Abstract
The SARS-CoV-2 P.1 variant of concern (VOC) was first identified in Brazil and is now spreading in European countries. It is characterized by the E484K mutation in the receptor-binding domain, which could contribute to the evasion from neutralizing antibodies. In Italy, this variant was first identified in January 2021. Here, we report an autochthonous outbreak of SARS-CoV-2 P.1 variant infections in southern Italy in subjects who had not travelled to endemic areas or outside the Apulia region. The outbreak involved seven subjects, three of whom had received a COVID-19 vaccine (one had received two doses and two had received one dose). Four patients had a mild clinical presentation. Laboratory investigations of nasopharyngeal swabs revealed that all strains were S-gene target failure-negative and molecular tests revealed they were the P.1 variant. Whole-genome sequencing confirmed that five subjects were infected with closely related strains classified as the P.1 lineage. The circulation of VOCs highlights the importance of strictly monitoring the spread of SARS-CoV-2 variants through genomic surveillance and of investigating local outbreaks. Furthermore, public health measures including social distancing, screening, and quarantine for travelers are key tools to slow down the viral transmission and to contain and mitigate the impact of VOC diffusion, and rapid scaling-up of vaccination is crucial to avoid a possible new epidemic wave.
Keywords: outbreak, P.1 variant, Gamma variant, SARS-CoV-2 infection, COVID-19, whole-genome sequencing
1. Introduction
In December 2020, the European Center for Disease Control and Prevention (ECDC) first reported the spread of a new SARS-CoV-2 variant of concern (VOC) characterized by multiple spike protein mutations and mutations in other genomic regions, called VOC 202012/01—lineage B.1.1.7, in the UK, and labeled Alpha variant by the World Health Organization [1,2]. A few weeks later, a new ECDC risk assessment described the emergence of two new VOCs, namely, the 501Y.V2 variant (Beta variant), which was isolated in South Africa, and the P.1 variant (Gamma variant), which was identified in Brazil, mostly in the Amazonas state [3]. The overall risk associated with the introduction and community spread of these VOCs was assessed as being high/very high [3]. In May 2021, the SARS-CoV-2 B.1.617.2 Delta variant emerged in India and has spread all over the world [4]. At the time of writing, all these lineages seem to have almost replaced the previous circulating viruses in the geographic regions and the Delta VOC, in particular, shows very high probability of becoming the dominant circulating strain in the EU/EAA [4,5,6,7]. The spread of VOCs with a high transmission potential poses a serious risk in terms of virulence, potential reinfections, and antibody responses to and efficacies of vaccines [4,8].
The P.1 variant is characterized by 11 amino acid changes in the spike protein, three of which are located in the receptor-binding domain (RBD) [3]. These amino acid changes are L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, H655Y, T1027I, and V1176F [3]. The 501Y.V2 and P.1 variants are both characterized by the E484K mutation in the RBD, which could contribute to the evasion from neutralizing antibodies [9,10]. Cases of reinfection caused by SARS-CoV-2 strains carrying the E484K mutation have been described [11]. The P.1 variant was first identified in Japan in four travelers from Brazil, but there was no indication it was associated with more severe disease [8]. However, recent studies reported evidence that disease severity is increased with this variant [12]. Moreover, an impact on transmissibility has also been demonstrated [13]. Retrospective analyses of samples collected in Manaus (Brazil) demonstrated the presence of the P.1 variant from November 2020, when case numbers of COVID-19 were high, and a rise of this variant from 0% to 87% in 7 weeks [13]. Moreover, a statistically significant association between P.1 infection and a lower Cycle threshold (Ct) value in real-time PCR, which is an indirect index of viral load in different specimens [14], was reported [13].
In Europe, infections of the P.1 variant, as well as the B.1.1.7, B.1.351, and B.1.617.2 VOCs, have been associated with a higher risk of hospitalization and intensive care unit admission [4,12]. In Italy, the P.1 variant was first reported in January 2021 in three patients returning from Brazil [15]. Monthly national flash surveys conducted in Italy to estimate the prevalence of VOCs from February 2021, reported that the estimated prevalence of the P.1 variant increased from 0% in February 2021 to 11.8% in June 2021 [16]. In the Apulia region, the estimated prevalence of the P.1 variant remained below 1% [16].
Here, we report an autochthonous outbreak of SARS-CoV-2 P.1 variant infections in southern Italy occurred in April 2021, when strict non-pharmaceutical interventions (NPI) were mandatory and travels outside the regions were forbidden.
