Abstract
Streptomycetes are sessile bacteria that produce metabolites that impact the behavior of microbial communities. Emerging studies have demonstrated that Streptomyces spores are distributed through various mechanisms, but it remains unclear how spores are transported to their preferred microenvironments, such as plant roots. Here, we show that Streptomyces spores are capable of utilizing the motility machinery of other soil bacteria. Motility assays and microscopy studies reveal that Streptomyces spores are transported to plant tissues by interacting directly with the flagella of both gram-positive and gram-negative bacteria. Genetics experiments demonstrate that this form of motility is facilitated by structural proteins on the spore coat. These results demonstrate that nonmotile bacteria are capable of utilizing the motility machinery of other microbes to complete necessary stages of their lifecycle.
Subject terms: Bacteria, Cellular microbiology
Introduction
Bacteria belonging to the genus Streptomyces are an integral component of diverse ecosystems and are well-known to produce chemically diverse metabolites, including the vast majority of all clinically relevant antibiotics [1, 2]. Soil-dwelling Streptomycetes, such as Streptomyces coelicolor (Sc), colonize plant roots and provide the associated plant protection from potential phytopathogens through antibiotic secretion [1]. The symbiosis of Streptomycetes with their plant hosts has been shown to improve plant health and productivity, and thereby provides a potential sustainable solution to increase crop yields [2–5]. The lifecycle of Streptomycetes is complex and involves stages of aerial hyphae formation on the soil surface to produce spores, and spore germination on plant roots to produce filamentous colonies [1]. Immotile Streptomyces bacteria distribute their spores over long distances through attachment to insects and nematodes, but it is unclear how they relocate over short distances to their preferred microenvironments such as plant root systems [6, 7].
While Streptomyces are nonmotile, many other soil microbes, such as Bacillus subtilis (Bs) and Pseudomonas fluorescens (Pf), are motile and can move through their environment by regulating the rotation of flagella [8]. On solid surfaces, flagellar rotation enables cell swarming. During swarming, cells are densely packed and continuously move outward toward unoccupied areas. In liquid media, flagella enable cell swimming where the cells move independently and can rapidly change swimming direction. In addition to flagellar motility, some bacteria can also move through a passive diffusion on surfaces called sliding [8]. Sliding does not involve flagella but occurs when cells are pushed through the forces generated by the outward growing colony.
Recent reports have revealed that microbe transport by inter-species interactions can occur between motile and immotile microbes [9]. These studies demonstrate that inter-microbial transport occurs among organisms natively found on abiotic surfaces [10, 11], plant surfaces [12], and within the soil [13, 14]. In some instances, the transportation of human pathogens are facilitated on abiotic surfaces, including nonmotile Staphylococcal species that directly adhere to their mobile partners [10], Aspergillus fumigatus (Af) spores that interact with the flagella of motile bacteria [14], various nonmotile human microbiome bacteria that are carried by Capnocytophaga gingivalis [15], and Legionella pneumophila that is transported internally by their amoebae hosts [16].
Here, we demonstrate that spores of the sessile Streptomycetes, such as Sc, are transported by Bs to their preferred microenvironment. Sc and Bs are both soil-dwelling bacteria that utilize plant root exudate as a nutritive source [1, 17]. Using microscopy methods, motility assays, and genetics approaches, we demonstrate that Bs transports Sc spores via direct attachment to Bs flagella, a mode of transportation we call “hitchhiking”. Hitchhiking is dependent on the conserved rodlin proteins, which form a fibrous outer layer on the spore coat of almost all Streptomycetes, but with a hitherto unclear function [18, 19]. These results exemplify that nonmotile bacteria are capable of utilizing the motility machinery of other microbes to occupy advantageous environments, and that this mode of transport may be widespread in nature.
Results
Bacillus subtilis disperses Streptomyces coelicolor spores
Transportation of Sc spores by Bs was demonstrated by mixing isolated Sc spores with a liquid culture of Bs followed by inoculation onto an agar swarm plate and incubation. After 5 days, Sc colonies are visible on the plate and are only dispersed in the presence of Bs in all samples tested (n = 13) (Fig. 1A). Spore dispersal by Bs occurs across the entire surface of the plate and to the plate’s edge (4.5 cm from the inoculation point). To demonstrate that the Sc spores are being moved by Bs cells and not merely “floating” in the expanding Bs colony, we conducted identical assays but inoculated the Sc spores and Bs culture separately and onto different areas of the plate. The resulting Sc colonies form streaks across the plate that emanate from the Bs inoculation site in a predictable manner in all samples tested (n = 10) (Fig. 1B). This experiment was repeated on 12 cm plates and demonstrate that Sc spores are dispersed to the edge of the plate, which is 10 cm from the spore innoculation point (n = 3) (Fig. S1A). To determine the effeciency of transport, this experiment was conducted with dilutions of Sc spores so that the total number of Sc colonies would germinate without overlap and could be individually counted. The results demonstrate that 86 ± 5.6% of the apparent colonies are located outside of the initial innoculation point under these conditions (n = 4) (Fig. S1B).
Fig. 1. S. coelicolor spores are transported by B. subtilis.
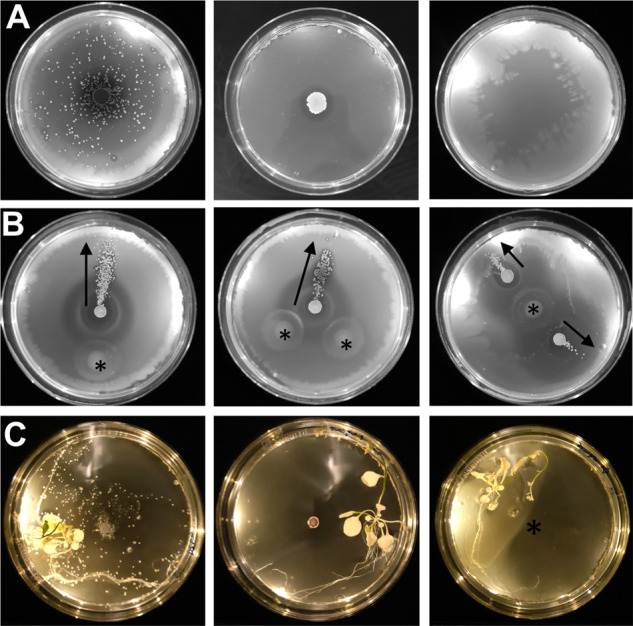
A When Sc and Bs are innoculated on the center of a swarm plate, visible Sc colonies (white dots) are apparent and are only dispersed in the presence of motile Bs. Left: Sc with Bs. Middle: Sc alone. Right: Bs alone. B When Sc and Bs are innoculated in different positions on swarm plates, the Sc colonies are dispersed in the swarming direction of the Bs cells (black arrows). Asterisks denote the Bs innoculation sites. C Bs moves spores toward plant tissues. Left: Sc with Bs. Middle: Sc alone. Right: Bs alone, asterisk denotes the Bs innoculation site.
