cells present) or 25°C (<99% of germ cells). Data is representative of 2 biologically independent experiments. f, Control, rde-8(tm2252) or mut-16(pk710) animals (all rde-1(ne219); oma-1(zu405ts) background) were injected with oma-1 pUG RNAs (n=9, 12 and 8, respectively) and % embryos hatched scored at 20°C. n=3 for all no injection. Error bars: s.d. of the mean.
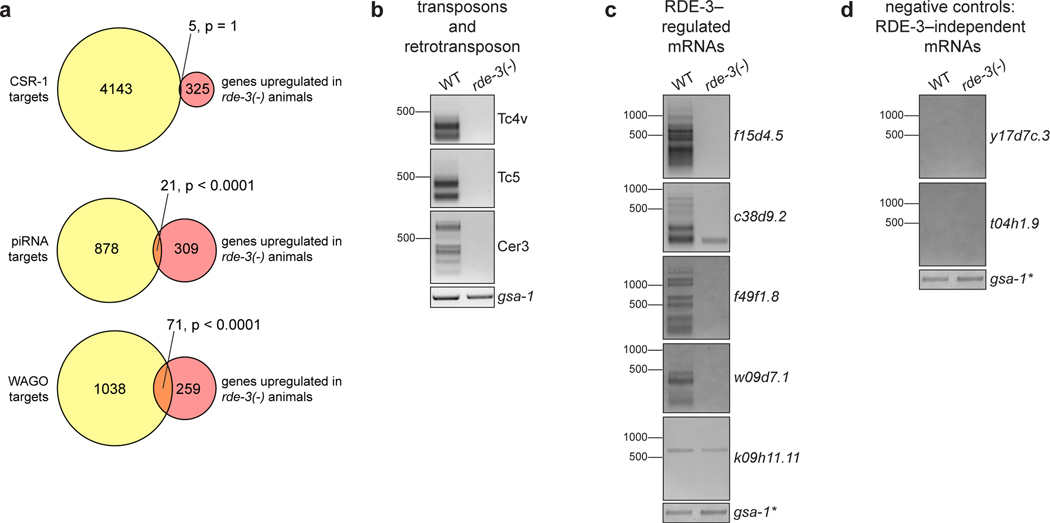
To identify additional targets of pUGylation, we conducted mRNA-seq on wild-type and rde-3(−) animals and identified 346 RNAs that were upregulated in rde-3(−) animals (Supplementary Table 2, adjusted p value <0.05 and log2 fold change >1.5), including Tc1 RNA1, as well as six other DNA transposons (Tc1A, Tc4, Tc5, MIRAGE1, CEMUDR1, Chapaev-2), several LTR retrotransposons (Cer3, Cer9, Cer13), and 294 predicted protein-coding RNAs (Supplementary Table 2, Extended Data Fig. 5a). Directed pUG PCR analyses confirmed that Tc4v, Tc5, Cer3, and four of five genes tested from amongst our list of the top 25 most RDE-3–regulated mRNAs were pUGylated in an RDE-3–dependent manner (Extended Data Fig. 5b,c). pUG tails were not detected on RNAs whose expression is unchanged in rde-3 mutants, including oma-1, gfp, dpy-11 (Extended Data Fig. 2a,b), and two additional genes selected at random (Extended Data Fig. 5d). We conclude that RDE-3 adds pUG tails to endogenous RNAs in C. elegans, which include, but are not limited to, transposon RNAs.
pUG RNAs localize to germ granules.
Germ granules are liquid-like condensates that form near the outer nuclear membrane in most animal germ cells and likely promote germ cell totipotency by concentrating germline determinants, including maternal RNAs and proteins, into developing germline blastomeres16. C. elegans RDE-3 localizes to perinuclear germ granules termed Mutator foci17. RNA fluorescence in situ hybridization (RNA FISH) using a fluorescently labeled p(AC)9 probe to detect pUG RNAs (pUG FISH) showed that pUG RNAs localized to perinuclear puncta in germ cells of wild-type, but not rde-3(−), animals (Fig. 3c). pUG FISH coupled with immunofluorescence (IF) to detect a GFP- and degron-tagged RDE-3 showed that pUG RNA foci co-localized with RDE-3 and, therefore, Mutator foci (Fig. 3d). These data suggest that pUG RNAs are produced, function, and/or are stored in Mutator foci in the C. elegans germline. Indeed, glp-1(q224) animals, which lack ≅99% of their germ cells when grown at 25°C (hereafter, glp-1(ts))18, failed to produce detectable Tc1 pUG RNAs (Fig. 3e) or oma-1 dsRNA-induced oma-1 pUG RNAs (Extended Data Fig. 6a) when grown at 25°C, confirming that pUG RNAs are produced or stored in germ cells. Incidentally, when glp-1(ts) animals were treated with dsRNA targeting the hypodermis-expressed dpy-11 gene13, dpy-11 pUG RNAs were detected in somatic cells (Extended Data Fig. 6a), consistent with previous reports showing that RDE-3 promotes RNAi within and between cells in the C. elegans soma17,19. Hereafter, this work focuses on the biogenesis and function of pUG RNAs in the germline.
To explore further how germline pUG RNAs and Mutator foci might relate, we asked if the glutamine/asparagine (Q/N) motif–rich protein MUT-16, which is required for Mutator focus assembly in germ cells17, was needed for pUG RNA biogenesis or function. mut-16(pk710) animals, which harbor a nonsense mutation in mut-16, produced elevated levels of oma-1 pUG RNAs in response to oma-1 dsRNA (Extended Data Fig. 6b,c) and decreased levels of Tc1 pUG RNAs (Extended Data Fig. 6d), suggesting that Mutator foci help to coordinate pUG RNA biogenesis. mut-16(pk710) animals were completely defective for silencing oma-1 after an oma-1 pUG RNA injection (Fig. 3f), indicating that Mutator foci are required for pUG RNA–based gene silencing, downstream of pUG RNA biogenesis.
pUG RNAs are templates for RdRPs.
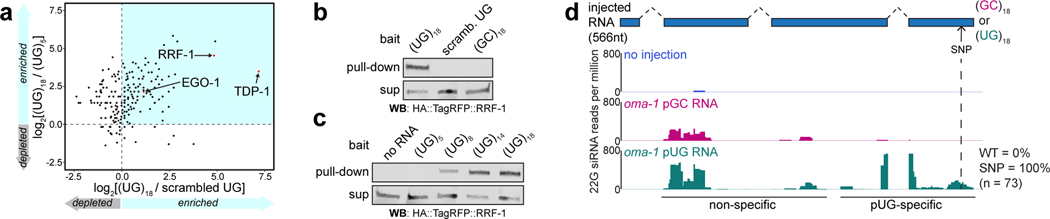
To understand how pUG tails might convert RNAs into agents of gene silencing, we sought to identify pUG tail–binding proteins. We conjugated 5’ biotinylated RNA oligonucleotides (oligos) consisting of 18 UG repeats, which conferred gene silencing activity onto oma-1 and gfp mRNA fragments (Fig. 2), to streptavidin beads. Beads were incubated with wild-type C. elegans extracts and bound proteins were analyzed with liquid chromatography–tandem mass spectrometry (LC-MS/MS). Beads conjugated to oligos consisting of 5 UG repeats and 36 scrambled UGs, neither of which conferred gene silencing activity (Fig. 2c), served as controls. This analysis identified 54 proteins that were enriched ≥2-fold in our (UG)18 RNA pull-down versus both control pull-downs (Fig. 4a, Supplementary Table 3). Amongst these proteins were TDP-1, the C. elegans ortholog of the mammalian UG-binding protein TDP-4320,21, as well as the RNA-dependent RNA polymerases (RdRPs) EGO-1 and RRF-1 (Fig. 4a, Supplementary Table 3). Current models posit that, during RNAi, RdRPs: 1) are recruited to mRNAs by siRNAs generated from dsRNA, and 2) use these mRNAs as templates to synthesize additional siRNAs, termed secondary (2°) siRNAs, which carry out gene silencing22–25. Interestingly, RRF-1, one of four C. elegans RdRPs, localizes to Mutator foci17. Thus, pUG tails may promote gene silencing by recruiting RdRPs, such as RRF-1, to pUG RNAs, which could then act as templates for siRNA synthesis.
Figure 4. pUG RNAs are templates for RdRPs.
