Abstract
Understanding the effects of changing climate and long-term human activities on soil organic carbon (SOC) and the mediating roles of microorganisms is critical to maintain soil C stability in agricultural ecosystem. Here, we took samples from a long-term soil transplantation experiment, in which large transects of Mollisol soil in a cold temperate region were translocated to warm temperate and mid-subtropical regions to simulate different climate conditions, with a fertilization treatment on top. This study aimed to understand fertilization effect on SOC and the role of soil microorganisms featured after long-term community incubation in warm climates. After 12 years of soil transplantation, fertilization led to less reduction of SOC, in which aromatic C increased and the consumption of O-alkyl C and carbonyl C decreased. Soil live microbes were analyzed using propidium monoazide to remove DNAs from dead cells, and their network modulization explained 60.4% of variations in soil labile C. Single-cell Raman spectroscopy combined with D2O isotope labeling indicated a higher metabolic activity of live microbes to use easily degradable C after soil transplantation. Compared with non-fertilization, there was a significant decrease in soil α- and β-glucosidase and delay on microbial growth with fertilization in warmer climate. Moreover, fertilization significantly increased microbial necromass as indicated by amino sugar content, and its contribution to soil resistant C reached 22.3%. This study evidentially highlights the substantial contribution of soil microbial metabolism and necromass to refractory C of SOC with addition of nutrients in the long-term.
Subject terms: Microbial ecology, Biodiversity
Introduction
Soil organic carbon (SOC) is the largest carbon pool in terrestrial ecosystems, and its decomposition and accumulation directly affect the global carbon balance [1]. Soil carbon (C) and nitrogen (N) cycles are closely coupled, and are greatly affected by changing climate and human activities. Long-term anthropogenic N inputs (such as fertilization) increase soil nutrient availability and substantially affect soil C dynamics [2–5]. Meta-analyses indicated that N addition replenished SOC pool by increasing the input of root exudates (exogenous C) [6], and increased soil recalcitrant C by 22% [7]. Although N application has the potential to increase soil total C storage, the long-term stability of soil C depends on future changes in climates [8]. The response of soil C to long-term fertilization in term of slow- and active-cycling SOC pools is affected by temperature and precipitation. The interactive influence of soil nutrients and climate factors determine the largest part of the SOC variance (32%) based on the long-term field observations and modeling in agricultural ecosystem [9], which is closely related to the microbial metabolism, enzyme activities, and necromass turnover [10–12].
Soil microorganisms can not only decompose and mineralize soil organic matter [13], but also metabolize plant residues or form necromass through anabolism to stabilize C in soil [14]. Microbial residues are usually closely bound to the surface of soil minerals and are considered to be relatively stable [15, 16]. The framework of soil microbial C pump indicates that microorganisms regulate soil C accumulation through ex vivo modification (reconstruction of organic C molecules by microbial extracellular enzymes) and in vivo turnover (resynthesis of C molecules by microbial necromass) [17]. It means that microbial metabolic products or necromass can contribute substantially to SOC as refractory C sources, and the addition of nutrients or easily degradable C can also increase soil organic matter in the long-term. This process can be greatly affected by multifactorial environmental changes [18–20]. Therefore, it requires further studies to elucidate the physiological metabolism of soil microorganisms and the contribution of necromass to soil C under long-term fertilization in different climatic conditions for better understanding the accumulation and stability of organic C in agricultural soils.
However, as a common practice, most studies extract DNA of total soil microbes, making it difficult to distinguish and quantify the roles of live and dead microbes in mediating SOC [21, 22]. Propidium monoazide (PMA) is a membrane-impermeant dye that selectively penetrates cells with compromised membranes [23]. PMA intercalates into DNA from cells and can be covalently cross-linked to it, which strongly inhibits PCR amplification. As a result, live microbes can theoretically be distinguished from total communities [23]. The combination of PMA method, metabolic activity analysis of living bacteria, and necromass analysis may be an effective way to discerning the biological mechanism of soil C changes by fertilization.
Long-term soil transplantation experiments are considered as an intuitive and effective method to simulate the integrated changes of various environmental factors under natural conditions [24, 25]. A soil transplantation experiment was carried out since 2005 by translocation large transects of Mollisols soil from northern China (cold temperate zone) to central China (warm temperate zone) and southern China (middle subtropical zone), simulating multifactorial environmental changes in the field. The microbial biomass, diversity, community structure, and succession rates were significantly changed after years of translocation [26–28]. At present, more than 10 years of prolonged community incubation under different environmental conditions makes it a microbial adaptation experiment, with on top of fertilization treatment. Here, we hypothesize that after 12 years of soil transplantation, the addition of fertilizer will select for a specific microbial community with contribution to the build of SOC. The refractory C pool in fertilized soil will be supplemented by the increase of microbial necromass. In addition, fertilization will lead to less reduction of labile C of SOC due to less priming effect. To test this hypothesis, we used solid-state 13C nuclear magnetic resonance (NMR) analysis to track the molecular fingerprint of SOC. The PMA method and single-cell Raman spectroscopy combined with heavy water (Raman-D2O) were performed to target live soil bacterial structure and physiology. Meanwhile, amino sugar content as an indicator for microbial necromass was also measured. Our results indicated that in agriculture soils, fertilization regulated microbial metabolism and necromass for the build of soil resistant C pool under warm climate conditions.
Materials and methods
Research sites and sample collection
Soil transplantation experiment is carried out at three sites in the National Field Science Research Stations of the Chinese Academy of Sciences [29]: Hailun station in Heilongjiang Province of northern China (N site, E126°38′ and N47°26′, cold temperate climate zone with soil type of Mollisol), Fengqiu station in Henan Province of central China (C site, E114°24′ and N35°00′, warm temperate climate zone with soil type of Inceptisol) and Yingtan station in Jiangxi Province of south China (S site, E116°55′ and N28°15′, subtropical climate zone with soil type of Ultisol). In each experimental station, 18 blocks with a size of 1.4 m in length × 1.2 m in width × 1.0 m in depth were set up in October 2005. The blocks were surrounded by 20-cm cement mortar brick walls and were paved underneath with quartz sand (3 cm in thickness) [30]. To ensure an intact soil matrix, the soil was stratified every 20 cm per layer during excavation. Six Mollisol soil blocks were excavated but remained in place to serve as in situ. Other 12 blocks were transferred to C site and S site, respectively (transplanted samples were thereby designated as TransS1 and TransS2). The mean annual temperature (MAT) and precipitation (MAP) is 1.5 °C and 550 mm in situ, 13.9 °C and 605 mm in C site, and 17.6 °C and 1795 mm in S site. Maize was planted every year since the spring of 2006 with non-fertilization (control) and regular fertilization, 150 kg/hm2 N, 75 kg/hm2 P and 60 kg/hm2 K in the forms of urea, (NH4)2HPO4 and KCl, respectively. All P and all K fertilizers and half of the N fertilizer were applied before maize cropping. The other half of the N fertilizer was applied as a top dressing at the large trumpet stage of maize growth. Three biological triplicates were performed for each treatment.
Twelve years after initiating the soil transplantation experiment, 18 Mollisol soil samples (in situ, TransS1 and TransS2) were collected for the analysis of C molecular groups, live and total microbial communities, single-cell Raman metabolic activity, C utilization capacity and necromass content in August to September 2017 after crop harvest. In addition, 6 local Inceptisol (C site) and 6 local Ultisol (S site) soil samples were taken simultaneously to track the entry of local microbes to the transplanted Mollisol soils. Ten soil cores with a diameter of 2 cm and a depth of 0–20 cm were collected from each block and mixed thoroughly to make one soil sample, sealed in polyethylene wrapper, stored on ice, and transported to the laboratory. Soils for geochemical analyses were stored at 4 °C, and soils for DNA extraction were stored at −80 °C.
