Abstract
Candidatus phylum Eremiobacterota (formerly WPS-2) is an as-yet-uncultured bacterial clade that takes its name from Ca. Eremiobacter, an Antarctic soil aerobe proposed to be capable of a novel form of chemolithoautotrophy termed atmospheric chemosynthesis, that uses the energy derived from atmospheric H2-oxidation to fix CO2 through the Calvin-Benson-Bassham (CBB) cycle via type 1E RuBisCO. To elucidate the phylogenetic affiliation and metabolic capacities of Ca. Eremiobacterota, we analysed 63 public metagenome-assembled genomes (MAGs) and nine new MAGs generated from Antarctic soil metagenomes. These MAGs represent both recognized classes within Ca. Eremiobacterota, namely Ca. Eremiobacteria and UBP9. Ca. Eremiobacteria are inferred to be facultatively acidophilic with a preference for peptides and amino acids as nutrient sources. Epifluorescence microscopy revealed Ca. Eremiobacteria cells from Antarctica desert soil to be coccoid in shape. Two orders are recognized within class Ca. Eremiobacteria: Ca. Eremiobacterales and Ca. Baltobacterales. The latter are metabolically versatile, with individual members having genes required for trace gas driven autotrophy, anoxygenic photosynthesis, CO oxidation, and anaerobic respiration. UBP9, here renamed Ca. Xenobia class. nov., are inferred to be obligate heterotrophs with acidophilic adaptations, but individual members having highly divergent metabolic capacities compared to Ca. Eremiobacteria, especially with regard to respiration and central carbon metabolism. We conclude Ca. Eremiobacterota to be an ecologically versatile phylum with the potential to thrive under an array of “extreme” environmental conditions.
Subject terms: Soil microbiology, Microbial ecology
Introduction
Candidatus phylum Eremiobacterota is an as-yet-uncultured bacterial lineage, with the type genus Ca. Eremiobacter based on a metagenome-assembled genome (MAG) from Antarctic soil [1]. Ca. Eremiobacter contains genes encoding ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) type 1E and high-affinity Group 1h [NiFe]- hydrogenase genes [2, 3]. Together, these genes are indicative of a novel form of chemolithoautotrophy called atmospheric chemosynthesis [4, 5]. Discovered in cold desert soils of Antarctica, bacteria genetically capable of this process use high-affinity Group 1h [NiFe]-hydrogenases to oxidize trace levels of hydrogen gas at below atmospheric levels [1, 6]. The energy and reductant derived from trace gas oxidation allow these chemolithoautotrophs to fix CO2 through the Calvin–Benson–Bassham (CBB) cycle [1, 2]. This process is one strategy for survival of bacteria in nutrient-poor desert soils [5], and is a major advance in our understanding of the potential ecological importance of members of Ca. Eremiobacterota. However, Ca. Eremiobacter is just one example of the potential ecological and metabolic diversity of the entire phylum Ca. Eremiobacterota.
The first 16S rRNA gene sequence of Ca. Eremiobacterota (originally candidate phylum WPS-2) was identified from a soil contaminated with polychlorinated biphenyl, with the designation of WPS-2 derived from “Wittenberg-polluted soil” [7]. As part of the rare biosphere (relative abundances usually <0.1%) [8, 9], growing evidence suggests a global distribution of Ca. Eremiobacterota, with 16S rRNA gene sequences reported from diverse terrestrial environments. Improved sequencing efforts have led to Ca. Eremiobacterota 16S rRNA gene sequences being recovered from environmental and animal sources: peatlands [9]; permafrost [10]; mosses in boreal forests [11, 12]; acidic, polluted soil [7]; bare, unvegetated soil [13]; canine and human oral microbiomes [14, 15]; fecal samples [16]; industrial waste processes [16]; and Antarctic desert soils [17–19].
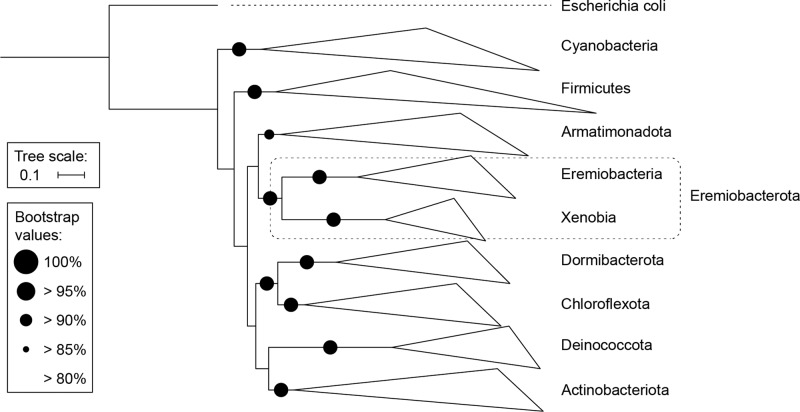
Whole-genome phylogenetic analysis has revealed phylum Ca. Eremiobacterota to be closest to the phyla Armatimonadota and Chloroflexota [1, 16]. Ca. Eremiobacterota is composed of two class-level lineages, Ca. Eremiobacteria and UBP9 (formerly SHA-109) [12, 16]. In recent years, Ca. Eremiobacteria MAGs have been extensively recovered from natural terrestrial environments [1, 10, 11]; UBP9 MAGs have been recovered from baboon feces and industrial waste [16]. Ca. Eremiobacteria is composed of two candidate orders, Ca. Eremiobacterales (type genus Ca. Eremiobacter) [1] and Ca. Baltobacterales (formerly UBP12) [12]. Ca. Baltobacterales MAGs have originated from forests and peatlands, with certain members inferred to be facultative aerobes capable of bacteriochlorophyll-based anoxygenic photosynthesis [12], whereas others were predicted to be aerobic organoheterotrophs [1, 13].
