Abstract
Posttranslational modification of a protein by ubiquitin usually results in rapid degradation of the ubiquitinated protein by the proteasome. The transfer of ubiquitin to substrate is a multistep process. Cdc4p is a component of a ubiquitin ligase that tethers the ubiquitin-conjugating enzyme Cdc34p to its substrates. Among the domains of Cdc4p that are crucial for function are the F-box, which links Cdc4p to Cdc53p through Skp1p, and the WD-40 repeats, which are required for binding the substrate for Cdc34p. In addition to Cdc4p, other F-box proteins, including Grr1p and Met30p, may similarly act together with Cdc53p and Skp1p to function as ubiquitin ligase complexes. Because the relative abundance of these complexes, known collectively as SCFs, is important for cell viability, we have sought evidence of mechanisms that modulate F-box protein regulation. Here we demonstrate that the abundance of Cdc4p is subject to control by a peptide segment that we term the R-motif (for “reduced abundance”). Furthermore, we show that binding of Skp1p to the F-box of Cdc4p inhibits R-motif-dependent degradation of Cdc4p. These results suggest a general model for control of SCF activities.
An important mechanism of regulating protein abundance in eukaryotes is the ubiquitin (Ub)-proteasome degradation pathway. Ub is a member of a family of conserved polypeptides that are covalently attached to protein substrates (reviewed in reference 16). Multiple rounds of modification create a poly(Ub) chain on the substrate that targets the substrate for degradation by the proteasome (reviewed in references 4, 12, 14, 15, 42, and 51). The transfer of free Ub onto a protein substrate is a multistep process. E1 activates free Ub at the expense of ATP. Ub is then transferred to an E2 (or ubiquitin protein-conjugating enzyme). Based on sequence comparison, yeast has 11 E2s, and it is believed that each E2 is responsible for ubiquitinating distinct substrates. Although a free E2 enzyme may directly transfer Ub onto a substrate in a purified system, this reaction is promoted by additional proteins referred to as E3s or ubiquitin protein ligases. Some E3s act as intermediary Ub carriers in the transfer of Ub from E2 to substrate (47). Other E3s act as adapters, tethering E2 to its substrate (8, 49). It is emerging that a variety of structurally distinct E3 proteins each serve to regulate the interaction between E2 proteins and various distinct substrates.
The ubiquitin-proteasome degradation pathway regulates two major cell cycle events: entry into S phase and entry into anaphase (reviewed in references 5, 22, and 38). In yeast, cell cycle progression is mediated by the activity of the cyclin-dependent kinase (CDK) Cdc28p (reviewed in references 26, 34, and 35). The activation state and specificity of Cdc28p are determined by cyclins and CDK inhibitors (reviewed in references 17, 26, 34, 36, and 43). Association with cyclins Cln1 to Cln3 activates Cdc28p during G1, while the mitotic cyclins, Clb1 to Clb6, are required for the S through M phases. Proteins regulating mitosis, including mitotic cyclins, are targeted for degradation by a cell cycle-regulated E3 complex, the anaphase-promoting complex. During G1, the CDK inhibitor Sic1p acts to inhibit CDK-Clb complex formation and prevent the initiation of S phase. Entry into S phase requires degradation of Sic1p by the ubiquitin-proteasome pathway (48). We and others have identified components of an E2-E3 complex that is necessary to target Sic1p for degradation (1, 10, 30).
Cells lacking the gene encoding the E2 enzyme Cdc34p remain in G1, develop multiple elongated buds, and fail to separate duplicated spindle pole bodies (10). This phenotype is consistent with failure to activate the Clb-CDK complexes because of the inability to degrade Sic1p (32, 48). Mutations in two other genes, CDC4 and CDC53, cause phenotypes indistinguishable from mutations in CDC34 (30). Cdc53p is a member of a family of proteins termed cullins (24, 30, 52). Cdc4p is a member of a family of proteins that are characterized by the presence of WD-40 repeats (37, 53) and a second motif known as the F-box (1). The entry into S phase requires CDC34 to act in concert with CDC4 and CDC53 (30). Indeed, we have also shown that CDC34, CDC4, and CDC53 gene products form a complex in vivo and that complex formation is necessary for S phase entry (30, 31). A fourth gene, SKP1 (1, 3), encodes yet another member of this complex, and recombinant Cdc34p, together with Cdc4p, Cdc53p, and Skp1p produced in insect cells, ubiquitinates Sic1p in vitro (8, 49). Thus, the Cdc4p-Cdc53p-Skp1p complex is an E3 for Cdc34p.
In addition to Cdc4p, two other F-box-containing proteins in yeast, Met30p (39) and Grr1p (2, 23, 27, 39), have been proposed to interact with Skp1p and Cdc53p to form complexes referred as SCFs (for Skp1p-Cdc53p-F box) (8, 39, 40). Like Cdc4p, both Met30p (50) and Grr1p (9) contain repetitive domains, WD-40 repeats, and leucine-rich repeats, respectively (20), which potentially interact with distinct Cdc34p substrates. Thus, a family of potential E3 complexes have been identified that are distinguishable by a component that has been proposed to recognize distinct substrates.
It has been suggested that each SCF complex contains only one F-box protein (39, 40). Thus, Cdc4p, Met30p, and Grr1p would compete for a common set of components, namely, Skp1p and Cdc53p. Therefore the relative abundances of F-box-containing proteins must be tightly regulated. The way in which the cell regulates SCF component abundance is not clear. Cdc34p is itself a substrate for ubiquitination, yet cdc34 mutants that are resistant to Cdc34p ubiquitination have no growth defect (11). Posttranslational modification of Cdc53p by the Ub-like protein Rub1p is stimulated when the abundance of Cdc4p and Cdc34p is altered (25). Furthermore, cells unable to modify Cdc53p by Rub1p attachment are sensitive to the abundance of Cdc34p and Cdc4p (25). In the course of performing genetic analysis of CDC4, we have identified Cdc4p domains that are responsible for regulating its abundance. We find that the normally low abundance and short half-life of Cdc4p are dependent on the presence of an R-motif that is located adjacent to the F-box. Furthermore, we show that the Skp1p–F-box interaction plays an important role in inhibiting the destabilizing effect of the R-motif. The results suggest a model whereby the decision to determine which SCF(s) will be present is regulated by stabilization of the F-box protein through Skp1p binding.
MATERIALS AND METHODS
Yeast strains and manipulations.
