Abstract
Hypertrophic growth of cardiac muscle cells is induced by a variety of physiological and pathological stimuli and is associated with a number of changes, including activation of genes such as atrial natriuretic factor. We found that two serum response element (SRE)-like DNA elements, one of which does not meet the consensus sequence and binds serum response factor (SRF) with low affinity, regulate the activity of this promoter. Surprisingly, the ability to induce the promoter by two different physiologic stimuli, as well as various activated transcription factors, including SRF-VP16, was primarily dependent upon the nonconsensus rather than the consensus SRE. This SRE controls the induction of gene expression via an unusual mechanism in that it is required to allow some, but not all, active transcription factors at unrelated sites on the promoter to stimulate gene expression. Thus, in addition to regulation of SRF activity by growth stimuli, regulation of a low-affinity SRE element controls inducible gene expression by modulating the ability of other transcription factors to stimulate the transcription machinery.
Postnatal growth of cardiac muscle cells occurs by hypertrophy rather than by cell division and is associated with a number of phenotypic changes, including increased expression of a number of cardiac muscle cell-specific genes such as atrial natriuretic factor (ANF) (11). Induction of hypertrophy and activation of the ANF promoter is achieved by many kinds of stimuli, including growth factors that bind to tyrosine kinase-linked receptors (34), cytokines that activate gp130-linked receptors (35), agonists such as phenylephrine or angiotensin II that activate G-protein-coupled receptors (27, 42, 46), mechanical stretch (43, 58), increased muscle cell contraction rate (32), and activators of protein kinase C, such as phorbol esters (3, 13). Given the diverse stimuli that activate this promoter, ANF is a good model for studying mechanisms that regulate inducible gene expression.
The ANF promoter contains two serum response element (SRE)-like DNA elements that are thought to be important for expression (48). Serum response factor (SRF) is one of the best-studied inducible transcription factors, with most of our information coming from the analysis of the c-Fos SRE. As its name suggests, the SRE element in the c-Fos promoter is stimulated by serum. SRF is also activated by other growth factors and quite different stimuli, such as alterations in the cytoskeleton. In some cases, activation of SRE-driven gene expression is achieved through accessory factors. The best-understood such mechanism involves the activation of ternary complex factors (TCFs), such as Elk-1 or SAP-1, that are recruited by SRF and are activated as transcription factors by phosphorylation by mitogen-activated protein (MAP) kinases (21, 25, 26, 30, 57). TCFs such as Elk-1 interact with DNA via an Ets site adjacent to the SRF-binding site, and activators that induce MAP kinase activity require an intact TCF site to stimulate the c-Fos promoter. Other stimuli, such as lysophosphatidic acid (LPA) or Rho family GTPases, are able to directly activate SRF (21, 22). Rho-dependent activation of SRF is not mediated through MAP kinases, does not appear to require TCFs, and involves as-yet-unidentified Rho effector pathways (22, 44).
Many and perhaps all of the stimuli that lead to cardiac hypertrophy and expression of ANF cause activation of one or more MAP kinase cascades (5–8, 38, 40, 41, 47, 52, 59). The role of MAP kinases in the regulation of this promoter has been confusing, with conflicting data regarding the roles of the ERK and SAPK-JNK pathways in gene expression (17, 18, 33, 37, 49, 51, 52, 56). Recently, more-consistent results from a number of groups have demonstrated that activation of the p38 pathway is sufficient to stimulate the ANF promoter (33, 55, 59).
We wished to examine the mechanism by which different stimuli are able to activate the ANF promoter. We first assessed the role of p38 in ANF induction by two different physiological stimuli: the alpha-1 adrenergic agonist phenylephrine and electrical-pacing-induced contraction. Electrical-pacing-induced, but not phenylephrine-induced, expression requires p38, and this is achieved in part through a cyclic AMP response element (CRE)-like DNA element in the promoter. In addition, both of these p38-dependent and p38-independent stimuli also require input from the SRE elements. Both SRE elements are important for the regulation of the basal activity of the promoter, but only the nonconsensus SRE regulates induction by phenylephrine, electrical pacing, and even overexpression of activated forms of SRF and GAL4-transcription factor fusion proteins. We find that hypertrophic stimuli activate SRE-regulated expression in a manner distinct from activation of SRF itself by serum, Rho, or LPA (22). Rather, the nonconsensus SRE is required to allow some, but not all, activated transcription factors at other sites on the promoter to stimulate gene expression. This mechanism therefore represents a method of inducible gene regulation where a regulated DNA element works by discriminating between activated transcription factors at other sites on the promoter and controlling their ability to induce the basal transcription machinery.
MATERIALS AND METHODS
Cell culture and transfections.
Cardiac muscle cells were cultured and transfected as previously described (24, 49, 51) except that for the electrical pacing experiments, the cells were plated at a higher density (2,000/mm2) than for the other treatments (260/mm2); this increased density is required for efficient electrical pacing in vitro. All transfections were conducted in duplicate or in triplicate in 3.5-cm tissue culture dishes by using the calcium phosphate precipitation method. Prior to transfection, all plasmids were purified by alkaline lysis followed by polyethylene glycol precipitation. Luciferase and β-galactosidase activity were assayed on a Dynatec MLX luminometer 48 h after transfection by harvesting the cells in reporter lysis buffer (Promega, Madison, Wis.) and incubation with the appropriate light emission accelerator (Tropix, Bedford, Mass.; Promega) reaction buffer. The data shown for each experiment represents the mean ± The Standard error of the mean for a single experiment that was representative of at least three repeated experiments.
Cell treatments.
Cells were treated as required with phenylephrine (5 to 100 μM) (Sigma, St. Louis, Mo.) and p38 inhibitor SB203580 (20 μM) (Calbiochem, La Jolla, Calif.) for 48 h. The media with and without inhibitors was replaced every 24 h for all cultures. For electrical stimulation to induce muscle cell contraction, the cells were plated in 24-well plates that were electrically connected via an agarose salt bridge and stimulated with a custom-built stimulator. In this case controls were from the same plate of cells but from wells that were not electrically paced.
Plasmids.
Expression plasmids encoding SRF and SRF-VP16 were provided by Art Alberts and Richard Treisman (ICRF, London, United Kingdom). GAL4-VP16 was provided by Don Ayer (University of Utah). The MEK6 expression plasmid was provided by Bernd Stein (Signal Pharmaceuticals, La Jolla, Calif.), and the MEKK1 expression plasmid was provided by Melanie Cobb (University of Texas Southwestern, Dallas, Tex.). The Raf:ER expression plasmid (50) was based on a molecule provided by Martin McMahon (University of California at San Francisco). GAL4-ATF2, GAL4-Jun, and GAL4-Elk expression plasmids were obtained from Stratagene (La Jolla, Calif.). All luciferase reporter plasmids were compared to a cotransfected Rous sarcoma virus-LacZ plasmid that was provided by Michael Kapiloff (OHSU, Portland, Oreg.). Promoter mutations were constructed by site-directed mutagenesis with the Quickchange Kit (Stratagene) in a −638 ANF luciferase plasmid, which was constructed in pGL3basic (Promega), or in the −132 ANF-luciferase, which is a HindIII truncation of the same plasmid. Mutations were constructed as follows: SRE1, bases −114 to −104 (CTTTAAAAGG) mutated to CGCGGATCCG; SRE2, bases −406 to −396 (CCTTATTTGG) mutated to CGCGGATCG; SRE1 → SRE2, bases −114 to −104 (CTTTAAAAGG) mutated to CCTTATTTGG; SRE2 → SRE1, bases −406 to −396 (CCTTATTTGG) mutated to CTTTAAAAGG; SRE1 → cFos, bases −114 to −104 (CTTTAAAAGG) mutated to CCATATTAGG; SRE1 → SREP, bases −114 to −104 (CTTTAAAAGG) mutated to ACATATTAGT; SRE1T → C, bases −114 to −104 (CTTTAAAAGG) mutated to CCTTAAAAGG; SRE1 ↔ SRE2, bases −114 to −104 (CTTTAAAAGG) mutated to CCTTATTTGG and bases −406 to −396 (CCTTATTTGG) mutated to CTTTAAAAGG; and mutCRE, bases −601 to −593 (TGACTTCA) mutated to GGATCCCA. Reporter plasmids were also constructed by using the core SRE elements with flanking KpnI sites cloned into a basal promoter (pGL3prl, provided by Michael Kapiloff) that contains 72 bp of the prolactin promoter, including the TATA box. The SRE2 element was also ligated to the end of the −132 promoter to create −132 SRE by using these same KpnI sites. In addition, a 300-bp piece of DNA was amplified from the kanamycin gene and fused to the −132 reporter, and then three copies of the SRE2 element were fused to this molecule. Finally, two GAL4 sites were fused to the −132 ANF to create −132GAL4. All of the promoter constructs were checked by sequencing. In some cases the mutant promoters were repaired by further mutagenesis to recreate the wild-type sequence as a control for mutagenesis specificity.
In vitro DNA binding.
