Abstract
Splicing enhancers are RNA sequences required for accurate splice site recognition and the control of alternative splicing. In this study, we used an in vitro selection procedure to identify and characterize novel RNA sequences capable of functioning as pre-mRNA splicing enhancers. Randomized 18-nucleotide RNA sequences were inserted downstream from a Drosophila doublesex pre-mRNA enhancer-dependent splicing substrate. Functional splicing enhancers were then selected by multiple rounds of in vitro splicing in nuclear extracts, reverse transcription, and selective PCR amplification of the spliced products. Characterization of the selected splicing enhancers revealed a highly heterogeneous population of sequences, but we identified six classes of recurring degenerate sequence motifs five to seven nucleotides in length including novel splicing enhancer sequence motifs. Analysis of selected splicing enhancer elements and other enhancers in S100 complementation assays led to the identification of individual enhancers capable of being activated by specific serine/arginine (SR)-rich splicing factors (SC35, 9G8, and SF2/ASF). In addition, a potent splicing enhancer sequence isolated in the selection specifically binds a 20-kDa SR protein. This enhancer sequence has a high level of sequence homology with a recently identified RNA-protein adduct that can be immunoprecipitated with an SRp20-specific antibody. We conclude that distinct classes of selected enhancers are activated by specific SR proteins, but there is considerable sequence degeneracy within each class. The results presented here, in conjunction with previous studies, reveal a remarkably broad spectrum of RNA sequences capable of binding specific SR proteins and/or functioning as SR-specific splicing enhancers.
Pre-mRNA splicing enhancer elements are short RNA sequences capable of activating weak splice sites in nearby introns (73; see references 2, 18, 27, 45, and 71 for recent reviews). These elements can activate heterologous pre-mRNAs (66, 68, 73) and can function at a distance as great as 500 nucleotides (nt) from the affected intron (70). Both constitutive (38, 60, 62) and regulated (69) splicing enhancers contain binding sites for SR proteins, a family of essential splicing factors containing one or more RNA recognition motifs (RRM) and an arginine/serine (RS)-rich domain (77; for reviews, see references 18 and 45). The RRM is required for RNA binding, and the RS domain is required for protein-protein interactions (4, 34, 74, 75). Mechanistic studies of splicing enhancer function are consistent with a recruitment model in which SR proteins activate splicing by binding to enhancers and recruiting the splicing machinery to the adjacent intron (21, 27, 69, 72, 78).
Because of the critical role of SR proteins in the activation of splicing enhancers, in vitro selection methods have been used to identify SR protein binding sites. For example, distinct high-affinity binding sites were identified for the human SR proteins SF2/ASF, SRp40, and SC35 by SELEX (systematic evolution of ligands by exponential enrichment). Multiple copies of the SF2/ASF and SRp40 RNA binding sites, but not the SC35 RNA binding sites, could function as splicing enhancers in HeLa cell nuclear extracts (63, 64) and function with specificity as SR-dependent enhancers in complementation assays (33, 63–65). Using a similar approach, Shi et al. identified a high-affinity RNA binding site for the Drosophila SR protein B52 but did not determine whether this sequence could function as an RNA splicing enhancer (59). An in vitro binding site selection with the Drosophila protein RBP1 (24), which identified an RNA sequence similar to a sequence found in several copies in the doublesex (dsx) splicing enhancer, showed specific cross-linking to the predicted RNA binding site in Drosophila Kc nuclear extracts, using a pre-mRNA containing a single site-specific label (42). In general, the selection of high-affinity RNA binding sites alone may not be sufficient for identifying the full array of sequences capable of functioning as SR protein-specific splicing enhancers (for a discussion, see reference 39).
An alternative approach to the identification of splicing enhancers is to use a functional in vitro (67) or in vivo (14) selection from a pool of randomized RNA sequences. An in vitro selection was used to identify RNA sequences capable of promoting exon inclusion in a substrate containing competing 3′ splice sites. A randomized cassette of RNA sequences was inserted into a pre-mRNA substrate such that the presence of a functional enhancer resulted in exon inclusion. Most of the enhancer elements obtained contained extended purine-rich sequences, but a second, novel class of sequences lacking stretches of purines was also identified (67). An in vivo selection was performed for RNA sequences that stimulated exon inclusion (14). In addition to isolation of purine-rich sequences, a novel class of A/C-rich splicing enhancers was isolated and characterized. Both of these selection approaches are based on altering the splice site utilization between competing splice sites, and the trans-acting factors that function in conjunction with these sequence motifs remain unknown. A recent in vitro functional selection was performed to identify SR protein-specific splicing enhancers capable of functioning downstream of an enhancer-dependent pre-mRNA (39). This report demonstrated that the SR proteins SF2/ASF, SRp40, and SRp55 could recognize significantly more degenerate consensus sequences than the high-affinity RNA binding sites identified by SELEX for SF2/ASF, SRp40, and B52/SRp55 (59, 63, 64).
We have made use of the well-characterized Drosophila dsx enhancer-dependent splicing substrate to select for functional splicing enhancer sequences. Alternative splicing of the Drosophila dsx pre-mRNA is positively regulated by a splicing enhancer located 300 nt downstream of a weak, female-specific 3′ splice site (for reviews, see references 5, 44, 53, 54, and 61). In females, a protein complex consisting of the Drosophila splicing regulators Transformer (Tra), Transformer-2 (Tra-2), and a specific SR protein (3, 42, 47, 68, 69) is assembled on the dsx repeat element (30, 49, 56, 68). This complex then recruits the splicing machinery to the dsx repeat element that resides downstream of the female-specific 3′ splice site (21, 27, 28, 69, 72, 78). A dsx pre-mRNA substrate containing the female-specific 3′ splice site and the dsx repeat element splicing enhancer is not efficiently spliced in human HeLa cell nuclear extracts unless the splicing reaction mixture is supplemented with recombinant Tra and/or Tra-2 (68). This splicing activity is lost when the dsx repeat element splicing enhancer is deleted, but splicing can be rescued by the insertion of a constitutive splicing enhancer downstream from the female-specific 3′ splice site (28, 43, 57, 60, 66, 70, 73). Thus, the dsx pre-mRNA provides a useful substrate for studying heterologous splicing enhancers and for the selection of novel constitutive splicing enhancers.
In this paper, we report the selection, identification, and characterization of novel splicing enhancer elements by using the dsx pre-mRNA. The selected enhancer sequences were separated into six different classes based on shared sequence similarities and then compared to sequences found in previously described splicing enhancers. Certain of the selected splicing enhancers display significant similarity to naturally occurring splicing enhancers or exonic splicing enhancer sequences found in human β-globin (hβ-globin) exons, whereas others appear to be novel. Individual selected enhancer sequences were analyzed for the ability to be activated by specific SR proteins in an S100 complementation assay (36). We found that individual selected enhancers could be differentially activated by individual SR proteins in complementation assays. Using a biotinylated RNA affinity technique, we also show that different selected enhancer RNA sequences can be bound by distinct SR proteins. Based on these functional and RNA binding assays, we identify distinct degenerate RNA consensus sequences for the SR proteins SC35, 9G8, and SRp20.
MATERIALS AND METHODS
DNA oligonucleotides.
The following oligonucleotides were used in the selection scheme: 1A and 1B (the randomized N18 and polypurine [AAG]6 oligonucleotides, respectively, from Oligos Etc.), 5′ TGCGGGTTCGAAATGACTCTCAGCAT(NNNNNNNNNNNNNNNNNN)AGTCGATCGATAAGCTTGGATCCGGAGAG 3′ (the Sa element [74] is in italics, the BstBI and HindIII sites are in boldface, and positions of the randomized nucleotides are indicated by N; the oligonucleotide for the synthetic dsx-[AAG]6 construct was isogenic except that the 18 randomized nucleotides were changed to six consecutive repeats of the AAG trinucleotide known to function as a constitutive splicing enhancer [67]); 2 (reverse transcription [RT] primer), 5′ CTCTCCGGATCCAAGCTTATGCATCGACT 3′; 3, (bridging), 5′ ATGCTGAGAGTCATTTCGAACCCGCAGCTCACCCCCGTCATAGATA 3′; 4 (T7 primer), 5′ TGTAATACGACTCACTATA 3′; 5 (dsx splice junction primer), 5′ TCGAAGAGGGCCAATACG 3′; 6 (Sa PCR primer), 5′ TGCGGGTTCGAAATGACTCTCAGCAT 3′; and 7 (dsx exon 4 sequencing primer), 5′ GCCAATACGTTGTGAATGAG 3′.
dsx-N18 pool construction.
The dsx-N18 construct was generated in three steps. In step 1, a 72-mer DNA oligonucleotide encoding a randomized cassette was generated between defined DNA sequences containing convenient restriction sites and primer hybridization sites to facilitate subcloning and PCR amplification of selected sequences. The randomized cassette in oligonucleotide 1A was flanked by well-characterized exon-derived sequence known to have no splicing enhancer activity on the dsx 3′ splice site (Sa element [73]) and some polylinker-derived sequence used for hybridization to the RT primer. The polypurine cassette encoding the repeated AAG trinucleotide (oligonucleotide 1B) was analogously designed. The 72-mers were amplified by PCR for 15 cycles using an oligonucleotide primer (RT primer; oligonucleotide 2) to yield a double-stranded DNA pool (round 0) PCR product.
In step 2, a PCR primer (bridging primer; oligonucleotide 3) was designed so that the 3′ half would be complementary to dsx exon 4 and the 5′ half would be complementary to the sense strand of the 72-mer containing the randomized pool (oligonucleotide 1A). The T7 promoter primer (oligonucleotide 4) and the bridging primer (oligonucleotide 3) were used to PCR amplify an FspI/BstBI fragment from plasmid pdsx(RI/FspI)T7 (see “Plasmid constructions” below). The resulting PCR product contains a T7 promoter upstream of the dsx sequence encoding exon 3, intervening sequence 3 (IVS3), 65 nt of exon 4, and a sequence complementary to the first 26 nt of oligonucleotide 1A (and oligonucleotide 1B).
In step 3, the PCR product encoding the dsx substrate was annealed with the randomized pool (round 0) in another PCR to generate the dsx-N18 transcription template for round 1. The resulting dsx-N18 transcription template has the first randomized nucleotide beginning at +90 relative to the dsx 3′ splice site. Using the dsx-N18 PCR product as a transcription template, 32P-labeled, capped pre-mRNAs were transcribed by using T7 RNA polymerase at a specific activity ratio of 160:1 (unlabeled to labeled UTP [800 Ci/mmol]). The 390-nt-long dsx-N18 pre-mRNA substrate consists of exon 3 (141 nt), IVS3 (114 nt), and exon 4/randomized pool (135 nt).
dsx-N18 selection and amplification.
