Abstract
Addition of purified human topoisomerase I (topo I) to simian virus 40 T antigen-driven in vitro DNA replication reactions performed with topo I-deficient extracts results in a greater than 10-fold stimulation of completed molecules as well as a more than 3-fold enhancement of overall DNA replication. To further characterize this stimulation, we first demonstrate that bovine topo I but not Escherichia coli topo I can also enhance DNA replication. By using several human topo I mutants, we show that a catalytically active form of topo I is required. To delineate whether topo I influences the initiation or the elongation step of replication, we performed delayed pulse, pulse-chase, and delayed pulse-chase experiments. The results illustrate that topo I cannot promote the completion of partially replicated molecules but is needed from the beginning of the reaction to initiate replication. Competitive inhibition experiments with the topo I binding T antigen fragment 1-246T and a catalytically inactive topo I mutant suggest that part of topo I’s stimulation of replication is mediated through a direct interaction with T antigen. Collectively, our data indicate that topo I enhances the synthesis of fully replicated DNA molecules by forming essential interactions with T antigen and stimulating initiation.
A major area of interest under investigation is the molecular mechanism involved in the early stages of mammalian cell DNA replication. However, neither the mammalian initiator protein(s) nor the helicase(s) specific for DNA replication has been definitively identified (23, 25). Moreover, the exact origins of mammalian DNA replication are also not defined and appear to reside in initiation zones rather than consist of precise DNA consensus sequences (27, 29). To circumvent these obstacles and still obtain valuable information regarding the early stages of mammalian cell DNA replication, we are exploiting the simian virus 40 (SV40) model of DNA replication in vitro, which depends on a well-defined origin of replication and a single initiator protein and DNA helicase, the virally encoded T antigen (10, 35, 59). All essential replication machinery components except T antigen are provided by permissive cellular extracts (34, 35, 59, 68). Further studies have led to the establishment of reconstituted systems from purified proteins that support SV40 DNA replication (26, 64, 65, 69). Use of these model systems has provided a wealth of information regarding the functions of proteins involved in eukaryotic DNA replication (recently reviewed in references 5 and 20). However, there are still significant gaps in our understanding of the exact mechanisms involved in eukaryotic DNA replication, in particular with respect to (i) the composition and geometry of the initiation and elongation complexes, (ii) the interactions among replication factors, and (iii) how replication is regulated at various stages.
T antigen provides the helicase activity by forming a double hexamer over the SV40 origin of replication in the presence of ATP (3, 11, 38). In assays using either cell extracts, purified proteins, or partially purified multienzyme complexes (1, 33, 37), the cellular proteins found to participate with this T-antigen double hexamer include DNA polymerase α-primase (12, 33, 39, 42, 43, 64, 69), replication protein A (14, 43, 64, 69, 70), proliferating cell nuclear antigen (24, 37, 43, 45, 46, 47, 64), DNA polymerase δ (40, 43, 46, 47, 64), topoisomerase I (topo I) and/or topo II (28, 33, 37, 64, 69, 72), replication factor C (43, 62, 63, 64, 65), 5′-3′ exonuclease (MF-1) (26, 33, 64), DNA ligase (26, 33, 37, 64), and RNase H (26, 33, 37, 64). Details of the temporal role of each of these proteins as well as the mechanism by which each functions in vivo remain to be determined.
Models like the SV40 system described above have been devised to isolate and study specific events in the complex mechanism of eukaryotic DNA replication. Of particular interest, the requirements and specific roles of type I and type II topoisomerases have yet to be clearly determined. Type II topoisomerases function enzymatically by producing double-strand breaks to relieve torsionally strained replicated molecules (7). Yang et al. (72) have demonstrated that topo II is required to separate catenated SV40 daughter molecules (60, 61). Eukaryotic type I topoisomerases generate single-stranded nicks in double-stranded DNA by forming a reversible 3′-phosphate bond and introducing or relieving either positive or negative supercoils (7, 55, 57). Either topo I or topo II can stimulate the early stages of DNA replication (72). Evidence for the presence of topo I in isolated multiprotein replication complexes has been obtained by several labs (1, 33, 37). Topo I has also been found associated with SV40 DNA replicative intermediates (RIs) (6, 7). Several groups have also shown that topo I can stimulate in vitro SV40 T-antigen-mediated DNA replication (69, 71).
T antigen and human or bovine topo I can associate in vitro (51). There is evidence that this interaction is functionally significant (44, 48, 48a, 52, 53). First, topo I interferes with T antigen’s ability to unwind DNA from nonorigin sites, implying that the specificity of origin unwinding is increased (53). Second, T antigen can reverse the formation of a covalent 3′-phosphate bond between topo I and the DNA in the presence of camptothecin, indicating that the activity of topo I can be modulated by T antigen (44). Third, T antigen activates topo I nicking activity and directs the enzyme to cleave specific sites near the origin of replication (52). We have postulated (52) that a complex of a double hexamer of T antigen and a single molecule of topo I unwinds and relaxes origin-containing DNA. Fourth, a complex of T antigen and topo I appears to be required to partially unwind circular origin-containing DNA (48, 48a). Fifth, topo I significantly increases T-antigen mediated DNA replication and generates much larger amounts of completed molecules (53).
Evidence from several labs demonstrates that the T-antigen double hexamer unwinds origin-containing closed circular DNA in the presence of at least RPA and DNA polymerase α-primase (8, 13, 39, 41, 43) and possibly a topoisomerase (10, 11, 48). As the origin opens up, torsional strain would significantly increase ahead of the replication forks unless a topoisomerase is recruited to relieve this strain. To explain the effects of topo I on SV40 DNA replication in vitro, it might be argued that the enzyme functions at replication forks to promote DNA synthesis and permit partially replicated molecules to go to completion. To gain a better understanding of topo I’s involvement in eukaryotic DNA replication, we established a modified in vitro system which dissects replication into two stages, initiation and elongation. The results of this study demonstrate that, contrary to expectations, topo I is involved in the initiation reaction and is unable to convert RIs to completed molecules.
MATERIALS AND METHODS
Plasmids.
The SV40 origin-containing plasmids pSKori (53) and pSV011+ (46, 58) have been described elsewhere.
Cells.
Sf9 insect cells were routinely maintained in spinner flasks, transferred to T150 flasks, and infected by standard protocols (PharMingen). 293 cells (16) were grown in Dulbecco modified Eagle medium containing 10% fetal bovine serum in spinner flasks in a 5% CO2 humidified incubator. Cytosolic 293 cell extracts were prepared as described previously (58, 59) and screened for topo I by Western blotting (ECL kit; Amersham), using the topo I monoclonal antibody 8G6 (53a).
Recombinant baculoviruses.
The construction of SV40 wild-type (WT) T antigen (32) as well as C-terminal (51) T-antigen deletion mutant-expressing baculoviruses has been described elsewhere. Stewart et al. (57) generated the baculovirus expressing human topo I used in this study.
Protein purification.
Human topo I was purified by column chromatography as described by Stewart et al. (57) and estimated to be about 90% pure (53). Purified Escherichia coli topo I was a generous gift from Ken Marians (Sloan-Kettering Institute), and purified bovine topo I was a generous gift from Tom Melendy (University of Buffalo). Purified Y723F, topo58, and topo12 were generous gifts from Jim Champoux and Lance Stewart (University of Washington).
