Abstract
Transcriptional regulation by transforming growth factor β (TGF-β) is a complex process which is likely to involve cross talk between different DNA responsive elements and transcription factors to achieve maximal promoter activation and specificity. Here, we describe a concurrent requirement for two discrete responsive elements in the regulation of the c-Jun promoter, one a binding site for a Smad3-Smad4 complex and the other an AP-1 binding site. The two elements are located 120 bp apart in the proximal c-Jun promoter, and each was able to independently bind its corresponding transcription factor complex. The effects of independently mutating each of these elements were nonadditive; disruption of either sequence resulted in complete or severe reductions in TGF-β responsiveness. This simultaneous requirement for two distinct and independent DNA binding elements suggests that Smad and AP-1 complexes function synergistically to mediate TGF-β-induced transcriptional activation of the c-Jun promoter.
Transforming growth factor β (TGF-β) is a multifunctional cytokine with a wide range of physiological as well as pathological effects (reviewed in references 31 and 42). Its physiological roles include inhibition of the proliferation of a variety of cell types, negative regulation of the immune system, and positive regulation of extracellular matrix deposition. Dysregulation of these processes can result in various fibrotic as well as malignant diseases. Indeed, many late stage cancers have lost expression of TGF-β receptors, which renders them resistant to TGF-β-mediated growth inhibition (19, 29, 36, 38, 50, 55); restoration of TGF-β pathways in these cells can often restore growth inhibition and decrease the malignant phenotype. TGF-β-mediated immune system suppression and stimulation of extracellular matrix (ECM) production may also contribute to tumor-promoting effects.
Regulation of transcription of specific sets of genes by TGF-β mediates many of these physiological roles. Upregulation of two cyclin-dependent kinase inhibitor genes, p21 and p15, has been shown to mediate TGF-β-induced growth arrest in certain cell types (7, 12, 41), while upregulation of ECM genes, including plasminogen activator inhibitor 1 (PAI-1), fibronectin, and collagen genes, may mediate other effects of TGF-β. However, many of the genes regulated by TGF-β are also regulated by a variety of other signals, including some signals which appear to play very distinct roles at the physiological level. Of particular note is a subset of TGF-β immediate-response target promoters, including the TGF-β1 ligand gene and most of the TGF-β-responsive extracellular matrix genes, in which AP-1 binding sites have been found to be involved in mediating the TGF-β signal (4, 22, 51). The use of AP-1 sites in TGF-β-dependent transcription has been particularly puzzling, given the extensively described mitogenic signaling pathways which also activate transcription through AP-1; the mechanism by which TGF-β regulates these promoter sequences has not been clarified. An additional level of complexity is introduced by the regulation by TGF-β of the expression of AP-1 family members themselves. This suggests that there can be both primary and secondary effects on transcription through AP-1 by TGF-β.
The regulation of AP-1 transcription factors by TGF-β varies with the specific family member and with cell type. The upregulation of c-Jun transcript occurs in a wide range of cell lines derived from both normal and transformed cells. This response to TGF-β is early and immediate, with mRNA induced within 15 to 30 min. While cycloheximide studies have been inconclusive, due to the inducing effects of the cycloheximide itself on c-jun transcription, the time course of induction strongly suggests that this gene could be a primary target of TGF-β (24, 26, 39), which is supported by the current study describing specific promoter elements capable of mediating TGF-β’s induction of c-Jun.
The model for TGF-β activation of transcription continues to undergo rapid development. The Smads are a recently identified family of proteins which operate downstream of various members of the TGF-β superfamily (reviewed in references 13, 14, 23, 30, and 37). Smad2 and Smad3 are downstream effectors of the TGF-β signaling pathway. Upon ligand binding, they are phosphorylated by the TGF-β type I receptor kinase and translocate to the nucleus in a complex with Smad4 (28, 35, 59). Recent work has identified a potential consensus Smad3-Smad4 DNA binding site, GTCTAGAC (58), by random oligonucleotide screening, as well as similar sequences in the PAI-1 promoter (9), the engineered TGF-β-responsive reporter construct, p3TP-lux promoter (57), the JunB promoter (18), and the COL7A1 collagen promoter (54). It was found that four copies of the oligonucleotide consensus site or nine copies of the PAI-1 site could confer TGF-β responsiveness on a minimal promoter. In addition, mutation of all three putative Smad3-Smad4 binding sites in the PAI-1 promoter could eliminate TGF-β responsiveness of that promoter in HepG2 cells.
Although these studies demonstrate the importance of Smad3-Smad4 binding sites in the mediation of TGF-β responsiveness, they do not fully address the issue of whether binding elements for other transcription factors are also required for TGF-β-mediated transcriptional activation of target promoters. Biochemical and overexpression studies have demonstrated that Smads are capable of functional interaction with Sp1 (33) and with AP-1; in fact, direct physical interaction between Smads and AP-1 family members has been demonstrated in model systems (27, 60). Cooperation between Smad2-Smad4 complexes and FAST-1 has been demonstrated at an activin responsive Xenopus promoter (2, 3). Finally, a very recent study reports that a binding site for the transcription factor muE3 (TFE3), as well as one for Smad3 and Smad4, is required for TGF-β-mediated transcription of a reporter controlled by a specific region of the PAI-1 promoter (16).
While the TGF-β-responsive elements in the c-Jun promoter have not previously been characterized, extensive work has established the importance of two AP-1/CRE sequences in the c-Jun promoter in regulation by phorbol-12-myristate-13-acetate (TPA), serum, UV, E1A, and interleukin 1 (IL-1) (1, 15, 34, 43, 53). Furthermore, a reporter construct controlled by the −79 to +170 sequence of the c-Jun promoter, which contains only the more proximal AP-1/CRE site (−71 to −64), has proved sufficient for a maximal response to most of these signals. Interestingly, none of these stimuli appears to change the occupancy of any identified binding sites in the c-Jun promoter. Thus, the prevailing model of activation by these other signals is thought to be through modification of a constitutively promoter-bound complex, in most cases c-Jun-ATF-2.
Here, we identify two DNA binding elements within this −79 to +170 region which are indispensable in TGF-β-mediated induction of c-Jun: the proximal AP-1/CRE site known to be important for the response to several other signals, and a novel Smad3/Smad4 binding site. Mutation of either site alone is found to abolish or severely reduce promoter upregulation by TGF-β, despite the presence of the remaining element. Our results suggest that the two complexes can cooperate synergistically in activating TGF-β-mediated transcription of this c-Jun promoter region.
MATERIALS AND METHODS
Antibodies and reagents.
Human TGF-β1 was from R&D Systems. Rabbit polyclonal antisera recognizing Smad3 and Smad4 were generated in this lab. Smad3 antiserum was raised against a specific Smad3 peptide (DAGSPNLSPNPMSPAHNNLD), while Smad4 antiserum was raised against full-length human glutathione S-transferase–Smad4.
Cell culture.
Mink lung epithelial cells and primary mouse embryo fibroblasts (MEFs) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), nonessential amino acids, penicillin, and streptomycin. Immortalized human keratinocyte cells (HaCaT) were grown in MEM supplemented with 10% FBS, penicillin and streptomycin, and 20 mM l-glutamine. Primary fibroblasts were harvested from day-14 embryos. Embryos were mechanically disrupted by passage through an 18-gauge needle and plated on gelatin-coated 10-cm-diameter plates in DMEM with 20% heat-inactivated FBS, penicillin, streptomycin, and gentamicin (Gibco BRL, Gaithersburg, Md.). When confluent, cells were trypsinized and further maintained in DMEM with 10% FBS. The targeted disruption of the Smad3 allele in these mice and the characterization of their phenotype are described elsewhere (6).
