Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for the pandemic of coronavirus disease (COVID-19). Whereas in most cases COVID-19 is asymptomatic or pauci-symptomatic, extremely severe clinical forms are observed. In this case, complex immune dysregulations and an excessive inflammatory response are reported and are the main cause of morbidity and mortality. Natural killer cells are key players in the control of viral infection, and their activity is regulated by a tight balance between activating and inhibitory receptors; an alteration of NK activity was suggested to be associated with the development of severe forms of COVID-19. In this study, we analyzed peripheral NK cell subpopulations and the expression of activating and inhibitory receptors in 30 patients suffering from neurological conditions who recovered from mild, moderate, or severe SARS-CoV-2 infection, comparing the results to those of 10 SARS-CoV-2-uninfected patients. Results showed that an expansion of NK subset with lower cytolytic activity and an augmented expression of the 2DL1 inhibitory receptor, particularly when in association with the C2 ligand (KIR2DL1-C2), characterized the immunological scenario of severe COVID-19 infection. An increase of NK expressing the ILT2 inhibitory receptor was instead seen in patients recovering from mild or moderate infection compared to controls. Results herein suggest that the KIR2DL1-C2 NK inhibitory complex is a risk factor toward the development of severe form of COVID-19. Our results confirm that a complex alteration of NK activity is present in COVID-19 infection and offer a molecular explanation for this observation.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12035-021-02517-4.
Keywords: NK, KIR, COVID-19, SARS-CoV-2, Innate immunity
Introduction
The COVID-19 is provoked by the Coronaviridae SARS-CoV-2, a single strain RNA virus characterized by an extremely high infectivity but a relatively low pathogenicity. Thus, while the majority of patients experience mild symptoms, around 20% of them develop a multi organ condition that, in the worst cases, results in a severe and potentially lethal acute respiratory distress syndrome (ARDS) [1]. The pathogenesis of COVID-19 is still unclear, but a growing number of evidences indicate that an excessive and dysregulated immune response is the main cause of morbidity and mortality [2]. Such immune response is associated with massive inflammation and an increased production of inflammatory cytokines, as well as with quantitative and qualitative alterations that affect the immune cells involved both in innate and acquired immune responses [3].
As is the case with all pathogens, and in particular with human respiratory RNA viruses, the establishment of a successful infection by SARS-CoV-2 is contingent upon the activation of mechanisms that bypass or suppress innate immune responses [4, 5]. Innate immunity, indeed, is the first line of defense against infections and acts prior to the protective responses mediated by the adaptive immune system. Among the actors of innate immunity, a pivotal role is played by natural killer (NK) cells, a population of immune cells that is also extremely important in integrating innate and adaptive immune responses [6, 7]. NK cells include a number of different subpopulations that are associated with diverse cytotoxic abilities [8–10]. NK cell activation, and the triggering of their cytotoxic abilities, is an active process that depends on the interaction between receptor molecules on the surface of NK cells and ligands on target cells. The killer cell immunoglobulin (Ig)-like receptors (KIRs) and the inhibitory receptors immunoglobulin-like transcript ILT-2 are the best described of such receptors, and ligate shared allelic determinants of HLA class I molecules as well as HLA-G on target cells [11]. These interactions regulate NK activity, as NK cell activation or lack thereof is the consequence of a delicate balance between signals that are generated from activating and inhibitory receptors belonging to many families.
Analyses focusing on NK cells in SARS-CoV-2 infection showed that reduced NK-cell counts and impaired cytolytic activity associate with the development of a severe COVID-19–related inflammation [12]. Other results indicated that an upregulation of NK-inhibitory receptors is present as well in SARS-CoV-2-infection. Thus, in patients with severe COVID-19 disease, a reduction of NK antiviral activity, characterized by an increase of the NKG2A and KIR2DL1 inhibitory receptor, and a reduced ability to express CD107 and to produce IFN-γ, IL-2, granzyme B, and TNF-α were shown [13, 14]. Taken together, these results indicate that NK cells might be exhausted in severe COVID-19.
In this study, we analyzed peripheral NK subpopulation and the expression of activating and inhibitory receptors in patients affected by neurological conditions who recovered from mild, moderate, or severe SARS-CoV-2 infection. Analyses were performed on samples obtained 45–60 days after SARS-CoV-2 infection onset, and results were compared to those of patients with similar neurological conditions that were never SARS-CoV-2-infected. Data herein confirm that COVID-19 results in a complex alteration of NK cell subsets and functions that is more marked in those individuals who underwent severe disease.
Materials and Methods
Patients and Controls
Forty patients admitted from March 2020 to June 2020 to the Fondazione Don Carlo Gnocchi Onlus in patient rehabilitation program (Milan, IRCCS Santa Maria Nascente and Rovato, Centro Spalenza) were enrolled in the study. All patients were undergoing intensive rehabilitation due to the disability caused by a neurological disease, and all of them received at least one SARS-CoV-2 RNA test on their nasopharyngeal swab (NPS) or bronchial aspirate. Thirty patients contracted SARS-CoV-2 infection, while ten other patients were repeatedly negative on their NPSs (control group).
Patients were divided into three diagnostic groups: brain injury causing a disorder of consciousness (DOC), injury of the central nervous system (CNS) without DOC history, and disease of the peripheral nervous system (PNS).
