Summary
Background
COVID-19 has the potential to cause outbreaks in hospitals. Given the comorbid and elderly cohort of patients hospitalized, hospital-acquired COVID-19 infection is often fatal. Pathogen genome sequencing is becoming increasingly important in infection prevention and control (IPC).
Aim
To inform the understanding of in-hospital SARS-CoV-2 transmission in order to improve IPC practices and to inform the future development of virological testing for IPC.
Methods
Patients detected COVID-19 positive by polymerase chain reaction on Ward A in April and May 2020 were included with contact tracing to identify other potential cases. Genome sequencing was undertaken for a subgroup of cases. Epidemiological, genomic, and cluster analyses were performed to describe the epidemiology and to identify factors contributing to the outbreak.
Findings
Fourteen cases were identified on Ward A. Contact tracing identified 16 further patient cases; in addition, eight healthcare workers (HCWs) were identified as being COVID-19 positive through a round of asymptomatic testing. Genome sequencing of 16 of these cases identified viral genomes differing by two single nucleotide polymorphisms or fewer, with further cluster analysis identifying two groups of infection (a five-person group and a six-person group).
Conclusion
Despite the temporal relationship of cases, genome sequencing identified that not all cases shared transmission events. However, 11 samples were found to be closely related and these likely represented in-hospital transmission. This included three HCWs, thereby confirming transmission between patients and HCWs.
Keywords: COVID-19, Hospital-acquired infection, Outbreak, Genome sequencing
Introduction
The COVID-19 pandemic has caused significant disruption and mortality globally. Up to April 11th, 2021, there have been 150 million cases of COVID-19 worldwide and more than three million deaths [1]. There is growing evidence that hospital-acquired infections are occurring and that they carry a significant mortality risk [[2], [3], [4]]. The sequencing of pathogen genomes has proven fruitful in previous communicable disease outbreaks and is already demonstrating utility in responding to the COVID-19 pandemic [[5], [6], [7], [8]].
In this report, the authors investigate an outbreak of COVID-19 on an elderly care ward at a hospital in the UK using epidemiological and genomic methods. The aim of this study was to better understand in-hospital SARS-CoV-2 transmission in order to improve immediate infection prevention and control (IPC) and to explore the potential use of virological testing for IPC purposes.
Methods
The investigation was retrospective, did not alter the management of the patients involved, and is reported in line with the ORION checklist for reporting an outbreak [9].
Ethics statement
Informed consent was not gained from patients involved in this outbreak. All patients were managed according to clinical judgement and infection control practices in order to treat them and control the outbreak according to local guidelines. Patients did not undergo randomization or intervention for the purpose of this report. Data have been analysed and presented anonymously.
Healthcare setting and affected patient population
Ward A is a 17-bed all-male elderly medicine ward at the Royal Sussex County Hospital (RSCH), a tertiary centre in East Sussex, UK. At the start of the COVID-19 pandemic, Ward A was designated for patients with no clinical, radiological, or polymerase chain reaction (PCR) evidence of COVID-19 infection (hereinafter referred to as ‘Green’). Ward A has two bays, connected with a corridor, but due to the shared facilities (toilets, kitchen, and staff) it was considered as one area in this analysis.
All patients presenting to the hospital were assessed for clinical signs of COVID-19. Patients were admitted to Green wards only if a specialty registrar or consultant deemed that COVID-19 infection was unlikely. The decision was based primarily upon the absence of symptoms or radiographic features of COVID-19 as SARS-COV-2 PCR tests took >24 h to return a result. Until April 27th, 2020, PCR testing for SARS-COV-2 was reserved for patients displaying symptoms of COVID-19 (shortness of breath, hypoxia, fever, or cough). However, after that time, hospital policy advised that all patients undergo an admission PCR test. Patients without COVID-19 symptoms could be moved to a Green ward before their PCR result was available.
Testing and laboratory methods
Testing for SARS-CoV-2 was performed by real-time reverse transcriptase (RT)–PCR method in the department of microbiology and infection (RSCH). The targets for the test include ORF1ab and E genes. The manufacturer reported 100% specificity and 95% sensitivity (with analytical detection limit of 25 copies/rxn). A random and limited number of positive samples in this study were then identified and assigned unique and anonymized COG IDs. A minimum of 25 μL of RNA extract was prepared for each participant sample in this study and then sent for sequencing to the research laboratory at Queen Alexandra Hospital in Portsmouth as per COG UK Sample Transfer Standard Operating Procedure.