Due to the phylogenetic diversity, potential metabolic disparity, and ecological importance of Ca. Eremiobacterota, we wish to elucidate the phylogeny, environmental distribution, and ecological roles of this clade. To do so, we performed a meta-analysis by collecting 16S rRNA gene amplicon sequencing data together with environmental metadata from cold and temperate terrestrial environments. We also comparatively analyzed 72 Ca. Eremiobacterota MAGs (including UBP9) to assess the metabolic potential of this clade. Finally, we used catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH) to elucidate the morphology of members of Ca. Eremiobacteria inhabiting Antarctic desert soil.
Materials and methods
16S rRNA gene sequence diversity, associated metadata, and multivariate analysis
Australian temperate and polar soil data were obtained from the Biome of Australia Soil Environments (BASE) project [19], and the Polar Soil Archive (PSA) biodiversity survey [20]. The former contains amplicon sequencing data across a range of terrestrial ecosystems within Australia (1641 samples, accessed on 15 November 2018), while the latter contains data spanning 223 soils from eight locations across the High Arctic and East Antarctica. The 16S rRNA gene sequence fragment in both datasets were amplified using primers 27F and 519R [19, 21]. The PCR amplicon in the BASE dataset was sequenced on either Illumina or the 454 platforms [19], while the latter was sequenced on the 454 platform only [22]. The raw sequences for the two datasets were processed using the Mothur pipeline and OTUs were identified at 96% or 97% sequence identity using UCLUST, respectively. Both datasets were not subsampled, as the read distribution was uneven across samples due to the mixed usage of Illumina and 454 sequencing platforms. Analysis has been performed previously that compared a subsampled and non-subsampled version of the dataset [22]. The results suggested that subsampling had no significant effect on the outcome of the diversity analysis and we therefore elected to leave the dataset intact. Both datasets contain comprehensive soil physicochemical data, comprising 64 measured chemicals including pH, moisture, total carbon, nitrogen, and phosphorus, and 67 microclimate factors.
Random Forest modeling was used to estimate the significance of soil physicochemical factors on the number of Ca. Eremiobacteria operational taxonomic units (OTUs) recovered (richness) and their relative abundance in Australian and polar soils datasets, separately. The importance of each predictor variable was determined by evaluating the decrease in prediction accuracy averaged over the forest (5000 trees), using the rfPermute package [23] under the R-environment. The significance of the model and the cross-validated R2 were assessed with 1000 permutations of each predictor. The influence (promoting or inhibiting) of these predictors was assessed based on Pearson correlation analysis using SPSS (version 22; IBM, 2013). The relationships between pH and Ca. Eremiobacteria were visualized using Origin (version b9.5.1.195; OriginLab Corporation, 2018).
Sequencing, metagenome assembly, and binning
Three Antarctic soil samples were collected from Mitchell Peninsula in Eastern Antarctica [20]. DNA extraction and shotgun sequencing were performed as described [1]. Raw low-quality reads were identified and processed with Trimmomatic (ver. 0.36) for adapter removal and quality filtering [24]. Quality-controlled reads were then assembled using MetaSPAdes (ver. 3.11.1) with default parameters [25]. Contigs whose length was <2000 bp were removed using BBMap (ver. 36.92) [26]. All samples were also co-assembled by combining their respective quality-controlled reads and repeating the assembly and contig filtering steps. Quality-controlled reads for each sample, or combined samples, were mapped onto their respective (co-)assemblies using BamM “make” (ver 1.73, M. Imelfort, unpublished, https://github.com/minillinim/BamM). Assemblies were binned by providing the contigs for each and mapped reads (as BAM files) as input to UniteM (ver. 0.0.15, D. Parks, unpublished, https://github.com/dparks1134/UniteM) and using GroopM (ver. 2.0.0) [27], Maxbin (ver. 2.2.4) [28], MetaBAT (ver. 0.32.5) [29], and MetaBAT2 (ver. 2.12.1) binning methods [27, 29–31].
Ca. Eremiobacterota MAGs retrieval, taxonomic classification, and analysis
Sixty-three MAGs classified as Ca. Eremiobacterota were retrieved from previous studies (Supplementary Table S1) [1, 10, 16]. Taxonomic assignment of MAGs, including identification of novel taxa, was performed using the Genome Taxonomy Database Toolkit (GTDB-Tk) (ver. 0.3.2; with reference to GTDB R04-R89), which provides an objective taxonomic classification of prokaryote genomes (including MAGs) by placing them into concatenated protein reference trees, using relative evolutionary divergence and average nucleotide identity to establish taxonomic ranks [32, 33]. MAG completeness and contamination were evaluated on the 63 retrieved MAGS and nine new MAGs using CheckM (ver. 1.0.12) [34]. Novel Candidatus taxa were named according to the proposed genomic standards [35]. Open reading frame (ORF) and gene functional prediction were performed using Prokka [36], with the annotations of all proteins discussed here manually confirmed using ExPASY BLAST, including RuBisCO large subunit (CbbL), based on BLAST searching against previously reported sequences [1, 37, 38]. Potential carbon monoxide dehydrogenase (CODH) large subunit and CbbL amino acid sequences were extracted from the respective genomes and aligned using MAFFT together with their respective reference sequences. Phylogenetic trees were generated using IQ-Tree with an approximate-maximum likelihood method and visualized using iTOL [39]. Genome analysis methods including CAZy (for carbohydrate-active enzymes) and genome-based phylogenetics are described in Supplementary Methods. Protein sequences identified as hydrogenases based on catalytic domains were classified further using the hydrogenase classifier HydDB [40].