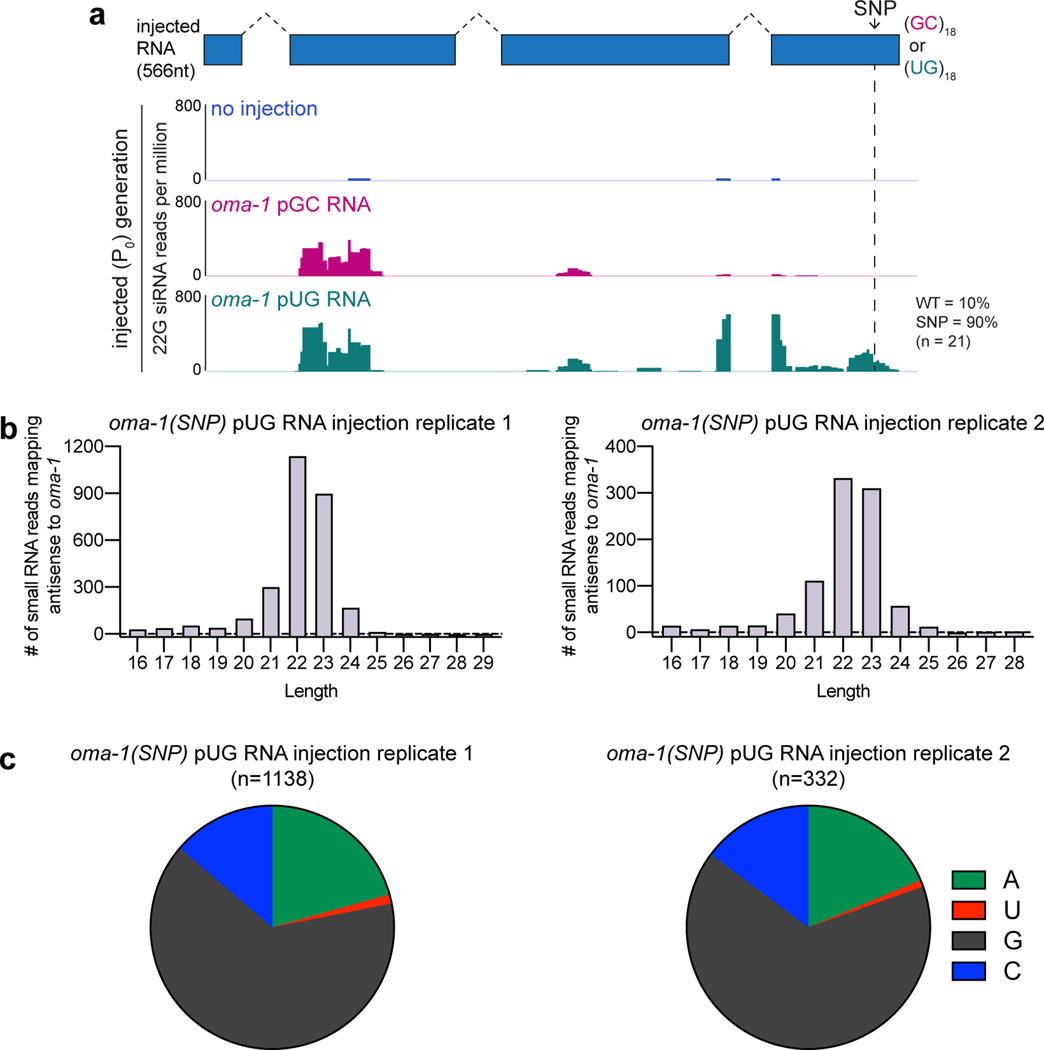
a, LC-MS/MS was performed on proteins that bound to 5’ biotinylated RNA oligos [(UG)5, (UG)18 or 36 scrambled UGs] conjugated to streptavidin beads. Shown is a scatter plot of log2-transformed fold enrichment in (UG)18 vs. scrambled UG pull-down (x-axis) and (UG)18 vs. (UG)5 pull-down (y-axis). Proteins enriched ≥2-fold in (UG)18 vs. beads-only pull-down are plotted. b-c, Indicated 5’ biotinylated RNA oligos were conjugated to streptavidin beads and incubated with extracts from animals expressing HA::tagRFP::RRF-1. Bead-bound material (pull-down) and supernatant (sup) were subjected to ɑ-HA immunoblotting. Data is representative of 2 biologically independent experiments. d, rde-1(ne219); oma-1(zu405ts) animals were injected with SNP-containing (dotted line) oma-1 (oma-1(SNP)) pUG or pGC-tailed RNAs and collected 1–4 hours later; small RNAs (20–30nts) were sequenced. Distribution of 22G siRNAs mapping antisense to oma-1 is shown, with 22G siRNA reads normalized to reads per million total reads. pUG-specific: 22G siRNAs observed only after oma-1(SNP) pUG RNA injection; non-specific: 22G siRNAs observed after oma-1(SNP) pUG and pGC RNA injections. See Extended Data Fig. 7 for biological replicate and details about sequenced small RNAs.
To confirm and expand upon potential pUG tail and RRF-1 interactions, we incubated beads conjugated to RNA oligos consisting of 5, 8, 14, or 18 UG repeats; 18 GC repeats; or 36 scrambled UGs with extracts from animals expressing HA- and TagRFP-tagged RRF-1. ɑ-HA immunoblotting showed that HA::TagRFP::RRF-1 interacted with (UG)18, but not scrambled UG or (GC)18, RNA oligos (Fig. 4b). Additionally, HA::Tag::RFP::RRF-1 precipitated strongly with (UG)14 and (UG)18 RNAs, weakly with a (UG)8 RNA, but not with a (UG)5 RNA (Fig. 4c). Together, these data show that the RdRP RRF-1 interacts with UG repeat RNAs and that the sequence determinants of this interaction largely mirror those required for pUG tail–mediated gene silencing in vivo (Fig. 2c).
To determine whether pUG RNAs act as templates for RdRPs in vivo, we sequenced small (20–30nt) RNAs from animals injected with either an oma-1 pGC or pUG RNA engineered to contain a single-nucleotide polymorphism (SNP) not present in the genomic copy of oma-1 (Fig. 4d, termed oma-1(SNP) RNAs). This SNP enabled differentiation of siRNAs templated from genomically encoded oma-1 mRNAs versus those templated from injected oma-1(SNP) pGC or pUG RNAs. In C. elegans, RdRP-derived (2°) siRNAs are also known as 22G siRNAs as they are typically antisense, 22nt in length and begin with a guanosine26. Small RNA sequencing showed that injection of the oma-1(SNP) pUG RNA, but not the oma-1(SNP) pGC RNA, triggered the synthesis of oma-1 22G siRNAs mapping near (≅100bp upstream) the site where the pUG tail was appended (Fig. 4d, Extended Data Fig. 7, Supplementary Table 4)27. For unknown reasons, both oma-1(SNP) pUG and pGC RNAs triggered non-specific siRNA synthesis ≅0.4kb upstream of where the tails were appended. Importantly, most (90–100%) pUG-specific 22G siRNAs antisense to the region of oma-1 containing the engineered SNP encoded the complement of the SNP (Fig. 4d, Extended Data Fig. 7a), indicating that these siRNAs were templated from the injected oma-1(SNP) pUG RNA. We conclude that one function of a pUG tail is to convert RNAs into templates for RdRPs.
pUG RNAs are vectors for TEI.
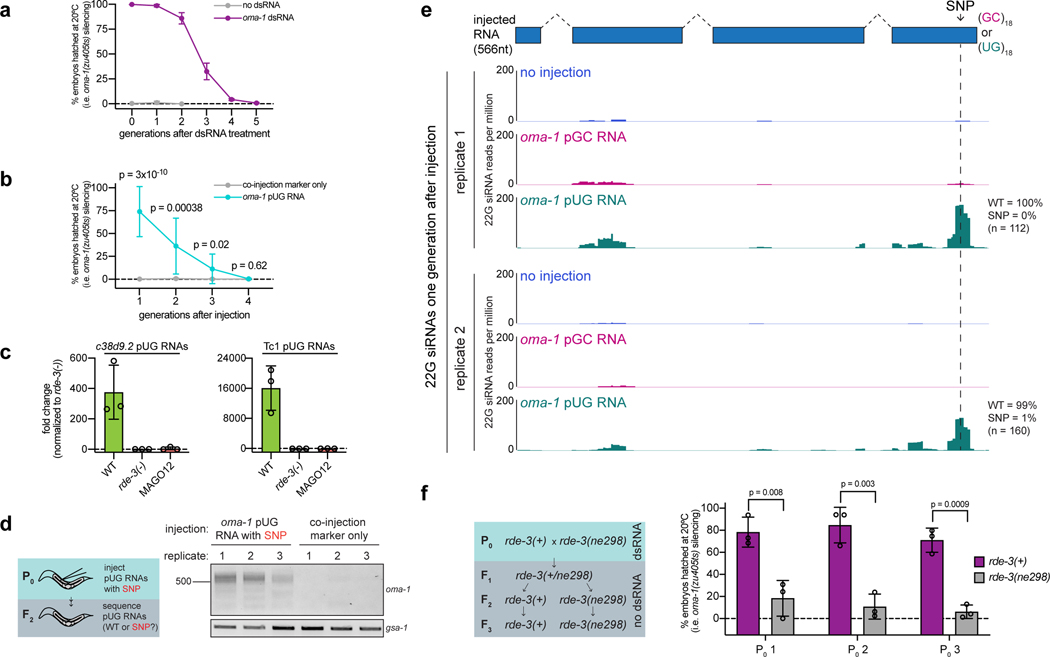
RNAi-triggered gene silencing can be inherited for multiple generations in C. elegans, making RNAi inheritance a robust and dramatic example of transgenerational epigenetic inheritance (TEI)28–33. Interestingly, a one-time exposure of animals to oma-1 dsRNA not only initiated the production of oma-1 pUG RNAs (Fig. 1a,b), but also caused oma-1 pUG RNAs to be expressed for four additional generations (Fig. 5a), concomitant with oma-1 gene silencing (Extended Data Fig. 8a), suggesting that pUG RNAs may contribute to TEI. To test this idea, we injected animals with gfp or oma-1 pUG RNAs and monitored gfp or oma-1 silencing over generations. gfp or oma-1 pUG RNAs were sufficient to silence gfp (Fig. 5b) or oma-1 (Extended Data Fig. 8b), respectively, for multiple generations. We conclude that pUG RNAs are sufficient to induce TEI.
Figure 5. pUG RNA/siRNA cycles drive heritable gene silencing.
a,oma-1 pUG PCR performed on total RNA from descendants of oma-1 dsRNA-treated animals. b, rde-1(ne219); gfp::h2b animals were injected with gfp pUG RNA, and gfp expression was monitored for six generations. n=3 (no injection), 9 (gfp pUG RNA). Error bars: s.d. of the mean. c, oma-1 pUG PCR performed on total RNA from oma-1 dsRNA-treated (P0) animals of indicated genotypes and their progeny (F1). Note: pUG RNAs appear longer in MAGO12 animals (Extended Data Fig. 10). d, pUG RNAs were Sanger sequenced from F2 progeny of rde-1(ne219); oma-1(zu405ts) animals injected with oma-1(SNP) pUG RNA. e, oma-1(zu405ts) hermaphrodites were fed oma-1 dsRNA and crossed to rde-3(ne298); oma-1(zu405ts) males (3 biologically independent crosses). oma-1 pUG PCR was performed on total RNA from rde-3(+) or rde-3(ne298) F3 progeny. a, c, d, e. Data is representative of 3 biologically independent experiments.