For sequencing, viral RNA samples were processed using the ARTIC nCoV-2019 sequencing protocol (GunIt) V.2 [10]. Full details of the laboratory methods and sequencing protocol can be found in the Appendix [[11], [12], [13]].
Analytical methods
Definitions
‘Primary cases’ were any patient detected as SARS-CoV-2 PCR positive while on Ward A. Patients were ‘contacts’ if they shared Ward A with a primary case in the 14 days (the incubation period of SARS-CoV-2) before or after the primary case's positive result. Contacts with a positive SARS-CoV-2 PCR result within 21 days of the primary case were considered involved in the outbreak and defined as ‘secondary cases’. The timeframe of 21 days was selected to capture the incubation period of SARS-CoV-2 with some additional range given the relatively low availability of testing.
The symptoms of COVID-19 investigated in this study were: new, persistent cough; hypoxia; shortness of breath; fever >37.5°C and anosmia.
Case-finding methods
All patients detected as COVID-19 positive on Ward A in March–May 2020 were identified through the hospital COVID-19 database. The clinical history and locations were extracted for these patients from their clinical notes and the electronic bed management system (Medway). Electronically stored patient lists were used to identify contacts. The clinical history of contacts was evaluated to identify any evidence of COVID-19 infection within 21 days of contact. If positive, their locations were extracted, and their patient contacts traced. This process was repeated until there were no further contacts.
Although not routine, all healthcare workers (HCWs) who worked on Ward A on May 29th and 30th, 2020, underwent an ad-hoc round of PCR testing on those days as part of the outbreak investigation and the results are included here.
Data analysis
Data were extracted in June 2020 and sequencing performed in August 2020. Data processing was performed in R (v1.2.5033), visualization with ggPlot2 (vs3.3.2), and iGraph (v1.2.6). The package transcluster (v0.1.0) in R was used to analyse the relatedness of case sequence data [14]. Based on previously published work, the serial transmission interval (β) was set at 5 days, the viral mutation rate (λ) at two mutations per month, and a probability threshold of 80% [5,6,14]. The number of inferred transmission events (T) was varied from one to 10. This allowed for the possibility that there were undetected cases in the clusters that contributed to the transmission chains. As a comparison, given the relatively low mutation rate of SARS-CoV-2, samples differing by two single nucleotide polymorephisms (SNPs) or fewer were considered related in a second analysis [5,15].
A descriptive root-cause analysis was subsequently undertaken.
Results
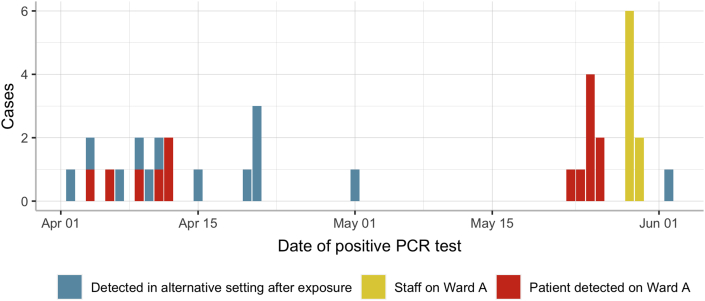
During the first wave of the COVID-19 pandemic, 14 cases of COVID-19 infection were detected on Ward A. Six of the cases were detected between April 4th and 12th, 2020, following which there was a 41-day period with no COVID-19 cases on Ward A. Another case was detected on the May 23rd, 2020, with seven patients subsequently testing positive for SARS-CoV-2 on Ward A in the week following. The two groupings of cases are subsequently referred to as Cluster One (April 4th to April 12th) and Cluster Two (May 23rd to May 26th) (Table I). Figure 1 displays the epidemiological curve of cases found on Ward A (red bars).
Table I.