B. subtilis transports S. coelicolor spores to plant tissues
In nature, Sc and Bs thrive near plant roots that excrete exudates but only Bs can move toward the root systems. We conducted assays to determine if Bs can transport Sc spores to plant tissues. Assays with the Bs strain alone demonstrate that the plates become “cloudy” with Bs cells in areas around plant tissues, perhaps due to the presence of nutritive plant exudates that facilitates bacterial growth (Fig. 1C). Like previous experiments, the Sc spores alone do not exhibit movement unless they are co-inoculated with Bs cells, and the dispersed spores preferentially establish colonies near plant tissues in all instances (n = 5) (Fig. 1C).
Spore dispersal occurs with swarming and sliding Bs strains
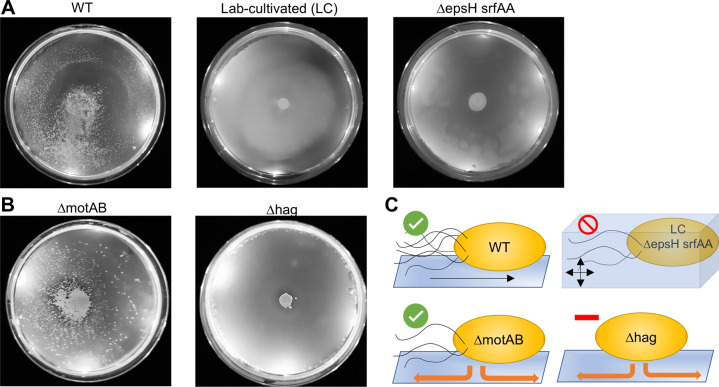
Bs has two modes of flagellar-mediated motility, swimming and swarming, that occur in liquid environments and on solid surfaces, respectively. When Bs senses that it is on a solid surface, it will differentiate into a swarmer cell that has a significant increase in the number of flagella and produces hydrophobic surfactants, such as surfactin, to efficiently spread across the surface [20–22]. In our experiments, we utilized an undomesticated strain of Bs (NCIB3610) that can swarm, unlike common laboratory strains that lack the ability to differentiate into swarmer cells and fail to produce surfactin to undergo swarming motility (Table S1) [20, 22–24]. To determine if the Sc spores are transported by both swarming and swimming motilities, we repeated the experiments with a laboratory-cultivated Bs strain that is incapable of swarming, and spore transport does not occur on the agar swimming plates (0.27% agar) (Figs. 2A and S2) [20]. Importantly, the laboratory-cultivated strain has accumulated many genetic defects in addition to defective swarming capabilities. To ensure that the decrease in spore dispersal can be attributed to limitations of bacterial swimming, we utilized a Bs strain with an undomesticated genetic background but has been genetically altered so that it is only capable of swimming motilities (Δepsh srfAA, DK1484). Like the laboratory-cultivated strain, this strain does not disperse Sc spores on swim plates (Figs. 2A and S2). Therefore, we conclude that spore transport can be accomplished by swarming motility but not swimming motility.
Fig. 2. B. subtilis can transport spores via swarming and sliding.
A The WT (swarming strain) can transport spores on agar plates (0.27–0.5%) (n = 13), but swimming only strains (laboratory-cultivated WT strain or ΔepsH srfAA) cannot (0.25–0.3% agar) (n = 6 and n = 8, respectively). Quantification of the results is shown in Fig. S2. B A Bs ΔmotAB strain that possesses flagella but lacks flagellar motility can disperse spores via sliding at WT levels (n = 6). A Bs Δhag strain that does not possess flagella cannot disperse the spores via sliding at WT levels (n = 6). Quantification of results is shown in Fig. S2. All Δhag plates are shown in Fig. S3. C In summation, spores are dispersed by swarming (WT) and sliding (ΔmotAB) Bs cells in a flagella-dependent manner. However, spores are not dispersed via swimming (lab-cultivated or ΔepsH srfAA), and are significantly less dispersed via sliding in the absence of flagella (Δhag).
In addition to flagellar motilities, Bs can also move through sliding on surfaces. To determine if spores can be dispersed through sliding, we utilized a Bs mutant that has flagella but the flagella cannot rotate (ΔmotAB, DS222) [25], and a mutant that does not possess flagella (Δhag, DS1677) [26]. The ΔmotAB strain moves spores over the entire surface and to the edge of the agar plates in all samples (4.5 cm from the inoculation point) (n = 6) (Figs. 2B and S2). The number of dispersed spores cannot be ascertained due to overlapping colonies that result in apparent smears across the plates, but each plate contains over 100 dispersed Sc colonies. The Δhag strain shows severely reduced dispersal, where an average of 3.2 spores are transported with an average maximum distance of 0.95 ± 0.85 cm from the inoculation point (n = 6) (Figs. 2B, S2 and S3). Therefore, we surmise that spore dispersal can also occur through sliding but is facilitated by the presence of flagella (Fig. 2C). As an additional control, we also conducted these experiments by first spreading the Bs cells across the surface of the plate and then inoculating the Sc spores in the center. Spore dispersal was significantly reduced for both the Bs WT (2.06 ± 0.05 cm) and ΔmotAB (1.25 ± 0.16 cm) strains when compared to co-inoculation in the center of the plate (Figs. S2 and S4). These data indicate that the Sc spores are transported by Bs cells that continuously move away from the inoculation point and are not transported after the motile Bs has covered the surface of the plate.
