Abstract
The recently sequenced Saccharomyces cerevisiae genome was searched for a gene with homology to the gene encoding the major human AP endonuclease, a component of the highly conserved DNA base excision repair pathway. An open reading frame was found to encode a putative protein (34% identical to the Schizosaccharomyces pombe eth1+ [open reading frame SPBC3D6.10] gene product) with a 347-residue segment homologous to the exonuclease III family of AP endonucleases. Synthesis of mRNA from ETH1 in wild-type cells was induced sixfold relative to that in untreated cells after exposure to the alkylating agent methyl methanesulfonate (MMS). To investigate the function of ETH1, deletions of the open reading frame were made in a wild-type strain and a strain deficient in the known yeast AP endonuclease encoded by APN1. eth1 strains were not more sensitive to killing by MMS, hydrogen peroxide, or phleomycin D1, whereas apn1 strains were ∼3-fold more sensitive to MMS and ∼10-fold more sensitive to hydrogen peroxide than was the wild type. Double-mutant strains (apn1 eth1) were ∼15-fold more sensitive to MMS and ∼2- to 3-fold more sensitive to hydrogen peroxide and phleomycin D1 than were apn1 strains. Elimination of ETH1 in apn1 strains also increased spontaneous mutation rates 9- or 31-fold compared to the wild type as determined by reversion to adenine or lysine prototrophy, respectively. Transformation of apn1 eth1 cells with an expression vector containing ETH1 reversed the hypersensitivity to MMS and limited the rate of spontaneous mutagenesis. Expression of ETH1 in a dut-1 xthA3 Escherichia coli strain demonstrated that the gene product functionally complements the missing AP endonuclease activity. Thus, in apn1 cells where the major AP endonuclease activity is missing, ETH1 offers an alternate capacity for repair of spontaneous or induced damage to DNA that is normally repaired by Apn1 protein.
Base excision repair is a DNA repair pathway that has been characterized in the bacterium Escherichia coli, the yeast Saccharomyces cerevisiae, and human cells (reviewed in reference 46). This DNA repair pathway is responsible for relieving the burden of approximately 10,000 abasic (apurinic/apyrimidinic [AP]) sites estimated to arise each day by spontaneous hydrolysis in the genome of a mammalian cell (24). Base excision repair also repairs damaged bases resulting from endogenous chemical reactions. If this naturally occurring damage is not repaired, genomic stability is compromised (16, 27). Alkylation of some bases changes the pairing preference for an incoming nucleotide during replication. To cope with the potential for mutagenesis due to such alkylation damage, specific DNA glycosylases recognize altered bases and cleave the N-glycosylic bond to result in AP sites (14). These AP sites, in turn, are incised by an AP endonuclease or lyase and further processed for removal of 5′ or 3′ blocking sugar fragments, respectively, thus allowing a DNA polymerase to replace the missing nucleotide (reviewed in reference 49). DNA polymerases may stall during replication upon encountering an AP site (27) or, for some DNA and RNA polymerases, may bypass the AP site (19). AP endonucleases also participate in the repair of DNA strand break damage caused by ionizing agents, hydrogen peroxide, and radiomimetic agents such as bleomycin (7, 55).
Base excision repair is an essential biochemical pathway. Deletion of the gene encoding the major AP endonuclease, DNA polymerase β, or XRCC1 protein in a mouse by targeted homologous recombination results in the absence of viable homozygous mice (embryonic lethal phenotype [25]). Mice deficient in 3-methyladenine DNA glycosylase have been engineered and are viable (9, 10, 20), presumably because the central part of the base excision repair pathway in these animals is still intact.
With the rapidly expanding genetic information available for diverse bacteria, archaea, yeasts, and other eukaryotic species, one can find open reading frames that encode putative proteins displaying homology to known proteins. So far, the genomes of 21 organisms have been completely sequenced (13, 23, 33, 48; see www-c.mcs.anl.gov/home/gaasterl/genomes.html and www.kegg.com). At the amino acid level, all of these organisms have many different conserved DNA repair proteins for diverse DNA repair pathways, which indicates that DNA repair processes evolved very early (4). However, there are some striking oddities. One of these is that E. coli has two distinct AP endonucleases. The first AP endonuclease in E. coli was discovered as an exonuclease and 3′ phosphatase and hence acquired the name of exonuclease III (40). Later, the enzyme was shown to possess an AP endonuclease activity that is about equal to its exonuclease activity (53). Another AP endonuclease was discovered and named endonuclease IV (26); it is induced by the redox-active compound paraquat, among others (5). These enzymes exhibit some differences in substrate preferences (50). Exonuclease III and endonuclease IV belong to separate AP endonuclease families (reviewed in reference 7) with representatives in all domains of life.
The budding yeast, S. cerevisiae, was previously shown to possess an AP endonuclease (encoded by APN1) that is 41% identical to E. coli endonuclease IV over a 280-residue segment (35). S. cerevisiae strains with APN1 deleted exhibit a remarkable sensitivity to oxidizing DNA-damaging agents such as hydrogen peroxide, t-butylhydroperoxide, and the DNA-alkylating agent methyl methanesulfonate (MMS) but not to bleomycin (38). Cell-free lysates from apn1 yeast lose approximately 97% of all AP endonuclease and DNA 3′ repair diesterase activities compared to wild-type cell-free lysates (35). In this report, I provide suggestive evidence for a second AP endonuclease activity in S. cerevisiae that is encoded by ETH1 (open reading frame YBL019W) (11, 17). The gene product is distinct from the yeast AP endonuclease encoded by APN1 and is a homolog of E. coli exonuclease III. S. cerevisiae is the first eukaryote known to possess homologs of each of the AP endonucleases of E. coli. I have also identified an exonuclease III homolog in the fission yeast, Schizosaccharomyces pombe, and propose the same name, eth1+, for the open reading frame SPBC3D6.10.
MATERIALS AND METHODS
Strains, media, and reagents.