SRF protein was prepared by coupled in vitro transcription-translation of a T7-driven SRF plasmid in reticulocyte lysate by using the TNT kit (Promega). Then, 1 μl of lysate programmed with either SRF or luciferase (provided by Promega) cDNAs was incubated with the appropriate double-stranded DNA probe for 30 min at room temperature in 10 mM HEPES (pH 7.9), 50 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, and 10% glycerol, along with 2 μl of poly(dI-dC) per 20-μl reaction. DNA-protein complexes were resolved on a 5% acrylamide gel in 0.5× Tris-borate buffer. All probes were double stranded and were radiolabeled with [γ−32P]ATP. The following are single-stranded sequences of the probes, with the core SREs underlined: SRE1(114) short, GGCTATACTTTAAAAGGCCGATAT; SRE2(406) short, GGCTATACCTTATTTGGCCGATAT; SRE1(−114), TCGCTGGACTGATAACTTTAAAAGGGCATCTTCTCCTGGC; and SRE2(−406), TGCCTCTCCTCCCGCCCTTATTTGGAGCCCCTGACAGCTG. For competition assays, 5×, 10×, 100×, or 1,000× cold double-stranded SRE1(114) short or SRE2(406) short was coincubated in reactions with double-stranded radiolabeled SRE2(406)-short probe as described above.
RESULTS
p38-dependent activation of ANF gene expression.
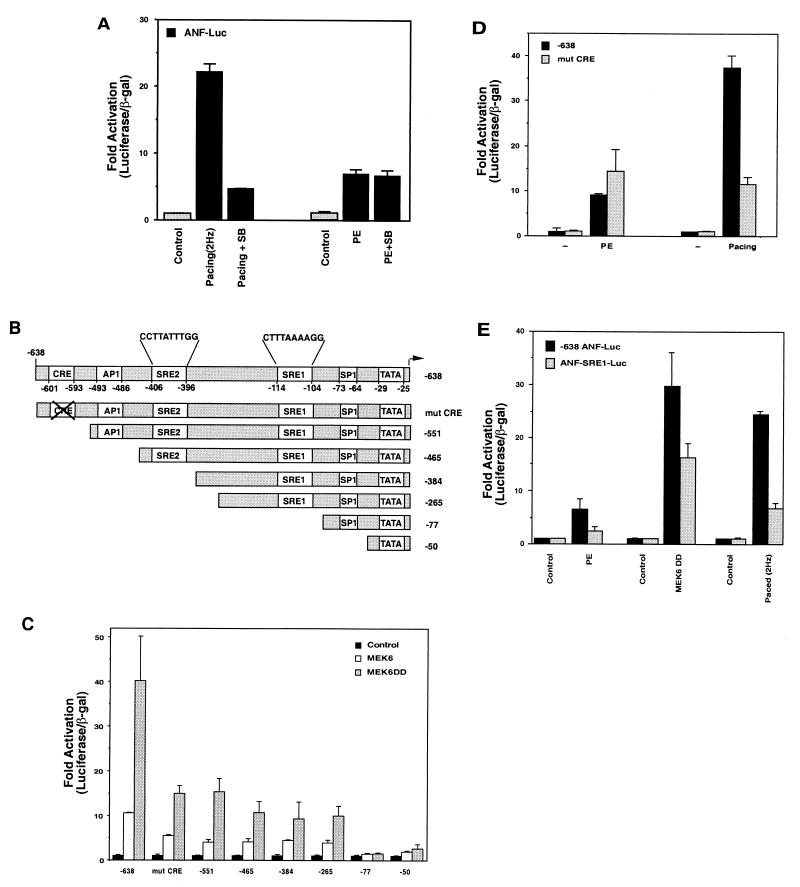
To first determine if diverse hypertrophic stimuli require the p38 MAP kinase, we used the p38 inhibitor SB203580 in transient-transfection experiments with ANF-luciferase reporter plasmids in neonatal rat ventricular myocytes. The myocytes were then stimulated by two different physiological stimuli: phenylephrine, which activates the α-adrenergic receptor, and electrical pacing to increase the contraction rate. Figure 1A shows that ANF-luciferase expression is increased in paced cells (2.5 Hz) compared to unpaced cells and that this activation is inhibited by the p38 inhibitor. In the same experiment, the p38 inhibitor failed to reduce phenylephrine-induced expression. We conclude that the basic signaling mechanisms that are used by phenylephrine and electrically stimulated contraction to induce ANF-luciferase expression differ in their requirement for p38 in these culture conditions.
FIG. 1.
Pacing- but not phenylephrine-induced ANF expression requires p38. (A) Cells were transfected with ANF-luciferase and then treated with phenylephrine (PE) or electrically paced to induce contraction at 2 Hz in the presence or absence of the p38 inhibitor SB 203580 (SB). Both electrical pacing and phenylephrine-induced ANF-luciferase expression, but inhibition of p38 only reduced pacing-induced expression. (B) Schematic of truncation mutants of the −638 ANF promoter, indicating putative transcriptional elements. (C) Cells were transfected with control vectors, expression vectors for wild-type or activated MEK6, and various truncations of the ANF promoter driving luciferase or a mutation in a CRE-like element. Note that MEK6 activity induces the ANF promoter and that this is partially inhibited by mutation or truncations that remove the CRE-like element. Complete inhibition of activation occurs when an SRE-like element is deleted (−77). (D) Cells were transfected with the wild-type −638 promoter, or one which contains the CRE mutation, and then stimulated by phenylephrine or pacing. Both stimuli induced the wild-type promoter and while the CRE mutation significantly inhibited pacing-induced expression, it had no effect on phenylephrine-stimulated expression. (E) Cells were transfected with the −638 promoter or a mutant promoter in the SRE1 sequence (ANF-SRE1-Luc) and treated with phenylephrine, cotransfected with active MEK6, or electrically paced. Activation of the SRE1 mutant promoter is compromised for all stimuli.
MAP kinase-induced gene expression is achieved through direct phosphorylation of transcription factors, leading to increased transcriptional activity. The ANF promoter contains a number of DNA elements that could potentially regulate induction (Fig. 1B). To begin to identify the transcription factors that mediate regulation of the ANF promoter in response to these diverse signaling pathways, we performed a deletion analysis to determine the sites that are responsible for MEK6 (and thus p38)-dependent activation of this gene. Figure 1C shows the results of a deletion analysis through a portion of the ANF promoter that contains 638 bp of promoter sequence where expression was induced by overexpression of wild-type MEK6 or a constitutively active MEK6 molecules (MEK6DD), i.e., treatments that should lead only to p38 activation. We found two truncations (−551) and (−77) that had a strong inhibitory effect on MEK6-induced ANF-luciferase expression. The first deletion reduced induction by about 50% and removed a CRE-like element. We therefore mutated this element (mutCRE) and tested the induction of this promoter with active MEK6. Mutation or deletion of the CRE element both led to similar 50% reductions in MEK6-induced gene expression. We conclude that this element is a target for p38-dependent signals, presumably via transcription factors such as ATF2 that can be activated by the stress-induced MAP kinases and that can bind to CRE elements (19, 53). If this DNA element is actually the target for p38-dependent signaling, we might expect that the mutCRE promoter molecule would be less sensitive to phenylephrine induction than to pacing induction. Figure 1D shows that this is indeed the case when direct comparison of induction between the wild-type promoter and the CRE mutant are examined in response to either phenylephrine or pacing.
We noted that while the CRE deletion or mutation had a significant effect on MEK6-induced gene expression, it did not completely abolish transactivation. The truncation that completely abolished MEK6-induced gene expression removed a DNA element with the sequence CTTTAAAAGG. This sequence, which we denote as SRE1, is similar to the consensus binding sequence for SRF (CCA/T6GG). To determine whether the SRE was important for the induction of ANF gene expression, we mutated the SRE1 and tested whether this affected gene expression stimulated by MEK6, phenylephrine, or pacing. Figure 1E shows that mutation of this SRE-like element inhibited expression by all of the stimuli that were tested including phenylephrine, which is not affected by the p38 inhibitor. Thus, in contrast to the CRE mutation, the SRE1 element was a target for both p38-dependent and p38-independent signals. This observation led us to further investigate the role of the SREs in regulating the induction of the ANF promoter.
SRF binds preferentially to SRE2 compared to SRE1.