Approximately four complete pools of the randomized 390-nt dsx-N18 pre-mRNA substrate were spliced in the first round in four large-scale (200-μl) in vitro splicing reactions, each performed with approximately 200,000 cpm (128 fmol) of 32P-UTP-labeled RNA. Similarly, four 200-μl reactions were performed in round 2, and two 200-μl reactions were performed in rounds 3 through 6. Splicing reactions otherwise were performed according to standard procedures (77), using 40% (vol/vol) HeLa cell nuclear extract. After a 2-h incubation at 30°C, the RNAs were deproteinized (proteinase K), phenol-chloroform extracted, and ethanol precipitated. The total RNA was reverse transcribed by using Superscript II reverse transcriptase as specified by the manufacturer (Life Technologies) but scaled up proportionately to a 50-μl reaction volume. One RT reaction using 50 pmol of RT primer (oligonucleotide 2) was performed for every 100-μl volume of in vitro splicing reaction mixture. The first-strand cDNA reaction was used to seed (20%, vol/vol) a 100-μl PCR to select for the dsx-N18 spliced products that are faithfully spliced in vitro. The selection for spliced products was accomplished via a sensitive PCR using oligonucleotide 2 and a dsx splice junction primer (oligonucleotide 5) which hybridizes across the splice junction generated by the correct ligation of exon 3 to exon 4.
The resulting splice junction PCR product theoretically contains functional splicing enhancers that are able to activate splicing of the weak dsx 3′ splice site in this chimeric dsx pre-mRNA. To regenerate a transcription template for subsequent rounds of selection, the splice junction must be eliminated and the selected enhancers must be retained. To accomplish this goal, enhancers were amplified by using oligonucleotide 2 and an Sa element primer (oligonucleotide 6) that hybridizes to the Sa element immediately upstream of the randomized residues. The Sa element PCR product containing the splicing enhancers was reinserted as a cassette into a new dsx pre-mRNA transcription template by performing another PCR with the T7-bridging dsx PCR product in a manner identical to that used for construction of the round 1 pool from round 0 (see “dsx-N18 pool construction” above).
Plasmid constructions.
For splicing and sequencing of the individual round 3 and round 6 clones, the splice junction PCR product was digested with BstBI and BamHI and cloned into the corresponding sites in plasmid pdsx(RI/FspI)T7. The net result is subcloning the enhancers back into the genetic context in which they underwent selection. Dideoxy sequencing of both strands was performed with a primer that hybridizes to exon 4 sequences (oligonucleotide 7) and an SP6 promoter primer. Some clones were sequenced and shown to have 16, 17, or 19 nt within the N18 template randomized after dideoxy sequencing. The individual enhancer clones were linearized at the HindIII site to eliminate the weak, intrinsic enhancer activity of the polylinker 3′ of the HindIII site.
For the biotinylated RNA experiments, individual splicing enhancers were sequentially subcloned to remove all dsx-derived sequences and as much polylinker sequence as possible. The BstBI/BamHI restriction fragments containing the individual splicing enhancers were subcloned into the corresponding sites in pGEM-7Z. A SmaI/BamHI fragment from the resulting construct was subcloned into the EcoRV/BamHI sites of SP73. The resulting SP6 transcription template allows the transcription of a splicing enhancer element in the absence of any dsx-derived sequences and a minimal amount of polylinker-derived sequences. The transcription templates were digested with HindIII before transcription.
In vitro splicing assays.
Pre-mRNA substrates were assayed for splicing activity by using complete premixed nuclear extract splicing reaction mixtures or complete premixed S100 complementation reaction mixtures requiring only the addition of the individual pre-mRNA substrate. For each nuclear extract splicing assay, the nuclear extract (40%, vol/vol) plus the basic components of the splicing reaction were premixed before addition of 10 to 20 fmol of a 32P-UTP-labeled gel-isolated pre-mRNA substrate.
The S100 extracts were prepared essentially as described elsewhere (1), with two modifications: phenylmethylsulfonyl fluoride was omitted from the dialysis buffer, and the 100,000 × g centrifugation was performed in a 70Ti fixed-angle rotor (Beckman). S100 complementation reactions were performed essentially as described previously (77), using ice-cold reagents: 40% (vol/vol) HeLa cell S100 extract in buffer D (15), 2.6% (vol/vol) polyvinyl alcohol (Sigma P-8136), 3.2 mM MgCl2, 20 mM creatine phosphate, 1.5 mM ATP, and 0.25 U of rRNasin (Promega) per μl. The order of addition of the reaction components was S100 premixed with cofactors followed by buffer D or the recombinant SR protein prediluted in buffer D. These premixed complementation reaction mixtures were aliquoted into individual reaction tubes, and the 32P-UTP-labeled pre-mRNA (10 to 20 fmol) was added to complete the reaction. S100 reaction and nuclear extract reaction mixtures were incubated for 3 h at 30°C. RNAs were deproteinized, extracted, and precipitated before being resolved on a 10% denaturing polyacrylamide (19:1)–7 M urea–1× Tris-borate-EDTA gel so that lariat-exon 4 intermediates could be resolved from the spliced product. RNAs were visualized by autoradiography.
The recombinant SR proteins SC35 and SF2/ASF were expressed and purified from baculovirus-infected cell lysates under native conditions as described elsewhere (69). The identities and phosphorylation states of the SR proteins were confirmed (data not shown) by their immunoreactivity with anti-SC35 and anti-9G8 monoclonal antisera (provided by Renate Gattoni and James Stévenin), anti-SF2/ASF monoclonal antisera (provided by Adrian Krainer), and the phosphoepitope-specific monoclonal antibody (MAb) 104 (provided by Mark Roth).
Biotinylated RNA experiments.
The enhancer RNAs were transcribed in the presence of biotin-21-UTP so that on average two biotin moieties are incorporated per enhancer RNA molecule. The biotinylated enhancer RNAs were trace labeled with [α-32P]UTP to identical specific activities. The protocol was performed essentially as described previously (76). Individual biotinylated enhancer RNAs were incubated in a nuclear extract (40%, vol/vol) splicing reaction mixture under splicing conditions for 10 min at 30°C. The biotinylated RNAs and associated proteins were purified under detergent and high-salt conditions as described elsewhere (76). The proteins associated with the RNAs were analyzed by Western blotting with MAb 104 (55).
RESULTS
In vitro selection of splicing enhancer sequences.
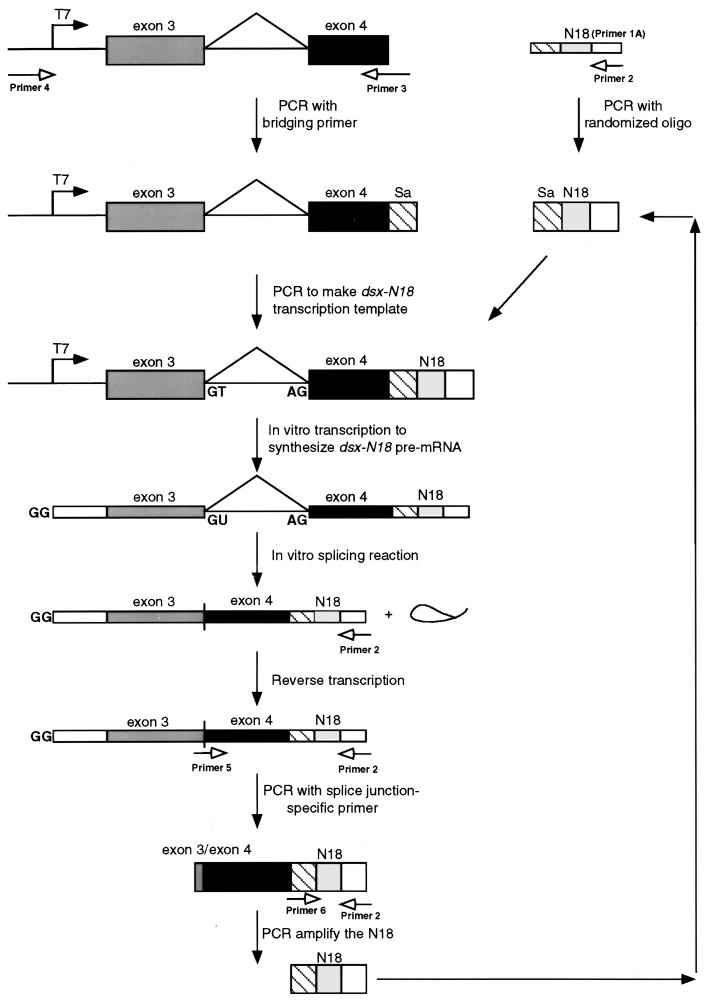
An in vitro selection was performed to identify short RNA sequences capable of functioning as pre-mRNA splicing enhancers in mammalian HeLa cell nuclear extracts. A diagram of the in vitro selection approach used to isolate functional exonic splicing enhancers capable of activating the enhancer-dependent 3′ splice site of the Drosophila dsx intron 3 is shown in Fig. 1. PCR was used to generate a transcription template that contained 18 consecutive randomized nucleotides, yielding a pool complexity of approximately 6.9 × 1010 sequences, positioned 90 nt downstream of the dsx 3′ splice site. The resulting DNA population was transcribed to generate a pool of dsx pre-mRNA substrates containing 18 consecutive randomized nucleotides (dsx-N18) in place of a splicing enhancer. The pre-mRNA substrates encoding the randomized pool were incubated in nuclear extracts under splicing conditions, and the spliced products containing the functional splicing enhancers were amplified by RT and selective PCR amplification of the spliced products using a splice junction-specific oligonucleotide primer. A second PCR was used to amplify the splicing enhancer elements. The isolated splicing enhancers were then reincorporated into new transcription templates for subsequent rounds of in vitro splicing, RT and PCR, thereby eliminating the possibility of accumulating Taq polymerase-mediated up-mutations that render the dsx substrate enhancer independent (70). The splicing enhancers recovered after three and six rounds of selection were subcloned and sequenced, and their splicing activities were assayed.
FIG. 1.