Demographic, epidemiological, and genomic features of the Ward A outbreak
| Variable | Cluster One | Cluster Two | Total |
|---|---|---|---|
| Size of epidemiological outbreak | |||
| Total | 21 | 17 | 38 |
| Patients | 21 | 9 | 30 |
| Healthcare workers | 0 | 8 | 8 |
| Patients | N = 21 | N = 9 | N = 30 |
| Demographics | |||
| Median age (years) | 82.5 | 83.7 | 82.5 |
| Female | 2 (9.5%) | 0 | 2 (6.7%) |
| Context of positive result | |||
| Before transferring to Ward A | 3 (14.3%) | 0 | 3 |
| On Ward A | 6 (28.6%) | 8 (88.9%) | 14 |
| On another ward | 1 (4.8%) | 0 | 1 |
| On admission to hospital post-exposure | 11 (52.4%) | 1 (11.1%) | 12 |
| Reason for testing | |||
| Admission testing on Ward A | 0 | 1 (11.1%) | 1 |
| Admission testing in A&E | 3 (14.3%) | 0 | 3 |
| Developed symptoms on Ward A | 6 (28.6%) | 0 | 6 |
| Screening on Ward A post-exposure | 0 | 7 (77.8%) | 7 |
| Screening in A&E post-exposure | 11 (52.4%) | 1 (11.1%) | 12 |
| Screening to facilitate discharge | 1 (4.8%) | 0 | 1 |
| Public Health England definitionsa | |||
| ‘definite healthcare-associated’ | 1 | 1 | 1 |
| ‘probable healthcare-associated’ | 3 | 3 | 6 |
| ‘indeterminate healthcare-associated’ | 3 | 3 | 6 |
| ‘community-acquired’ | 0 | 1 | 1 |
| No. of patients readmitted | |||
| After contact on Ward A | 9 | 1 | 10 |
| After household contact | 2 | 0 | 2 |
| Time from Ward A discharge to positive PCR | 12.9 | 7.2 | |
| 30-day outcome | |||
| Death | 4 (19.0%) | 1 (12.5%) | 5 (16.7%) |
| Staff | N = 0 | N = 8 | N = 8 |
| No. of asymptomatic PCR-positive staff | 0 | 5 | 5 |
| Genomic investigation | |||
| No. of samples sequenced | 13 | 6 | 19 |
| Sample source | All patients | 2 patients 4 staff |
15 patients 4 staff |
| Median no. of SNPs different | 9 | 0 | 11 |
| No. of identical genomes | 2 | 5 | 7 |
| No. of genomes ≤2 SNPs different | 10b | 6 | 16 |
| No. of samples clustered at T = 3 | 6 | 5 | 11 (6 + 5) |
PCR, polymerase chain reaction; SNP, single nucleotid polymorphism.
Public Health England definitions for ‘healthcare-associated infections’ were applied to patients detected to be SARS-COV-2 positive during their admission (‘definite’: first positive >15 days after admission; ‘probable’: first positive 8–14 days after admission; ‘indeterminate’: first positive 2–7 days after admission).
Comprising two groups of seven and three persons.
Figure 1.
The epidemic curve of 27 cases on Ward A (April 1st to June 2nd). Colours indicate the category of case. Patients 7–9 were COVID-19 positive prior to admission on to Ward A and are therefore not included in the epidemic curve.
Cluster One
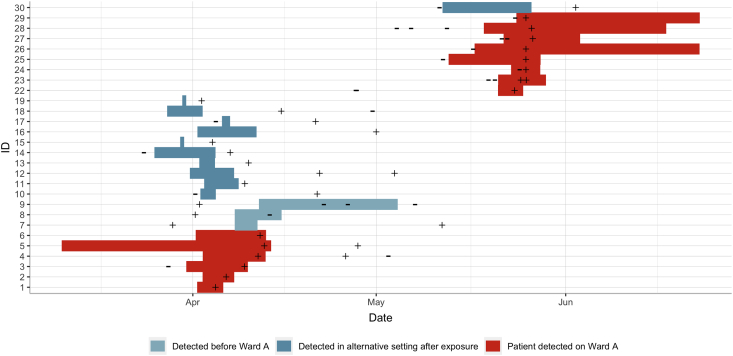
On April 4th, 2020 (three days into their Ward A admission), while displaying symptoms of a respiratory infection, Patient 1 was tested for SARS-CoV-2 and returned a positive result. In the following eight days, five inpatients on Ward A developed respiratory symptoms and tested positive for SARS-CoV-2 (Patients 2–6). They had been on Ward A for between four and 32 days prior to their COVID-19 diagnoses. Only one of the patients had previously undergone SARS-CoV-2 PCR testing (Patient 3, prior to their admission to Ward A; Figure 2; Appendix).
Figure 2.
Timeline plot of patients 1–19 and 22–30. Bars illustrate time spent on Ward A. Colours indicate category of patient. ‘–’, date/time of negative PCR test; ‘+’, date/time of positive PCR test.