The yeast strains used in this study are as follows: A2.7.A3p (MATa cdc4-3 leu2-3,112 his3-11,15), Y382 (MATα ade2 ade3 ura3 leu2 trp1) (kindly provided by A. Bender), YPH1172 (MATa ura3-52 trp1-Δ63 his3-Δ200 leu2-Δ1 ade2-101 skp1Δ1::TRP skp1-3::LEU2), YPH1161 (MATa ura3-52 trp1-Δ63 his3-Δ200 leu2-Δ1 ade2-101 skp1Δ1::TRP skp1-4::LEU2) (both kindly provided by P. Hieter), MHY803 (MATα ura3-52 trp1-1 his3-Δ200 leu2-3,112 lys2-801 Δdoa3::HIS3 [YCplac22 DOA3]), and MHY792 (MATα ura3-52 trp1-1 his3-Δ200 leu2-3,112 lys2-801 Δdoa3::HIS3 [YCplac22 doa3-1]) (both kindly provided by M. Hochstrasser). Standard rich (YPD) and defined minimal SD medium were prepared as described previously (44). Transformations were carried out as described previously (7). For plasmid selection, yeast cells were grown on defined minimal medium supplemented with the appropriate amino acids. For galactose induction, cells were grown to early logarithmic phase in minimal medium containing sucrose instead of dextrose, galactose-containing medium (2%) was added, and the culture was incubated for a further 2 to 3 h. To measure protein half-lives, GST-Cdc4p fusions were transiently induced from the GAL1-10 promoter for 30 min. Glucose (2%) and cycloheximide (1 mg/ml) were added to repress transcription and translation, respectively. For complementation experiments, patches derived from single colonies were grown under permissive conditions (23°C) and then replica plated and incubated further under nonpermissive conditions (37°C).
Plasmid constructions.
Escherichia coli DH5α was used to propagate plasmids. Plasmid manipulations were performed by standard methods (46). Plasmid pSJ4101 has been described previously (30). The vector pEG(KG) was used for the expression of glutathione S-transferase (GST) fusion proteins (33). Expression of the GST fusion proteins was from the GAL1-10 promoter. All GST-CDC4 fusion constructs were created by cloning PCR-generated DNA fragments with plasmid-borne CDC4 DNA as template by methods as described previously (29). GST-MET30 fusion constructs were created by cloning PCR-generated DNA fragments from yeast genomic DNA as the template. Primers annealing at the 5′ end of either CDC4 or MET30 were designed to incorporate either an XbaI or a BamHI restriction site, and primers annealing at the 3′ end of either CDC4 or MET30 were designed to incorporate either a SalI or an AvrII restriction site. The PCR products were cleaved with the appropriate enzymes and ligated into pEG(KG), which had previously been digested with the appropriate enzymes. Table 1 lists primers used in this study. Table 2 lists the constructs derived from using different sets of primers, which are numbered to indicate the amino acids from the complete Cdc4p protein that are encoded. pGST-CDC4(Δ272–318) was generated by cloning a PCR fragment generated by CDC48aAvr and NM202b into pGSTCDC4(1–269)(Bam-Xba). This construct replaces residues 272 to 318 with a glycine residue pGST-CDC4(Δ321–341) was generated by cloning a PCR fragment generated by CDC43aAvr2 and NM202b into pGSTCDC4(1–319). This construct replaces residues 321 to 341 with a glycine residue. pGST-CDC4(L278R) was generated by cloning a PCR fragment generated by CDC4L278R2a and NM202b into pGSTCDC4(1-L278R). To clone the R-motif, two complementary oligonucleotides encoding amino acids 320 to 341 were annealed. BamHI and SalI restriction sites were incorporated into these oligonucleotides, and following restriction with these enzymes, the fragment was ligated into pEG(KG) vector previously cleaved with the same restriction enzymes. A PCR-generated SKP1 fragment obtained with primers 5′-AAAGGATCCATGGTGACTTCTAATGTTGTC-3′ and 5′-GGGGTCGACCTAACGGTCTTCAGCCCATTC-3′ was produced. BamHI and SalI restriction sites were incorporated into these oligonucleotides, and following restriction with these enzymes, the fragment was ligated into p424ADH vector (American Type Culture Collection) cleaved with the same restriction enzymes. This cloning placed SKP1 under the control of the ADH1 promoter and allowed for SKP1 overexpression.
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| F-BOX1A | GGGGGATCCCACATGATCAAGATCGACTTC |
| F-BOX1B | AAATCTAGAGGATCCTTTCCAAGGTCGCGTAGTTTG |
| CDC41aXba2 | AAATCTAGACATGGGGTCGTTTCCCTTAGCTGAG |
| CDC42b | AAAGTCGACGGCAAAGACGTTATTAGGTCCCTC |
| CDC43b | TGAGTCGACCTACTAAGACGATCTTGTTGTGAGAG |
| CDC45aAvr2 | GGCCCTAGGCAATTTAAAGAGGGACCTAATAACG |
| NM202b | AACGTCGACTCATGGTATTATAGTTGTCCTCG |
| CDC43aAvr | GGCCCTAGGCTACCCAAAACTCTCACAACAAG |
| CDC41aBam | AAAGGATCCATGGGGTCGTTTCCCTTAGCTGAG |
| CDC47bXba | GGGTCTAGATTATCCTTGATTAAAGTCCCCAAG |
| CDC48aAvr | AAGCCTAGGCCTTCTGATATCGGAAAATTTTGTG |
| CDC4L278R2a | GGGCCTAGGCCTTTTGAAATAAGTTTG |
| CDC49bXba | GGGTCTAGAAGTTTTTTCCACAACGATGTAGATTTTC |
| CDC4L278R1b | GGGTCTAGACGTTATTAGGTCCCTC |
TABLE 2.
GST-CDC4 fusion constructs made during this study
| Construct | Primers used |
|---|---|
| pGST-CDC4(1–278) | CDC41aXba2 and CDC42b |
| pGST-CDC4(1–351) | CDC41aXba2 and CDC43b |
| pGST-CDC4(269–779) | CDC45aAvr and NM202b |
| pGST-CDC4(341–779) | CDC43aAvr2 and NM202b |
| pGST-CDC4(269–341) | CDC45aAvr and CDC43b |
| pGST-CDC4(1–319) | CDC41aBam and Cdc49bXba |
| pGST-CDC4(279–779) | CDC4L278R2a and NM202b |
| pGST-CDC4(319–779) | CDC48aAvr and NM202b |
| pGST-CDC4(1–269)(Bam-Xba) | CDC41aBam and CDC47bXba |
| pGST-CDC4(1–277) | CDC4aBam and CDC4L278R1b |
| pGST-Met30(176–275) | F-BOX1A and F-BOX2B |
Protein preparation and Western immunoblot techniques.
Yeast lysate preparations and Western immunoblot techniques were carried out as described previously (29). Antibodies raised against GST were purchased from Sigma.
RESULTS
The Cdc4p F-box and WD-40 repeats are required for Cdc4p in vivo activity.