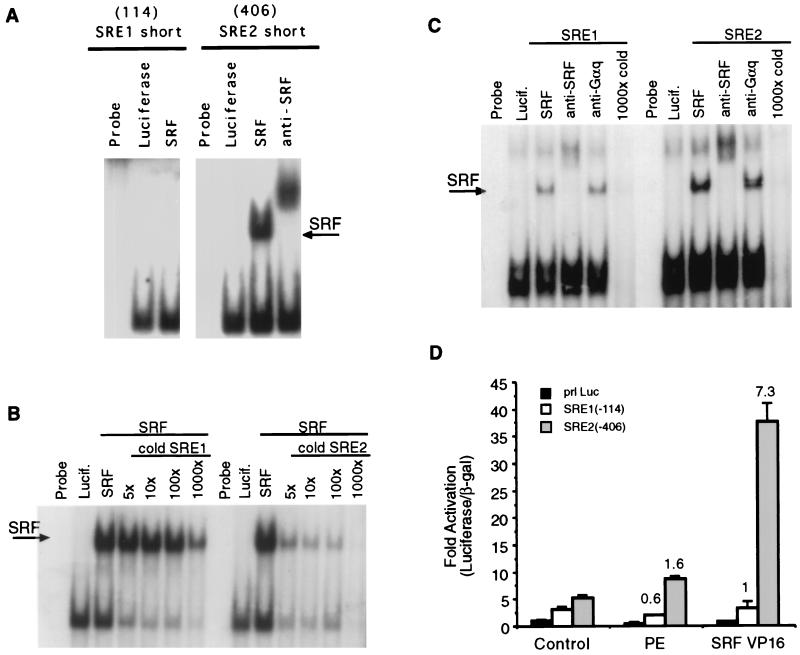
Since several diverse stimuli appeared to require the same DNA element for maximal induction (SRE1), we focused our subsequent studies on the mechanism of action at the ANF SRE elements. The ANF promoter contains two SRE-like elements; SRE2 matches the consensus binding sequence, while SRE1 does not. To test whether SRF could bind to these sequences, we performed binding studies by using in vitro-translated SRF and the core SRE elements. Figure 2A shows that SRF can bind efficiently to the core SRE2 element but that binding to the core SRE1 element was barely detectable. SRF bound with similar efficiency to the core c-Fos SRE as to SRE2 (data not shown). Figure 2B shows competition assays with SRE2 as probe, which was coincubated with excess cold SRE1 or SRE2. It was evident that these SREs have different affinities for SRF, since 5× cold SRE2 was more effective at competition than 1,000× cold SRE1. The nonspecific band below the shifted complex is competed off almost equally well by both competitors. To test whether SRF could bind if we included more flanking sequences, we repeated experiments shown in Fig. 1A with 40-mer oligonucleotides containing either the SRE1 sequence or the SRE2 sequence in the middle of native flanking ANF sequences. Figure 2C indicates that SRF can in fact bind to both SRE1 and SRE2 when longer oligonucleotides are used. However, binding was considerably stronger to the consensus SRE2 sequence than to the nonconsensus SRE1 sequence. A nonspecific band occurs in all lanes in a position near, but distinct from, the supershifted complex. Cardiac nuclear extracts contain a protein that binds to these sequences and which can be shifted by anti-SRF antibodies (reference 48 and data not shown). We found that this band was stronger when the consensus SRE2 element rather than the nonconsensus SRE1 element is used in the binding assay (data not shown).
FIG. 2.
SRF binding to SRE1 and SRE2. (A) In vitro binding reactions with SRF or luciferase from reticulocyte lysate in the presence of anti-SRF antibody as indicated, along with the core SRE1 (SRE1 short) and SRE2 (SRE2 short) probes. SRF binds more efficiently to SRE2 than to SRE1. (B) Competition assay with SRE2 as a probe along with excess cold SRE1 or SRE2. (C) Binding reactions similar to those in panel A but with longer oligonucleotides that contained 15 bp of the native ANF flanking sequence on either side of the two core SRE elements and the indicated antibodies. Although both SREs can bind to SRF, SRE2 binds much more efficiently than did SRE1. (D) Minimal promoters driven by single copies of SRE1 or SRE2 were transfected into cardiac muscle cells along with SRF-VP16 or empty vector and treated 100 μM phenylephrine (PE). The SRE2-luciferase plasmid is stimulated by SRF-VP16 but not by phenylephrine. SRE1-luciferase is not significantly stimulated by either phenylephrine or SRF-VP16.
To test whether SRF could bind to these SREs in intact heart cells, we constructed reporter plasmids that consist of a minimal promoter fused to the core SRE1 or SRE2 sequences. Figure 2D shows an experiment where an expression plasmid encoding a full-length SRF cDNA fused to the transactivation domain from the strong viral transcription factor VP16 was transfected along with SRE1 or SRE2 reporters. SRF-VP16 activated the reporter plasmids that contain the consensus SRE2 sequence but not the SRE1 sequences. These data indicate that SRF-VP16 can bind more efficiently to the SRE2 sequence than to the SRE1 sequence in cardiac muscle cells. We also tested whether phenylephrine treatment or electrical pacing stimulated gene expression that was controlled by these elements. The SRE2-driven reporter was modestly activated by less than 50% by phenylephrine, while the SRE1-driven reporter was not stimulated at all by this agonist. Electrical pacing also failed to stimulate expression from these constructs (data not shown). These data suggest that phenylephrine activates SRF differently than does serum or the Rho family GTPases, where SRF bound to a minimal SRE is a more effective transcription factor in the presence of the stimulus (2, 21, 22). Taken together, these experiments produce the expected result: the SRE2 element that matches the consensus binding sequence functions as an effective SRF binding element, while the nonconsensus SRE1 element does not, and the two SRE elements are therefore not equivalent to each other. In addition, the SRE1 sequence, while critical for induction of the intact promoter by phenylephrine or pacing (Fig. 1E), does not appear to be an independent regulatory element that is sufficient for induction in isolation.
Induction and basal activity of the ANF promoter is dependent on the SRE elements.
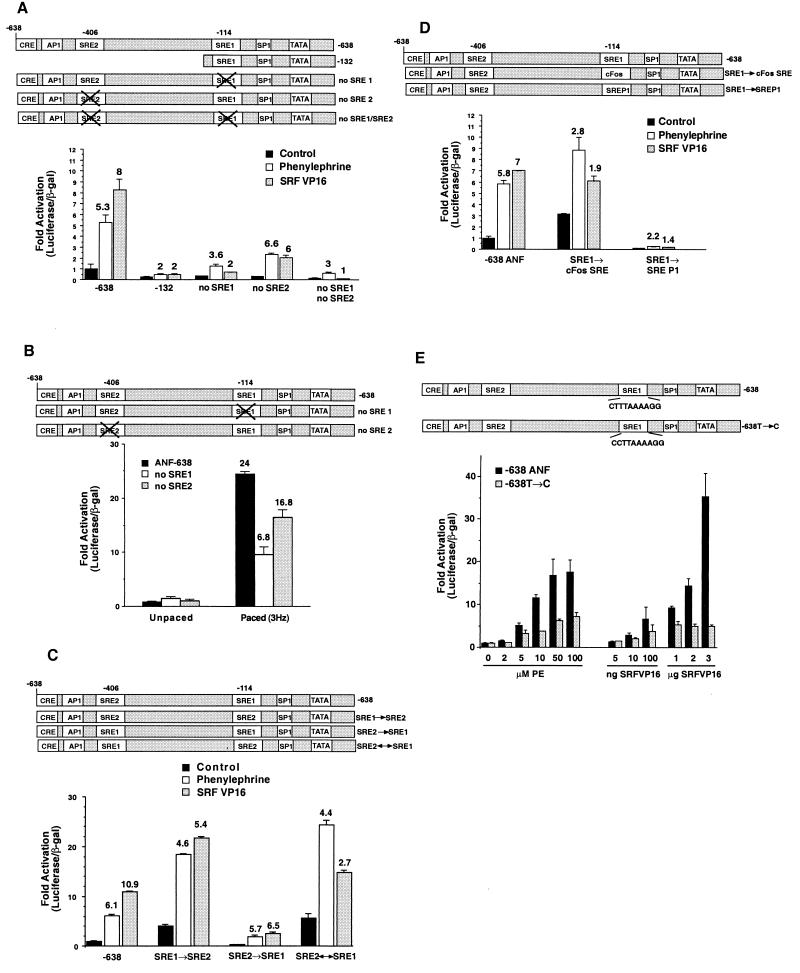
The activity of a gene such as ANF is a product of both the basal activity of the promoter and the level of induction by stimuli such as phenylephrine, electrical pacing, or expression of activated transcription factors such as SRF-VP16. The data shown in Fig. 1 indicate that the SRE1 element plays a role in the regulation of ANF induction by several stimuli despite the fact that these stimuli may utilize different signaling pathways. To more completely examine the role of the SRE elements in both basal and stimulated gene expression, we constructed a series of mutant promoters. Figure 3A shows a series of experiments with the wild-type promoter (−638), a truncated promoter that contained the SRE1 element but not the SRE2 element (−132), the −638 promoter containing mutations in SRE1 (no SRE1), mutations in SRE2 (no SRE2), or mutations in both SREs (no SRE1, no SRE2). We monitored the effects on the basal activity of the various promoters and the level of induction by phenylephrine or SRF-VP16. The wild-type promoter had a reasonable basal activity and was induced 5.3-fold by phenylephrine and about 8-fold by SRF-VP16. This result indicates that SRF-VP16 can bind to one or both of the SRE elements in their native context in the ANF promoter. In contrast, the truncated promoter (−132) that contains only SRE1 but not SRE2 had reduced basal activity and was only modestly induced (about twofold) by either phenylephrine or SRF-VP16. A similar result was obtained with the mutant −638 promoter that contained an intact SRE2 but a mutant SRE1 element. That is, the basal activity was reduced compared with the wild-type, whereas phenylephrine and SRF-VP16 induced expression only 3.6- and 2-fold, respectively. The mutant promoter that contained an intact SRE1 element but a mutant SRE2 element also showed reduced basal activity but was still efficiently induced (about sixfold) by both phenylephrine and SRF-VP16. The double mutations in both SREs produced even lower basal levels of gene expression and again reduced the induction by both stimuli. These data indicate that the basal activity of the ANF promoter is dependent upon both SRE1 and SRE2 elements; however, induction by phenylephrine and SRF-VP16 is primarily dependent upon SRE1 but not SRE2. This result is particularly surprising in the case of SRF-VP16-induced expression. Although SRF binds to SRE2 better than to SRE1 and although SRF-VP16 can activate SRE2- but not SRE1-driven expression in cells (Fig. 2), the ability to induce the ANF promoter by SRF-VP16 is primarily dependent upon the nonconsensus SRE element that cannot efficiently bind to SRF. A similar result was obtained when we examined activation of the mutant promoters by electrical pacing (Fig. 3B), although in this case there was also some role for the SRE2 element in induction.