WT and most mutant T antigens were immunoaffinity purified from Sf9 cells with monoclonal antibody PAb101 (17) as described by Simanis and Lane (50). 1-246T was purified with monoclonal antibody PAb419 (19) as described by Mastrangelo et al. (38). The T antigens prepared for use in in vitro DNA replication assays were dialyzed against buffer F [10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 7), 5 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride, 10% (vol/vol) glycerol (50)] and stored in aliquots at −80°C. The WT T antigen used in the enzyme-linked immunosorbent assay (ELISA) in Fig. 1B was purified in the same manner (50) with monoclonal antibody PAb416 (19), dialyzed against storage buffer (10 mM Tris [pH 8], 1 mM EDTA, 100 mM NaCl, 1 mM DTT, 50% [vol/vol] glycerol [38]), and stored at −20°C. Silver staining of 10% Laemmli gels (30) allowed for the estimation of the concentration of the purified proteins relative to a phosphorylase b standard.
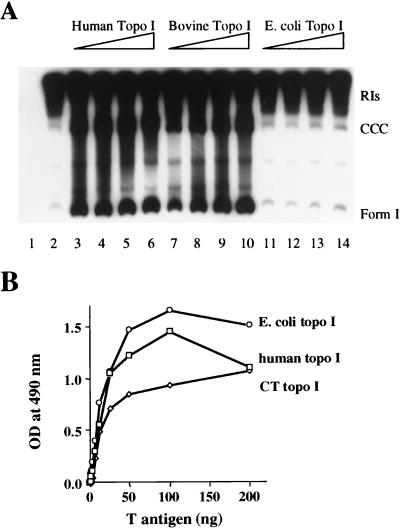
FIG. 1.
Comparison of the activities of topo I isolated from different species. (A) In vitro DNA replication assays were performed with topo I-deficient 293 cell extracts supplemented with 200 ng of pSKori, 1 μCi of [α-32P]dATP, and 2 μg of T antigen in the absence (lane 2) or presence of increasing amounts of human, bovine, or E. coli topo I (50 ng [lane 3, 7, or 11], 100 ng [lane 4, 8, or 12], 200 ng [lane 5, 9, or 13], and 300 ng [lane 6, 10, or 14]. As a control, neither T antigen nor topo I was added to lane 1. After purification, the labeled replicated DNA was analyzed by gel electrophoresis on a 1.5% agarose gel. The positions of RIs, CCC relaxed DNA, and form I DNAs are shown. (B) ELISAs were performed by adsorbing 100 ng of human, bovine (CT), or E. coli topo I to the wells of a microtiter plate and reacting them with increasing quantities (0, 0.75, 1.5, 3, 6.25, 12.5, 25, 50, 100, and 200 ng) of T antigen. The specific complexes formed were detected by using biotinylated PAb101, streptavidin-conjugated HRP, and o-phenylenediamine substrate solution. Absorbance at (optical density [OD]) 490 nm was measured with a Dynex MRX plate reader.
ELISAs.
ELISAs were performed as detailed by Simmons et al. (51), with 100 ng of either human, bovine, or E. coli topo I as the first protein. Increasing quantities of immunoaffinity-purified WT T antigen were allowed to react with the bound topo I. After washing, the bound protein complexes were incubated with 300 ng of biotin-conjugated PAb101. The wells were washed, streptavidin-conjugated horseradish peroxidase (HRP; Sigma) was added, and substrate was introduced after another wash. The reactions were stopped with H2SO4, and absorbance at 490 nm was read on a Dynex MRX plate reader. A parallel series of reactions lacking topo I was performed as a background control, and their values were subtracted from those of the corresponding topo I-containing samples.
In vitro DNA replication assays with a 60-min label.
Standard DNA synthesis reaction mixes were prepared similarly to those described by Stillman and Gluzman (59) (see Fig. 3A), using replication buffer A (30 mM HEPES-KOH [pH 7.5], 7 mM MgCl2, 0.5 mM DTT, 4 mM ATP, 200 μM CTP, GTP, and UTP, 100 μM dTTP, dGTP, and dCTP, 25 μM dATP, 40 mM creatine phosphate, 20 ng of creatine phosphokinase per μl [final concentrations]). Unless otherwise indicated, 2 μg of T antigen, 200 to 300 μg of topo I-deficient 293 cell extract, 236 ng of pSV011+, and 1 μCi [α-32P]dATP were used in each 50-μl reaction. Purified topo I, topo I mutant, or 1-246T was added when indicated. After the DNA was synthesized for 1 h at 37°C, the reactions were terminated by the addition of stop buffer (21 mM EDTA, 0.42% sodium dodecyl sulfate, 0.21 μg of proteinase K per μl [final concentrations]). After digestion for 30 min at 37°C, the labeled DNA was sequentially extracted with phenol, chloroform-isoamyl alcohol, and ether and precipitated with ethanol before being analyzed on a 1.5% agarose gel. After the gel was dried, the labeled replicated DNA products were quantitated on a Molecular Dynamics PhosphorImager unless otherwise indicated and exposed to film.
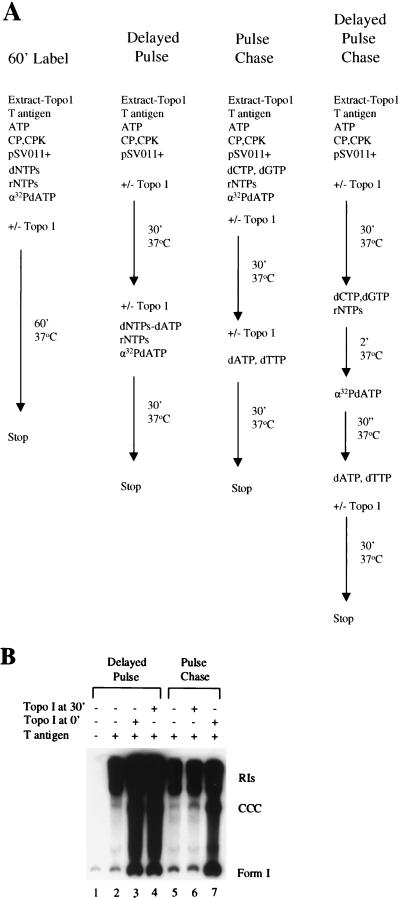
FIG. 3.
Delayed-pulse and pulse-chase in vitro DNA replication assays. (A) Schematic diagram of the order of addition of the components in the procedures used throughout this study. CP, creatine phosphate; CPK, creatine phosphokinase. (B) In the delayed pulse experiment, replication reactions were begun with ATP but not the other nucleotides. At 30 min, 1 μCi of [α-32]dATP, along with cold dTTP, dCTP, dGTP, CTP, GTP, and UTP, was added, and DNA synthesis proceeded for 30 min at 37°C. In the pulse-chase assays, all ribonucleotides and deoxyribonucleotides including 1 μCi of [α-32]dATP were added from the beginning. After 30 min, the label was chased with an excess of cold dATP for 30 min. Human topo I (100 ng) was added either from the beginning (topo I at 0′) or at 30 min (topo I at 30′). After the reactions were stopped, the labeled replicative products were purified and analyzed as described for Fig. 1A.
Delayed pulse replication assays.