Bacterial strain XL1Blue or DH5α was used in all cloning steps. The yeast strains used in this study are presented in Table 1. E. coli was grown in antibiotic-supplemented modified Luria broth (LB; 1% Bacto Tryptone, 0.75% yeast extract, 0.5% sodium chloride); yeast was grown in rich medium (2% Bacto Peptone, 1% yeast extract) supplemented with 80 mg of adenine per liter and 2% (vol/vol) glucose (YPAD). For selection of plasmids, yeast was grown in complete minimal medium (2) lacking uracil but with 2% galactose (CM−ura+gal) or 2% glucose (CM−ura+glu). Rich medium lacking adenine (YPD) was used for growth on plates. A special CM−ura+gal that is limiting in either adenine (0.75 μg/ml) or lysine (1 μg/ml) was used for fluctuation test experiments (see below) (52). All restriction enzymes and DNA-modifying enzymes were obtained from New England Biolabs. MMS was purchased from Sigma, hydrogen peroxide was purchased from Fluka, and phleomycin D1 was purchased from Invitrogen. Radionuclides for Northern analysis were purchased from Dupont NEN.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| DBY747 | MATa leu2-3,112 his3-Δ1 trp1-289am ura3-52 gal2 | Stock of B. Demple laboratory (35) |
| DRY370 | DBY747 apn1-Δ1::HIS3 | Stock of B. Demple laboratory (38) |
| MKP-o | MATα can1-100 ade2-1 lys2-1 ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 | Stock of B. Demple laboratory (34) |
| DRY373 | MKP-o apn1-Δ1::HIS3 | Stock of B. Demple laboratory (38) |
| RBY1, RBY3, RBY4 | MKP-o eth1Δ1::hisG URA3 hisG | This study |
| RBY5, RBY7, RBY8 | DRY373 eth1Δ1::hisG URA3 hisG | This study |
| RBY31 | MKP-o eth1Δ1::hisG | This study |
| RBY71 | DRY373 eth1Δ1::hisG | This study |
Cloning of ETH1.
ETH1 was cloned from DBY747 (35) genomic DNA by PCR with oligonucleotides obtained from Operon. 5′-CAGTCTGCAGTTATTATTGCTGGC and 5′-CAGTCTCGAGTCTCAACTACCGAAG specify the sequences of the oligonucleotides. Underlined bases indicate yeast genomic sequences 5′ and 3′ to the ETH1 open reading frame. Pfu DNA polymerase and the preceding oligonucleotide primers were used to amplify ETH1 from yeast genomic DNA. The PCR product was purified with a PCR purification kit (Qiagen) and subsequently cleaved with PstI and XhoI at sites introduced into the PCR product by the oligonucleotides. Likewise, Bluescript II KS (Stratagene) was cleaved with PstI and XhoI. The PCR product was ligated into Bluescript II KS plasmid under standard conditions, resulting in plasmid BlueETH (2). Plasmid clones were sequenced to verify that the PCR product matched the reported sequence (GenBank accession no. Z35780). A yeast expression plasmid was constructed by subcloning a BamHI-XhoI fragment of BlueETH into pYES2 (Invitrogen), resulting in pETH1.
Disrupting ETH1 in wild-type and apn1 yeast.
BlueETH was digested with BglII to release a 1.38-kb internal piece of ETH1. A 3.84-kb BamHI-BglII fragment, containing one Salmonella hisG sequence on either side of S. cerevisiae URA3, from pNKY51 (a gift of N. Kleckner), was then ligated into the BglII sites in BlueETH to create a deletion and insertion in ETH1. The ETH1 disruption plasmid was named BlueETH::hisG URA3 hisG. Yeast strains MKP-o and DRY373 were transformed (44) with a SacII-XhoI fragment containing the disrupted ETH1 gene, and Ura+ transformants were selected on CM−ura+glu plates. Deletion-insertion mutants were verified by Southern analysis (2). Next, Ura− strains were selected by allowing growth in YPAD overnight followed by growth on 5-fluoroorotic acid plates (2). PCR across the mutated ETH1 locus and subsequent restriction analysis of the PCR product verified that the locus was eth1Δ1::hisG.
Gradient plate analysis.
The technique of constructing a gradient plate has been described previously (6). Briefly, different gradients of MMS were obtained by adding various amounts of MMS to 30 ml of YPD, CM−ura+gal, or CM−ura+glu to create the bottom layer of the plate. After solidification of the bottom layer, a top layer consisting of 30 ml of the corresponding medium alone was added. The plates were dried in a 37°C incubator for 30 min prior to use. A 150-μl aliquot of cells was placed onto a sterile microscope slide. The edge of another sterile microscope slide was then dipped into the cells and stamped onto the gradient plate parallel to the gradient. The plates were incubated upright at 30°C for 3 days.
Toxicity measurements.
Exponential-phase cultures were treated with various concentrations of hydrogen peroxide or phleomycin D1 for 1 h at 30°C with shaking (250 rpm) as in the reported method (38). In other experiments, cells were exposed to MMS as previously described (38). Surviving cells were determined by plating various dilutions on YPD plates and scoring colonies after 2 to 3 days of growth at 30°C. The percentage of cells surviving was calculated from the number of viable colonies per number of cells plated.
Northern analysis.
A single yeast colony was inoculated into 4 ml of YPAD and allowed to grow overnight at 30°C. This culture was then diluted in 10 ml of YPAD to an optical density at 600 nm (OD600) of 0.04 and allowed to grow at 30°C to an OD600 of 0.5. Each 10-ml culture was next treated with 0.1% or 0.2% (vol/vol) MMS for various times. Total RNA was isolated by the published method (45).
Spontaneous mutation rate assay.
Reversion of ochre alleles in lys2-1 and ade2-1 was measured as described previously (38, 52). Briefly, 1,000 cells were plated per well into at least 96 wells for each experiment and strain. Following 10 days of growth at 30°C, the fluctuation test, using the method of P0, was employed to calculate the spontaneous mutation rate (52).
Complementation in bacteria.
E. coli BW287 (Hfr KL16 dut-1 xthA3) (6, 51) was transformed with expression vector pKEN2 or pKENETH, which contains an XbaI-PstI fragment of ETH1 from pT7-7ETH. An expression vector containing the human AP endonuclease cDNA, pKENAPE, was also transformed into BW287 for a positive control. The strain is temperature sensitive; therefore, transformation of competent cells (2) was carried out for 30 min on ice followed by 45 s at 42°C and 2 h at room temperature in 10 volumes of LB prior to plating on Luria agar base supplemented with 75 μg of ampicillin per ml and 125 μg of thymidine per ml. Colonies were visible after 2 days of growth at room temperature (21 to 24°C). Colonies were randomly chosen with a sterile toothpick and transferred onto a new plate by streak-dilution to test for their ability to form single colonies with or without 750 μM isopropyl-β-d-thiogalactopyranoside (IPTG) when incubated at room temperature or 30, 32, 34, 35, 37, or 42°C.
RESULTS
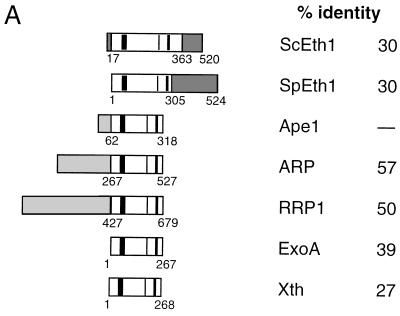
To determine if S. cerevisiae has an exonuclease III homolog, several searches were performed with the complete sequences of exonuclease III and human AP endonuclease by using the program BLASTP (Washington University). These searches revealed no matches. However, when the yeast database (11, 17) was searched with a highly conserved core of 19 amino acids of human AP endonuclease (amino acids 300 to 318 [8, 41, 47]), open reading frame YBL019W (GenBank accession no. Z35780) appeared as a potential match with a segment-pairing BLAST score of P = 0.70. All other potential matches had P > 0.99, indicating a worse match. Direct comparisons of residues 17 to 363 of the yeast open reading frame revealed 30% identity to the major human AP endonuclease, 29% identity to exonuclease III, and 34% identity to a Schizosaccharomyces pombe open reading frame (SPBC3D6.10) (Fig. 1A). Multiple sequence alignments place the predicted Eth1 protein within the exonuclease III family (Fig. 1). Despite amino acid substitutions and additions throughout the yeast proteins compared to other AP endonucleases, all proposed catalytic residues are conserved in both S. cerevisiae and S. pombe Eth1 proteins (Fig. 1B) (35).