It is well known that COVID-19 severity can be highly variable, with the disease ranging from mild flu-like symptoms to severe interstitial pneumonia. In line with Chen and colleagues [15], COVID severity was graded on three levels:
Mild: asymptomatic or flu-like symptoms without pneumonia manifestation
Moderate: pneumonia manifestation in imaging without respiratory distress
Severe: pneumonia with respiratory distress treated with respiratory support (from continuous positive airway pressure up to oro-tracheal intubation or tracheostomy)
The study was approved by the local ethics committee (Comitato Etico, IRCCS Fondazione Don Gnocchi, Milano), and patients or guardians provided written informed consent.
Of interest for the current study, we recently described the spread of the SARS-CoV-2 infection and the clinical course of the COVID-19 in a cohort of DOC patients who have been exposed to the virus [16]. In that work, we detail in full the course of several clinical and laboratory abnormalities (e.g. C-reactive protein, white blood cell) related to the COVID-19 and the management of the infection in this unique patients’ population. To note, some of the DOC patients recruited in our previous study have been also recruited here. Therefore, by adding clinical details about the disease in some of the patients recruited here, our recent work [16] should actually be considered a companion of the current one.
Blood Sample Collection, Immunofluorescent Staining, and Analysis by Flow Cytometry
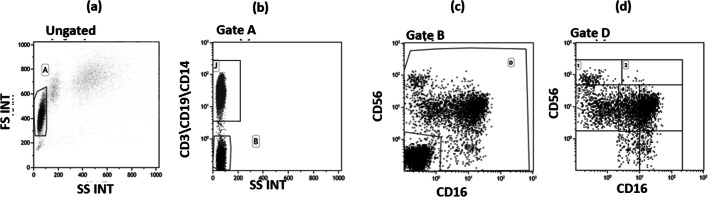
Whole blood was collected in vacutainer tubes containing ethylenediamine tetra-acetic acid (EDTA) (Becton Dickinson & Co., Rutherford, NJ), cell counts were performed using an XN 1000 Sysmex hematology analyzer (Dasit Group, Italy), and viability was evaluated by the automated cell counter ADAM-MC (Digital Bio, NanoEnTek Inc., Korea), after red cell lysis. Immunophenotypic analyses were performed on 600 μl of EDTA peripheral blood incubated for 30 min at 4 °C with the fluorochrome-labeled monoclonal antibodies. Erythrocyte lysis was obtained with the Immuno-Prep Epics Kit and Q-Prep Work Station (Beckman-Coulter Brea, CA, USA). The analyses were performed using a Beckman-Coulter GALLIOS flow cytometer equipped with a 22 mW blue solid-state diode laser operating at 488 nm and with a 25 mW red solid-state diode laser operating at 638 nm and interfaced with Kaluza analysis software. For each analysis, 20,000 events were acquired and gated on forward and side scatter properties for lymphocyte and on the CD3-CD19-CD14- and side scatter properties to exclude T, B, and monocyte cells; the remaining triple-negative cells were analyzed in a CD56 versus CD16 dot plot to identify the natural killer (NK) cell subsets, CD56brightCD16−, CD56brightCD16dim, CD56dimCD16bright, CD56dimCD16dim, CD56dimCD16−, and CD56 − CD16bright, considering isotype background (Fig. 1). The expression of KIR receptors was performed on NK subsets. The flow cytometry compensation was performed, using the fluorescence minus one (FMO) control approach. Briefly, all antibody conjugates in the experiment are included except the one that is controlled for. The FMO measures the spread of fluorescence from the other staining parameters into the channel of interest, determining the threshold for positive staining.
Fig. 1.
Flow cytometry dot plots indicating the gate strategy used to identify natural killer cell subsets. a Lymphocyte selected by forward (FS) and side scatter (SS) properties (gate A). b The CD3 + CD19 + CD14+ vs the SS dot plot allows the discrimination of monocytes and T and B lymphocytes (gate J); the remaining triple negative cells (gate B) were analyzed within a CD56 vs CD16 dot plot (c) leading to the identification of natural killer cells (gate D). d Natural killer subsets were selected in the CD56 vs CD16 dot plot as follows: CD56brightCD16− (1), CD56brightCD16dim (2), CD56dimCD16-(3), CD56dimCD16dim(4), CD56dimCD16bright (5), and CD56 − CD16bright(6)
Monoclonal Antibodies (mAbs)
The following mAbs were used: anti-CD3 phycoerythrin-cyanine 7 (PE-Cy7) (Mouse IgG1, clone: UCHT1) (Beckman-Coulter); anti-CD19 PC-7 (Mouse IgG1, clone: J3–119) (Beckman-Coulter); anti-CD14 PC- 7 (IgG2a Mouse, clone: RMO52) (Beckman-Coulter); anti-CD4 phycoerythrin (PE) (Mouse Ig1; clone 13B8.2); anti-CD8 phycoerythrin-cyanine 5 (PE-Cy5) (Mouse IG1; clone B9.11); anti-CD16 fluorescein (FITC) or PE-Cy5 (Mouse IgG1, clone: 3G8) (Beckman-Coulter); anti-CD56 PE (Mouse IgG1, clone: N901) (NKH-1) (Beckman-Coulter); anti-human KIR2DL1/CD158a FITC (Mouse IgG1, clone: 143211) (R&D Systems, Minneapolis, MN, USA); anti-human KIR2DS4/CD158i allophycocyanin (APC) (Mouse IgG2a, clone: 179315) (R&D Systems); anti-human ILT2/CD85j APC (Mouse IgG1, clone: 292305) (R&D Systems); anti-human KIR2DS1/CD158h Alexa Fluor 700 (Rabbit IgG, clone: 1127B) (R&D Systems); and anti-anti-KIR2DS2/CD158b polyclonal antibody FITC (Rabbit IgG aa39–65) (LSBio, Seattle, WA, USA).