Solid-state 13C nuclear magnetic resonance (NMR) analysis
Hydrofluoric (HF) acid was used in a pretreatment step to prevent interference from Fe3+ and Mn2+ ions in the soil. After pretreatment, soil sample was subjected to solid-state magic-angle spinning NMR measurement (AVANCE II 300 MH). The chemical shift in the main 13C signal of SOC corresponded to the following C structures [31]: 0–45 ppm alkyl C, 45–95 ppm O-alkyl C, 95–110 ppm acetal C, 110–160 ppm aromatic C, and 160–220 ppm carbonyl C. In general, SOC can be classified as labile C (LC) with molecular groups of O-alkyl, methoxy and carbonyl, and resistant C (RC) with alkyl, aromatic and phenol groups [32, 33].
High-throughput sequencing analysis of live and total microbes
PMA method was used to extract DNA from soil live microbes [34]. PMA is a membrane-impermeant dye that selectively penetrates dead cells. Once inside the cells, PMA intercalates into the DNA and can be covalently cross-linked to it, which strongly inhibits PCR amplification. By using PCR after PMA treatment, it is theoretically possible to obtain cells with intact cell membranes only. For the soil total microbes, microbial genomic DNA was extracted from soil samples using the MoBio Kit in combination with liquid nitrogen freeze-milling. The concentration and purity of the extracted DNA were tested using a NanoDrop 2000 (Thermo Fisher Scientific, USA).
Extracting bacteria by Nycodenz density-gradient separation (NDGS)
The NDGS method described in a previous study [35] was used to extract live bacterial cells from soil samples from in situ, TransS1, and TransS2. The sieved soils (1 g, 0.6 mm sieve) were homogenized in 5 mL of phosphate-buffered saline (PBS, 8 g L−1 NaCl, 0.2 g L−1 KCl, 1.44 g L−1 Na2HPO4, 0.24 g L−1 KH2PO4) amended with 25 μL of Tween 20. After vigorous vortexing for 30 min, the soil-associated bacteria were detached. To separate bacteria from soil particles, the suspension was gently added to 5 mL of Nycodenz (y98%, Aladdin) solution (1.42 g/mL), dissolved in 5 mL of sterile water, and then centrifuged at 14,000 × g for 90 min at 4 °C. After centrifugation, the middle layer containing bacteria was collected into a new tube containing PBS. The microorganisms inside were collected by centrifugation at 5000 rpm for 10 min at room temperature and washed twice with ultrapure water to remove residual PBS and other traces of reagents. Then, the soil bacteria extracted by NDGS were inoculated into starch culture and cellulose culture amended with 50% D2O and incubated at 28 °C for 24 h prior to Raman analysis.
Single-cell Raman measurements and Raman mapping analysis
All bacterial samples were washed three times with ultrapure water to remove residual culture base. An aliquot of 2 μL of the as-prepared bacterial solution was loaded on Al foil and air-dried at room temperature prior to Raman measurements [36]. Raman spectra and Raman mapping were acquired from a LabRAM Aramis confocal Raman microscope (HORIBA Jobin-Yvon) equipped with a 532 nm Nd:YAG laser (Laser Quantum), 300 g/mm grating and a ×100 objective (Olympus, NA = 0.09). The Raman band of a silicon wafer at 520.6 cm−1 was used to calibrate the Raman spectrometer before each measurement. Fifty individual cells from each sample were randomly selected for single-cell Raman measurements. Two independent experiments were performed. The spectra were acquired in the range of 500–3200 cm−1 at an acquisition time of 15 s for each point. Raman imaging was acquired at a step size of 1 μm, and a pseudocolored Raman image was generated on the basis of the C–H and C–D bands. All spectra were preprocessed by baseline correction and normalization to the C–H bands via LabSpec6 software (HORIBA Jobin-Yvon). Peak intensity was quantified by calculating the peak area. To indicate the degree of D substitution in C–H bonds, the intensities of the C–D peak (2040–2300 cm−1) and the C–H peak (2800–3100 cm−1) were used to calculate the C–D ratio of CD/(CD + CH). Raman spectra and the C–D ratios were obtained in R language software (version 3.4.4). Correlation and variance analyses were performed using R language software.
Carbon degradation experiment (starch and cellulose degradation)
To determine the degradation capacity of soil live microbes, starch, and cellulose were added to the inorganic salt base medium (100 mL to obtain a concentration of 1%) and inoculated with the soil microbial suspension at a concentration of 2% [37]. A tube containing 10 mL of 0.1 m NaOH was suspended in the culture bottle, and the system was sealed with silica gel. The above devices were cultured at a constant temperature of 28 °C and kept away from light. Twenty-four hours later, the device was opened, and the tube was removed. The volume of CO2 generated in the system was determined by hydrochloric acid titration. Three replicates were performed for each treatment.
Soil extracellular enzyme activity assays
Soil α-glucosidase and β-glucosidase were measured using 4-methylumbelliferyl substrate, which is split into high-fluorescence cleavage products upon hydrolysis [38]. Two enzyme assays were set up in 96-well microplates. Sixteen replicate wells were set up for each sample and each standard concentration. The assay plate was incubated in the dark at 25 °C for 3 h to mimic the average soil temperature. Enzyme activities were corrected using fluorescence quenching. Fluorescence was measured using a microplate reader (EnSpire 2300 Multilabel Reader, Perkin Elmer, Waltham, MA, USA) with 355-nm excitation and 460-nm emission filters. The activities were expressed as μmol d−1 g−1 dry soil.
Growth curve measurements
To further understand the growth of live microbes in different carbon sources, the growth curves of microorganisms were measured. Based on the above C source degradation experiments, 18 soil suspensions were cultured in two culture media (inorganic salt + starch medium and inorganic salt + cellulose medium). Suspensions were added to the two different culture media, and nine replicates were performed for each sample. The experiment was performed in 96-well plates. The inoculation amount of soil was 10% (270 µL of medium + 30 µL of suspension). Then, the plates were put into a Bioscreen C (Type: FP-1100-C, Oy Growth Curves AB, USA), incubated for 5 days at 28 °C with a wavelength of 600 nm, and measured every hour during incubation.
Microbiological determination of dead microbes (amino sugar experiment)
The content of amino sugars was measured as tracers for microbial necromass. The amino sugars in the soil samples were hydrolyzed, purified, and derived by gas chromatography (Agilent 6890, Agilent Technologies, USA) [39]. Briefly, soil samples (containing ~0.4 mg N) were hydrolyzed with 10 mL of 6 M HCl at 105 °C for 8 h. The cooled sample was added to 100 μg of myoinositol as a surrogate standard, filtered, evaporated to dryness at 40 °C under reduced pressure and redissolved in 20 mL of Milli-Q water with the pH adjusted to 6.6–6.8 using 0.4 M potassium hydroxide. Precipitates were discarded after centrifugation (3000 × g, 10 min), and the supernatant was freeze-dried. Amino sugars were redissolved in methanol and separated from salts by centrifugation. After the addition of a quantitative standard (methyl-glucamine), amino sugars were transformed into aldononitrile derivatives by heating in 0.3 mL of a derivatization reagent (containing 32 mg/mL hydroxylamine hydrochloride and 40 mg/mL 4-(dimethylamino) pyridine in a pyridine and methanol mixture (4:1; v/v)) at 75–80 °C for 30 min. The derivatives were further acetylated with 1 mL of acetic anhydride for 20 min and mixed with 1.5 mL of dichloromethane after cooling. Excess derivatization reagents were removed by extraction with 1 M HCl (added once) and Milli-Q water, while the dichloromethane phase containing amino sugar derivatives was dried under nitrogen before quantification.