Plasmid constructs.
Flag-tagged human Smad4 was a generous gift from Rik Derynck. Human pCGN Smad3 was described previously (57). The c-Jun luciferase reporter containing the −79 to +170 sequence of the human c-Jun promoter was generously provided by Bin Su (48). The rest of the promoter mutants and 3′ deletion constructs were made by PCR mutagenesis using the following primer sets: as 5′ primers, wild type, 5′CCC AAG CTT GGC CTT GGG GTG ACA TCA TGG GC3′; AP-1/CRE mutant, 5′CCC AAG CTT GGC CTT GGG GAT CCA CCA TGG GCT ATT TTT AGG GG3′; and as 3′ primers, wild type, 5′AAA CTG CAG GCC GAC CTG GCT GGC TGG CTG TGT CTG TCT GTC3′; mutant, 5′AAA CTG CAG GCC GAC CTG GCT GGC TGG CTG TTC CAA GCT CCT TGC CTG ACT CCG3′. A HindIII site was engineered into the 5′ end of each PCR product, and a PstI site was engineered into the 3′ end of each PCR product. PCR products were subcloned into pGEMT (Promega, Madison, Wis.), and then the HindIII/PstI fragments were purified on an agarose gel, extracted with a QIAEX II gel extraction kit (Qiagen Inc., Santa Clarita, Calif.), and subcloned back into the HindIII and PstI sites flanking the 5′ and 3′ ends, respectively, of the −79 to +170 sequence insert in the −79 to +170 luciferase reporter construct. Constructs were verified by restriction digestion with HindIII/PstI and by sequencing.
Transfection and luciferase assays.
Transient transfections were performed with the standard DEAE-dextran method and the luciferase activity was measured 24 h after the addition of 100 pM human TGF-β1 as described previously (8). For all experiments, 3 μg of the indicated luciferase reporter and, when indicated, 1 μg of Smad3 expression vector were used (57). Total DNA was kept constant by using empty pCGN vector. All transfections were normalized to β-galactosidase activity by cotransfection of 0.5 μg of a β-galactosidase (pCMV-β-Gal) expression vector. The luciferase data shown are representative of experiments performed in duplicate in at least three independent experiments.
Nuclear extracts.
Nuclear lysates were prepared from control and TGF-β1-treated cells. Briefly, confluent cells from 10-cm-diameter dishes were washed twice with phosphate-buffered saline. After washing, 5 ml of ice-cold hypotonic lysis buffer was added (20 mM HEPES [pH 7.6], 20% glycerol, 10 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1% Triton X-100, 25 mM NaF, 25 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, 1 mM dithiothreitol, and protease inhibitors). The cells were allowed to swell on ice for 5 min before they were scraped and collected. Nuclei were pelleted by centrifugation at 500 rpm in a Beckman swinging-bucket tabletop centrifuge for 5 min and resuspended in 100 to 200 μl of nuclear extraction buffer (hypotonic buffer plus 500 mM NaCl). After incubation and rocking at 4°C, the lysates were cleared of debris by centrifugation.
Western blot analysis.
Western blot analysis for c-Jun was performed on nuclear lysates prepared from MEFs. Prior to treatment with TGF-β1 for the indicated times, cells were serum starved for 12 h in DMEM–0.2% FBS. Equal protein amounts were resolved by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis, and Western blotting was performed with a 1:1,000 dilution of the rabbit polyclonal antibody α-c-Jun (9162) from New England Biolabs, Inc. (Beverly, Mass.).
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed by using 1 to 3 μg of nuclear extracts prepared from untreated cells or cells treated with 100 pM TGF-β1 for 1 h and probes derived from a SacI/BamHI fragment of luciferase construct containing the c-Jun sequence from −79 to +170. The digest produced two fragments of the c-Jun promoter that consist of the sequences from −79 to −19 and −18 to +170. Gel shift conditions were exactly as previously described (57). For supershift analysis of Smads, 2 μl of Smads 3 and 4 immune-phase and preimmune-phase antisera and 2 μg of Smad2 (S-20-X) or Smad4 (C-20-X) antibodies from Santa Cruz Biotechnology, Inc. (Santa Cruz, Calif.), were used. For other supershifts, 2 μg of anti-c-Jun (KM-1-X), ATF-2 (FRBR-1-X and C-19-X), and CREB (C21-X and 24H4B-X) from Santa Cruz Biotechnology, Inc., were used. The sequences of the competitor oligonucleotides used to identify the Smad binding site are shown in Fig. 3A. The sequence of the competitor oligonucleotide containing a CREB/ATF binding site (CRE) was 5′-AGA GAT TGC CTG ACG TCA GGA GCT AG-3′ and its complementary strand. The sequence of the mutated CRE site was 5′-AGA GAT TGC CTG TGG TCA GAG AGC TAG-3′. Where results for only one lysate are shown, similar results were obtained for both HaCaT lysates and mink lung lysates.
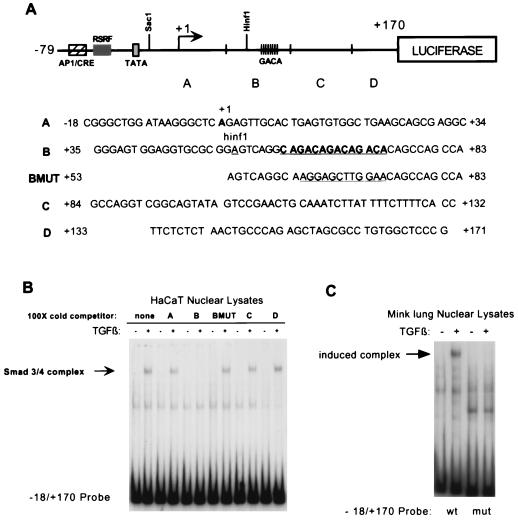
FIG. 3.
The Smad3-Smad4 binding site in the human c-Jun promoter is identified as a CAGA triplet located 3′ of the TATA box. (A) Schematic diagram of the −79 to +170 region of the c-Jun promoter. Four oligonucleotide sequences, named A through D, were designed to span the −18 to +170 region of the promoter. An additional oligonucleotide bearing a mutation in a CAGA triplet from +62 to +73 (BMUT) is also diagrammed (see text for additional discussion). The mutation changed the sequence from GACAGACAGACA to AGGAGCTTGCAA. (B) EMSA was performed by using the same −18 to +170 probe and HaCaT lysates as described for Fig. 2. A 100-fold molar excess of unlabeled oligonucleotides was incubated with the nuclear lysates before addition of radiolabeled probe, in order to compete with binding. The induced Smad3-Smad4 binding complex is indicated with an arrow. (C) EMSA was performed by using nuclear lysates from untreated mink lung cells or mink lung cells treated with TGF-β1 for 1 h and the same −18 to +170 probe. Radiolabeled probe was either the wild-type sequence from −18 to +170 or the mutated sequence from +62 to +73 (the CAGA triplet).
RESULTS
TGF-β treatment induces DNA binding of a Smad3- and Smad4-containing complex to a sequence in the 3′ region of the c-Jun promoter.