Delayed pulse experiments were carried out as described previously (15, 59) (see Fig. 3A). Topo I-deficient 293 cell extracts were incubated with T antigen and pSV011+ in replication buffer B (40 mM HEPES-KOH [pH 7.5], 8 mM MgCl2, 0.5 mM DTT, 3 mM ATP, 40 mM creatine phosphate, 20 to 40 ng of creatine phosphokinase per μl [final concentrations]) at 37°C for 30 min. To initiate DNA synthesis, GTP, CTP, and UTP (0.2 mM each), dGTP, dCTP, and dTTP (0.1 mM each), and 1 μCi of [α-32P]dATP (3,000 Ci/mmol) were added with or without topo I, and DNA synthesis continued at 37°C for 30 min. The reaction products were purified and analyzed as described above. In some cases, after separation of the replication products on a 1.5% agarose gel, the completed molecules were visualized by ethidium bromide, excised from the gel, Cerenkov counted, purified by GeneClean (Bio 101), digested with BstUI and SspI (New England Biolabs), and applied to a 2% agarose gel.
Pulse-chase replication assays.
Pulse-chase assays were performed by a modification of the procedures described by Fotedar et al. (15) (see Fig. 3A). Specifically, T antigen, topo I-deficient 293 cell extracts, pSV011+, 0.2 mM each CTP, GTP, and UTP, 0.1 mM dCTP, 0.1 mM dGTP, and 1 μCi of [α-32P]dATP (3,000 Ci/mmol) were incubated in replication buffer B for 30 min at 37°C to allow for the formation of primed initiation complexes. At 30 min, the label was chased with 0.1 mM cold (unlabeled) dATP (15,000-fold excess) and 0.1 mM dTTP was added. After 30 min at 37°C, the reactions were stopped and the newly synthesized DNA was extracted and analyzed as before.
Delayed pulse-chase replication assays.
Topo I-deficient extracts were incubated under the initial conditions described above for the delayed pulse replication assays (see Fig. 3A). At 30 min, the complexes were primed at 37°C by the addition of 0.2 mM each CTP, GTP, and UTP, 0.1 mM dCTP, and 0.1 mM dGTP; 1 μCi of [α-32P] dATP (3,000 Ci/mmol) was then added to allow for a limited elongation reaction in the absence of dTTP for 30 s at 37°C. A 15,000-fold excess of cold dATP (0.1 mM dATP) was added along with 0.1 mM dTTP, and the reaction was incubated at 37°C for 30 min before the addition of stop buffer. Topo I was added either at the beginning of the reaction or with the chase.
RESULTS
Human and bovine topo I stimulate SV40 DNA replication.
We have previously demonstrated that human topo I promotes the synthesis of fully replicated SV40 DNA in vitro (53). The mechanism by which DNA replication is stimulated is not known. In an effort to characterize this reaction further, we first asked whether topo I isolated from other species could substitute for the human enzyme. Figure 1A demonstrates that human and bovine but not E. coli topo I stimulates the production of completed circular plasmid DNA containing the entire SV40 replication origin. Replication reactions were carried out with 293 cell extracts deficient in topo I as determined by Western blotting (data not shown). In the absence of added topo I, T-antigen-mediated replication resulted in the formation of unfinished RIs (Fig. 1A, lane 2) as previously shown (53). The addition of even the lowest quantity (50 ng) (Fig. 1A, lane 3) of human topo I increased the formation of completed molecules (covalently closed circular [CCC] DNA, form I DNA, and all topoisomers migrating in between) approximately 9.3-fold while increasing total DNA synthesis approximately 2.3-fold. Maximal stimulation was observed with 100 ng of human topo I (11.6-fold for completed molecules and 2.5-fold for overall), and higher concentrations reduced the stimulation (Fig. 1A, lanes 4 to 6). Bovine topo I also significantly increased the formation of completed molecules (Fig. 1A, lanes 7 to 10) (11-fold with 300 ng), although a 3-fold-higher concentration was needed to obtain maximal stimulation. In contrast, E. coli topo I had no effect on T-antigen-mediated DNA replication (Fig. 1A, lanes 11 to 14). The E. coli topo I used in these experiments was shown to be catalytically active in a DNA nicking reaction (data not shown).
We previously showed that human and bovine topo I can bind T antigen in vitro (51). To determine if the failure of E. coli topo I to stimulate DNA replication was due to an inability to interact with T antigen, we measured T antigen-topo I binding with an ELISA (Fig. 1B). Purified topo I (100 ng) from each species was adsorbed to the surface of the wells of a microtiter plate, and then increasing quantities (0 to 200 ng) of WT T antigen were allowed to bind to the attached topo I. Protein-protein complexes were reacted with biotinylated PAb101, an anti-T monoclonal antibody, and detected with streptavidin-conjugated HRP and its substrate. E. coli topo I bound T antigen at least as well as human or bovine topo I (Fig. 1B). To ensure that binding was due to direct protein-protein interactions and was not mediated by contaminating DNA, the reactions were repeated in the presence of ethidium bromide (100 μg/ml) (18, 31); the same results were obtained (data not shown). Therefore, E. coli topo I’s inability to stimulate DNA replication cannot be explained by a simple lack of binding.
Topo I catalysis is required for stimulation of DNA replication.
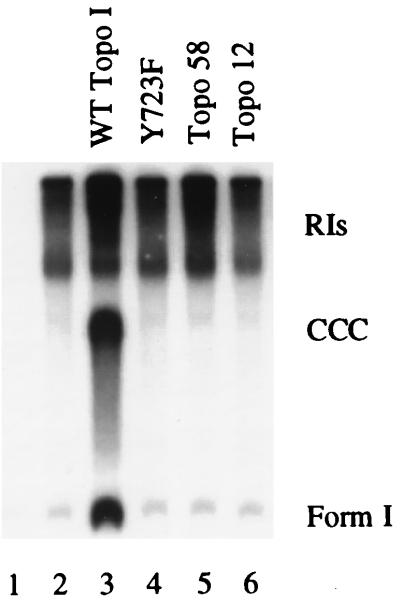
It is well established that topoisomerases (I or II) are required for SV40 DNA replication (69, 71, 72) and presumably must also be necessary for cellular DNA replication. The dependence for topo I in the SV40 system has also been demonstrated by the inhibition of DNA replication with the topo I poison camptothecin (49, 54, 68). Camptothecin interferes with the second step in catalysis, the religation of nicked strands (2, 9, 21). To determine if nicking activity is also required to promote T-antigen-mediated DNA replication, we tested the ability of various human topo I mutants to stimulate replication (Fig. 2). These include Y723F (a catalytic site point mutant [57]) and deletion mutants topo58 (amino acids 175 to 659 [55, 57]) and topo12 (amino acids 658 to 765 [56]), which are also catalytically inactive (56). Y723F binds T antigen normally in an ELISA, whereas topo58 binds less well and topo12 is inactive (data not shown). Replication reactions were carried out in the absence or presence of 100 ng of full-length topo I or the molar equivalent of the deletion mutants (Fig. 2). In this assay, none of the mutants stimulated DNA replication (Fig. 2; compare lanes 4 through 6 with lane 3). These results provide evidence that topo I catalytic activity is required for stimulation of DNA replication.
FIG. 2.
Effect of catalytically inactive topo I on in vitro DNA replication. Replication reactions were carried out in the absence (lanes 1 and 2) or in the presence of various forms of topo I as indicated above the lanes. Lane 1 contained no T antigen as a negative control. The labeled replicative products were analyzed as described for Fig. 1A.
Topo I stimulates DNA replication from the point of initiation.