Ca. Eremiobacteria fluorescence in situ hybridization (FISH) probe design
Near-complete SSU RNA gene sequences (>1300 bp) classified as Ca. Eremiobacteria were retrieved from the SILVA databases (n = 185) [41]. The retrieved sequences were used to design a class-level Ca. Eremiobacteria-specific FISH probe (Erem-289-Cy3, 5′- TCGCTCTCTCAAACCAGC[CY3]-3′) using the ARB probe design function [42], ensuring the site of hybridization was carefully selected based on the SSU rRNA accessibility map [43]. The specificity of the Ca. Eremiobacteria probe was tested in silico using testprobe 3.0 against the nonredundant SILVA Reference dataset [41]. Due to the lack of pure or enriched Ca. Eremiobacteria culture for use as positive controls, the Erem-289 FISH probe was optimized and validated using clone-FISH [44], with further details on the design of positive and negative control clones provided in the Supplementary Methods, Supplementary Fig. S1, and Supplementary Table S2.
CARD-FISH
Bacterial cells were extracted from 0.25 g of soil collected from Mitchell Peninsula using a Nycodenz density gradient medium, in triplicate [45] (see Supplementary Methods). Isolated bacterial cells were filtered onto a 0.2-µm polycarbonate membrane (Millipore, Australia) using a vacuum manifold and washed twice with 100-µL sterile Milli Q water [46]. The membranes were embedded in 0.1% (w/v) low melting temperature agarose and cells fixed with 100 µL of 4% paraformaldehyde at 4 °C for 24 h, followed by a second fixation step in 100-µL ethanol and PBS (1:1) at 4 °C for 24 h. Hybridization was performed overnight as described [46]. The CARD reaction was performed by incubation of the membrane in 100 µL of the tyramide-Cy3 working solution from the TSA Plus Fluorescence Kit (Perkin Elmer, Melbourne, Australia), for 10 min at RT. Membranes were then washed in 100 µL of 1x PBS, followed by 100-µl sterile water (1 min), before dehydration in 100-µL 96% ethanol, and counterstaining with 4′,6-diamidino-2-phenylindol (DAPI; Thermo Fisher, Australia) at a concentration of 2 µg/mL. Cells were visualized using a BX61 motorized epifluorescence microscope with an LED light source and a DP71 digital camera (Olympus), using appropriate filters for the emission of Cy3 (excitation at 531 nm, emission at 593 nm), Cy5 (excitation at 628 nm, emission at 692 nm), and DAPI (excitation at 377 nm, emission at 447 nm). Soil samples, positive and negative clone-FISH controls were prepared and analysed in triplicate (Supplementary Information). In total, 65 Cy3 and DAPI-positive cells were identified between three replicate soil samples.
Results and discussion
Ca. Eremiobacterota: a taxonomic and phylogenetic overview
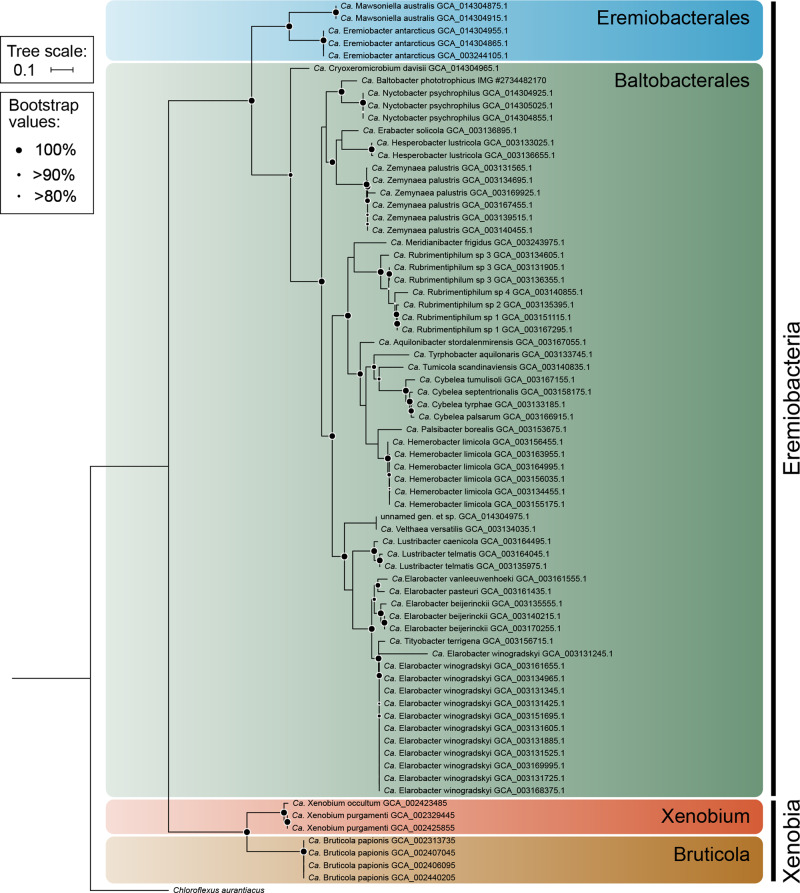