FIG. 1.
(A) Protein alignment for selected AP endonucleases of the exonuclease III family. Gray shading indicates sequences that do not have sequence homology to exonuclease III. The percent amino acid identity relative to human AP endonuclease (Ape1) is given for the recombination protein (ARP) of Arabidopsis, the recombination repair protein (RRP1) of Drosophila melanogaster, exonuclease A (ExoA) of Streptococcus pneumoniae, exonuclease III (Xth) of Escherichia coli, the exonuclease III homolog (ScEth1) of Saccharomyces cerevisiae, and the exonuclease III homolog (SpEth1) of Schizosaccharomyces pombe. The vertical bars in the AP endonuclease domain (open box) show highly conserved residues encompassing the proposed catalytic amino acids; other conserved short regions exist but are not indicated (see panel B for details). (B) Partial amino acid sequence alignment of nine selected exonuclease III family members over the DNA repair domains of these proteins. Asterisks indicate proposed catalytic amino acids (18, 29–31). Dots indicate gaps in the sequences. A black background indicates amino acid identity, while gray shading indicates amino acid similarity. Pileup was used to align the sequences (15).
To understand if ETH1 is involved in a physiological response in yeast, deletions in ETH1 were engineered in MKP-o (wild type) and the isogeneic strain DRY373, which carries a deletion and insertion at the APN1 locus (Table 1). The general method used (targeted gene replacement with selection) has been described previously (1). Southern analysis of genomic DNA established that the gene was deleted and replaced by the URA3 cassette at the correct locus (data not shown) in both single- and double-mutant strains. Three independent eth1 single-mutant strains and three independent apn1 eth1 double-mutant strains were isolated, and the eth1Δ1::hisG (Ura−) derivatives, RBY31 and RBY71, were obtained from RBY3 and RBY7 by selection on 5-fluoroorotic acid medium (2) (Table 1).
Role of ETH1 in base excision repair.
Alkylating agents such as MMS add their reactive alkyl groups to nucleophiles in the bases of DNA. These alkylated bases are removed by specific DNA glycosylases, leaving an abasic site. Because the Eth1 protein belongs to the exonuclease III family of AP endonucleases, I hypothesized that yeast strains deficient in this gene would be sensitive to MMS or other DNA-damaging agents and would be similar to an apn1 strain (38). Therefore, initial gradient plate assay experiments were designed to test the sensitivity of these strains to MMS and various DNA-damaging agents. The most striking difference was seen with MMS in the apn1 eth1 strain (RBY71). These strains were also tested for their sensitivity to irradiation by 254-nm light, which causes pyrimidine dimer formation (14). Mutant strains showed no change in sensitivity to this DNA-damaging agent compared to wild-type strains over a range of 10 to 240 J/m2 irradiation (data not shown). These initial results suggested that ETH1 plays some role at least in the response to alkylation damage but not in the response to UV damage.
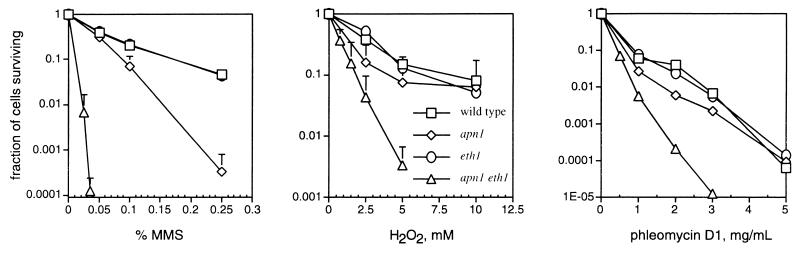
Thus, since there was a phenotypic difference between the double-mutant strain and the single-mutant strain (eth1 strain RBY31) or a wild-type strain when challenged with MMS, a more quantitative investigation of the phenotype was needed. A study of the surviving fraction of cells following acute exposure to MMS, hydrogen peroxide, or phleomycin D1 was undertaken to quantify better the sensitivities of these strains to these agents.
Figure 2 shows the MMS sensitivities of the wild-type and single- and double-mutant strains. Disruption of ETH1 in a wild-type background produced no difference in the number of cells surviving an acute exposure to MMS. The apn1 strain was 3.3-fold more sensitive to MMS than was its wild-type parent, as calculated from the estimated D10 (concentration of agent where 10% of cells survive exposure), in agreement with an earlier report (38). Disruption of ETH1 in an apn1 strain resulted in a dramatic killing effect by MMS. At 0.035% MMS, ∼0.01% of the double-mutant cells survived the challenge whereas ∼50% of apn1 and eth1 single-mutant and wild-type cells remained viable. An estimated D10 value for the double-mutant strain, RBY71, shows that this strain is 20 times more sensitive to MMS than are the wild-type and eth1 strains; this result is therefore in agreement with pilot gradient plate results (data not shown).
FIG. 2.
MMS, hydrogen peroxide, and phleomycin D1 toxicity. Strains are as follows: □, MKP-o; ◊, DRY373; ○, RBY31; ▵, RBY71. The genotypes of these strains are given in the figure. All data are plotted as means and standard deviations from at least three independent experiments. For the phleomycin data, the standard deviations are smaller than the size of the data points. For the limit of detection for this assay, more than 10 colonies had to survive from 2 × 106 cells plated.
Because substrates other than AP sites have relevance in the base excision repair pathway, these yeast strains were exposed to two other agents that cause strand breaks with 3′-blocking deoxyribose fragments (14) and/or oxidized AP sites that are also repaired by this pathway (55; reviewed in reference 7). Treatment of strain DRY373 with hydrogen peroxide revealed a biphasic survival curve. At low concentrations of hydrogen peroxide, apn1 cells were moderately more sensitive than wild-type cells; however, at 5 mM or higher, apn1 cells exhibited a survival similar to wild-type cells (Fig. 2). RBY31 was no more sensitive to hydrogen peroxide than was the wild-type strain. RBY71 revealed a fivefold-greater sensitivity to hydrogen peroxide challenge relative to wild-type cells and was ca. two- to threefold more sensitive to hydrogen peroxide than were single mutant apn1 cells based on the estimated D10 (Fig. 2).