KIR and HLA Ligand Genotyping
Genomic DNA was isolated from peripheral blood by phenol-chloroform extraction using standard procedures. Molecular genotyping of KIRs and HLA-Bw4+/Bw4- C1/C2 Kir ligands was performed by PCR on genomic DNA using sequence specific primers (SSP) according to the manufacturer’s instructions (BAG, Lich, Germany; Astra Formedic, Milan, Italy). Allele detection was done after amplification in a GeneAmp PCR 9700 thermocycler (Applied Biosystems, Foster City, CA, USA) by gel electrophoresis on 2% agarose gel.
KIR-HLA complexes were defined as follows:
Statistical Analyses
The normality of distribution of continuous variables was evaluated using the Kolmogorov-Smirnov test. Quantitative data were defined normally or not normally distributed (Shapiro-Wilk test) and are therefore summarized as median and standard deviation or median and interquartile range (IQR; 25th and 75th percentiles) respectively. Comparisons between groups were performed using a t-test or two-tailed Mann-Whitney test for independent samples. Kruskal-Wallis analysis of variance was utilized for each variable. Chi-square analysis was used to evaluate KIR and HLA genetic distribution between groups. Data analysis was performed using the MEDCALC statistical package (MedCalc Software bvba, Mariakerke, Belgium).
Results
Patients’ clinical characteristics are given in Table 1. COVID-19 respiratory involvement ranged from mild to severe, and, about the lungs’ modifications on the chest CT scan, eight patients infected by SARS-CoV-2 showed ground glass opacities, six showed consolidations, and three showed both ground glass opacities and consolidations. No abnormalities on the chest CT scan were found in negative patients.
Table 1.
Clinical characteristics of COVID-19 convalescent patients and healthy controls. CNS central nervous system, PNS peripheral nervous system, DOC disorder of consciousness, Q1 and Q3 first and third quartile, respectively. For each diagnostic group, the most common diagnosis is given (in brackets) as well as the COVID severity (1, mild; 2, moderate; 3, severe). Duration of infection: days between the first positive and the first negative SARS-CoV-2 test on nasopharyngeal swab (NPS) or bronchial aspirate
| COVID-19 patients | No COVID | |
|---|---|---|
| Age, median (Q1–Q3), years | 66 (58–76) | 65 (58–75) |
| Gender, male/female | 19/11 | 7/3 |
|
Brain injury with DOC history (hemorrhagic stroke) |
6 (4) | 6 (4) |
| COVID severity, median (range) | 2 (1–2) | – |
|
Injury of the CNS without DOC history (stroke and cerebrovascular disease) |
15 (8) | 3 (2) |
| COVID severity, median (range) | 2 (1–3) | – |
|
Disease of the PNS (peripheral neuropathy) |
9 (7) | 1 (1) |
| COVID severity, median (range) | 3 (1–3) | – |
| Duration of infection, median (Q1–Q3), days | 42 (32.5–58) | – |
Regarding long-term comorbidities (i.e., those present before being exposed to the virus), patients were affected on median by one disease each (Table 2), with hypertension being the most common one.
Table 2.
Patients’ comorbidity. N number of patients affected by the disease before the diagnosis of COVID-19. These diseases are known to be associated with poor prognosis in SARS-CoV-2 infection. Each patient was diagnosed as being affected by at least one of these conditions
| Disease | N |
|---|---|
| Hypertension | 25 |
| Diabetes mellitus | 8 |
| Ischemic heart disease | 6 |
| Active smoking | 5 |
| Chronic obstructive pulmonary disease | 4 |
| Malignancy | 4 |
| Obstructive sleep apnea syndrome | 2 |
Analysis of Peripheral T, B, and NK Subsets
Thirty COVID-19 convalescent patients who had been diagnosed as being affected by either mild, moderate, or severe COVID-19 disease (10/group) as well as 10 SARS-CoV-2-uninfected controls were analyzed. The following NK cell subsets were analyzed in peripheral blood of all the individuals enrolled in the study: (1) CD56brightCD16−; (2) CD56brightCD16dim; (3) CD56dimCD16−; (4) CD56dimCD16dim; (5) CD56dimCD16bright; and (6) CD56 − CD16bright. The expression of the activating receptors KIR2DS1, KIR2DS2, and KIR2DS4 as well as that of the inhibitory receptors KIR2DL1 and KIRILT-2 was analyzed as well in all patients and controls.
Percentage and absolute number of T, B, and NK lymphocyte subsets were analyzed in all the individuals enrolled in the study. Results did not show the presence of any significant differences in any of the immune populations when COVID-19 patients were compared to controls or when they were divided up based on disease severity (Table 3).
Table 3.