S. coelicolor spores attach to B. subtilis flagella
We utilized several microscopy methods to elucidate a mechanism for Sc spore dispersal by Bs. Fluorescently labeled Sc spores were imaged under a fluorescence microscope and were immotile as expected (Supplementary Movie 1). However, when Bs cells were added to the fluorescent spores, the spores localize near the Bs cell poles (Supplementary Movie 2 and Fig. 3A). In some instances, the spores are stationary on the surface of the glass slide and an associated Bs cell is seen rotating around the spore (Movie 1). The observed Bs cell rotation in these assays is reminiscent of rotations seen in Bs cells that have their flagella chemically tethered to a solid surface, whereby the torque generated by the immobilized flagella induces rotation of the cell body [27]. This observation suggests that the Sc spores adhere directly to the Bs flagella, and therefore effectively mimic a flagellar tether in these instances. To verify that the Sc spores do not directly interact with the Bs cell body, we imaged a mixture of Sc spores and Bs cells with a cryo-electron microscope. Like the fluorescence microscopy images, the Sc spores are localized near the Bs cell poles but do not make direct contact with the Bs cell body (Figs. 3B and S5A). In total, ~77% of the spores are located within 1 μm of a Bs cell poll (n = 35 spores). To determine if the spores interact with Bs flagella, we utilized a Bs “mini-cell” strain (minD::TnYLB) that lacks the excreted material inhibiting their direct visualization. Indeed, when mixed with Sc spores, the flagella can be seen co-localizing with the spores in two-dimensional cryo-EM images (Figs. 3C and S5B).
Fig. 3. Microscopy methods indicate that Sc spores directly adhere to Bs flagella.
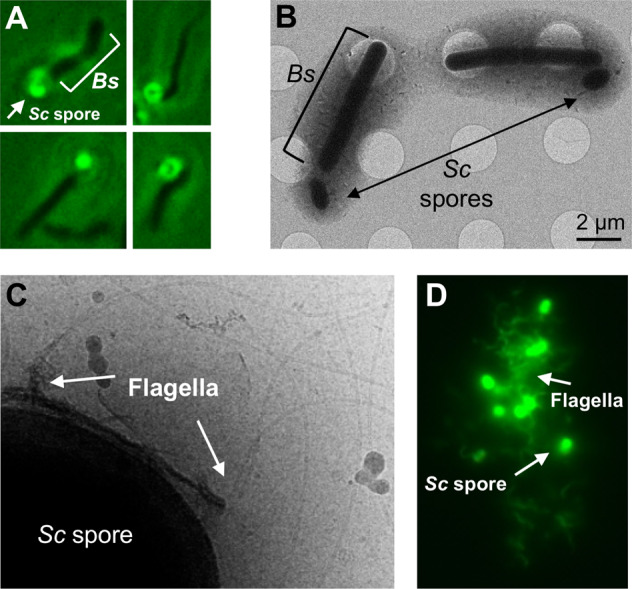
A Fluorescence microscopy of dye-labeled spores with unlabeled Bs cells demonstrate that the spores localize to the cell poles of Bs. B Cryo-electron microscopy samples of mixed Sc spores and Bs cells reveal that the spores do not directly adhere to the Bs cell body. C Cryo-EM shows the Bs flagella colocalize with the Sc spore coat. D Fluorescence microscopy of dye-labeled sheared flagella and spores demonstrate that spores directly interact with Bs flagella to form extended associations of both components.
Sc spore adherence to Bs flagella was further confirmed by visualization of a mixture of Sc spores with sheared flagella via fluorescence microscopy. Fluorescent labeling of Bs flagella was accomplished using a Bs strain (DS1919) that is mutated in a single flagellin residue (T209C) [28]. The surface-exposed thiol allows for direct labeling with dyes that possess a reactive maleimide group. This dye also labels proteins on the surface of the Sc spore coat. Indeed, when sheared Bs flagella are isolated and mixed with spores, and unassociated flagella are washed from the mixture, a majority of spores still retain associated flagella. In total, ~64% of the spores are associated with flagella (n = 130 spores). In some instances, large “clumps” of spores entangled in flagella are observed (Figs. 3D and S5C). In comparison, the flagella without spores added do not form aggregates and are randomly dispersed (Fig. S5C).
Hitchhiking is conserved in Streptomycetes
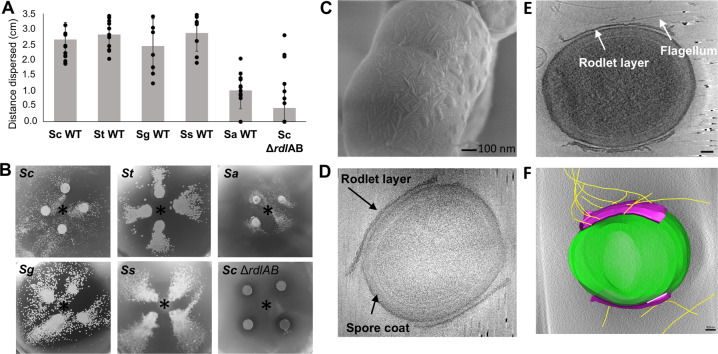
To determine if spore dispersal by Bs also occurs in other Streptomyces species, we conducted Bs swarm plate assays with Streptomyces tendae (St), Streptomyces griseus (Sg), Streptomyces scabies (Ss), and Streptomyces avermitilis (Sa). To quantify spore dispersal, we prepared swarm plate assays where Bs cells were inoculated at the center of the plate (9 cm in diameter) and the isolated spores are inoculated in four equidistant positions around the Bs inoculation site. The maximum dispersal distance, which is the distance from the center of the spore inoculation site to the most distant dispersed colony, was measured for each of the four spore samples. The wild-type (WT) Sc spores are dispersed by Bs in 100% of samples and are moved an average maximum distance of 2.67 ± 0.43 cm from the initial inoculation point (n = 20). Likewise, these assays demonstrate that St (n = 12), Sg (n = 8), and Ss (n = 8) spores are dispersed in 100% of samples to similar distances as WT Sc spores. However, the Sa spores are dispersed at significantly shorter distances (n = 12) (Fig. 4A, B) and are dispersed in 83% of the samples. As the last common ancestor of Sc and Sg existed more than 200 million years ago and both species are capable of hitchhiking, these data suggest that the ancestor also possessed this dispersal mechanism and it remained conserved.
Fig. 4. Hitchhiking of Streptomyces spores is facilitated by the presence of rodlin proteins.
ABs swarm assays with both wild-type (WT) Streptomyces spores (S. coelicolor n = 20, S. tendae n = 12, S. griseus n = 8, S. scabies n = 8, S. avermitilis n = 12) and Sc spores lacking the rodlin proteins (Sc ΔrdlAB n = 24) demonstrate that spores are dispersed in all tested WT species but with S. avermitilis dispersed the shortest distance, and dispersal is abrogated by the loss of the rodlins in Sc. Results are expressed as the mean of dispersal distance ± standard error of the mean. Differences for Sc ΔrdlAB and Sa dispersal compared to WT Sc, St, Sg, and Ss strains are statistically significant using a two-tailed null hypothesis significance test (p < 0.05). B Representative images of Bs/spore swarm plates from A. The Bs innoculation site is denoted with an asterisk. C SEM of WT Sc spores shows the rodlet layer with pairwise rodlets of 20 nm spacing. D A representative cryo-ET image of isolated Sc WT spores shows that the rodlet layer does not cover the entire spore but leaves the poles exposed (n = 22). E Cryo-ET reconstructions show that flagella preferentially interact with the rodlet layer (n = 12). Scale bar 100 nm. F Segmentation of the reconstruction from E clearly demonstrates the flagella:rodlin interaction. Spore body: green, rodlet layer: purple, flagella: yellow. Scale bar 100 nm (Color figure online).