Phleomycin D1 is a glycopeptide antibiotic, closely related in structure to the bleomycins, that produces oxidized AP sites and strand breaks with 3′ phosphoglycolate termini. The accumulation of unrepaired double-strand breaks is believed to be the reason for cytotoxicity produced by bleomycin and, by inference, phleomycin D1 (56). Because phleomycin was reported to be more active at releasing nucleosomes and degrading chromosomes in S. cerevisiae than was bleomycin (32), phleomycin D1 was chosen as the agent in experiments to investigate whether the eth1 single mutant (RBY31) is more sensitive to this compound than is a wild-type strain. An earlier report investigating the cellular role of Apn1 protein stated that apn1 cells were not more sensitive to bleomycin or ionizing radiation than were wild-type cells (38). That finding was surprising since Apn1 was predicted to be essential for removal of 3′ phosphoglycolate moieties resulting from bleomycin-induced DNA damage. That Apn1-deficient cells were not more sensitive to bleomycin than were wild-type cells suggested that there was another mechanism for the repair of bleomycin-induced DNA damage. Therefore, the authors of that report (38) speculated that there might be another AP endonuclease with an associated 3′ DNA repair diesterase activity. In this study, I show that apn1 cells were slightly more sensitive to phleomycin D1 than were wild-type cells and eth1 cells. Again, the eth1 strain was not different from the wild-type strain; however the double-mutant strain, RBY71, was two- to threefold more sensitive to this agent than were wild-type cells as estimated from D10 values (Fig. 2). The results of experiments with the two oxidizing agents, hydrogen peroxide and phleomycin D1, are consistent with a role for Eth1 protein in removing 3′ blocking groups from DNA during base excision repair. However, biochemical experiments with specific substrates and purified protein are needed to confirm this hypothesis.
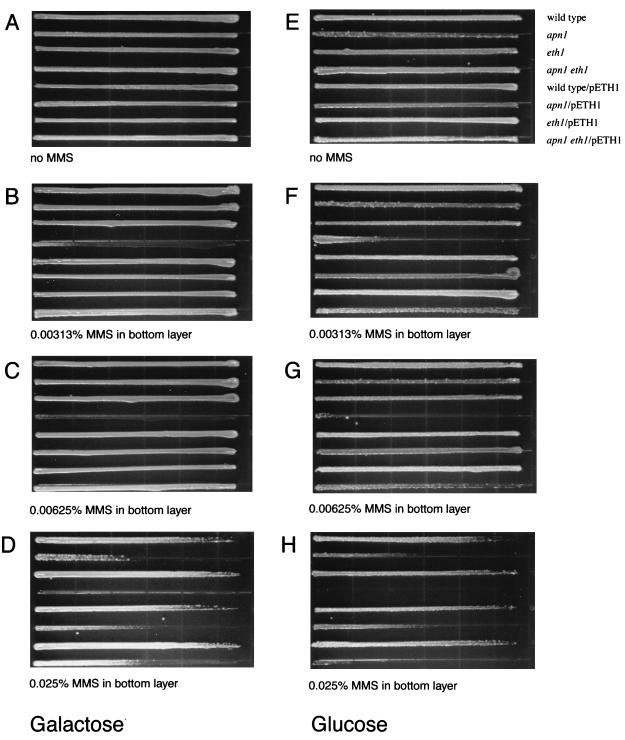
Lastly, I confirmed that the observed sensitivities to MMS were due to the lack of a functional ETH1 gene with galactose-dependent ETH1 expression from pETH1. By gradient plate analysis, induced expression of Eth1 protein clearly reversed the killing effect by MMS (Fig. 3). Growth of RBY71 was completely prevented at concentrations of MMS greater than 0.00313% if the strain contained pYES2 vector. However, RBY71 grew almost as well as an Apn1-deficient strain when complemented with pETH1 and grown on galactose. It should be noted that galactose-dependent ETH1 expression from pETH1 does not provide increased resistance to MMS in any strain except the double-mutant strain. If Apn1 has a broader substrate range for repairing damage caused by MMS than Eth1 does, an increased resistance to MMS would not be seen when Eth1 is expressed in an apn1 strain; alternatively, the endogenous ETH1 mRNA is sufficient for the amount of Eth1 protein and extra mRNA from pETH1 does not result in an increased amount of Eth1 protein. This finding is consistent with complementation of other AP endonucleases in E. coli (36, 37) or yeast (28, 54).
FIG. 3.
Replacement of ETH1. MKP-o (wild type), DRY373 (apn1), RBY31 (eth1), and RBY71 (apn1 eth1) contain a yeast expression vector, pYES2 (top four strains in each panel), or the ETH1 expression vector, pETH1 (bottom four strains in each panel). The growth medium used for the gradient plate is CM−ura+gal (selection for the plasmid and inducing conditions [A to D]) or CM−ura+glu (selection for the plasmid and noninducing conditions [E to H]). The bottom layer does not contain MMS (A and E) or contains 0.00313% MMS (B and F), 0.00625% MMS (C and G), or 0.025% MMS (D and H).
Eth1 protein contributes to genomic stability.
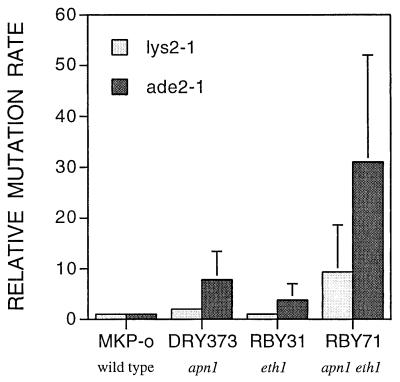
The spontaneous mutation reversion rate of the lys2-1 or ade2-1 ochre mutations was calculated by fluctuation analysis in wild-type and mutant strains. Apn1-deficient strains revealed a spontaneous mutation rate that was twofold higher for lysine prototrophy and about eightfold for adenine prototrophy than that of the wild-type strain, MKP-o. The mutation rate of the eth1 mutant strain, RBY31, showed no difference at lys2-1 but was elevated 3.8-fold at the ade2-1 locus compared to the wild-type strain. This elevation in the spontaneous mutation rate was the only observed phenotypic difference observed for an eth1 strain relative to the wild type. However, in the apn1 eth1 double mutant, RBY71, reversion to lysine or adenine prototrophy was 9- and 31-fold greater, respectively, than the reversion rates of MKP-o at these loci (Fig. 4). Preliminary experiments investigating forward mutation to canavanine resistance indicate that the apn1 eth1 strains display a hypermutator phenotype (data not shown). When the deleted gene was returned to the yeast on an expression plasmid, the increased spontaneous mutation rates returned to wild-type levels (data not shown). Spontaneous mutations giving rise to prototrophy may occur by ochre suppression or true reversion. True reversions accounted for about 10 to 20% of the total revertants, while the majority of revertants (80 to 90%) were likely results of extragenic mutations (suppressors).