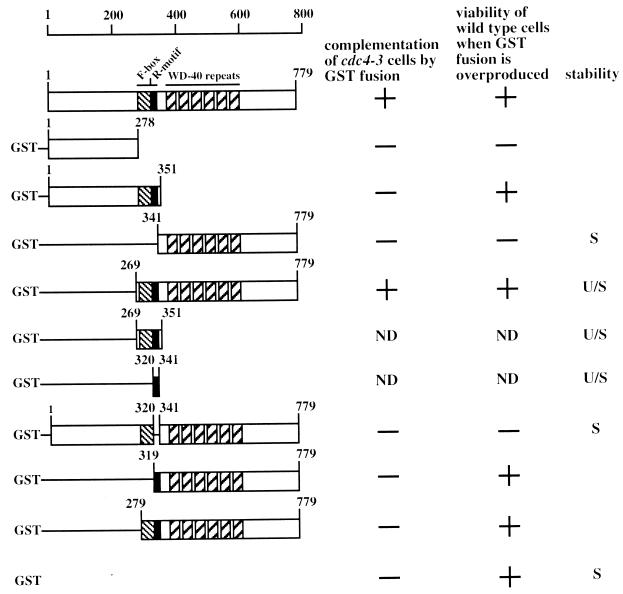
Cdc4p is a component of a multiprotein complex that is required for the degradation of two CDK inhibitors, Sic1p (48) and Far1p (13), as well as Cdc6p (6, 41), which regulates the formation of prereplicative complexes at origins of DNA replication. The most prominent cdc4 defect is failure to degrade Sic1p, which prevents the usual cell cycle rise in Clb-associated CDK activity (48). Cells lacking cdc4 activity arrest prior to S phase with characteristic multielongated bud morphology. Two domains have been characterized in Cdc4p that are required for the interaction of Cdc4p with other members of the complex and for substrate recognition. The Cdc4p F-box has been proposed to interact with other members of the complex, which include Skp1p and Cdc53p (39). The Cdc4p WD-40 repeats have been proposed to bind the substrate for ubiquitination (49). To achieve a further dissection of the functional domains in Cdc4p, we constructed GST fusion constructs including various portions of CDC4 (Fig. 1). To assess the function of the resulting fusion proteins, each construct, as well as the empty vector and plasmid encoding untagged full-length Cdc4p, was transformed into strain A2.7.A3p, which contains the temperature-sensitive cdc4-3 allele. Isolated transformants were patched onto dextrose medium lacking leucine to select for the presence of the plasmid and then incubated at 23°C, the permissive temperature for cells containing cdc4-3. These patches were replica plated to the same media and incubated at 37°C to test the ability of each GST fusion protein to rescue the temperature-sensitive phenotype of A2.7.A3p (Fig. 2). Plasmids encoding full-length untagged Cdc4p [Cdc4(1–779)p] and GST-Cdc4(269–779)p, which includes both the F-box and the WD-40 repeats, allowed A2.7.A3p cells to grow at the nonpermissive temperature. Neither the plasmid encoding only GST nor plasmids encoding the other GST-Cdc4p fusion proteins permitted growth at the nonpermissive temperature (Fig. 2), and microscopic examination showed that these cells displayed the multielongated bud phenotype typical of loss of CDC4 activity. These findings demonstrate that the F-box and WD-40 repeat sequences are necessary and sufficient to complement a cdc4 temperature-sensitive mutant. Furthermore, it is clear that the Cdc4p amino-terminal segment (residues 1 to 278) is not required for the essential Cdc4p activity in vivo.
FIG. 1.
Cdc4p and GST-Cdc4p deletion panel. A schematic diagram illustrating Cdc4p and the GST-Cdc4p fusion constructs used in this study is presented. The positions of the F-box, R-motif, and WD-40 repeat motifs are as indicated. Amino acid residues are numbered. Also indicated is a summary of the biological activities of the different GST-Cdc4p fusion constructs. The ability of each construct to complement (+) or failure to complement (−) cdc4-3 cells is indicated. The viability (+) or inviability (−) of Y382 cells when each construct is overproduced is indicated. The stability of the indicated construct, as determined by measurement of the half-life of the protein, is represented as stable (S) or unstable (U/S). ND, not determined.
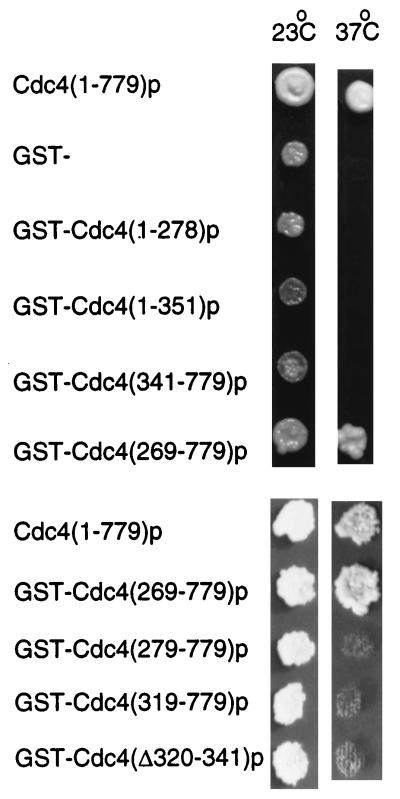
FIG. 2.
Functional Cdc4p domains. A.2.7.A3p cells containing the cdc4-3 temperature-sensitive mutation were transformed with a plasmid that allowed the production of the indicated proteins. Replica patches were made, and cells were incubated at the indicated temperature for 3 days.
Identification of dominant lethal and dominant negative Cdc4p domains.
The plasmids used in the above experiments expressed CDC4 and GST-CDC4 genes from a galactose-inducible promoter, GAL1-10. Cells grown in the presence of dextrose weakly express GAL1-10-controlled genes, whereas cells grown in the presence of galactose strongly express GAL1-10-controlled genes. The fact that Cdc4p and GST-Cdc4(269–779)p do not have to be overproduced to rescue the cdc4-3 mutant suggests that only a low level of full-length Cdc4p is required in the cell. Indeed, we have estimated that cell viability is maintained when Cdc4p is present at only five copies per cell (19). Increasing the abundance of one F-box protein might be expected to have a detrimental effect on the cell, since it might titrate common SCF components away from other essential F-box proteins. There is much genetic evidence indicating that the abundance of SCF components must be balanced to allow normal cell growth (23, 25, 27, 30, 39). However, overexpression of genes encoding F-box proteins only has a detrimental effect on cell growth when the cell also contains mutations in other SCF components or other F-box proteins. Furthermore, increased CDC4 transcription does not adversely affect cell growth (see below) (31). These data suggests that the abundance of F-box proteins may be regulated in some manner. However, overexpressing different portions of CDC4 may be detrimental to the cell, since these Cdc4p fragments may lack potentially self-regulating sequences. To assess whether the overproduction of the GST-Cdc4 fusion proteins is toxic to cells containing normal SCF components, Y382 cells were transformed with each construct described above. Isolated transformants were grown on sucrose medium lacking leucine to select for the presence of the plasmid and then transferred to medium containing galactose as the sole carbon source to induce the expression of each fusion protein and assess its potential toxicity (Fig. 3A).