FIG. 3.
SRE1 and SRE2 regulate basal activity of the ANF promoter but only SRE1 regulates induction. (A) Activation by phenylephrine or SRF-VP16 was determined for the −638 promoter, a truncated promoter (−132), or the −638 promoter with mutations in SRE1, SRE2, or both SRE1 and SRE2. All promoters were normalized to unstimulated wild-type or mutant promoter. The numbers above the bars represent the fold activation by phenylephrine or SRF-VP16 for each individual promoter compared to untreated cells. Mutations in either SRE reduced the basal activity of the promoter; however, only mutation in the SRE1 element significantly reduced induction by phenylephrine or SRF-VP16. (B) Wild-type −638 promoter or the SRE1 or SRE2 mutation were assayed in control cells or in cells that were stimulated by electrical pacing. Contraction-induced gene expression was significantly inhibited by the SRE1 mutation and only moderately inhibited by the SRE2 mutation. (C) Activation by phenylephrine or SRF-VP16 was determined for the wild-type promoter or for mutant promoters where the SRE1 element was mutated so that it was identical to SRE2 (SRE1 → SRE2), the SRE2 element was mutated to be identical to SRE1 (SRE2 → SRE1), or where the SREs were reversed (SRE2 ↔ SRE1). The basal activity was increased for the SRE1 → SRE2 mutant; however, induction by phenylephrine or SRF-VP16 was reduced. The mutant containing two SRE1 elements had slightly reduced induction by SRF-VP16 but virtually no reduction in phenylephrine induction. The SRE2 ↔ SRE1 had an expression level similar to that of SRE1 → SRE2. (D) Activation by phenylephrine or SRF-VP16 of wild-type −638 promoter or mutant promoters, where SRE1 was mutated to the c-Fos SRE sequence or the low-affinity SREP1 sequence (23). Mutation of SRE1 to the c-Fos SRE increased the basal promoter activity but reduced the induction by either phenylephrine or SRF-VP16. Mutation to the poorly binding SREP1 sequence reduced both basal activity and induction. (E) Point mutation of the SRE1 element to more closely match a consensus SRE sequence (638T → C) increased the basal activity of the promoter 6.6-fold but reduced induction by phenylephrine or SRF-VP16 at all doses of the two agonists. Note that the data for this set of transfections was normalized to each unstimulated promoter.
We performed further mutagenesis studies to change the sequence of the two SRE elements. We constructed a promoter that contained either a mutant SRE1 element that was identical to SRE2 (SRE1 → SRE2), a molecule that contains mutations to convert the SRE2 sequence to SRE1 (SRE2 → SRE1), or a construct that reverses the two SRE elements (SRE2 ↔ SRE1). These molecules were again compared to the wild-type molecule that contained one copy of each SRE element in their correct positions. Figure 3C shows that mutation of the SRE1 element to create a promoter with two consensus SRE2 elements resulted in a significant (fourfold) increase in basal activity but reduced the induction of the promoter by either phenylephrine or SRF-VP16. Conversely, mutation of the SRE2 element to create a promoter with two copies of the nonconsensus SRE1 element caused a reduction in basal activity and SRF-VP16 induction but no significant effect on induction by phenylephrine (5.7-fold). Reversal of SRE1 and SRE2 (SRE2 ↔ SRE1) had a phenotype similar to the construct that had two consensus SRE2 elements (SRE1 → SRE2) and was activated by both phenylephrine and SRF-VP16 to a lesser extent than was the wild-type promoter.
We then constructed promoters that contain well-characterized SREs in place of the SRE1 element. The c-Fos SRE binds SRF with high affinity, while SREP binds SRF with very low affinity (23). Again these promoters were compared to the wild-type molecule. Figure 3D shows that the high-affinity cFos SRE in the place of the SRE1 caused an increase in basal activity but a significant reduction in the level of induction by either phenylephrine or SRF-VP16. Mutation of SRE1 so that it was identical to the SREP element caused reduced basal activity and reduced induction by both phenylephrine and SRF-VP16. Finally, we made a point mutation in the promoter to change the sequence of the SRE1 element from CTTTAAAAGG to CCTTAAAAGG, thus making the endogenous SRE1 element match a consensus SRF binding sequence (638T → C). Figure 3E shows that this point mutation also reduced the fold induction by either phenylephrine or SRF-VP16 at all of the concentrations of the two agonists. The mutation also caused a fivefold increase in basal activity compared to the wild-type promoter (data not shown).
Taken together, these data suggest that the inducibility of the promoter is dependent upon the presence of the low-affinity SRE1 element at its normal position in the promoter. Artificially increasing the likelihood of SRF binding at the SRE1 element by mutation to a higher-affinity site increased the basal activity but reduced induction. These data suggest that induction of ANF gene expression is achieved by starting with a DNA sequence that confers low basal activity on the promoter.
Cooperation between SRE1 and SRE2.
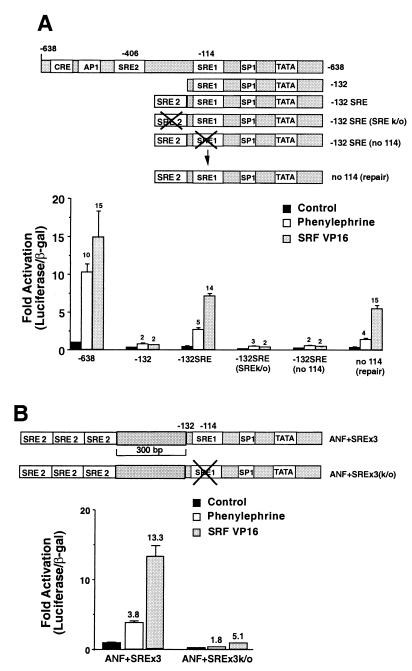
To further analyze the mechanism of gene regulation via the SRE elements, we constructed a series of molecules by using the truncated −132 promoter. These molecules were compared to the wild-type −638 promoter. Figure 4A again shows that the truncated −132 promoter had reduced basal activity and very little induction by either phenylephrine or SRF-VP16. Therefore, although the SRE1 element is an important sequence for induction by these stimuli and the −132 promoter has an intact SRE1, other sequences in the −638 promoter are clearly required for induction. Fusion of a single copy of the SRE2 element to the end of the −132 promoter (−132SRE) slightly rescued the basal activity, led to significant (but lower than for wild-type) activation by phenylephrine, and completely rescued the ability of SRF-VP16 to induce the promoter. Induction of the −132SRE compared to the −132 promoter construct by SRF-VP16 might be interpreted as meaning that SRF-VP16 was working via the high-affinity SRE2 element, i.e., as in the situation with a minimal promoter containing either SRE1 or SRE2 (Fig. 2D). Such a conclusion might be supported by the observation that mutation of the SRE2 element in this plasmid (−132SRE, SRE2k/o) abolished activation by SRF-VP16 (or phenylephrine). However, this interpretation is incorrect, since we found that mutation of the SRE1 element in this plasmid (−132SRE, SRE1k/o) also abolished induction. Thus, a molecule that contains an intact SRE2 element that efficiently binds SRF is not induced by SRF-VP16 unless the low-affinity SRE1 element downstream is also intact. To exclude the possibility that this result was due to inadvertent mutation of other sequences in the plasmid (e.g., in the luciferase gene) during the mutagenesis, we performed further mutagenesis to repair the SRE1 knockout in this plasmid (SRE1 repair). This repaired molecule regained the ability to be stimulated by SRF-VP16. We conclude that the SRE2 element cooperates with the SRE1 element but that the SRF-VP16-dependent induction of gene expression is still highly dependent upon the SRE1 element. The plasmid containing the SRE2 mutation in the context of the −638 promoter is still significantly activated by SRF-VP16 (Fig. 3A). We conclude from this result that other sequences in the −638 promoter may be able to function like the SRE2 element fused to the end of the −132 promoter, perhaps to promote SRF-VP16 binding at SRE1.
FIG. 4.