FIG. 4.
Spontaneous reversion rates for lys2-1 and ade2-1. The relative increase in mutation rate is plotted with respect to that of the wild-type strain MKP-o, which is defined as 1. Data are means and standard deviations from three independent experiments.
Complementation of an exonuclease III temperature-sensitive mutant by ETH1—evidence for AP endonuclease activity.
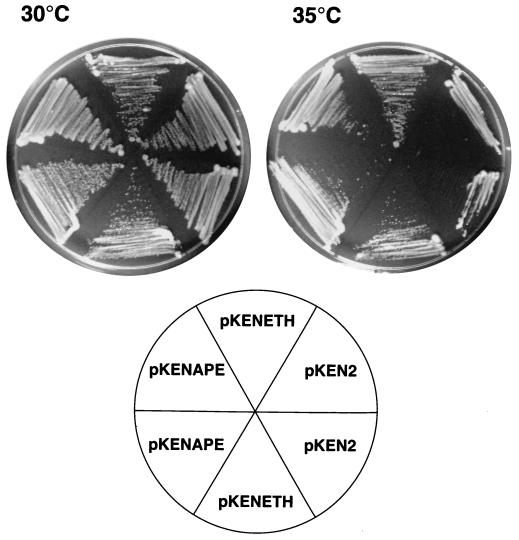
E. coli dut mutants are deficient in dUTPase and incorporate large amounts of dUMP into DNA from large intracellular dUTP pools (6). Because a dut strain is wild type with respect to its uracil-DNA N-glycosylase activity, many AP sites are created in the chromosome. Survival of this strain is drastically reduced if it is then made deficient for AP endonuclease activity. However, a temperature-sensitive mutant that grows at room temperature but not at 37°C was isolated. (Survival is less than 10−4 at temperatures greater than or equal to 37°C.) This dut-1 xthA3 strain (BW287) (6, 51) can therefore be used as a tool to test whether a gene expressed in trans can complement the missing AP endonuclease (exonuclease III). The conditional lethal phenotype suggests that exonuclease III is essential for the repair of AP sites created within DNA by uracil DNA N-glycosylase. IPTG-inducible expression of ETH1 from pKENETH in BW287 resulted in a runny and wet colony phenotype. Fewer colonies arose on plates that contained IPTG than on plates that did not (data not shown). These two observations suggest a toxic effect of Eth1 in E. coli and may explain why attempts at expression of protein have failed with E. coli. It should be noted that many codons in ETH1 are rare codons for E. coli, which could be the most important reason why full-length protein was not obtained with bacterial expression systems. Expression of ETH1 in yeast has not resulted in sufficient quantity of protein either. Therefore, genetic complementation of an exonuclease III-deficient E. coli strain became a working alternative to establish whether Eth1 protein has nicking activity. When pKENETH was transformed into BW287, Eth1 demonstrably rescued the temperature-sensitive phenotype of this strain and thus evidently functions to replace the missing exonuclease III activity. The complementation does not determine if the nicking occurred on the 5′ side (AP endonuclease) or the 3′ side (AP lyase) of the AP site, but it seems unlikely that Eth1 would have the mechanism of a lyase, since it does not have any conserved sequences that look like those of lyases. Also, none of the known exonuclease III family members have lyase activity. The genetic complementation was obtained under the restrictive conditions of no IPTG (leaky expression) and 35°C and with freshly transformed cells from plates incubated at room temperature (Fig. 5).
FIG. 5.
Complementation of E. coli BW287. Three plasmids were transformed into BW287: pKEN2 (vector control), pKENETH (ETH1 expression), and pKENAPE (human AP endonuclease protein expression). The plates were incubated at the indicated temperatures for 24 h.
ETH1 transcription.
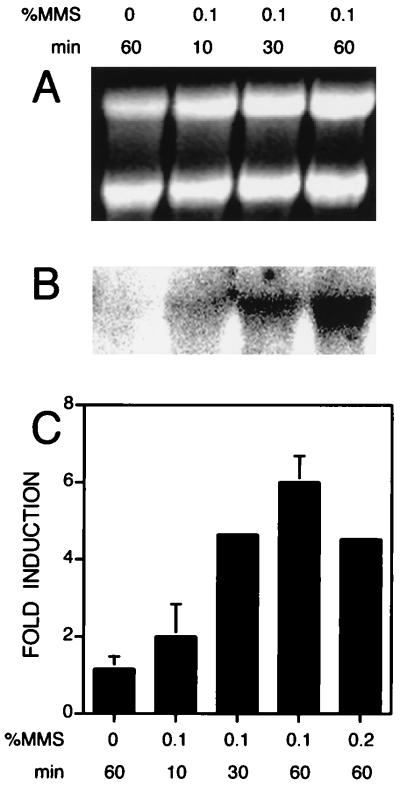
During the course of the characterization of the phenotype of an eth1 strain and an apn1 eth1 strain, I tested a wild-type strain for the possibility that ETH1 is regulated at the transcriptional level by the alkylating agent MMS, which followed from an initial finding that ETH1 mRNA might be induced by MMS as assayed by gene chip technology (20a). The wild-type strain, DBY747, was grown to mid-log phase (OD600 = 0.5) and then challenged with different concentrations of MMS and times of exposure (Fig. 6). ETH1 mRNA is induced as much as sixfold in a time-dependent manner by MMS. This new finding implies that ETH1 plays an important role in a stress response program that is initiated upon exposure to an agent that causes methylation of DNA and protein species. Many genes are induced in response to MMS, but not many are induced at the fivefold level or higher. In fact, it is remarkable that ETH1 is inducible whereas APN1 mRNA is only slightly induced (2.4-fold under the same conditions) (21). The identical experiment was performed with an apn1 strain (DRY370). The expression of ETH1 mRNA in the apn1 strain increased with the same kinetics and to the same level with the same concentration of MMS used in experiments with the wild-type strain, DBY747 (data not shown). Thus, the absence of APN1 does not affect the expression of ETH1.
FIG. 6.
Northern blot analysis of ETH1. Total RNA was harvested from DBY747, and 20 μg was loaded into each lane. Following electrophoresis and transfer to a positively charged nylon membrane (Boehringer Mannheim), the RNA was hybridized to a radioactive internal BglII fragment of ETH1. (A) 28S and 18S rRNA revealed by ethidium bromide staining. (B) Hybridization intensity of the radiolabeled probe to ETH1 mRNA revealed by phosphorimager analysis (Bio-Rad) of RNA from the gel shown in panel A. (C) Fold induction shown from Northern analysis of three independent measurements from two experiments. To correct for RNA-loading differences, the phosphorimager data in panel B are normalized to the quantified intensities of the 28S and 18S rRNA shown in panel A. Data are presented as means and standard deviations (n = 3).