2. Results
Of the seven patients involved in the outbreak, six were members of the same family and one was a friend of the index case. The demographic and clinical characteristics of the patients are shown in Table 1.
Table 1.
Demographic and clinical characteristics of seven cases of SARS-CoV-2 P.1 variant infections.
| Patient Number | Relationship with the Index Case | Age (Years) | Sex | Comorbidities | Date of Onset of Symptoms | Date of Diagnosis | Clinical Presentation | Vaccinated |
|---|---|---|---|---|---|---|---|---|
| 1 | Index case | 45 | Male | No | 20 April 2021 | 21 April 2021 | Mild | No |
| 2 | Wife | 31 | Female | No | - | 29 April 2021 | Asymptomatic | No |
| 3 | Father-in-law | 73 | Male | Diabetes, Hypertension | 28 April 2021 | 28 April 2021 | Mild | Yes, BNT162b2 (second dose on 30 March 2021) |
| 4 | Mother-in-law | 69 | Female | No | - | 28 April 2021 | Asymptomatic | Yes, BNT162b2 (first dose on 19 April 2021) |
| 5 | Brother-in-law | 53 | Male | No | 21 April 2021 | 22 April 2021 | Mild | No |
| 6 | Nephew | 16 | Male | No | - | 12 May 2021 | Asymptomatic | No |
| 7 | Friend | 37 | Male | No | 26 April 2021 | 27 April 2021 | Mild | Yes, ChAdOx1-S (first dose on 29 March 2021) |
The index case (patient 1) was a healthy 45-year-old man who resided in a small town with about 13,000 inhabitants in the province of Bari, Italy, and presented with high-grade fever (>39 °C), sore throat, diarrhea, anosmia, and ageusia on 20 April 2021. He reported no history of travel in any area endemic for the SARS-CoV-2 P.1 variant or travel out of the Apulia region because travel was forbidden by national health authorities, nor contacts with any SARS-CoV-2 positive case. The patient tested positive for SARS-CoV-2 on 21 April 2021. His wife and other household members also tested positive for SARS-CoV-2 and were promptly quarantined. Contact tracing revealed that 30 subjects were contacts of the index case and his household members. All of them were tested for SARS-CoV-2 infection. Among these contacts, only a friend of the index case who showed symptoms on 26 April 2021, tested positive (patient 7). Epidemiological investigation revealed that the contact with the index case occurred on 20 April 2021 during football training. Patients 3, 5, and 7 had mild clinical presentation, with symptoms developing between 21 and 26 April 2021. Patient 3 was fully vaccinated with two doses of the BNT162b2 COVID-19 vaccine. The first dose was administered on 8 March 2021, and the second dose was administered on 30 March 2021, in accordance with the recommended schedule. Patients 4 had received one dose of the BNT162b2 COVID-19 vaccine on 19 April 2021, and patient 7 had received one dose of the ChAdOx1-S COVID-19 vaccine on 29 March 2021 (Table 1).
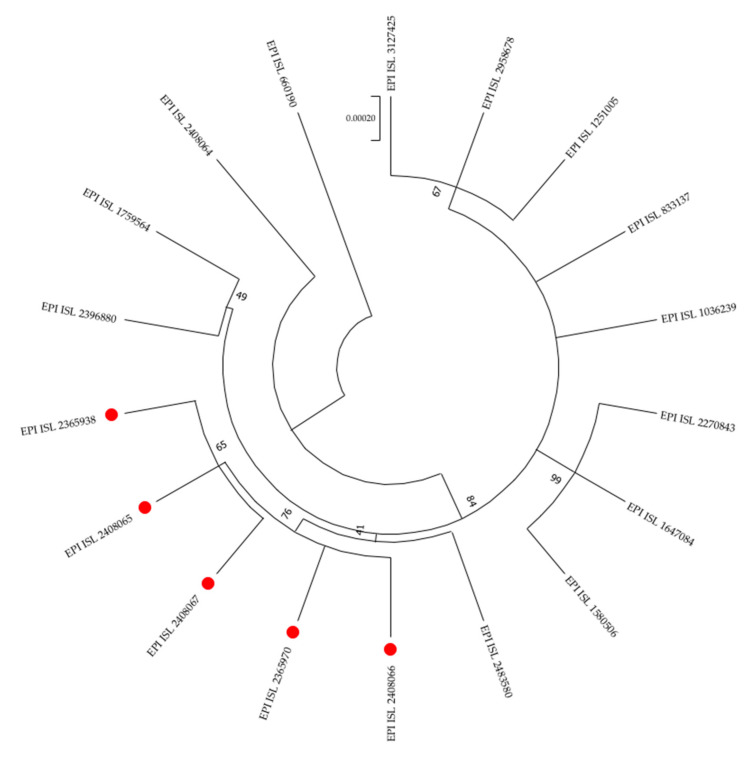
The seven patients involved in the outbreak tested positive for SARS-CoV-2 by real-time PCR and all were S-gene target failure (SGTF)-negative. Therefore, samples were subjected to molecular screening for variants and designated the P.1 variant because of the presence of the K417T, E484K, and N501Y spike mutations. Whole-genome sequencing was performed of all seven strains, but high-quality SARS-CoV-2 genome sequences were only obtained for patients 1, 3, 4, 5, and 7. These were imputed into the PANGOLIN tool for lineage classification [17] and classified as the P.1 lineage. The sequences were deposited in the GISAID database (www.gisaid.com, accessed on 11 August 2021). The accession numbers are reported in Figure 1. The phylogenetic tree showed closely related strains, thus suggesting a common source of exposure (Figure 1).