An increase in c-Jun mRNA level has been previously observed within 15 to 30 min of TGF-β treatment in a variety of cell types (24, 39, 49). In order to confirm the induction of endogenous c-Jun by TGF-β, we performed Northern analysis of RNA and Western analysis of nuclear extracts isolated from similarly treated cells. In both mink lung epithelial cells (Mv1Lu) and HaCaT cells, the level of c-Jun transcript increased within 1 h of TGF-β treatment and protein levels were dramatically increased within 2 h of TGF-β treatment (data not shown), confirming that the induction of c-Jun by TGF-β occurs in these cells and is likely to be an early response. The induction by TGF-β was most evident in Mv1Lu cells if the cells were serum starved overnight before addition of TGF-β, since the c-Jun transcript is upregulated by serum.
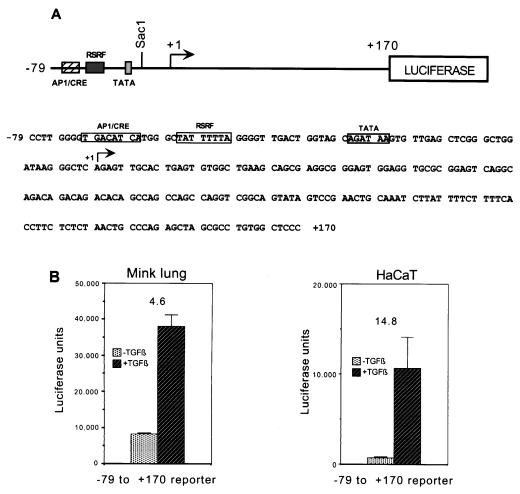
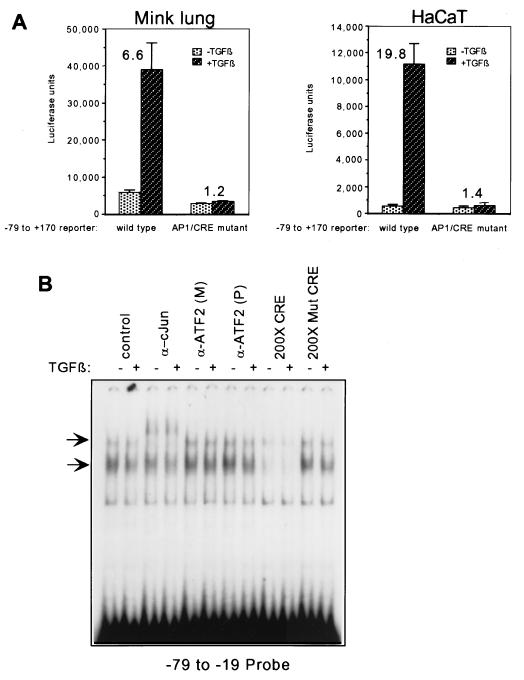
To aid in defining TGF-β responsive elements in the human c-Jun promoter, we next obtained a luciferase reporter construct under control of the sequence from −79 to +170 of the c-Jun promoter (48). This region, diagrammed in Fig. 1A, contains the proximal AP-1/CRE site and the adjacent AT-rich sequence (a putative RSRF [related to serum response factor] site) which is important in epidermal growth factor (EGF) induction of c-Jun, as well as the native TATA box and approximately 170 bp of the sequence 3′ of the start site. As mentioned above, this region was sufficient to convey maximal responsiveness to UV, TPA, EGF, and serum. We transiently transfected this construct into Mv1Lu and HaCaT cells, and measured luciferase activity after TGF-β treatment. As shown in Fig. 1B, the construct was highly responsive to TGF-β, giving 4.6-fold induction in Mv1Lu cells and 14.8-fold induction in HaCaT cells.
FIG. 1.
The −79 to +170 region of the human c-Jun promoter is sufficient to convey TGF-β and Smad3 responsiveness to a luciferase reporter. (A) Schematic representation of the −79 to +170 luciferase reporter. (B) The reporter was transiently transfected into Mv1Lu or HaCaT cells, and TGF-β-induced luciferase activity was measured in relative light units (luciferase units). Fold inductions are indicated above the bars and were calculated by comparing the luciferase activities of cells treated with TGF-β and those of untreated controls.
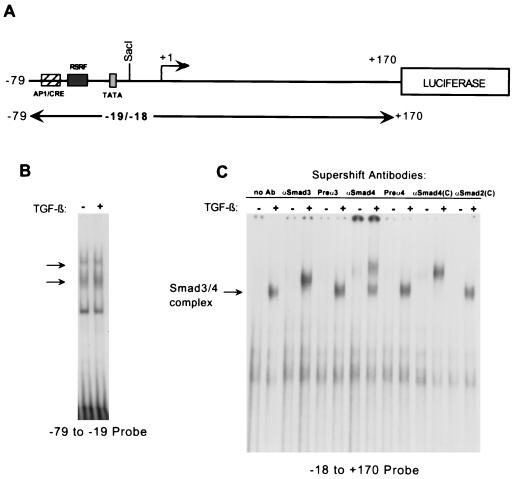
Having determined that the −79 to +170 portion of the c-Jun promoter was sufficient to convey TGF-β responsiveness, we next examined whether the mechanism of activation might involve induction of Smad DNA binding to a site in this region. We performed an EMSA by using a 5′ portion or a 3′ portion of the −79 to +170 region as a probe (Fig. 2A) and nuclear extracts from HaCaT cells treated for 1 h with TGF-β. The −79 to −19 probe bound two complexes (small arrows), and no change was observed upon TGF-β treatment (Fig. 2B). On the other hand, the −18 to +170 probe bound a complex that was strongly induced by TGF-β treatment (Fig. 2C). This induced complex appeared within 30 min of TGF-β treatment and was still present at 2 h (data not shown). Using an antiserum specific to Smad3 as well as an antiserum and commercial antibody specific to Smad4, we were able to supershift the induced complex, indicating the presence of both Smad3 and Smad4 in the complex. No supershift was seen with the corresponding preimmune-phase antisera, and a commercially available Smad2 antibody also failed to cause a supershift (Fig. 2C). Similar results were obtained with nuclear extracts from Mv1Lu cells (data not shown).
FIG. 2.
EMSAs showing induced binding of a Smad3- and Smad4-containing complex to the 3′ region of the human c-Jun promoter. (A) Schematic representation of the −79 to +170 region of the c-Jun promoter showing the probes used for EMSAs. (B) EMSA was performed by using a radiolabeled restriction fragment spanning the −79 to −19 region of the c-Jun promoter and nuclear lysates from either untreated HaCaT cells or HaCaT cells treated with TGF-β1 for 1 h. Two constitutively binding complexes are indicated with arrows. (C) EMSA was performed by using a radiolabeled restriction fragment spanning the −18 to +170 region of the c-Jun promoter and the same HaCaT lysates. A complex that shows binding induced by TGF-β treatment is indicated with an arrow. Supershifts were performed using antiserum against Smad3 or Smad4, shown with their corresponding preimmune-phase antiserum (Preα3 and Preα4) or with commercial antibodies against Smad4 [αSmad4(C)] and Smad2 [αSmad2(C)].
These results establish the existence of a Smad3-Smad4 binding site contained within the −18 to +170 region of the c-Jun promoter. The binding of Smad3-Smad4 is rapidly induced upon TGF-β treatment, with a time course consistent with that of Smad phosphorylation and subsequent translocation to the nucleus (see references 14 and 23 for reviews). In contrast, the pattern of binding to the −79 to −19 region of the promoter is unchanged upon TGF-β treatment.