How might pUG RNAs drive TEI? We speculated that if pUG RNA–templated siRNAs (Fig. 4d, Extended Data Fig. 7a) could direct de novo mRNA pUGylation, then generationally repeated cycles of pUG RNA–templated siRNA synthesis and siRNA-directed pUG RNA biogenesis could be maintained in the absence of initiating dsRNA triggers and, thus, propagate gene silencing across generations. Three lines of evidence support this “pUG/siRNA cycling” model for RNAi-directed TEI. First, RdRP-derived 2° siRNAs in C. elegans can engage twelve AGO proteins (termed WAGOs) to mediate gene silencing34. MAGO12 animals, which harbor deletions in all twelve WAGOs, produced oma-1 pUG RNAs after oma-1 RNAi (Fig. 5c). Progeny of RNAi-treated MAGO12 animals, however, failed to produce oma-1 pUG RNAs (Fig. 5c). Thus, the 2° siRNA system is needed to maintain pUG RNAs specifically during the inheriting generations of TEI, consistent with a pUG/siRNA cycling model for TEI. Interestingly, pUG RNAs derived from endogenous pUGylation targets c38d9.2 and Tc1 were also dependent upon the WAGOs (Extended Data Fig. 8c), suggesting that the endogenous targets of RDE-3 also undergo heritable silencing via pUG/siRNA cycling.
Second, when we injected animals with an oma-1(SNP) pUG RNA, oma-1 pUG RNAs were detectable in subsequent generations (Extended Data Fig. 8d), but did not contain the engineered SNP (Fig. 5d). Similarly, <1% of siRNAs sequenced from progeny of oma-1(SNP) pUG RNA injected animals possessed the SNP complement (Extended Data Fig. 8e). Combined, these data show de novo pUGylation events occur during the inheriting generations of RNAi-directed TEI and these newly derived pUG RNAs become templates for further siRNA synthesis, supporting the idea that repeated pUG/siRNA cycling mediates TEI.
Third, we conducted a genetic analysis (Extended Data Fig. 8f) that showed that these de novo pUGylation events in inheriting generations are required for TEI. We crossed oma-1 RNAi–treated wild-type hermaphrodites with rde-3(ne298) males, isolated rde-3(+) and rde-3(ne298) F2 progeny, and then assayed the F3 generation of this cross (Fig. 5e) for oma-1 pUG RNA expression and oma-1 gene silencing. rde-3(ne298) animals lacked oma-1 pUG RNAs (Fig. 5e) and failed to silence the oma-1 locus (Extended Data Fig. 8f) during the inheriting generations of TEI, supporting the idea that pUG RNA biogenesis and, therefore, pUG/siRNA cycling in progeny is necessary for TEI maintenance. We conclude that pUG tails convert otherwise inert RNA fragments into drivers of an RNA-based memory system, which is likely propagated across generations via iterative cycles of sense pUG RNA and antisense siRNA biogenesis.
Discussion
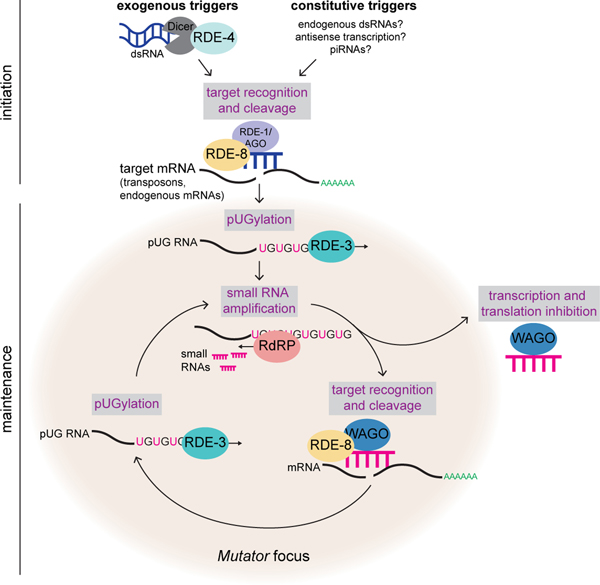
Here we show that RDE-3 adds pUG tails to germline- and soma-expressed RNAs in C. elegans and also demonstrate a role for this modification in transposon silencing and TEI. We find that RdRPs are recruited, either directly or indirectly, to pUG tails and use pUG RNAs as templates for siRNA synthesis. Assemblage of RDE-317 and other proteins, like the endonuclease RDE-812 and the RdRP RRF-117, into germline condensates termed Mutator foci likely coordinates RNA target recognition, cleavage, pUGylation, and siRNA amplification (Extended Data Fig. 9). We find that functional pUG tails consist of more than eight pairs of perfect or near-perfect 3’ UG repeats. These precise length and sequence requirements for pUG tail function hint that long pUG tails may impart stability upon mRNA fragments and/or form a structure which helps to recruit, and possibly prime, RdRPs, similar to the proposed role for poly(U)-tailing in small RNA–based gene silencing in Tetrahymena35. Additionally, our data show that proteins other than RdRPs, such as TDP-1, the C. elegans ortholog of the mammalian UG-binding protein TDP-4320,21, also interact with pUG repeats. We speculate that these other proteins may regulate the localization, stability, or function of pUG-tailed RNAs. Note: Extended Data Fig. 9 relates our findings to a previous report suggesting RDE-3 may uridylate targets of RNAi12.
Further, our data show that pUG RNAs act as informational vectors for TEI when they engage in feed-forward amplification cycles with RdRP-generated 2° siRNAs (Extended Data Fig. 9). These pUG/siRNA cycles, we speculate, allow C. elegans to remember past gene silencing events and inoculate progeny against expressing unwanted and/or dangerous genetic elements. Experimental RNAi-initiated pUG/siRNA cycles perdure for several generations (Fig. 5a), but are not permanent, suggesting that C. elegans possesses systems to prevent pUG/siRNA cycles from propagating in perpetuity. Interestingly, we find that RNAi-initiated pUG RNAs shorten progressively during TEI, suggesting that pUG RNA shortening, which may be an inevitable consequence of RdRP-based 2° siRNA synthesis (Extended Data Fig. 10), could function as one such brake on TEI. In contrast, the natural targets of pUGylation, such as transposons, are constitutively silenced by the pUG/siRNA system, suggesting that genetic systems, such as genomically encoded PIWI-interacting RNAs or endogenous dsRNAs, likely reinforce and refocus epigenetic pUG/siRNA silencing at these loci each generation (Extended Data Fig. 9).
The logic of sense pUG RNA/antisense siRNA cycling resembles that of fly and mammalian piRNA “ping-pong” systems in which iterative base-pairing between genomically encoded sense/antisense transposon RNAs, and piRNAs derived from these RNAs, mediates stable transposon silencing36. We speculate that related sense/antisense RNA systems could contribute to other biological processes for which long-term memories of past expression states are needed, such as antiviral immunity, development, or inheritance of environmentally acquired traits. Finally, our data show that long non-templated and non-homopolymeric tracts of ribonucleotides can be appended to, and confer novel functions to, RNAs in C. elegans. It will be of obvious interest to ask whether pUG-tailed RNAs, or RNAs bearing other unexpected tails, are restricted to C. elegans or are, instead, emissaries of a new class of eukaryotic RNA.
Methods
Genetics.
C. elegans culture and genetics were performed as described previously37. Unless otherwise noted, all C. elegans strains (Supplementary Table 5) were maintained at 20°C on NGM growth media and fed OP50 E. coli bacteria.
RNAi.
To perform RNAi experiments, embryos were obtained via hypochlorite treatment (egg prep) of gravid adult hermaphrodites and dropped onto RNAi plates (standard NGM plates with 1mM isopropyl β-D-1-thiogalactopyranoside and 25μg/ml carbenicillin) seeded with HT115 E. coli bacteria expressing either L4440 (Addgene, #1654) empty vector control or L4440 carrying inserts to trigger the production of dsRNA against a gene of interest. To perform pUG RNA analysis after RNAi treatment, gravid adults were washed off plates after 3–4 days using M9 + Triton X-100 buffer, collected in TRIzol, flash frozen in liquid nitrogen and stored at −80°C until total RNA extraction (see below). To look at oma-1 pUG RNAs across generations after RNAi, embryos were dropped onto plates seeded with HT115 bacteria expressing either empty vector control or dsRNA of interest. Some gravid adults were collected for the P0 generation sample and the remaining were egg prepped onto plates without dsRNA every generation, for the indicated number of generations. Each generation, some adult animals were collected while some were egg prepped to obtain the next generation. pUG RNAs were then detected as described below. To measure % embryos hatched after oma-1 RNAi, embryos obtained from animals harboring the oma-1(zu405ts) allele14 were dropped onto plates seeded with HT115 bacteria expressing either empty vector control or oma-1 RNAi and grown at 20°C. 6 adults were then singled per treatment for each strain/genotype and allowed to lay embryos overnight. The total number of embryos laid was counted, and then embryos were allowed to hatch for 24 hours, after which the total number of embryos that hatched was counted. For transgenerational RNAi experiments, empty vector control-treated and oma-1 RNAi–treated adults were egg prepped onto plates without dsRNA every generation and % embryos hatched was counted as just described until embryos no longer hatched. The dpy-11 RNAi clone and the oma-1 RNAi clone used, unless noted below, came from the C. elegans RNAi collection (Ahringer lab). The second oma-1 RNAi clone (referred to as pAS74 and used for Fig. 5a and Extended Data Figs. 1b, 8a, 10a, 10b) was a custom clone made to target exon 6 of oma-1. The gfp RNAi clone was obtained from the Fire lab.
pUG PCRs and qRT-PCRs.