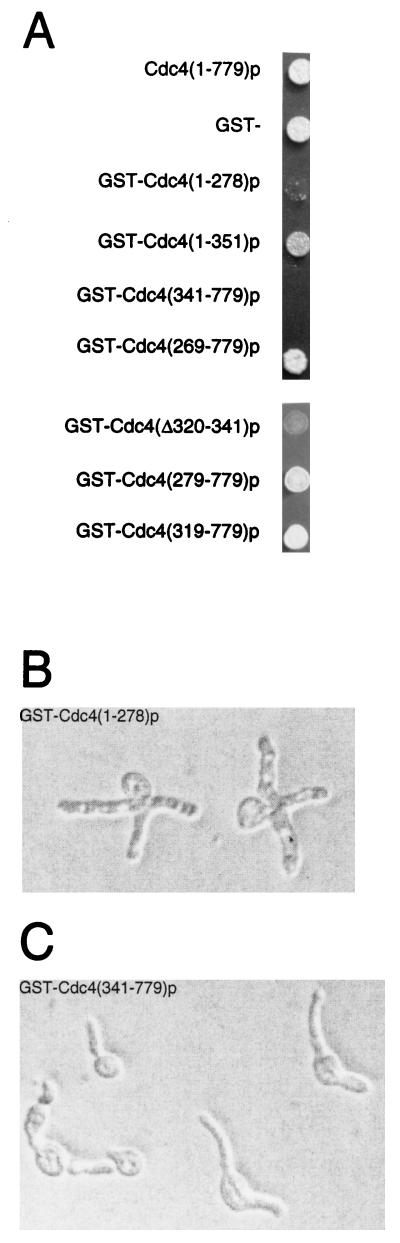
FIG. 3.
Identification of dominant lethal and dominant negative Cdc4p domains. (A) Y382 cells, wild type for CDC4, were transformed with the plasmid that allowed the production of the indicated protein in the presence of galactose. Transformants were grown in the presence of sucrose, patched onto medium containing galactose as the sole carbon source, and incubated for 3 days. (B) Terminal phenotype of GST-CDC5(1–278) overexpression. (C) Terminal phenotype of GST-CDC5(341–779) overexpression.
Overexpression of genes encoding full-length Cdc4p, GST, GST-Cdc4(1–351)p, and GST-Cdc4(269–779)p did not inhibit the growth of Y382 cells (Fig. 3A). However, overexpression of genes encoding GST-Cdc4(1–269)p and GST-Cdc4(341–779)p did inhibit the growth of Y382 cells (Fig. 3A). Therefore, overexpressed portions of CDC4 which encode residues 269 to 341 are not toxic to Y382 cells. Cdc4p residues 269 to 341 contain the F-box, a Skp1p binding polypeptide sequence (1). We next wanted to distinguish whether the F-box sequence (residues 278 to 319) or the sequence contiguous with the F-box (residues 320 to 341) was responsible for alleviating the toxicity when fragments of Cdc4p were overproduced. Overexpression of genes encoding GST-Cdc4(279–779)p, which contains a small deletion in the F-box, and GST-Cdc4(319–779)p, which completely lacks the F-box, did not inhibit Y382 cell growth. Therefore, residues 320 to 341, and not the F-box, are necessary to permit cell growth when a potentially toxic fragment of Cdc4p is overproduced. Finally, we wanted to test whether residues 320 to 341 were necessary to prevent full-length CDC4 from being toxic when it is overexpressed. A GST-CDC4 gene overexpressing GST-Cdc4(Δ320–341)p inhibited cell growth. Therefore, we have identified a small polypeptide region in Cdc4p, residues 320 to 341, that is both necessary and sufficient to prevent cell death when CDC4 is overexpressed.
To begin to understand why overexpression of different regions of CDC4 inhibited cell growth, we carried out microscopic examination of cells which overproduced GST-Cdc4(1–278)p, GST-Cdc4(341–779)p, and GST-Cdc4(Δ320–341)p. Cells overproducing GST-Cdc4(Δ320–341)p displayed no cell cycle arrest. Therefore, it is unlikely that overproduction of this protein interferes with Sic1p uniquitination and degradation, since cells unable to degrade Sic1p have a characteristic multielongated bud morphology. The cell death induced by GST-Cdc4(Δ320–341)p overproduction may have been due to loss of essential SCF activities which contain other F-box proteins.
Microscopic examination showed that cells overexpressing GST-CDC4(1–278) displayed the multielongated bud phenotype typically seen with loss of CDC4 activity (Fig. 3B). Thus, overproduction of GST-Cdc4(1–278)p generated a dominant negative phenotype. Curiously, this amino-terminal domain is nonessential for Cdc4p activity in vivo (Fig. 2). A possible explanation for why a nonessential domain generates a dominant lethal phenotype when overproduced is that some component of the SCFCdc4p complex partially associates with this region and is thus titrated away from its essential role in SCFCdc4p function. However, neither CDC34 nor CDC53 overexpression relieved the toxicity or terminal phenotype induced by GST-CDC4(1–278) overexpression (data not shown), suggesting that other SCFCdc4p components that remain unidentified may interact with Cdc4(1–278)p. Alternatively, residues 1 to 278 may act as a negative regulator of SCFCdc4p activity.
Overproduction of GST-Cdc4(341–779)p also inhibited the growth of Y382 cells. Microscopic examination showed that these cells also generated an elongated bud morphology but one that was distinct from the phenotype seen for loss of CDC4 activity (Fig. 3C). In some cases buds formed at opposite poles of the elongated cell, whereas cdc4 cells form multiple buds close to the same site. Cdc4p residues 341 to 779 contain WD-40 repeats almost exclusively and have been proposed to interact with SCFCdc4p substrates, which include Sic1p, Cdc6p, and Far1p. Why overproduction of this Cdc4p motif is toxic to cells is unclear. Nevertheless, it is clear that overproducing either fragments of Cdc4p or full-length Cdc4p that lack residues 320 to 341 is toxic to cells. Thus, residues 320 to 341 may regulate Cdc4p abundance or activity.
Cdc4p residues 320 to 341 act as a transferable destabilizing signal.