DISCUSSION
In this report, I describe the function of a previously unknown damage-inducible gene in S. cerevisiae which, in the absence of APN1, is responsible for limiting the occurrence of spontaneous mutations and for protection against the cytotoxic effects of three different DNA-damaging agents, MMS, hydrogen peroxide, and phleomycin D1, whose damages are processed by the base excision repair pathway. The effect of another DNA-damaging agent, 254-nm light (up to a dose of 240 J/m2), on the growth of mutant strains was the same as that on the wild-type strain. This result is not surprising since base excision repair has not been associated with repair of UV-induced DNA damage in yeast. However, I would predict that apn1 eth1 or eth1 cells would display sensitivity to ionizing radiation or t-butylhydroperoxide if Eth1 were responsible for repairing 3′ blocking fragments from DNA strand breaks or repair of oxidized AP sites, since these agents also cause similar, if not identical, damage to that caused by hydrogen peroxide or phleomycin D1. Such experiments will indicate whether the DNA damage produced by these agents (presumably DNA strand breaks with 3′ phosphoglycolate or 3′ phosphate groups) is a substrate for repair by Eth1 in vivo. In contrast to xth E. coli strains, yeast strains deficient in Eth1 show only a modest increase in their sensitivity to hydrogen peroxide, and this small hypersensitivity is observed only when they are already deficient for the major yeast AP endonuclease encoded by APN1. The major human AP endonuclease, when expressed in yeast, is also only modestly able to confer added resistance to hydrogen peroxide, which suggests that Eth1 protein may be more like other eukaryotic AP endonucleases than like bacterial forms of this enzyme (54).
That yeast and E. coli are opposite with respect to the ratios of each type of AP endonuclease also suggests some specialized niche selected for the prevalence of one type of enzyme over the other. E. coli may encounter agents that result in damage that is more efficiently repaired by exonuclease III, but S. cerevisiae requires mostly the endonuclease IV-like Apn1 protein for the damage it most often encounters. It is possible that the subtle differences in substrates, i.e., AP sites generated from spontaneous hydrolysis of the N-glycosylic bond and from glycosylase-mediated chemistry versus the oxidized AP sites and breaks containing fragmented sugars at the 3′ end, dictate the necessity for the two different AP endonucleases. However, both types of enzymes cleave the AP site, resulting in an identical product.
Another comparison can be made among the AP endonucleases of S. cerevisiae and E. coli. The major AP endonucleases of both organisms, Apn1 and exonuclease III, respectively, are seemingly noninducible or mildly inducible. The minor AP endonuclease, endonuclease IV of E. coli or Eth1 of S. cerevisiae, is the AP endonuclease that is inducible by agents that tax the ability of the cell to grow. The major AP endonucleases are needed at relatively high levels and at all times in an aerobic environment to repair the numerous spontaneously arising abasic sites. MMS alkylates bases in DNA that are processed into AP sites that look no different from a spontaneously arising AP site. Apn1 therefore repairs these AP sites as well. However, MMS causes other damage and initiates a global stress response that results in the change in copy number of hundreds of mRNAs (21). ETH1 is one of the many genes that are induced by this program. The minor AP endonucleases are thus available for recruitment when there is a particular need during certain stress conditions to repair a specific subset of damages or to provide backup DNA repair capacity.
Since S. pombe has no detectable AP endonuclease activity and yet has homologs of both APN1 (39; GenBank accession no. U33625) and ETH1 (this work), future experiments with S. pombe are needed to determine whether its eth1 and/or apn1 mRNA is also inducible and how exactly the organism survives a challenge by MMS or oxidative DNA damage. S. pombe may use its endonuclease III protein to remove AP sites (22, 42).
Attempts have been made to express and purify Eth1 protein for biochemical studies of its enzymatic activity on various DNA substrates. However, finding an expression system that produces sufficient quantity of protein has been unsuccessful. Although the current data indicate that ETH1 encodes a protein with AP endonuclease and DNA 3′-repair diesterase activities, future biochemical experiments with purified protein would be helpful in confirming the results presented here. Cell-free lysates of apn1 eth1 strains expressing ETH1 from a galactose-inducible promoter in the high-copy-number pETH1, however, exhibit neither AP endonuclease nor DNA 3′-repair diesterase activity despite the presence of abundant relevant mRNA (data not shown). Therefore, either these cell-free lysates lack a necessary cofactor or Eth1 is so labile that all activity is lost during the preparation of the lysate. Alternatively, the lack of detectable protein in yeast lysates may result from tightly controlled expression in yeast. This possibility is supported by two observations: (i) cells growing in rich medium show little or no detectable ETH1 mRNA, but mRNA is found in cells exposed to 0.1% MMS and increases in amount over time; and (ii) the high level of ETH1 mRNA from pETH1 does not lead to a proportionately high level of Eth1 protein (data not shown). However, Sander and Ramotar were successful in partially purifying an AP endonuclease activity from apn1 cells (43). Whether this activity is the same as the activity of the ETH1 gene product remains to be proven.
Two other eukaryotic AP endonucleases complement E. coli genetically (37, 56), and so this approach was used to establish whether Eth1 has AP endonuclease activity in vivo. ETH1 was able to complement the exonuclease III deficit in strain BW287 to allow growth at a restrictive temperature, but not without an adverse effect on E. coli. Complementation was observed only at 35°C and with or without IPTG. Although expression and complementation was observed when IPTG was added, the number of colonies formed was smaller than if it were absent from the medium.
Some comments regarding the structural features of these enzymes are warranted. Figure 1B shows a partial alignment of nine different AP endonucleases belonging to the exonuclease III family. A larger family is possible if DNase I and poly(A) elements like human L1 endonuclease are included (12). DNase I and the retrotransposons such as L1 endonuclease have conserved amino acids surrounding the active-site residues in the AP endonucleases; however, L1 endonuclease and DNase I do not cleave AP sites (12). L1 endonuclease cleaves preferentially at DNA sites containing L1-like DNA sequences, and DNase I nicks DNA nonspecifically (12). The nonspecific nicking activity may be the result of three helical sequences present among the AP endonucleases but absent in DNase I and L1 endonuclease (18). Another feature of the exonuclease III family of proteins is that of the C-terminal sequences of 160 and 220 amino acids in Eth1 for S. cerevisiae and S. pombe, respectively. Only these organisms have such sequences, and yet these organisms diverged from each other evolutionarily almost as long ago as humans diverged from them (3). The yeast-specific C-terminal domains contain short runs of basic amino acids that are similar to the nuclear targeting sequences identified for Apn1 (36). Perhaps these extra C-terminal amino acids are required for nuclear targeting of these proteins.
The use of future genetic crosses of RBY71 with the many available mutations in DNA repair genes, as well as screening for extrageneic suppressors, will be invaluable in elucidating the physiological role of the ETH1 gene product and its contribution to overall base excision DNA repair pathways. Currently, no AP endonuclease-deficient mammalian cell lines have been isolated for studies of the mechanisms of base excision repair, despite attempts to generate them. Here I describe the first eukaryote having deletions in two genes coding for AP endonuclease activity, which offers a valuable tool for investigating eukaryotic base excision repair.