Contact tracing identified 114 patients who could have plausibly been exposed to COVID-19 on Ward A or have been the source of infection. Thirteen met the definition for secondary cases, as they had had a positive SARS-CoV-2 PCR result in the 21 days before or after their contact on Ward A.
Three of the secondary cases had been diagnosed with COVID-19 earlier in their hospital admission and were transferred to Ward A from a COVID-19 ward 10 days after their positive PCR result as they were no longer deemed infectious (Patients 7–9). As they were only present on Ward A when not infectious, these cases were considered not to be related to the others in Cluster One (Figure 2).
The remaining 10 cases had their positive SARS-CoV-2 PCR result after contact on Ward A. Patient 19 had tested positive on a different ward after transferring from Ward A whereas the remaining nine patients were discharged from hospital, and then all tested positive in accident and emergency on readmission into hospital (Patients 10–18). Out of the 10, only three had undergone SARS-CoV-2 PCR testing during their previous admission (all of which were negative). In addition, case note review identified that household members of both patients 10 and 13 were also admitted to hospital and tested positive (Patients 20 and 21, respectively).
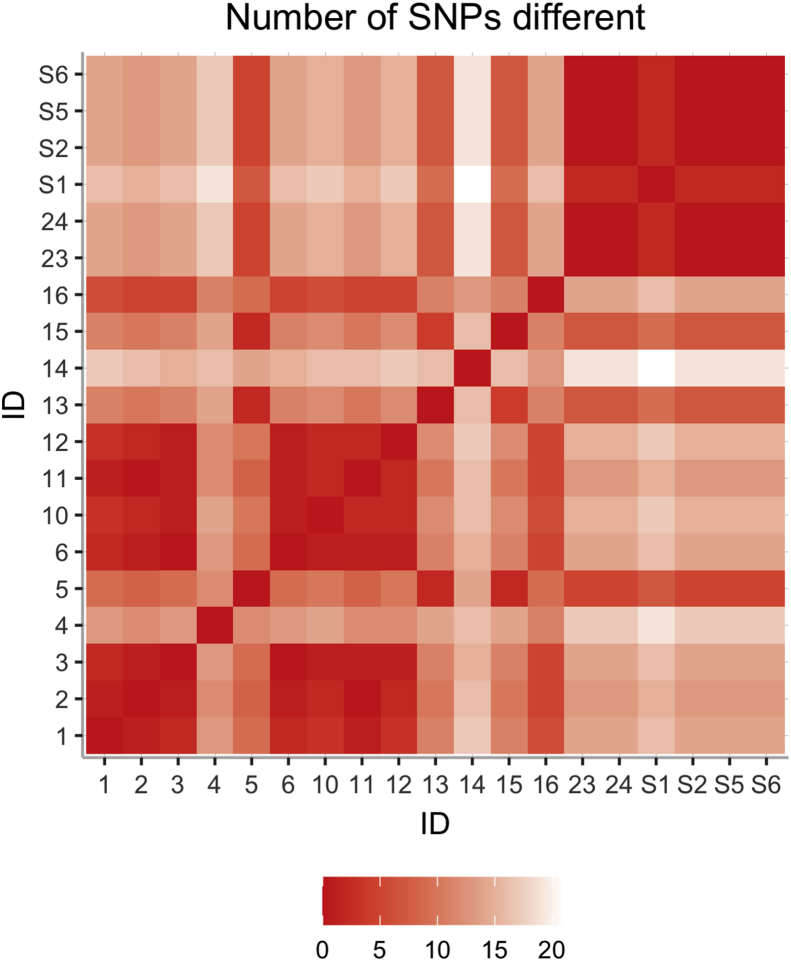
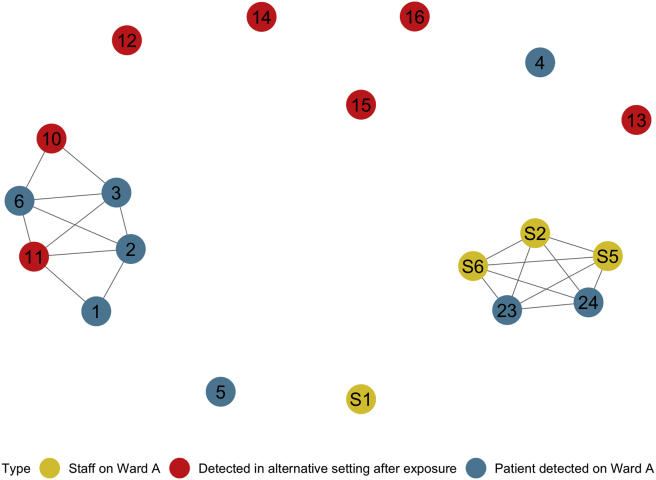
Contact tracing broadened the scope of the investigation from the six cases on Ward A to 18 positive cases in total. Thirteen samples were sequenced (Patients 1–6 and 10–16), and their SNP differences are illustrated in Figure 3. Ten samples were fewer than two SNPs different to others and formed seven- and three-sample groups (Appendix). Due to the low proportion of samples sequenced and the time elapsed in the outbreak, cluster analysis was performed allowing for three intermediate transmissions (T ≤ 3). This revealed six samples compatible with in-hospital transmission and suggests that seven of the cases were unrelated to others in the outbreak (Figure 4). The six-patient group included Patient 10, whose household member (Patient 21) was also subsequently admitted positive for SARS-CoV-2.