It has long been assumed that a topoisomerase is required for DNA replication for the purpose of relieving the torsional strain imposed on the unreplicated portions of the molecule. Our observation (reference 53 and this report) that topo I primarily stimulates the production of completed molecules was in agreement with this hypothesis and suggested that the enzyme acts during elongation to relax DNA ahead of replication forks. To investigate this further, we first asked whether topo I had to be added at the same time as the T-antigen initiator to stimulate DNA replication. A delayed pulse reaction was carried out to answer this question (Fig. 3). Topo I-deficient 293 cell extracts were first incubated with T antigen and SV40 origin-containing DNA under replication conditions in the presence of ATP but not the other nucleoside triphosphates (NTPs) to form a preinitiation complex. After 30 min, the remaining rNTPs and all dNTPs (including 32P-labeled dATP) were added to permit initiation and subsequent elongation. In the absence of added topo I, the primary labeled products were RIs, just as in the 60-min label reaction (Fig. 3B, lane 2). Whether topo I was added with the T antigen from the beginning of the reaction (Fig. 3B, lane 3) or 30 min later with the label (Fig. 3B, lane 4), there was an 11-fold stimulation in the formation of completed molecules. Moreover, there was no decrease in the amounts of labeled RIs (1.6-fold stimulation) with the addition of topo I, implying that topo I was not functioning solely in elongation. Therefore, topo I can stimulate the synthesis of completed molecules even when added after the binding of T antigen to the DNA.
In an effort to distinguish between an effect of topo I on initiation and elongation, we performed a pulse-chase experiment (Fig. 3B, lanes 5 to 7). Here, all nucleotides and the labeled dATP were present from the beginning of the reaction and a 15,000-fold excess of cold dATP (0.1 mM) was added at 30 min (Fig. 3A). Once again, topo I was added either from the beginning or at 30 min with the chase. If topo I functions during elongation, it would promote the formation of completed molecules from RIs, and the labeled reaction products generated when topo I is present during the chase would be mostly completed molecules and very few RIs. If topo I is needed for the initiation reaction alone or during initiation and elongation, one would expect to observe a significant stimulation of completed molecules when topo I is added from the beginning. Figure 3B (lanes 5 and 6) shows that there is no effect on replication when topo I is added with the chase, demonstrating that topo I is unable to promote the completion of partially replicated DNAs. There was a 2.9-fold stimulation of overall replication and a 7.2-fold stimulation of completed molecules when topo I was present from the beginning of the reaction (Fig. 3B, lane 7), indicating that it influences initiation.
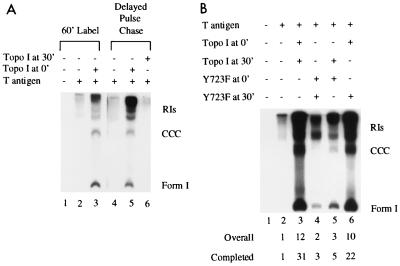
To obtain additional support for this interpretation, we did a delayed pulse-chase experiment (Fig. 4A). In this reaction, topo I-deficient extracts were incubated with T antigen, origin-containing DNA, and ATP for 30 min to permit T antigen to bind the origin (Fig. 3A). At 30 min, the DNA was pulse-labeled for 30 seconds with [α-32P]dATP in the presence of all four rNTPs plus dCTP and dGTP. The label was then chased for 30 min with excess cold dATP (15,000-fold) in the presence of the fourth dNTP (dTTP). The elimination of dTTP from the initiation reaction allowed us to clearly discriminate between effects on the two phases of DNA replication (15), and the brief pulse permitted us to observe the effects on only the molecules that are primed to initiate replication. Topo I was added from the beginning of the reaction (Fig. 4A, lane 5) or with the chase (Fig. 4A, lane 6). Results were compared to those obtained with a standard 60-min label DNA synthesis reaction in the absence (Fig. 4A, lane 2) or presence (Fig. 4A, lane 3) of topo I. As long as topo I was present when the initiation complex formed, it was able to significantly stimulate the reaction, but it was unable to convert preformed RIs to completed molecules. Therefore, topo I does not function solely during elongation.
FIG. 4.
Delayed pulse-chase assays. (A) For lanes 1 to 3, reaction mixtures contained 1 μCi of [α-32P]dATP for 1 h in the absence (lane 2) or presence (lane 3) of 100 ng of human topo I; lane 1 was a control without T antigen or topo I. The delayed pulse-chase reactions were set up like the delayed pulse experiments for the first 30 min, at which time the rest of the rNTPs, dGTP, and dCTP were added. Then 1 μCi [α-32P]dATP was added to each sample for 30 s at 37°C, after which time dTTP and excess cold dATP were added for an additional 30 min. The labeled products were analyzed as described for Fig. 1A; however, the gel was exposed only one-fifth as long. (B) Delayed pulse-chase reactions were set up as for panel A except that 50 ng of either WT topo I or Y723F was added at the beginning (0′). Another 50 ng of either WT topo I or Y723F was added with the chase (30′) as indicated above the lanes. The amount of replication relative to that with T antigen alone is indicated below the lanes.
The above experiments could be explained by the requirement for topo I in the formation of an initiation complex at the origin of replication. The existence of such a complex is substantiated by the ability of T antigen to activate topo I to nick origin DNA at sites different from the ones the enzyme selects on its own (52). We reasoned that if topo I associates early in the formation of the replication complex, insertion of the catalytically inactive form of the enzyme at that point would generate a defective complex that could not be rescued with WT topo I. To evaluate this possibility, we performed another delayed pulse-chase experiment (Fig. 4B) in which 50 ng of either WT topo I or mutant Y723F was added from the beginning of the reaction. After the brief pulse, 50 ng of either WT topo I or Y723F was added at the beginning of the chase period. The amount of replication relative to that with T antigen alone is indicated below the lanes. WT topo I was unable to stimulate elongation when Y723F was present during initiation (Fig. 4B, lane 5). However, if WT topo I was present from the start of the reaction, it was capable of stimulating the formation of completed molecules even when Y723F was added during elongation (Fig. 4B, lane 6). These results suggest that once formed, an initiation complex consisting of WT or mutant topo I becomes irreversibly programmed in the replication pathway. This experiment also substantiates our conclusion that a catalytically active topo I is essential during the initiation reaction to stimulate the formation of completed molecules.
Competition by mutant T antigen or mutant topo I.