Cooperation between SRE2 and SRE1. (A) The wild-type −638 promoter or the −132 truncation (−132), the −132 promoter with a functional SRE2 element fused to the end (−132 SRE), −132 mutants containing mutated SRE2 (SREk/o) or SRE1 (no114), or repairs of the mutated sequence (no 114 repair) were induced by phenylephrine or SRF-VP16. All transfections were normalized to the untreated controls for each promoter. The fold activation for each promoter (from its unstimulated control) is given by the numbers above the bars. Note that the −132 promoter is not significantly activated by either phenylephrine or SRF-VP16, while the promoter with a single copy of the SRE2 element fused to the end of −132 regains wild-type activation by SRF-VP16 and partially rescues activation by phenylephrine. Mutation of either SRE element reduces activation by both stimuli, while remutation to repair the SRE1 mutant rescues activation by both stimuli. (B) Three SRE2 elements were fused to the end of a 300-bp DNA fragment from the Kan gene on the end of the −132 promoter. The SRE1 element was mutated in this construct, and stimulation was assessed after treatment with phenylephrine or SRF-VP16. Note that mutation of the SRE1 element reduces basal promoter activity and induction by both phenylephrine and SRF-VP16.
We were concerned that the altered spacing between the elements might have affected our results. To address this question, we constructed a molecule that contained 300 bp of irrelevant DNA sequence from the kanamycin gene between the −132 position and a cloning site and inserted three copies of the high-affinity SRE2 element to maximize the likelihood of SRF binding at this site. This molecule therefore has multiple high-affinity SRF-binding sequences that are correctly spaced relative to the SRE1 element. Figure 4B shows that this molecule is efficiently induced by SRF-VP16 but that induction is compromised by mutation of the SRE1 element. Thus, the presence of high-affinity SRF-binding elements at the end of the truncated −132 promoter is not sufficient to cause efficient induction of gene expression by SRF-VP16 unless the SRE1 element is intact.
An intact SRE1 element is required for transcriptional activation by chimeric transcription factors.
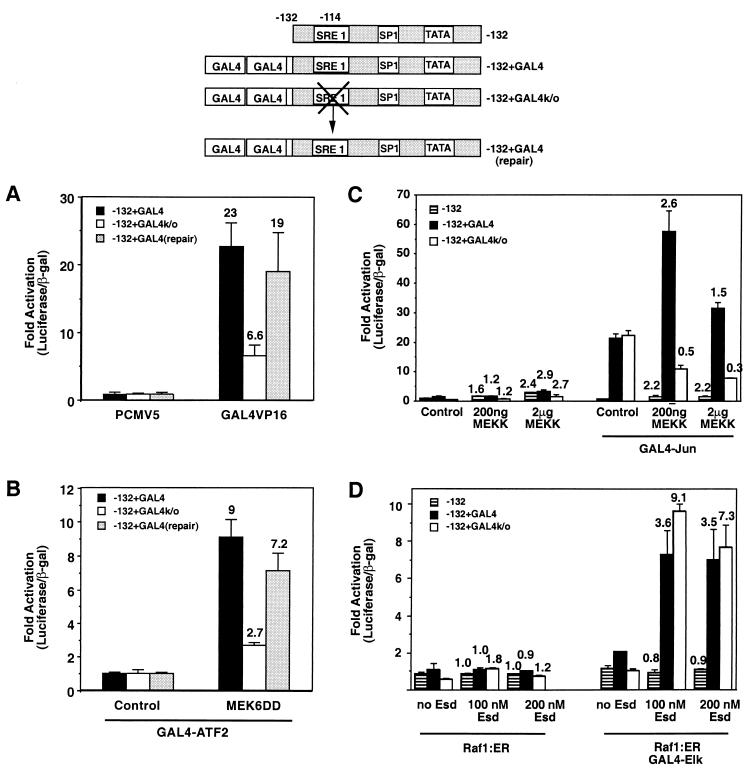
We considered two possible explanations for the results shown in Fig. 4. First, we considered the possibility that perhaps SRF-VP16 was only able to bind to either SRE element in the ANF promoter in a strictly cooperative manner. This model seemed unlikely since it did not account for our results with plasmids that contained multiple SRE2 elements and a mutated SRE1 element (Fig. 4B). This model was also difficult to reconcile with the data presented in Fig. 2D, where a single isolated SRE2 element in a basal promoter was efficiently activated by SRF-VP16. We therefore developed a second hypothesis to explain the data: in the absence of binding at SRE1, transcription induced by factors bound at other sites is abrogated. According to this view, it did not matter whether the activation site was the SRE2 element or a different DNA element that was bound to an active transcription factor, there would be minimal gene expression unless SRE1 was also functional. To test this hypothesis, we constructed a reporter plasmid with two GAL4 binding sites fused to the end of the −132 promoter containing an intact or a mutated SRE1 element. Figure 5A shows that a GAL4-VP16 molecule was able to efficiently transactivate this reporter when the SRE1 element was intact but not when it was mutated. To exclude the possibility of this result being due to another mutation that arose during the mutagenesis, we repaired the SRE1 element in this molecule. This plasmid was activated to wild-type levels by GAL4-VP16. We conclude that in the absence of the intact SRE1 element, stimulation of the ANF promoter is compromised even when stimulation is achieved by a strong artificial transcription factor such as GAL4-VP16. Thus, part of the mechanism of activation of this gene is by regulating interaction at the SRE1 element, and only when this interaction occurs can other active transcription factors induce expression.
FIG. 5.
Activation of transcription by a heterologous transcription factor requires SRE1 activity. (A) Two consensus GAL4-binding sequences were fused to the end of the −132 promoter (−132+GAL4), to a version with the SRE1 element mutated (−132+GAL4k/o), or to a repaired version of this molecule with a reconstructed wild-type SRE1 (repair). Cells were transfected with control vector or a GAL4-VP16 expression plasmid. Note that GAL4-VP16 activates the parental plasmid but that mutation of the SRE1 element in this molecule reduces activation, while a repaired version of this molecule has rescued induction. (B) The GAL4-containing promoters were cotransfected with an expression plasmid encoding GAL4-ATF2, along with a control plasmid or an activated MEK6 expression plasmid, which phosphorylates ATF2. MEK6 induces GAL4-ATF2-dependent gene expression in an SRE1-dependent manner. (C) −132-Luc, GAL4-132-Luc, or the GAL4-132-Luc molecule with mutated SRE1 (−132+GAL4k/o) were transfected with increasing amounts of a MEKK1 expression plasmid with or without a GAL4-Jun expression plasmid. Note that GAL4-Jun increased the expression of the GAL4-containing promoters even in the absence of MEKK1 activity and that this occurs irrespective of the presence or absence of an intact SRE1 element. Expression of active MEKK1 further stimulated GAL4-Jun-dependent expression; however, this stimulation was compromised when the SRE1 element was mutated. (D) Luciferase reporters as in panel C were transfected with a Raf1:ER expression plasmid and with or without a GAL4-Elk expression plasmid. Raf activity was stimulated by adding increasing doses of estradiol to activate GAL4-Elk. In this case, mutation of the SRE1 element had no effect on the level of gene expression induced by the active GAL4-Elk protein.
To create a simple model for how regulated transcription factors at other sites might have their activity controlled by the SRE1 element; we transfected cells with the GAL4-132 promoter (or the SRE1 mutant) along with GAL4-binding transcription factors that were activated by MAP kinase-dependent phosphorylation. Figure 5B shows that MEK6 can induce expression from the GAL4-containing reporter driven by activated GAL4-ATF2. This activation is inhibited by the SRE1 mutation, indicating that the activity of a regulated transcription factor binding to an unrelated DNA sequence is dependent upon the SRE1 element. Figure 5C shows the results obtained when we compared activation of the −132 promoter or the GAL4-132 promoter with an intact or mutated SRE1 element after stimulation of GAL4-Jun. Activation of GAL4-Jun was achieved by stimulating the JNK-SAPK pathway with active MEKK1. Mutation of SRE1 also inhibited the ability of activated GAL4-Jun to induce the promoter. Figure 5D shows the results obtained when we activated GAL4-Elk with an estrogen-induced Raf molecule (ΔRaf1:ER) that leads to ERK activation in transfected cells (45, 50). The data shown in Fig. 5C and D demonstrate two further aspects of this regulation. First, we noted that simply expressing GAL4-Jun with no MEKK1 stimulation was sufficient to increase expression from the Gal4-containing promoters (i.e., GAL4-Jun has some ability to increase transcription even without increased JNK activity). Interestingly, this activity was not affected by the SRE1 mutation, while the further stimulation upon MEKK1 transfection (i.e., JNK-dependent phosphorylation of the transcription factor to increase its activity) was abrogated by the SRE1 mutation. Thus, transcription that is stimulated by JNK-dependent phosphorylation of the transcription factor requires the activity at SRE1, while the transcriptional activity of the unphosphorylated GAL4-Jun molecule does not. Conversely, active and phosphorylated GAL4-Elk does not require the presence of the SRE1 element in order to stimulate transcription. Indeed, the fold induction of the SRE1 mutant promoter was greater than that of the wild-type promoter upon Raf activation. Thus, the regulation of the promoter via SRE1 is dependent upon which particular transcription factor is bound to the promoter. SRE1 can control the activity of VP16-, phosphorylated-ATF2-, and phosphorylated-Jun-dependent gene transcription, but it does not control transcription that is activated by unphosphorylated Jun or by phosphorylated Elk.