Figure 1.
Phylogenetic tree of 18 SARS-CoV-2 full genome sequences, including the five genomes examined in this study (red dots). The reference SARS-CoV-2 genome (GISAID accession number: EPI_ISL_660190) was included to root the tree. P.1 reference strain has been used to construct the tree (GISAID accession numbers: EPI_ISL_833137). Other Italian P.1 SARS-CoV-2 sequences have been used for comparison (GISAID accession numbers: EPI_ISL_2408064, EPI_ISL_1759564, EPI_ISL_2396880, EPI_ISL_2483580, EPI_ISL_1580506, EPI_ISL_1647084, EPI_ISL_2270843, EPI_ISL_1036239, EPI_ISL_1251005, EPI_ISL_2958678, EPI_ISL_3127425). Evolutionary analyses were conducted in MEGAX by using the maximum likelihood method and Tamura-Nei model. The robustness of branching pattern was tested by 1000 bootstrap replications. Bootstrap values are reported. The scale bar indicates nucleotides substitutions per site.
3. Discussion
Virus importation associated with travel has been the key driver of viral spread since the first wave of the COVID-19 pandemic in Italy [18]. Cases of imported SARS-CoV-2 variant P.1 infection were first reported on 7 January 2021 in Central Italy in travelers returning from Brazil, and this variant was also subsequently identified in northern and southern Italy [15]. All direct flights from Brazil to Italy were cancelled on 16 January 2021. Another case of P.1 infection was identified in a traveler returning from Brazil on 17 January 2021, thus confirming the risk of introducing variants via indirect flights [19]. An in-depth molecular epidemiological analysis performed by Di Giallonardo et al. showed intensive local transmission of the P.1 variant in Italy after its travel-linked introduction [20]. In the Apulia region, the P.1 variant was first identified through a flash national survey on 20 April 2021 [21]. It was identified in a patient without risk factors for P.1 variant infection (i.e., travel or contact with a P.1 variant-positive case). Thereafter, we documented the circulation of the P.1 variant in two more provinces of the Apulia region (data not published). The present study reports an outbreak of the SARS-CoV-2 P.1 VOC in southern Italy. Among cases, no subjects with known travel history, nor contacts with other SARS-CoV-2 positive subjects were identified. Therefore, despite lockdown restrictions being imposed in Italy from December 2020, the cluster described here is concerning and suggests a local ongoing transmission, although at a low level, of this variant in the Apulia region.
The symptomatic cases of the outbreak here described showed a mild clinical presentation. Recently, de Siqueira et al. also reported a familial cluster of five cases, with three having severe disease, one of whom died [22]. We could speculate that the difference in clinical presentation between this previous study and the cases described here could be related to the fact that three of our seven cases had received at least one dose of a COVID-19 vaccine, which may have protected them against severe disease. Of note, patient 4 received the first vaccine dose only a few days before being diagnosed and was asymptomatic, while patient 3 was fully vaccinated and received the second dose 1 month before the clinical onset of symptoms. As previously reported for the B.1.1.7 lineage VOC in the Apulia region [23], the P.1 variant raises concerns about such strains causing possible symptomatic post-vaccination infections. Some studies demonstrated that samples of vaccinated and convalescent people exhibit lower neutralization activity against SARS-CoV-2 strains harboring the E484K spike mutation [24], showing the need to induce the highest neutralization titers through vaccination. According to the European approach, the Italian national guidelines indicate that the booster vaccination should be postponed to provide more subjects with a first vaccination [25]. However, this approach will result in a lower level of neutralizing antibodies and could leave some vaccinees unprotected in the context of the rising spread of SARS-CoV-2 variants [24,26].