Phylogenetic analysis of MAGs (Figs. 1 and 2) and 16S rRNA gene sequences (Supplementary Fig. S2 and Supplementary Table S3) support the subdivision of Ca. Eremiobacterota into two class-level lineages, Ca. Eremiobacteria and UBP9. Within Ca. Eremiobacteria, two order-level clades were resolved, corresponding to Ca. Eremiobacterales and Ca. Baltobacterales [1, 12], each of which contained a single family-level clade, Ca. Eremiobacteraceae fam. nov. and Ca. Baltobacteraceae, respectively (Fig. 2 and Supplementary Table S1). Ca. Eremiobacterales include Ca. Eremiobacter and Ca. Mawsoniella gen. nov. (Table 1 and Supplementary Table S1). Ca. Baltobacterales include the type genus Ca. Baltobacter, which was inferred to be capable of both heterotrophy and anoxygenic photosynthesis [12]; and Ca. Rubrimentiphilum, which was predicted to be an obligate heterotroph (Fig. 2 and Table 1) [13]. An additional 59 Ca. Eremiobacteria MAGs were found to be representatives of Ca. Baltobacterales [10]; based on GTDB taxonomy and phylogenetic analysis, these were resolved into 16 novel genus-level taxa (Fig. 2, Table 1, and Supplementary Table S1). Of these, Ca. Cryoxeromicrobium gen. nov. is resolved as the most basal known genus within Ca. Baltobacterales, and Ca. Nyctobacter gen. nov. is the sister taxon of Ca. Baltobacter (Fig. 2). These genera and Ca. Erabacter gen. nov., Ca. Hesperobacter gen. nov., and Ca. Zemynaea gen. nov. form a single cluster within Ca. Baltobacterales. A second Ca. Baltobacterales cluster includes Ca. Rubrimentiphilum and Ca. Meridianibacter gen. nov. (Bin 23) [1, 13]. A third cluster comprises five genera (Ca. Aquilonibacter gen. nov., Ca. Tyrphobacter gen. nov., Ca. Tumulicola gen. nov., Ca. Cybelea gen. nov., Ca. Palsibacter gen. nov., Ca. Hemerobacter gen. nov.). The fourth Ca. Baltobacterales cluster comprises four genera (Ca. Velthaea gen. nov.; Ca. Lustribacter gen. nov.; Ca. Elarobacter gen. nov.; Ca. Tityobacter gen. nov.) and tended to contain the largest genomes within Ca. Eremiobacteria (~4.5–5 Mbp) (Table 1 and Supplementary Table S1).
Fig. 1. Phylogenetic tree based on 15 concatenated marker protein sequences, which demonstrates the relationship of Candidatus Eremiobacterota to closely related bacterial phyla.
Escherichia coli (Proteobacteria) was set as the outgroup.
Fig. 2. Phylogenetic tree of Candidatus Eremiobacterota based on 15 concatenated ribosomal protein sequences.
Phylum Ca. Eremiobacterota is composed of two classes, Ca. Eremiobacteria and Ca. Xenobia class. nov. Ca. Eremiobacteria contain two orders, Ca. Eremiobacterales and Ca. Baltobacterales. Ca. Eremiobacter (Ga011786) [1] is the type genus of family Ca. Eremiobacteraceae fam. nov., order Ca. Eremiobacterales class Ca. Eremiobacteria, and phylum Ca. Eremiobacterota. Ca. Baltobacter (WPS2_44; IMG accession #2734482170) is the type genus of family Ca. Baltobacteraceae and order Ca. Baltobacterales [12]. Note that because genome completeness was <70% for GCA_014304975.1, no genus or species name was proposed for this metagenome-assembled genome. Ca. Xenobia class. nov. contains a single order Ca. Xenobiales ord. nov., a single family Ca. Xenobiaceae fam. nov., and two genera Ca. Xenobium gen. nov. and Ca. Bruticola gen. nov. Chloroflexus aurantiacus (Chloroflexota) was set as the outgroup. Information for the MAGs discussed in this study, including sampled location, is presented in Supplementary Table S1.
Table 1.
Predicted physiological and metabolic traits inferred for Candidatus Eremiobacterota genera, for the three known orders Ca. Eremiobacterales, Ca. Baltobacterales, and Ca. Xenobiales ord. nov.