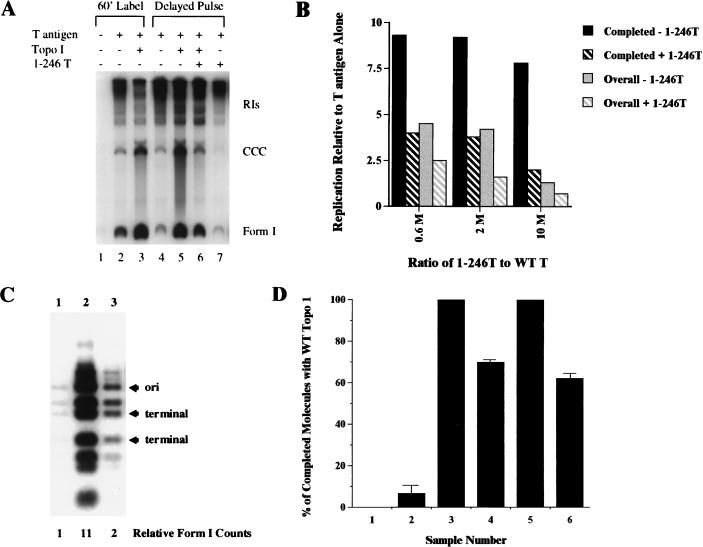
Previous work from our lab has shown that an interaction between T antigen and topo I is likely involved in the ability of topo I to inhibit unwinding of nonorigin DNA (53) and to partially unwind circular origin-containing DNA (48a). These conclusions were based on the inhibitory effects of T-antigen fragments that bind to topo I. We used a similar approach here and determined whether the T-antigen deletion mutant 1-246T interferes with topo I’s stimulation of replication. This fragment was chosen because it binds topo I as well as WT T antigen (51), does not bind DNA (unpublished data), and does not interfere with T antigen’s unwinding activity (53). A delayed pulse experiment was carried out as illustrated in Fig. 3A except that topo I was added only at 30 min in the presence or absence of a 10-fold molar excess of 1-246T relative to the amount of full-length T antigen (Fig. 5A). Due to the very low levels of incorporation into completed molecules in the absence of topo I, addition of 1-246T had a slight stimulatory or slight inhibitory (Fig. 5A, lane 7) effect depending on the experiment; however, this was not significant. In the presence of topo I, 1-246T always partially inhibited the stimulation of replication by topo I alone (Fig. 5A; compare lanes 5 and 6). In this experiment, 1-246T reduced topo I’s stimulation of completed molecules to only 2-fold, compared to 7.8-fold with topo I alone (Fig. 5A; Fig. 5B, 10 M). To verify this partial inhibition, the experiment was repeated with different concentrations of 1-246T under delayed pulse and delayed pulse-chase conditions. Figure 5B quantifies the stimulation of DNA replication by topo I alone and by a combination of topo I and 1-246T. Even with 0.6 M equivalents of 1-246T relative to full-length T antigen, the stimulation by topo I was reduced from 9.3- to 4-fold for completed molecules and from 4.5- to 2.5-fold for overall replication. A 2 M excess of 1-246T resulted in an intermediate effect (Fig. 5B).
FIG. 5.
Competitive in vitro DNA replication reactions. (A) Competition with 1-246T. For lanes 1 to 3, 1 μg of immunoaffinity-purified T antigen was used to program an in vitro DNA replication reaction of 236 ng of pSV0II+ and 1 μCi of [α-32P]dATP in the absence (lanes 1 and 2) or presence (lane 3) of 100 ng of human topo I; lane 1 is a negative control without T antigen. For lanes 4 to 7, T-antigen-mediated in vitro DNA replication was initiated without label for 30 min. Then 1 μCi of [α-32P]dATP, 100 ng of human topo I (lanes 5 and 6), and a 10 M excess (over WT T antigen) of 1-246T (lanes 6 and 7) were added, and the reaction continued for an additional 30 min. The labeled DNA was purified, analyzed on a 1.5% agarose gel, dried, and exposed to film. The radioactive products were excised and subjected to Cerenkov counting. (B) Stimulation of completed molecules and overall DNA replication in the presence of topo I (−1-246T) or in the presence of topo I and 1-246T (+1-246T) DNA replication in the presence of T antigen alone is given a value of 1, and fold stimulation is calculated for each molar excess of 1-246T over WT T antigen and depicted graphically. The experiment containing 0.6 M equivalents of 1-246T (relative to WT T antigen) was done with a delayed pulse-chase assay. Experiments containing 2 and 10 M excesses of 1-246T were performed with delayed pulse assays; 10 M excess represents the experiment illustrated in panel A. (C) Restriction enzyme digestion of the form I molecules obtained during a delayed pulse competition reaction. A competition reaction with a 10 M excess of 1-246T was carried out as for panel A. After separation of the replication products on a 1.5% agarose gel, the completed molecules were visualized by ethidium bromide, excised from the gel, Cerenkov counted, purified by GeneClean (Bio 101), digested with BstUI and SspI (New England Biolabs), and applied to a 2% agarose gel. The 739-bp fragment contains the origin of replication (ori), and the 493- and 330-bp fragments are on the opposite side close to the site of termination. The numbers below the lanes represent relative incorporation into form I DNA. (D) Competition with the catalytic site topo I mutant Y723F. An in vitro DNA replication assay was performed in the presence or absence of various quantities of WT human topo I and mutant topo I Y723F. The DNA was synthesized in the presence of label for 1 h and analyzed on a 1.5% agarose gel. Replicated completed molecules (from CCC to form I on the gel) were quantitated, and the results (from two independent experiments) are expressed as a percentage of the values obtained with WT topo I alone. Samples: 1, no T antigen (negative control); 2, T antigen alone; 3, 100 ng of human topo I; 4, 50 ng of topo I and 50 ng of Y723F; 5, 150 ng of topo I; 6, 50 ng of topo I and 100 ng of Y723F. Samples 1, 2, and 4 were normalized to sample 3, and sample 6 was normalized to sample 5.
If topo I stimulates DNA replication by participating in the formation of an initiation complex, the addition of 1-246T should interfere with the initiation of replication. We confirmed this prediction by performing another delayed pulse experiment in the presence or absence of topo I and 1-246T. After DNA synthesis, the products were separated by gel electrophoresis and the form I bands were excised and subjected to restriction enzyme digestion (Fig. 5C). The two enzymes used (BstUI and SspI) cut the plasmid DNA into nine fragments and permitted us to analyze the incorporation of label into different portions of the replicating plasmid. 1-246T decreased the stimulation of labeled form I DNA by topo I from 11- to 2-fold (Fig. 5C). This significant reduction in incorporation of label was equal thoughout the plasmid DNA; all of the restriction fragments showed proportional decreases in labeling (Fig. 5C, lanes 2 and 3). The reduction in labeling of the fragments derived from the replication termination region was equivalent to that of the origin containing fragment. If 1-246T were interfering with topo I’s activity in elongation, one would predict it to preferentially diminish the incorporation of label into the fragments farthest from the origin. These results provide additional evidence for our interpretation that topo I participates with T antigen in the initiation reaction.
If this partial inhibition of topo I activity by 1-246T is due to a block in the formation of WT T antigen-topo I complexes, it should also be possible to partially abrogate topo I’s stimulation of replication with a functionally inactive mutant form of topo I. We therefore investigated the ability of Y723F to compete with WT topo I and reduce its effect on DNA replication. Since high concentrations of WT topo I interfere with its stimulation (Fig. 1A), we determined the effect of Y723F by comparing reactions containing equal amounts of total (WT and mutant) topo I. In the presence of 50 ng each of topo I and Y723F (1:1), the stimulation of completed molecules was 70% ± 1% of that with 100 ng of WT topo I (Fig. 5D, samples 3 and 4). When 100 ng of Y723F was included with 50 ng of topo I (2:1), the stimulation was 62% ± 3% of that with 150 ng of WT topo I (Fig. 5D, samples 5 and 6). One would predict that, at maximum, the 1:1 and 2:1 combinations would reduce the completed molecules to 50 and 33%, respectively. This partial inhibition is consistent with the interpretation that a portion of topo I’s stimulation of DNA replication is due to a direct interaction with T antigen.
DISCUSSION
The primary effect of adding purified human topo I to a DNA replication reaction utilizing topo I-deficient extracts is to stimulate the production of completed DNA molecules (reference 53 and this report). Specifically, the average levels of stimulation of T-antigen-mediated DNA replication by human topo I from 27 experiments were 11.5 ± 4.0-fold for completed molecules and 3.6 ± 1.0 fold for overall replication. This result suggested that topo I acts mostly at the elongation stage of DNA synthesis to relieve torsional strain ahead of the replication forks. However, we demonstrated here that under a number of different experimental conditions, topo I is required during or prior to initiation of DNA replication. This finding suggests that topo I is a component of the initiation complex.