Spore dispersal by B. subtilis is facilitated by the rodlin proteins
The outer surface of most Streptomyces spores is characterized by a fibrillar rodlet layer, which is a striated pattern of pairwise aligned rodlets composed of the rodlin proteins [18, 19]. Scanning electron microscopy (SEM) images of Sc and Ss spores show the striated rodlet layer (Fig. 4C). In previous studies, the rodlets of Sc, S. lividans, St, Sg, and Ss were visually indistinguishable [19, 29]. Using electron microscopy images from this and previous studies, we measured the spacing of the rodlets in these species, which is highly conserved and around ~20 nm (when measured from the center of the rodlet fibers) (Table S2). Furthermore, the rodlin proteins from Sc, St, and Sg have ~34% sequence identity despite the species’ distant evolutionary relation (Fig. S6) [19].
Intriguingly, Sa spores are less widely dispersed than the other Streptomyces species and it is the only tested species that natively lacks rodlin proteins [19]. In agreement, an Sc mutant strain that lacks the rodlin proteins (ΔrdlAB) abrogates hitchhiking by Bs (n = 3, Figs. S2 and S7). Importantly, previous studies demonstrate that the Sc ΔrdlAB strain is not delayed in germination, and does not exhibit any behavioral or phenotypic change compared to the WT strain with the exception of the rodlet layer [18, 19, 30]. In contrast, Sc mutants that lack proteins which produce polysaccharides found on the surface of Streptomycetes (ΔcslA and ΔmatAB) are unaffected (n = 3, Figs. S2 and S7) [31]. However, spore dispersal was not completely abolished in the ΔrdlAB strain. Using identical swarm plate assays described in the section above, the ΔrdlAB strain is dispersed with an average maximum distance of 0.46 ± 0.82 cm from the initial inoculation point and dispersal occurs in 33% of the samples (n = 24) (Fig. 2).
To characterize how rodlins interact with flagella in three dimensions, we conducted cryo-ET experiments of samples containing Bs minicells and Sc spores. Reconstructions show that the Sc spores are oval shaped and possess a thick coat. The rodlet layer can be seen as a sheath around the lateral sides of the spore with frayed edges, leaving the poles exposed, and suggest that the rodlet sheath easily peels away from the cell body (n = 22) (Fig. 4D). A similar spore morphology has also been observed in Streptomyces albus [32]. Bs flagella accumulate around and directly interact with the rodlet layer (n = 12) (Fig. 4E, F and Movie 2). However, due to the thickness of the spores the resolution is limited and we could not deduce if the flagella preferentially bind specific features of the rodlet layer. Collectively, these data suggest that the rodlet layer facilitates spore dispersal by interacting directly with flagella.
Hitchhiking of S. coelicolor spores is not limited to Bacillus
Although B. subtilis is ubiquitous in soil, other genera are also flagellated and may also contribute to dispersal of Sc spores. We therefore conducted swarm plate assays with a P. fluorescens WT strain (R1SS101), which is also natively associated with plant roots [33]. Importantly, Pf disperses spore similarly to Bs, but Sc colonies appear in patterns that are reminiscent of Pseudomonas swarm patterns [34] (n = 6) (Fig. S8). These data demonstrate that hitchhiking is a widespread mechanism that allows Streptomyces spores to disperse at cm-scales (Fig. 5).
Fig. 5. An overview of the hitchhiking model.

Aerial Streptomyces spores are transported on the cm-scale to plant root systems by directly adhering to the flagella of motile bacteria (inset). Here, the spores germinate and produce antibiotics (red gradient) to ward off microbial competitors (Color figure online).
Discussion
Sessile Streptomycetes have a complex lifecycle that involves formation of aerial hyphae that differentiate into spores. The spores of some Streptomyces species, including Sc, are dispersed over long distances by direct attachment to insects and nematodes [6]. Intriguingly, recent reports identify that specific volatile metabolites secreted by Streptomycetes attract arthropods as a mechanism for spore dispersal [7], and they can induce the formation of Streptomyces “explorer cells”, while simultaneously starving microbial competitors [35]. However, it is unclear how the spores are dispersed specifically at the centimeter scale to plant root microenvironments. Here, we demonstrate that Streptomyces spores are able to utilize the motility machinery of motile soil microbes by directly attaching to their flagella. While these experiments demonstrate that Sc spores are dispersed by Bs and Pf regardless of their destination, these motile bacteria are also known to associate with plant roots. Therefore, this mechanism of dispersal, called hitchhiking, may provide Streptomyces spores a mechanism for translocation to beneficial environments. Indeed, assays with A. thaliana plants demonstrate that Bs can transport Sc spores to plant tissues. This allows spores to germinate near nutrient-rich plant exudate to generate filamentous colonies that produce antibiotics, thereby protecting the plant from potential phytopathogens.
Hitchhiking is facilitated by two spore coat proteins, RdlA and RdlB, which are conserved in most Streptomycetes. These proteins assemble into pairwise aligned filaments, called rodlets, on the outer surface of the spores and are spaced ~20 nm apart (Table S2). Until now, the function of the rodlets has remained elusive [30]. Interestingly, the diameter of the bacterial flagellar filament is also ~20 nm [22, 36]. Therefore, it is possible that the rodlet layer provides a gripped surface for the flagella, which become “wrapped” in the grooves made by the rodlin proteins and thereby facilitates spore transport. However, our cryo-ET data can not support such speculations due to the limited resolution of the whole-cell reconstructions. Therefore, it is still unclear what properties of the rodlet layer encourage interactions with flagella.