| Candidatus genus | Candidatus species | Source | Physiology and metabolism (inferred from MAGs) | |
|---|---|---|---|---|
| Eremiobacterales |
Eremiobacter Ji et al. [1] |
E. antarcticus sp. nov. | Soil |
Heterotroph and autotroph. Hydrogenotrophic chemolithoautotrophy using Group 1h [NiFe] hydrogenase and CBB cycle; capable of using atmospheric H2. Organic substrates include peptides, amino acids, carboxylates, acetate, sarcosine, formate, methanol, poly- and oligosaccharides, sugars, catechol, 4-hydroxybenzoate. Glycogen storage. |
| Mawsoniella gen. nov. | M. australis sp nov. | Soil |
Obligate heterotroph. Hydrogen oxidation using Group 1h [NiFe] hydrogenase. Organic substrates include peptides, amino acids, carboxylates, sarcosine, oligosaccharides, sugars, catechol, 4-hydroxybenzoate. PHA storage. Polyphosphate storage. |
|
| Baltobacterales | Cryoxeromicrobium gen. nov. | C. davisii sp. nov. | Soil |
Obligate heterotroph. Organic substrates include peptides, amino acids, carboxylates, sarcosine, poly- and oligosaccharides, sugars. Polyphosphate storage. |
| Nyctobacter gen. nov. | N. psychrophilus sp. nov. | Soil |
Heterotroph and autotroph. Hydrogenotrophic chemolithoautotrophy using Group 1h [NiFe] hydrogenase and CBB cycle; capable of using atmospheric H2. Organic substrates include peptides, amino acids, carboxylates, poly- and oligosaccharides, sugars. PHA storage; glycogen storage. Polyphosphate storage. |
|
| Erabacter gen. nov. | E. solicola sp. nov. | Palsa |
Obligate heterotroph. Organic substrates include peptides, amino acids, carboxylates, sarcosine, poly- and oligosaccharides, sugars. PHA storage. |
|
| Hesperobacter gen. nov. | H. lustricola sp. nov | Bog |
Heterotroph and autotroph. Hydrogenotrophic chemolithoautotrophy using Group 1h [NiFe] hydrogenase and CBB cycle; capable of using atmospheric H2. Photoautotrophy; photoreceptors. CO oxidation. Organic substrates include peptides, amino acids, carboxylates, glycerol, taurine, cyanate, acetate, sarcosine, alcohols (including ethanol), poly- and oligosaccharides, sugars, fluoroacetate. PHA storage. Polyphosphate storage. |
|
| Zemynaea gen. nov. | Z. palustris sp. nov. | Palsa |
Obligate heterotroph. Anaerobic respiration: nitrate. Organic substrates include peptides, amino acids, carboxylates, sarcosine, formate, poly- and oligosaccharides, sugars. PHA storage. |
|
| Meridianibacter gen. nov. | M. frigidus sp. nov. | Soil |
Obligate heterotroph. Organic substrates include peptides, amino acids, carboxylates, poly- and oligosaccharides, sugars. Polyphosphate storage. |
|
| Aquilonibacter gen. nov. | A. stordalenmirensis sp. nov. | Palsa |
Obligate heterotroph. CO oxidation. Organic substrates include peptides, amino acids, carboxylates, glycerol, alcohols (including ethanol, PVA), arylsulfates, poly- and oligosaccharides, sugars, fluoroacetate. PHA storage. Polyphosphate storage. Motile by flagella. |
|
| Tyrphobacter gen. nov. | T. aquilonaris sp. nov. | Palsa |
Obligate heterotroph. Organic substrates include peptides, amino acids, carboxylates, methanol, poly- and oligosaccharides, sugars. PHA storage. Polyphosphate storage. |
|
| Tumulicola gen. nov. | T. scandinaviensis sp. nov. | Palsa |
Obligate heterotroph. CO oxidation. Organic substrates include peptides, amino acids, carboxylates, taurine, acetate, sarcosine, arylsulfates, poly- and oligosaccharides, sugars. PHA storage. |
|
| Cybelea gen. nov. | C. septentrionalis sp. nov.; C. tumulisoli sp. nov.; C. tyrphae sp. nov.; C. palsarum sp. nov. | Palsa |
Obligate heterotroph. Hydrogen oxidation using Group 1h [NiFe] hydrogenase (C. septentrionalis only). Organic substrates include peptides, amino acids, carboxylates, taurine, urea, sarcosine, PVA, arylsulfates, poly- and oligosaccharides, sugars, fluoroacetate (substrates vary for individual spp.). PHA storage. Polyphosphate storage. |
|
| Palsibacter gen. nov. | P. borealis sp. nov. | Palsa |
Heterotroph and autotroph. Hydrogenotrophic chemolithoautotrophy using Group 1h [NiFe] hydrogenases and CBB cycle; capable of using atmospheric H2. CO oxidation Organic substrates include peptides, amino acids, carboxylates, glycerol, taurine, urea, cyanate, acetate, sarcosine, alcohols (including methanol, ethanol), arylsulfates, poly- and oligosaccharides, sugars. PHA storage; glycogen storage. Polyphosphate storage. Bidirectional [NiFe] hydrogenase (Group 3d). |
|
| Hemerobacter gen. nov. | H. limicola sp. nov. | Bog |
Heterotroph and autotroph. Hydrogenotrophic chemolithoautotrophy using Group 1f and/or 2a [NiFe] hydrogenase and CBB cycle; capable of using atmospheric H2. CO oxidation. Photoautotrophy; carboxysomes. Anaerobic respiration: nitric oxide. Organic substrates include peptides, amino acids, carboxylates, sarcosine, arylsulfates, poly- and oligosaccharides, sugars. PHA storage. Polyphosphate storage. |
|
| Velthaea gen. nov. | V. versatilis sp. nov. | Bog |
Heterotroph and autotroph. Hydrogenotrophic chemolithoautotrophy using Group 1h [NiFe] hydrogenase and CBB cycle; capable of using atmospheric H2. Photoautotrophy; carboxysomes; photoreceptors. CO oxidation. Anaerobic respiration: sulfoxides. Assimilatory nitrate reduction. Organic substrates include peptides, amino acids, carboxylates, glycerol, taurine, urea, cyanate, sarcosine, alcohols (including methanol, ethanol), poly- and oligosaccharides, sugars, catechol, propane. PHA storage; glycogen storage. Motile by flagella (including phototaxis). Polyphosphate storage. Bidirectional [NiFe] hydrogenase (Group 3b). |