Data processing and statistical analysis
Pearson’s correlation coefficient analysis and other analyses were conducted to analyze changes in SOC and its molecular groups by SPSS statistical software (SPSS Inc., Chicago, IL, USA). Moreover, these methods were performed to identify the phyla of bacteria associated with the LC and RC components. In addition, structural equation modeling was used to simultaneously test the direct, indirect and total effects of temperature, soil nutrients and total microbes on the LC and RC components. The major criteria used to evaluate the model were as follows [40]: (1) a chi-square value-to-degrees of freedom ratio (chi-square/df) less than or equal to 2 and a chi-square test p value greater than 0.05, (2) a goodness-of-fit index value greater than 0.90, and (3) a mean square and variance (RMSEA) of progressive residual error less than 0.05. Before statistical analysis, the normality test was carried out, and all the data were in accordance with the normal variance test. The following analyses were performed in R software (version 3.4.4; http://www.r-project.org/). The α- and β-diversity (including Shannon index, richness, canonical correspondence analysis ordination, nonparametric statistical multivariate analyses, and Mantel analysis) of the live and total microbial communities were compared [41–43]. Partial redundancy analysis (pRDA) was used to evaluate the effects of explanatory variables, including soil geochemical attributes (NO3−–N, NH4+–N, pH, SOC, AN, AP), live bacteria (the DNA of live bacteria) and dead bacteria (muramic acid from dead bacteria in soil amino sugars), on the LC and RC components. The contributions of microbial phyla in different modules to the SOC components were predicted using a random forest model [44]. Moreover, cooccurrence networks were constructed using the “CoNet” plugin in Cytoscape software (version 3.6.1; http://www.cytoscape.org/), and Gephi software (version 0.9.2; http://www.gephi.org/) was used to modularize the OTUs of microbial communities significantly related to LC and RC components [45]. All of the above analyses were performed using 18 soil samples (six samples each from in situ, TransS1 and TransS2). Moreover, for 12 samples from the C site and S site, microbial communities were used for SourceTracker analysis [46] in R language (version 3.4.4).
Results
Effect of fertilization on amount and quality of SOC in different climate conditions
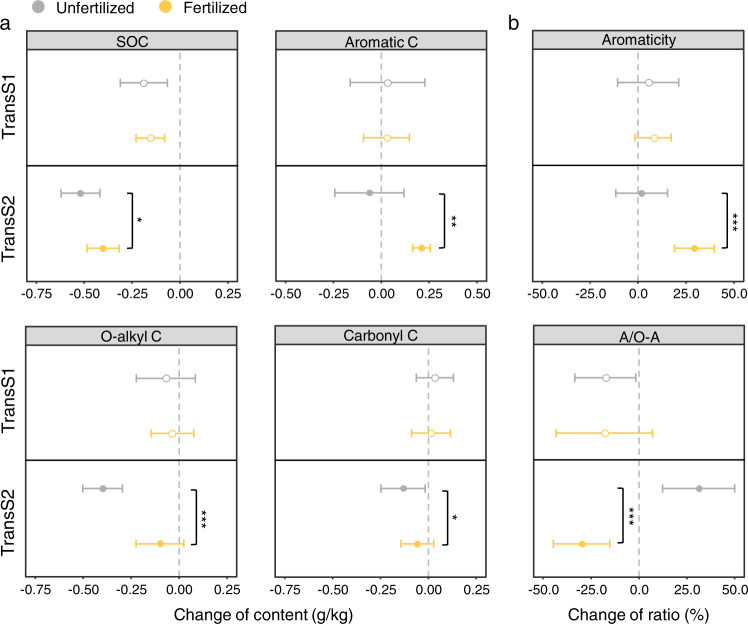
After 12 years of soil transplantation, the loss of SOC was significantly less in the warmer climate (TransS2 treatment) (Fig. 1). We further used NMR technology to determine the changes of SOC molecular groups (Fig. 1, Fig. S1). Fertilization significantly increased the aromatic C in soil RC in TransS2. The aromaticity in fertilized soils in TransS2 was 2.7 times that of TransS1, indicating an increased humification with fertilization under warmer climate conditions. For soil LC, O-alkyl C and carbonyl C decreased in unfertilized soils, while fertilization significantly mitigated the loss of LC in warmer climate. The decrease in the ratio of alkyl/O-alkyl by fertilization in TransS2 indicated a potential inhibition of the consumption of easily decomposable small-molecule C. Fertilization and soil translocation also changed some other soil properties, such as pH and soil nutrients (Table S1).
Fig. 1. Effects of fertilization on SOC and its molecular groups 12 years after soil transplantation (in different climates).
a Effects of fertilization on SOC and its molecular groups (alkyl C, O-alkyl C, acetal C, aromatic C, and carbonyl C) in different climates. Only SOC molecular groups with significant changes are shown, and the rest are shown in Fig. S1. b A/O-A ratio (%) = [alkyl C peak area (0–45 ppm)]/[O-alkyl C peak area (45–90 ppm)] ×100; Aromaticity (%) = [Aromatic C peak area (110–160 ppm)]/[Total peak area (0–160 ppm)] ×100. Data are the means ± SD (n = 9). Significance analysis is performed between unfertilized and fertilized soils by one-way ANOVA, *p < 0.05, **p < 0.01, ***p < 0.001.
Changes of the composition and structure of live and total microbial communities
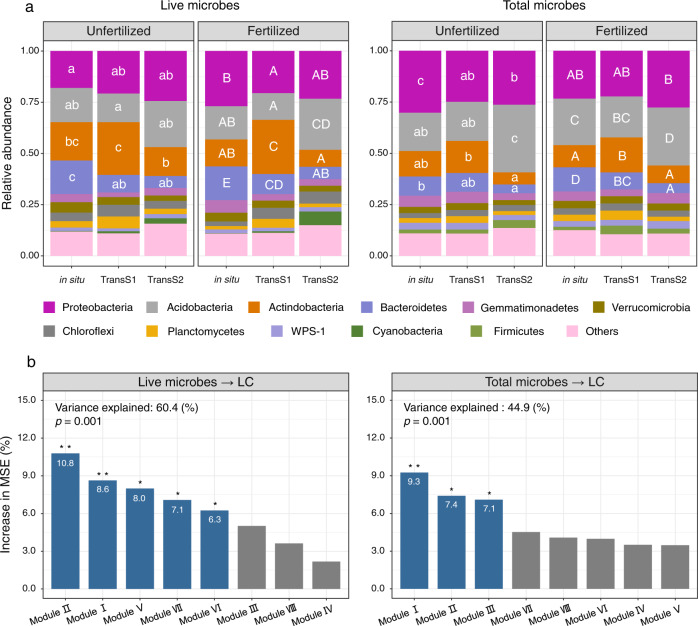
Rarefaction analysis indicated that the sampling efforts captured most of the live and total microbiota members (Fig. S2). A Bayesian approach was employed to identify potential sources of bacterial communities in Mollisol soil after transplantation at the OTU level (Fig. S3). The greatest potential source of soil microorganisms after transplantation to TransS1 was in situ Mollisol, accounting for 69%, followed by 21% of unknown sources and only 10% from local soil (Inceptisol). Similar results were obtained for TransS2. These results indicated that only a small proportion of the microbes came from the local soil (Inceptisol and Ultisol) (Fig. S3, Table S2).
Fertilization significantly changed the composition and structures of bacterial community in different climate conditions (Fig. 2a, Fig. S4, Table S3), increased the α-diversity of live microbes, and decreased the α-diversity of total microbes (Fig. S5). Mantel and partial Mantel tests were used to determine how environmental factors affected the live and total bacterial communities (Table 1). In comparison to the effects of soil geochemical attributes, fertilization and climatic factors (MAT and MAP) were the dominant factor in shifting the community structures of live and total microbes (p < 0.05).
Fig. 2. Composition and modulization of live and total microbial communities.
a Comparison of composition of live and total bacterial community. Only the top ten phyla with the highest relative abundance are shown, and the others are represented by “others” in the figure. Lowercase letters indicate significant differences between live and total microbes under non-fertilization, and capital letters indicate that under fertilization. Statistical significance is performed by one-way ANOVA and Tukey’s HSD test. b Contribution of the top eight network modules in live and total microbes to the changes in soil labile carbon (LC) components by the random forest model. The data of each module in the histogram are standardized by Z-score. Detailed microbial network module is shown in Figs. S6 and S7. The prediction of changes in soil resistant carbon (RC) by live microbes and total microbes is shown in Fig. S9. The blue bars indicate significance (*p < 0.05, **p < 0.01), and gray bars indicate no significant difference.
Table 1.