The role of topo I in initiating replication was elucidated by varying the time of its addition relative to the other components of the replication reaction. Delayed pulse assays showed that stimulation took place even when it was introduced 30 min after the addition of T antigen but before the onset of replication (Fig. 3B). In these reactions, topo I was able to promote the synthesis of completed molecules without diminishing the amounts of RIs. Labeled completed molecules generated under these delayed pulse conditions showed both uniform stimulation of incorporation in all fragments in the presence of topo I as well as a uniform reduction upon the addition of the competitor, 1-246T (Fig. 5C). Further evidence was obtained from the pulse-chase experiments (Fig. 3B), which illustrated that topo I must be present from the beginning of the reaction and cannot stimulate the completion of preformed RIs. The delayed pulse-chase experiments (Fig. 4A) clearly distinguished between the two phases since preinitiation complexes were stalled by a lack of nucleotides, and after priming of these complexes, the brief label without the fourth deoxynucleotide limited elongation. Once again, topo I was able to stimulate replication when added before the reaction but not after the labeling period. Furthermore, a catalytically active topo I is required during initiation to promote the formation of completed molecules (Fig. 4B). Collectively, these results lead to the conclusion that topo I must act before or during initiation, but they do not rule out the likely possibility that it also functions during elongation.
Camptothecin, an inhibitor of the religation activity of topo I, is capable of inhibiting overall SV40 DNA replication in vitro and in vivo (49, 54, 68). Replication would cease presumably because the replication complex at the fork would be stalled by an unsealed nick. One might anticipate that topo I nicking is also required since the torsional strain created ahead of the forks would prevent further elongation. The demonstration that catalytically inactive topo I mutants (Y723F, topo58, and topo12 [56, 57]) are unable to stimulate DNA replication (Fig. 2) is consistent with this hypothesis. Therefore, a catalytically active topo I is required to promote the formation of completed replicated DNA molecules.
Evidence of a direct T antigen-topo I functional complex comes from our competition experiments with 1-246T and Y723F. These mutants should theoretically compete with their WT counterparts to limit the formation of active T antigen-topo I complexes. There was substantial inhibition of topo I’s ability to stimulate overall DNA replication and to form completed molecules by a small amount of 1-246T, and this inhibition was more pronounced, although not complete, in the presence of a large excess of 1-246T (Fig. 5A to C). A similar effect was observed with Y723F, although the reduction in stimulation was about half of that anticipated (Fig. 5D). Since both types of competition experiments resulted in only partial inhibition, it is conceivable that topo I stimulates DNA replication via both T-antigen-dependent and T-antigen-independent mechanisms.
In contrast to human and bovine topo I, E. coli topo I has no effect on T-antigen-mediated circular unwinding (48, 48a) or in vitro DNA replication (this report). However, it has been established that T antigen binds human, bovine (51), and now E. coli topo I, so binding per se was not sufficient to allow for biological activity. Others have demonstrated that E. coli topo I binds to the 5′-phosphate at the site of the nick whereas the eukaryotic topo I binds through the 3′-phosphate (reference 36 and references therein). E. coli topo I also differs from the eukaryotic topo I in its inability to relax positively supercoiled DNA (66, 67). It is conceivable that one or more of these mechanistic differences prohibits a functional interaction between E. coli topo I and the helicase in our viral model of eukaryotic DNA replication.
It is interesting to speculate on the nature of the interaction between topo I and the T-antigen helicase. We (51) have shown that there are at least two topo I binding sites on T antigen, one between amino acids 82 and 246 and the other between amino acids 246 and 708. Furthermore, it has recently been demonstrated that topo I possesses two T antigen binding sites, one between amino acids 1 and 139 and one between amino acids 383 and 765 (18). It has been proposed that the nonfunctional N-terminal end of topo I may contain cellular protein binding domains (56). One could hypothesize that T antigen or a cellular helicase and topo I must bind through both sites to produce a functional complex during DNA replication. We have previously proposed (52) that this complex consists of a single molecule of topo I bound to a double hexamer of T antigen. If true, this stoichiometry would imply that the topo I contacts at most two T-antigen monomers and that it is associated with DNA on only one side of the origin. Our observation (Fig. 3B) that topo I can stimulate replication after T antigen has bound to the origin in the presence of ATP implies that topo I can functionally interact with a preformed double hexamer. Furthermore, the inability of WT topo I to displace a catalytically inactive topo I on the initiation complex indicates that once formed, the complex is stable. It will be interesting to determine at what point topo I becomes associated with the initiation complex relative to other replication factors.
DNA replication requires both an origin of replication as well as an initiator protein that recognizes and acts on this DNA sequence. During SV40 DNA replication, T antigen serves the dual role of an initiator protein and a helicase. In yeast, these two functions appear to be provided by separate proteins. Autonomously replicating sequences have been defined as origins of replication in yeast and an initiator protein complex, the origin recognition complex has been shown to bind to the autonomously replicating sequences in an ATP-dependent manner (reference 73 and references therein). However, this binding is not sufficient to initiate DNA replication. Initiation also requires the recruitment of several additional proteins including Cdc6p, Cdc45p, and the Mcm (minichromosome maintenance) proteins, after which the origin recognition complex can be released without affecting initiation (references 22 and 73 and references therein). The helicase specifically used during DNA replication in yeast has yet to be definitively identified, although Dna2p is a likely candidate (4). Homologs for the yeast proteins are being found in mammalian cells (23, 25). One study in search of the cellular helicase counterpart to SV40 T antigen has identified fractions from FM3A mouse cells which exhibit helicase activity, although it is not yet known if either of the two proteins found actually plays a role in DNA replication, repair, or recombination (23). A second report provides evidence that a hexamer comprised of two MCM4-MCM6-MCM7 complexes from HeLa cells exhibits helicase and ATPase activities (25). Further study of the function of these helicases will have to await the establishment of an in vitro replication assay. Based on the findings described in this paper, we speculate that the cellular helicase would also require an interaction with topo I during the initiation and elongation phases of DNA replication.
We have previously determined that a complex of T antigen and topo I is likely involved in DNA replication by simultaneously melting, unwinding, and relaxing superhelical DNA ahead of replication forks (52, 53). In this study, we provide additional evidence suggesting that a catalytically active human or bovine, but not E. coli, topo I is required early in replication. The results presented here are consistent with the interpretation that a T antigen-topo I complex is an integral part of the initiation machinery and that this complex moves with replication forks during DNA replication.
ACKNOWLEDGMENTS
This work was supported by PHS grant CA36118 to D.T.S.
We thank Pat Hearing for the 293 cells, Lance Stewart and Jim Champoux for the baculovirus expressing full-length human topo I and for purified Y723F, topo58, and topo12, Ken Marians for purified E. coli topo I, and Tom Melendy for purified bovine topo I.