DISCUSSION
Like many inducible genes, the ANF promoter is a target for diverse stimuli that are likely to work through different signaling pathways. We focused here on two different physiological stimuli: activation of the α-adrenergic receptor by phenylephrine and electrical pacing to increase the muscle cell contraction rate (to mimic the increased workload that is a major inducer of hypertrophy in response to pathological stimuli such as aortic stenosis). The hypertrophic stimuli that have been tested to date activate various MAP kinase cascades, but the mechanism of gene activation by these signaling pathways is still unclear. Our data indicate that a common mechanism may regulate gene activation by many signaling pathways. While investigating the targets of transcriptional regulation of the ANF promoter by p38 and electrical pacing, we found that a nonconsensus SRE appears to regulate induction by multiple stimuli and active transcription factors.
p38-dependent activation of the ANF promoter.
The experiments here showed that electrical-pacing-induced ANF expression is regulated at least in part through p38-dependent signals. In contrast, phenylephrine, which works through a Gq-coupled receptor, was not significantly inhibited when we treated cells with a p38 inhibitor. These data indicate that different kinds of stimuli can use different signaling mechanisms to stimulate ANF expression. We note that these data contradict results from other investigators who showed that phenylephrine-induced ANF expression could be inhibited by the p38 inhibitor (59). We have excluded trivial explanations for the difference between our results (such as our inhibitor being inactive). Possibilities could include different culture conditions used by various laboratories or the various inhibitor incubation periods, which have been shown to have varied effects on cell morphology (12). We recently found that, in dense cultures, phenylephrine causes an increased rate of contraction of the cells that is associated with p38 activation (24a). Thus, one explanation for the discrepancy is that a contraction-induced component of the ANF activation was being detected in the experiments where phenylephrine-induced expression was affected by the p38 inhibitor. Irrespective of the reason for these differences, an important point arising from our work is that, under the conditions that we used here, phenylephrine-induced ANF expression is not sensitive to a p38 inhibitor, whereas contraction-induced expression is sensitive to this inhibitor.
p38-dependent activation of the gene was partially dependent upon a CRE-like element that is presumably a target for an activated transcription factor, such as a member of the ATF-2 family. We found that pacing-induced expression but not phenylephrine-induced expression was affected by mutation of this sequence. Work from another group has shown that pressure overload-induced ANF expression in transgenic animals is not affected by mutation of the CRE-like element (54). Pressure overload-induced hypertrophy is dependent upon Gq signaling (1); thus, our results are consistent with these data in implying that this DNA element is not required for Gq-dependent signaling to this promoter.
In addition to stimulus-specific activator sequences such as the CRE, two SRE elements (the consensus SRE2 element and the nonconsensus SRE1 element) in the ANF promoter are important for regulation of the gene. As expected, the consensus SRE2 was able to bind to SRF with higher affinity than was the nonconsensus SRE1 element. The difference in binding was most apparent for the core SRE elements and occurred both in vitro and in cells transfected with a constitutively active form of SRF (SRF-VP16). The SRE1 sequence is also similar to a MEF2 binding site (36) and, since MEF2C is known to be a target for p38-dependent signals (20), we tested whether MEF2C could bind to the SRE1 element. We have been unable to show any significant binding of MEF2C to SRE1 in vitro (data not shown).
Multiple agonists and transcription factors require SRE1.
Mutation of the SRE1 reduced induction by physiological stimuli such as phenylephrine and electrical pacing. Surprisingly, mutations in this element (but not the consensus SRE2 element) also had a major effect on induction by completely artificial stimuli such as SRF-VP16, GAL4-VP16, GAL4-ATF2, and GAL4-Jun. Expression of an inducible gene such as ANF is a product of both the amount of induction and the basal level of expression. When we tried to separate these two activities we found that the consensus SRE2 element played a role in the regulation of basal levels of gene expression but had a minor role in induction. Importantly, simply making the nonconsensus SRE1 element more closely match a consensus SRF binding site actually reduced the amount of induction that was achievable by various stimuli. A similar result was obtained when we retained both SRE elements but switched their positions in the promoter. In these cases, the unstimulated basal rate of expression was increased without a proportional increase in the stimulated samples. These data suggest that an essential aspect of the regulation of induction of the ANF promoter is achieved by creating a low basal state, because SRF can bind only inefficiently to the native SRE1 element. Thus, if stimuli promote SRF binding at this site, we will see increased gene expression; in this view, SRF is responsible for the regulation of induction of this promoter, but the mechanism of induction is via regulation of SRF’s ability to interact with its binding site. Such a model is similar to many inducible transcription factors, including SRF itself, which can show altered DNA binding to consensus SREs when it is phosphorylated (31, 39). However, if hypertrophic stimuli simply increased the binding affinity of SRF for the SRE1 sequence, thus bringing an active transcription factor to the promoter, we would expect to see an increase in expression from the reporters that contain this element in isolation. In the case of SRF at the c-Fos SRE element, this is clearly what occurs (although perhaps by several different mechanisms, depending on the stimulus). We do not observe significant activation of the isolated SRE1 element (or the SRE2 element) by hypertrophic stimuli. Therefore, we conclude that the mechanism of gene induction, while dependent upon the SRE1 site, is not simply achieved by increased recruitment of active SRF at this element. Our data also do not support a model where SRF that is already bound to DNA is “activated” to become a more effective transcription factor by hypertrophic stimuli, e.g., as it may be by Rho-dependent signals (22).
SRF controls transforming growth factor β (TGF-β)-dependent expression of the skeletal α-actin gene in cardiac muscle cells (29). In this case, the mechanism involves SRF and YY1 binding to overlapping sites at a consensus SRE. SRF binding promotes gene expression, while YY1 binding causes repression. While there are obvious similarities with our situation, we do not think that this mechanism explains the regulation of the ANF promoter. First, the flanking region of the nonconsensus SRE1 element, unlike the consensus SRE in the skeletal α-actin promoter, does not have the correct sequence for YY1 binding. Second, the observations in the skeletal α-actin promoter are qualitatively different from our results in that the isolated skeletal α-actin SRE element was induced by TGF-β, whereas hypertrophic stimuli do not themselves activate SRE1-dependent gene expression. However, regulation of basal promoter activity by the SRE elements does seem to be similar between ANF and skeletal α-actin. In both cases (see Fig. 3 and reference 29) increasing the likelihood of SRF binding at the SRE increases the basal activity of the gene. In this regard, the nonconsensus sequence of the SRE1 element in the ANF promoter could be the functional equivalent of the overlapping YY1-binding site in the skeletal α-actin promoter, which inhibits SRF binding and is able to keep the basal level of gene expression low.
A number of different transcription factor binding sites have been identified in the ANF promoter (see, for example references 4, 48, and 54) and include the homeodomain protein Csx/NKX2.5 and the zinc finger protein GATA4 (15, 28). These two factors cooperate to regulate tissue specificity of this promoter (14, 28). Such cooperativity also occurs between NKX2.5 and SRF on the cardiac α-actin promoter (10). Such results have led to the suggestion that the ANF promoter might represent an enhanceosome-like structure (16) similar to the enhanceosome that controls beta interferon expression (9). The beta interferon enhanceosome requires proper spacing between cooperative DNA elements for it to function. Therefore, our data showing that the low-affinity SRE1 element must be positioned at the correct site in the promoter to achieve efficient induction by the stimuli that we tested supports an enhanceosome-type model.
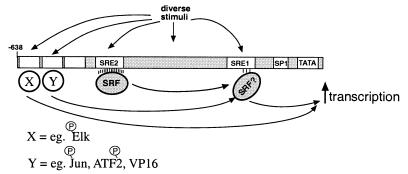
Our data also indicate that part of the mechanism of activation of the ANF promoter is because the SRE1 element is required to allow transcription factors that are present at different sites on the promoter to induce gene expression. This is most clearly demonstrated in the experiments where the ability of active GAL4-binding transcription factors to induce gene expression can be compromised by mutations in the SRE1 element. One mechanism through which the SRE1 element could be required for induction by factors that bind to other sites could be through a cooperative interaction with other transcription factors, i.e., in a manner analogous to the TCF binding at the c-Fos SRE or NKX2.5-SRF interaction at the cardiac α-actin promoter (10). However, such a model seems unlikely to completely explain our data since it is difficult to imagine how SRF could assist in the recognition of the GAL4 DNA binding domain to its binding site. In addition, the idea that the effects that we observe are due to an increase in cooperative DNA binding is difficult to reconcile with the observation that not all activated GAL4-binding transcription factors are affected by the SRE1 mutation. An alternative model is that the SRE1 element acts as an essential mediator of transcriptional regulation to allow communication between some active transcription factors and the basal transcription apparatus (Fig. 6). Such a model implies that the diverse stimuli that induce the ANF promoter will activate different subsets of transcription factors that bind to different sites on the promoter but that the requirement for SRE1 function will be common to many of these transcription factors and thus many stimuli. If SRE1 is part of an enhanceosome, this structure must be required to allow efficient activation by some, but not all, transcription factors at other positions on the promoter. SRF can interact with components of the basal transcription machinery (60); one possibility, therefore, is that SRF at the SRE1 sequence functions as an “activator bridge” to promote productive interactions between activated transcription factors and the basal transcription machinery. In this regard it should be noted that while we know that SRF can bind to this element, we cannot exclude the possibility that the effect on gene expression is achieved through another protein that is also able to recognize the SRE1. Our data indicate that the SRE1 sequence is only required for some transcription factors, implying that the underlying mechanism allows discrimination between different activation domains. Further analysis of the mechanism of regulation through this DNA element should allow us to characterize such an activity.