The Smad3-Smad4 binding site in the c-Jun promoter is a CAGA triplet located 3′ of the TATA box.
In order to identify the Smad3-Smad4 binding site within the −18 to +170 region, four oligonucleotides scanning this sequence (Fig. 3A) were used as cold competitors in the EMSA. Only the +35 to +83 region was found to compete with the binding of the induced complex (Fig. 3B). When the oligonucleotide for this region was cut at a convenient HinfI site and the two halves were compared, binding could be further localized to the +53 to +83 region. Three mutant competitor oligonucleotides of the +53 to +83 region were then designed. We had noted a sequence in the middle of this region, ACAGACAGACAGACACAG, which bore great similarity to repeats of the Smad box as identified by previous studies (9, 57, 58) and was recently confirmed by the crystal structure of MH1-Smad3 bound to the CAGA box (47). Therefore, we made mutations to disrupt either this potential Smad binding site or the sequence 5′ or 3′ of it within the +53 to +83 region. Of the three, only the CAGA mutant oligonucleotide had lost its ability to compete with binding of the induced complex (Fig. 3B and data not shown), indicating that this mutation had disrupted the Smad binding site. Confirming this, a −18 to +170 probe containing mutated CAGA sequence was shown to no longer bind the induced Smad3-Smad4 complex (Fig. 3C). From these experiments we concluded that the Smad3-Smad4 binding site was located at the CAGA repeats within the +62 to +73 region of the c-Jun promoter. These results also established that no other sequences in the −18 to +170 region are absolutely required for DNA binding of the induced complex containing Smad3-Smad4.
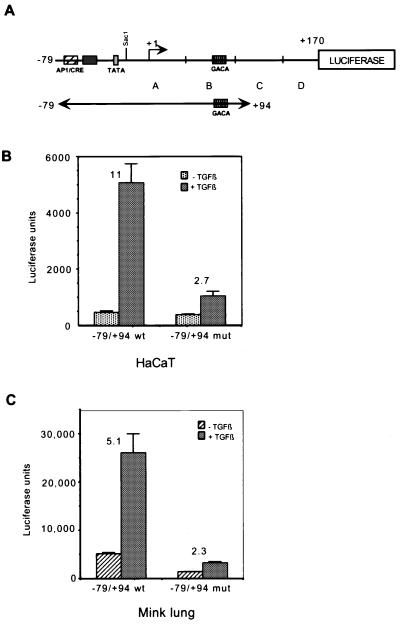
Mutation of the Smad3-Smad4 binding site in the c-Jun promoter abrogates responsiveness to TGF-β.
Having identified the Smad3-Smad4 binding site in the c-Jun promoter, we set out to determine its importance in mediating the TGF-β response. Using PCR mutagenesis, we created a −79 to +94 wild-type reporter and corresponding −79 to +94 mutant reporters (Fig. 4A). We found that the wild-type −79 to +94 reporter was induced by TGF-β 11-fold and 5.1-fold in HaCaT cells and Mv1Lu cells, respectively. Mutation of the CAGA sequences reduced the response to 2.7-fold and 2.3-fold, respectively (Fig. 4B and C). This demonstrates that the Smad3-Smad4 binding site is important for response to TGF-β in the context of this c-Jun promoter construct.
FIG. 4.
Mutation of the Smad3-Smad4 binding site abrogates responsiveness to TGF-β. (A) Diagram of new reporter constructs created by PCR mutagenesis. Two reporters for the region from −79 to +94 of the c-Jun promoter were created, i.e., one with wild-type sequence and the other mutated at the Smad3-Smad4 binding site (CAGA triplet) from +62 to +73. (B) The −79 to +94 wild-type and −79 to +94 mutant reporters were transfected into HaCaT cells. Cells were treated with TGF-β1 for 24 h before harvesting for luciferase assays. Fold inductions were calculated by comparing the luciferase activities of TGF-β-treated cells and untreated control cells. (C) The procedures used were the same as described for panel B except that Mv1Lu cells were used instead of HaCaT cells.
Although these findings indicate that the identified Smad3-Smad4 binding site is critical in conferring a complete response to TGF-β, there is a small degree of responsiveness which remains after mutation of the Smad3-Smad4 binding site. It is possible that the remaining TGF-β responsiveness is mediated through the AP-1/CRE site at the −71 to −64 region of the c-Jun promoter in a manner similar to that observed in our previous study of the 4XTRE reporter (57).
TGF-β induction and induced complex binding are lost in Smad3-deficient MEFs.
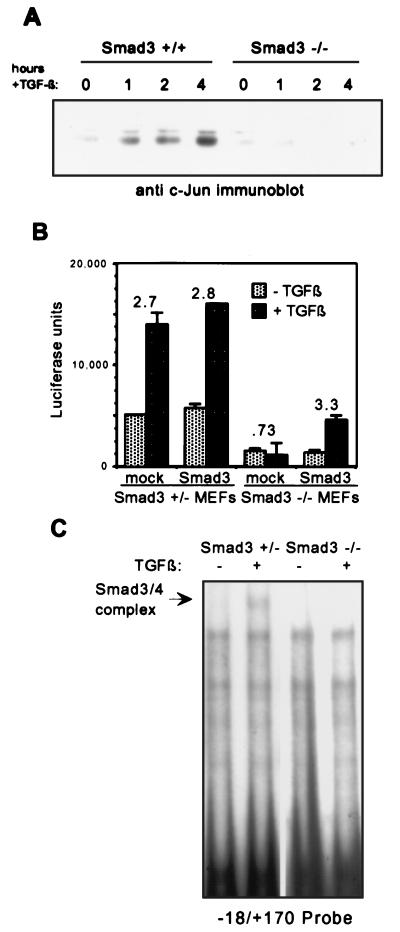
The recent creation of Smad3-deficient mice (6) has introduced a powerful new tool for studying the functional importance of Smad3 in isolation. We first compared induction by TGF-β of endogenous c-Jun in primary MEFs established from Smad3+/+ and Smad3−/− mice. Primary MEFs were serum starved for 12 h and treated with TGF-β for 4 h, and then nuclear lysates were prepared. As shown by Western blot analysis, induction by TGF-β of total c-Jun protein levels is lost in Smad3−/− MEFs whereas that in Smad3+/+ MEFs is intact (Fig. 5A).
FIG. 5.
Induction of c-Jun by TGF-β is lost in Smad3 null fibroblasts. (A) Western blotting was performed by using nuclear lysates from Smad3+/+ or Smad3−/− primary MEFs treated with TGF-β1 for 0, 1, 2, or 4 h. MEFs were serum starved for 12 h in 0.2% serum before treatment. (B) The −79 to +170 reporter was transfected into Smad3+/− or Smad3−/− MEFs with empty expression vector (mock) or Smad3 expression vector (Smad3). Cells were treated with TGF-β1 for 24 h before harvesting for luciferase assays. Fold induction by TGF-β1 is indicated over the bars. (C) EMSA was performed by using the −18 to +170 probe and nuclear lysates from untreated Smad3+/− MEFs or Smad3−/− MEFs or cells of the same types treated with TGF-β1 for 1 h. The induced Smad3-Smad4 complex is indicated with an arrow.