This study has some limitations. First, only five in seven samples were successfully subjected to WGS. However, the molecular screening for variants and the epidemiological linkage suggested the presence of SARS-CoV-2 P.1 lineage in all cases. Second, serum samples after vaccination and before symptom onset for anti-spike IgG detection were not available. Nevertheless, due to the incomplete vaccination schedule for two subjects, we could hypothesize a low humoral response to vaccination.
In Italy, where the vaccination campaign is accelerating but has not yet reached sufficient coverage, the spread of variants with higher transmissibility may have a significant impact, also in the light of the reduction of NPI. The current scenario characterized by the circulation of multiple VOCs and, in particular, of the Delta VOC [4], highlights the importance of closely monitoring the spread of SARS-CoV-2 variants through genomic surveillance and of investigating local outbreaks. Furthermore, maintaining public health measures including social distancing, screening, and quarantine for travelers are key tools to slow down the viral transmission and to contain and mitigate the impact of VOCs diffusion on the National Health Service. Finally, rapid up scaling of vaccination in Italy is crucial to avoid a possible new epidemic wave, particularly in younger people who have not yet been vaccinated.
4. Materials and Methods
Seven patients were involved in the outbreak. Their clinical presentations were classified according to the National Institute of Health (NIH) clinical staging of COVID-19 disease [27]. Nasopharyngeal swabs were collected from all patients at the Local Health Unit of Bari (Italy) and were processed at the Laboratory of Molecular Epidemiology and Public Health of the Hygiene Unit (A.O.U.C. Policlinico Bari), which is the coordinator of the Regional Laboratory Network for SARS-CoV-2 diagnosis in the Apulia region. RNA was extracted using a MagMAX Viral/Pathogen Nucleic Acid Isolation Kit (Thermo Fisher Scientific, Waltham, MA, USA). The molecular test was performed using a three-target commercial multiplex real-time PCR assay targeting the N, ORF1ab, and S genes (TaqPath RT-PCR COVID-19 Assay; Thermo Fisher Scientific). SGTF was assessed to rule out the B.1.1.7 lineage VOC because SGTF can be considered a robust proxy of VOC 202012/01 [26,28]. SGTF-negative samples were screened for the presence of notable types of spike protein mutations (HV 69-70 deletion, N501Y, K417N, E484K, and K417T) using a commercial multiplex real-time PCR kit (Seegene Allplex SARS-CoV-2 Variants I Assay, Arrows Diagnostics, Genova, Italy). Whole-genome sequencing was performed using the Ion Torrent platform (Thermo Fisher Scientific, Monza, Italy). The library was prepared using an Ion AmpliSeq Library Kit Plus according to the manufacturer’s instructions and the Ion AmpliSeq SARS-CoV-2 RNA custom primer panel (Thermo Fisher Scientific, Monza, Italy). Quality control of AmpliSeq reads and their alignment to the complete genome of the SARS-CoV-2 Wuhan-Hu-1 isolate were performed using the Torrent Server of the Ion Torrent S5 sequencer with default settings. The aligned reads were utilized for both reference-guided assembly and variant calling. The quality metrics for the reference-based assemblies are as follows: 784,477 sequence reads (average length 149 bp), 674,160 mapped reads, GC% 40, and an average base coverage depth of 3222. The total genome size was 29,780 bp. Assembly was performed using the Iterative Refinement Meta-Assembler (IRMA) v.1.3.0.2, which produced a consensus sequence for the sample using a cut-off of >50% for calling single nucleotide polymorphisms. The whole-genome sequences have been deposited in the GISAID database (https://www.gisaid.com, accessed on 11 August 2021). Phylogenetic analysis was performed using MEGAX software.
Author Contributions
Conceptualization, M.C. and D.L.; methodology, A.S. (Anna Sallustio); formal analysis, A.S. (Anna Sallustio) and M.A.; investigation, M.M.F. and A.S. (Antonio Sanguedolce); data curation, F.C. and D.C.; writing—original draft preparation, D.L. and M.A.; writing—review and editing, M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Institutional Review Board Statement
Ethical review and approval were waived for this study because all the activities were conducted as part of the legislated mandate of the Health Promotion and Public Health Department of Apulia. All procedures were carried out in accordance with the Declaration of Helsinki, as revised in 2013, for human subjects.