|
| Lustribacter gen. nov. | L. caenicola sp. nov.; L. telmatis sp. nov. | Bog |
Obligate heterotroph. Hydrogen oxidation using Group 1h [NiFe] hydrogenase. Anaerobic respiration: sulfoxides (L. caenicola), urocanate (L. telmatis). Organic substrates include peptides, amino acids, carboxylates, taurine, urea, cyanate, acetate, formate, alcohols (including methanol, ethanol), arylsulfates, alkanesulfonates, oligosaccharides, sugars (substrates vary for individual spp.), fluoroacetate, halobenzoate, phenoxypropionate, ethylbenzene, 4-hydroxybenzoate, 4-sulfocatechol (L. telmatis only). PHA storage; glycogen storage. Motile by flagella. Polyphosphate storage. Bidirectional [NiFe] hydrogenase (Group 3d) (L. caenicola only). |
|
| Elarobacter gen. nov. | E. winogradskyi sp. nov.; E. vanleeuwenhoeki sp. nov.; E. pasteuri sp. nov.; E. beijerinckii sp. nov. | Palsa |
Obligate heterotroph. CO oxidation. Organic substrates include peptides, amino acids, carboxylates, glycerol, urea, acetate, sarcosine, formate, ethanol, oligosaccharides, sugars (substrates vary for individual spp.), fluoroacetate. PHA storage; glycogen storage. Polyphosphate storage. Motile by flagella. |
|
| Tityobacter gen. nov. | T. terrigena sp. nov. | Palsa |
Heterotroph and autotroph. CO oxidation. Hydrogenotrophic chemolithoautotrophy using Group 1h [NiFe] hydrogenase and CBB cycle; capable of using atmospheric H2. Organic substrates include peptides, amino acids, carboxylates, glycerol, taurine, acetate, alcohols (including methanol, ethanol, polyvinyl alcohol), aldehydes, poly- and oligosaccharides, sugars, fluoroacetate, catechol. Glycogen storage. Polyphosphate storage. BMC for sequestering toxic metabolites. |
|
| Xenobiales | Xenobium gen. nov. | X. occultum sp. nov.; X. purgamenti sp. nov. | Tailing pond (X. occultum); palm oil mill effluent and dechlorination bioreactor (X. purgamenti) |
Obligate heterotroph. Microaerobic; anaerobic respiration by DNRA. Fermentative. Organic substrates include peptides, amino acids, carboxylates, glycerol, poly- and oligosaccharides, sugars, 4-hydroxybenzoate. PHA storage; glycogen storage. BMC for sequestering toxic metabolites. Bidirectional Fe-only hydrogenases (for redox balance?, H2 sensor?). |
| Bruticola gen. nov. | B. papionis sp. nov. | Baboon feces |
Obligate heterotroph. Fermentative (no respiration). Organic substrates include peptides, amino acids, citrate, starch, maltodextrin, glucose. Bidirectional Fe-only hydrogenases (for redox balance?). Glycogen storage. |
Full species names, types, and locality data are presented in Supplementary Table S1.
BMC bacterial microcompartment, CBB cycle Calvin–Benson–Bassham cycle, DNRA dissimilatory nitrate reduction to ammonium, MAG metagenome-assembled genome, PHA polyhydroxyalkanoate, RuBisCO ribulose-1,5-bisphosphate carboxylase/oxygenase.
Ca. Eremiobacterota also comprises the candidate class UBP9, the first genomic representatives of clade SHA-109 (Fig. 2 and Supplementary Table S1) [16]. UBP9 is here renamed Ca. Xenobia class. nov. and was determined to include a single order and family, with the type genus Ca. Xenobium gen. nov. recovered from samples sourced from industrial processes, and containing the largest estimated genome size for Ca. Eremiobacterota (~5 Mbp) (Supplementary Table S1). A second genus, Ca. Bruticola gen. nov., which was recovered from baboon fecal samples, has a genome almost half the size of that of Ca. Xenobium, with a much lower % GC content (Ca. Bruticola, ~43–44%; Ca. Xenobium, ~67–68%). The % GC content of the Ca. Xenobium genome is comparable to that of genomes from Ca. Eremiobacteria (~58–77%) (Supplementary Table S1).
Identification of environmental determinants of Ca. Eremiobacteria
Ca. Eremiobacteria 16S rRNA gene sequences were identified in 163 of the 223 samples in the PSA dataset, with an average relative abundance of 5.8% and an average of 16 OTUs identified (Fig. 3 and Supplementary Fig. S3). Two soils containing the most abundant and diverse Ca. Eremiobacteria community were identified from Mitchell Peninsula (Antarctica), with 66 OTUs comprising up to 25% of the relative abundance. In contrast, 1440 out of the 1641 samples in the Australian biome dataset contained Ca. Eremiobacteria OTUs, with an average relative abundance of only 0.5%, and an average richness of 27 OTUs (Supplementary Fig. S3). In general, Ca. Eremiobacteria showed higher relative abundances and richness in polar soils that had an organic carbon content of 0.1–1%, but this relationship was not evident for Australian soils (Supplementary Fig. S4 and Supplementary Fig. S5). In the Australian soils, Ca. Eremiobacteria were more abundant in soils from Rutherglen (Victoria, Australia) with a relative abundance of 7.2%. Soils from Booderee (NSW, Australia) exhibited the highest richness with 252 OTUs identified from a single sample. This demonstrates that while the Polar soils exhibit a greater relative abundance of Ca. Eremiobacteria, richness in these soils was lower compared to Australian temperate samples.
Fig. 3. Scatter plot showing the significant correlations between Candidatus Eremiobacteria relative abundance and pH in Australian and Polar soils.