Mantel and partial Mantel test on the relationships between bacterial community composition and soil geochemical properties (pH, SOC, TN, TP, NO3−–N, NH4+–N), warming climatic regimes and fertilization.
| Correlation between bacterial community composition and: | Controlling for | Live bacteria | Total bacteria | ||
|---|---|---|---|---|---|
| Rm | pm | Rm | pm | ||
| All samples | |||||
| All variables | 0.815*** | <0.001 | 0.816*** | <0.001 | |
| Warming climatic regimes | Soil and fertilization | 0.819*** | <0.001 | 0.821*** | <0.001 |
| Fertilization | Fertilization and soil | 0.280** | 0.015 | 0.333** | 0.004 |
| Soil geochemical properties | Warming climatic regimes and fertilization | −0.023ns | 0.560 | 0.065ns | 0.250 |
Bold value indicates p < 0.05. (*<0.05, **<0.01, ***<0.001, ns non-significant at 5%).
Relationship between live/total microbes and soil LC and RC components
Pearson correlation analysis indicated that there were 1148 and 874 OTUs of live and total microbes that significantly correlated with LC (p < 0.05), and 134 from live bacteria and 120 from total bacteria correlated with RC (Table. S4). Network modularization analysis indicated that LC-related OTUs parsed into eight top modules that accounted for 82.0% and 88.4% of the whole networks of live and total microbes, respectively (Fig. S6, Table. S4). There were five modules from live microbes and three modules from total microbes that significantly contributed to the changes in LC by random forest models, with 60.4% and 44.9% explanation, respectively (Fig. 2b, Figs. S7, S8). The modularization of RC-related OTUs was simpler than that of LC-related OTUs (Fig. S9, Table. S4). The total microbes explained more of the variation in RC (78.81%) than the live microbes (70.69%) and showed a positive contribution to RC (Fig. S10).
Microcosm experiments on the contributions of live microbes to C components
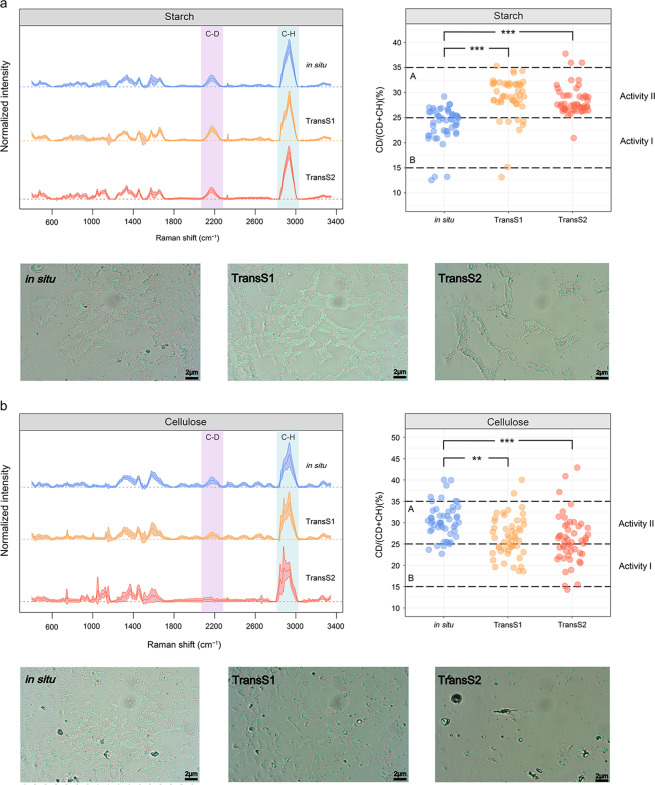
The metabolic activities of live bacteria were studied by Raman-D2O to confirm their degradation capacity after soil transplantation (Fig. 3). The bacterial cultures using D2O in its substrate metabolism showed obvious C–D Raman bands in the range of 2040–2300 cm−1 (Fig. 3a). The ratio of C–D to (C–D + C–H) in TransS1 and TransS2 was significantly higher than that in situ with starch as C source, but the opposite was true in the case with cellulose as C source (Fig. 3b).
Fig. 3. D2O isotope labeling combined with single-cell Raman spectroscopy to study the metabolic activities of soil bacteria with different carbon sources (starch and cellulose) in fertilized soils.
a Raman spectra (left), C–D ratios (C–D to (C–D + C–H)) (right) (n = 50), and Raman mapping images based on C–D bands of soil bacteria incubated in inorganic salt + starch medium with 50% D2O for 24 h. b Raman spectra, C–D ratios (n = 50), and Raman mapping images of soil bacteria incubated in inorganic salt + cellulose medium. Significance analysis is performed by one-way ANOVA and Tukey’s HSD test, **p < 0.01, ***p < 0.001.
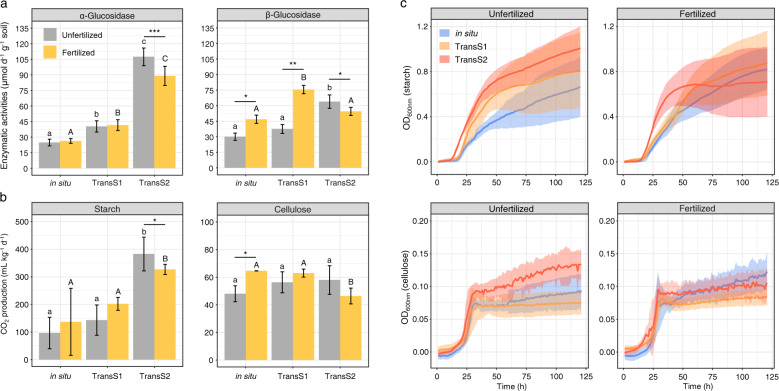
Fertilization resulted a significant decrease in soil α- and β-glucosidase activity in TransS2 (Fig. 4a). The ability of live microbes to decompose different C sources (starch and cellulose) was tested by inoculation of soil suspensions (Fig. 4b). Our results showed that fertilization significantly reduced the CO2 produced by the decomposition of starch and cellulose. These result indicated that the loss of unstable and stable C was reduced with fertilization in TransS2. Additionally, the growth curves of live bacteria in starch and cellulose were determined separately (Fig. 4c). The growth activity of bacteria in fertilized soils was lower than that in unfertilized soils in warmer climate. Especially in the starch C source, the bacteria in the fertilized soil accelerated to enter the stationary phase, while the bacteria in the unfertilized soils continued to grow.
Fig. 4. The metabolic capacity and growth of soil microorganisms on different C sources with fertilization in different climates.
a Soil enzymatic activities (α- and β-glucosidase). b Microcosm experiments on the microbial degradation capacity of starch and cellulose (CO2 production). The data of each bar are expressed as the mean ± SD (n = 3). c Growth curve of live soil microbes on starch and cellulose with inoculation of soil suspension. The growth curve is the average of nine replicates. The shaded area denotes the standard deviations of the average. Statistical analysis is performed among in situ, transS1 and transS2 by one-way ANOVA with Tukey’s HSD test and expressed with letters. Significance between unfertilized and fertilized soils is expressed with asterisk. (*p < 0.05, **p < 0.01, and ***p < 0.001, one-way ANOVA).