Total RNA was extracted using TRIzol Reagent (Life Technologies, 15596018). 5ug of total RNA and 1pmol of reverse transcription (RT) oligo was used to generate first-strand cDNA using the Superscript III First-Strand Synthesis System (Invitrogen, 18080051). Note: total RNA was heated with dNTPs and RT oligo to 65°C for 5 mins and immediately chilled on ice before proceeding with remaining cDNA synthesis steps. 1ul of cDNA was used for the first PCR (20ul volume) performed with Taq DNA polymerase (New England BioLabs, M0273) and primers listed in Supplementary Table 5. First PCR reactions were diluted 1:100, and then 1ul was used for a second PCR (50ul volume) using primers listed in Supplementary Table 5. gsa-1, which has an 18nt long genomically encoded pUG repeat in its 3’UTR, served as a control for all pUG PCR analyses. PCR reactions were then run on agarose gels. For Sanger sequencing, lanes of interest were cut out from agarose gels and gel extracted using a QIAquick Gel Extraction Kit (Qiagen, 28706). 3ul of gel extracted PCR product was used for TA cloning with the pGEM-T Easy Vector System (Promega, A1360) according to manufacturer’s instructions. Ligation reactions were incubated overnight at 4°C. Transformations were performed with 5-alpha Competent E. coli cells (NEB, C2987H) and plated on LB/ampicillin/IPTG/X-gal plates (according to pGEM-T Easy Vector System manufacturer’s instructions). On the next day, white colonies were selected and inoculated, and then liquid cultures were miniprepped using QIAprep Spin Miniprep Kit (Qiagen, 27106). Plasmids were Sanger sequenced using a universal SP6 primer (5’-CATACGATTTAGGTGACACTATAG-3’) (Dana-Farber/Harvard Cancer Center DNA Resource Core, Harvard Medical School). qRT-PCRs were performed using 2ul of 1:100 diluted first PCRs as a template with qPCR primers (Supplementary Table 5) and iTaq Universal SYBR Green Supermix (Bio-Rad) according to manufacturer’s instructions.
MiSeq.
oma-1 pUG PCRs were sequenced on an Illumina MiSeq from animals fed HT115 bacteria expressing L4440 empty vector control plasmid (2 biological replicates), oma-1 RNAi clone from the Ahringer RNAi library or our custom oma-1 RNAi clone (pAS74). F1 to F4 descendants from pAS74-fed animals were obtained as described above and also sequenced (1 replicate each generation). A first round of PCR was performed with the same primers as described above (Supplementary Table 5). Primers were modified for the second PCR to contain Illumina p5 and p7 sequences, read 1 and 2 sequencing primers, a unique index (reverse primer only) for multiplexing and unique molecular identifiers (NNN) (Supplementary Table 5). PCR reactions were then pooled, run on an agarose gel and gel purified as described above. Gel purified DNA was sequenced on an Illumina MiSeq (Biopolymers Facility, Harvard Medical School) to obtain paired-end reads (67bp for Read 1, 248bp for Read 2).
MiSeq sequencing analysis.
First, unique molecular identifiers (UMIs) were removed from each read pair and appended to the end of the read name using UMI-tools v.1.0.038. Then, Cutadapt v2.5 was used for the following: 1) low-quality bases (quality score < 20) were trimmed from the 3’ ends of reads; 2) read pairs containing the inline portion of the 5’ adapter (AACAACGAGAAGATCGATGA) in Read 1 were selected for and then trimmed; 3) Read pairs containing the inline portion of the 3’ adapter (GGCGTCGCCATATTCTACTTACACACACACACACACAC) in Read 2 were selected for and trimmed; and 4) If the 5’ adapter was present in any Read 2 sequences, the adapter was trimmed from those sequences39. After adapter trimming, Read 2 sequences were screened for additional pACs at the 5’ end: reads that did not contain additional pACs (and therefore did not have a pUG tail longer than the adapter) were discarded; reads that did contain additional pACs were retained, and the pACs were trimmed using Cutadapt v2.5 (pAC and pCA sequences were provided as non-internal 5’ adapters)39. After pAC trimming, Read 2 sequences shorter than 5 nucleotides were discarded. The remaining Read 2 sequences were aligned to the C. elegans genome (WormBase release WS260) using STAR v2.7.0f40. SAM and BED files of unique alignments were generated using SAMtools v1.9 and BEDtools v2.27.1 and then imported into R for subsequent analyses41–43. Alignments were deduplicated based on the combination of the UMI and end coordinate. Alignments that mapped to the “+” strand and/or to coordinates outside of the oma-1 gene were discarded.
To systematically define the “oma-1” and “pUG” portions of each read, the pre-pAC-trimmed version of the read was reverse-complemented and then split as follows. By default, the aligned portion of the read was designated as “oma-1”, and any sequence downstream of the aligned portion was designated as the “pUG.” Then, the “oma-1” portion was matched to an oma-1 reference sequence (spliced + UTRs) using Biostrings v2.50.244. If the first 1–6 nucleotides that occurred 3’ of the match were the same in the oma-1 reference as they were in the read prior to pAC trimming (and therefore had the potential to be templated), then those nucleotides were reassigned to the “oma-1” portion of the read. End coordinates of the alignments were adjusted accordingly. A small portion of reads (<15%) were misannotated with the above approach, largely due to soft-clipping at the 3’ end during alignment. To systematically filter out such reads, reads for which the annotated “pUG” started with a base other than “U” or “G” and/or contained 2 or more bases other than “U” or “G” within the “pUG” sequence were discarded. A list of oma-1 pUG RNA reads can be found in Supplementary Table 1. The abundance of each pUGylation site (Extended Data Fig. 1a) was plotted in R using Sushi v1.20.045 for pUG RNAs sequenced from wild-type animals fed the oma-1 RNAi clone from the RNAi collection. To generate the pUG site logos shown in Extended Data Fig. 1b, a list of unique pUG sites was combined for pUG RNAs sequenced from wild-type animals fed the oma-1 RNAi clone from the RNAi collection and pAS74 (our custom oma-1 RNAi clone). This combined list was sorted by the last nucleotide of the “oma-1” portion and then plotted in R using ggseqlogo v0.146.
pUG RNA injections.
For gfp and oma-1 pUG RNA injections, pUG RNAs were synthesized in vitro using MEGAscript T7 Transcription Kit (Invitrogen, AM1334). DNA templates for in vitro transcription reactions were gel purified PCR products (150ng per in vitro transcription reaction) amplified using primers listed in Supplementary Table 5. Reactions were incubated overnight at 37°C. in vitro transcribed RNA was purified using TRIzol Reagent (Life Technologies, 15596018) and stored at −80°C. Injection mix consisted of 0.5pmol/ul in vitro transcribed RNA and 2.5ng/ul co-injection marker (pmyo-2::mCherry::unc-54 3’UTR) plasmid pCFJ90 (Addgene, plasmid #19327), dissolved in water. Animals expressing the co-injection marker show mCherry expression in the pharynx. For gfp pUG RNA injections, mjIs31 (gfp::h2b); rde-1(ne219) animals were injected in the germline and allowed to lay a brood at 20°C. Progeny of injected animals (F1 generation) were washed off plates using M9 + Triton X-100 buffer, mounted onto slides and 12–20 mCherry-expressing progeny per injected animal were scored for gfp expression using the Plan-Apochromat 20 × /0.8 M27 objective on an Axio Observer.Z1 fluorescent microscope (Zeiss). Images were taken with the Plan-Apochromat 63 × /1.4 Oil DIC M27 objective. For transgenerational inheritance experiments, 5 F1 progeny expressing mCherry per injected animal were picked under an Axis Zoom.V16 fluorescent dissecting microscope using a PlanNeoFluar Z 1x/0.25 FWD 56mm objective to a new plate to lay an F2 brood. Five random F2 animals were picked to a new plate to lay F3 progeny, while the remaining F2 adults (~40/injected animal) were scored for gfp expression as described above, but without regard for mCherry expression. This process was continued for several generations until 100% of animals expressed gfp. For all experiments, no injection control animals or animals injected with other RNA species were scored as described for pUG RNA injected animals. For oma-1 pUG RNA injections, oma-1(zu405ts); rde-1(ne219) were injected in the germline and allowed to recover at 15°C for two days. oma-1(zu405ts) is a gain-of-function temperature-sensitive allele of oma-1. oma-1(zu405ts) animals lay arrested embryos at 20°C, unless oma-1(zu405ts) is silenced47. Two days after injections, injected animals were shifted to 20°C. To measure oma-1(zu405ts) silencing, 5 adult mCherry-expressing progeny (F1 generation) per injected animal were picked as described above and transferred to a new plate. Animals were removed after laying 50–100 embryos, and oma-1(zu405ts) silencing was measured as percentage of embryos hatched (# hatched embryos/total number of embryos laid). For transgenerational inheritance experiments, F2 progeny that hatched were picked to new plates and allowed to grow to adults at 20°C. Five adult F2 animals per injected animal were picked to a new plate and % embryos hatched was scored as described above. This process was continued for several generations until 100% of embryos failed to hatch at 20°C. No injection control animals were maintained at 15°C. Every generation, 5 animals were shifted to 20°C before reaching adulthood and % embryos hatched was scored once they were adults. For Tc1 pUG RNA injections, T7 in vitro transcription was performed as described above to synthesize a Tc1 pUG RNA consisting of a 36nt pUG tail appended to a 338nt long fragment of Tc1 RNA (see Supplementary Table 5 for primers used). 18 and 22 rde-3(−); unc-22::tc1 animals were injected with a Tc1 pUG RNA + co-injection marker or co-injection marker alone, respectively. unc-22(st136) animals have a Tc1 DNA transposon insertion in the unc-22 gene, resulting in paralysis. mCherry-expressing progeny of Tc1 pUG RNA + co-injection marker or co-injection marker only injected animals were picked at the L4 stage and randomly pooled (25 animals per pool) onto 10cM NGM plates and allowed to lay a brood. The number of mobile adult progeny in each pool was counted 6–7 days later.
RNA FISH + Immunofluorescence.