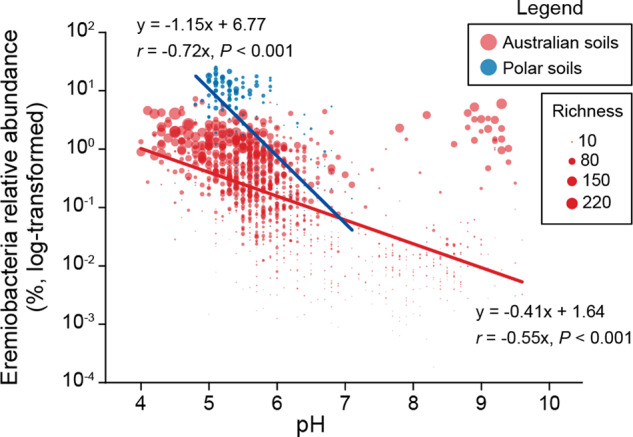
Dot size represents the richness (number of OTUs) of Ca. Eremiobacteria.
Random Forest and Spearman correlations identified pH and moisture (p < 0.001) (Fig. 3, Supplementary Fig. S4, and Supplementary Table S4) as two of the strongest determinants for Ca. Eremiobacteria, with Ca. Eremiobacteria richness and relative abundance negatively correlated with increased pH [47]. Ca. Eremiobacteria is acidotolerant, with a preference for pH < 6, which is consistent with previous work [12]. Of the 195 soil samples with a relative abundance of Ca. Eremiobacteria >1%, the majority (n = 154) were identified from soil samples with acidic pH (pH < 6) (Fig. 3). However, 19 samples with alkaline pH (pH > 8) also exhibited Ca. Eremiobacteria relative abundances >1%; these were from a single locality (Yellabinna Regional Reserve, NSW, Australia) which was very low in organic carbon content (0.24–0.6%) (Supplementary Table S4). Further analysis revealed that 16S rRNA gene sequences from the Yellabinna locality were dispersed within the Ca. Eremiobacteria tree, rather than constituting a separate Ca. Eremiobacteria clade (Supplementary Fig. S6). As pH appears to be the major determinant of Ca. Eremiobacteria distribution, it is possible that Ca. Eremiobacteria abundance at this alkaline locality was influenced more by their capacity to survive in dry, carbon-limited soils, such as by scavenging atmospheric gases as energy and carbon sources [1, 5].
Predicted metabolic properties of Ca. Eremiobacteria
Autotrophy and trace gas oxidation
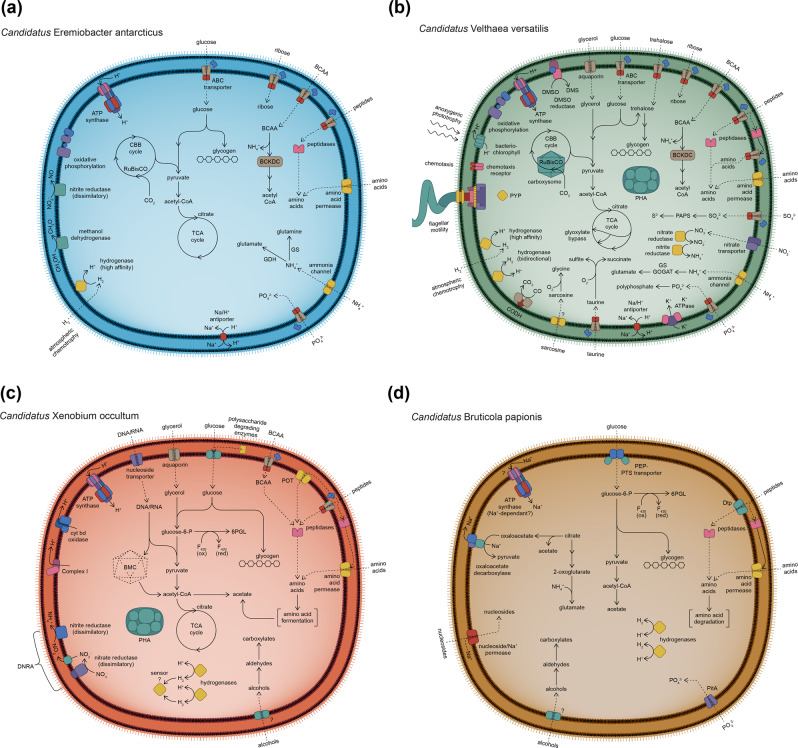
The genetic capacity for trace gas oxidation and carbon fixation through the CBB cycle [1] was represented among Ca. Eremiobacteria genomes. Ca. Eremiobacterales genera (Ca. Eremiobacter and Ca. Mawsoniella) and six Ca. Baltobacterales genera (Ca. Nyctobacter, Ca. Hesperobacter, Ca. Cybelea, Ca. Palsibacter, Ca. Velthaea, and Ca. Tityobacter) encode the high-affinity Group 1h [NiFe]-hydrogenase (hhyL) (Table 1, Supplementary Table S5, and Supplementary Table S6). Of these, five encode RuBisCO (cbbL) type IE (Ca. Eremiobacter, Ca. Nyctobacter, Ca. Palsibacter, Ca. Velthaea, and Ca. Tityobacter) (Table 1, Supplementary Table S5, and Supplementary Fig. S7). Together, these two genes indicate the potential to use atmospheric H2-oxidation to drive carbon fixation (“atmospheric chemosynthesis”) (Fig. 3) [1, 5]. In addition, Ca. Hemerobacter encodes both Group 1f and 2a [NiFe] hydrogenases, as well as RuBisCO (both type 1A and 1E), suggesting a capacity to use atmospheric H2-oxidation to drive carbon fixation; bacteria with Group 1f and 2a [NiFe] hydrogenases have previously been found to be capable of H2 uptake at sub-atmospheric concentrations [48, 49]. One proposed explanation for different atmospheric H2-oxidizing hydrogenases is that Group 2a is adapted for exponential phase growth, whereas Group 1h is linked to energy conservation during persistence [50]. Group 1f hydrogenases have also been accorded a function in protection against reactive oxygen species [3, 51]. Given that two genera (Ca. Hemerobacter and Ca. Lustribacter) that encode Group 1f hydrogenases are inferred to be capable of anaerobic respiration (see Respiration, below) and both were recovered from bog soil samples [10], we propose that Group 1f hydrogenases might be employed by Ca. Eremiobacteria for H2-oxidation under anaerobic conditions. The distribution of genes required for atmospheric H2-oxidation and carbon fixation across Ca. Eremiobacteria taxa is best explained by horizontal gene transfer (HGT) rather than these inferred abilities being ancestral for this phylum. For other [NiFe] hydrogenases found in Ca. Eremiobacteria, Group 3b (Ca. Velthaea) and Group 3d (Ca. Palsibacter and Ca. Lustribacter) hydrogenases are bidirectional, and may help to maintain redox balance within the cell (Table 1 and Supplementary Table S6) [3, 52].