Emerging studies have demonstrated that flagella preferentially interact with hydrophobic surfaces and flagellin can undergo methylation to increase flagella hydrophobicity [37–39]. This increase in hydrophobicity allows pathogenic bacteria to adhere to host cells [37–39], and flagellar adherence to plant cells has also been implicated in establishing colonization [40, 41]. Flagella hydrophobicity may facilitate interactions with hydrophobic spores and may account for spore transport that is seen in the absence of rodlins, given that the spore surface without rodlins remains hydrophobic (in Sa WT and Sc ΔrdlAB strains) [18, 42, 43]. Therefore, the same flagellar interactions that facilitate adherence to plant roots may also contribute to adherence to spores, and necessitate that motile bacteria not evolve to eliminate this interaction. Since spore:flagella interactions are in-part facilitated by hydrophobic interactions, this mode of transport may be influenced by environmental factors that impact Van der Waals screening distances such as salt concentrations and pH.
Although spore hitchhiking seems disadvantageous to the motile partner, previous research has shown that fungal hyphae can form so-called “fungal highways” that serve as bridges for motile bacteria over air gaps (such as air gaps found in soil) [14, 44, 45]. Like fungi, Steptomycetes form aerial hyphae that are structurally similar to fungal hyphae [46]. Therefore, Streptomycetes may be able to form “bacterial bridges” for their motile partner, but such interactions have not been reported yet. If such “bacterial bridges” do form, this would supply the system with a synergistic transport that has been previously observed between nonmotile fungal spores and motile bacteria [14].
The hitchhiking model is supported by a previous study that examines the interaction of two other root-colonizing microbes: the immotile fungus Af and the motile bacterium Paenibacillus vortex (Pv) [14]. Af spores are demonstrated to be dispersed by Pv in a swarming-dependent manner via direct attachment to flagella; dispersal is abrogated by the addition of excess purified Pv flagella or perturbations to the Af spore coat [14]. Furthermore, scanning EM micrographs show direct contact between Pv flagella and Af spores [14]. Although this study does not identify the spore coat component(s) responsible for adherence to flagella, Aspergillus spores also possess a rodlet layer [47]. Additionally, this study demonstrates that some Penicillium species are also transported by Pv, and these fungi possess a rodlet layer [48]. Collectively, these data may suggest that hitchhiking of spores onto motile bacteria via the formation of a striated rodlet layer is a dispersal mechanism that convergently evolved in both domains of life.
The colonization of plant roots by some Streptomycetes improves plant health and performance in a natural and sustainable manner [2–5]. Therefore, our data are applicable to industrial initiatives that aim to improve soil conditions for Streptomyces root colonization. Likewise, many Aspergillus fungi, like Af and A. niger, are human and plant pathogens. Therefore, insights into hitchhiking of these sessile organisms may elucidate unknown infection mechanisms.
Methods and materials
Streptomyces spore isolation
The following Streptomyces strains were used in this study: S. coelicolor M145 [49], S. coelicolor ΔrdlAB6 [42], S. tendae Tü901/8c [50], S. griseus (ATCC 13273), S. avermitilis (ATCC 31267), and S. scabies ISP5078. Spores were harvested from MS agar plates and quantified as described before [49].
Bacillus subtilis cultivation
The undomesticated Bs strain NCIB3610 [23] 25% glycerol stock was placed into 5 ml of LB and grown overnight at 30 °C. After 16 h of growth, 100 μl of the overnight culture was diluted into 5 ml of LB and grown at 37 °C to an O.D. of 0.4–0.5.
Pseudomonas fluorescens cultivation
The Pf strain R1SS101 25% glycerol stock was placed into 5 ml of 50% TB and grown overnight at 30 °C. After 16 h of growth, 100 μl of the overnight culture was diluted into 5 ml of 50% TB and grown at 37 °C to an O.D. of 0.4–0.5.
Swarm and swim plate assays
Swarm and swim plates were conducted on nutrient broth plates (0.5% peptone, 0.3% yeast extract, 0.5% NaCl) containing specific amounts of agar (0.27–0.5%). All components were mixed, autoclaved for 20 min, and 30 ml of the media was poured into a plastic petri dish with a 9 cm diameter. The plates were cooled for 30 min in a sterile fume hood and then stored in a 4 °C fridge for a maximum of 1 week. To determine if Sc spores are dispersed by Bs, 3 μl of Bs cells are inoculated onto the plate and 3 μl of Sc spore stocks are either added on the Bs inoculation site or to a separate inoculation site. The plates are incubated at 30 °C for 5 days and imaged on a light box.
For plates that first had Bs cells spread across the surface, 100 μl of Bs cells at an O.D. 0.4–0.5 were pipetted onto the plate (0.5% agar) and then spread over the surface of the plate using an L-spreader. Then 3 μl of Sc spore stock was added to the center of the plate. Plates were incubated at 30 °C for 5 days and imaged on a light box.
For all plate images, the distance of spore dispersal was determined using Image J bundled with Java 1.8.0_172 software. To measure the distances, the diameter of the petri dish (9 cm) was used as a reference scale. Then, a line was drawn from the inoculation point of the spores to the Streptomyces colony that is furthest from the inoculation point. The distance of the line was calculated based on the value of the reference scale.
Fluorescence microscopy
For fluorescent labeling of spores with unlabeled B. subtilis, 10 μl of Sc spore stock was added to 1 ml of iced LB. The 1 μl of the fluorescent styryl dye, FM2–10 (Thermo Fisher Scientific), was added to the 1 ml solution and inverted to mix. Excess dye was removed by rinsing the spores 4X with 1 ml of iced LB via centrifugation and decanting. After the final decantation, 1 ml of Bs cells with an O.D. of 0.4 were added to the spores, mixed via pipetting, and incubated at ambient temperatures for 5 min. Immediately before imaging, 5 μl of the samples were placed on a glass slide and a glass coverslip was placed on top. The sample was imaged on a Zeiss Axioscope A1 fluorescent microscope scope equipped with an Axiocam Mrc5 camera (Zeiss) in the Institute of Biology Microscopy Unit using a GFP filter. Images were collected and processed using Axiovision software (Zeiss).
For fluorescent labeling of sheared B. subtilis flagella with spores, B. subtilis (strain Δhag amyE::Phag-hagT209C spec) was grown to an O.D. of 0.6, and 2 ml of the cells were pelleted via centrifugation and resuspended in 1 ml of PBS buffer pH 7.5. The 1 ml suspension was passed back-and-forth between two 5 ml syringes with 21 gauge needles that were connected by a plastic tube (10 cm long with an inner diameter of 0.58 mm). The cell suspension was gently passed back and forth between the syringes 50 times, with 1 min pauses every ten passes. The cells were removed from the mixture via centrifugation at 5000 × g for 5 min. The supernatant containing the flagella were then centrifuged once again to remove any residual cells. In total, 5 μg/ml Alexa Fluor 488 C5 maleimide dye was added to the suspension and incubated for 5 min at room temperature. In total, 10 μl of Sc WT spore stock was then added to the mixture. The tube was gently inverted 50 times and the spores with associated flagella were pelleted via centrifugation at 5000 × g for 5 min. The pellet was washed with 1 ml PBS buffer pH 7.5 and centrifuged once again. The resulting pellet was resuspended in 50 μl PBS buffer pH 7.5. The sample was imaged on a Zeiss Axioscope A1 fluorescent microscope scope equipped with an Axiocam Mrc5 camera (Zeiss) in the Institute of Biology Microscopy Unit using a GFP filter. Images were collected and processed using Axiovision software (Zeiss).