Approximately 30 animals were dissected in 15 μl of 1X egg buffer (25 mM HEPES (pH 7.3), 118 mM NaCl2, 48 mM KCl, 2 mM CaCl2, 2 mM MgCl2) on SuperFrost Plus Adhesion slides (VWR, 631–0448) to isolate gonads. A coverslip was placed on top of dissected tissue, excess buffer was soaked up using a Kimwipe and slides were placed onto a metal block pre-chilling on dry ice for 10 min. Coverslips were popped off and slides were submerged in methanol at −20°C for 10 min. Slides were then washed twice, 5 min per wash in 1X PBS + 0.1% Tween-20 (PBSTW). Samples were then fixed with 4% paraformaldehyde solution in 1X PBS for 20 min, followed by two 5 min washes in PBSTW. Samples were then incubated at 37°C for 6 hours in a humid chamber with a 1:50 dilution of fluorescent RNA FISH probe in hybridization buffer (10% formamide, 2X SSC, 10% dextran sulfate (w/v)). The RNA FISH probe to detect pUG RNAs (/5Alexa647/CACACACACACACACACACA) was ordered from Integrated DNA Technologies (IDT) and stored at a stock concentration of 100uM at −20°C. The RNA FISH probe to detect ama-1 mRNA was ordered from Stellaris (SMF-6011–1). After 6 hours, slides were washed twice, 10 min per wash, in FISH Wash Buffer (2X SSC, 10% formamide, 0.1% Tween-20). Samples were then washed for 5 min in 2X SSC. Slides were sealed using 15ul of VECTASHIELD Antifade Mounting Medium (H-1000) with 4′,6-diamidino-2-phenylindole (DAPI). For experiments in which RNA FISH and immunofluorescence were combined, RNA FISH was first performed as above. After the final 2X SSC wash, slides were washed once with PBST for 5 min, samples were incubated overnight at room temperature in a humid chamber with a 1:1000 dilution of GFP antibody (Abcam, ab290) in PBSTW. Slides were then washed three times, 10 min per wash, in PBSTW and incubated in a 1:100 dilution (in PBSTW) of Goat anti-Rabbit secondary antibody, Alexa Fluor 555 (Invitrogen, A-21429) for 2 hours at room temperature in a humid chamber. Slides were next washed three times, 10 min per wash, in PBSTW and then sealed with 15ul of VECTASHIELD Antifade Mounting Medium (H-1000) with DAPI. All imaging was performed on an Axio Observer.Z1 fluorescent microscope (Zeiss) using the Plan-Apochromat 63 × /1.4 Oil DIC M27 objective. All image processing was done using Fiji48.
RNA-seq.
Total RNA was extracted using TRIzol Reagent (Life Technologies, 15596018). RNA quality (RIN) and quantity were assessed on the TapeStation 2200 (Agilent). Two rounds of mRNA purification were performed on 1ug total RNA using the Dynabeads mRNA DIRECT Kit (Invitrogen, 61011). First-strand cDNA was generated using the Superscript III First-Strand Synthesis System (Invitrogen, 18080051), followed by second-strand synthesis using DNA polymerase I (Invitrogen, 11917010). cDNA libraries were prepared using the Nextera XT DNA Library Preparation Kit (Illumina, FC-131–1024). Libraries were sequenced on the Illumina NextSeq500 platform (Biopolymers Facility, Harvard Medical School) and 75bp paired-end reads were obtained.
RNA-seq analysis.
Reads were trimmed to remove sequencing adapters and low-quality bases using Trim Galore version 0.4.4_dev (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Trimmed reads were then aligned to the C. elegans genome (UCSC ce11/WBcel235) using STAR version 2.7.0a49. Differential expression analysis of genes and repeat elements was performed using the TEtranscripts package in TEToolkit version 2.0.350. Gene annotations were obtained from Ensembl (WormBase release WS260)51. Repeat annotations were obtained from UCSC by downloading the RepeatMasker (rmsk) table in the Table Browser program. The table was reformatted to a GTF file using the Perl script makeTEgtf.pl (http://labshare.cshl.edu/shares/mhammelllab/www-data/TEToolkit/TE_GTF/). Features with an adjusted p value of < 0.05 and a log2 fold change > 1.5 were reported. Overlap (Extended Data Fig. 5a) was determined between mRNAs upregulated in rde-3(−) animals and published lists of: (1) RNAs targeted by CSR-1–bound endo-siRNAs52, (2) piRNA-targeted mRNAs (http://cosbi6.ee.ncku.edu.tw/piRTarBase/, Stringent and CLASH list)53, and (3) WAGO-class mRNAs26.
CRISPR.
The CRISPR strategy described previously54 was used to revert the missense mutation in rde-3(ne298) animals to wild-type and to tag the N-terminus of rrf-1 with ha::tagRFP. SapTrap cloning55,56 and the selection-based CRISPR strategy described previously57 was used to tag rde-3 at the N-terminus with gfp::degron and to introduce 3xflag::rde-3 (with 2kb upstream of the ATG and 2kb downstream of the stop codon) at the LGII MosSCI site ttTi560558 into rde-3(ne3370) animals. All guide RNAs were designed using the guide RNA selection tool CRISPOR59.
Small RNA sequencing.
rde-1(ne219); oma-1(zu405) animals were injected with an oma-1 pUG or pGC RNA (oma-1 mRNA fragment with (UG)18 or (GC)18 tail) in which the oma-1 sequence (the first 566nt of oma-1 mRNA) was modified to contain a SNP in exon 4 (ATTCATCCCG A>T TCATGGACCA). Injection mix was prepared as described above. For P0 analysis (Fig. 4d and Extended Data Fig. 7a), ~100 rde-1(ne219) animals were injected per experiment. After recovering for 1–4 hours at room temperature, injected animals were collected for total RNA extraction. For F1 analysis (Extended Data Fig. 8e), ~20 rde-1(ne219) animals were injected per experiment. Injected animals recovered at 15°C for two days and were returned to room temperature. ~500 adult co-injection marker-expressing progeny of injected animals were collected for total RNA extraction. Small RNAs were size-selected, cloned and sequenced as described previously60. Libraries were sequenced on the Illumina NextSeq500 platform (Biopolymers Facility, Harvard Medical School) to obtain 50bp single-end reads. Note: the same SNP-containing oma-1 pUG RNA was injected for the experiment described in Fig. 5d and Extended Data Fig. 8d. For this experiment, rde-1(ne219); oma-1(zu405ts) animals were injected with either co-injection marker only or the oma-1(SNP) pUG RNA with co-injection marker. Injected animals were allowed to recover at 15°C for ~4 days, after which 5–8 co-injection marker-expressing progeny from 5 injected animals were picked (as described above), pooled onto a 10cM plate for each replicate (3 replicates total) and allowed to lay a brood, which was then collected in TRIzol and oma-1 pUG PCR analysis was performed as described above.
Small RNA sequencing analysis.
A custom Python script was used to select reads starting with the last 4 nucleotides of the 5’ adaptor (either AGCG or CGTC). Cutadapt 1.1439 was then used to trim the 3’ adaptor (CTGTAGGCACCATCAATAGATCGGAAGAGCAC) and the in-line portion of the 5’ adaptor (AGCG and CGTC) (both with a minimum phred score = 20), allowing only sequences >= 16nt after trimming to pass (cutadapt -q 20 -m 16 -u 4 -a CTGTAGGCACCATCAATAGATCGGAAGAGCAC --discard-untrimmed). The quality of the trimming was assessed with FastQC 0.11.561. For downstream analysis, custom Python scripts were used to select reads that were 22nt in length and began with a G (22G siRNA reads). Tophat 2.1.162 was then used to map 22G siRNA reads to the C. elegans genome (WBcel235). Gene annotations were obtained from Ensembl51 (WormBase release WS269) and custom shell scripts were used to select protein-coding genes only. One mismatch was allowed to identify 22G siRNAs with SNPs. Using Samtools v0.1.19, only uniquely mapping sequences were retained. See Supplementary Table 4 for a list of all small RNA reads mapping to oma-1 and antisense 22G siRNAs mapping to oma-1. 22G siRNA pileup figures were generated as follows: first, bam files generated from Tophat v2.1.162 were normalized by DeepTools v3.0.263 based on counts per million and only antisense reads were kept for further analysis (bamCoverage -bs 2 --normalizeUsing CPM -samFlagExclide 16). Then, the normalized antisense 22G small RNA sequences (bedGraph files) were visualized using Sushi 1.20.045 in R. The number of reads mapping antisense to each gene was calculated by featureCounts 1.6.064 (featureCounts -s 2 -a *.gtf -t exon -g gene_name ). All custom scripts used in this section are available at: https://github.com/Yuhan-Fei/pUG-analysis.
pUG RNA chromatography.
Wild-type adult animals (~1–2 full 10cm plates per experiment) were frozen in liquid nitrogen as small droplets and ground into powder with a mortar and pestle. Powder was dissolved in lysis buffer (5mM HEPES-NaOH(pH7.5), 50mM NaCl, 5mM MgCl2, 0.5mM EDTA (pH8.0), 5% glycerol, 0.25% Triton X-100, 0.5mM DTT, 1mM PMSF, 1 tablet of cOmplete protease inhibitor (Roche, 11697498001)) and rotated for 30 min at 4°C. The resulting lysate was centrifuged at top speed for 10 min at 4°C. Supernatant was distributed evenly among experiments, and RNaseOUT recombinant ribonuclease inhibitor (Invitrogen, 10777019) was added to lysate (1ul per 100ul lysate). For each experiment, 160pmol of biotinylated RNA was conjugated to 400ug Dynabeads MyOne Streptavidin beads (Invitrogen, 65001) as per manufacturer’s instructions. Beads were added to lysates and rotated at room temperature for 1 hour. Beads were separated from supernatant on a magnetic rack, and the supernatant was collected and saved (“sup” fraction). Beads were washed 3 times with lysis buffer and rotated for 5 min at 4°C in lysis buffer. To perform liquid chromatography with tandem mass spectrometry (LC/MS/MS), beads were incubated in 500mM NH4OH and shaken at 37°C for 20 min after the last wash with lysis buffer. Beads were then pelleted using a magnetic rack and the supernatant was removed and vacuum dried until NH4OH had completely evaporated. 100ul of 100% TCA and 400ul of pre-chilled water was added to each sample, followed by a 15 min incubation on ice. Samples were spun at top speed at 4°C for 20 min. The supernatant was removed and the previous step was repeated using 1mL of 10% TCA solution. Samples were then washed two times with acetone and centrifuged for 10 min at 4°C after each wash. Acetone was removed and samples were dried in a speed vacuum to remove all residual acetone. Samples were analyzed using liquid chromatography-tandem mass spectrometry (LC-MS/MS, Taplin Mass Spectrometry Facility, Harvard Medical School). Note: a beads-only pull-down served as a control for this experiment. To analyze the LC-MS/MS data, 1 peptide count was assigned to all proteins with 0 peptide counts and the peptide counts for each pull-down sample were then normalized by the total number of peptides identified for that sample (Supplementary Table 3). Only proteins with normalized peptide counts that were ≥2-fold more enriched in the (UG)18 pull-down versus the beads-only control pull-down were kept for further analysis (Fig. 4a and Supplementary Table 3). For gel electrophoresis and Western blotting experiments, lysates were made from animals harboring rrf-1 tagged with ha and TagRFP at the endogenous locus. pUG RNA chromatography was performed as above. After the last wash with lysis buffer, beads (“pull-down” fraction) and “sup” were dissolved in 2x Laemmli sample buffer (Biorad, 1610737, final concentration=1x) with 5% 2-mercaptoethanol and heated for 5 min at 95°C and chilled on ice. “Pull-down” and “sup” fractions were loaded into 4–15% Mini-PROTEAN TGX Precast protein gels (Biorad, 4561086) and run in Tris-glycine running buffer (25mM Tris, 192mM glycine, 0.1% SDS). Proteins were then transferred to nitrocellulose membrane (BioRad) at 100V for 1 hour in electrotransfer buffer (50mM Tris, 40mM glycine, 9% methanol, 0.2% SDS). Blotted membranes were blocked with 5% milk in PBST (phosphate-buffered saline, 1.0% Tween-20) for 1 hour at room temperature and probed with primary antibody (1:1000 HA-Tag Rabbit mAb, Cell Signaling, #3724, in 5% milk) overnight at 4°C. After washing with PBST 3 times, membrane was probed with secondary antibody (1:10000 IRDye 800CW Goat anti-Rabbit IgG, LI-COR, 926–32211, in 5% milk) for 1 hour at room temperature. Membrane was washed with PBST 3 times before imaging using Odyssey Fc Dual-Mode Imaging System (LI-COR).