Percentage (%) and absolute number of peripheral blood lymphocyte subsets in convalescent COVID-19 patients who suffered from mild, moderate, or severe disease and in healthy controls. Median and interquartile range are indicated. n.s. Not significant
| No-COVID | Mild | Moderate | Severe | ||
|---|---|---|---|---|---|
| p Value | |||||
| CD3% | 83.5 | 79 | 76 | 80 | n.s. |
| (79–88.7) | (73–85) | (74.5–77) | (74.2–81.5) | ||
| CD3 | 1320 | 1270 | 1079 | 1409 | |
| (1011–2222) | (763–2060) | (751–1296) | (1103–1854) | ||
| CD4% | 53.5 | 48 | 50 | 44 | n.s. |
| (48–57) | (34–53) | (45–55) | (38–54) | ||
| CD4 | 895 | 713 | 681 | 852 | |
| (802–1577) | (398–1088) | (440–828) | (690–1171) | ||
| CD8% | 25 | 30 | 24 | 30 | n.s. |
| (22–32) | (24–35) | (23–30) | (23–38) | ||
| CD8# | 448 | 435 | 357 | 512 | |
| (278–840) | (282–666) | (247–432) | (330–737) | ||
| CD16% | 8 | 7 | 11 | 7 | n.s. |
| (5–14) | (4–12) | (5–13) | (5–15) | ||
| CD16 | 177 | 98 | 117 | 143 | |
| (66–251) | (88–153) | (97–168) | (78–198) | ||
| CD19% | 5 | 6 | 10 | 5 | n.s. |
| (4–10) | (5–9) | (7–12) | (4–8) | ||
| CD19 | 179 | 103 | 133 | 113 | |
| (58–241) | (70–128) | (64–151) | (75–156) | ||
| CD3 + DR+ % | 13 | 22 | 17 | 23 | n.s |
| (12–16) | (13–23) | (10–20) | (11–31) | ||
| CD3 + DR+ | 290 | 252 | 186 | 492 | |
| (190–391) | (180–390) | (94–350) | (109–630) | ||
| CD4 + DR+ % | 4 | 5 | 4 | 5 | n.s. |
| (3–6) | (4–7) | (3–5) | (4–9) | ||
| CD4 + DR+ | 64 | 92 | 68 | 117 | |
| (38–95) | (74–107) | (30–116) | (70–160) | ||
| CD8 + DR+ % | 8 | 14 | 5 | 11 | n.s. |
| (5–10) | (5–17) | (3–12) | (8–18) | ||
| CD8 + DR+ | 166 | 178 | 72 | 226 | |
| (118–240) | (111–222) | (31–171) | (88–316) | ||
| CD16 + DR+ % | 1 | 1 | 1 | 2 | n.s. |
| (0–3) | (0–2) | (0–2) | (0–4) | ||
| CD16 + DR+ |
14 (13–54) |
16 (5–23) |
10 (5–30) |
34 (2–63) |
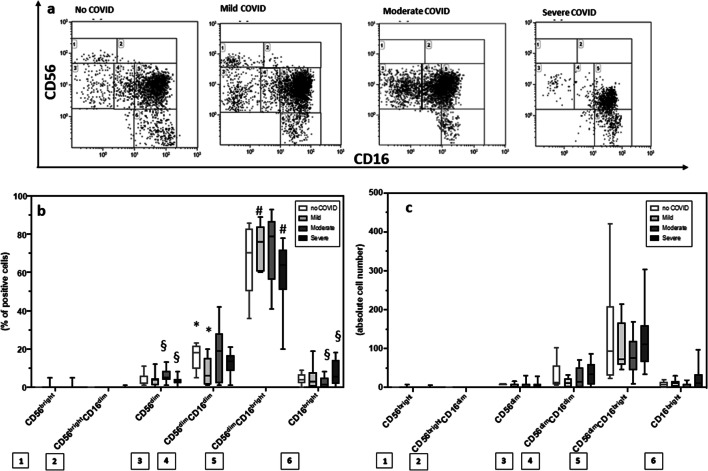
Results showed that the percentage of (1) CD56dimCD16bright and CD56dimCD16− cells was reduced in severe (median = 60% and 3% respectively) compared to mild (CD56dimCD16bright median = 76%, p = 0.03) and moderate (CD56dimCD16− median = 6%, p = 0.01) COVID-19 patients; (2) CD56 − CD16bright cells was increased in severe (median = 7.4%) compared to mild (median = 5%) and moderate (median = 3%, p = 0.04) COVID-19 patients and controls (median = 4%); and (3) CD56dimCD16dim cells was reduced in mild (median = 8%) COVID-19 patients compared to SARS-CoV-2-uninfected controls (median = 16%, p = 0.04) (Fig. 2). Notably, no differences were observed between the analyzed groups when the NK subset absolute numbers were analyzed.
Fig. 2.
COVID-19 is associated with dysregulated NK cell subsets. (a) CD56 vs CD16 dot plot identifying natural killer cell subsets. Summary results of median percentage (b) and of absolute numbers (c) of NK subsets relative to total NK population in SARS-CoV-2-uninfected healthy controls (white bar) and in convalescent COVID-19 patients who suffered from mild (light gray bar), moderate (gray bar), or severe (black bar) disease are shown. The boxes stretch from the 25th to the 75th percentile. The lines across the boxes indicate the median values. The lines stretching from the boxes indicate extreme values. Statistical significance is shown: *comparisons mild vs no COVID; #comparisons mild vs severe; §comparisons moderate vs severe
Analysis of KIR Receptor Expression
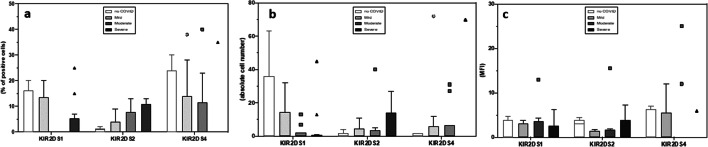
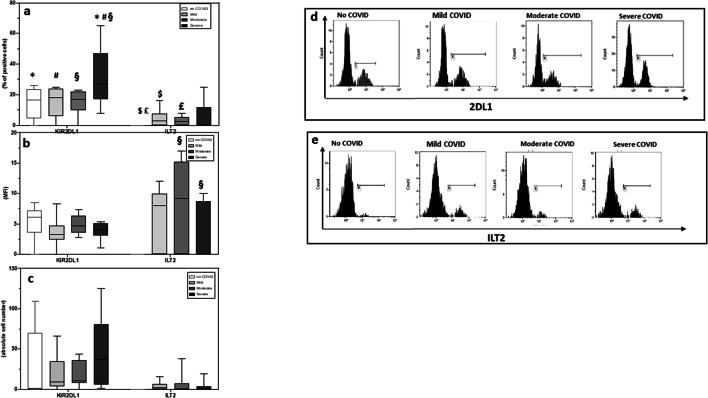
The expression of KIR activating and inhibitory receptors was analyzed next on NK CD56dimCD16bright cells, the major representative subset in peripheral blood. Results showed that the percentage and absolute number of NK expressing the activating 2DS1, 2DS2, and 2DS4 receptors were similar in all groups analyzed (Fig. 3). In contrast with these data, the percentage of NK cells expressing the inhibitory receptor 2DL1 was significantly increased in severe (median = 30%) compared to mild (median = 15%) (p = 0.03) or moderate (median = 14%) (p = 0.02) COVID-19 patients and SARS-CoV-2-uninfected controls (median = 14%) (p = 0.02). No differences were observed upon analyses of MFI of these receptors.