FIG. 6.
Model for transcriptional induction of the ANF promoter by diverse stimuli. The SRE1 sequence in the ANF promoter could function as an “activator bridge” to promote productive interactions between activated transcription factors at other sites and the basal transcription machinery.
ACKNOWLEDGMENTS
We are grateful to the various colleagues who provided plasmids that were used in this work. We thank Jamie Coombs for help with several of these experiments and Don Ayer and Steve Prescott for comments on the manuscript.
This research was supported by NIH grant HL 52010, the Thomas D. Dee Fellowship in Human Genetics (W.A.H.), and funds from the Huntsman Cancer Institute and was completed in partial fulfillment of the Ph.D. degree in Human Genetics (W.A.H.).
REFERENCES
- 1.Akhter S A, Luttrell L M, Rockman H A, Iaccarino G, Lefkowitz R J, Koch W J. Targeting the receptor-Gq interface to inhibit in vivo pressure overload myocardial hypertrophy. Science. 1998;280:574–577. doi: 10.1126/science.280.5363.574. [DOI] [PubMed] [Google Scholar]
- 2.Alberts A S, Geneste O, Treisman R. Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell. 1998;92:475–487. doi: 10.1016/s0092-8674(00)80941-1. [DOI] [PubMed] [Google Scholar]
- 3.Allo S N, McDermott P J, Carl L L, Morgan H E. Phorbol ester stimulation of protein kinase C activity and ribosomal DNA transcription: role in hypertrophic growth of cultured cardiomyocytes. J Biol Chem. 1991;266:22003–22009. [PubMed] [Google Scholar]
- 4.Ardati A, Nemer M. A nuclear pathway for α1-adrenergic receptor signaling in cardiac cells. EMBO J. 1993;12:5131–5139. doi: 10.1002/j.1460-2075.1993.tb06208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogoyevitch M A, Gillespie-Brown J, Ketterman A J, Fuller S J, Ben-Levy R, Ashworth A, Marshall C J, Sugden P H. Stimulation of the stress-activated mitogen-activated protein kinase subfamilies in perfused heart. Circ Res. 1996;79:162–173. doi: 10.1161/01.res.79.2.162. [DOI] [PubMed] [Google Scholar]
- 6.Bogoyevitch M A, Glennon P E, Andersson M B, Clerk A, Lazou A, Marshall C J, Parker P J, Sugden P H. Endothelin-1 and fibroblast growth factors stimulate the mitogen-activated protein kinase signaling cascade in cardiac myocytes. J Biol Chem. 1994;269:1110–1119. [PubMed] [Google Scholar]
- 7.Bogoyevitch M A, Glennon P E, Sugden P H. Endothelin-1, phorbol esters and phenylephrine stimulate MAP kinase activities in ventricular cardiomyocytes. FEBS Lett. 1993;317:271–275. doi: 10.1016/0014-5793(93)81291-7. [DOI] [PubMed] [Google Scholar]
- 8.Bogoyevitch M A, Marshall C, Sugden P H. Hypertrophic agonists stimulate the activities of the protein kinases c-Raf and A-Raf in cultured ventricular myocytes. J Biol Chem. 1995;270:26303–26310. doi: 10.1074/jbc.270.44.26303. [DOI] [PubMed] [Google Scholar]
- 9.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 10.Chen C Y, Schwartz R J. Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac α-actin gene transcription. Mol Cell Biol. 1996;16:6372–6384. doi: 10.1128/mcb.16.11.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chien K R, Knowlton K U, Zhu H, Chien S. Regulation of cardiac gene expression during myocardial growth and hypertrophy: molecular studies of an adaptive response. FASEB J. 1991;5:3037–3046. doi: 10.1096/fasebj.5.15.1835945. [DOI] [PubMed] [Google Scholar]
- 12.Clerk A, Michael A, Sugden P H. Stimulation of the p38 mitogen-activated protein kinase pathway in neonatal rat ventricular myocytes by the G protein-coupled receptor agonists, endothelin-1 and phenylephrine: a role in cardiac myocyte hypertrophy? J Cell Biol. 1998;142:523–535. doi: 10.1083/jcb.142.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunnmon P, Iwaki K, Henderson S, Sen A, Chien K R. Phorbol esters induce immediate-early genes and stimulate cardiac gene transcription in neonatal rat myocardial cells. J Mol Cell Cardiol. 1990;22:901–910. doi: 10.1016/0022-2828(90)90121-h. [DOI] [PubMed] [Google Scholar]
- 14.Durocher D, Charron F, Warren R, Schwartz R J, Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durocher D, Chen C Y, Ardati A, Schwartz R J, Nemer M. The atrial natriuretic factor promoter is a downstream target for Nkx-2.5 in the myocardium. Mol Cell Biol. 1996;16:4648–4655. doi: 10.1128/mcb.16.9.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durocher D, Nemer M. Combinatorial interactions regulating cardiac transcription. Dev Genet. 1998;22:250–262. doi: 10.1002/(SICI)1520-6408(1998)22:3<250::AID-DVG7>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 17.Gillespie-Brown J, Fuller S J, Bogoyevitch M A, Cowley S, Sugden P H. The mitogen-activated protein kinase kinase MEK1 stimulates a pattern of gene expression typical of the hypertrophic phenotype in rat ventricular cardiomyocytes. J Biol Chem. 1995;270:28092–28096. doi: 10.1074/jbc.270.47.28092. [DOI] [PubMed] [Google Scholar]
- 18.Glennon P E, Kaddoura S, Sale E M, Sale G J, Fuller S J, Sugden P H. Depletion of mitogen-activated protein kinase using an antisense oligodeoxynucleotide approach downregulates the phenylephrine-induced hypertrophic response in cardiac myocytes. Circ Res. 1996;78:954–961. doi: 10.1161/01.res.78.6.954. [DOI] [PubMed] [Google Scholar]
- 19.Gupta S, Campbell D, Derijard B, Davis R J. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 20.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 21.Hill C S, Treisman R. Differential activation of c-fos promoter elements by serum, lysophosphatidic acid, G proteins and polypeptide growth factors. EMBO J. 1995;14:5037–5047. doi: 10.1002/j.1460-2075.1995.tb00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill C S, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 23.Hill C S, Wynne J, Treisman R. Serum-regulated transcription by serum response factor (SRF): a novel role for the DNA binding domain. EMBO J. 1994;13:5421–5432. doi: 10.1002/j.1460-2075.1994.tb06877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hines W A, Thorburn A. Ras and rho are required for Gαq-induced hypertrophic gene expression in neonatal rat cardiac myocytes. J Mol Cell Cardiol. 1998;30:485–494. doi: 10.1006/jmcc.1997.0613. [DOI] [PubMed] [Google Scholar]
- 24a.Hines, W. A., J. Thorburn, and A. Thorburn. Submitted for publication.