Figure 3.
Matrix showing the number of single nucleotide polymorphisms different between samples.
Figure 4.
Network diagram illustrating the grouping of samples after genomic and cluster analysis. Colours represent category of sample (as per Figure 1).
Cluster Two
On May 23rd, 2020, the clinical team identified that Patient 22 had been transferred to Ward A without having the (at that time) mandatory admission SARS-CoV-2 PCR test. They were asymptomatic at the time, were tested, and returned a positive result. All of the other patients on the ward underwent asymptomatic screening for COVID-19 infection, with seven testing positive by May 26th (Patients 23–29). Forty-one contacts were traced, one testing positive on readmission to hospital post-discharge on June 2nd (Patient 30). Except for patient 22, all of the positive cases in Cluster Two had a negative SARS-CoV-2 PCR test earlier in their hospital admission.
Given the developing outbreak, all HCWs working on Ward A on May 29th and 30th underwent an ad-hoc round of asymptomatic SARS-CoV-2 PCR testing. Eight out of 27 HCWs tested positive, with only three of them going on to develop symptoms.
In summary, between May 23rd and June 2nd, nine patients and eight HCWs tested positive for COVID-19. Six of these samples (Patients 23 and 24, and Staff 1, 2, 5, and 6) were sequenced. The samples from the two clusters were not closely related, suggesting that there was no cryptic or undetected transmission event on Ward A connecting the two clusters. From Cluster Two, five of the samples were identical, with the remaining sample (Staff 1) different by just one SNP. Cluster analysis confirmed that the five identical samples are related, and, although not related in transcluster analysis, Staff 1 is likely to be part of the group given the single SNP difference.
Discussion
This report describes the experiences on an elderly care ward in April and May 2020. Thirty-eight PCR-confirmed cases of COVID-19 were related to Ward A during this time-period: 30 patients and eight staff. Genomic sequencing of 19 samples has identified 16 cases that were related to others by two SNPs or fewer with cluster analysis identifying two discrete groups of infection comprising 11 cases (six and five patients each).
This analysis builds upon previously published work demonstrating the utility of genome sequencing in complementing conventional epidemiological investigation to elucidate the epidemiology of COVID-19 nosocomial outbreaks [5,15].
First, robust contact tracing of Cluster One identified nine patients who were on Ward A, discharged to the community, and then readmitted COVID-19 positive. All of these patients were exposed to SARS-CoV-2 in hospital but were not part of routine contact tracing at the time. Our analysis found no evidence of transmission for between two and five of these samples depending on whether two SNPs or transcluster was used to determine transmission plausibility. In this context, genome sequencing provided evidence to help rule out nosocomial infection.
Second, despite the above point, genome analysis confirms that two patients (Patients 10 and 11) who were discharged from Ward A during Cluster One had viral sequences compatible with hospital-acquired infection. This has important implications as patients recently discharged from hospital are not captured in conventional definitions of hospital-acquired COVID-19 infection. Interestingly, Patient 10 was readmitted 17 days after discharge from Ward A with asymptomatic COVID-19 infection, highlighting that time from discharge to positive test cannot always be relied upon to exclude nosocomial infection. Moreover, genome sequencing further highlighted that Patients 1 and 2, although testing positive early into their hospital admission (four and five days respectively), were related.