Fig. 3.
NK cell expressing activating receptors are similar in all groups analyzed. Summary results of median percentage (a), absolute numbers (b), and MFI (c) of NK cells expressing the 2DS1, 2DS2, and 2DS4 activating KIR receptor in SARS-CoV-2-uninfected (white bar) and in convalescent COVID-19 patients who suffered from mild (light gray bar), moderate (gray bar), or severe (black bar)
The expression and MFI of ILT-2, another inhibitory receptor, was analyzed next on in all patients and controls. Results showed that ILT-2 expression was increased in all groups of COVID-19 patients compared to SARS-CoV-2-uninfected controls. These differences reached statistical significance in mild (median = 4.3%) and moderate (median = 3.2%) COVID-19 patients compared to SARS-CoV-2-uninfected controls (median = 0.2%) (p = 0.04 and p = 0.03 respectively vs controls). ILT-2 MFI was similarly increased in all groups of COVID-19 patients compared to SARS-CoV-2-uninfected controls, but these differences were not statistically significant (Fig. 4). Results of further analyses showed that in all groups of COVID-19 patients, 85% of CD56-CD16bright cells express ILT2 (Supplemental Fig. 1).
Fig. 4.
NK cell expressing inhibitory receptors are expanded in COVID-19 patients. Summary results of median percentage (a), absolute numbers (b), and MFI (c) of NK expressing the 2DL1 and ILT2 inhibitory KIR receptors in SARS-CoV-2-uninfected healthy controls (white bar) and in convalescent COVID-19 patients who suffered from mild (light gray bar), moderate (gray bar), or severe (black bar) disease. The percentage of NK cell percentage expressing the 2DL1 (d) and ILT2 (e) inhibitory KIR receptors are also shown. The boxes stretch from the 25th to the 75th percentile. The lines across the boxes indicate the median values. The lines stretching from the boxes indicate extreme values. Statistical significance is shown. *Comparisons no COVID vs severe; #comparisons mild vs severe; §comparisons moderate vs severe; $comparisons no COVID vs mild; £comparisons no COVID vs moderate
KIR and HLA Ligand Distributions
The genetic pattern of distribution of all analyzed KIRs and HLA-C, HLA Bw4 ligands, as well as the number of activating and inhibitory KIR genes and of KIR-HLA activating and inhibitory complexes is reported in Supplementary Table 1. No statistical differences were observed between groups.
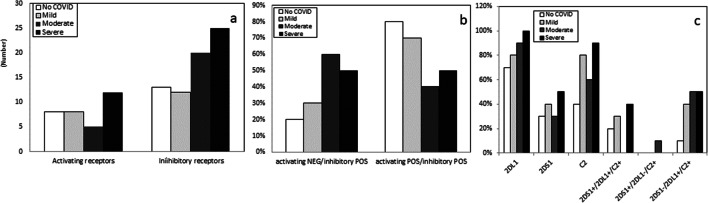
However, KIR-HLA inhibitory complexes (2DL1-C2, 2DL2-C1, 3DL3-C1, 3DL1-Bw4) were more frequently present in patients who developed either a moderate (absolute number of KIR-HLA inhibitory complex: N = 20) or a severe (N = 25) form of COVID-19 compared to the situation observed in individuals with mild disease (N = 12) or SARS-CoV-2-uninfected controls (N = 13) (Table 1S and Fig. 5a). These differences approached but did not reach statistical significance, possibly because of the limited number of analyzed individuals.
Fig. 5.
Absolute number of activating and inhibitory KIR receptors in SARS-CoV-2-uninfected healthy controls (white bar) and in convalescent COVID-19 patients who suffered from mild (light gray bar), moderate (gray bar), or severe (black bar) disease (a). Percentage of subjects carrying inhibitory but not activating KIR or carrying both inhibitory and activating KIR (b). Percentage of subjects carrying KIR2DS1, KIR2DL1, and HLA-C2 ligands and of subjects expressing different KIR2DS1/2DL1-HLA-C2 functional complexes (c): 2DS1 + C2+/2DL1 + C2+ (both inhibitory and activating receptor are functional); 2DS1 + C2+/2DL1-C2+ (only activating receptor is functional)
2DS1-C2+/2DL1 + C2+ (only inhibitory receptor is functional).