ADDENDUM
During the review of this work, another name (APN2) was chosen by Louise Prakash to describe the gene identified and characterized in this report. ETH1 was registered as the name for this gene with the curators of the Saccharomyces Genome Database on 17 March 1998.
ACKNOWLEDGMENTS
I thank Bernard Weiss for providing bacterial strains for genetic complementation experiments, David M. Wilson III for construction of pKENAPE, and Yuji Masuda, Yong-jie Xu, Elisabeth M. Bailey, Brian J. Glassner, Scott A. Jelinsky, Bruce Demple, and Donny Wong for discussions and critical reading of the manuscript. Special recognition and thanks are given to Scott A. Jelinsky for the initial observation that ETH1 mRNA is induced by exposure of yeast to MMS and to Bruce Demple for providing financial support and encouragement.
This work was supported by NIH grants GM40000, ES03926, and CA71993 to B. Demple. R.A.O.B. is a Charles A. King Trust Fellow and is partially funded by Fleet Investment Management, Trustee of the Charles A. King Trust.
ADDENDUM IN PROOF
Bernard A. Connolly and his group in the United Kingdom recently demonstrated the conversion of bovine pancreatic DNase I to a DNA repair AP endonuclease. The recombinant protein has 14 amino acids, of which 10 are amino acids of an α-helix in exonuclease III that is not present in the natural form of DNase I, inserted at a position on the surface of DNase I corresponding to the location of the extra α-helix in exonuclease III (S. Cal, K. L. Tan, A. McGregor, and B. A. Connolly, EMBO J. 17:7128–7138, 1998).
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 3.Berbee M L, Taylor J W. Dating the evolutionary radiations of the true funghi. Can J Bot. 1993;71:1114–1127. [Google Scholar]
- 4.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Chan E, Weiss B. Endonuclease IV of Escherichia coli is induced by paraquat. Proc Natl Acad Sci USA. 1987;84:3189–3193. doi: 10.1073/pnas.84.10.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham R P, Saporito S M, Spitzer S G, Weiss B. Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol. 1986;168:1120–1127. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 8.Demple B, Herman T, Chen D S. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc Natl Acad Sci USA. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elder R H, Weeks R J, Willington M A, Cooper D P, Rafferty J A, Deans B, Hendry J H, Margison G P. Meeting of the American Association for Cancer Research. San Diego, Calif: American Association for Cancer Research; 1997. Alkylpurine-DNA-N-glycosylase (APNG) knockout mice, abstr. 2411. [Google Scholar]
- 10.Engelward B P, Weeda G, Wyatt M D, Broekhof J L, de Wit J, Donker I, Allan J M, Gold B, Hoeijmakers J H, Samson L D. Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc Natl Acad Sci USA. 1997;94:13087–13092. doi: 10.1073/pnas.94.24.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldmann H, Aigle M, Aljinovic G, Andre B, Baclet M C, Barthe C, Baur A, Becam A M, Biteau N, Boles E, et al. Complete DNA sequence of yeast chromosome II. EMBO J. 1994;13:5795–5809. doi: 10.1002/j.1460-2075.1994.tb06923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Q, Moran J V, Kazazian H H, Jr, Boeke J D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 13.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Venter J C, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 14.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C: ASM Press; 1995. [Google Scholar]
- 15.Genetics Computer Group. Program manual for the Wisconsin Package. 8th ed. Madison, Wis: GCG Inc.; 1994. [Google Scholar]
- 16.Gentil A, Cabral-Neto J B, Mariage-Samson R, Margot A, Imbach J L, Rayner B, Sarasin A. Mutagenicity of a unique apurinic/apyrimidinic site in mammalian cells. J Mol Biol. 1992;227:981–984. doi: 10.1016/0022-2836(92)90513-j. [DOI] [PubMed] [Google Scholar]
- 17.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, Louis E J, Mewes H W, Murakami Y, Philippsen P, Tettelin H, Oliver S G. Life with 6000 genes. Science. 1996;274:546. doi: 10.1126/science.274.5287.546. , 563–567. [DOI] [PubMed] [Google Scholar]
- 18.Gorman M A, Morera S, Rothwell D G, de La Fortelle E, Mol C D, Tainer J A, Hickson I D, Freemont P S. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J. 1997;16:6548–6558. doi: 10.1093/emboj/16.21.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halas A, Baranowska H, Policinska Z, Jachymczyk W J. Involvement of the REV3 gene in the methylated base-excision repair system. Cooperation of two DNA polymerases, delta and Rev3p, in the repair of MMS-induced lesions in the DNA of Saccharomyces cerevisiae. Curr Genet. 1997;31:292–301. doi: 10.1007/s002940050208. [DOI] [PubMed] [Google Scholar]
- 20.Hang B, Singer B, Margison G P, Elder R H. Targeted deletion of alkylpurine-DNA-N-glycosylase in mice eliminates repair of 1,N6-ethenoadenine and hypoxanthine but not of 3,N4-ethenocytosine or 8-oxoguanine. Proc Natl Acad Sci USA. 1997;94:12869–12874. doi: 10.1073/pnas.94.24.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Jelinsky, S. A. Personal communication.