Heterozygous Experiment.
To perform the experiment in Fig. 5e and Extended Data Fig. 8f, embryos were obtained via hypochlorite treatment of oma-1(zu405ts) gravid adult hermaphrodites and dropped onto RNAi plates seeded with HT115 bacteria expressing dsRNA targeting oma-1. L4 hermaphrodites were then transferred, along with rde-3(ne298); oma-1(zu405ts) males, onto RNAi plates seeded with 25ul (small area of food to encourage mating) of oma-1 dsRNA-expressing bacteria. Once hermaphrodites were adults, they were singled onto NGM plates seeded with OP50 and allowed to lay F1 progeny. 12–15 F1s were singled from 3 independently mated hermaphrodites and genotyped to ensure that they were heterozygous for rde-3(ne298). To obtain F3 animals, 12–15 F2s per F1 (verified to be heterozygous for rde-3(ne298)) were singled to 15°C (so as to avoid embryonic arrest due to temperature) and allowed to lay a brood. F2s were then single worm genotyped to identify rde-3(+) and rde(ne298) homozygous animals. Then, % embryonic arrest was calculated by pooling 5 L4 stage F3 animals per F2 at 20°C until they had laid a brood of 50–200 progeny and counting the # of embryos that were laid vs. hatched on the following day. rde-3(+) and rde(ne298) homozygous F3 broods were pooled for all plates that were derived from the same P0 and oma-1 pUG PCR was performed as described above.
Extended Data
Extended Data Fig. 1. Analysis of oma-1 pUGylation sites.
a, Illumina MiSeq was performed (n=1 biological experiment) on oma-1 pUG PCR products derived from WT and rde-3(−) animals, +/− oma-1 dsRNA. # of sequenced pUG RNAs (y-axis) mapping to each pUGylation site (x-axis) is shown. Inset: total number of sequenced oma-1 pUG RNAs from indicated samples and total number of these sequenced pUG RNAs in which the oma-1 sequence was spliced. b, MiSeq-sequenced oma-1 pUG RNAs were sorted into four groups based on the nucleotide (nt) at the last templated position (−1) of the oma-1 mRNA. The % of oma-1 pUG RNAs (MiSeq reads) with each nt in the −1 position is shown. Logo analysis was then performed on each of the four groups to determine the probability of finding each nucleotide at the first position of the pUG tail (+1), as well as at the second-to-last templated nucleotide of oma-1 (−2). This analysis showed that if the last templated nucleotide of the oma-1 mRNA fragment was an A or a C, then RDE-3 was equally likely to add a U or a G as the first nucleotide of an elongating pUG tail. If, however, the last templated nt was a U or G, then RDE-3 preferentially added a G or U, respectively, as the first nt in an elongating pUG tail. *Note: To perform the analyses in this figure, we assumed that if a U or G could have been genomically encoded, then it was. If, instead, RDE-3 added the U or G shown in the −1 position as the first nucleotide of the pUG tail, then these data show that the second nucleotide that RDE-3 prefers to add is a G after a U or a U after a G.
CCA-adding rNT enzymes modify the 3’ termini of tRNAs with non-templated CCA nts. The mechanism by which these enzymes add non-templated nonhomopolymeric stretches of nts is thought to involve allosteric regulation of the nt binding pocket by the 3’ nt of a substrate tRNA65. A similar mechanism may explain how RDE-3 can add pUG tails to its mRNA substrates. For instance, when the 3’ nt of an RDE-3 substrate is a U, the rNTP binding pocket of RDE-3 might adopt a structure that preferentially binds G and vice versa when 3’ nt of an RDE-3 substrate is a G. Such a model could explain how a single rNT enzyme adds perfectly alternating U and G nts to RNA substrates. There are also alternative models for how RDE-3 might add pUG tails to an RNA. These include: 1) the existence a poly(AC) nucleic acid template used by RDE-3 during pUG tail synthesis, 2) the existence of one or more rNTs that cooperate with RDE-3 to produce pUG tails, or 3) the possibility that RDE-3 binds and incorporates UG or GU dinucleotides. We disfavor the first two possibilities as these models are difficult to reconcile with the observation that RDE-3 adds UG repeats to tethered RNAs in yeast or in Xenopus oocytes5. The third proposed model may be true, but because our sequencing shows that pUG tails can initiate with either a U or G (this figure, Supplementary Table 1), then RDE-3 would need to be able to bind both UG and GU dinucleotides. Determining the mechanism by which RDE-3 adds pUG tails will likely involve structural studies and/or in vitro pUGylation assays using recombinant RDE-3 protein.
Extended Data Fig. 2. RNAi-triggered pUGylation and pUG RNA–directed gene silencing are general and sequence-specific.
a,gfp::h2b, rde-3(−); gfp::h2b and WT (no gfp::h2b) animals were fed E.coli expressing either empty vector control or gfp dsRNA. b, WT and rde-3(−) animals were fed E.coli expressing empty vector control and either oma-1 or dpy-11 dsRNA. For a and b, gfp, dpy-11 and oma-1 pUG RNAs were detected using the assay outlined in Fig. 1a. Data is representative of 3 biologically independent experiments. c, rde-1(ne219); oma-1(zu405ts) animals were injected with either an oma-1 (n=6) or gfp (n=10) pUG RNA. n=3 for no injection. % embryos hatched was scored for the progeny of injected animals. Inset: injected RNAs run on a 2% agarose gel to assess RNA integrity. Error bars: s.d. of the mean. d, rde-1(ne219); gfp::h2b animals were injected with either an oma-1 or gfp pUG RNA (n=10 for both, 3 for no injection). Mean % progeny with gfp::h2b silenced is indicated ± standard deviation (s.d.). For c and d, all pUG tails were 36nt in length.
Extended Data Fig. 3. RDE-3–mediated pUGylation is necessary for RNAi.
a, Animals of the indicated genotypes (all harboring the oma-1(zu405ts) mutation) were treated +/− oma-1 dsRNA. For each experiment, % embryos hatched was scored at 20°C and averaged for 6 individual animals per treatment for each genotype. rde-1(ne219) mutants, which cannot respond to dsRNA3, serve as a control for this experiment. Error bars: s.d. of the mean for 3 biologically independent experiments. b, Control or rde-3(ne298) animals (all rde-1(ne219); oma-1(zu405ts) background) were injected with oma-1 pUG RNAs and % embryos hatched was scored at 20°C. n=10 noninjected and 16 injected animals for control. n=8 noninjected and 14 injected animals for rde-3(ne298). Error bars: s.d. of the mean.
Extended Data Fig. 4. pUG tails must be appended to sense RNAs of >50nt for functionality.
rde-1(ne219); oma-1(zu405ts) animals were injected with: a, an oma-1 pUG RNA consisting of the sense or antisense strand of the same 541nt long oma-1 mRNA fragment (beginning at the aug) with a 36nt 3’ pUG tail (n=9 for both; n=3 for no injection). b, oma-1 pUG RNAs consisting of oma-1 mRNA fragments of varying lengths (with position 1 starting at the aug of the oma-1 mRNA sequence) all appended to a 36nt pUG tail. n=6 (no injection), 10 (1–50), 17 (1–100), 8 (1–270), 9 (271–541) and 15 (1–541). For a and b, % embryonic arrest was scored at 20°C. Error bars: s.d. of the mean.
Extended Data Fig. 5. Endogenous targets of pUGylation in C. elegans.
a, mRNAs upregulated in rde-3(−) mutants (Supplementary Table 2) were compared to published lists of: (1) RNAs targeted by CSR-1–bound endo-siRNAs52, (2) piRNA-targeted mRNAs (based on predictive and experimental approaches)53, and (3) WAGO-class mRNAs26. p-values were generated using a one-sided Fisher’s exact test. This analysis showed statistically significant overlap between the mRNAs upregulated in rde-3(−) mutants and both piRNAs targets and WAGO-class mRNAs. b-d, Total RNA was extracted from WT or rde-3(−) animals. The assay outlined in Fig. 1a was used to detect pUG RNAs for b, two DNA transposons (Tc4v and Tc5) and a retrotransposon (Cer3) that were significantly upregulated in rde-3(−) animals; c, predicted protein-coding mRNAs that were significantly upregulated in rde-3(−) animals; and d, two randomly selected mRNAs whose expression does not change in rde-3(−) mutants. Data is representative of 3 biologically independent experiments. *Note: the same RT samples were used for panels c and d and, therefore, the gsa-1 loading control is the same for both panels.