- 25.Hipskind R A, Baccarini M, Nordheim A. Transient activation of Raf-1, MEK, and ERK2 coincides kinetically with ternary complex factor phosphorylation and immediate-early gene promoter activity in vivo. Mol Cell Biol. 1994;14:6219–6231. doi: 10.1128/mcb.14.9.6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hipskind R A, Buscher D, Nordheim A, Baccarini M. Ras/MAP kinase-dependent and -independent signaling pathways target distinct ternary complex factors. Genes Dev. 1994;8:1803–1816. doi: 10.1101/gad.8.15.1803. [DOI] [PubMed] [Google Scholar]
- 27.Knowlton K U, Barrachini E, Ross R S, Harris A N, Henderson S A, Evans S M, Glembotski C C, Chien K R. Co-regulation of the atrial natriuretic factor and cardiac myosin light chain-2 genes during α-adrenergic stimulation of neonatal rat ventricular cells. J Biol Chem. 1991;266:7759–7768. [PubMed] [Google Scholar]
- 28.Lee Y, Shioi T, Kasahara H, Jobe S M, Wiese R J, Markham B E, Izumo S. The cardiac tissue-restricted homeobox protein Csx/Nkx2.5 physically associates with the zinc finger protein GATA4 and cooperatively activates atrial natriuretic factor gene expression. Mol Cell Biol. 1998;18:3120–3129. doi: 10.1128/mcb.18.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacLellan W R, Lee T-C, Schwartz R J, Schneider M D. Transforming growth factor-β response elements of the skeletal a actin gene. J Biol Chem. 1994;269:16754–16760. [PubMed] [Google Scholar]
- 30.Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- 31.Marais R M, Hsuan J J, McGuigan C, Wynne J, Treisman R. Casein kinase II phosphorylation increases the rate of serum response factor-binding site exchange. EMBO J. 1992;11:97–105. doi: 10.1002/j.1460-2075.1992.tb05032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonough P M, Glembotski C C. Induction of atrial natriuretic factor and myosin light chain-2 gene expression in cultured ventricular myocytes by electrical stimulation of contraction. J Biol Chem. 1992;267:11665–11668. [PubMed] [Google Scholar]
- 33.Nemoto S, Sheng Z, Lin A. Opposing effects of Jun kinase and p38 mitogen-activated protein kinases on cardiomyocyte hypertrophy. Mol Cell Biol. 1998;18:3518–3526. doi: 10.1128/mcb.18.6.3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker T G, Packer S E, Schneider M D. Peptide growth factors can provoke “fetal” contractile protein gene expression in rat cardiac myocytes. J Clin Invest. 1990;85:507–514. doi: 10.1172/JCI114466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pennica D, Ling K L, Shaw K J, Luis E, Rullamas J, Luoh S-H, Darbonne W C, Knutzon D S, Yen R, Chien K R, Baker J B, Wood W I. Expression cloning of cardiotrophin 1, a cytokine that induces cardiac myocyte hypertrophy. Proc Natl Acad Sci USA. 1995;92:1142–1146. doi: 10.1073/pnas.92.4.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pollock R, Treisman R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 1991;5:2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- 37.Post G R, Goldstein D, Thuerauf D J, Glembotski C C, Brown J H. Dissociation of p44 and p42 mitogen-activated protein kinase activation from receptor-induced hypertrophy in neonatal rat ventricular myocytes. J Biol Chem. 1996;271:8452–8457. doi: 10.1074/jbc.271.14.8452. [DOI] [PubMed] [Google Scholar]
- 38.Ramirez M T, Sah V P, Zhao X-L, Hunter J J, Chien K R, Brown J H. The MEKK-JNK pathway is stimulated by α-adrenergic receptor and Ras activation and is associated with in vitro and in vivo cardiac hypertrophy. J Biol Chem. 1997;272:14057–14061. doi: 10.1074/jbc.272.22.14057. [DOI] [PubMed] [Google Scholar]
- 39.Rivera V M, Miranti C K, Misra R P, Ginty D D, Chen R H, Blenis J, Greenberg M E. A growth factor-induced kinase phosphorylates the serum response factor at a site that regulates its DNA-binding activity. Mol Cell Biol. 1993;13:6260–6273. doi: 10.1128/mcb.13.10.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadoshima J, Izumo S. The heterotrimeric Gq protein-coupled angiotensin II receptor activates p21ras via the tyrosine kinase-Shc-Grb2-Sos pathway in cardiac myocytes. EMBO J. 1996;15:775–787. [PMC free article] [PubMed] [Google Scholar]
- 41.Sadoshima J, Izumo S. Mechanical stretch rapidly activates multiple signal transduction pathways in cardiac myocytes: potential involvement of an autocrine/paracrine mechanism. EMBO J. 1993;12:1681–1692. doi: 10.1002/j.1460-2075.1993.tb05813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadoshima J, Izumo S. Molecular characterization of angiotensin II-induced hypertrophy of cardiac myocytes and hyperplasia of cardiac fibroblasts. Circ Res. 1993;73:413–423. doi: 10.1161/01.res.73.3.413. [DOI] [PubMed] [Google Scholar]
- 43.Sadoshima J, Jahn L, Takahashi T, Kulik T J, Izumo S. Molecular characterization of the stretch-induced adaptation of cultured cardiac cells. J Biol Chem. 1992;267:10551–10560. [PubMed] [Google Scholar]
- 44.Sahai E, Alberts A S, Treisman R. RhoA effector mutants reveal distinct effector pathways for cytoskeletal reorganization, SRF activation and transformation. EMBO J. 1998;17:1350–1361. doi: 10.1093/emboj/17.5.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samuels M L, Weber M J, Bishop J M, McMahon M. Conditional transformation of cells and rapid activation of the mitogen-activated protein kinase cascade by an estradiol-dependent human Raf-1 protein kinase. Mol Cell Biol. 1993;13:6241–6252. doi: 10.1128/mcb.13.10.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sei C A, Irons C E, Sprenkle A B, McDonough P M, Brown J H, Glembotski C C. The α-adrenergic stimulation of atrial natriuretic factor expression in cardiac myocytes requires calcium influx, protein kinase C, and calmodulin-regulated pathways. J Biol Chem. 1991;266:15910–15916. [PubMed] [Google Scholar]
- 47.Sheng Z, Knowlton K, Chen J, Hoshijima M, Brown J H, Chien K R. Cardiotrophin 1 (CT-1) inhibition of cardiac myocyte apoptosis via a mitogen-activated protein kinase-dependent pathway. J Biol Chem. 1997;272:5783–5791. doi: 10.1074/jbc.272.9.5783. [DOI] [PubMed] [Google Scholar]
- 48.Sprenkle A, Murray S F, Glembotski C C. Involvement of multiple cis elements in basal- and α-adrenergic agonist-inducible atrial natriuretic factor transcription. Circ Res. 1995;77:1060–1069. doi: 10.1161/01.res.77.6.1060. [DOI] [PubMed] [Google Scholar]
- 49.Thorburn J, Carlson M, Mansour S J, Chien K R, Ahn N G, Thorburn A. Inhibition of a signaling pathway in cardiac muscle cells by active mitogen-activated protein kinase kinase. Mol Biol Cell. 1995;6:1479–1490. doi: 10.1091/mbc.6.11.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thorburn J, McMahon M, Thorburn A. Raf-1 kinase activity is necessary and sufficient for gene expression changes but not sufficient for cellular morphology changes associated with cardiac myocyte hypertrophy. J Biol Chem. 1994;269:30580–30586. [PubMed] [Google Scholar]
- 51.Thorburn J, Xu S, Thorburn A. MAP kinase- and Rho-dependent signals interact to regulate gene expression but not actin morphology in cardiac muscle cells. EMBO J. 1997;16:1888–1900. doi: 10.1093/emboj/16.8.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thorburn J S, Frost J A, Thorburn A M. Mitogen-activated protein kinases mediate changes in gene expression, but not cytoskeletal organization associated with cardiac muscle cell hypertrophy. J Cell Biol. 1994;126:1565–1572. doi: 10.1083/jcb.126.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Dam H, Wilhelm D, Herr I, Steffen A, Herrlich P, Angel P. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 1995;14:1798–1811. doi: 10.1002/j.1460-2075.1995.tb07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Harsdorf R, Edwards J G, Shen Y-T, Kudej R K, Dietz R, Leinwand L A, Nadal-Ginard B, Vatner S F. Identification of a cis-acting regulatory element conferring inducibility of the atrial natriuretic factor gene in acute pressure overload. J Clin Invest. 1997;100:1294–1304. doi: 10.1172/JCI119643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y, Huang S, Sah V P, Ross J, Brown J H, Han J, Chien K. Cardiac muscle cell hypertrophy and apoptosis induced by distinct members of the p38 mitogen-activated protein kinase family. J Biol Chem. 1998;273:2161–2168. doi: 10.1074/jbc.273.4.2161. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, Su B, Sah V P, Brown J H, Han J, Chien K R. Cardiac hypertrophy induced by mitogen-activated protein kinase kinase 7, a specific activator for c-Jun NH2-terminal kinase in ventricular muscle cells. J Biol Chem. 1998;273:5423–5426. doi: 10.1074/jbc.273.10.5423. [DOI] [PubMed] [Google Scholar]
- 57.Whitmarsh A J, Shore P, Sharrocks A D, Davis R J. Integration of MAP kinase signal transduction pathways at the serum response element. Science. 1995;269:403–407. doi: 10.1126/science.7618106. [DOI] [PubMed] [Google Scholar]
- 58.Yamazaki T, Tobe K, Hoh E, Meamura K, Kaida T, Komuro I, Tamemoto H, Kadowski T, Nagai R, Yazaki Y. Mechanical loading activates mitogen-activated protein kinase and S6 peptide kinase in cultured rat cardiac myocytes. J Biol Chem. 1993;268:12069–12076. [PubMed] [Google Scholar]
- 59.Zechner D, Thuerauf D J, Hanford D S, McDonough P M, Glembotski C C. A role for the p38 mitogen-activated protein kinase pathway in myocardial cell growth, sarcomeric organization and cardiac-specific gene expression. J Cell Biol. 1997;139:115–127. doi: 10.1083/jcb.139.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu H, Joliot V, Prywes R. Role of transcription factor TFIIF in serum response factor-activated transcription. J Biol Chem. 1994;269:3489–3497. [PubMed] [Google Scholar]