Cryo-electron microscopy
B. subtilis cells were grown to an O.D. of 0.5 and 1 ml of B. subtilis cells were mixed with 5 μl S. coelicolor spores glycerol stock and incubated at ambient temperatures for 5 min. Cells were concentrated by centrifugation and 3 μl aliquots of the cell suspension are applied to glow-discharged R2/2 200 mesh copper Quantifoil grids (Quantifoil Micro Tools), the sample was pre-blotted for 30 s, and then blotted for 2 s. Grids were pre-blotted and blotted at 20 °C and at 95% humidity. The grids were plunge frozen in liquid ethane using an automated Leica EM GP system (Leica Microsystems) and stored in liquid nitrogen. The grids were imaged on a 120 kV Talos L120C cryo-electron microscope (Thermo Fisher Scientific) at the Netherlands Center for Electron Nanoscopy.
Cryo-electron tomography
B. subtilis mini-cell strain was grown from a 20% glycerol stock to an O.D. of 0.6 in 50 ml of LB. The cells were centrifuged at 8000 × g for 30 min. The supernatant was collected and then centrifuged at 12,000 × g for 20 min. The resulting cell pellet was resuspended in 20 μl of LB and 8 μl of WT Sc spore stock was added to the cell mixture. A 1/10 dilution of protein A- treated 10-nm colloidal gold solution (Cell Microscopy Core, Utrecht University, Utrecht, The Netherlands) was added to the mixture and mixed by pipetting. The grids were prepared using an automated Leica EM GP system (Leica Microsystems) with the sample chamber set at 20 °C and at 95% humidity. In total, 3 μl of the sample mixture was applied to a freshly glow-discharged copper R2/2 200 grid (Quantifoil Micro Tools), pre-blotted for 30 s, and then blotted for 2 s. The grid was plunge frozen in liquid ethane and stored in liquid nitrogen.
Images were recorded with a Gatan K3 Summit direct electron detector equipped with a Gatan GIF Quantum energy filter with a slit width of 20 eV. Images were taken at a magnification of ×19,500, which corresponds to a pixel size of 4.4 Å. Tilt series were collected using SerialEM with a bidirectional dose-symmetric tilt scheme (−60° to 60°, starting from 0°) with a 2° increment. The defocus was set to −12 μm and the cumulative exposure per tilt series was 160 e-/A2. Bead tracking-based tilt series alignment and drift correcting were done using IMOD [51] and CTFplotter was used for contrast transfer function determination and correction [52]. Tomograms were reconstructed using simultaneous iterative reconstruction with iteration set to 4. Segmentation was done in IMOD.
Plant growth
Arabidopsis thaliana Col-0 strain was grown from sterilized seedlings on sterilized plant MS agar media. Harvested A. thaliana seeds were sterilized in a sterile fume hood by incubation in 10% bleach for 30 min, washed with sterile water, and then incubated in 70% ethanol for 5 min. The seeds were then washed 6X with sterile water, placed on sterile filter paper, and placed in a dark 4 °C fridge for 3–4 days in a sterile and parafilm-sealed petri dish. Plant agar media plates were prepared by autoclaving Murashige and Skoog (MS) media (0.22% MS media with vitamins, 1.2% plant agar, 0.5% sucrose, pH 5.8) and pouring 100 ml of the media into square petri dishes with 12 cm length. The plates were allowed to cool for 1 h in a sterile fume hood. A. thaliana seeds were manually placed on the surface of the plates 1 cm apart by picking up the seeds with sterilized wooden picks. The plates were sealed with parafilm and placed in a climate-controlled plant growth chamber at a 20° angle so the plant roots grew on the surface of the media. The plant chamber was kept at 21 °C with a 16-h light cycle. The plants were allowed to grow for 1 month before use in chemoattraction assays (below).
Chemotaxis attractant assays with plant roots
Chemoattraction of Bs cells to plant roots in the presence and absence of Sc spores was conducted on minimal media plates with 0.25% agar. The media was prepared according to previous methods [53] in round petri dishes with 9 cm diameter. One month old sterile A. thaliana plants were removed from their sterile media and placed on the edge of the minimal media plates. In total, 3 μl of B. subtilis culture was placed to the center of the plate, and then 3 μl of the isolated spore stock was also added to the center. Controls of each bacteria by itself were also prepared. The plates were incubated for 16 h at 30 °C and then placed in a climate-controlled plant growth chamber for 2 weeks. The plant chamber was kept at 21 °C with a 16-h light cycle. After Sc colonies were visible, the plates were imaged on a light box.
Supplementary information
Acknowledgements
We thank Dr. Daniel Kearns for his guidance and advice during the preparation of this manuscript. We also thank him for the following B. subtilis strains: DK605, DS1677, DS222, DS1919, and DK1484. Mark Ladinsky at Caltech for the 3D segmentation of the Sc spores with Bs minicells, Dr. Jos Raaijmakers for the P. fluorescens strain R1SS101 and the A. thaliana Col-0 strain, Dr. Chris Rao for the undomesticated B. subtilis strain NCIB3610, Dr. Rose Loria for the S. scabies ISP5078 strain, and Dr. Joost Willemse for assistance with the fluorescence microscopy experiments. We also thank Dr. Jos Raaijmakers and Dr. Gilles van Wezel for their on-going support and advice with this project. We thank the Netherlands Centre for Electron Nanoscopy (NeCEN) for access to cryo-EM data collection and processing facilities, and the Institute of Biology Microscopy Unit at Leiden University for access to and training with light and fluorescence microscopes. This work is part of the research program National Roadmap for Large-Scale Research Infrastructure 2017–2018 with project number 184.034.014, which is financed in part by the Dutch Research Council (NWO). This project was funded by the European Union under a Marie-Sklodowska-Curie COFUND LEaDing fellowship to ARM, and an NWO Talent Programme Veni grant to ARM.
Author contributions
ARM, DC, and AB designed research; ARM and DC conducted experiments; ARM and DC analyzed data; ARM, DC, and AB wrote the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-00952-8.