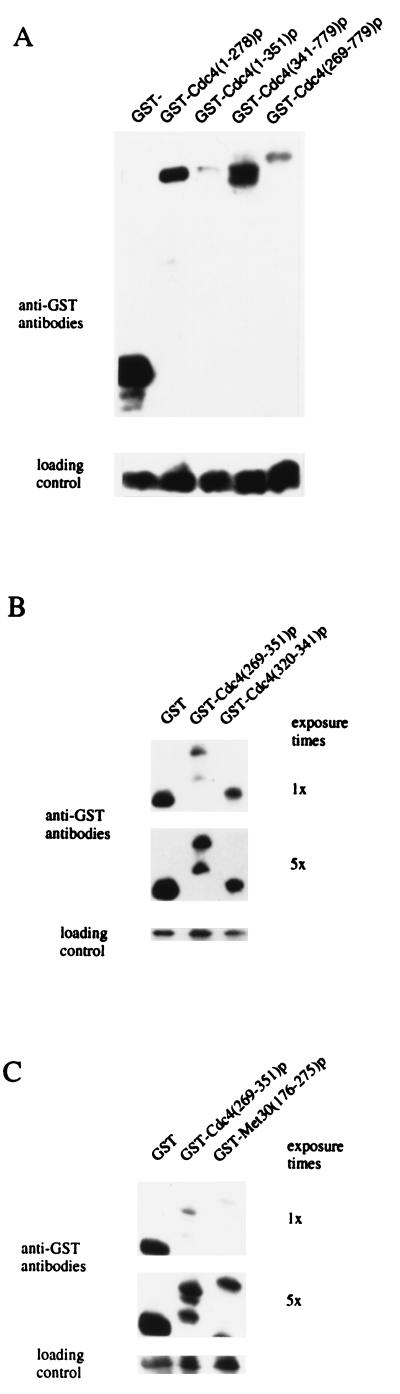
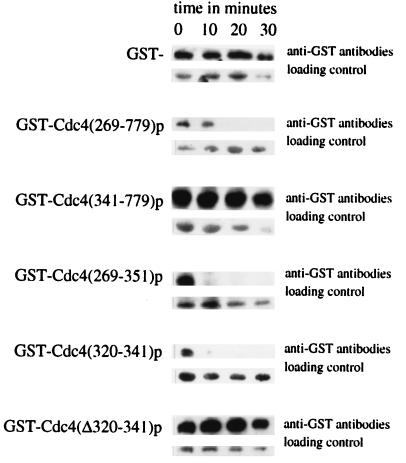
Since the toxicity of overexpressing CDC4 truncations was ameliorated by the presence of Cdc4p residues 320 to 341, we sought to establish the function of this sequence. Growth inhibition induced by overproducing different portions of Cdc4p may be induced by titrating SCF components into a nonfunctioning complex. Because a number of F-box proteins function in concert with a common set of SCF components (27, 39, 40), titrating these components into a nonfunctional complex would result in loss of activity for a number of SCF complexes. Therefore, one function of residues 320 to 341 could be to reduce Cdc4p abundance. Thus, suspecting an effect on protein abundance, we carried out Western blot analysis with anti-GST antibodies to determine the steady-state abundance of different GST-Cdc4p fusion proteins (Fig. 4A). We found a clear correlation between the steady-state abundance of different GST-Cdc4p fusion proteins and their ability to inhibit cell growth when overproduced (Fig. 4A). GST-Cdc4(1–278)p and GST-Cdc4(341–779)p, which inhibited cell growth (Fig. 3A), were both produced to a relatively high abundance with respect to GST-Cdc4p fusions proteins that were not toxic to cells when overproduced. Both GST-Cdc4(1–351)p and GST-Cdc4(269–779)p, which do not inhibit cell growth when overproduced, are low-abundance proteins. When fused to GST, the region common to these low-abundance proteins, residues 269 to 351, also reduced the steady-state abundance of GST (Fig. 4B). We concluded that residues 320 to 341 functioned to reduce the toxicity of various GST-Cdc4p fusions. When fused to GST, residues 320 to 341 functioned to lower the steady-state abundance of the fusion protein (Fig. 4B). We named Cdc4p residues 320 to 341 the R-motif, since this domain is necessary and sufficient to reduce the abundance of GST-Cdc4p fusions.
FIG. 4.
Cdc4p residues 320 to 341 reduce the steady-state abundance of GST-Cdc4p fusion proteins. Panels A through C show the anti-GST Western blot of soluble protein extracts from cells expressing the indicated GST-fusion proteins. A cross-reacting band was used as a loading control.
The R-motif, LLISENFVSPKGFNSLNLKLSQ, appears to be a unique sequence, since we were unable to find significant matches in any database. However, in Cdc4p it is located adjacent to the F-box, which appears to be conserved throughout eukaryotes (2, 18, 28, 29, 40, 45). We next investigated whether an F-box-containing sequence from another protein might act in a similar fashion. A GST fusion protein was constructed which contained a 100-amino-acid segment of Met30p that includes the Met30p F-box (Fig. 4C). Western blot analysis showed that the addition of this sequence to GST had an effect similar to that of the corresponding segment of CDC4, severely reducing the steady-state abundance of GST (Fig. 4C). Fusion of the same Met30p sequence onto the segment of CDC4 that encodes the WD-40 repeats suppressed the toxicity of the Cdc4p carboxyl terminal when overproduced in wild-type cells (data not shown). We conclude that sequences near the F-box from either protein are capable of regulating the abundance of Cdc4p in a cis-acting manner.
One mechanism by which the R-motif may reduce GST-Cdc4p steady-state abundance is to target the protein for degradation. To explore this possibility, we sought to measure the half-lives of different GST-Cdc4p fusions by promoter shutoff experiments (Fig. 5). GST and GST-Cdc4(341–779)p were shown to be stable during the time course. However, GST-Cdc4(269–779)p had a much shorter half-life than GST-Cdc4(341–779)p. This indicated that residues 269 to 351 were responsible for the instability of GST-Cdc4(269–779)p. Indeed, GST-Cdc4(269–351)p, which contains the R-motif, was also shown to have a short half-life. When fused to GST, the R-motif residues (residues 320 to 341) were sufficient to decrease the half-life of GST (Fig. 5). To investigate whether the R-motif is necessary for Cdc4p degradation, the half-life of a GST-Cdc4p fusion that lacked these residues was measured. GST-Cdc4(Δ320–341)p and was stable over the course of the experiment. Thus, we have shown that GST-Cdc4p fusions containing the R-motif are rapidly degraded and that the R-motif is necessary and sufficient for this degradation.
FIG. 5.
The R-motif targets Cdc4p for degradation. Time course experiments to measure the stability of various GST-Cdc4p fusions after promoter shutoff. The GST-Cdc4p fusions tested are as indicated. The same cross-reacting band was used as a loading control in each case.
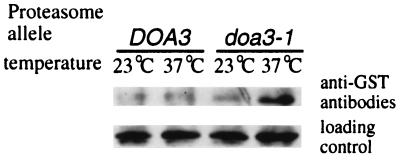
Finally, we wanted to investigate the mechanism by which Cdc4p may be degraded. One potential mechanism by which Cdc4p is degraded is by the ubiquitin-proteasome pathway. To investigate whether Cdc4p is degraded in a proteasome-dependent fashion GST-CDC4(269–779) was overexpressed in isogenic cells that either were wild type or contained a temperature-sensitive allele of an essential proteasome subunit, doa3-1. GST-Cdc4(269–779)p was produced at the permissive temperature, 23°C, or the restrictive temperature, 36°C, for doa3-1 in both cell types. In cells containing the wild-type DOA3 allele, the abundance of GST-Cdc4(269–779)p remained the same at either temperature (Fig. 6). However the abundance of GST-Cdc4(269–779)p increased in doa3-1 cells incubated at the restrictive temperature. Thus, Cdc4p abundance appears to be controlled by a proteasome-dependent pathway.
FIG. 6.
Proteasome-dependent degradation of GST-Cdc4(269–779)p. Cells containing the indicated DOA3 allele were incubated at the indicated temperature. The steady-state abundance of GST-Cdc4(269–779)p was monitored by Western blot analysis with anti-GST antibodies.
The F-box–Skp1p interaction regulates GST-Cdc4p steady-state abundance.