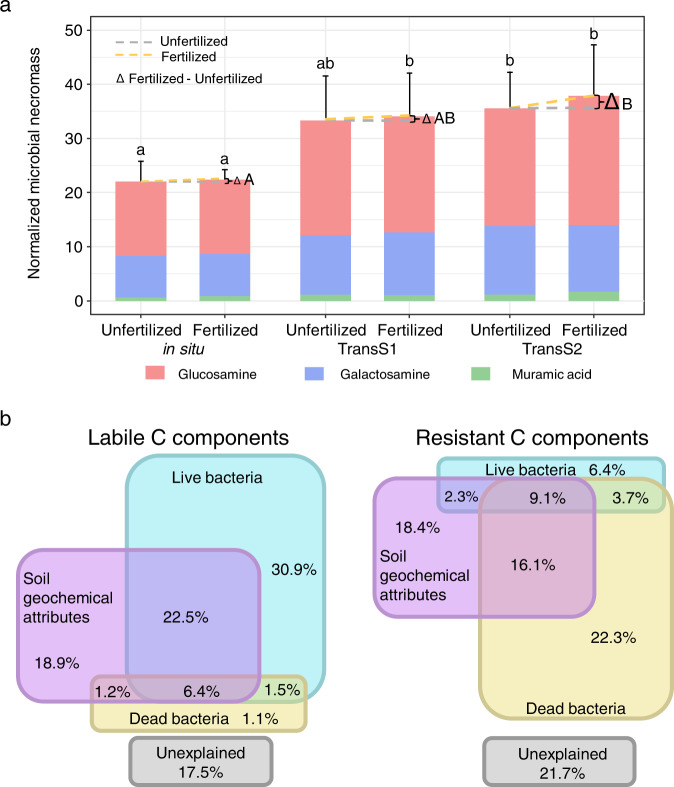
Contributions of microbial necromass to SOC by soil amino sugars analysis
Soil amino sugars were measured as a tracker for microbial necromass. Our results showed a significant increase in the amounts of microbial necromass with fertilization in TransS2 (Fig. 5a). Furthermore, the accumulation of microbial necromass was enhanced 12 years after transplantation. The contributions of live and dead microbes and soil geochemical attributes to the LC and RC components were estimated using the partial RDA method (Fig. 5b). The contribution of live microbes to LC components was 30.9%, much higher than the contribution of dead microbes (1.1%). For RC components, the contribution of dead microbes was 22.3%, greater than that of live microbes (6.4%). This result indicated that dead microbes may be the main contributor to soil RC.
Fig. 5. Contributions of microbial necromass to SOC.
a Soil amino sugar content as necromass biomarker, including glucosamine from dead fungi and aminogalactose and muramic acid from dead bacteria. Data are presented as the means ± SD (n = 3). Δ means the difference between fertilization and non-fertilization (F–U). Capital letters indicate significant differences of fertilization effect in different climates, and lowercase letters indicate significant differences between in situ and transplantation (one-way ANOVA with Duncan’s multiple range test at p < 0.005). b Partial redundancy analysis (pRDA) differentiating the effects of soil geochemical attributes, live bacterial biomass, and dead bacterial amino sugar content on soil LC and RC.
Discussion
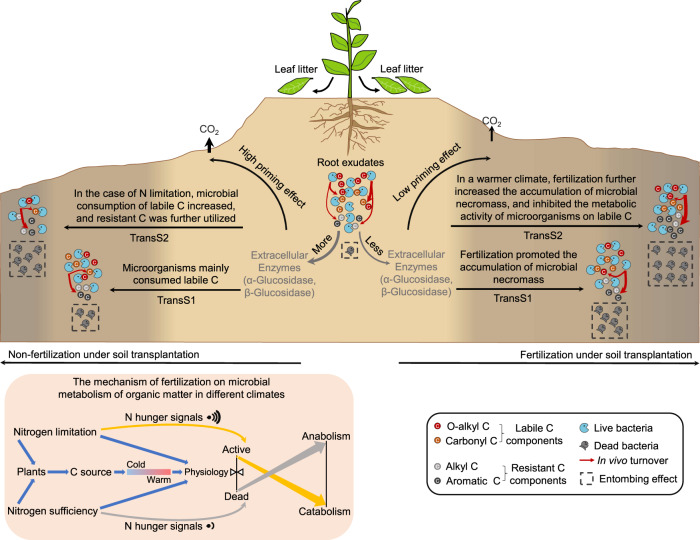
Under the joint influence of climate change and human activities, the regulation of SOC by N addition in the terrestrial biosphere has attracted much attention [47–49]. In general agricultural management, the strategy of replenishing SOC usually comes from the input of refractory C sources such as straw or compost, which are considered as the best option to increase RC in soil. However, these approaches do not adequately reflect current views of SOC sequestration and neglect the role of microbial biomass. Based on the theory of microbial C pump, microbial metabolic products or necromass can contribute substantially to RC-SOC, emphasizing the regulation of microbes in C sequestration [17, 50]. Hence, it is necessary to further reveal the microbial mechanism of long-term fertilization effect on SOC and test this under different climatic conditions by combining field and cultivation experiments. Our study evidentially demonstrated that under warm climate, soil microbial necromass played an important role in the build of RC-SOC. Fertilization changed the composition and metabolic characteristics of soil microbial communities, resulting in more accumulation of microbial necromass for long-term soil C sequestration. Here, we constructed a conceptual framework to reveal the potential mechanisms to explain the regulatory effect of microbial physiological metabolism and necromass on SOC sequestration in the cases of N deficiency and sufficiency (Fig. 6). Under warm climate, fertilization can supplement soil refractory C (aromatic C) by promoting the accumulation of microbial necromass. On the other hand, live microorganisms consumed less unstable C component of SOC (such as O-alkyl C, carbonyl C, etc.) due to less priming effect.
Fig. 6. Schematic diagram of potential regulation mechanism of soil microorganisms on SOC molecular groups in fertilized and non-fertilized soils under different climates.
The soil color gradient in the above figure reflects the changes of LC and RC components, and the height represents the content of SOC. The lower left figure shows the effect mechanism of nitrogen limitation or sufficiency on microbial metabolism of SOC under different climatic conditions. Without fertilization, due to nitrogen limitation, plants and microorganisms may send higher signals of nitrogen starvation, and microorganisms produce more extracellular enzymes to decompose SOC to obtain nitrogen source. In addition, nitrogen deficiency reduced the input of plant root exudates (easily degradable C source), warming accelerated the consumption of unstable C in soil, further stimulated the respiration of stable C in soil by microorganisms, resulting in the decrease of SOC. In the case of fertilization, the increase of soil microbial necromass supplemented soil unstable C, and low N starvation signal reduced the consumption of soil stable C by microorganisms.
Microorganisms can accumulate metabolites or microbial residues (intact cell or after lysis and fragmentation) iteratively and stabilize them within or on soil particles [17]. Here, we found that the total microbes positively contributed to RC and explained the changes of RC under combination effects of fertilization and soil transplantation. The biomass of live microbes only accounts for less than 5% of SOC [51], and it is generally considered that their contribution to soil C storage is negligible [52]. However, based on the absorbing Markov chain approach, it is estimated that the size of soil microbial necromass C pool is about 40 times larger than that of soil microbial biomass [52]. On average, the contribution of microbial necromass accounts for 55.6% of total SOC for temperate agricultural soils based on a global analysis [53]. We used amino sugar as biomarker, and found that the contribution of microbial necromass to the stable C component (22.3%) was higher than that of live microbes (6.4%), which showed that microbial necromass can contribute substantially to resistant SOC. The accumulation of microbial necromass is controlled by environmental factors, which affects the process of soil C sequestration. For example, recent study indicates the enhanced physical preservation of SOC under winter warming and emphasizes the role of soil microorganisms in aggregate life cycles [54]. Therefore, we infer that the contribution of microorganisms to soil stable C pool is mainly reflected in the increase of necromass under the combined effects of changing climate and fertilization.
The accumulation of microbial residues in soil is of great significance to the conservation and sequestration of soil C [55–58]. In this study, we found that fertilization significantly increased the accumulation of microbial neromass in warmer climate (Fig. 5a), which may be the direct evidence of increasing soil stable C and delaying the loss of SOC. This may be explained by the following reasons: (1) Fertilization would make the microorganisms with rich resource acquisition strategy become the dominant species [20], increase the biomass of these species in warming climate condition, and directly promote the accumulation of microbial residues. (2) Increasing input of root exudates (exogenous C) of aboveground plants by fertilization can regulate soil C/N and indirectly increase soil microbial necromass [59]. (3) In climates with higher rainfall and temperatures, microbial residue stimulated by fertilization can be closely combined with the minerals on the soil surface to form a stable organic matter with longer turnover time, and is not easy to be decomposed through mineral protection [59–61]. Our research showed that under long-term climate change, the microbial necromass increased by fertilization contributed significantly to the build of SOC, which provides solid evidence to support the viewpoint that microbial anabolism is a major contributor to the stable SOC pool.