The presence/absence of activating and inhibitory KIR-HLA complexes was finally evaluated. The phenotype characterized by the simultaneous absence of activating and the presence of inhibitory KIR-HLA complexes was, once again, more frequently detected in patients who developed moderate (60%) and severe (50%) forms of COVID-19 compared to the situation seen in individuals with mild disease (30%) or in SARS-CoV-2-uninfected controls (20%) (Fig. 5b). In particular, the genetic KIR2DS1-C2 (activatory)-negative/KIR2DL1-C2 (inhibitory)-positive complex was more frequently observed in patients with moderate (50%) and severe (50%) COVID-19 than in those with mild (40%) disease or in controls (10%) (Fig. 5c). These results were confirmed by the observation that cells of individuals with mild COVID-19 and SARS-CoV-2-uninfected more frequently carried both activatory and inhibitory KIRs (70% and 80%, respectively) (Fig. 5b).
Discussion
We performed an analysis of innate immune responses in a group of patients affected by neurological conditions who either were recovering from SARS-CoV-2 infection or were never infected by the virus, with a focus on NK cells. Results of phenotypic analyses showed that patients recovering from severe COVID-19 are characterized by a peculiar skewing in NK cell subpopulations. Thus, in these patients, CD56-CD16bright were increased, whereas CD56dimCD16− and CD56dimCD16bright NK cells were reduced in comparison to patients who suffered from either mild or moderate COVID-19.
An expansion of CD56 − CD16 bright NK cells was previously described in chronic HCV infection [19] and in viremic HIV-infected patients, but not in those individuals in whom antiretroviral therapy resulted in viremia suppression [20]. Even more recently the same phenomenon was described in Cytomegalovirus and Epstein-Barr virus co-infected elderly individuals [21]. Importantly, results obtained in chronic HIV-1 and HCV infections showed that expansion of CD56neg NK cells is accompanied by decreases in numbers of CD56dim NK cells [19, 22]. CD56neg NK cells were observed to be phenotypically more closely related to CD56dim NK cells than CD56bright NK cells. Thus, results obtained by principal component analysis (PCA) [23] and by studies focusing on epigenetic modifications, [24, 25] showed that CD56neg cells display a transcriptional feature more similar to that of CD56dim than to that of CD56bright NK cells.
CD56 − CD16 bright NK cells are characterized by a significantly lower cytolytic activity and a scarce ability to secrete cytokines [26]; we observed that this NK subset also expresses the inhibitory ILT2 receptor in SARS-CoV-2-infected patients. CD56dimCD16− cells are instead endowed with a potent antiviral activity mediated both by cytolytic mechanisms and by the production of high amounts of IFNγ [27, 28]. These cells were recently shown to be decreased in COVID-19 patients with a diagnosis of ARDS and who required mechanical ventilation [29]. A higher expression of ILT2 and 2DL2 on CD56- NK was shown to associate with an increased receptor-specific inhibition of CD16-mediated cytolytic activity in HIV infection [20]. The observation that this same cell population is expanded in COVID-19 patients indicates that similar alterations of NK cell populations are present in both diseases. The additional observation that the CD56dimCD16− and the CD56dimCD16bright NK cell subsets were reduced in patients recovering from severe COVID-19 reinforces the idea that NK cell-mediated immune responses are impaired in severe SARS-CoV-2 infection. CD56dimCD16bright NK cells, finally, besides being characterized as well by a potent cytolytic activity, were also shown to be particularly apt at activating antibody-dependent cellular cytotoxicity (ADCC) in response to viruses, including influenza virus, herpes simplex virus type 1, and HCMV [30–32]. NK-mediated ADCC was recently suggested to contribute to viral control in COVID-19 patients, as antibodies elicited toward the SARS-CoV-2 S glycoprotein (S309 and S306)-transfected cells could efficiently trigger ADCC [33].
The Don Gnocchi Foundation, where the study was conducted, is a rehabilitation clinical center; as such, we did not have access to patients with ongoing, acute COVID-19 infection who died from their disease. Other authors have nevertheless analyzed NK cell subsets in these patients. Results showed that an increase of exhausted CD56dimCD16bright and of CD57+ mature cells [34, 35] and a decrease of CD56bright NK characterize patients suffering from extremely severe COVID-19 disease [34]. These results could be seen as being in disagreement with ours, but once the subset of patients who did survive extremely severe infection was analyzed alone, results showed that, similarly to what we observed, exhausted CD56dimCD16bright NK cells were diminished. Taken together these results allow the speculation that the extensive viral replication and the proinflammatory milieu seen in severe COVID-19 results in the increase of CD56dimCD16bright and of CD57+ mature NK cells, which are characterized by impaired cytolytic functions, and the reduction of CD56bright cells, which are known to be endowed with a potent antiviral activity.
NK cells are activated following the interaction between HLA molecules and KIR activating and inhibitory receptors, and the net imbalance between these two families of qualitatively antagonist receptors dictates whether NK cell will or will not be activated. Analyses of KIR activating and inhibitory receptors on NK CD56dimCD16bright cells, the major representative subset in peripheral blood, showed that, whereas no differences could be seen in the expression the activating 2DS1, 2DS2, and 2DS4 receptors, the percentage of NK cells expressing the inhibitory receptor 2DL1 was significantly increased in severe COVID-19 patients. The skewing toward a preferential expression of inhibitory NK cell-associated molecules in COVID-19 patients was further supported by results showing that ILT2 expression, another inhibitory receptor, was augmented as well on NK cells of all COVID-19 patients. Our results also suggest that the presence of the inhibitory 2DL1complex, particularly when in association with the C2 protein (KIR2DL1-C2), is a risk factor toward more severe form of COVID-19 development.