Informed Consent Statement
Informed written consent was obtained from all subjects involved in the study.
Data Availability Statement
All data regarding the patients and laboratory tests are available from the corresponding author by e-mail request.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.European Center for Disease Control and Prevention Threat Assessment Brief: Rapid Increase of a SARS-CoV-2 Variant with Multiple Spike Protein Mutations Observed in the United Kingdom. [(accessed on 30 May 2021)];Risk Assessment. 2020 Available online: https://www.ecdc.europa.eu/en/publications-data/threat-assessment-brief-rapid-increase-sars-cov-2-variant-united-kingdom.
- 2.World Health Organization Tracking SARS-CoV-2 Variants. [(accessed on 15 July 2021)];2021 Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
- 3.European Center for Disease Control and Prevention Risk Related to the Spread of New SARS-CoV-2 Variants of Concern in the EU/EEA—First Update. [(accessed on 28 January 2021)];2021 Available online: https://www.ecdc.europa.eu/en/publications-data/covid-19-risk-assessment-spread-new-variants-concern-eueea-first-update.
- 4.European Center for Disease Control and Prevention Risk Assessment: Implications for the EU/EEA on the Spread of the SARS-CoV-2 Delta (B.1.617.2) Variant of Concern. [(accessed on 15 July 2021)];2021 Available online: https://www.ecdc.europa.eu/en/publications-data/threat-assessment-emergence-and-impact-sars-cov-2-delta-variant.
- 5.O’Toole Á., Hill V., Pybus O.G., Watts A., Bogoch I.I., Khan K., Messina J.P., Tegally H., Lessells R.R., Giandhari J., et al. Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2. Wellcome Open Res. 2021;6:121. doi: 10.12688/wellcomeopenres.16661.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walensky R.P., Walke H.T., Fauci A.S. SARS-CoV-2 variants of concern in the United States—Challenges and opportunities. JAMA. 2021;325:1037. doi: 10.1001/jama.2021.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sabino E.C., Buss L.F., Carvalho M.P.S., Prete C.A., Crispim M.A.E., Fraiji N.A., Pereira R.H.M., Parag K.V., Peixoto P.D.S., Kraemer M.U.G., et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397:452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujino T., Nomoto H., Kutsuna S., Ujiie M., Suzuki T., Sato R., Fujimoto T., Kuroda M., Wakita T., Ohmagari N. Novel SARS-CoV-2 variant in travelers from Brazil to Japan. Emerg. Infect. Dis. 2021;27:1243–1245. doi: 10.3201/eid2704.210138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. bioRxiv. 2020 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greaney A.J., Loes A.N., Crawford K.H., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nonaka C.K.V., Franco M.M., Gräf T., Barcia C.A.L., Mendonça R.N., de Sousa K.A.F., Neiva L.M.C., Fosenca V., Mendes A.V.A., de Aguiar R.S., et al. Genomic evidence of SARS-CoV-2 reinfection involving E484K spike mutation, Brazil. Emerg. Infect. Dis. 2021;27:1522–1524. doi: 10.3201/eid2705.210191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Funk T., Pharris A., Spiteri G., Bundle N., Melidou A., Carr M., Gonzalez G., Garcia-Leon A., Crispie F., O’Connor L., et al. Characteristics of SARS-CoV-2 variants of concern B.1.1.7, B.1.351 or P.1: Data from seven EU/EEA countries, weeks 38/2020 to 10/2021. Eurosurveillance. 2021;26:2100348. doi: 10.2807/1560-7917.ES.2021.26.16.2100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T., et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romero-Gómez M.P., Gómez-Sebastian S., Cendejas-Bueno E., Montero-Vega M.D., Mingorance J., García-Rodríguez J. Ct value is not enough to discriminate patients harbouring infective virus. J. Infect. 2020;82:e35–e37. doi: 10.1016/j.jinf.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Compagni E.D., Jurisic L., Caporale M., Bacà F., Scialabba S., Fanì S., Perullo A., Toro M., Marchegiano A., Martino M., et al. Genome sequences of three SARS-CoV-2 P.1 strains identified from patients returning from Brazil to Italy. Microbiol. Resour. Announc. 2021;10 doi: 10.1128/mra.00177-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Istituto Superiore di Sanità Prevalenza delle Varianti VOC (Variant of Concern) Appartenenti ai Lignaggi B.1.1.7, P.1, B.1.617.2 e B.1.351 e di Altre Varianti in Italia. [(accessed on 10 July 2021)];Indagine del 22/06/2021. Available online: https://www.epicentro.iss.it/coronavirus/pdf/sars-cov-2-monitoraggio-varianti-indagini-rapide-22-giugno-2021.pdf.