Extended Data Fig. 6. Mutator foci likely coordinate pUG RNA biogenesis within germ cells.
a,dpy-11 and oma-1 pUG PCR (Fig. 1a) were performed on total RNA from glp-1(q224/ts) animals grown at 15°C (germ cells present) or 25°C (<99% of germ cells), +/− oma-1 and dpy-11 dsRNA. Data is representative of 2 biologically independent experiments. *Note: the samples in a are the same as those used in Fig. 3e and, therefore, the gsa-1 loading control is the same. b, oma-1 pUG PCR was performed on total RNA extracted from wild-type, rde-3(−), and mut-16(pk710) animals, +/− oma-1 dsRNA. Data is representative of 4 biologically independent experiments. c, qRT-PCR was used to quantify levels of oma-1 pUG RNAs in wild-type, rde-3(−), and mut-16(pk710) animals, +/− oma-1 dsRNA. Data is represented as fold change in the levels of oma-1 pUG RNAs +/− oma-1 dsRNA (y-axis) for each strain (x-axis). n=3 biologically independent samples per treatment for each strain. Error bars: s.d. of the mean. d, qRT-PCR was used to quantify levels of Tc1 pUG RNAs in wild-type, rde-3(−), and mut-16(pk710) animals. Note: the RNA samples used for d are the same as those used in c, except that the data for +/− oma-1 dsRNA samples were pooled for each strain. n=6 biologically independent samples for each strain. Error bars: s.d. of the mean. The analyses in c and d showed that mut-16 mutant animals produced more oma-1, but fewer Tc1, pUG RNAs, than wild-type animals. The increased levels of oma-1 pUG RNAs in mut-16(pk710) animals was also suggested by the gel in b. Together, these data suggest that Mutator foci likely have an important role in coordinating pUG RNA biogenesis in germ cells, as pUG RNA levels become misregulated in mut-16(pk710) mutants.
Extended Data Fig. 7. pUG RNAs are templates for RdRPs.
a, A biological replicate of the experiment shown in Fig. 4d was performed. oma-1(SNP) pUG or pGC RNAs were injected into rde-1(ne219); oma-1(zu405ts) germlines. SNP location is indicated with the dotted line. Injected animals were collected 1–4 hours after injection, total RNA was isolated and small RNAs (20–30nt) were sequenced. Distribution of 22G siRNAs mapping antisense to oma-1 is shown, with 22G siRNA reads normalized to reads per million total reads. oma-1 pUG (but not pGC) RNA injection triggered 22G siRNA production near the site of the pUG tail (“pUG-specific” 22G siRNAs). For unknown reasons, both pUG and pGC RNA injections triggered small RNA production ≅400bp 5’ of either tail. b, Length distribution of small RNA reads mapping antisense to oma-1 is shown for small RNAs sequenced after oma-1(SNP) pUG RNA injections (Fig. 4d and a). c, Proportion of 22nt long small RNAs mapping antisense to oma-1 containing 5’ adenine, uracil, guanine, or cytosine is shown.
Extended Data Fig. 8. De novo pUGylation events in progeny are required for TEI.
a,oma-1(zu405ts) animals were fed bacteria expressing empty vector control or oma-1 dsRNA and % embryos hatched at 20°C was scored for 6 generations. Error bars represent s.d. of the mean of three biologically independent experiments. For each experiment, % embryos hatched at 20°C was averaged for 6 individual animals per treatment for each genotype. b, rde-1(ne219); oma-1(zu405ts) animals were injected with co-injection marker alone (n=12) or co-injection marker + oma-1 pUG RNA (n=19) and % embryos hatched at 20°C was scored for four generations in lineages of animals established from injected parents (see Methods for details of experimental setup). Error bars: s.d. of the mean. p-values: two-tailed unpaired Student’s t-test. c, c38d9.2 and Tc1 pUG RNA expression quantified in embryos harvested from wild-type, rde-3(−) or MAGO12 animals using qRT-PCR. Fold change normalized to rde-3(−). Each point (n) represents a biologically independent replicate, n=3 independent replicates/strain. Error bars: s.d. of the mean. d, Same experiment as Fig. 5d. rde-1(ne219); oma-1(zu405ts) animals were injected with an oma-1(SNP) pUG RNA or with co-injection marker only. Co-injection marker-expressing F1 progeny were picked and allowed to lay their F2 broods. oma-1 pUG PCR was performed on total RNA from F2 progeny. Shown is data from three biological replicates. e, Two biological replicates of small RNAs sequenced from the progeny of rde-1(ne219); oma-1(zu405ts) animals injected with oma-1(SNP) pUG or pGC RNAs are shown. Dotted line indicates the location of the SNP incorporated into oma-1. Distribution of 22G siRNAs mapping antisense to oma-1 is shown, with 22G siRNA reads normalized to reads per million total reads. In Fig. 4d and Extended Data Fig. 7a, small RNAs were sequenced 1–4 hours after injection and 100% of 22G siRNAs antisense to the region of the engineered SNP in oma-1 were found to encode the complement of the SNP. Shown here, <1% of 22G siRNAs from progeny of injected animals encoded the SNP complement. Note: siRNAs mapping near the pUG tail were observed only after oma-1(SNP) pUG RNA injection (pUG-specific siRNAs). For unknown reasons, both oma-1(SNP) pUG and pGC RNAs triggered small RNA production 5’ of the pUG-specific siRNAs. It is possible that these siRNAs were triggered by systems that respond to foreign RNAs, such as the piRNA system. Further work will be needed to ascertain the etiology of these siRNAs. f, Same experiment as Fig. 5e. oma-1(zu405ts) hermaphrodites were fed oma-1 dsRNA and crossed to rde-3(ne298); oma-1(zu405ts) males. F2 progeny from this cross were genotyped for rde-3(ne298). WT and rde-3(ne298) homozygous F3 progeny were phenotyped for % embryonic arrest at 20°C. 3 biologically independent crosses (P0 1–3) were performed. Error bars: s.d. of the mean. p-values: two-tailed unpaired Student’s t-tests.
Extended Data Fig. 9. Working model for pUG RNA/siRNA cycling during RNAi.
Initiation: exogenous and constitutive (i.e. genomically encoded such as dsRNA, piRNAs) triggers direct RDE-3 to pUGylate RNAs previously fragmented by factors in the RNAi pathway. Maintenance: pUG RNAs are templates for 2° siRNA synthesis by RdRPs. Argonaute proteins (termed WAGOs) bind 2° siRNAs and: 1) target homologous RNAs for transcriptional and translational silencing29,34,66,67, as well as 2) direct the cleavage and de novo pUGylation of additional mRNAs. In this way, cycles of pUG RNA-based siRNA production and siRNA-directed mRNA pUGylation maintain silencing over time and across generations. This model shows germline perinuclear condensates termed Mutator foci as the likely sites of pUG RNA biogenesis in germ cells for several reasons. RDE-3 localizes to Mutator foci17 and we show in Fig. 3d that endogenous pUG RNAs localize to Mutator foci. The fact that enzyme and enzyme product both localize to Mutator foci suggests that Mutator foci may be sites of RNA pUGylation. In addition, while pUG RNAs are still made in mut-16 mutants (Extended Data Fig. 6b-d), which lack Mutator foci, the levels of both dsRNA-triggered and endogenous pUG RNAs are misregulated. Thus, while RDE-3 still has enzymatic activity in the absence of Mutator foci, these perinuclear condensates are likely coordinating target recognition and pUGylation in wild-type animals. Indeed, both the endonuclease RDE-8, which cleaves mRNAs targeted by dsRNA12, and the RdRP RRF-117 also localize to Mutator foci, further suggesting that pUG RNA/siRNA cycling occurs in Mutator foci.
Previous studies have shown that animals lacking RDE-3 still produce some 22G endo- siRNAs, including 22G siRNAs that associate with the Argonaute CSR-1 and whose biogenesis depends upon the RdRP EGO-126,68. Thus, EGO-1 may also produce some 22G siRNAs via a pUG RNA-independent mechanism.
A previous study showed that, in rrf-1 mutants that lack germlines, sel-1 RNAi causes a small fraction of sel-1 mRNA fragments to be uridylated in a largely RDE-3–dependent manner in the soma12. This data suggests that, in somatic tissues, RDE-3 may add non-templated Us to the 3’ termini of mRNA fragments generated during RNAi. It was proposed that this uridylation may be important for turnover or decay of RNAi targets12. Our work, combined with this earlier data about RDE-3–dependent uridylation12, suggests two models. First, RDE-3 may possess two distinct catalytic activities: uridylation and pUGylation. According to this model, RDE-3 might add Us or UGs depending on context (e.g. cell/tissue-type or developmental timing). Alternatively, the mRNA uridylation observed in the soma could depend upon RDE-3 and the pUGylation system, but may be mediated by another, currently unknown, poly(U) polymerase.
Extended Data Fig. 10. pUG RNA shortening may act as a brake on TEI.
a, The gel shown is the same as in Fig. 5a, except that oma-1 pUG RNAs from the P0 generation are included for WT and rde-3(−) animals. Data is representative of 3 biologically independent experiments. b, oma-1 pUG RNA reads from MiSeq (n=1 biological experiment) were mapped to oma-1 and the length of the oma-1 mRNA portion of each pUG RNA read was determined (y-axis). Shown is a Box and Whisker plot representing the interquartile range (IQR, box) and median (line in the box) of lengths at the indicated generations after dsRNA treatment. The y-axis starts at the aug of the oma-1 mRNA. The whiskers extend to values below and above 1.5*IQR from the first and third quartiles, respectively. Data beyond the end of the whiskers are outliers and plotted as points. These data support the gel in a, showing that pUG RNAs get shorter in each generation during RNAi-triggered TEI. c, A “ratchet” model to explain pUG RNA shortening. pUG RNA shortening may be due to the 3’→5’ directionality of RdRPs, which, during the maintenance phase of pUG/siRNA cycling (see model in Extended Data Fig. 9), causes each turn of the pUG/siRNA cycle to trigger cleavage and pUGylation of target mRNAs at sites more 5’ than in the previous cycle. Eventually, pUG RNAs are too short to act as RdRP templates, cycling cannot be maintained and silencing ends. Additional support for the ratchet model comes from Fig. 5c, which shows that RNAi-triggered pUG RNAs are longer in MAGO12 mutant animals than in wild-type animals. Note: the P0 generation animals in Fig. 5c were exposed to dsRNA continuously from embryos to adulthood, when they were harvested. These longer pUG RNAs are likely due to continued initiation of pUGylation triggered by the exogenously provided dsRNA without downstream pUG/siRNA cycling. In the absence of this cycling, pUG RNA shortening does not occur. Finally, a number of recent studies in C. elegans have reported transgenerational inheritance of acquired traits, which lasts 3–4 generations69–74. As shown in a, the expression of oma-1 RNAi–directed pUG RNAs also perdures for 3–4 generations. These shared generational timescales of inheritance hint that the inheritance of acquired traits in C. elegans may be mediated by pUG RNAs whose generational “half-life” is limited to 3–4 generations due to the built-in brake on TEI provided by pUG RNA shortening.