Under the warm climate conditions, the content of SOC in Mollisol soil decreased significantly, mainly due to the reduction of LC in SOC. Increasing temperature was reported to increase the abundance of microbial functional genes encoding LC degrading enzymes [62]. Similarly, the metabolic activity of live microorganisms to degrade unstable C sources increased after soil transplantation in this study. However, we found that the magnitude of this increase was less under fertilization conditions in warmer climates (Fig. 4). The microbial necromass C source may mainly come from root exudates or plant residuals instead of SOC under fertilization. This is consistent with that compared with non-fertilization, the loss of LC in fertilized soil was significantly less, which may be explained by N-Mining hypothesis [63, 64]—the priming effect of accelerating SOM decomposition for N-supply. With increasing amount of fresh root exudate input, microorganisms consume SOM for acquisition of N to meet their nutrient demands. Especially, in the unfertilized soil, crop growth needs a lot of nutrients, and trade between plants and microorganisms will be enhanced [64], whereby microorganisms obtain C from roots to exchange N (and P) released from SOM for root uptake [65]. In the case of fertilization, microorganisms will not enhance the N acquisition of SOM in order to meet their own or crop nutrient needs [66]. Second, energy-induced exoenzyme synthesis is proposed for explaining this negative mineral N-induced priming effects [67]. Microbial extracellular enzymes directly catalyze the decomposition of SOC [68, 69]. In our study, we found that in warmer climate, compared with non-fertilization, the extracellular enzymes of microorganisms in the fertilized soil were decreased, and the respiration of using starch as easily decomposed C source was weakened, suggesting reduced C mineralization capacity. This may indicate a low energy cellular energy status that would not signal the potential for growth, thus reducing unnecessary energy expenditure [70]. It can be confirmed by our study that the growth curve of bacteria in fertilized soil was lower than that in unfertilized soil in warmer climate. Third, N addition significantly decreased soil pH. Low soil pH would reduce decomposition rates and promote soil C storage [71, 72].
Overall, this study highlights the important contribution of microbial metabolism and necromass to SOC sequestration under the joint effects of fertilization and different climatic conditions. Facing the intense debate that is going on C sequestration, our findings provide new insights for agricultural management on the build of SOC under warm climate conditions by understanding microbial metabolism and necromass response to N fertilization. In contrast to the general opinion, the addition of nutrients or easily degradable C can also increase soil organic matter in the long-term, which is critically important for predicting soil C pool dynamics and promoting the C sequestration in response to climate change. In agricultural management, it is a long-term process for straw and other refractory C sources to form soil organic matter components. Integrating the continual microbial transformation of organic C from labile to persistent anabolic forms is practical for SOC storage in agricultural ecosystem.
Supplementary information
Acknowledgements
The authors are grateful to the Editor and anonymous referees whose constructive comments substantially improved the manuscript. We would like to thank Professer Xudong Zhang for helping the amino sugar experiment, and Dr. Anning Zhu, Jianbo Fan in Fengqiu and Yingtan Research Stations for experiment management and sampling assistance. This study was supported by National Natural Science Foundation of China (41530856), Strategic Priority Research Program of the Chinese Academy of Sciences (XDA24020104), National Natural Science Foundation of China (41622104, 41877060), Scholar Program of the Jiangsu Province (BRA2019333), Youth Innovation Promotion Association of Chinese Academy of Sciences (2016284) and Top‐Notch Young Talents Program of China (W03070089).
Author contributions
All authors contributed intellectual input and assistance to this study and manuscript. B.S. and Y.L. developed the original framework. Y.S. contributed field experiment maintenance and sampling. X.J. and H.N. contributed to single-cell Raman spectroscopy analysis. Y.L., H.N., X.X., N.Z., and X.W. contributed experiment and data analysis. Y.L. and H.N. wrote the paper.
Data availability
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number CRA001931, which are publicly accessible at http://bigd.big.ac.cn/gsa.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-00950-w.
References
- 1.Sakschewski B, von Bloh W, Boit A, Poorter L, Peña-Claros M, Heinke J, et al. Resilience of Amazon forests emerges from plant trait diversity. Nat Clim Change. 2016;6:1032–6. doi: 10.1038/nclimate3109. [DOI] [Google Scholar]
- 2.Gruber N, Galloway JN. An Earth-system perspective of the global nitrogen cycle. Nature. 2008;451:293–6. doi: 10.1038/nature06592. [DOI] [PubMed] [Google Scholar]
- 3.Kicklighter DW, Melillo JM, Monier E, Sokolov AP, Zhuang Q. Future nitrogen availability and its effect on carbon sequestration in Northern Eurasia. Nat Commun. 2019;10:1–19. doi: 10.1038/s41467-019-10944-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L, Greaver TL. A global perspective on belowground carbon dynamics under nitrogen enrichment. Ecol Lett. 2010;13:819–28. doi: 10.1111/j.1461-0248.2010.01482.x. [DOI] [PubMed] [Google Scholar]
- 5.Maaroufi NI, Nordin A, Hasselquist NJ, Bach LH, Palmqvist K, Gundale MJ. Anthropogenic nitrogen deposition enhances carbon sequestration in boreal soils. Glob Chang. Glob Change Biol. 2015;21:3169–80. doi: 10.1111/gcb.12904. [DOI] [PubMed] [Google Scholar]
- 6.Lu M, Zhou X, Luo Y, Yang Y, Fang C, Chen J, et al. Minor stimulation of soil carbon storage by nitrogen addition: a meta-analysis. Agr Ecosyst Environ. 2011;140:234–44. doi: 10.1016/j.agee.2010.12.010. [DOI] [Google Scholar]
- 7.Chen J, Luo Y, van Groenigen KJ, Hungate BA, Cao J, Zhou X, et al. A keystone microbial enzyme for nitrogen control of soil carbon storage. Sci Adv. 2018;4:q1689. doi: 10.1126/sciadv.aaq1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cusack DF, Torn MS, Mcdowell WH, Silver WL. The response of heterotrophic activity and carbon cycling to nitrogen additions and warming in two tropical soils. Glob Change Biol. 2010;16:2555–72. [Google Scholar]
- 9.Zhang J, Balkovic J, Azevedo LB, Skalsky R, Bouwman AF, Xu G, et al. Analyzing and modelling the effect of long-term fertilizer management on crop yield and soil organic carbon in China. Sci Total Environ. 2018;627:361–72. doi: 10.1016/j.scitotenv.2018.01.090. [DOI] [PubMed] [Google Scholar]
- 10.Bending GD, Putland C, Rayns F. Changes in microbial community metabolism and labile organic matter fractions as early indicators of the impact of management on soil biological quality. Biol Fertil Soils. 2000;31:78–84. doi: 10.1007/s003740050627. [DOI] [Google Scholar]
- 11.Kallenbach CM, Frey SD, Grandy AS. Direct evidence for microbial-derived soil organic matter formation and its ecophysiological controls. Nat Commun. 2016;7:13630. doi: 10.1038/ncomms13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leinweber P, Jandl G, Baum C, Eckhardt K, Kandeler E. Stability and composition of soil organic matter control respiration and soil enzyme activities. Soil Biol Biochem. 2008;40:1496–505. doi: 10.1016/j.soilbio.2008.01.003. [DOI] [Google Scholar]