REFERENCES
- 1.Applegren N, Hickey R J, Kleinschmidt A M, Zhou Q, Coll J, Wills P, Swaby R, Wei Y, Quan J Y, Lee M Y W T, Malkas L H. Further characterization of the human cell multiprotein DNA replication complex. J Cell Biochem. 1995;59:91–107. doi: 10.1002/jcb.240590111. [DOI] [PubMed] [Google Scholar]
- 2.Been M D, Burgess R R, Champoux J J. DNA strand breakage by wheat germ type 1 topoisomerase. Biochim Biophys Acta. 1984;782:304–312. doi: 10.1016/0167-4781(84)90066-6. [DOI] [PubMed] [Google Scholar]
- 3.Borowiec J, Hurwitz J. ATP stimulates the binding of simian virus 40 (SV40) large tumor antigen to the SV40 origin of replication. Proc Natl Acad Sci USA. 1988;85:64–68. doi: 10.1073/pnas.85.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Budd M E, Choe W C, Campbell J L. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J Biol Chem. 1995;270:26766–26769. doi: 10.1074/jbc.270.45.26766. [DOI] [PubMed] [Google Scholar]
- 5.Bullock P A. The initiation of simian virus 40 DNA replication in vitro. Crit Rev Biochem Mol Biol. 1997;32:503–568. doi: 10.3109/10409239709082001. [DOI] [PubMed] [Google Scholar]
- 6.Champoux J J. Topoisomerase I is preferentially associated with isolated replicating simian virus 40 molecules after treatment of infected cells with camptothecin. J Virol. 1988;62:3675–3683. doi: 10.1128/jvi.62.10.3675-3683.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Champoux J J. Topoisomerase I is preferentially associated with normal SV40 replicative intermediates, but is associated with both replicating and nonreplicating SV40 DNAs which are deficient in histones. Nucleic Acids Res. 1992;20:3347–3352. doi: 10.1093/nar/20.13.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins K, Kelly T J. Effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase α-primase. Mol Cell Biol. 1991;11:2108–2115. doi: 10.1128/mcb.11.4.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Covey J M, Jaxel C, Kohn K W, Pommier Y. Protein-linked DNA strand breaks induced in mammalian cells by camptothecin, an inhibitor of topoisomerase I. Cancer Res. 1989;49:5016–5022. [PubMed] [Google Scholar]
- 10.Dean F B, Bullock P, Murakami Y, Wobbe C R, Weissbach L, Hurwitz J. Simian virus 40 (SV40) DNA replication: SV40 large T antigen unwinds DNA containing the SV40 origin of replication. Proc Natl Acad Sci USA. 1987;84:16–20. doi: 10.1073/pnas.84.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean F B, Dodson M, Echols H, Hurwitz J. ATP-dependent formation of specialized nucleoprotein structure by simian virus 40 (SV40) large tumor antigen at the SV40 replication origin. Proc Natl Acad Sci USA. 1987;84:8981–8985. doi: 10.1073/pnas.84.24.8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dornreiter I, Copeland W C, Wang T S F. Initiation of simian virus 40 DNA replication requires the interaction of a specific domain of human DNA polymerase α with large T antigen. Mol Cell Biol. 1993;13:809–820. doi: 10.1128/mcb.13.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dornreiter I, Eerdile L F, Gilbert I U, von Winkler D, Kelly T J, Fanning E. Interaction of DNA polymerase α-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fairman M P, Stillman B. Cellular factors required for multiple stages of SV40 DNA replication in vitro. EMBO J. 1988;7:1211–1218. doi: 10.1002/j.1460-2075.1988.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fotedar A, Cannella D, Fitzgerald P, Rousselle T, Gupta S, Doree M, Fotedar R. Role for cyclin A-dependent kinase in DNA replication in human S phase cell extracts. J Biol Chem. 1996;271:31627–31637. doi: 10.1074/jbc.271.49.31627. [DOI] [PubMed] [Google Scholar]
- 16.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 17.Gurney E G, Harrison R O, Fenno J. Monoclonal antibodies against simian virus 40 T antigens: evidence for distinct subclasses of large T antigen and for similarities among nonviral T antigens. J Virol. 1980;34:752–763. doi: 10.1128/jvi.34.3.752-763.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haluska P, Jr, Saleem A, Edwards T K, Rubin E H. Interaction between the N-terminus of human topoisomerase I and SV40 large T antigen. Nucleic Acids Res. 1998;26:1841–1847. doi: 10.1093/nar/26.7.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harlow E, Crawford L V, Pim D C, Williamson N M. Monoclonal antibodies specific for simian virus 40 tumor antigens. J Virol. 1981;39:861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hickey R J, Malkas L H. Mammalian cell DNA replication. Crit Rev Eukaryotic Gene Expr. 1997;7:125–157. doi: 10.1615/critreveukargeneexpr.v7.i1-2.80. [DOI] [PubMed] [Google Scholar]
- 21.Hsiang Y H, Hertzberg R, Hecht S, Liu L F. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 22.Hua X H, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J Cell Biol. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes P, Baldacci G. A DNA helicase purified by replication protein (RPA) affinity chromatography from mouse FM3A cells. Nucleic Acids Res. 1997;25:3881–3888. doi: 10.1093/nar/25.19.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurwitz J, Dean F B, Kwong A D, Lee S H. The in vitro replication of DNA containing the SV40 origin. J Biol Chem. 1990;265:18043–18046. [PubMed] [Google Scholar]
- 25.Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J Biol Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 26.Ishimi Y, Claude A, Bullock P, Hurwitz Complete enzymatic synthesis of DNA containing the SV40 origin of replication. J Biol Chem. 1988;263:19723–19733. [PubMed] [Google Scholar]
- 27.Ishimi Y, Matsumoto K, Ohba R. DNA replication from initiation zones of mammalian cells in a model system. Mol Cell Biol. 1994;14:6489–6496. doi: 10.1128/mcb.14.10.6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishimi Y, Sugasawa K, Hanaoka F, Eki T, Hurwitz J. Topoisomerase II plays an essential role as a swivelase in the late stage of SV40 chromosome replication in vitro. J Biol Chem. 1992;267:462–466. [PubMed] [Google Scholar]
- 29.Kobayashi T, Rein T, DePamphilis M. Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol Cell Biol. 1998;18:3266–3277. doi: 10.1128/mcb.18.6.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Lai J-S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lanford R E. Expression of simian virus 40 T antigen in insect cells using a baculovirus expression vector. Virology. 1988;167:72–81. doi: 10.1016/0042-6822(88)90055-4. [DOI] [PubMed] [Google Scholar]
- 33.Li C, Cao L G, Wang Y L, Baril E F. Further purification and characterization of a multienzyme complex for DNA synthesis in human cells. J Cell Biochem. 1993;53:405–419. doi: 10.1002/jcb.240530418. [DOI] [PubMed] [Google Scholar]
- 34.Li J J, Kelly T J. Simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1984;81:6973–6977. doi: 10.1073/pnas.81.22.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J J, Kelly T J. Simian virus 40 DNA replication in vitro: specificity of initiation and evidence for bidirectional replication. Mol Cell Biol. 1985;5:1238–1246. doi: 10.1128/mcb.5.6.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madden K R, Champoux J J. Overexpression of human topoisomerase I in baby hamster kidney cells: hypersensitivity of clonal isolates to camptothecin. Cancer Res. 1992;52:525–532. [PubMed] [Google Scholar]
- 37.Malkas L H, Hickey R J, Li C, Pederson N, Baril E F. A 21S enzyme complex from HeLa cells that functions in simian virus 40 DNA replication in vitro. Biochemistry. 1990;29:6362–6374. doi: 10.1021/bi00479a004. [DOI] [PubMed] [Google Scholar]