We next investigated the ability of exogenous Smad3 expression to rescue c-Jun reporter induction in Smad3 null fibroblasts. Smad3+/− and Smad3−/− MEFs were transfected with −79 to +170 reporter with empty vector or with a Smad3 expression vector. Note that Smad3+/− MEFs express Smad3 and that they activate representative responses to TGF-β to an extent similar to Smad3+/+ MEFs (6). The c-Jun promoter was induced approximately threefold by TGF-β treatment in Smad3+/− fibroblasts (Fig. 5B), which is comparable to the fold induction of other TGF-β-responsive promoters examined in these cells (6). However, in Smad3−/− MEFs, TGF-β failed to induce reporter activity (Fig. 5B). Although the uninduced overall activity is lower in the null cells, the full threefold induction by TGF-β was restored upon cotransfection with Smad3. This establishes the absence of Smad3 as the defect responsible for loss of c-Jun promoter activation in these cells, and this result demonstrates that Smad3 is absolutely and specifically required for c-Jun promoter regulation by TGF-β.
Finally, we looked at DNA binding to the Smad3-Smad4 site in the absence of Smad3, to determine whether Smad3 was indeed required for binding of the TGF-β-induced complex. An EMSA was performed by using the wild-type −18 to +170 probe containing the Smad3-Smad4 binding site (see Fig. 2A). Induced complex binding was observed in Smad3+/− fibroblasts, but no induced complex was seen in Smad3−/− fibroblasts (Fig. 5C). This suggests that Smad3 is not only present in but also critical to the formation of the DNA binding complex which is induced upon TGF-β treatment. The correlation between loss of the induced complex and loss of endogenous c-Jun induction and c-Jun reporter activation further supports the importance of the induced Smad3-Smad4 binding complex to TGF-β regulation of c-Jun transcription, as well as firmly establishing the requirement for Smad3 in this process.
Mutation of an AP-1/CRE site can independently abrogate TGF-β responsiveness of the c-Jun promoter.
The AP-1/CRE site at −71 to −64 has previously been shown to be important for induction of c-Jun by other signals (1, 15, 34, 53). A consensus AP-1 site was also shown to be not only necessary but also sufficient for TGF-β and Smad responsiveness in the context of a multimerized TRE reporter (57), and mutation of the Smad3-Smad4 binding site in the c-Jun promoter eliminated nearly all but not all TGF-β responsiveness (Fig. 4). While a recent study by Dennler et al. (9) established the importance of three Smad3-Smad4 binding sites in TGF-β regulation of the PAI-1 promoter, it did not address whether the AP-1-like sites present in the promoter (21) may also be important for TGF-β regulation in that context. In order to investigate the importance of the AP-1/CRE site in induction by TGF-β of this c-Jun promoter region, we used PCR mutagenesis to mutate this site in the −79 to +170 reporter. Mutating the AP-1/CRE site abrogated all transcriptional induction of the reporter by TGF-β (Fig. 6A), despite the fact that the Smad3-Smad4 site identified as described above (Fig. 4) remained intact. This suggests that a synergistic functional cooperation exists between Smads and AP-1/CRE complexes in the context of TGF-β-induced transcriptional activation of this c-Jun promoter region.
FIG. 6.
An AP-1/CRE site is also required for TGF-β and Smad responsiveness of the c-Jun promoter. (A) A −79 to +170 luciferase reporter carrying a mutation in the AP-1/CRE site induced by PCR mutagenesis was transfected into HaCaT cells alongside the wild type −79 to +170 reporter. The mutation changed the sequence from TGACATCA to ATCCACCA. Fold induction was calculated by comparing TGF-β-treated cells to untreated control cells. Cells were treated with TGF-β1 for 24 h before harvesting for luciferase assays. (B) EMSA was performed as described in the legend for Fig. 2B. Attempts to perform supershifts were made using a monoclonal antibody against c-Jun (α-cJun) and polyclonal [α-ATF2(P)] and monoclonal [α-ATF2(M)] antibodies against ATF-2 (third through eighth lanes). Competition with 200-fold molar excess of wild-type CRE consensus site oligonucleotide (200× CRE) or 200-fold molar excess of mutated CRE site oligonucleotide (200× Mut CRE) is illustrated in the last four lanes.
We next sought to identify which proteins bind to this AP-1/CRE site in our system. We performed an EMSA using the −79 to −19 probe diagrammed in Fig. 2, where we had observed that there was no change in the pattern of binding to this sequence upon TGF-β addition. Since previous studies (15) had identified c-Jun and ATF-2 as the components constitutively bound to this site, we attempted to supershift the bound complexes with antibodies against these two transcription factors. An antibody specific to c-Jun caused a supershift of the slower-migrating complex, confirming the presence of c-Jun (Fig. 6B). However, we did not see a supershift on this probe when we used two commercially available antibodies specific to ATF-2 (Fig. 6B), which had been successfully used to supershift ATF-2-containing complexes in a previous study (11). Additionally, several commercial antibodies against CREB were unable to supershift this complex (data not shown). Nonetheless, we were able to compete away binding of the faster- and slower-migrating complexes using unlabeled consensus CRE site oligonucleotide in 200× molar excess, whereas the same molar excess of unlabeled mutant CRE oligonucleotides did not compete with the binding. This suggests that a component of the bound complexes is a CRE binding protein. These results demonstrate that a constitutively bound complex containing c-Jun, either as a homodimer or in combination with a yet unknown CRE binding partner, is required in conjunction with the Smad complex in mediating the TGF-β activation of this promoter region.
DISCUSSION
We identify here a novel Smad3-Smad4 binding site in the 5′ untranslated region (UTR) of the c-Jun promoter and introduce evidence for the simultaneous requirement for two different responsive elements in mediating TGF-β-induced c-Jun transcription. The first is a Smad3-Smad4 binding site, and the second is a spatially distinct AP-1/CRE binding site. The two elements are capable of binding their corresponding transcription factor complexes independently. Importantly, mutation of either element alone severely diminishes TGF-β responsiveness, suggesting that the two elements have a functionally synergistic relationship. This notion is supported by the fact that, in an additive system, mutation of either element would result in only a partial loss of TGF-β response; a complete loss of TGF-β response would require the simultaneous mutation of all contributing elements because each element could function alone to mediate a partial response. On the other hand, in a synergistic relationship such as the one we have identified, neither element is capable of mediating a vigorous transcriptional response in the absence of the other, so the effect of the two elements acting together is greater than the sum of the effects of each element alone.
These findings introduce important nuances into the developing model of Smad-mediated transcriptional regulation and offer an illustration to support aspects of Smad function predicted by biochemical and structural observations. They suggest that synergy between Smads and other transcription factors could be an important mechanism for mediating both the specificity and the responsiveness to cross talk of the TGF-β transcriptional activation signal.
Sequence comparison of Smad3-Smad4 binding sites.
Numerous studies have identified Smad3-Smad4 DNA binding sites using various approaches. As seen in Table 1, the sequences found by various groups are essentially identical; regardless of whether one defines a Smad3-Smad4 binding site as the palindrome AGACGTCT, as the CAGA box, or as repeats of GACA, all of the identified sites contain the Smad box, 5′-GTCT-3′, or its reverse complement, 5′-AGAC-3′ (9, 57, 58). Most recently, another Smad3-Smad4-responsive site, CAGACAGtCTGTCTG in the junB promoter, was identified (18). Only the COL7A1 promoter presents a discrepancy, in that the deletions which abrogate Smad binding do not directly disturb the Smad box-like sequences (54). It may be, as the authors suggest, that the small deletions at the ends of their binding element disrupted binding in a non-sequence-specific manner.
TABLE 1.