- 13.Roth V, Lange M, Simon C, Hertkorn N, Bucher S, Goodall T, et al. Persistence of dissolved organic matter explained by molecular changes during its passage through soil. Nat Geosci. 2019;12:755–61. doi: 10.1038/s41561-019-0417-4. [DOI] [Google Scholar]
- 14.Kallenbach CM, Grandy AS, Frey SD, Diefendorf AF. Microbial physiology and necromass regulate agricultural soil carbon accumulation. Soil Biol Biochem. 2015;91:279–90. doi: 10.1016/j.soilbio.2015.09.005. [DOI] [Google Scholar]
- 15.Chen L, Liu L, Qin S, Yang G, Fang K, Zhu B, et al. Regulation of priming effect by soil organic matter stability over a broad geographic scale. Nat Commun. 2019;10:5112. doi: 10.1038/s41467-019-13119-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sollins P, Kramer MG, Swanston C, Lajtha K, Filley T, Aufdenkampe AK, et al. Sequential density fractionation across soils of contrasting mineralogy: evidence for both microbial- and mineral-controlled soil organic matter stabilization. Biogeochemistry. 2009;96:209–31. doi: 10.1007/s10533-009-9359-z. [DOI] [Google Scholar]
- 17.Liang C, Schimel JP, Jastrow JD. The importance of anabolism in microbial control over soil carbon storage. Nat Microbiol. 2017;2:17105. doi: 10.1038/nmicrobiol.2017.105. [DOI] [PubMed] [Google Scholar]
- 18.Ding X, Chen S, Zhang B, Liang C, He H, Horwath WR. Warming increases microbial residue contribution to soil organic carbon in an alpine meadow. Soil Biol Biochem. 2019;135:13–9. doi: 10.1016/j.soilbio.2019.04.004. [DOI] [Google Scholar]
- 19.Ni X, Liao S, Tan S, Peng Y, Wang D, Yue K, et al. The vertical distribution and control of microbial necromass carbon in forest soils. Glob Ecol Biogeogr. 2020;29:1829–39. doi: 10.1111/geb.13159. [DOI] [Google Scholar]
- 20.Shao P, Lynch L, Xie H, Bao X, Liang C. Tradeoffs among microbial life history strategies influence the fate of microbial residues in subtropical forest soils. Soil Biol Biochem. 2021;153:108112. doi: 10.1016/j.soilbio.2020.108112. [DOI] [Google Scholar]
- 21.Ma T, Zhu S, Wang Z, Chen D, Dai G, Feng B, et al. Divergent accumulation of microbial necromass and plant lignin components in grassland soils. Nat Commun. 2018;9:3480. doi: 10.1038/s41467-018-05891-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malik AA, Puissant J, Buckeridge KM, Goodall T, Jehmlich N, Chowdhury S, et al. Land use driven change in soil pH affects microbial carbon cycling processes. Nat Commun. 2018;9:3591. doi: 10.1038/s41467-018-05980-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nocker A, Sossa-Fernandez P, Burr MD, Camper AK. Use of propidium monoazide for live/dead distinction in microbial ecology. Appl Environ Microbiol. 2007;73:5111–7. doi: 10.1128/AEM.02987-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castro HF, Classen AT, Austin EE, Norby RJ, Schadt CW. Soil microbial community responses to multiple experimental climate change drivers. Appl Environ Microbiol. 2010;76:999–1007. doi: 10.1128/AEM.02874-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rinnan R, MichelsenI A, Bååth E, Jonasson S. Fifteen years of climate change manipulations alter soil microbial communities in a subarctic heath ecosystem. Glob Change Biol. 2007;13:28–39. doi: 10.1111/j.1365-2486.2006.01263.x. [DOI] [Google Scholar]
- 26.Liang Y, Xiao X, Nuccio EE, Yuan M, Zhang N, Xue K, et al. Differentiation strategies of soil rare and abundant microbial taxa in response to changing climatic regimes. Environ Microbiol. 2020;22:1327–40. doi: 10.1111/1462-2920.14945. [DOI] [PubMed] [Google Scholar]
- 27.Wang M, Ding J, Sun B, Zhang J, Wyckoff KN, Yue H, et al. Microbial responses to inorganic nutrient amendment overridden by warming: consequences on soil carbon stability. Environ Microbiol. 2018;20:2509–22. doi: 10.1111/1462-2920.14239. [DOI] [PubMed] [Google Scholar]
- 28.Zhao M, Xue K, Wang F, Liu S, Bai S, Sun B, et al. Microbial mediation of biogeochemical cycles revealed by simulation of global changes with soil transplant and cropping. ISME J. 2014;8:2045–55. doi: 10.1038/ismej.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang Y, Jiang Y, Wang F, Wen C, Deng Y, Xue K, et al. Long-term soil transplant simulating climate change with latitude significantly alters microbial temporal turnover. ISME J. 2015;9:2561–72. doi: 10.1038/ismej.2015.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun B, Wang F, Jiang Y, Li Y, Dong Z, Li Z, et al. A long-term field experiment of soil transplantation demonstrating the role of contemporary geographic separation in shaping soil microbial community structure. Ecol Evol. 2014;4:1073–87. doi: 10.1002/ece3.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez I, Álvarez-González JG, Carrasco B, Ruíz-González AD, Cabaneiro A. Post-thinning soil organic matter evolution and soil CO2 effluxes in temperate radiata pine plantations: impacts of moderate thinning regimes on the forest C cycle. Can J Res. 2012;42:1953–64. doi: 10.1139/x2012-137. [DOI] [Google Scholar]
- 32.Baldock JA, Oades JM, Waters AG, Peng X, Vassallo AM, Wilson MA. Aspects of the chemical structure of soil organic materials as revealed by solid-state 13C NMR spectroscopy. Biogeochemistry. 1992;16:1–42. doi: 10.1007/BF02402261. [DOI] [Google Scholar]
- 33.Coxall HK, Wilson PA, Pälike H, Lear CH, Backman J. Rapid stepwise onset of Antarctic glaciation and deeper calcite compensation in the Pacific Ocean. Nature. 2005;433:53–7. doi: 10.1038/nature03135. [DOI] [PubMed] [Google Scholar]
- 34.Carini P, Marsden PJ, Leff JW, Morgan EE, Strickland MS, Fierer N, et al. is abundant in soil and obscures estimates of soil microbial diversity. Nat Microbiol. 2016;2:16242. doi: 10.1038/nmicrobiol.2016.242. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Bi Q, Yang K, Zheng B, Pu Q, Cui L. D2O-isotope-labeling approach to probing phosphate-solubilizing bacteria in complex soil communities by single-cell raman spectroscopy. Anal Chem. 2019;91:2239–46. doi: 10.1021/acs.analchem.8b04820. [DOI] [PubMed] [Google Scholar]
- 36.Cui L, Butler HJ, Martin-Hirsch PL, Martin FL. Aluminium foil as a potential substrate for ATR-FTIR, transflection FTIR or Raman spectrochemical analysis of biological specimens. Anal Methods. 2016;8:481–7. doi: 10.1039/C5AY02638E. [DOI] [Google Scholar]
- 37.Bååth E, Pettersson M, Söderberg KH. Adaptation of a rapid and economical microcentrifugation method to measure thymidine and leucine incorporation by soil bacteria. Soil Biol Biochem. 2001;33:1571–4. doi: 10.1016/S0038-0717(01)00073-6. [DOI] [Google Scholar]
- 38.Trivedi P, Delgado-Baquerizo M, Trivedi C, Hu H, Anderson IC, Jeffries TC, et al. Microbial regulation of the soil carbon cycle: evidence from gene-enzyme relationships. ISME J. 2016;10:2593–604. doi: 10.1038/ismej.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Amelung W. Gas chromatographic determination of muramic acid, glucosamine, mannosamine, and galactosamine in soils. Soil Biol Biochem. 1996;28:1201–6. doi: 10.1016/0038-0717(96)00117-4. [DOI] [Google Scholar]
- 40.Schermelleh-Engel K, Moosbrugger H. Evaluating the fit of structural equation models: tests of significance and descriptive goodness-of-fit measures. Methods Psychological Res Online. 2003;8:23–74. [Google Scholar]
- 41.Clarke KR. Non-parametric multivariate analyses of changes in community structure. Austral Ecol. 1993;18:117–43. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 42.Kruskal JB. Nonmetric multidimensional scaling: a numerical method. Psychometrika. 1964;29:115–29. doi: 10.1007/BF02289694. [DOI] [Google Scholar]
- 43.Legendre P, Legendre L. Numerical Ecology, 3rd English Edition. Oxford, UK: Elsevier; 2012.