An imbalance in the ratio of NK inhibitory and activating receptors is present in a number of diseases [36], and it is known that KIR/HLA interactions influence both susceptibility and protection toward infective diseases [37–41]. Within the scenario of infectious diseases, an increased expression of inhibitory NK cell receptor on CD56 − CD16 + NK cells was shown to be present in HIV infection and to result in the inhibition of CD16-induced cytotoxicity [18]. Notably, recent results showed that the density of 2DL1 is higher on NK cells of COVID-19 patients with a diagnosis of acute respiratory distress syndrome (ARDS) [14]. This result, thus, further supports the hypothesis that NK function is defective in patients who suffer from severe forms of SARS-CoV-2 infection within an extremely complex scenario of immune impairment that involves multiple cell types. These alterations can be summarized as an altered distribution of NK subsets and a preferential expression of NK inhibitory receptor, which is particularly evident in convalescent COVID-19 patients who suffered from a more severe form of infection.
The limit of the study is the small number of patients. Ampler cohorts of individuals, possibly followed within a longitudinal study, will be needed to better understand the different mechanisms implemented by the virus to evade the immune system during infection, and the immune defense strategies put into play to counter and eliminate the virus.
Supplementary Information
(PNG 678 kb)
(DOCX 22 kb)
Author Contribution
MS, DT, and MC conceived and designed the research; MS, DT, IM, FLR, EB, MZ, and FP performed the experiments; AC, AL, LB, PB, and JN are responsible for the clinical cohorts of patients; MS, DT, FRG, and MC analyzed the data and prepared the manuscript.
Funding
The study was supported by Italian Ministry of Health (RC 2020) and by COVID Regione Lombardia, Italy project “Risposta immune in pazienti con COVID-19 e comorbidità.”
Availability of Data and Materials
Data generated during the current study are available from the corresponding author on reasonable request.
Declarations
Consent to Participate
Informed consent was obtained from all individual participants included in the study. The informed consent was approved by the Ethic committee of the IRCCS Fondazione Don Gnocchi Milano.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Marina Saresella and Daria Trabattoni contributed equally to this work.
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, et al. China novel coronavirus investigating and research seam. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2019;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan L, Wang Q, Zhang D, Ding J, Huang Q, et al. Lymphopenia predicts disease severity of COVID-19: A descriptive and predictive study. Signal Transduct Target Ther. 2020;5(1):33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin C, Zhou L, Hu Z, Zhang S, Yang S, et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelemans T, Kikkert M. Viral innate immune evasion and the pathogenesis of emerging RNA virus infections. Viruses. 2019;11:961. doi: 10.3390/v11100961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 6.Marcenaro E, Carlomagno S, Pesce S, Moretta A, Sivori S. Bridging innate NK cell functions with adaptive immunity. Adv Exp Med Biol. 2011;780:45–55. doi: 10.1007/978-1-4419-5632-3_5. [DOI] [PubMed] [Google Scholar]
- 7.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331(6013):44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michel T, Poli A, Cuapio A, Briquemont B, Iserentant G, et al. Human CD56bright NK cells: An update. J Immunol. 2016;196(7):2923–2931. doi: 10.4049/jimmunol.1502570. [DOI] [PubMed] [Google Scholar]
- 9.Poli A, Michel T, Theresine M, Andres E, Hentges F, et al. CD56bright natural killer (NK) cells: An important NK cell subset. Immunology. 2009;126:458–465. doi: 10.1111/j.1365-2567.2008.03027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633–640. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 11.Clements CS, Kjer-Nielsen L, Kostenko L, Hoare HL, Dunstone MA, et al. Crystal structure of HLA-G: A nonclassical MHC class I molecule expressed at the fetal-maternal interface. Proc Natl Acad Sci U S A. 2005;A102(9):3360–3365. doi: 10.1073/pnas.0409676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osman M, Faridi RM, Sligl W, Shabani-Rad MT, Dharmani-Khan P, et al. Impaired natural killer cell counts and cytolytic activity in patients with severe COVID-19. Blood Adv. 2020;4(20):5035–5039. doi: 10.1182/bloodadvances.2020002650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng M, Gao Y, Wang G, Song G, Liu S, et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17(5):533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demaria O, Carvelli J, Batista L, Thibult ML, Morel A, et al. Identification of druggable inhibitory immune checkpoints on natural killer cells in COVID-19. Cell Mol Immunol. 2020;17(9):995–997. doi: 10.1038/s41423-020-0493-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen T, Wu D, Chen H, Yan W, Yang D, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: Retrospective study. BMJ. 2020;368:m1091. doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caronni A, Liaci E, Bianchi A, Viganò A, Marenco F, et al. Clinical course of SARS-CoV-2 infection in patients with severe acquired brain injury and a disorder of consciousness: An observational study. Brain Inj. 2021;35(5):520–529. doi: 10.1080/02699052.2021.1887937. [DOI] [PubMed] [Google Scholar]