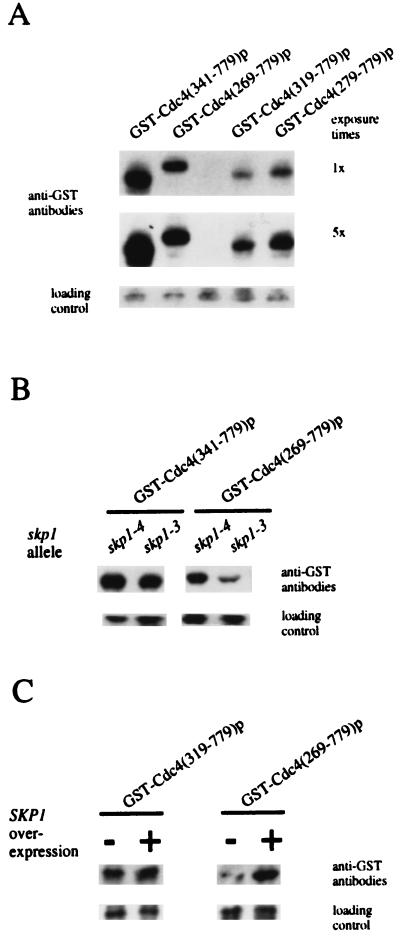
We have demonstrated that the R-motif is necessary and sufficient to target GST-Cdc4p for proteasome-dependent degradation. Thus, the R-motif is a negative regulatory sequence for Cdc4p function. Curiously, the R-motif lies adjacent to the F-box. The F-box is a polypeptide sequence necessary for Skp1p binding, and the F-box–Skp1p interaction has been previously noted to stimulate Cdc4p activity (1). We therefore wanted to investigate whether the function of the F-box affected R-motif activity. Accordingly, we constructed specific mutations within the F-box to observe their effects on GST-Cdc4p abundance (Fig. 7A). We made two GST-Cdc4p fusions that contained mutations within the F-box: one lacked the first residue of the F-box, GST-Cdc4(279–779)p, and the second lacked the F-box, GST-Cdc4(319–779)p. The steady-state abundance of these GST-Cdc4p fusions was compared with that of similar constructs that contained wild-type F-box GST-Cdc4(269–779)p. Western blot analysis (Fig. 7A) showed that the abundance of GST-Cdc4p fusion was lower in cells containing mutations in the Cdc4p F-box than in cells containing a wild-type Cdc4p F-box. Thus, the effect of the R-motif is enhanced when mutations are made within the F-box.
FIG. 7.
The F-box–Skp1p interaction affects the abundance of GST-Cdc4p fusions. Panels A through C show the anti-GST Western blot analysis of soluble protein extracts in cells expressing the indicated GST-CDC4 genes. (B) Extracts were prepared from cells containing the indicated SKP1 allele. (C) Extracts were prepared from cells overexpressing (+) or not overexpressing (−) SKP1.
Because the F-box is necessary for Skp1p binding in a variety of proteins (1, 23, 39), the above results suggest that the F-box–Skp1p interaction masks the effect of the R-motif. Above, we have shown that mutating the F-box, which eliminates the ability of Cdc4p to bind Skp1p, has the effect of reducing the steady-state abundance of GST-Cdc4p. As a second means of testing the significance of the F-box–Skp1p interactions, we measured the GST-Cdc4p fusion abundance when Skp1p activity was limiting. Two temperature-sensitive mutant skp1 alleles that have different terminal morphologies at the nonpermissive temperature have been described (3). skp1-3 causes cells to have a cdc34-like phenotype. The second mutant allele, skp1-4, generates cells that arrest after S phase and have a chromosome missegregation phenotype, consistent with a role for SKP1 in kinetochore function (3, 21). Isogenic strains containing either skp1-3 or skp1-4 were transformed with GST-CDC4 fusion constructs encoding either residues 341 to 779 or 269 to 779, and the relative steady-state abundance of these proteins was measured by Western blot analysis. We found that GST-Cdc4(341–779)p, which lacks the F-box, is produced at very high abundance in the presence of either skp1 allele. However, the abundance of GST-Cdc4(269–779)p, which contains the F-box, differed between strains containing two different skp1 alleles. Those containing skp1-4 produced GST-Cdc4(269–779)p to an abundance equivalent to cells that contain wild-type SKP1, while skp1-3 cells produced GST-Cdc4(269–779)p to a much lower abundance (Fig. 7B). The abundance of GST-Cdc4(269–779)p was further reduced when the cultures were transferred to the nonpermissive temperature, 37°C, for skp1 alleles (data not shown). Thus, the integrity of Skp1p in the cell affects the abundance of an F-box-containing protein. Furthermore, we noted that many skp1-3 cells became arrested with multiple elongated buds, consistent with the expectation that they were compromised for SCFCdc4p activity because of low Cdc4p abundance. Clearly, both Skp1p and the site to which it is known to bind, the F-box are important in regulating the abundance of Cdc4p.
Finally, we wanted to test whether promoting the Skp1p–F-box interaction would increase the steady-state abundance. To do this, we cooverexpressed SKP1 with GST-CDC4 genes that produced GST-Cdc4p fusions containing a wild-type F-box, GST-Cdc4(269–779)p, or lacking the F-box, GST-Cdc4(319–779)p. We observed that the steady-state abundance of GST-Cdc4(269–779)p was increased when it was co-overexpressed with SKP1 whereas the steady-state abundance of GST-Cdc4(319–779)p was unaffected when it was co-overproduced with Skp1p (Fig. 7C). Therefore, the F-box–Skp1p interaction acts to suppress R-motif mediated degradation.
DISCUSSION
We have defined domains in Cdc4p that are required for its function in an active SCF complex. By testing the effects of overproduction, we have identified domains that cause dominant defects in cell cycle progression. The presence of a small polypeptide sequence relieves the toxicity of these domains, and we have also shown that this sequence negatively regulates Cdc4p abundance and targets Cdc4p for proteasome-dependent degradation. Finally, we have demonstrated that the F-box–Skp1p interaction is necessary to stabilize Cdc4p.
Two domains on Cdc4p are well characterized (Fig. 1). The first is the F-box, which links Cdc4p to Cdc53p through Skp1p and is required for the formation of the SCFCdc4p complex. The second consists of the WD-40 repeats, which are necessary to bind Cdc34p substrates for ubiquitination. This complex targets Cdc34p substrates for ubiquitin-dependent degradation. By monitoring complementing activities of a Cdc4p deletion panel, we have shown that the F-box and WD-40 repeats are both necessary and sufficient to complement a cdc4 temperature-sensitive mutant (Fig. 2).
By overproducing the panel of Cdc4p deletions, we identified fragments of Cdc4p that inhibit cell growth (Fig. 3A). We have identified a short polypeptide region in Cdc4p, which we have termed the R-motif, that is necessary and sufficient to relieve this toxicity (Fig. 3A). Western blot analysis demonstrated that the R-motif is necessary and sufficient to reduce Cdc4p abundance and shorten the half-life of a protein (Fig. 4 and 5). Thus, one function of the R-motif is to reduce the Cdc4p abundance in the cell. Curiously, the R-motif is essential for Cdc4p in vivo activity (Fig. 2). Possibly, the R-motif binds additional factors necessary for SCFCdc4p activity. Alternatively, the R-motif may confer important structural features to Cdc4p.