Schematic diagram of the in vitro selection strategy used to identify splicing enhancer sequences. The diagram illustrates the strategy used to construct the dsx-N18 transcription template, transcribe the dsx-N18 pre-mRNA, splice the dsx-N18 substrate in vitro, isolate the functional N18 splicing enhancers, and regenerate the transcription template for subsequent rounds of selection (see Materials and Methods for details). Boxes represent exon sequences, and horizontal lines represent intron sequences (including the branched, excised lariat). Thick boxes represent double-stranded nucleic acids (PCR template, transcription templates, and PCR products), and thin boxes represent single-stranded nucleic acids (DNA oligonucleotides, RNAs, and cDNAs). Solid arrows represent sequential steps in the construction, processing, and regeneration of the DNA and RNA molecules used in the dsx-N18 selection. Unfilled arrows indicate PCR primers (see Materials and Methods for sequences) and their sites of hybridization. In the dsx-N18 construct, the N18 enhancer is positioned at +90 relative to the dsx weak 3′ splice site. The 5′ cap, 5′ splice site, and 3′ splice site in the pre-RNAs are indicated by GG, GU, and AG, respectively. The line connecting the exons indicates the splicing pattern of the female-specific dsx IVS3 minigene upon activation by a splicing enhancer.
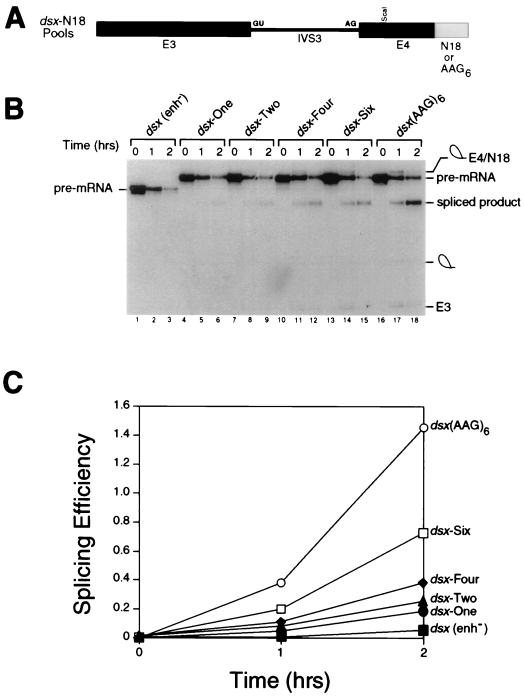
Several large-scale in vitro splicing reactions were assembled so that at least four complete pools of randomized sequences (shown schematically in Fig. 2A) were sampled for enhancer activity in the initial round of selection. Low levels of splicing could be observed in the second round of selection, and the efficiency and rate of splicing increased with each subsequent round of selection and amplification (Fig. 2B and C). The efficiency of splicing (defined as the ratio of spliced product to precursor; see the legend to Fig. 2) of the round 6 pool (Fig. 2B, lanes 13 to 15) was approximately 50% of that observed with a well-characterized strong splicing enhancer (the repeated polypurine sequence AAG [66]) (Fig. 2B, lanes 16 to 18; see Fig. 2C for quantitation). The selection was terminated after six rounds to examine the largest possible collection of functional splicing enhancers for common sequence motifs and to avoid the dilemma of isolating a single “winner” sequence.
FIG. 2.
Evolution of the dsx-N18 pool. (A) The dsx-N18 and dsx-(AAG)6 constructs are shown schematically. Exon 3, intron 3, exon 4, and the enhancer(s) are indicated by E3, IVS3, E4, and N18 or AAG6, respectively. The 5′ and 3′ splice sites are indicated by GU and AG, respectively. (B) Kinetic analysis showing in vitro splicing assays performed with HeLa cell nuclear extracts and uniformly labeled pre-mRNA splicing substrates comprising the total pool of dsx-N18 pre-mRNAs after various rounds of the selection (rounds 1, 2, 4, and 6 are shown in lanes 4 to 6, 7 to 9, 10 to 12, and 13 to 15, respectively). The negative control pre-mRNA (lanes 1 to 3) is an dsx pre-mRNA lacking an enhancer [dsx(enh−)]. The positive control pre-mRNA (lanes 16 to 18) is a dsx pre-mRNA activated by six consecutive copies of a multimerized AAG trinucleotide splicing enhancer (modeled after a synthetic polypurine splicing enhancer in reference 66) that is otherwise isogenic to the dsx-N18 construct. In the kinetic analysis shown, the reaction mixtures were incubated for the number of hours indicated at the top, and positions of the precursors, intermediates, and products of the splicing reaction are indicated to the left and right. The RNAs were analyzed on a 10% denaturing gel in order to resolve the lariat-exon 4 intermediate from the spliced product. (C) Quantitation of the in vitro splicing reactions in panel B. The splicing efficiency (ratio of spliced product to precursor) is calculated from quantitation of individual bands after subtraction of background using a BAS2000 phosphorimager.
Analysis of selected splicing enhancer sequences.
A total of 84 individual clones recovered after rounds 3 and 6 of selection were sequenced and analyzed for in vitro splicing efficiency. The splicing efficiencies of the control pre-mRNAs varied from essentially zero for individual dsx pre-mRNAs lacking an enhancer to 81% conversion to spliced product for an hβ-globin pre-mRNA splicing substrate (Table 1, controls). Several of the individual round 6 clones had splicing efficiencies greater than 50%, which compares favorably with the dsx–avian sarcoma-leukemia virus (ASLV) pre-mRNA’s splicing efficiency of 58%. The round 3 and round 6 clones were divided into seven classes based on their sequences. Class I and class II clones (Table 1) are sequences that are predominantly purine rich and pyrimidine rich, respectively; class III to VI clones (Table 2) encode recurring sequence motifs found in clones that have strong splicing enhancer activities; and class VII clones (Table 2) consist of clones isolated only once that have particularly potent splicing enhancer activities. Note that some potent splicing enhancers are represented in more than one class, consistent with the notion of a composite splicing enhancer (16, 39). For example, clone 6-38 has two nonoverlapping sequences representative of both class III and class V consensus sequence motifs (Table 2).
TABLE 1.
Purine- and pyrimidine-rich sequences in the selected splicing enhancers
| Clone no. | Splicing enhancer sequencea | Splicing efficiencyb (%) |
|---|---|---|
| Class I, purine-rich enhancers (≥65% purine content) | ||
| Motif A, GGGGA | ||
| 3-7 | GCAACGGGGACGCGGC | 40 |
| 3-1 | AGCGGUCGCGGUUGGGGGag | 32 |
| 6-43 | GCGGAGGAGGCCCGUGGGag | 50 |
| Motif B, GGAGGA | ||
| 6-43 | GCGGAGGAGGCCCGUGGGag | 50 |
| 6-19 | GCCAGCGGAGGAUGCGG | 53 |
| Motif C, GGAGA | ||
| 3-35 | CUGGAAUACGGAGACCGG | 36 |
| 6-40 | GGUGAGCGGAGAUGCUGC | 31 |
| Others | ||
| 3-36 | GGACCUAGAGGUGGCGAC | 40 |
| 6-29 | GACCGUCGGACAGGAGC | 36 |
| Class II, pyrimidine-rich enhancers (≥67% pyrimidine content) | ||
| Motif D, UCUCC | ||
| 6-13 | auCUCCACGUCGCCUGCUGC | 38 |
| 6-16 | auCUCCACGUCGCCUGCUGC | 37 |
| 6-24 | UUUGCGGUCUCCGGCCUCC | 56 |
| Motif E, UCUUC | ||
| 6-5 | UGCCACCCGCGGUCUUCC | 26 |
| 6-12 | UCGUCGUCUUCGCGGCCC | 49 |
| 3-32 | CCUGCUGCGUCUUUGUCC | 27 |
| Motif F, UCCUC | ||
| 6-7 | CCUGUCCUCGGUGUUGC | 36 |
| 6-22 | CGUCCUCGUGUCACCGCC | 37 |
| 6-6 | GGUUCCUGUCGCCGCCCC | 41 |
| Controls (reference) | ||
| dsx, enhancerless | ≤1 | |
| hβ-globin (51) | 81 | |
| dsx-ASLV (60) | 58 |
Boldface letters indicate purine- and pyrimidine-rich tracts consisting of five or more consecutive nucleotides; uppercase letters indicate splicing enhancer sequences specific to the indicated clone; lowercase letters indicate sequences common to all clones; underlining indicates the position of sequence deviation from the motif. The splicing enhancers are in the sequence context gcauNNNNNNNNNNNNNNNNNNaguc. Note that some of the enhancers contain 16, 17, or 19 nt.
Quantitated as described in the legend to Fig. 2 according to formula [S][S + P]−1 × 100, where S is the spliced product and P is the precursor RNA.
TABLE 2.
Recurring motifs in selected splicing enhancers and other strong enhancers
| Clone | Selected enhancer sequencea | Splicing efficiencyb (%) |
|---|---|---|
| Class III, enhancers containing permutations of the sequence (U)GGACCNG | ||
| 6-14 | GCCGCCGCUUCGUGGACCag | 53 |
| 6-25 | CACGCUCCUCGCUGGACCag | 53 |
| 6-38 | GCCGCCGUGGUGGACCGGag | 50 |
| 6-26 | CCGAGCUACAGGACCGGag | 35 |
| 6-29 | GACCGUCGGACAGGAGC | 36 |
| 3-35 | CUGGAAUACGGAGACCGGag | 36 |
| 3-36 | uGGACCUAGAGGUGGCGAC | 40 |
| 6-9 | uGGACCGCCCUGCCAUACC | 34 |
| 3-3 | CAGGCGGGACCGCGACG | 17 |
| Class IV, enhancers containing the sequence (C)CACC(C) | ||
| 6-28 | CCGGAGCCACCCGGUACC | 29 |
| 6-5 | UGCCACCCGCGGUCUUCC | 26 |
| 6-2 | CGUCGCACCCUGUCUGCC | 29 |
| 6-22 | CGUCCUCGUGUCACCGCC | 37 |
| 6-35 | UCCUGGCGUCACCGUAC | 27 |
| Class V, enhancers containing the sequence YGCCGCC | ||
| 6-14 | uGCCGCCGCUUCGUGGACC | 53 |
| 6-38 | uGCCGCCGUGGUGGACCGG | 50 |
| 6-45 | uGCCGCCGCGAGUUGGGGC | 32 |
| 6-8 | GCCAGUAGUUGCCGCCGC | 24 |
| 6-6 | GGUUCCUGUCGCCGCCCC | 41 |
| 6-1 | GGACACCUGUGCGCCGCCag | 43 |
| Class VI, enhancers containing the sequence RGAACYU | ||
| 3-25 | CCACGUGGAACCUCGUCC | 35 |
| 6-44 | ACGGCGCGGAACCUUUCC | 47 |
| 6-23 | GCCCGAGAACUUCUUGCC | 40 |
| Class VII, other strong enhancers | ||
| 6-18 | CCGACGCCAUGGACGACGag | 55 |
| 6-3 | GGCUGCCAGUCGGAAUUGG | 52 |
| 6-47 | CCGUGACAGCAUCGGCGG | 50 |
| 3-23 | CGUCGGCAGGUGGUCCCG | 47 |
| 6-39 | UCUGGAUCCUGCGGAUGG | 44 |
(i) Enhancers with purine-rich sequences.