- 17.Middleton D, Williams F, Halfpenny IA. KIR genes. Transpl Immunol. 2005;14:135–142. doi: 10.1016/j.trim.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31(4):429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez VD, Falconer K, Björkström NK, Blom KG, Weiland O, et al. Expansion of functionally skewed CD56-negative NK cells in chronic hepatitis C virus infection: Correlation with outcome of pegylated IFN-alpha and ribavirin treatment. J Immunol. 2009;183(10):6612–6618. doi: 10.4049/jimmunol.0901437. [DOI] [PubMed] [Google Scholar]
- 20.Mavilio D, Benjamin J, Daucher M, Lombardo G, Kottilil S, et al. Natural killer cells in HIV-1 infection: Dichotomous effects of viremia on inhibitory and activating receptors and their functional correlates. Proc Natl Acad Sci U S A. 2003;100(25):15011–15016. doi: 10.1073/pnas.2336091100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller-Durovic B, Grählert J, Devine OP, Akbar AN, Hess C. CD56-negative NK cells with impaired effector function expand in CMV and EBV co-infected healthy donors with age. Aging. 2019;11:724–740. doi: 10.18632/aging.101774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alter G, Teigen N, Davis BT, Addo MM, Suscovich TJ, et al. Sequential deregulation of NK cell subset distribution and function starting in acute HIV-1 infection. Blood. 2005;106:3366–3369. doi: 10.1182/blood-2005-03-1100. [DOI] [PubMed] [Google Scholar]
- 23.Ishiyama K, Kitawaki T, Otsuka Y, Takaori-Kondo A, Kadowaki N. Programmed cell death 1-expressing CD56-negative natural killer (NK) cell expansion is a hallmark of chronic NK cell activation during dasatinib treatment. Cancer Sci. 2021;112:523–536. doi: 10.1111/cas.14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith SL, Kennedy PR, Stacey KB, Worboys JD, Yarwood A, et al. Diversity of peripheral blood human NK cells identified by single-cell RNA sequencing. Blood Adv. 2020;4:1388–1406. doi: 10.1182/bloodadvances.2019000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang C, Siebert JR, Burns R, Gerbec ZJ, Bonacci B, et al. Heterogeneity of human bone marrow and blood natural killer cells defined by single-cell transcriptome. Nat Commun. 2019;10:3931. doi: 10.1038/s41467-019-11947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mavilio D, Lombardo G, Benjamin J, Kim D, Follman D, et al. Characterization of CD56-CD16+ natural killer (NK) cells: A highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proc Natl Acad Sci U S A. 2005;102:2886–2891. doi: 10.1073/pnas.0409872102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilk AJ, Blish CA. Diversification of human NK cells: Lessons from deep profiling. J Leukoc Biol. 2018;103:629–641. doi: 10.1002/JLB.6RI0917-390R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martínez-Colón GJ, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26(7):1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J, Zhang T, Hwang I, Kim A, Nitschke L, et al. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity. 2015;42(3):431–442. doi: 10.1016/j.immuni.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang T, Scott JM, Hwang I, Kim S. Cutting edge: Antibody dependent memory-like NK cells distinguished by FcRγ deficiency. J Immunol. 2013;190:1402–1406. doi: 10.4049/jimmunol.1203034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costa-Garcia M, Vera A, Moraru M, Vilches C, López-Botet M, et al. Antibody-mediated response of NKG2Cbright NK cells against human cytomegalovirus. J Immunol. 2015;194(6):2715–2724. doi: 10.4049/jimmunol.1402281. [DOI] [PubMed] [Google Scholar]
- 33.Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583(7815):290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 34.Varchetta S, Mele D, Oliviero B, Mantovani S, Ludovisi S, et al. Unique immunological profile in patients with COVID-19. Cell Mol Immunol. 2021;18(3):604–612. doi: 10.1038/s41423-020-00557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu C, Martins AJ, Lau WW, Rachmaninoff N, Chen J, et al. Time-resolved systems immunology reveals a late juncture linked to fatal COVID-19. Cell. 2021;184(7):1836–1857.e22. doi: 10.1016/j.cell.2021.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guerini FR, Clerici M. NK cells in human disease: An evolving story. Clin Immunol. 2012;43:203–206. doi: 10.1016/j.clim.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Dizaji Asl K, Velaei K, Rafat A, Tayefi Nasrabadi H, Movassaghpour AA, et al. The role of KIR positive NK cells in diseases and its importance in clinical intervention. Int Immunopharmacol. 2021;92:107361. doi: 10.1016/j.intimp.2020.107361. [DOI] [PubMed] [Google Scholar]
- 38.Wawina-Bokalanga T, Vanmechelen B, Lhermitte V, Martí-Carreras J, Vergote V, et al. Human diversity of killer cell immunoglobulin-like receptors and human leukocyte antigen class I alleles and Ebola virus disease outcomes. Emerg Infect Dis. 2021;27(1):76–84. doi: 10.3201/eid2701.202177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guerini FR, Mancuso R, Agostini S, Agliardi C, Zanzottera M, et al. Activating KIR/HLA complexes in classic Kaposi's sarcoma. Infect Agent Cancer. 2012;7:9. doi: 10.1186/1750-9378-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soria A, Guerini FR, Bandera A, Bolognesi E, Uglietti A, et al. KIR-HLA genotypes in HIV-infected patients lacking immunological recovery despite effective antiretroviral therapy. PLoS One. 2011;6(11):e27349. doi: 10.1371/journal.pone.0027349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guerini FR, Lo Caputo S, Gori A, Bandera A, Mazzotta F, et al. Under representation of the inhibitory KIR3DL1 molecule and the KIR3DL1+/BW4+ complex in HIV exposed seronegative individuals. J Infect Dis. 2011;203(9):1235–1239. doi: 10.1093/infdis/jir020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PNG 678 kb)
(DOCX 22 kb)
Data Availability Statement
Data generated during the current study are available from the corresponding author on reasonable request.