Third, those patients who acquired COVID-19 in hospital and were discharged have the potential to transmit in the community. Patient 10 was readmitted after discharge from Ward A with a genome sequencing suggesting hospital acquisition. It is likely that there was onward community transmission as when Patient 10 was readmitted, a member of their household (Patient 21) was also admitted and tested positive for COVID-19. Similarly, one patient was diagnosed with COVID-19 infection after being transferred to another ward from Ward A, demonstrating the potential hospital-wide effect of undetected cases.
Fourth, analysis of Cluster Two confirms transmission between patients and staff (although the directionality is uncertain) as samples from two patients and three HCWs were identical. This is in keeping with other published literature and raises important IPC questions.
Finally, as has been reported in other studies, there was a high level of asymptomatic infection detected in both patients and HCWs in Cluster Two, illustrating the importance of screening exposed individuals to identify cases early [16,17]. Interestingly, all of the cases in Cluster One were symptomatic and this likely reflects the availability of SARS-CoV-2 PCR testing, which at that time was largely reserved for those with symptoms. It is thus plausible that asymptomatic infections (either acquired in the hospital or the community) may not have been identified in this investigation.
This study has several limitations to discuss. First, since not all exposed patients underwent testing, it is likely that some infections were undetected. This is particularly relevant during Cluster One and is noteworthy in light of the high rate of asymptomatic infection reported here. Second, due to limitations on resources, only half of the positive samples (sampled randomly) were sequenced, which limits our understanding of the outbreaks. Third, samples from the clustered and non-clustered samples differ by only one or two SNPs and therefore we cannot completely exclude community acquisition in some of the cases. Finally, contact tracing was performed using ward handover lists and therefore it is possible that some patients were missed.
The utility of the COG-UK network in providing whole genome sequencing context to aid in the understanding of hospital outbreak clusters highlights the benefit that such technology might have in the future for understanding pathogens in acute NHS care.
Recommendations
-
–
Test all patients exposed to COVID-19 in hospital regardless of symptoms.
-
–
Test staff exposed to COVID-19 on non-COVID wards.
-
–
Use genome sequencing to generate and test hypotheses to improve IPC practices.
-
–
Consider patients readmitted to hospital with COVID-19 after recent discharge as nosocomial infection.
Author contributions
R.D. Wenlock: conceptualization, data curation, formal analysis, investigation, methodology, project administration, resources, software, supervision, validation, visualization, roles/writing – original draft, writing – review and editing; M. Tausan: conceptualization, data curation, formal analysis, investigation, methodology, project administration, resources, software, supervision, validation, visualization, roles/writing – original draft, writing – review and editing; R. Mann: conceptualization, data curation, investigation, methodology, writing – review and editing; W. Garr: conceptualization, data curation, investigation, methodology, writing – review and editing; R. Preston: conceptualization, data curation, investigation, methodology, writing – review and editing; A. Arnold: conceptualization, data curation, investigation, methodology, writing – review and editing, J. Hoban: conceptualization, data curation, investigation, methodology, writing – review and editing; L. Webb: conceptualization, data curation, investigation, methodology, writing – review and editing; C. Quick: conceptualization, data curation, investigation, methodology, writing – review and editing; A. Beckett: data curation, formal analysis, investigation, methodology, project administration, resources, software, writing – review and editing; K. Loveson: data curation, formal analysis, investigation, methodology, project administration, resources, software, writing – review and editing; S. Glaysher: data curation, formal analysis, investigation, methodology, project administration, resources, software, writing – review and editing; S. Elliott: data curation, formal analysis, investigation, methodology, project administration, resources, software, writing – review and editing; C. Malone: data curation, formal analysis, investigation, methodology, project administration, resources, software, writing – review and editing; B. Cogger: data curation, formal analysis, investigation, methodology, project administration, resources, software, writing – review and editing; L. Easton: data curation, formal analysis, investigation, methodology, project administration, resources, software, writing – review and editing; The COVID-19 Genomics UK (COG-UK) consortium: S.C. Robson: conceptualization, data curation, formal analysis, investigation, methodology, project administration, resources, software, supervision, validation, visualization, roles/writing – original draft, writing – review and editing; M.O. Hassan-Ibrahim: investigation, methodology, project administration, resources, software, supervision, writing – review and editing; C. Sargent: conceptualization, data curation, formal analysis, investigation, methodology, project administration, resources, software, supervision, validation, visualization, roles/writing – original draft, writing – review and editing.
Conflict of interest statement
None declared.