Class I sequences are splicing enhancers that are more than 65% purine rich (Table I). A notable feature of the selected sequences was the low frequency of extended tracts, operationally defined as five or more consecutive nucleotides, of either purines or pyrimidines. The purine-rich clones were further organized into three purine tract-containing sequence motifs (motifs A to C). Two round 6 clones, 6-19 and 6-43, share the purine-rich motif GGAGGA (motif B) that was earlier characterized as a synthetic (GGA)8 purine-rich enhancer capable of activating the dsx 3′ splice site (66). Clones 6-19 and 6-43 share the sequence submotif GAGGA, which is found in one copy in the fibronectin EDIIIA splicing enhancer (46) and in two copies each in the β-tropomyosin (25) and the cardiac troponin T (cTNT) (50) splicing enhancers (Fig. 3, comparison A). Substitution mutations within this sequence element in the cTNT enhancer severely affect the inclusion of the cTNT exon 5 (Fig. 3, comparison A; see also references 12, 13, and 50). This GAGGA sequence motif was also recovered in a previous in vitro selection for RNA sequences capable of stimulating inclusion of an internal exon (i.e., clones 5.8, 7.16, 7.19, 7.23, and 7.24 in reference 67) and in SELEX experiments performed on SF2/ASF and SC35 (clones A39, A48, A53, S31, and S38 in reference 64). Two of the purine-rich selected clones have the sequence YGGAGA (3-35 and 6-40; Table 1, class I, motif C). Two sequences matching this consensus were also recovered in an earlier functional selection (i.e., clones 5.9 and 5.42 in reference 67).
FIG. 3.
Sequence comparison of individual dsx-N18 enhancers to other naturally occurring splicing enhancer sequences characterized previously by point mutation(s), block substitutions, or deletion mutations. Sequence homology is indicated by boxes. Individual dsx-N18 clone sequences and consensus motifs are designated as in Tables 1 and 2. The ASLV-6U mutant (mut.) shown has no loss of splicing enhancer function compared to the wild type (wt) yet shows an altered pattern of cross-linked factors (60). The third of the six point mutations in the ASLV-6U mutant generates a better match to the class V consensus motif. All other mutants result in at least some loss of splicing enhancer function in vitro or in vivo. Sequence polymorphisms, point mutations, and substitution mutations are underlined. Deletions are indicated by Δ. Due to the large size of some deletions, some sequence information inclusive of the deletion is omitted and indicated by an ellipsis.
(ii) Enhancers with pyrimidine-rich sequences.
Class II sequences are splicing enhancers that are more than 67% pyrimidine rich (Table 1). Interestingly, sequences that are predominantly pyrimidine rich were not recovered in any of the previous selections for functional splicing enhancers. The class II sequences were further organized into three pyrimidine tract-containing sequence motifs based on sequence homology within tracts of pyrimidines (motifs D through F). Two of the pyrimidine-rich clones, 6-13 and 6-16, are independent isolates of the same enhancer sequence (as the consequence of either separate isolates of two different molecules or a single isolate yielding two bacterial colonies). Splicing enhancer sequences that are as much as 78% pyrimidine rich, such as clones 6-13 and 6-16, are capable of functioning as strong splicing enhancers. A pyrimidine tract within these two pyrimidine-rich splicing enhancers (motif D) contains an excellent match to the 5′ half of the dsx repeat element consensus, UC(U/A)(U/A)C (8, 49). Motifs D, E, and F are all highly homologous to the 5′ half of the dsx repeat element, a sequence which is phylogenetically conserved (26), cross-links a specific SR protein in extracts (42), and is required for enhancer-dependent splice site activation (31). The most potent class II enhancer, 6-24, which is 74% pyrimidine rich and shares motif D with 6-13 and 6-16, is activated by specific SR proteins in splicing complementation assays (see below), thus demonstrating a role for pyrimidine-rich sequences in splice site activation in addition to the more typical purine-rich sequences.
(iii) Enhancers that are similar to sequences in naturally occurring exons.
Clone 3-36 contains a 13-nt sequence which is nearly identical to a naturally occurring splicing enhancer that we have identified in hβ-globin exon 2 (Fig. 3, comparison B) (57). This 13-nt sequence is completely conserved between human and rabbit β-globin genes over four wobble positions in the protein coding sequence, and a specific double-point mutation of this sequence inactivates its splicing enhancer function (Fig. 3, comparison B) (57). This sequence motif is very similar to a degenerate sequence motif that was independently isolated nine times in our selection (Table 2, class III). Clone 6-29 contains one permutation to the class III motif consensus GGACCNG (Table 2) and actually shares a larger region of homology to a naturally occurring sequence in the splicing enhancer in the mouse immunoglobulin M (IgM) exon 2 (73) (Fig. 3, comparison C). In addition, the class III clones 6-26 and 6-38 each share an extended region of homology with both clone 6-29 and mouse IgM splicing enhancer (Fig. 3, comparison C).
Class VII clone 6-18 (Table 2), one of the most efficiently spliced clones isolated in our selection, contains significant sequence homology to the splicing enhancer in cTNT exon 5 (Fig. 3, comparison D). Mutation of this particular sequence within the cTNT splicing enhancer compromises its splicing enhancer efficiency (13). Notably, this sequence is virtually identical to one of the consensus high-affinity binding sites identified by SELEX for the SR protein 9G8 (8a; see also Discussion). Another sequence motif with homology to naturally occurring enhancers was isolated six times independently in our selection (Table 2, class V). A similar sequence motif is found naturally occurring in both the bovine growth hormone (bGH) and ASLV splicing enhancers (Fig. 3, comparison E). A deletion (23) and a sextuple point mutant (60) overlapping this sequence (Fig. 3, comparison E) have been analyzed in the bGH and ASLV enhancers, respectively, but the individual sequence element remains uncharacterized (see also the legend to Fig. 3).
(iv) Other enhancer sequence motifs.
Several other sequence motifs were recovered several times in our screen for splicing enhancers. Several clones contain the sequence (C)CACC(C) (6-2, 6-5, 6-22, 6-28, and 6-35 [Table 2, class IV]). This sequence most likely corresponds to a class of A/C-rich splicing enhancers recovered in a previous selection for functional enhancers (14). Finally, the two class VI clones 3-25 and 6-44 have a splicing efficiency of greater than 35% and share sequence motif (Table 2). A third class VI clone, 6-23, shows a more limited homology (Table 2) with these two clones. To our knowledge, no previously characterized splicing enhancers contain this sequence motif.
Individual SR proteins specifically activate the splicing of pre-mRNA substrates containing the selected enhancers.
Previous studies have demonstrated that some splicing enhancers bind SR proteins (38, 50, 60, 62, 69). Certain pre-mRNAs can also be differentially committed to the splicing pathway by SR proteins (10, 17, 69), but the specific sequence elements responsible for this function remain uncharacterized. In addition, two purine-rich splicing enhancer sequences isolated by SELEX can function as splicing enhancers in nuclear extracts (63, 64) and SF2/ASF- or SRp40-specific enhancers in S100 complementation reactions (33, 63, 65). We have used the S100 complementation assay (35) to determine whether pre-mRNA splicing substrates containing our enhancers selected in nuclear extracts can be activated by specific recombinant SR proteins, such as those recently characterized for SF2/ASF, SRp40, and SRp55 (39). The individual recombinant SR proteins 9G8, SC35, and SF2/ASF were tested with dsx premRNAs containing the selected enhancers or other well-characterized splicing enhancers (28, 43, 57) as positive controls.
(i) SC35.
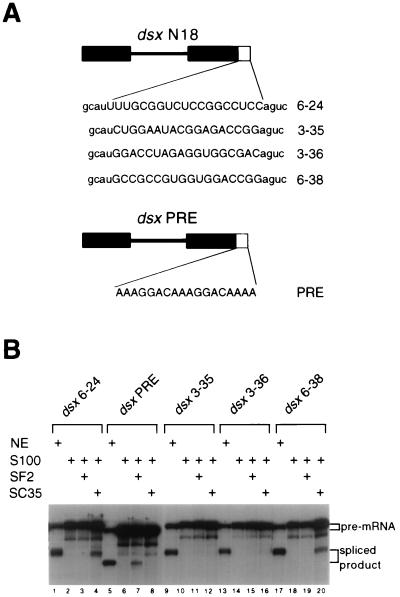
The individual dsx pre-mRNAs containing the selected enhancer clones (Fig. 4A) were tested in S100 assays for complementation by the recombinant SR proteins SF2/ASF and SC35 (19, 20, 37). Our initial result was that one of the pyrimidine-rich splicing enhancers, 6-24, was efficiently and specifically complemented by the SR protein SC35 but not SF2/ASF (Fig. 4B, compare lanes 4 and 3). The dsx-PRE construct (28) was used as a specificity control for SF2/ASF, as it is efficiently complemented by SF2/ASF but not SC35 (Fig. 4B; compare lanes 7 and 8). To determine whether the ability to be complemented by SC35 is a general property of pyrimidine-rich enhancers, selected clones containing pyrimidine-rich enhancers (Table 1, class II) were tested in SC35 complementation reactions. Selected clones containing non-pyrimidine-rich enhancers, such as 3-35, 3-36, and 6-38 (Fig. 4B, lanes 9 to 20), were used as specificity controls. Even though some of the pre-mRNAs containing pyrimidine-rich enhancers tested (such as 3-32, 6-5, and 6-12 [data not shown]) showed modest levels of SC35-dependent complementation, pyrimidine richness does not seem to be an intrinsic property of SC35-dependent enhancers. The two most potent SC35-dependent enhancers were 6-24 and 6-38, a pyrimidine-rich enhancer and a non-pyrimidine-rich enhancer, respectively (Fig. 4B, lanes 4 and 20). In parallel complementation reactions, dsx-N18 clones 6-24 and 6-38 show efficient complementation with SC35 but not SF2/ASF (Fig. 4B; compare lanes 4 and 3 and lanes 20 and 19). The otherwise isogenic clones 3-35 and 3-36 show levels of splicing enhancer activity similar to those of 6-24 and 6-38 in nuclear extracts (Fig. 4B, compare lanes 1, 9, 13, and 17), but clones 3-35 and 3-36 are complemented neither by SC35 nor by SF2/ASF (Fig. 4B, lanes 11 and 12 and lanes 15 and 16, respectively). Both clones 3-35 and 6-38 are members of the class III sequence motif (Table 2) and fortuitously share the identical RGACCGG sequence at the 3′ half of their splicing enhancers (positions 13-18 within their 18-nt splicing enhancers; Fig. 4A). Thus, clones 3-35 and 6-38 differ from each other at only 13 of 390 positions within their pre-mRNAs, yet they differ dramatically in the ability to be complemented by SC35 (Fig. 4B; compare lanes 12 and 20). This directly implicates the 5′ half of the 6-38 enhancer in SC35-dependent activation. Clones 6-38 and 6-14 have the class V sequence motif at identical positions at the 5′ end of their enhancers, and clone 6-14 can be complemented by SC35, albeit less strongly than 6-38 (data not shown). 6-24 and the 5′ half of 6-38 show sequence homology to each other as well as to an SC35-dependent splicing enhancer derived from hβ-globin exon 2. The hβ-globin exon 2 SC35-dependent splicing enhancer has been characterized by detailed mutagenesis and site-specific cross-linking studies (57). Each of these SC35-dependent splicing enhancers has the sequence UGCNGYY, a sequence motif not found in the PRE, 3-35, or 3-36 splicing enhancer (see also Discussion). We conclude that otherwise isogenic dsx pre-mRNA substrates can be differentially complemented by the SR protein SC35, and these SC35-responsive clones share sequence homology with each other. In addition, we directly demonstrate that splicing enhancers with pyrimidine-rich sequence compositions or with balanced pyrimidine-purine sequence compositions can function as SR-specific splicing enhancers in S100 assays.