There are several possible explanations why overexpressing cdc4 mutants may cause cell death. It is possible that these portions of Cdc4p have unregulated ubiquitinating activities and thus target essential proteins for degradation. However, we favor an alternative argument. Considerable genetic evidence has indicated that regulating the abundance of F-box-containing proteins is essential for cell viability (23, 25, 27, 30, 39) because such proteins are in competition with one another for binding common components. Additional evidence that altering the abundance of F-box proteins affects the activities of other SCFs is seen in the fact that overexpressing GRR1 exacerbates the growth defect of skp1-11 cells, presumably due to titration of Skp1p away from Cdc4p and Met30p (27). GRR1 overexpression also retards the growth of cdc34 and cdc53 mutants (23, 39). However, MET30 overexpression only weakly affects temperature-sensitive cdc34 and cdc53 strains and has no effect on skp1 mutants (39). The reasons for this remain obscure, but Met30p may be more difficult to overexpress than GRR1. Conversely, loss of GRR1 suppresses cdc34-1 sic1 cells (23), which arrest at a later stage in the cell cycle than cdc34 mutants (48). The reason for this suppression may be due to the relative abundance of different F-box-containing proteins. Since cdc34-1 sic1 cells have a common terminal morphology with cdc4 sic1 cells, it has been suggested that SCFCdc4p is also required later in the cell cycle (48). Thus, one means of suppressing cdc34-1 sic1 cells is that the efficiency of SCFCdc4p is increased. The absence of a nonessential F-box protein would increase the pool of Skp1p, Cdc53p, and Cdc34p and thereby allow greater SCFCdc4p efficiency for an uncharacterized G2/M function (48).
Clearly, regulating the relative abundances of different F-box-containing proteins is essential for maintaining cell viability. However, the data above has been generated by overproducing one F-box protein while another SCF component was encoded by a temperature-sensitive allele. Simply overproducing an F-box protein is not detrimental to the cell (Fig. 3A). Therefore, it is likely that the abundance of an F-box protein is self-regulated. Indeed, Cdc4p has recently been shown to be an unstable protein (54). In this paper, we have identified a motif on one F-box protein, Cdc4p, that regulates its abundance. Overexpressing CDC4 that lacks the R-motif is toxic to cells, although other SCF components are wild type (Fig. 3A).
It seems unlikely that R-motif activity is unregulated. Unregulated R-motif activity would probably result in insufficient Cdc4p to maintain cell viability. During the course of our work, we noted that mutations in the Cdc4p F-box that presumably disrupt the F-box–Skp1p interaction reduced the abundance of the GST-Cdc4p fusions (Fig. 7A). A skp1 allele that specifically hinders SCFCdc4p function also decreases the abundance of only F-box-containing GST-Cdc4p fusions (Fig. 7B). Finally, overexpressing SKP1 increased the abundance of only GST-Cdc4p that contained an F-box (Fig. 8C). Taken together, these data demonstrate the importance of the F-box–Skp1p interaction as a positive regulator of Cdc4p abundance. This conclusion is supported by other genetic evidence, as shown by the original isolation of SKP1 as a high-copy-number suppressor of cdc4 (1). Increasing the abundance of Skp1p increases the abundance of Cdc4p, thereby relieving cdc4 temperature sensitivity.
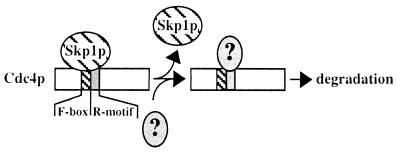
FIG. 8.
Skp1p and the Cdc4p F-box act in concert to regulate Cdc4p abundance. A schematic diagram illustrating the action of two Cdc4p domains is presented. Skp1p binding to the F-box masks the effect of the adjacent R-motif. Release of Skp1p exposes the R-motif, which may recruit proteins that will subsequently target Cdc4p for degradation.
Accordingly, the Skp1p–F-box interaction plays an important role in regulating Cdc4p abundance and hence SCFCdc4p activity (Fig. 8). We favor the model that the binding of Skp1p to the F-box confers stability by preventing the R-motif from binding other factors that destabilize this protein. Specifically, failure of the F-box–Skp1p interaction would expose residues 320 to 341, which would then be capable of binding these putative destabilizing factors. We note, however, that sequences similar to residues 320 to 341 do not appear to be present in other proteins, including other F-box-containing proteins. Therefore, Cdc4p may interact with specific destabilization factors which target Cdc4p for removal from the cell without compromising the abundances of other F-box-containing proteins. Although Met30p similarly lacks any sequence resembling the Cdc4p R-motif, we have demonstrated that a sequence near or within the Met30p F-box also decreases protein abundance (Fig. 4C), and so the model described in Fig. 8 may hold true for Met30p. Like Cdc4p, Met30p is also decreased in its abundance in skp1 mutants (39), suggesting that the Met30p–F-box–Skp1p interaction similarly modulates Met30p abundance. Our data for Cdc4p also parallels that for another yeast protein, Ctf13p, which is involved in kinetochore activity (21). Ctf13p contains an F-box (40), its half-life is decreased in skp1 mutants, and it is also targeted for degradation by the ubiquitin-proteasome pathway (21). Like Cdc4p, Ctf13p activity requires Skp1p binding (21). Potentially, binding of Skp1p to F-box proteins may not be necessary for F-box protein biochemical activity but may be needed to prevent F-box protein degradation. The abundance of an F-box/leucine-rich repeat protein in humans, Skp2, has recently been shown to oscillate during the cell cycle (28). Whether F-box and R-motif-like activities are involved in changes in Skp2 abundance has yet to be explored.
Regulating the abundance of F-box proteins is critical for normal cellular functions. In this paper, we have defined a domain, the R-motif, that is important in regulating the abundance of Cdc4p. Overexpression of Cdc4p lacking this region leads to inviability, even though the cell is wild type for all other SCF components. The precise mechanism by which the R-motif induces a decrease in Cdc4p steady-state abundance remains unknown. However, we have provided evidence that Cdc4p abundance is dependent on the proteasome. Thus, it is likely that the abundance of an ubiquitin ligase might itself be regulated by ubiquitin-dependent degradation pathway.
ACKNOWLEDGMENTS
We thank Alan Bender, Phil Hieter, and Mark Hochstrasser for generously providing strains. We thank Doug Lammer, Peter Roach, and Ron Wek for stimulating discussions and Doug Lammer and Frank Li for critical reading of the manuscript.
This work was supported by National Science Foundation grant MCB-9728069 to M.G.