Comparison of Smad3-Smad4 binding sites
| Reference | Location | Sequence |
|---|---|---|
| This study | c-Jun, +62 to +73 | CAGACAGACAGACACA |
| 9 | PAI-1 | |
| −730 | AGCCAGACA | |
| −580 | AGACAGACA | |
| −280 | AGACAGACA | |
| 58 | Oligo screen | GTCTAGAC |
| 57 | 2X TRE (from p3TP-lux) | TGAGTCAGACA (21 bp) TGAGTCAGACA |
In agreement with these other studies, the novel Smad3-Smad4 binding site identified in the c-Jun 5′ UTR consists of three Smad boxes in a row. Although it is unusual to find enhancing elements in the 5′ UTR, it is not unprecedented. Transcriptional activators with binding sites in the 5′ UTR of promoters or in intronic sequences are hypothesized to function transiently, i.e., during the establishment of the initiation of transcription (5, 32, 44). The molecular mechanism for Smad-mediated activation of transcription is not yet well defined, but a transient role of Smads in transcriptional initiation, through their binding to the sequence in the 5′ UTR of the c-Jun gene, would be consistent with the transient presence of Smads in the nucleus after TGF-β stimulation.
The Smad consensus binding site, or Smad box, has now been confirmed by the elucidation of the crystal structure of Smad3 bound to DNA (46). A single Smad3 MH1 binds asymmetrically through a novel DNA binding β-hairpin structure to a 4-bp Smad box (CAGA) with sequence-specific interactions (9, 58). Note that in vivo Smads exist as homo- and hetero-oligomers (20, 25, 46, 56, 61), which would explain why more than one 4-bp repeat has been found to be required for binding of natural Smad complexes in the studies discussed above.
Synergy between Smads and AP-1 family members.
Our results further demonstrate that while the Smad3-Smad4 site is important for TGF-β induction of c-Jun, an AP-1/CRE site is also required for TGF-β regulation of the c-Jun promoter. Mutation of either site in the context of the −79 to +170 region of this promoter eliminated the ability of TGF-β to elicit maximal induction of the c-Jun promoter. There are several possible mechanisms by which such synergy may be achieved, and elucidating the mechanism for this synergistic cooperation is an important area for future investigations.
The first possibility is that direct physical interaction between Smads and AP-1 family members is responsible for mediating the functional cooperation. Recent studies have described an interaction between Jun family members and Smad3 (27, 60). In fact, Smad3 and Smad4 have both been found to interact with all members of the Jun family to varying degrees. The Jun family members interact with Smads at a small C-terminal domain which is highly conserved among Jun proteins. While the interaction between Jun and Smads is direct, the involvement of this protein-protein interaction in transcriptional activation of the c-Jun promoter is unclear. Although it is a strong possibility, direct protein interaction is certainly not the only possible explanation for the observed functional cooperation seen in the c-Jun promoter between Smads and AP-1.
Another possible mechanism for functional synergy is cooperative DNA binding. We do not know whether AP-1/CRE complexes and Smad3-Smad4 complexes may cooperatively bind their corresponding sites in vivo, even though they clearly can strongly bind their corresponding c-Jun promoter sites independently in vitro. It is possible that the interactions of each complex with DNA in vivo may be enhanced by cooperative recruitment and stabilization or by an alteration in local DNA structure which is fostered by the binding of both complexes at once.
Synergy is a functional cooperation that can also be independent of any physical interaction. It is possible that Smads and AP-1 may cooperate by contributing complementary but necessary subfunctions of transcriptional activation, for instance by recruiting different required members of the basal transcriptional machinery. The location of the Smad binding site 3′ in relation to the TATA box in the c-Jun promoter strongly suggests that the role of Smads is transient and limited to the start of transcription, perhaps involving the establishment of the transcription initiation complex. AP-1 may contribute complementary functions to promote transcription.
Finally, it is possible that TGF-β signal transduction can directly affect the activity of AP-1 complex bound to the promoter element. It has been postulated that TGF-β may signal through the mitogen-activated protein kinase pathway and activate AP-1 through phosphorylation. This remains to be clearly shown and is currently under investigation. Such an activity would add yet another dimension to the cooperativity in the c-Jun promoter region demonstrated here.
Further implications.
The model of required synergistic cooperation may explain some discrepancies in our understanding of Smad function to date. A number of recent studies have established the abilities of Smad3 and Smad4 to interact and function synergistically with the transcriptional coactivator CREB binding protein/p300 (10, 17, 40, 45, 52). Although these findings suggest that a DNA-bound Smad3-Smad4 complex is able to independently recruit CREB binding protein/p300 and hence possibly initiate transcription on its own, it does not appear to do so. A close examination of studies on Smad3-Smad4 binding sites reveals that no single Smad3-Smad4 site has been found to be sufficient for TGF-β responsiveness. In all of these studies, multiple copies of the Smad binding site were found to be required to confer TGF-β responsiveness (9, 58). Perhaps a single Smad3-Smad4 complex is unable to successfully recruit the factors necessary to accomplish transcriptional activation on its own. Cooperation with another transcription factor, such as AP-1, Sp1 (33), or TFE3 (16), or collaboration between a number of Smad3-Smad4 binding sites is required to build strong enough interactions to activate transcription.
It is worth noting that while we have examined responsive elements and Smad binding in the −79 to +170 region of the c-Jun promoter, there may be additional Smad3-Smad4 binding sites, or other TGF-β responsive elements, elsewhere in the native c-Jun promoter sequence. These could in fact cooperate further with the Smad3-Smad4 binding site identified in this study to mediate c-Jun regulation in vivo.
Finally, an investigation into other examples of cooperating responsive elements could yield critical insight into TGF-β signaling specificity and cross talk with other signaling pathways. Given the description in the present study of a joint requirement for Smad3-Smad4 binding and an AP-1/CRE site, it may be interesting to look for additional required elements in other TGF-β-responsive promoters. The recent work by Hua et al. (16) revealed another important example of such cooperativity and lends further support to the possibility that similar modes of synergistic transcriptional activation may exist in the context of many Smad-responsive promoters.
We have identified in these studies a functional cooperation between a novel Smad3-Smad4 site and an AP-1/CRE binding site within the −79 to +170 region of the c-Jun promoter, which functions in transcriptional activation by TGF-β. These findings not only solidify the role of Smad3 as an intracellular effector for the TGF-β signal but also support a new and more complex model of Smad3-Smad4 transcriptional regulation, i.e., one which involves cooperation with neighboring response elements and may allow coordination of other interacting pathways with the TGF-β signal. The synergistic interaction between TGF-β-specific effectors and other transcription factors proposed in this model could mediate the activation of different subsets of target genes in different cell types and physiological states, translating into the diversity of physiological and pathological roles played by TGF-β in different tissue types, stages of development, and disease states.
ACKNOWLEDGMENTS
C.W. and E.M.R.-C. contributed equally to this work.
We thank Bing Su for graciously providing the human c-Jun −79 to +170 luciferase reporter, Rik Derynck for his generous gifts of human Smad2 and Smad3, Yong Yu for technical assistance, and Patrick Hu and Xing Shen for their valuable discussions and assistance with experiments.
This work was supported by grant DK45756 from the National Institutes of Health to X.-F.W. and U.S. Army Breast Cancer Research grants BC962225 and BC971814 to E.M.R.-C. and J.P.F., respectively. N.T.L. was supported by a predoctoral fellowship from the National Science Foundation. X.-F.W. is a Leukemia Society Scholar.