- 38.Mastrangelo I A, Hough P V C, Wall J S, Dodson M, Dean F B, Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature (London) 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 39.Matsumoto T, Eki T, Hurwitz J. Studies on the initiation and elongation reactions in the simian virus 40 DNA replication system. Proc Natl Acad Sci USA. 1990;87:9712–9716. doi: 10.1073/pnas.87.24.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melendy T, Stillman B. Purification of DNA polymerase δ as an essential simian virus 40 DNA replication factor. J Biol Chem. 1991;266:1942–1949. [PubMed] [Google Scholar]
- 41.Melendy T, Stillman B. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J Biol Chem. 1993;268:3389–3395. [PubMed] [Google Scholar]
- 42.Murakami M, Wobbe C R, Weissbach L, Dean F B, Hurwitz J. Role of DNA polymerase α and DNA primase in simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1986;83:2869–2873. doi: 10.1073/pnas.83.9.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murakami Y, Hurwitz J. Functional interactions between SV40 T antigen and other replication proteins at the replication fork. J Biol Chem. 1993;268:11008–11017. [PubMed] [Google Scholar]
- 44.Pommier Y, Kohlhagen G, Wu C, Simmons D T. Mammalian DNA topoisomerase I activity and poisoning by camptothecin are inhibited by simian virus 40 large T antigen. Biochemistry. 1998;37:3818–3823. doi: 10.1021/bi972067d. [DOI] [PubMed] [Google Scholar]
- 45.Prelich G, Kostura M, Marshak D R, Mathews M B, Stillman B. The cell-cycle regulated proliferating cell nuclear antigen is required for SV40 DNA replication in vitro. Nature (London) 1987;326:471–475. doi: 10.1038/326471a0. [DOI] [PubMed] [Google Scholar]
- 46.Prelich G, Stillman B. Coordinated leading and lagging strand synthesis during SV40 DNA replication in vitro requires PCNA. Cell. 1988;53:117–126. doi: 10.1016/0092-8674(88)90493-x. [DOI] [PubMed] [Google Scholar]
- 47.Prelich G, Tan C K, Kostura M, Mathews M B, So A G, Downey K M, Stillman B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-δ auxiliary protein. Nature (London) 1987;326:517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- 48.Roberts J M. Simian virus 40 (SV40) large tumor antigen causes stepwise changes in SV40 origin structure during initiation of DNA replication. Proc Natl Acad Sci USA. 1989;86:3939–3943. doi: 10.1073/pnas.86.11.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48a.Roy, R., et al. Unpublished data.
- 49.Shin C G, Snapka R M. Patterns of strongly protein-associated simian virus 40 DNA replication intermediates resulting from exposures to specific topoisomerase poisons. Biochemistry. 1990;29:10934–10939. doi: 10.1021/bi00501a011. [DOI] [PubMed] [Google Scholar]
- 50.Simanis V, Lane D P. An immunoaffinity purification procedure for SV40 large T antigen. Virology. 1985;144:88–100. doi: 10.1016/0042-6822(85)90308-3. [DOI] [PubMed] [Google Scholar]
- 51.Simmons D T, Melendy T, Usher D, Stillman B. Simian virus 40 large T antigen binds to topoisomerase I. Virology. 1996;222:365–374. doi: 10.1006/viro.1996.0433. [DOI] [PubMed] [Google Scholar]
- 52.Simmons D T, Roy R, Chen L, Gai D, Trowbridge P W. The activity of topoisomerase I is modulated by large T antigen during unwinding of the SV40 origin. J Biol Chem. 1998;273:20390–20396. doi: 10.1074/jbc.273.32.20390. [DOI] [PubMed] [Google Scholar]
- 53.Simmons D T, Trowbridge P W, Roy R. Topoisomerase I stimulates SV40 T antigen-mediated DNA replication and inhibits T antigen’s ability to unwind DNA at nonorigin sites. Virology. 1998;242:435–443. doi: 10.1006/viro.1997.9024. [DOI] [PubMed] [Google Scholar]
- 53a.Simmons, D. T., et al. Unpublished data.
- 54.Snapka R M. Topoisomerase inhibitors can selectively interfere with different stages of simian virus 40 DNA replication. Mol Cell Biol. 1986;6:4221–4227. doi: 10.1128/mcb.6.12.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stewart L, Ireton G C, Champoux J J. The domain organization of human topoisomerase I. J Biol Chem. 1996;271:7602–7608. doi: 10.1074/jbc.271.13.7602. [DOI] [PubMed] [Google Scholar]
- 56.Stewart L, Ireton G C, Champoux J J. Reconstitution of human topoisomerase I by fragment complementation. J Mol Biol. 1997;269:355–372. doi: 10.1006/jmbi.1997.1056. [DOI] [PubMed] [Google Scholar]
- 57.Stewart L, Ireton G C, Parker L H, Madden K R, Champoux J J. Biochemical and biophysical analyses of recombinant forms of human topoisomerase I. J Biol Chem. 1996;271:7593–7601. doi: 10.1074/jbc.271.13.7593. [DOI] [PubMed] [Google Scholar]
- 58.Stillman B, Gerard R D, Guggenheimer R A, Gluzman Y. T antigen and template requirements for SV40 DNA replication in vitro. EMBO J. 1985;4:2933–2939. doi: 10.1002/j.1460-2075.1985.tb04026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stillman B W, Gluzman Y. Replication and supercoiling of simian virus 40 DNA in cell extracts from human cells. Mol Cell Biol. 1985;5:2051–2060. doi: 10.1128/mcb.5.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sundin O, Varshavsky A. Terminal stages of SV40 DNA replication proceed via multiply intertwined catenated dimers. Cell. 1980;21:103–114. doi: 10.1016/0092-8674(80)90118-x. [DOI] [PubMed] [Google Scholar]
- 61.Sundin O, Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell. 1981;25:659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- 62.Tsurimoto T, Stillman B. Multiple replication factors augment DNA synthesis by the two eukaryotic DNA polymerases, α and δ. EMBO J. 1989;8:3883–3889. doi: 10.1002/j.1460-2075.1989.tb08567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase α and δ during initiation of leading and lagging strand synthesis. J Biol Chem. 1991;266:1961–1968. [PubMed] [Google Scholar]
- 64.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 65.Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature (London) 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 66.Wang J C. Interaction between DNA and an Escherichia coli protein ω. J Mol Biol. 1971;55:523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- 67.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- 68.Wobbe C R, Dean F, Weissbach L, Hurwitz J. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc Natl Acad Sci USA. 1985;82:5710–5714. doi: 10.1073/pnas.82.17.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wobbe C R, Weissbach L, Borowiec J A, Dean F B, Murakami Y, Bullock P, Hurwitz J. Replication of simian virus 40 origin-containing DNA in vitro with purified proteins. Proc Natl Acad Sci USA. 1987;84:1834–1838. doi: 10.1073/pnas.84.7.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wold M S, Kelly T. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc Natl Acad Sci USA. 1988;85:2523–2527. doi: 10.1073/pnas.85.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wold M S, Weinberg D H, Virshup D M, Li J J, Kelly T J. Identification of cellular proteins required for simian virus 40 DNA replication. J Biol Chem. 1989;264:2801–2809. [PubMed] [Google Scholar]
- 72.Yang L, Wold M S, Li J J, Kelly T J, Liu L F. Roles of DNA topoisomerases in simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1987;84:950–954. doi: 10.1073/pnas.84.4.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zou L, Mitchell J, Stillman B. CDC445, a novel yeast gene that functions with the origin recognition complex and Mcm proteins in initiation of DNA replication. Mol Cell Biol. 1997;17:553–563. doi: 10.1128/mcb.17.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]