Funding sources
S.R. and A.B. are part-funded from Research England’s Expanding Excellence in England (E3) Fund. The sequencing costs were funded by the COVID-19 Genomics UK (COG-UK) Consortium which is supported by funding from the Medical Research Council (MRC) part of UK Research & Innovation (UKRI), the National Institute for Health Research (NIHR) and Genome Research Limited, operating as the Wellcome Sanger Institute. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.infpip.2021.100165.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.World Health Organization . 2020. Coronavirus disease (COVID-19) Available at: https://covid19.who.int [last accessed 11 April 2020]. [Google Scholar]
- 2.Taylor J., Rangaiah J., Narasimhan S., Clark J., Alexander Z., Manuel R. Nosocomial COVID-19: experience from a large acute NHS Trust in South-West London. J Hosp Infect. 2020;106:621–625. doi: 10.1016/j.jhin.2020.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jewkes S.V., Zhang Y., Nicholl D.J. Nosocomial spread of COVID-19: lessons learned from an audit on a stroke/neurology ward in a UK district general hospital. Clin Med. 2020;20:e173–e177. doi: 10.7861/clinmed.2020-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lessells R., Moosa Y., De Oliveira T. KwaZulu-Natal Research Innovation and Sequencing Platform (KRISP); 2020. Report into a nosocomial outbreak of coronavirus disease 2019 (COVID-19) at Netcare St. Augustine’s Hospital; p. 37.https://www.krisp.org.za/manuscripts/StAugustinesHospitalOutbreakInvestigation_FinalReport_15may2020_comp.pdf Available at: [last accessed July 2021] [Google Scholar]
- 5.Meredith L.W., Hamilton W.L., Warne B., Houldcroft C.J., Hosmillo M., Jahun A.S. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect Dis. 2020;20:1263–1271. doi: 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamilton W.L., Tonkin-Hill G., Smith E., Houldcroft C.J., Warne B., Meredith L.W. COVID-19 infection dynamics in care homes in the East of England: a retrospective genomic epidemiology study. medRxiv. 2020 2020.08.26.20182279. [Google Scholar]
- 7.Reuter S., Ellington M.J., Cartwright E.J., Köser C.U., Török M.E., Gouliouris T. Rapid bacterial whole-genome sequencing to enhance diagnostic and public health microbiology. JAMA Intern Med. 2013;173:1397–1404. doi: 10.1001/jamainternmed.2013.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Köser C.U., Ellington M.J., Cartwright E.J., Gillespie S.H., Brown N.M., Farrington M. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stone S.P., Cooper B.S., Kibbler C.C., Cookson B.D., Roberts J.A., Medley G.F. The ORION statement: guidelines for transparent reporting of outbreak reports and intervention studies of nosocomial infection. J Antimicrob Chemother. 2007;59:833–840. doi: 10.1093/jac/dkm055. [DOI] [PubMed] [Google Scholar]
- 10.Quick J. 2020. nCoV-2019 sequencing protocol v2 (GunIt) V.2. [DOI] [Google Scholar]
- 11.Loman N.J., Quick J., Simpson J.T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods. 2015;12:733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- 12.Li H. Sequence analysis Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shu Y., McCauley J. GISAID: Global Initiative on Sharing all Influenza Data – from vision to reality. Eurosurveillance. 2017;22:2–4. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stimson J., Gardy J., Mathema B., Crudu V., Cohen T., Colijn C. Beyond the SNP threshold: identifying outbreak clusters using inferred transmissions. Mol Biol Evol. 2019;36:587–603. doi: 10.1093/molbev/msy242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rockett R.J., Arnott A., Lam C., Sadsad R., Timms V., Gray K.-A. Revealing COVID-19 transmission in Australia by SARS-CoV-2 genome sequencing and agent-based modeling. Nat Med. 2020;26:1398–1404. doi: 10.1038/s41591-020-1000-7. [DOI] [PubMed] [Google Scholar]
- 16.Rivett L., Sridhar S., Sparkes D., Routledge M., Jones N.K., Forrest S. Screening of healthcare workers for SARS-CoV-2 highlights the role of asymptomatic carriage in COVID-19 transmission. Elife. 2020;9 doi: 10.7554/eLife.58728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones N.K., Rivett L., Sparkes D., Forrest S., Sridhar S., Young J. Effective control of SARS-COV-2 transmission between healthcare workers during a period of diminished community prevalence of COVID-19. Elife. 2020;9 doi: 10.7554/eLife.59391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.