References
- 1.van der Meij A, Worsley SF, Hutchings MI, van Wezel GP. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol Rev. 2017;41:392–416. doi: 10.1093/femsre/fux005. [DOI] [PubMed] [Google Scholar]
- 2.Worsley SF, Newitt J, Rassbach J, Batey SFD, Holmes NA, Murrell JC, et al. Streptomyces endophytes promote host health and enhance growth across plant species. Appl Environ Microbiol. 2020;86:1–35. doi: 10.1128/AEM.01053-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vurukonda SSKP, Giovanardi D, Stefani E. Plant growth promoting and biocontrol activity of streptomyces spp. as endophytes. Int J Mol Sci. 2018;19:1–26. doi: 10.3390/ijms19040952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franco C, Michelsen P, Percy N, Conn V, Listiana E, Moll S, et al. Actinobacterial endophytes for improved crop performance. Australas Plant Pathol. 2007;36:524–31. doi: 10.1071/AP07067. [DOI] [Google Scholar]
- 5.Olanrewaju OS, Babalola OO. Streptomyces: implications and interactions in plant growth promotion. Appl Microbiol Biotechnol. 2019;103:1179–88. doi: 10.1007/s00253-018-09577-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruddick SM, Williams ST. Studies on the ecology of actinomycetes in soil V. some factors influencing the dispersal and adsorption of spores in soil. Soil Biol Biochem. 1972;4:93–103. doi: 10.1016/0038-0717(72)90046-6. [DOI] [Google Scholar]
- 7.Becher PG, Verschut V, Bibb MJ, Bush MJ, Molnár BP, Barane E, et al. Developmentally regulated volatiles geosmin and 2-methylisoborneol attract a soil arthropod to Streptomyces bacteria promoting spore dispersal. Nat Microbiol. 2020;5:821–9. doi: 10.1038/s41564-020-0697-x. [DOI] [PubMed] [Google Scholar]
- 8.Jarrell KF, McBride MJ. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol. 2008;6:466–76. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- 9.Muok AR, Briegel A. Intermicrobial Hitchhiking: how nonmotile microbes leverage communal motility. Trends Microbiol. 2020:1–9. [DOI] [PubMed]
- 10.Samad T, Billings N, Birjiniuk A, Crouzier T, Doyle PS, Ribbeck K. Swimming bacteria promote dispersal of non-motile staphylococcal species. ISME J. 2017;11:1933–7. doi: 10.1038/ismej.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiong L, Cao Y, Cooper R, Rappel WJ, Hasty J, Tsimring L. Flower-like patterns in multi-species bacterial colonies. elife. 2020;9:1–27. doi: 10.7554/eLife.48885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagai E, Dvora R, Havkin-Blank T, Zelinger E, Porat Z, Schulz S, et al. Surface-motility induction, attraction and hitchhiking between bacterial species promote dispersal on solid surfaces. ISME J. 2014;8:1147–51. doi: 10.1038/ismej.2013.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkelshtein A, Roth D, Ben JE, Ingham CJ. Bacterial swarms recruit cargo bacteria to pave the way in toxic environments. mBio. 2015;6:1–10. doi: 10.1128/mBio.00074-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inghama CJ, Kalismand O, Finkelshteind A, Ben-Jacob E. Mutually facilitated dispersal between the nonmotile fungus Aspergillus fumigatus and the swarming bacterium Paenibacillus vortex. Proc Natl Acad Sci USA. 2011;108:19731–6. doi: 10.1073/pnas.1102097108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shrivastava A, Patel VK, Tang Y, Yost SC, Dewhirst FE, Berg HC. Cargo transport shapes the spatial organization of a microbial community. Proc Natl Acad Sci USA. 2018;115:8633–8. doi: 10.1073/pnas.1808966115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol. 1980;33:1179–83. doi: 10.1136/jcp.33.12.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allard-massicotte R, Tessier L, Lécuyer F, Lakshmanan V, Lucier J. Bacillus subtilis early colonization of Arabidopsis thaliana roots. mBio. 2016;7:1–10. doi: 10.1128/mBio.01664-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang W, Willemse J, Sawyer EB, Lou F, Gong W, Zhang H, et al. The propensity of the bacterial rodlin protein RdlB to form amyloid fibrils determines its function in Streptomyces coelicolor. Sci Rep. 2017;7:1–13. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Claessen D, Stokroos L, Deelstra HJ, Penninga NA, Bormann C, Salas JA, et al. The formation of the rodlet layer of streptomycetes is the result of the interplay between rodlins and chaplins. Mol Microbiol. 2004;53:433–43. doi: 10.1111/j.1365-2958.2004.04143.x. [DOI] [PubMed] [Google Scholar]
- 20.Patrick JE, Kearns DB. Laboratory strains of Bacillus subtilis do not exhibit swarming motility. J Bacteriol. 2009;191:7129–33. doi: 10.1128/JB.00905-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Najafi J, Shaebani MR, John T, Altegoer F, Bange G, Wagner C. Flagellar number governs bacterial spreading and transport efficiency. Sci Adv. 2018;4:1–9. doi: 10.1126/sciadv.aar6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee S, Kearns DB. The structure and regulation of flagella in Bacillus subtilis. Annu Rev Genet. 2014;48:319–40. doi: 10.1146/annurev-genet-120213-092406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nye TM, Schroeder JW, Kearns DB, Simmons LA. Complete genome sequence of undomesticated Bacillus subtilis strain NCIB 3610. Genome Announc. 2017;5:12–3. doi: 10.1128/genomeA.00364-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kearns DB, Losick R. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol. 2003;49:581–90. doi: 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- 25.Chan JM, Guttenplan SB, Kearns DB. Defects in the flagellar motor increase synthesis of poly-γ-glutamate in Bacillus subtilis. J Bacteriol. 2014;196:740–53. doi: 10.1128/JB.01217-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukherjee S, Babitzke P, Kearns DB. FliW and flis function independently to control cytoplasmic flagellin levels in Bacillus subtilis. J Bacteriol. 2013;195:297–306. doi: 10.1128/JB.01654-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glekas GD, Cates JR, Cohen TM, Rao CV, Ordal GW. Site-specific methylation in Bacillus subtilis chemotaxis: Effect of covalent modifications to the chemotaxis receptor McpB. Microbiology. 