REFERENCES
- 1.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-Box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 2.Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- 3.Connelly C, Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coux O, Tanaka K, Goldberg A L. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 5.Deshaises R J. The self destructive personality of a cell cycle in transition. Curr Opin Cell Biol. 1995;7:781–789. doi: 10.1016/0955-0674(95)80061-1. [DOI] [PubMed] [Google Scholar]
- 6.Drury L S, Perkins G, Diffley J F. The Cdc4/34/53 pathway targets Cdc6p proteolysis in budding yeast. EMBO J. 1997;16:5966–5979. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elbe R. A simple and efficient procedure for transformation of yeasts. BioTechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- 8.Feldman R M, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/Cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sicp. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 9.Flick J S, Johnston M. Grr1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol Cell Biol. 1991;11:5101–5112. doi: 10.1128/mcb.11.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goebl M G, Yochem J, Jentsch S, McGrath J P, Varchavsky A, Byers B. The yeast cell cycle gene cdc34 encodes a ubiquitin conjugating enzyme. Science. 1988;241:1331–1334. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- 11.Goebl M G, Goetsch L, Byers B. The Ubc3 (Cdc34) ubiquitin-conjugating enzyme is ubiquitinated and phosphorylated in vivo. Mol Cell Biol. 1994;14:3022–3029. doi: 10.1128/mcb.14.5.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haas A L, Siepmann T J. Pathways of ubiquitin conjugation. FASEB J. 1997;11:1257–1268. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- 13.Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaises R J, Peter M. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 1997;11:3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilt W, Wolf D H. Proteasomes: destruction as a programme. Trends Biochem Sci. 1996;21:96–102. [PubMed] [Google Scholar]
- 15.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 16.Hochstrasser M. There’s the Rub: a novel ubiquitin-like modification linked to cell cycle regulation. Genes Dev. 1998;12:901–907. doi: 10.1101/gad.12.7.901. [DOI] [PubMed] [Google Scholar]
- 17.Hunter T. Braking the cycle. Cell. 1993;75:839–841. doi: 10.1016/0092-8674(93)90528-x. [DOI] [PubMed] [Google Scholar]
- 18.Ingram G C, Doyle S, Carpenter R, Schultz E A, Simon R, Coen E S. Dual role for fimbriata in regulating floral homeotic genes and cell division in Antirrhinum. EMBO J. 1997;16:6521–6534. doi: 10.1093/emboj/16.21.6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson S L. Structure and functional analysis of the yeast CDC4 gene product. Ph.D. dissertation. Seattle: University of Washington; 1991. [Google Scholar]
- 20.Kajava A V. Structural diversity of leucine-rich repeat proteins. J Mol Biol. 1998;277:519–527. doi: 10.1006/jmbi.1998.1643. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan K B, Hyman A A, Sorger P K. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell. 1997;91:491–500. doi: 10.1016/s0092-8674(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 22.King R W, Deshaises R J, Peters J-M, Kirschner M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 23.Kishi T, Seno T, Yamao F. Grr1 functions in the ubiquitin pathway in Saccharomyces cerevisiae through association with Skp1. Mol Gen Genet. 1998;257:143–148. doi: 10.1007/s004380050633. [DOI] [PubMed] [Google Scholar]
- 24.Kipreos E T, Lander L E, Wing J P, He W W, Hedgecock E M. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- 25.Lammer D, Mathias N, Laplaza J M, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lew D L, Reed S. Morphogenesis in the yeast cell cycle: regulation of Cdc28 by cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li F, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:5625–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lisztwan J, Marti A, Sutterluty H, Gstaiger M, Wirbelauer C, Krek W. Association of human CUL-1 and ubiquitin-conjugating enzyme CDC34 with the F-box protein p45(SKP2): evidence for evolutionary conservation in the subunit composition of the CDC34-SCF pathway. EMBO J. 1998;17:368–383. doi: 10.1093/emboj/17.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Mathias N, Steussy C N, Goebl M G. Intragenic suppression among CDC34 (UBC3) mutations defines a class of ubiquitin-conjugating catalytic domains. Mol Cell Biol. 1995;15:5635–5644. doi: 10.1128/mcb.15.10.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathias N, Johnson S L, Winey M, Adams A E M, Goetsch L, Pringle J R, Byers B, Goebl M G. Cdc53 acts in concert with Cdc4 and Cdc34 to control the G1 and S phase transition and identifies a conserved family of proteins. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathias N, Steussy C N, Goebl M G. An essential domain with Cdc34p is required for binding to a complex containing Cdc4p and Cdc53p in Saccharomyces cerevisiae. J Biol Chem. 1998;273:4040–4045. doi: 10.1074/jbc.273.7.4040. [DOI] [PubMed] [Google Scholar]
- 32.Mendenhall M D. An inhibitor of p34CDC28 protein kinase activity from Saccharomyces cerevisiae. Science. 1993;259:216–219. doi: 10.1126/science.8421781. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell D A, Marshall T K, Deschenes R J. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–723. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 34.Nasmyth K. Control of the yeast cell cycle by the Cdc28 protein kinase. Curr Opin Cell Biol. 1993;5:166–179. doi: 10.1016/0955-0674(93)90099-c. [DOI] [PubMed] [Google Scholar]
- 35.Nasmyth K. At the heart of the budding yeast cell cycle. Trends Genet. 1996;12:405–412. doi: 10.1016/0168-9525(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 36.Nasmyth K, Hunt T. Cell cycle. Dams and sluices. Nature. 1993;366:634–635. doi: 10.1038/366634a0. [DOI] [PubMed] [Google Scholar]
- 37.Neer E J, Schmidt C J, Nambudripad R, Smith T F. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 38.Pagano M. Cell cycle regulation by the ubiquitin pathway. FASEB J. 1997;11:1067–1075. doi: 10.1096/fasebj.11.13.9367342. [DOI] [PubMed] [Google Scholar]
- 39.Patton E E, Willems A R, Sa D, Kuras L, Thomas D, Craig K L, Tyers M. Cdc53 is a scaffold protein for multiple Cdc53/Skp1/F-box protein complexes that regulate cell division and methionine metabolism in yeast. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patton E E, Willems A R, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don’t Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 41.Piatti S, Bohm T, Cocker J H, Diffley J F, Nasmyth K. Activation of S-phase promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 42.Pickart C M. Targeting substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 43.Pines J. Arresting developments in cell-cycle control. Trends Biochem Sci. 1994;19:143–145. doi: 10.1016/0968-0004(94)90272-0. [DOI] [PubMed] [Google Scholar]
- 44.Rose M D, Winston F, Heiter P. Methods in yeast genetics: a laboratory course. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 45.Ruegger M, Dewey E, Gray W M, Hobbie L, Turner J, Estelle M. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 1998;12:198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Scheffner M, Nuber U, Huibregtse J M. Protein ubiquitination involving an E1-E2-E3 enzyme cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 48.Schwob E, Bohm T, Mendenhall M D, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 49.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 50.Thomas D, Kuras L, Barbey R, Cherest H, Blaiseau P-L, Surdin-Kerjan Y. Met30p, a yeast transcriptional inhibitor that responds to S-adenosylmethionine, is an essential protein with WD-40 repeats. Mol Cell Biol. 1995;15:6526–6534. doi: 10.1128/mcb.15.12.6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 52.Willems A R, Lanker S, Patton E E, Craig K L, Nason T F, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 53.Yochem J, Byers B. Structural comparison of the yeast cell division cycle gene CDC4 and a related pseudogene. J Mol Biol. 1987;195:233–245. doi: 10.1016/0022-2836(87)90646-2. [DOI] [PubMed] [Google Scholar]
- 54.Zhou P, Howley P. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligase. Mol Cell. 1998;2:1–20. doi: 10.1016/s1097-2765(00)80156-2. [DOI] [PubMed] [Google Scholar]