- 44.Liaw A, Wiener M. Classification and regression by randomForest. R N. 2002;2:18–22. [Google Scholar]
- 45.Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. 2012;8:e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu L, Ning D, Zhang B, Li Y, Zhang P, Shan X, et al. Global diversity and biogeography of bacterial communities in wastewater treatment plants. Nat Microbiol. 2019;4:1183–95. doi: 10.1038/s41564-019-0426-5. [DOI] [PubMed] [Google Scholar]
- 47.Crowther TW, Todd-Brown KEO, Rowe CW, Wieder WR, Carey JC, Machmuller MB, et al. Quantifying global soil carbon losses in response to warming. Nature. 2016;540:104–8. doi: 10.1038/nature20150. [DOI] [PubMed] [Google Scholar]
- 48.Hicks Pries CE, Castanha C, Porras RC, Torn MS. The whole-soil carbon flux in response to warming. Science. 2017;355:1420–3. doi: 10.1126/science.aal1319. [DOI] [PubMed] [Google Scholar]
- 49.Peñuelas J, Ciais P, Canadell JG, Janssens IA, Fernández-Martínez M, Carnicer J, et al. Shifting from a fertilization-dominated to a warming-dominated period. Nat Ecol Evol. 2017;1:1438–45. doi: 10.1038/s41559-017-0274-8. [DOI] [PubMed] [Google Scholar]
- 50.Sokol NW, Bradford MA. Microbial formation of stable soil carbon is more efficient from belowground than aboveground input. Nat Geosci. 2019;12:46–53. doi: 10.1038/s41561-018-0258-6. [DOI] [Google Scholar]
- 51.Trumbore SE, Chadwick OA, Amundson R. Rapid exchange between soil carbon and atmospheric carbon dioxide driven by temperature change. Science. 1996;272:393–6. doi: 10.1126/science.272.5260.393. [DOI] [Google Scholar]
- 52.Liang C, Cheng G, Wixon DL, Balser TC. An Absorbing Markov Chain approach to understanding the microbial role in soil carbon stabilization. Biogeochemistry. 2011;106:303–9. doi: 10.1007/s10533-010-9525-3. [DOI] [Google Scholar]
- 53.Liang C, Amelung W, Lehmann J, Kästner M. Quantitative assessment of microbial necromass contribution to soil organic matter. Glob Change Biol. 2019;00:1–13. doi: 10.1111/gcb.14781. [DOI] [PubMed] [Google Scholar]
- 54.Tian J, Zong N, Hartley IP. Microbial metabolic response to winter warming stabilizes soil carbon. Glob Change Biol. 2021;00:1–18. doi: 10.1111/gcb.15538. [DOI] [PubMed] [Google Scholar]
- 55.Coxall HK, Wilson PA, Pälike H, Lear CH, Backman J. Rapid stepwise onset of Antarctic glaciation and deeper calcite compensation in the Pacific Ocean. Nature. 2005;433:53–7. doi: 10.1038/nature03135. [DOI] [PubMed] [Google Scholar]
- 56.Ding X, Chen S, Zhang B, He H, Filley TR, Horwath WR. Warming yields distinct accumulation patterns of microbial residues in dry and wet alpine grasslands on the Qinghai-Tibetan Plateau. Biol Fertil Soils. 2020;56:881–92. doi: 10.1007/s00374-020-01474-9. [DOI] [Google Scholar]
- 57.Jia J, Feng X, He J, He H, Lin L, Liu Z. Comparing microbial carbon sequestration and priming in the subsoil versus topsoil of a Qinghai-Tibetan alpine grassland. Soil Biol Biochem. 2017;104:141–51. doi: 10.1016/j.soilbio.2016.10.018. [DOI] [Google Scholar]
- 58.Zhou X, Chen C, Wang Y, Xu Z, Duan J, Hao Y, et al. Soil extractable carbon and nitrogen, microbial biomass and microbial metabolic activity in response to warming and increased precipitation in a semiarid Inner Mongolian grassland. Geoderma. 2013;206:24–31. doi: 10.1016/j.geoderma.2013.04.020. [DOI] [Google Scholar]
- 59.Chen J, Ji C, Fang J, He H, Zhu B. Dynamics of microbial residues control the responses of mineral-associated soil organic carbon to N addition in two temperate forests. Sci Total Environ. 2020;748:141318. doi: 10.1016/j.scitotenv.2020.141318. [DOI] [PubMed] [Google Scholar]
- 60.Fontaine S, Barot S, Barré P, Bdioui N, Mary B, Rumpel C. Stability of organic carbon in deep soil layers controlled by fresh carbon supply. Nature. 2007;450:277–80. doi: 10.1038/nature06275. [DOI] [PubMed] [Google Scholar]
- 61.Wang C, Wang X, Pei G, Xia Z, Peng B, Sun L, et al. Stabilization of microbial residues in soil organic matter after two years of decomposition. Soil Biol Biochem. 2020;141:107687. doi: 10.1016/j.soilbio.2019.107687. [DOI] [Google Scholar]
- 62.Zhou J, Xue K, Xie J, Deng Y, Wu L, Cheng X, et al. Microbial mediation of carbon-cycle feedbacks to climate warming. Nat Clim Change. 2011;2:106–10. doi: 10.1038/nclimate1331. [DOI] [Google Scholar]
- 63.Chen R, Senbayram M, Blagodatsky S, Myachina O, Dittert K, Lin X, et al. Soil C and N availability determine the priming effect: microbial N mining and stoichiometric decomposition theories. Glob Change Biol. 2014;20:2356–67. doi: 10.1111/gcb.12475. [DOI] [PubMed] [Google Scholar]
- 64.Fontaine S, Henault C, Aamor A, Bdioui N, Bloor JMG, Maire V, et al. Fungi mediate long term sequestration of carbon and nitrogen in soil through their priming effect. Soil Biol Biochem. 2011;43:86–96. doi: 10.1016/j.soilbio.2010.09.017. [DOI] [Google Scholar]
- 65.Jones DL, Nguyen C, Finlay RD. Carbon flow in the rhizosphere: carbon trading at the soil–root interface. Plant Soil. 2009;321:5–33. doi: 10.1007/s11104-009-9925-0. [DOI] [Google Scholar]
- 66.Zang H, Wang J, Kuzyakov Y. N fertilization decreases soil organic matter decomposition in the rhizosphere. Appl Soil Ecol. 2016;108:47–53. doi: 10.1016/j.apsoil.2016.07.021. [DOI] [Google Scholar]
- 67.Mason-Jones K, Schmücker N, Kuzyakov Y. Contrasting effects of organic and mineral nitrogen challenge the N-Mining Hypothesis for soil organic matter priming. Soil Biol Biochem. 2018;124:38–46. doi: 10.1016/j.soilbio.2018.05.024. [DOI] [Google Scholar]
- 68.Carreiro MM, Sinsabaugh RL, Repert DA, Parkhurst DF. Microbial enzyme shifts explain litter decay responses to simulated nitrogen deposition. Ecology. 2000;81:2359–65. doi: 10.1890/0012-9658(2000)081[2359:MESELD]2.0.CO;2. [DOI] [Google Scholar]
- 69.Sinsabaugh RL, Hill BH, Follstad Shah JJ. Ecoenzymatic stoichiometry of microbial organic nutrient acquisition in soil and sediment. Nature. 2009;462:795–98. doi: 10.1038/nature08632. [DOI] [PubMed] [Google Scholar]
- 70.Burns RG, DeForest JL, Marxsen J, Sinsabaugh RL, Stromberger ME, Wallenstein MD, et al. Soil enzymes in a changing environment: current knowledge and future directions. Soil Biol Biochem. 2013;58:216–34. doi: 10.1016/j.soilbio.2012.11.009. [DOI] [Google Scholar]
- 71.Tian D, Niu S. A global analysis of soil acidification caused by nitrogen addition. Environ Res Lett. 2015;10:24019. doi: 10.1088/1748-9326/10/2/024019. [DOI] [Google Scholar]
- 72.Zhou Z, Wang C, Zheng M, Jiang L, Luo Y. Patterns and mechanisms of responses by soil microbial communities to nitrogen addition. Soil Biol Biochem. 2017;115:433–41. doi: 10.1016/j.soilbio.2017.09.015. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number CRA001931, which are publicly accessible at http://bigd.big.ac.cn/gsa.