Supplementary Material
Acknowledgements
We thank past and present members of the Kennedy, Butcher and Wickens labs for helpful discussions; the Biopolymers Facility at HMS for Illumina sequencing; the Dana-Farber/Harvard Cancer Center DNA Resource Core for Sanger sequencing; and the Taplin Mass Spectrometry Facility in the Cell Biology Department at HMS for performing LC-MS/MS. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). Some strains were provided by the Mitani laboratory through the National BioResource Project (Tokyo, Japan), which is part of the International C. elegans Gene Knockout Consortium. A.S. and J.Y. were supported by the Ruth L. Kirschstein T32 Predoctoral NRSA (T32GM096911) and NSF Graduate Research Fellowships (A.S.: DGE1144152, DGE1745303; J.Y.: DGE1745303). A.E.D. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2304-17). D.J.P. was supported by a Ruth L. Kirschstein National Research Service Award (1F32GM125345-01).
Footnotes
Code availability
A description of custom scripts used to analyze MiSeq and RNA-seq data is provided in the Methods section and scripts are available upon request. Custom Python scripts used to analyze small RNA sequencing data are deposited at: https://github.com/Yuhan-Fei/pUG-analysis.
Data availability
All DNA and RNA sequencing data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus75 and are accessible through GEO Series accession number GSE148134. Processed MiSeq and small RNA sequencing reads are also provided in Supplementary Tables 1 and 4, respectively.
Competing interests
M.W. has a patent (US20160145666A1) through Wisconsin Alumni Research Foundation (Madison, WI) for methods, kits, and compositions of matter relating to poly(UG) polymerases.
Main references
- 1.Collins J, Saari B. & Anderson P. Activation of a transposable element in the germ line but not the soma of Caenorhabditis elegans. Nature 328, 726–728 (1987). [DOI] [PubMed] [Google Scholar]
- 2.Ketting RF, Haverkamp THA, van Luenen HGAM & Plasterk RHA mut-7 of C. elegans, Required for Transposon Silencing and RNA Interference, Is a Homolog of Werner Syndrome Helicase and RNaseD. Cell 99, 133–141 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Tabara H. et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123–132 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Chen C-CG et al. A member of the polymerase beta nucleotidyltransferase superfamily is required for RNA interference in C. elegans. Curr. Biol 15, 378–383 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Preston MA et al. Unbiased screen of RNA tailing activities reveals a poly(UG) polymerase. Nat. Methods 16, 437–445 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fire A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Aravind L. & Koonin EV DNA polymerase beta-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 27, 1609–1618 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin G. & Keller W. RNA-specific ribonucleotidyl transferases. RNA 13, 1834–1849 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Detwiler MR, Reuben M, Li X, Rogers E. & Lin R. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev. Cell 1, 187–199 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Parrish S. & Fire A. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA 7, 1397–1402 (2001). [PMC free article] [PubMed] [Google Scholar]
- 11.Tabara H, Yigit E, Siomi H. & Mello CC The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109, 861–871 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Tsai H-Y et al. A ribonuclease coordinates siRNA amplification and mRNA cleavage during RNAi. Cell 160, 407–419 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ko FCF & Chow KL A novel thioredoxin-like protein encoded by the C. elegans dpy-11 gene is required for body and sensory organ morphogenesis. Development 129, 1185–1194 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Lin R. A gain-of-function mutation in oma-1, a C. elegans gene required for oocyte maturation, results in delayed degradation of maternal proteins and embryonic lethality. Dev. Biol 258, 226–239 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Fischer SEJ, Wienholds E. & Plasterk RHA Continuous exchange of sequence information between dispersed Tc1 transposons in the Caenorhabditis elegans genome. Genetics 164, 127–134 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voronina E, Seydoux G, Sassone-Corsi P. & Nagamori I. RNA granules in germ cells. Cold Spring Harb. Perspect. Biol 3, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips CM, Montgomery TA, Breen PC & Ruvkun G. MUT-16 promotes formation of perinuclear mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 26, 1433–1444 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Austin J. & Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 51, 589–599 (1987). [DOI] [PubMed] [Google Scholar]
- 19.Jose AM, Garcia GA & Hunter CP Two classes of silencing RNAs move between Caenorhabditis elegans tissues. Nat. Struct. Mol. Biol 18, 1184–1188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buratti E. & Baralle FE Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem 276, 36337–36343 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Kuo P-H, Doudeva LG, Wang Y-T, Shen C-KJ & Yuan HS Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 37, 1799–1808 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sijen T. et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107, 465–476 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Ambros V, Lee RC, Lavanway A, Williams PT & Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol 13, 807–818 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Sijen T, Steiner FA, Thijssen KL & Plasterk RHA Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315, 244–247 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Pak J. & Fire A. Distinct Populations of Primary and Secondary Effectors During RNAi in C. elegans. Science 315, 241–244 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Gu W. et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 36, 231–244 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Billi AC, Fischer SEJ & Kim JK Endogenous RNAi pathways in C. elegans. WormBook 1–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vastenhouw NL et al. Gene expression: long-term gene silencing by RNAi. Nature 442, 882 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Buckley BA et al. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489, 447–451 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashe A. et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150, 88–99 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shirayama M. et al. piRNAs Initiate an Epigenetic Memory of Nonself RNA in the C. elegans Germline. Cell 150, 65–77 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luteijn MJ et al. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 31, 3422–3430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sapetschnig A, Sarkies P, Lehrbach NJ & Miska EA Tertiary siRNAs mediate paramutation in C. elegans. PLoS Genet. 11, e1005078 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yigit E. et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127, 747–757 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Talsky KB & Collins K. Initiation by a eukaryotic RNA-dependent RNA polymerase requires looping of the template end and is influenced by the template-tailing activity of an associated uridyltransferase. J. Biol. Chem 285, 27614–27623 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czech B. et al. piRNA-Guided Genome Defense: From Biogenesis to Silencing. Annu. Rev. Genet 52, 131–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional references
- 37.Brenner S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith T, Heger A. & Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 27, 491–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011). [Google Scholar]
- 40.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Team RStudio. RStudio: Integrated Development for R. (2016). [Google Scholar]
- 44.Pagès H, Aboyoun P, Gentleman R. & DebRoy S. Biostrings: Efficient manipulation of biological strings. R package version 20.50.2 2, (2017). [Google Scholar]
- 45.Phanstiel DH, Boyle AP, Araya CL & Snyder MP Sushi.R: flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics 30, 2808–2810 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagih O. ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics 33, 3645–3647 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Alcazar RM, Lin R. & Fire AZ Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics 180, 1275–1288 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schindelin J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin Y, Tam OH, Paniagua E. & Hammell M. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 31, 3593–3599 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cunningham F. et al. Ensembl 2019. Nucleic Acids Res. 47, D745–D751 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Claycomb JM et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139, 123–134 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu W-S et al. piRTarBase: a database of piRNA targeting sites and their roles in gene regulation. Nucleic Acids Res. 47, D181–D187 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wan G. et al. Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature vol. 557 679–683 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwartz ML & Jorgensen EM SapTrap, a Toolkit for High-Throughput CRISPR/Cas9 Gene Modification in Caenorhabditis elegans. Genetics vol. 202 1277–1288 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dickinson DJ, Slabodnick MM, Chen AH & Goldstein B. SapTrap assembly of repair templates for Cas9-triggered homologous recombination with a self-excising cassette. MicroPublication Biol. Dataset 10, W2KT0N (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dickinson DJ, Pani AM, Heppert JK, Higgins CD & Goldstein B. Streamlined Genome Engineering with a Self-Excising Drug Selection Cassette. Genetics 200, 1035–1049 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frøkjaer-Jensen C. et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet 40, 1375–1383 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haeussler M. et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dodson AE & Kennedy S. Germ Granules Coordinate RNA-Based Epigenetic Inheritance Pathways. Dev. Cell 50, 704–715.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andrews S. & Others. FastQC: a quality control tool for high throughput sequence data. (2010). [Google Scholar]
- 62.Trapnell C, Pachter L. & Salzberg SL TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramírez F, Dündar F, Diehl S, Grüning BA & Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liao Y, Smyth GK & Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Xiong Y. & Steitz TA Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature 430, 640–645 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Guang S. et al. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321, 537–541 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guang S. et al. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature vol. 465 1097–1101 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang C. et al. mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A 108, 1201–1208 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Remy J-J Stable inheritance of an acquired behavior in Caenorhabditis elegans. Curr. Biol 20, R877–8 (2010). [DOI] [PubMed] [Google Scholar]
- 70.Rechavi O. et al. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell 158, 277–287 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schott D, Yanai I. & Hunter CP Natural RNA interference directs a heritable response to the environment. Sci. Rep 4, 7387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jobson MA et al. Transgenerational Effects of Early Life Starvation on Growth, Reproduction, and Stress Resistance in Caenorhabditis elegans. Genetics 201, 201–212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moore RS, Kaletsky R. & Murphy CT Piwi/PRG-1 Argonaute and TGF-β Mediate Transgenerational Learned Pathogenic Avoidance. Cell 177, 1827–1841.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Posner R. et al. Neuronal Small RNAs Control Behavior Transgenerationally. Cell 177, 1814–1826.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edgar R, Domrachev M. & Lash AE Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.