FIG. 4.
Functional characterization of SC35-dependent enhancers. (A) The dsx pre-mRNA substrates are indicated schematically, with the dsx portion shown in black. Sequences of the different splicing enhancers tested are in capital letters; sequences common to all dsx-N18 clones are in lowercase letters. The dsx-PRE construct is similar but not identical to the dsx-N18 constructs, as it does not contain an inert Sa element. (B) Two dsx-N18 clones that show sequence homology are specifically activated by SC35. The pre-mRNA used in the S100 complementation reactions is indicated above the autoradiogram. The presence of the indicated reaction component in splicing assays and complementation reactions is indicated by a plus sign above the appropriate reaction lane: HeLa cell nuclear extract (NE), HeLa S100 extract complemented with buffer D (S100), or the SR protein indicated by a plus sign. Amounts of SC35 and SF2/ASF used in the complementation assays were 400 and 200 ng, respectively. The dsx-N18 and dsx-PRE pre-mRNAs and spliced products are resolved on a 10% denaturing polyacrylamide gel; their positions are indicated to the right.
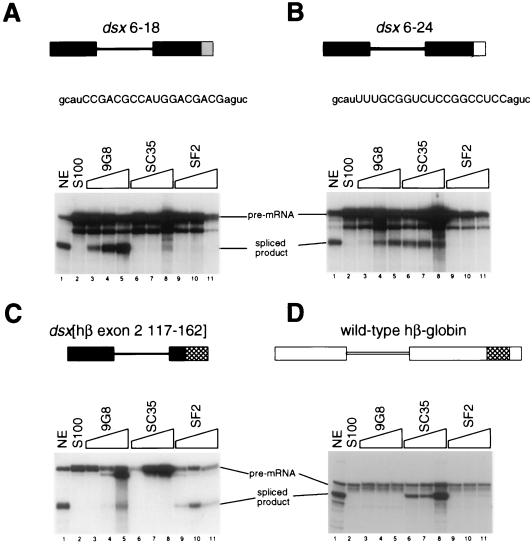
(ii) 9G8.
To determine whether the selected clones could be specifically complemented by SR proteins other than SC35, complementation reactions were performed with recombinant 9G8 (9). Class VII clone 6-18 is one of the most potent splicing enhancers in nuclear extracts (Table 2) and shows strong homology to a high-affinity 9G8 RNA binding site characterized by SELEX (8a) (see Discussion). Clones 6-18, 6-24 (an SC35-dependent enhancer [Fig. 4B]), a dsx pre-mRNA containing an hβ-globin-derived SF2/ASF-dependent enhancer (57), and the wild-type hβ-globin pre-mRNA were tested in parallel S100 assays in which the recombinant SR proteins 9G8, SC35, and SF2/ASF were each titrated. The selected clone 6-18 showed complementation at every concentration of 9G8 (Fig. 5A; compare lanes 2 to 5) tested in the titration. In contrast, clone 6-18 showed little or no activity in S100 extracts at any concentration of SC35 (Fig. 5A, lanes 6 to 8) or SF2/ASF (Fig. 5A, lanes 9 to 11) tested. By comparison, clone 6-24 showed efficient complementation at every concentration of SC35 tested (Fig. 5B, lanes 6 to 8). Surprisingly, clone 6-24 also showed efficient complementation at the two highest concentrations of 9G8 tested (Fig. 5B, lanes 3 to 5), but no complementation was observed at any concentration of SF2/ASF (Fig. 5B, lanes 9 to 11). The SR protein SF2/ASF did not function in complementation assays with either clone 6-18 (Fig. 5A, lanes 9 to 11) or clone 6-24 (Fig. 5B, lanes 9 to 11), but SF2/ASF showed complementation activity at every concentration tested (Fig. 5C, lanes 9 to 11) on a dsx chimeric pre-mRNA containing a β-globin-derived SF2/ASF-dependent splicing enhancer (57). This pre-mRNA was also efficiently complemented by 9G8 at the two highest concentrations tested (Fig. 5C, lanes 3 to 5), and no complementation was observed with SC35 at any concentration tested (lanes 6 to 8). The SF2/ASF-dependent enhancer has some sequence homology with a motif found in two copies in the dsx-PRE construct (57). Consistent with these observations, the dsx PRE can be efficiently complemented by both SF2/ASF and 9G8 (29) but not by SC35 (Fig. 4, lanes 7 and 8; see also reference 28). The wild-type β-globin substrate was efficiently complemented at every concentration of SC35 tested (Fig. 5D, lanes 6 to 8), modestly complemented at every concentration of SF2/ASF tested (lanes 9 to 11), and not complemented with 9G8 (lanes 3 to 5). Based on the findings that each SR protein was titrated within its linear range of complementation activity (data not shown) and that there is at least one pre-mRNA for which each SR protein gives efficient complementation at every concentration tested (and at least one pre-mRNA in which no complementation was observed at every concentration tested), we conclude that dsx chimeras containing different enhancer sequences can be differentially complemented by the SR protein 9G8, SC35 and SF2/ASF. Interestingly, the SR protein 9G8 can efficiently complement both clones 6-18 and 6-24, containing very disparate sequences, whereas SC35 efficiently complements clone 6-24 but not 6-18 (see Discussion).
FIG. 5.
Functional characterization of 9G8-dependent enhancers. (A) The dsx 6-18 pre-mRNA is activated efficiently by 9G8 in S100 complementation assays. The presence of the indicated reaction component in splicing assays and complementation reactions is indicated above the appropriate reaction lane: NE, HeLa cell nuclear extract (lane 1); S100, HeLa S100 extract complemented with buffer D (lane 2). The relative SR protein concentration for each series of SR protein titrations (indicated by a gradient above the lanes) increases by factors of 1.00, 1.50, and 2.25. The actual amounts of SR protein tested in the complementation assays were as follows: 9G8, 67 ng (lane 3), 100 ng (lane 4), and 150 ng (lane 5); SC35, 267 ng (lane 6), 400 ng (lane 7), and 600 ng (lane 8); SF2/ASF, 89 ng (lane 9), 133 ng (lane 10), and 200 ng (lane 11). These amounts were empirically determined to be within the linear complementation range of each SR protein’s specific activity (data not shown). The dsx RNA substrates are indicated schematically, and labeling is as in Fig. 3. The RNA substrates and splicing products were resolved on a 10% denaturing polyacrylamide gel. The reactions were performed in parallel with those in panels B to D. (B) The dsx 6-24 pre-mRNA is activated efficiently by both SC35 and 9G8. (C) The dsx[hβ 117-162] pre-mRNA (57) is activated efficiently by SF2/ASF and by 9G8. (D) The wild-type hβ-globin pre-mRNA (51) is activated efficiently by SC35.
Interaction of specific SR proteins with the selected splicing enhancers.
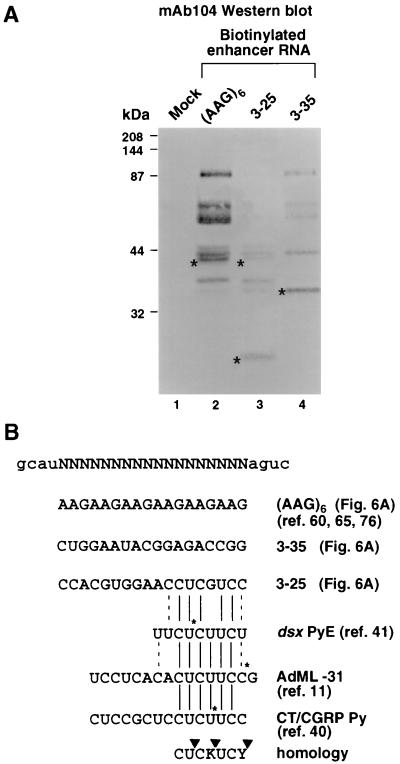
Strong splicing enhancers isolated in the selection were individually tested in the absence of any dsx sequences for the ability to interact with specific SR proteins. Biotinylated splicing enhancer RNAs were incubated in nuclear extracts under splicing conditions, isolated with strepavidin-agarose, and washed under high-salt and detergent conditions as previously described (76). The SR proteins that remain bound to the enhancers were analyzed by Western blotting with MAb which recognizes a phosphoepitope common to many SR proteins (55). Analysis of the SR protein composition of the N18 clones in Fig. 6 shows that some SR proteins bind to each of the enhancers shown in Fig. 6A, but other SR proteins bind specifically to individual enhancers. This procedure may not be generally applicable for all splicing enhancer RNAs, as some enhancers, such as 3-36 and 6-43, bind only the SR proteins common to all the enhancers shown in Fig. 6A (data not shown), and other enhancer RNAs, such as 6-24, show no bound SR proteins (data not shown). This could be a consequence of the high-salt/detergent conditions used in the washes or the fact that individual enhancers recruit a specific trans-acting factor not recognized by MAb 104.