REFERENCES
- 1.Angel P, Hattori K, Smeal T, Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP1. Cell. 1988;55:875–885. doi: 10.1016/0092-8674(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 2.Chen X, Rubock M J, Whitman M. A transcriptional partner for MAD proteins in TGF-beta signaling. Nature. 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- 3.Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- 4.Chung K-Y, Agarwal A, Uitto J, Mauviel A. An AP-1 binding sequence is essential for regulation of the human a2(I) collagen (COL1A2) promoter activity by transforming growth factor-β. J Biol Chem. 1996;271:3272–3278. doi: 10.1074/jbc.271.6.3272. [DOI] [PubMed] [Google Scholar]
- 5.Damert A, Leibiger B, Leibiger I B. Dual function of the intron of the rat insulin I gene in regulation of gene expression. Diabetologia. 1996;39:1165–1172. doi: 10.1007/BF02658502. [DOI] [PubMed] [Google Scholar]
- 6.Datto, M. B., J. P. Frederick, L. Pan, A. J. Borton, Y. Zhuang, and X.-F. Wang. Targeted disruption of Smad3 reveals an essential role in transforming growth factor β-mediated signal transduction. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 7.Datto M B, Li Y, Panus J, Howe D J, Xiong Y, Wang X-Y. TGF-β mediated growth inhibition is associated with induction of the cyclin-dependent kinase inhibitor, p21. Proc Natl Acad Sci USA. 1995;92:5545–5549. doi: 10.1073/pnas.92.12.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datto, M. B., Y. Yu, and X.-F. Wang. 1995. Functional analysis of the transforming growth factor β responsive elements in the WAF1/Cip1/p21 promoter. J. Biol. Chem. 270. [DOI] [PubMed]
- 9.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier J M. Direct binding of Smad3 and Smad4 to critical TGFb-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng X-H, Zhang Y, Wu R-Y, Derynck R. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for Smad3 in TGF-β-induced transcriptional activation. Genes Dev. 1998;12:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo Z, Du X, Iacovitti L. Regulation of tyrosine hydroxylase gene expression during transdifferentiation of striatal neurons: changes in transcription factors binding the AP-1 site. J Neurosci. 1998;18:8163–8174. doi: 10.1523/JNEUROSCI.18-20-08163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hannon G J, Beach D. p15INK4B is a potential effector of TGF-B-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 13.Hata A, Shi Y, Massague J. TGF-beta signaling and cancer: structural and functional consequences of mutations in Smads. Mol Med Today. 1998;4:257–262. doi: 10.1016/s1357-4310(98)01247-7. [DOI] [PubMed] [Google Scholar]
- 14.Heldin C-H, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 15.Herr I, van Dam H, Angel P. Binding of promoter-associated AP-1 is not altered during induction and subsequent repression of the c-jun promoter by TPA and UV irradiation. Carcinogenesis. 1994;15:1105–1113. doi: 10.1093/carcin/15.6.1105. [DOI] [PubMed] [Google Scholar]
- 16.Hua X, Liu X, Ansari D O, Lodish H F. Synergistic cooperation of TFE3 and Smad proteins in TGF-β induced transcription of the plasminogen activator inhibitor-1 gene. Genes Dev. 1998;12:3084–3095. doi: 10.1101/gad.12.19.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janknecht R, Wells N J, Hunter T. TGF-β-stimulated cooperation of Smad proteins with the coactivators CBP/p300. Genes Dev. 1998;12:2114–2119. doi: 10.1101/gad.12.14.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jonk L J C, Itoh S, Heldin C-H, ten Dijke P, Kruijer W. Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-β, activin, and bone morphogenetic protein-inducible enhancer. J Biol Chem. 1998;273:21145–21152. doi: 10.1074/jbc.273.33.21145. [DOI] [PubMed] [Google Scholar]
- 19.Kalkhoven E, Roelen B A, de Winter J P, Mummery C L, van den Eijnden-van Raaij A J, van der Saag P T, van der Burg B. Resistance to transforming growth factor beta and activin due to reduced receptor expression in human breast tumor cell lines. Cell Growth Differ. 1995;6:1151–1161. [PubMed] [Google Scholar]
- 20.Kawabata M, Inoue H, Hanyu A, Imamura T, Miyazono K. Smad proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. EMBO J. 1998;17:4056–4065. doi: 10.1093/emboj/17.14.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keeton M R, Curriden S A, Van Sonneveld A-J, Loskutoff D J. Identification of regulatory sequences in the type I plasminogen activator inhibitor gene responsive to transforming growth factor β. J Biol Chem. 1991;266:23048–23052. [PubMed] [Google Scholar]
- 22.Kim S-J, Angel P, Lafyatis R, Hattori K, Kim K Y, Sporn M B, Karin M, Roberts A B. Autoinduction of transforming growth factor β1 is mediated by the AP-1 complex. Mol Cell Biol. 1990;10:1492–1497. doi: 10.1128/mcb.10.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kretzschmar M, Massague J. SMADs: mediators and regulators of TGF-beta signaling. Curr Opin Genet Dev. 1998;8:103–111. doi: 10.1016/s0959-437x(98)80069-5. [DOI] [PubMed] [Google Scholar]
- 24.Lafon C, Mazars P, Guerrin M, Barboule N, Charcosset J-Y, Valette A. Early gene responses associated with transforming growth factor-β1 growth inhibition and autoinduction in MCF-7 breast adenocarcinoma cells. Biochim Biophys Acta. 1995;1266:288–295. doi: 10.1016/0167-4889(95)00023-l. [DOI] [PubMed] [Google Scholar]
- 25.Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Partnership between DPC4 and SMAD proteins in TGF-beta signaling pathways. Nature. 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 26.Li L, Hu J-S, Olson E N. Different members of the jun proto-oncogene family exhibit distinct patterns of expression in response to type β transforming growth factor. J Biol Chem. 1990;265:1556–1562. [PubMed] [Google Scholar]
- 27.Liberati, N. T., X. Shen, M. B. Datto, J. P. Frederick, and X.-F. Wang. Smads bind directly to the Jun family of AP-1 transcription factors. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 28.Macias-Silva M, Abdollah S, Hoodless P, Pirone R, Attisano L, Wrana J. MADR2 is a substrate of the TGF-β receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 29.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L Z, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, Brattain M, Willson J K V. Inactivation of the type II TGF-β receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 30.Massague J. TGF-β signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 31.Massague J. The transforming growth factor-β family. Annu Rev Cell Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 32.McCarthy T L, Thomas M J, Centrella M, Rotwein P. Regulation of insulin-like growth factor I transcription by cyclic adenosine 3′,5′-monophosphate (cAMP) in fetal rat bone cells through an element within exon 1: protein kinase A-dependent control without a consensus AMP response element. Endocrinology. 1995;136:3901–3908. doi: 10.1210/endo.136.9.7649098. [DOI] [PubMed] [Google Scholar]