- 21.Jelinsky, S. A., and L. D. Samson. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl. Acad. Sci., in press. [DOI] [PMC free article] [PubMed]
- 22.Karahalil B, Roldan-Arjona T, Dizdaroglu M. Substrate specificity of Schizosaccharomyces pombe Nth protein for products of oxidative DNA damage. Biochemistry. 1998;37:590–595. doi: 10.1021/bi971660s. [DOI] [PubMed] [Google Scholar]
- 23.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Venter J C, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 24.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 25.Lindahl T, Karran P, Wood R D. DNA excision repair pathways. Curr Opin Genet Dev. 1997;7:158–169. doi: 10.1016/s0959-437x(97)80124-4. [DOI] [PubMed] [Google Scholar]
- 26.Ljungquist S, Lindahl T, Howard-Flanders P. Methyl methane sulfonate-sensitive mutant of Escherichia coli deficient in an endonuclease specific for apurinic sites in deoxyribonucleic acid. J Bacteriol. 1976;126:646–653. doi: 10.1128/jb.126.2.646-653.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loeb L A, Preston B D. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 28.Masson J Y, Ramotar D. Normal processing of AP sites in Apn1-deficient Saccharomyces cerevisiae is restored by Escherichia coli genes expressing either exonuclease III or endonuclease III. Mol Microbiol. 1997;24:711–721. doi: 10.1046/j.1365-2958.1997.3841748.x. [DOI] [PubMed] [Google Scholar]
- 29.Masuda Y, Bennett R A O, Demple B. Dynamics of the interaction of human apurinic endonuclease (Ape1) with its substrate and product. J Biol Chem. 1998;273:30352–30359. doi: 10.1074/jbc.273.46.30352. [DOI] [PubMed] [Google Scholar]
- 30.Masuda Y, Bennett R A O, Demple B. Rapid dissociation of human apurinic endonuclease (Ape1) from incised DNA induced by magnesium. J Biol Chem. 1998;273:30360–30365. doi: 10.1074/jbc.273.46.30360. [DOI] [PubMed] [Google Scholar]
- 31.Mol C D, Kuo C F, Thayer M M, Cunningham R P, Tainer J A. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature. 1995;374:381–386. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- 32.Moore C W. Cleavage of cellular and extracellular Saccharomyces cerevisiae DNA by bleomycin and phleomycin. Cancer Res. 1989;49:6935–6940. [PubMed] [Google Scholar]
- 33.Pennisi E. Laboratory workhorse decoded. Science. 1997;277:1432–1434. doi: 10.1126/science.277.5331.1432. [DOI] [PubMed] [Google Scholar]
- 34.Pierce M K, Giroux C N, Kunz B A. Development of a yeast system to assay mutational specificity. Mutat Res. 1987;182:65–74. doi: 10.1016/0165-1161(87)90055-0. [DOI] [PubMed] [Google Scholar]
- 35.Popoff S C, Spira A I, Johnson A W, Demple B. Yeast structural gene (APN1) for the major apurinic endonuclease: homology to Escherichia coli endonuclease IV. Proc Natl Acad Sci USA. 1990;87:4193–4197. doi: 10.1073/pnas.87.11.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramotar D, Kim C, Lillis R, Demple B. Intracellular localization of the Apn1 DNA repair enzyme of Saccharomyces cerevisiae. Nuclear transport signals and biological role. J Biol Chem. 1993;268:20533–20539. [PubMed] [Google Scholar]
- 37.Ramotar D, Popoff S C, Demple B. Complementation of DNA repair-deficient Escherichia coli by the yeast Apn1 apurinic/apyrimidinic endonuclease gene. Mol Microbiol. 1991;5:149–155. doi: 10.1111/j.1365-2958.1991.tb01835.x. [DOI] [PubMed] [Google Scholar]
- 38.Ramotar D, Popoff S C, Gralla E B, Demple B. Cellular role of yeast Apn1 apurinic endonuclease/3′-diesterase: repair of oxidative and alkylation DNA damage and control of spontaneous mutation. Mol Cell Biol. 1991;11:4537–4544. doi: 10.1128/mcb.11.9.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramotar D, Vadnais J, Masson J Y, Tremblay S. Schizosaccharomyces pombe apn1 encodes a homologue of the Escherichia coli endonuclease IV family of DNA repair proteins. Biochim Biophys Acta. 1998;1396:15–20. doi: 10.1016/s0167-4781(97)00160-7. [DOI] [PubMed] [Google Scholar]
- 40.Richardson C, Kornberg A. A deoxyribonucleic acid phosphatase-exonuclease from Escherichia coli. I. Purification of the enzyme and characterization of the phosphatase activity. J Biol Chem. 1964;239:242–250. [PubMed] [Google Scholar]
- 41.Robson C N, Hickson I D. Isolation of cDNA clones encoding a human apurinic/apyrimidinic endonuclease that corrects DNA repair and mutagenesis defects in E. coli xth (exonuclease III) mutants. Nucleic Acids Res. 1991;19:5519–5523. doi: 10.1093/nar/19.20.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roldan-Arjona T, Anselmino C, Lindahl T. Molecular cloning and functional analysis of a Schizosaccharomyces pombe homologue of Escherichia coli endonuclease III. Nucleic Acids Res. 1996;24:3307–3312. doi: 10.1093/nar/24.17.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sander M, Ramotar D. Partial purification of Pde1 from Saccharomyces cerevisiae: enzymatic redundancy for the repair of 3′-terminal DNA lesions and abasic sites in yeast. Biochemistry. 1997;36:6100–6106. doi: 10.1021/bi970048y. [DOI] [PubMed] [Google Scholar]
- 44.Schiestl R H, Gietz R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 45.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seeberg E, Eide L, Bjoras M. The base excision repair pathway. Trends Biochem Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 47.Seki S, Hatsushika M, Watanabe S, Akiyama K, Nagao K, Tsutsui K. cDNA cloning, sequencing, expression and possible domain structure of human APEX nuclease homologous to Escherichia coli exonuclease III. Biochim Biophys Acta. 1992;1131:287–299. doi: 10.1016/0167-4781(92)90027-w. [DOI] [PubMed] [Google Scholar]
- 48.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Reeve J N, et al. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Srivastava D K, Vandeberg B J, Prasad R, Molina J T, Beard W A, Tomkinson A E, Wilson S H. Mammalian abasic site base excision repair—identification of the reaction sequence and rate-determining steps. J Biol Chem. 1998;273:21203–21209. doi: 10.1074/jbc.273.33.21203. [DOI] [PubMed] [Google Scholar]
- 50.Takeuchi M, Lillis R, Demple B, Takeshita M. Interactions of Escherichia coli endonuclease IV and exonuclease III with abasic sites in DNA. J Biol Chem. 1994;269:21907–21914. [PubMed] [Google Scholar]
- 51.Taylor A F, Weiss B. Role of exonuclease III in the base excision repair of uracil-containing DNA. J Bacteriol. 1982;151:351–357. doi: 10.1128/jb.151.1.351-357.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Von Borstel R C. Measuring spontaneous mutation rates in yeast. Methods Cell Biol. 1978;20:1–24. doi: 10.1016/s0091-679x(08)62005-1. [DOI] [PubMed] [Google Scholar]
- 53.Weiss B. Endonuclease II of Escherichia coli is exonuclease III. J Biol Chem. 1976;251:1896–1901. [PubMed] [Google Scholar]
- 54.Wilson D M, III, Bennett R A O, Marquis J C, Ansari P, Demple B. Trans-complementation by human apurinic endonuclease (Ape) of hypersensitivity to DNA damage and spontaneous mutator phenotype in apn1−yeast. Nucleic Acids Res. 1995;23:5027–5033. doi: 10.1093/nar/23.24.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Y-J, Kim E Y, Demple B. Excision of C4′-oxidized deoxyribose lesions from double-stranded DNA by human AP endonuclease (Ape1 protein) and DNA polymerase β. J Biol Chem. 1998;273:28837–28844. doi: 10.1074/jbc.273.44.28837. [DOI] [PubMed] [Google Scholar]
- 56.Yamamoto K, Hutchinson F. Response to bleomycin of E. coli mutants deficient in DNA repair. J Antibiot (Tokyo) 1979;32:1181. doi: 10.7164/antibiotics.32.1181. [DOI] [PubMed] [Google Scholar]