FIG. 6.
Binding of SR proteins to individual N18 enhancers. (A) Individual N18 enhancer RNAs were subcloned downstream of a T7 promoter, transcribed in the presence of biotin-21-UTP, and incubated in nuclear extracts by the method of Yeakley et al. (76). The RNA binding proteins were purified by using avidin-agarose and analyzed by Western blotting with MAb 104, which is immunoreactive to a phosphoepitope common to many SR protein family members. Mock refers to a control reaction without RNA included in the assay to determine the background levels of SR recruitment by the avidin-agarose resin in nuclear extracts. The 40-kDa SR protein (present in lanes 2 and 3 but absent in lane 4), the 20-kDa SR protein (present in lane 3 but absent in lanes 2 and 4), and the 35-kDa SR protein (present in lane 4 but absent in lanes 2 and 3) are indicated by asterisks. High-molecular-weight standards (prestained; Bio-Rad) are indicated to the left. (B) The 3-25 enhancer shows good homology to three sequences, dsx PyE (41), CT/CGRT Py (40), and AdML-31 (11), each of which contains a single site-specific label and can cross-link a 20-kDa protein. Solid lines indicate sequence identity, and dashed lines indicate conservative transition changes. Solid asterisks indicate positions of the engineered site-specific labels. The homology among the three sequences that bind SRp20 is CUCKUCY (where K is guanosine or uridine and Y is cytidine or uridine). Triangles integrate the positions of the three site-specific cross-linking experiments.
With these caveats in mind, the selected enhancer RNAs that can recruit a specific SR protein (as well as the SR proteins common to all clones) are shown in Fig. 6A. For example, a 40-kDa SR protein (Fig. 6A, lane 2; indicated with an asterisk), possibly one of the human Tra-2 homologues, binds to both the (AAG)6 enhancer as previously described (60, 65, 76) and the 3-25 enhancer (lane 3) but not the 3-35 enhancer (lane 4). Similarly, a 35-kDa SR protein of unknown identity binds specifically to the 3-35 enhancer (Fig. 6A, lane 4; indicated with an asterisk), and a 20-kDa SR protein (presumably SRp20) binds specifically to the 3-25 enhancer (lane 3; indicated with an asterisk). A mock incubation without RNA (Fig. 6A, lane 1) or with a nonbiotinylated RNA (i.e., not including biotin-21-UTP in the transcription reaction [data not shown]) results in no recruitment of SR proteins, not even those common to all clones. Because SR proteins are known to interact in vivo in the two-hybrid assay and are known to aggregate under low-salt conditions, one explanation for the above results is that one SR protein binds specifically to the RNA sequence and then nucleates the formation of an SR protein complex. Alternatively, the binding of multiple SR proteins to a single element may reflect the presence of multiple SR protein binding sites within the enhancer sequence. Interestingly, the 3-25 splicing enhancer shows significant sequence homology to exonic sequences shown in three earlier studies to cross-link a 20-kDa protein by using pre-mRNAs containing a single site-specific label (11, 40, 41).
A previous site-specific labeling experiment showed that a 20-kDa protein binds to an RNA sequence located upstream of the adenovirus major late (AdML) 5′ splice site in isolated E complex (11). The 20-kDa protein cross-linked to this RNA when a single 32P-labeled phosphate was located at −31 relative to the 5′ splice site (AdML −31 [Fig. 6B]), but not when the label was located at −26, −19, −15, or −3 relative to the 5′ splice site (11). A 20-kDa protein cross-links to an 8-nt spacer region (dsx PyE [Fig. 6B]) located between dsx repeats 3 and 4 in the D. melanogaster dsx repeat element containing a single site-specific label (41) but not to a similar single label positioned at various locations within the adjacent dsx repeat 4 (42). In addition, a recent study of SRp20’s effect on polyadenylation/exon inclusion showed that an SRp20-specific antibody immunoprecipitates SRp20 cross-linked to a CT/CGRP core enhancer element pyrimidine tract sequence (CT/CGRP Py [Fig. 6B]) containing a site-specific label that has strong homology to the 3-25 enhancer. Finally, we have detected the binding of a 20-kDa protein to an RNA containing the 3-25 enhancer and showed that it is an SR protein by virtue of its immunoreactivity with MAb 104 (Fig. 6A, lane 3). Significantly, all sequences implicated in binding the 20-kDa SR protein SRp20 share the sequence CUC(U/G)UCY (Fig. 6B).
DISCUSSION
We have used the enhancer-dependent dsx pre-mRNA substrate and an in vitro selection/amplification strategy to identify novel splicing enhancer sequences. Analysis of these sequences reveals that an extraordinarily large number of them can function as splicing enhancers. How can this sequence diversity be explained if all splicing enhancers are recognized by SR proteins? First, it is possible that there are numerous yet to be discovered SR proteins. Second, individual SR proteins may be capable of binding to relatively degenerate RNA sequences. Third, SR proteins may be capable of forming distinct SR protein complexes, each capable of recognizing a different sequence. There is evidence for each of these possibilities. The first possibility is supported by the observation that a number of MAbs raised against nuclear matrix proteins react with proteins shown to bear striking similarities with known SR proteins (7). The authors of that study speculate that there may be a large number of yet to be characterized SR proteins. However, additional studies will be required to determine whether these SR-like proteins are capable of functioning as splicing enhancer factors. The second possibility is supported by a recent analysis of SF2/ASF-dependent splicing enhancers identified by in vitro selection (39). These authors characterized several such enhancer elements, and the consensus sequence derived from the analysis of these elements is highly degenerate. Similarly, our studies on dsx-N18 pre-mRNAs that are complemented by SC35 are consistent with a degenerate consensus sequence of UGCNGYY (58), with the more potent clones having more homology to the UGCYGUU sequences (57) found in β-globin exons 1 and exon 2. The third possibility is consistent with the analysis of SR protein complexes that bind to individual dsx repeat elements (42). A heterotrimeric complex consisting of the Tra and Tra-2 proteins and a specific SR protein binds to each repeat. The SR and Tra-2 proteins contact the 5′ and 3′ ends, respectively, of each repeat, while Tra is required for the formation of the complex. Thus, all three possibilities may be responsible for the sequence diversity of splicing enhancers.
A distinguishing feature of the splicing enhancers recovered in our in vitro selection is the relative paucity of purine-rich sequences. With few exceptions, most previously characterized naturally occurring splicing enhancers are purine rich, as were most of the splicing enhancers isolated in a cis-competition screen (67). It should be noted that the identification of naturally occurring splicing enhancer elements has been biased toward those exonic elements found in alternatively spliced pre-mRNAs in which the alternative splicing enhancer switches splice site utilization to the weaker of two competing splice sites. Similarly, the purine-rich sequences isolated in the Tian and Kole in vitro selection (67) were performed as a cis-competition assay (52) against the constitutively spliced, full-length β-globin exon 2, an exon recently shown to have multiple SR-dependent splicing enhancers (57). By definition, these enhancers would have to be strong enough to outcompete the additive sum (28) of the multiple splicing enhancers present in the full-length β-globin exon 2 (57) in order to activate the internal 3′ splice site of two competing 3′ splice sites. These observations raise the interesting possibility that constitutively spliced exons utilize exon recognition elements that are mostly balanced or pyrimidine rich in nucleotide composition (such as those isolated in our selection), whereas the alternative splice site utilization may be controlled by factors which recognize the purine-rich elements typically found downstream of 3′ splice sites subject to alternative exon inclusion (such as those isolated in the Tian and Kole selection). Consistent with this notion that constitutive splicing enhancers may be mostly balanced or pyrimidine rich in composition, a number of short hβ-globin exon 2-derived splicing enhancers (for example, the exon 2-derived 13-24 and 63-80 sequences [57]), and a majority of enhancers isolated in our selection are balanced or pyrimidine rich in nucleotide composition (Tables 1 and 2). In theory, the strength of a strong composite enhancer might be the sum of the RNA binding affinities of the RNA binding domains, the intrinsic strength of activation of the SR domains (21), the additivity conferred by multiple enhancer complexes (28), the possible effects of cooperativity of binding (28) by protein-protein interactions (i.e., by Tra-like factors), and the distance from the regulated 3′ splice site (21). A strong composite enhancer containing one or more closely spaced (purine-rich) binding sites might now be sufficient to outcompete the multiple splicing enhancers (pyrimidine-rich or balanced nucleotide composition) found in a constitutive exon in a sensitive assay such as the cis competition. In addition to the general observation that splicing enhancers typically found in exons downstream of introns subject to alternative splicing are purine rich (for reviews, see references 6 and 45), two specific examples are consistent with this notion. Purine-rich sequences (as well as A/C-rich sequences [14]), were isolated in both in vitro (67) and in vivo (14) selection strategies based on alternative exon inclusion of an exon containing a randomized sequence that is normally skipped in a cis-competition assay against the constitutive β-globin exon 2.
Identification of degenerate RNA consensus sequences for SC35- and 9G8-mediated splice site activation.
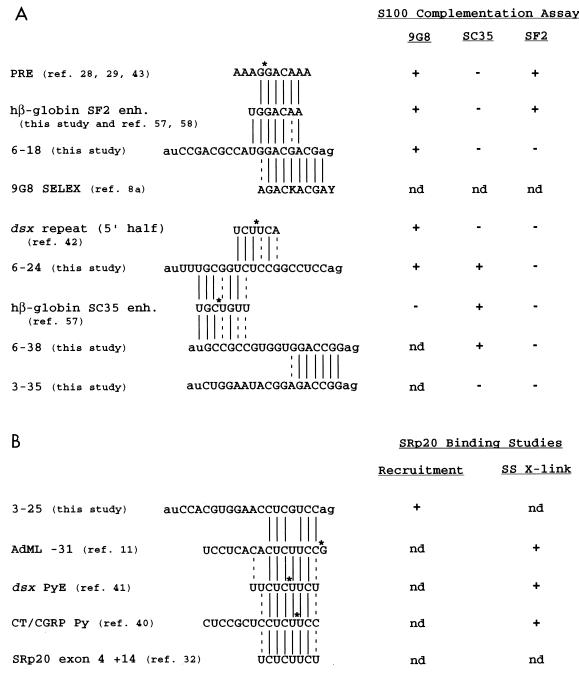
In this study, we characterized two different 18-nt enhancer sequences that could be specifically activated by 9G8 and two independent enhancer sequences that could be specifically activated by SC35. The 9G8-dependent enhancers show homology to two other sequences implicated in 9G8 binding studies (SELEX [8a] and site-specific cross-linking [42]), and the SC35-dependent enhancers show sequence homology with each other as well as other SC35-dependent enhancers (57). None of these enhancers could be efficiently activated by SF2/ASF, although two different control pre-mRNAs containing the PRE or nt 117 to 162 from hβ-globin exon 2 could be efficiently activated by SF2/ASF. These results are summarized in Fig. 7.