- 33.Moustakas A, Kardassis D. Regulation of the human p21/WAF1/Cip1 promoter in hepatic cells by functional interactions between Sp1 and Smad family members. Proc Natl Acad Sci USA. 1998;95:6733–6738. doi: 10.1073/pnas.95.12.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muegge K, Vila M, Gusella G L, Musso T, Herrlich P, Stein B, Durum S K. Interleukin 1 induction of the c-jun promoter. Proc Natl Acad Sci USA. 1993;90:7054–7058. doi: 10.1073/pnas.90.15.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda A, Tamaki K, Hanai J, Heldin C H, Miyazono K, ten Dijke P. TGFβ receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norgaard P, Damstrup L, Rygaard K, Spang-Thomsen M S, Poulsen H S. Growth suppression by transforming growth factor β1 of human small-cell lung cancer cell lines is associated with expression of the type II receptor. Br J Cancer. 1994;69:802–808. doi: 10.1038/bjc.1994.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Padgett R W, Das P, Krishna S. TGF-beta signaling, Smads, and tumor suppressors. Bioessays. 1998;20:382–390. doi: 10.1002/(SICI)1521-1878(199805)20:5<382::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 38.Park K, Kim S-J, Bang Y-J, Park J-G, Kim N K, Roberts A B, Sporn M B. Genetic changes in the transforming growth factor β (TGF-β) type II receptor gene in human gastric cancer cells: correlation with sensitivity to growth inhibition by TGF-β. Proc Natl Acad Sci USA. 1994;91:8772–8776. doi: 10.1073/pnas.91.19.8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pertovaara L, Sistonen L, Bos T J, Vogt P K, Keski-Oja J, Alitalo K. Enhanced jun gene expression is an early genomic response to transforming growth factor β stimulation. Mol Cell Biol. 1989;9:1255–1262. doi: 10.1128/mcb.9.3.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pouponnot C, Jayaraman L, Massague J. Physical and functional interaction of SMADs and p300/CBP. J Biol Chem. 1998;273:22865–22868. doi: 10.1074/jbc.273.36.22865. [DOI] [PubMed] [Google Scholar]
- 41.Reynisdottir I, Polyak K, Iavarone A, Massague J. Kip/Cip and Ink4 cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-beta. Genes Dev. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 42.Roberts A B, Sporn M B. The transforming growth factor β’s. In: Sporn M B, Roberts A R, editors. Peptides, growth factors and their receptors, part I. Berlin, Germany: Springer-Verlag; 1990. pp. 419–472. [Google Scholar]
- 43.Rozek D, Pfeifer G P. In vivo protein-DNA interactions at the c-jun promoter: preformed complexes mediate the UV response. Mol Cell Biol. 1993;13:5490–5499. doi: 10.1128/mcb.13.9.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schollen E, De Meirsman C, Matthijs G, Cassiman J J. A regulatory element in the 5′ UTR directs cell-specific expression of the mouse alpha 4 gene. Biochem Biophys Res Commun. 1995;211:115–122. doi: 10.1006/bbrc.1995.1785. [DOI] [PubMed] [Google Scholar]
- 45.Shen X, Hu P P-C, Liberati N T, Datto M B, Frederick J P, Wang X-F. TGF-β induced phosphorylation of Smad3 regulates its interaction with coactivator p300/CBP. Mol Biol Cell. 1998;9:3309–3319. doi: 10.1091/mbc.9.12.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shioda T, Lechleider R J, Dunwoodie S L, Li H, Yhata T, de Caestecker M P, Fenner M H, Roberts A B, Isselbacher K J. Transcriptional activating activity of Smad4: roles of SMAD heterooligomerization and enhancement by an associating transactivator. Proc Natl Acad Sci USA. 1998;95:9785–9790. doi: 10.1073/pnas.95.17.9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi Y, Wang Y-F, Jayaraman L, Yang H, Massague J, Pavletich N P. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA-binding in TGF-β signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- 48.Su B, Jacinto E, Hibi M, Kallunki T, Karin M, Ben-Neriah Y. JNK is involved in signal integration during costimulation of T lymphocytes. Cell. 1994;77:727–736. doi: 10.1016/0092-8674(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 49.Subramaniam M, Oursier M J, Rasmussen K, Riggs B L, Spelsberg T C. TGF-β regulation of nuclear proto-oncogenes and TGFβ gene expression in normal human osteoblast-like cells. J Cell Biochem. 1995;57:52–61. doi: 10.1002/jcb.240570107. [DOI] [PubMed] [Google Scholar]
- 50.Sun L, Wu G, Willson J K V, Zborowska E, Yang J, Rajkarunanayake I, Wang J, Gentry L E, Wang X-F, Brattain M G. Expression of transforming growth factor β type II receptor leads to reduced malignancy in human breast cancer MCF-7 cells. J Biol Chem. 1994;269:26449–26455. [PubMed] [Google Scholar]
- 51.Tang W, Yang L, Yan Y-C, Leng S X, Elias J A. Transforming growth factor-β stimulates interleukin-11 transcription via complex activating protein-1-dependent pathways. J Biol Chem. 1998;273:5506–5513. doi: 10.1074/jbc.273.10.5506. [DOI] [PubMed] [Google Scholar]
- 52.Topper J N, DiChiara M R, Brown J D, Williams A J, Falb D, Collins T, Gimbrone M A., Jr CREB binding protein is a required coactivator for Smad-dependent, transforming growth factor β transcriptional responses in endothelial cells. Proc Natl Acad Sci USA. 1998;95:9506–9511. doi: 10.1073/pnas.95.16.9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Dam H, Duyndam M, Rottier R, Bosch A, de Vries-Smits L, Herrlich P, Zantema A, Angel P, van der Eb A J. Heterodimer formation of cJun and ATF-2 is responsible for induction of c-jun by the 243 amino acid adenovirus E1A protein. EMBO J. 1993;12:479–487. doi: 10.1002/j.1460-2075.1993.tb05680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vindevoghel L, Kon A, Lechleider R J, Uitto J, Roberts A B, Mauviel A. Smad-dependent transcriptional activation of human type VII collagen gene (COL7A1) promoter by transforming growth factor-β. J Biol Chem. 1998;273:13053–13057. doi: 10.1074/jbc.273.21.13053. [DOI] [PubMed] [Google Scholar]
- 55.Wang J, Sun L Z, Myeroff L, Wang X-F, Gentry L E, Yang J, Liang J, Zborowska E, Markowitz S, Willson J K V, Brattain M G. Demonstration that mutation of the type II transforming growth factor β receptor inactivates its tumor suppressor activity in replication error-positive colon carcinoma cells. J Biol Chem. 1995;270:22044–22049. doi: 10.1074/jbc.270.37.22044. [DOI] [PubMed] [Google Scholar]
- 56.Wu R-Y, Zhang Y, Feng X-H, Derynck R. Heteromeric and homomeric interactions correlate with signaling activity and functional cooperativity of Smad3 and Smad4/DPC4. Mol Cell Biol. 1997;17:2521–2528. doi: 10.1128/mcb.17.5.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yingling J M, Datto M B, Wong C, Frederick J P, Liberati N T, Wang X-F. Tumor suppressor Smad-4 is a transforming growth factor β-inducible DNA binding protein. Mol Cell Biol. 1997;17:7019–7028. doi: 10.1128/mcb.17.12.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zawel L, Dai J L, Buckhaults P, Zhou S, Kinzler K W, Vogelstein B, Kern S E. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Feng X-H, Wu R-Y, Derynck R. Receptor-associated Mad homologues synergize as effectors of the TGF-β response. Nature. 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Feng X-H, Derynck R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGFbeta-induced transcription. Nature. 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Y, Musci T, Derynck R. The tumor suppressor Smad4/DPC4 as a central mediator of Smad function. Curr Biol. 1997;7:270–276. doi: 10.1016/s0960-9822(06)00123-0. [DOI] [PubMed] [Google Scholar]