2011;157:56–65. doi: 10.1099/mic.0.044685-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guttenplan SB, Shaw S, Kearns DB. The cell biology of peritrichous flagella in Bacillus subtilis. Mol Microbiol. 2013;87:211–29. doi: 10.1111/mmi.12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Berardo C, Capstick DS, Bibb MJ, Findlay KC, Buttner MJ, Elliot MA. Function and redundancy of the chaplin cell surface proteins in aerial hypha formation, rodlet assembly, and viability in Streptomyces coelicolor. J Bacteriol. 2008;190:5879–89. doi: 10.1128/JB.00685-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elliot MA, Karoonuthaisiri N, Huang J, Bibb MJ, Cohen SN, Kao CM, et al. The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 2003;17:1727–40. doi: 10.1101/gad.264403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Dissel D, Willemse J, Zacchetti B, Claessen D, Pier GB, van Wezel GP. Production of poly-β−1,6-N-acetylglucosamine by MatAB is required for hyphal aggregation and hydrophilic surface adhesion by Streptomyces. Micro Cell. 2018;5:269–79. doi: 10.15698/mic2018.06.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sexton DL, Tocheva EI. Ultrastructure of exospore formation in Streptomyces revealed by cryo-electron tomography. Front Microbiol. 2020;11:1–9. doi: 10.3389/fmicb.2020.581135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oku S, Komatsu A, Nakashimada Y, Tajima T, Kato J. Identification of Pseudomonas fluorescens chemotaxis sensory proteins for malate, succinate, and fumarate, and their involvement in root colonization. Microbes Environ. 2014;29:413–9. doi: 10.1264/jsme2.ME14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kollaran AM, Joge S, Kotian HS, Badal D, Prakash D, Mishra A, et al. Context-specific requirement of forty-four two-component loci in Pseudomonas aeruginosa swarming. iScience. 2019;13:305–17. doi: 10.1016/j.isci.2019.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones SE, Pham CA, Zambri MP, McKillip J, Carlson EE, Elliot MA. Streptomyces volatile compounds influence exploration and microbial community dynamics by altering iron availability. mBio. 2019;10:1–18. doi: 10.1128/mBio.00171-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imada K. Bacterial flagellar axial structure and its construction. Biophys Rev. 2018;10:559–70. doi: 10.1007/s12551-017-0378-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedlander RS, Vogel N, Aizenberg J. Role of flagella in adhesion of Escherichia coli to abiotic surfaces. Langmuir. 2015;31:6137–44. doi: 10.1021/acs.langmuir.5b00815. [DOI] [PubMed] [Google Scholar]
- 38.Horstmann JA, Lunelli M, Cazzola H, Heidemann J, Kühne C, Steffen P, et al. Methylation of Salmonella Typhimurium flagella promotes bacterial adhesion and host cell invasion. Nat Commun. 2020;11:1–11. doi: 10.1038/s41467-020-15738-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lillehoj EP, Kim BT, Kim KC. Identification of Pseudomonas aeruginosa flagellin as an adhesin for Muc1 mucin. Am J Physiol Lung Cell Mol Physiol. 2002;282:751–6. doi: 10.1152/ajplung.00383.2001. [DOI] [PubMed] [Google Scholar]
- 40.Rossez Y, Wolfson EB, Holmes A, Gally DL, Holden NJ. Bacterial flagella: twist and stick, or dodge across the kingdoms. PLoS Pathog. 2015;11:1–15. doi: 10.1371/journal.ppat.1004483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wheatley RM, Poole PS. Mechanisms of bacterial attachment to roots. FEMS Microbiol Rev. 2018;42:448–61. doi: 10.1093/femsre/fuy014. [DOI] [PubMed] [Google Scholar]
- 42.Claessen D, Wösten HAB, Van Keulen G, Faber OG, Alves AMCR, Meijer WG, et al. Two novel homologous proteins of Streptomyces coelicolor and Streptomyces lividans are involved in the formation of the rodlet layer and mediate attachment to a hydrophobic surface. Mol Microbiol. 2002;44:1483–92. doi: 10.1046/j.1365-2958.2002.02980.x. [DOI] [PubMed] [Google Scholar]
- 43.Claessen D, Rink R, De Jong W, Siebring J, De Vreugd P, Boersma FGH, et al. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 2003;17:1714–26. doi: 10.1101/gad.264303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kohlmeier S, Smits THM, Ford RM, Keel C, Harms H, Wick LY. Taking the fungal highway: Mobilization of pollutant-degrading bacteria by fungi. Environ Sci Technol. 2005;39:4640–6. doi: 10.1021/es047979z. [DOI] [PubMed] [Google Scholar]
- 45.Warmink JA, Nazir R, Corten B, van Elsas JD. Hitchhikers on the fungal highway: The helper effect for bacterial migration via fungal hyphae. Soil Biol Biochem. 2011;43:760–5. doi: 10.1016/j.soilbio.2010.12.009. [DOI] [Google Scholar]
- 46.Barka EA, Vatsa P, Sanchez L, Nathalie Gaveau-Vaillant CJ, Klenk H-P, Clément C, et al. Taxonomy, physiology, and natural products of Actinobacteria. Am Soc Microbiol. 2016;80:1–43. doi: 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hess WM, Stocks DL. Surface characteristics of Aspergillus conidia. Mycologia. 1969;61:560–71. doi: 10.1080/00275514.1969.12018769. [DOI] [PubMed] [Google Scholar]
- 48.Hess WM, Sassen MA, Remsen CC. Surface characteristics of Penicillium Conidia. Mycologia. 1968;60:290–303. doi: 10.1080/00275514.1968.12018570. [DOI] [PubMed] [Google Scholar]
- 49.Kieser T, Bibb MJ, Chater KF, Butter MJ, Hopwood DA, Chater KF, et al. Practical Streptomyces genetics. Norwich, UK: John Innes Foundation; 2000. p. 1–613.
- 50.Richter M, Willey JM, Süßmuth R, Jung G, Fiedler HP. Streptofactin, a novel biosurfactant with aerial mycelium inducing activity from Streptomyces tendae Tu 901/8c. FEMS Microbiol Lett. 1998;163:165–71. [Google Scholar]
- 51.Mastronarde DN. Dual-axis tomography: An approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–52. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- 52.Xiong Q, Morphew MK, Schwartz CL, Hoenger AH, Mastronarde DN. CTF determination and correction for low dose tomographic tilt series. J Struct Biol. 2009;168:378–87. doi: 10.1016/j.jsb.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pham HT, Parkinson JS. Phenol sensing by Escherichia coli chemoreceptors: a nonclassical mechanism. J Bacteriol. 2011;193:6597–604. doi: 10.1128/JB.05987-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.