FIG. 7.
Summary of SR protein-specific enhancer sequences. Splicing enhancers (enh.) show specificity in their binding of or activation by individual SR protein family members as characterized by RNA binding studies or S100 biochemical complementation assays. Results for S100 extracts complemented with recombinant SR protein 9G8, SC35, or SF2/ASF (A) and results of studies indicating specific binding of SRp20 (B) are shown. The recruitment assay is the biotinylated RNA affinity technique performed as described by Yeakley et al. (76), and the site-specific cross-linking (SS X-link) refers to the technique developed by Moore and Sharp (48) to incorporate a single 32P-labeled phosphate into a pre-mRNA substrate. Asterisks indicate positions of single-labeled phosphates positioned within functional pre-mRNAs of dsx PRE (42), hβ-globin SC35 enhancer (57), dsx PyE (41), and AdML (11) or of a site-specifically labeled CT/CGRP Py ribo-oligonucleotide (40). The dsx repeat element (5′ half) shown is an A-type repeat element that is found in three of the six D. melanogaster repeats (31) and all four of the Drosophila virilis repeats (26). Solid lines indicate sequence identity, and dashed lines indicate conservative transitions between splicing enhancer sequences. A putative SRp20 site, based on its sequence identity with the dsx PyE site, is located downstream of an autoregulated 3′ splice site in the SRp20 pre-mRNA (32). K, guanosine or uridine; nd, not determined.
Clones 6-18 and 6-24 contained 9G8-dependent splicing enhancers even though their enhancers show widely disparate primary sequences. Clone 6-18 contains the sequence GGACGACGAG, which shows strong homology to the 9G8 consensus sequence AGAC(G/U)ACGAY isolated by SELEX (Fig. 7 and reference 8a). The pyrimidine-rich clone 6-24 shows no sequence homology with the SELEX sequence but does show some sequence homology with the 5′ half of the dsx repeat element consensus (Fig. 7). The 5′ half of the dsx repeat element was previously shown to site specifically cross-link the 9G8 protein in HeLa cell nuclear extracts supplemented with Tra and Tra-2 (42). The binding of 9G8 with Tra and Tra-2 to the dsx repeat element is cooperative, and this cooperativity is presumably necessary for the stable complex formation required for activation at a distance (28). When the dsx repeat element is artificially positioned within 100 nt of the dsx 3′ splice site, it is Tra and Tra2 independent (70) and can be complemented in a dsx activation assay using S100 extracts supplemented with recombinant 9G8 (29). Thus, it is conceivable that 9G8 recognizes two very different, functionally relevant RNA sequences: one similar to the SELEX consensus sequence in clone 6-18 and one similar to the 5′ half of the dsx repeat element-like sequence in clone 6-24. The 9G8 protein has been proposed to have two (RRM-type and zinc knuckle-type [9]) RNA binding domains. Interestingly, a SELEX performed with a mutant zinc knuckle region of 9G8 identified a consensus sequence very similar to the dsx repeat element 5′ half (8a). It is possible that the two RNA binding domains operate independently of each other to recognize the disparate sequences in clones 6-18 and 6-24. A precedent for RNA binding domains operating independently of each other in an SR protein containing two RNA binding domains was suggested by the experiments of Chandler et al. (10). The authors showed that an SF2/ASF derivative lacking one of the RRMs, but not the reciprocal construct, could function comparably to the full-length SF2/ASF in a commitment complex functional assay.
Clones 6-24 and 6-38 contained SC35-dependent splicing enhancers, and the enhancer sequences show significant sequence homology with each other as well as with a recently identified SC35-dependent enhancer in hβ-globin exon 2. The β-globin exon 2-derived sequence UGCUGUU was subjected to mutagenesis and site-specific cross-linking studies for SC35-dependent activation and RNA binding (57). The most homologous sequences in the two strongest SC35-dependent enhancers identified among the dsx-N18 clones are the UGCGGUC sequence in 6-24 and the UGCCGCC sequence in 6-38. We propose the degenerate consensus RNA binding sequence UGCNGYY for SC35, a protein predicted to have only a single RRM-type RNA binding domain.
Identification of degenerate RNA binding sequence for SRp20.
We also identified another 18-nt enhancer capable of specifically recruiting the SR protein SRp20, using a biotinylated RNA affinity technique. This enhancer bears a strong sequence homology to the sequences implicated as SRp20 binding sites in three independent site-specific cross-linking studies. The functional significance of this sequence homology is that an exon whose inclusion is regulated by SRp20 shares multiple potential SRp20 sites, and a functional SRp20 binding site in the CT/CGRP enhancer enhances both polyadenylation and exon inclusion of an upstream exon. By using a biotinylated RNA affinity technique (76), the enhancer of clone 3-25 was shown to specifically bind a 20-kDa SR protein, whereas the enhancers of clones 3-35 and (AAG)6 did not bind a 20-kDa SR protein. No other enhancers tested with this assay were able to interact with a 20-kDa SR protein (data not shown). Clone 3-25 shows significant sequence homology with two other exonic sequences characterized by using site-specific cross-linking. A single-labeled phosphate was positioned upstream of the 5′ splice site in the AdML pre-mRNA and a 20-kDa protein specifically cross-linked at or near position −31 but not at adjacent positions (11). Similarly, a single-labeled phosphate was positioned in the 8-nt inter-repeat spacer between Drosophila melanogaster repeats 3 and 4 and shown to cross-link a 20-kDa protein (41). A recent study demonstrated that SRp20 binds to the CT/CGRP polyadenylation enhancer and increases an upstream exon’s inclusion in cultured cells transfected with SRp20 but not SF2/ASF, Drosophila SRp55, or an SRp20 derivative lacking an activation domain (40). This study was able to confirm the identity of SRp20 by immunoprecipitating a cross-linked RNA containing a site-specific label within this sequence by using an SRp20-specific antibody (40). The authors of each of these site-specific cross-linking studies (11, 40, 41) suggest that the 20-kDa protein might be SRp20. Here we show that a sequence with strong homology to these sequences interacts with a 20-kDa SR protein. Recent studies have shown that SRp20 pre-mRNA is autoregulated by its own gene product to encode a protein with a truncated SR domain (32), a domain necessary and sufficient to activate enhancer-dependent splicing of other pre-mRNAs containing weak 3′ splice sites (22). In cultured cells, transfected SRp20 is able to promote the inclusion of exon 4, which is usually skipped (32). We propose that the mechanism of activation might be due to one or more SRp20 binding site(s) within the regulated exon 4. There is a perfect match over eight consecutive nucleotides between the PyE site that was shown to cross-link SRp20 in dsx repeat element and a putative SRp20 site located within exon 4 at a position +14 relative to the weak 3′ splice site in intron 3 in the autoregulated SRp20 pre-mRNA. The sequence homology shared between the sequences implicated in SRp20 RNA binding is the degenerate sequence CUC(U/G)UCY. In total, the results of this work and previous studies suggest that both SELEX-isolated high-affinity consensus binding sites (8a, 33, 63–65) and more degenerate (and possibly lower-affinity) binding sites (this report and references 14, 39, and 67) can function as exonic splicing enhancers.
ACKNOWLEDGMENTS
We thank Brenton Graveley, Klemens Hertel, Jim Olesen, Jinghua Yang, and other members of the Maniatis lab, as well as Tom Cooper (Baylor University), Xiang-Dong Fu (University of California, San Diego), Kevin Jarrell (Boston University School of Medicine), Hong-Xiang Liu (Cold Spring Harbor Laboratory), Kristen W. Lynch (University of California, San Francisco), Robin Reed (Harvard Medical School), and Ming Tian (Harvard Medical School), for helpful discussions, encouragement, and critical comments on the manuscript. We are grateful to Jim Bruzik (Case Western Reserve University) for his S100 extract preparation protocol; Renate Gattoni and James Stévenin (CNRS, Strasbourg, France), Adrian Krainer (Cold Spring Harbor Laboratory), and Mark Roth (Fred Hutchinson Cancer Research Center) for MAbs and hybridomas; Klemens Hertel for permission to cite unpublished data; Cyril Bourgeois and J. Stévenin (CNRS, Strasbourg, France) for communicating results prior to publication; and Dave Smith (Harvard University Biological Laboratories Imaging Center) for help with preparation of figures.
This work was supported by a National Institutes of Health genetics training grant to T.D.S. and National Institutes of Health grant GM42231 to T.M.