- 17.Rambaut A., Holmes E.C., O’Toole A., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Giallonardo F., Duchene S., Puglia I., Curini V., Profeta F., Cammà C., Marcacci M., Calistri P., Holmes E.C., Lorusso A. Genomic epidemiology of the first wave of SARS-CoV-2 in Italy. Viruses. 2020;12:1438. doi: 10.3390/v12121438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maggi F., Novazzi F., Genoni A., Baj A., Spezia P.G., Focosi D., Zago C., Colombo A., Cassani G., Pasciuta R., et al. Imported SARS-CoV-2 variant, P.1 in traveler returning from Brazil to Italy. Emerg. Infect. Dis. 2021;27:1249–1251. doi: 10.3201/eid2704.210183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Giallonardo F., Puglia I., Curini V., Cammà C., Mangone I., Calistri P., Cobbin J., Holmes E., Lorusso A. Emergence and spread of SARS-CoV-2 lineages, B.1.1.7 and P.1 in Italy. Viruses. 2021;13:794. doi: 10.3390/v13050794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Istituto Superiore di Sanità Prevalenza delle VOC (Variant of Concern) del Virus SARS-CoV-2 in Italia: Lineage B.1.1.7, P.1 e B.1.351, e Altre Varianti (Variant of Interest, VOI) Tra cui Lineage P.2 e Lineage B.1.525. [(accessed on 28 May 2021)];Indagine del 20/4/2021. Available online: https://www.iss.it/documents/20126/0/Relazione+tecnica+indagine+rapida+varianti+SARS-COV-2.pdf/f425e647-efdb-3f8c-2f86-87379d56ce8d?t=1620232350272.
- 22.De Siqueira I.C., Camelier A.A., Maciel E.A., Nonaka C.K.V., Neves M.C.L., Macêdo Y.S.F., de Sousa K.A.F., Araujo V.C., Paste A.A., Souza B.S.D.F., et al. Early detection of P.1 variant of SARS-CoV-2 in a cluster of cases in Salvador, Brazil. Int. J. Infect. Dis. 2021;108:252–255. doi: 10.1016/j.ijid.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loconsole D., Sallustio A., Accogli M., Leaci A., Sanguedolce A., Parisi A., Chironna M. Investigation of an outbreak of symptomatic SARS-CoV-2 VOC 202012/01-lineage B.1.1.7 infection in healthcare workers, Italy. Clin. Microbiol. Infect. 2021;27:1174. doi: 10.1016/j.cmi.2021.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jangra S., Ye C., Rathnasinghe R., Stadlbauer D., Krammer F., Simon V., Martinez-Sobrido L., García-Sastre A., Schotsaert M., Alshammary H., et al. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe. 2021 doi: 10.1016/S2666-5247(21)00068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ministero della Salute Circolare 0019748-05/05/2021-DGPRE-DGPRE-P. Trasmissione Parere del CTS in Merito alla Estensione dell’Intervallo Tra le Due Dosi dei Vaccini a mRNAe alla Seconda Dose del Vaccino Vaxzevria. [(accessed on 30 May 2021)]; Available online: https://www.trovanorme.salute.gov.it/norme/renderNormsanPdf?anno=2021&codLeg=80236&parte=1%20&serie=null.
- 26.Loconsole D., Centrone F., Morcavallo C., Campanella S., Sallustio A., Accogli M., Fortunato F., Parisi A., Chironna M. Rapid spread of the SARS-CoV-2 variant of concern 202012/01 in southern Italy (December 2020–March 2021) Int. J. Environ. Res. Public Health. 2021;18:4766. doi: 10.3390/ijerph18094766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Institute of Health (NIH) Coronavirus Disease 2019 (COVID-19) Treatment Guidelines. [(accessed on 30 May 2021)]; Available online: https://files.covid19treatmentguidelines.nih.gov/guidelines/covid19treatmentguidelines.pdf. [PubMed]
- 28.Investigation of Novel SARS-CoV-2 Variant: Variant of Concern 202012/01—Technical Briefing 7. [(accessed on 15 March 2021)]; Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/968581/Variants_of_Concern_VOC_Technical_Briefing_7_England.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data regarding the patients and laboratory tests are available from the corresponding author by e-mail request.