REFERENCES
- 1.Abmayr S M, Workman J L. Preparation of nuclear and cytoplasmic extracts from mammalian cells. In: Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1987. pp. 12.1.1–12.1.9. [Google Scholar]
- 2.Adams M D, Rudner D Z, Rio D C. Biochemistry and regulation of pre-mRNA splicing. Curr Opin Cell Biol. 1996;8:331–339. doi: 10.1016/s0955-0674(96)80006-8. [DOI] [PubMed] [Google Scholar]
- 3.Amrein H, Gorman M, Nöthiger R. The sex-determining gene tra-2 of Drosophila encodes a putative RNA binding protein. Cell. 1988;55:1025–1035. doi: 10.1016/0092-8674(88)90247-4. [DOI] [PubMed] [Google Scholar]
- 4.Amrein H, Hedley M L, Maniatis T. The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by Transformer-2. Cell. 1994;76:735–746. doi: 10.1016/0092-8674(94)90512-6. [DOI] [PubMed] [Google Scholar]
- 5.Baker B S. Sex in flies: the splice of life. Nature. 1989;340:521–524. doi: 10.1038/340521a0. [DOI] [PubMed] [Google Scholar]
- 6.Black D L. Finding splice sites within a wilderness of RNA. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- 7.Blencowe B J, Nickerson J A, Issner R, Penman S, Sharp P A. Association of nuclear matrix antigens with exon-containing splicing complexes. J Cell Biol. 1994;127:593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burtis K C, Baker B S. Drosophila doublesex gene controls somatic sexual differentiation by producing alternative spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989;56:997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- 8a.Cavaloc, Y., C. F. Bourgeois, L. Kister, and J. Stévenin. RNA, in press. [DOI] [PMC free article] [PubMed]
- 9.Cavaloc Y, Popielarz M, Fuchs J-P, Gattoni R, Stévenin J. Characterization and cloning of the human splicing factor 9G8: a novel 35 kDa factor of the serine/arginine protein family. EMBO J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandler S D, Mayeda A, Yeakley J M, Krainer A R, Fu X-D. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc Natl Acad Sci USA. 1997;94:3596–3601. doi: 10.1073/pnas.94.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiara M D, Gozani O, Bennett M, Champion-Arnaud P, Palandjian L, Reed R. Identification of proteins that interact with exon sequences, splice sites, and the branchpoint sequence during each stage of spliceosome assembly. Mol Cell Biol. 1996;16:3317–3326. doi: 10.1128/mcb.16.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper T A. In vitro splicing of cardiac troponin T precursors. Exon mutations disrupt splicing of the upstream intron. J Biol Chem. 1991;267:5330–5338. [PubMed] [Google Scholar]
- 13.Cooper T A, Ordahl C P. Nucleotide substitutions within the cardiac troponin T alternative exon disrupt pre-mRNA alternative splicing. Nucleic Acids Res. 1989;17:7905–7921. doi: 10.1093/nar/17.19.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coulter L R, Landree M A, Cooper T A. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol Cell Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elrick L L, Humphrey M B, Cooper T A, Berget S M. A short sequence within two purine-rich enhancers determines 5′ splice site specificity. Mol Cell Biol. 1998;18:343–352. doi: 10.1128/mcb.18.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu X-D. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- 18.Fu X-D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 19.Fu X-D, Maniatis T. Isolation of a complementary DNA that encodes the mammalian splicing factor SC35. Science. 1992;256:535–538. doi: 10.1126/science.1373910. [DOI] [PubMed] [Google Scholar]
- 20.Ge H, Zuo P, Manley J L. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- 21.Graveley B R, Hertel K J, Maniatis T. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J. 1998;17:6747–6756. doi: 10.1093/emboj/17.22.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graveley B R, Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol Cell. 1998;1:765–771. doi: 10.1016/s1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- 23.Hampson R K, La Follette L, Rottman F M. Alternative processing of bovine growth hormone mRNA is influenced by downstream exon sequences. Mol Cell Biol. 1989;9:1604–1610. doi: 10.1128/mcb.9.4.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinrichs V, Baker B S. The Drosophila SR protein RBP1 contributes to the regulation of doublesex alternative splicing by recognizing RBP1 RNA target sequences. EMBO J. 1995;14:3987–4000. doi: 10.1002/j.1460-2075.1995.tb00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helfman D M, Ricci W M, Finn L A. Alternative splicing of tropomyosin pre-mRNAs in vitro and in vivo. Genes Dev. 1988;2:1627–1638. doi: 10.1101/gad.2.12a.1627. [DOI] [PubMed] [Google Scholar]
- 26.Hertel K J, Lynch K W, Hsiao E C, Liu E H, Maniatis T. Structural and functional conservation of the Drosophila doublesex splicing enhancer repeat elements. RNA. 1996;2:969–981. [PMC free article] [PubMed] [Google Scholar]
- 27.Hertel K J, Lynch K W, Maniatis T. Common themes in the function of transcription and splicing enhancers. Curr Opin Cell Biol. 1997;9:350–357. doi: 10.1016/s0955-0674(97)80007-5. [DOI] [PubMed] [Google Scholar]
- 28.Hertel K J, Maniatis T. The function of multisite splicing enhancers. Mol Cell. 1998;1:449–455. doi: 10.1016/s1097-2765(00)80045-3. [DOI] [PubMed] [Google Scholar]
- 29.Hertel, K. J., and T. Maniatis. Unpublished observations.
- 30.Hoshijima K, Inoue K, Higuchi I, Sakamoto H, Shimura Y. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science. 1991;252:833–836. doi: 10.1126/science.1902987. [DOI] [PubMed] [Google Scholar]
- 31.Inoue K, Hoshijima K, Higuchi I, Sakamoto H, Shimura Y. Binding of the Drosophila transformer and transformer-2 proteins to the regulatory elements of doublesex primary transcript for sex-specific RNA processing. Proc Natl Acad Sci USA. 1992;89:8092–8096. doi: 10.1073/pnas.89.17.8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jumaa H, Nielsen P J. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16:5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanopka A, Mühlemann O, Akusjärvi G. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature. 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- 34.Kohtz J D, Jamison S F, Will C L, Zuo P, Lührmann R, Garcia-Blanco M A, Manley J L. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 35.Krainer A R, Maniatis T. Multiple factors including the small nuclear ribonucleoproteins U1 and U2 are necessary for pre-mRNA splicing in vitro. Cell. 1985;42:725–736. doi: 10.1016/0092-8674(85)90269-7. [DOI] [PubMed] [Google Scholar]
- 36.Krainer A R, Maniatis T, Ruskin B, Green M R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- 37.Krainer A R, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 38.Lavigueur A, La Branche H, Kornblihtt A R, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 39.Liu H X, Zhang M, Krainer A R. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lou H, Neugebauer K M, Gagel R F, Berget S M. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol Cell Biol. 1998;18:4977–4985. doi: 10.1128/mcb.18.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lynch K W. Ph.D. thesis. Cambridge, Mass: Harvard University; 1996. [Google Scholar]
- 42.Lynch K W, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 43.Lynch K W, Maniatis T. Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes Dev. 1995;9:284–293. doi: 10.1101/gad.9.3.284. [DOI] [PubMed] [Google Scholar]
- 44.Maniatis T. Mechanisms of alternative pre-mRNA splicing. Science. 1991;251:33–34. doi: 10.1126/science.1824726. [DOI] [PubMed] [Google Scholar]
- 45.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 46.Mardon H J, Sebastio G, Baralle F E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987;15:7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKeown M, Belote J M, Boggs R T. Ectopic expression of the female transformer gene product leads to female differentiation of chromosomally male Drosophila. Cell. 1988;53:887–895. doi: 10.1016/s0092-8674(88)90369-8. [DOI] [PubMed] [Google Scholar]
- 48.Moore M J, Sharp P A. Site-specific modification of pre-mRNA: the 2′-hydroxyl groups at the splice sites. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- 49.Nagoshi R N, Baker B S. Regulation of sex-specific RNA splicing at the Drosophila doublesex gene: cis-acting mutations in exon sequences alter sex-specific RNA splicing patterns. Genes Dev. 1990;4:89–97. doi: 10.1101/gad.4.1.89. [DOI] [PubMed] [Google Scholar]
- 50.Ramchatesingh J, Zahler A M, Neugebauer K M, Roth M B, Cooper T A. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by directing interactions with an exonic enhancer. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reed R, Griffith J, Maniatis T. Purification and visualization of native spliceosomes. Cell. 1988;53:949–961. doi: 10.1016/s0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 52.Reed R, Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986;46:681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- 53.Rio D C. RNA binding proteins, splice site selection, and alternative pre-mRNA splicing. Gene Expr. 1992;2:1–5. [PMC free article] [PubMed] [Google Scholar]
- 54.Rio D C. Splicing of pre-mRNA: mechanism, regulation and role in development. Curr Opin Genet Dev. 1993;3:574–584. doi: 10.1016/0959-437x(93)90093-5. [DOI] [PubMed] [Google Scholar]
- 55.Roth M B, Murphy C, Gall J G. A monoclonal antibody that recognizes a phosphorylated epitope stains lampbrush chromosome loops and small granules in the amphibian germinal vesicle. J Cell Biol. 1990;111:2217–2223. doi: 10.1083/jcb.111.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ryner L C, Baker B S. Regulation of doublesex pre-mRNA processing by 3′ splice site activation. Genes Dev. 1991;5:2071–2085. doi: 10.1101/gad.5.11.2071. [DOI] [PubMed] [Google Scholar]
- 57.Schaal T D, Maniatis T. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol Cell Biol. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schaal, T. D., and T. Maniatis. Unpublished data.
- 59.Shi H, Hoffman B E, Lis J T. A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol Cell Biol. 1997;17:2649–2657. doi: 10.1128/mcb.17.5.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steinmann-Zwicky M, Amrein H, Nöthiger R. Genetic control of sex determination in Drosophila. Adv Genet. 1990;27:189–237. doi: 10.1016/s0065-2660(08)60026-7. [DOI] [PubMed] [Google Scholar]
- 62.Sun Q, Mayeda A, Hampson R K, Krainer A R, Rottman F M. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 63.Tacke R, Chen Y, Manley J L. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc Natl Acad Sci USA. 1997;94:1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tacke R, Manley J L. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tacke R, Tohyama M, Ogawa S, Manley J L. Human Tra-2 proteins are sequence-specific activators of pre-mRNA splicing. Cell. 1998;93:139–148. doi: 10.1016/s0092-8674(00)81153-8. [DOI] [PubMed] [Google Scholar]
- 66.Tanaka K, Watakabe A, Shimura Y. Polypurine sequences within a downstream exon function as a splicing factor. Mol Cell Biol. 1994;14:1347–1354. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tian H, Kole R. Selection of novel exon recognition elements from a pool of random sequences. Mol Cell Biol. 1995;15:6291–6298. doi: 10.1128/mcb.15.11.6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tian M, Maniatis T. Positive control of pre-mRNA splicing in vitro. Science. 1992;256:237–240. doi: 10.1126/science.1566072. [DOI] [PubMed] [Google Scholar]
- 69.Tian M, Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- 70.Tian M, Maniatis T. A splicing enhancer exhibits both constitutive and regulated activities. Genes Dev. 1994;8:1703–1712. doi: 10.1101/gad.8.14.1703. [DOI] [PubMed] [Google Scholar]
- 71.Wang J, Manley J L. Regulation of pre-mRNA splicing in metazoa. Curr Opin Genet Dev. 1997;7:205–211. doi: 10.1016/s0959-437x(97)80130-x. [DOI] [PubMed] [Google Scholar]
- 72.Wang Z, Hoffmann H M, Grabowski P J. Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA. 1995;1:21–35. [PMC free article] [PubMed] [Google Scholar]
- 73.Watakabe A, Tanaka K, Shimura Y. The role of exon sequences in splice site selection. Genes Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- 74.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 75.Xiao S H, Manley J L. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 76.Yeakley J M, Morfin J-P, Rosenfeld M G, Fu X-D. A complex of nuclear proteins mediates SR protein binding to a purine-rich splicing enhancer. Proc Natl Acad Sci USA. 1996;93:7582–7587. doi: 10.1073/pnas.93.15.7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zahler A M, Lane W S, Stolk J A, Roth M B. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 78.Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]