Aerobic CODH is also encoded across Ca. Baltobacterales (Supplementary Fig. S8) with five of these genera also containing RuBisCO (either type 1E or 1A) (Ca. Hesperobacter, Ca. Hemerobacter, Ca. Palsibacter, Ca. Velthaea, and Ca. Tityobacter) (Supplementary Fig. S7), suggesting that these genera can oxidize atmospheric CO to generate CO2 for autotrophic growth via the CBB cycle [1, 2]. In addition, for heterotrophic growth, aerobic atmospheric CO oxidation serves as a potential supplemental energy source to support survival during nutrient starvation [50].
Three Ca. Baltobacterales genera contain genes for RuBisCO type 1A, of which two (Ca. Hemerobacter and Ca. Velthaea) also contain RuBisCO type 1E (Table 1, Supplementary Table S5, and Supplementary Fig. S7). Many autotrophic bacteria improve CO2 fixation by sequestering RuBisCO into inclusions called carboxysomes [53]; these were encoded in Ca. Hemerobacter and Ca. Velthaea (Table 1 and Supplementary Table S5). Four Ca. Baltobacterales genera have the genetic potential for anoxygenic phototrophy using Type II reaction centers (Ca. Baltobacter, Ca. Hesperobacter, Ca. Hemerobacter, and Ca. Velthaea), as inferred previously for MAGs belonging to these genera [11, 12] (Table 1 and Supplementary Table S5). HGT has been proposed to explain the distribution of phototrophy-related genes within Ca. Eremiobacteria [12]. For bacteriochlorophyll synthesis, magnesium-protoporphyrin IX monomethyl ester cyclase exists as alternative aerobic (acsF) or anaerobic (bchE) enzymes, and both are encoded in Ca. Hemerobacter and Ca. Velthaea. This is consistent with an ability to grow phototrophically under both aerobic and anaerobic conditions; those genera that had bchE also have the genetic capacity for anaerobic respiration (see “Respiration,” below).
Ca. Velthaea has two photoreceptor genes: photoactive yellow protein (PYP) and bacteriophytochrome (Table 1, Fig. 3b, and Supplementary Table S5). Ca. Hesperobacter has a gene for PYP. Both PYP and bacteriophytochrome are light-detecting photoreceptors found across prokaryotes, and involved in cellular processes such as phototaxis and upregulation of hydrolytic enzymes involved in degradation of plant material [54, 55].
Respiration
Ca. Eremiobacteria are typically associated with oxygen-rich environments, including moss and surface soils [12, 56]. All genera examined here have the capacity for aerobic respiration using cytochrome c oxidase. Many species additionally encoded a cytochrome bd quinol oxidase, associated with aerobic respiration under O2-limited conditions (Supplementary Table S5) [57]. Several genera also encoded the capacity to use terminal electron acceptors other than oxygen, including nitrate (Ca. Zemynaea), nitric oxide (Ca. Hemerobacter), sulfoxides (possibly dimethylsulfoxide and/or trimethylamine N-oxide) (Ca. Velthaea and Ca. Lustribacter) [58], and urocanate (Ca. Lustribacter) [59] (Supplementary Table S5). Microaerobic and anaerobic capacities are consistent with low oxygen levels that prevail in peatlands, including palsa and bog anoxic layers [10]. There was no evidence of anaerobic respiration in any genus sourced from Antarctic soils (either Ca. Eremiobacterales or Ca. Baltobacterales), although Ca. Eremiobacter encodes a dissimilatory nitrite reductase (Fig. 4a), possibly for redox balance [60].
Fig. 4. Illustrations depicting the predicted diverse metabolic capacities within phylum Candidatus Eremiobacterota.
Class Candidatus Eremiobacteria a order Candidatus Eremiobacterales, represented by Candidatus Eremiobacter antarcticus sp. nov.; b order Candidatus Baltobacterales, represented by Candidatus Velthaea versatilis gen. et. sp. nov.; and class Candidatus Xenobia class. nov., represented by c Candidatus Xenobium occultum gen. et. sp. nov.; and d Candidatus Bruticola papionis gen. et. sp. nov. All cells are shown as diderm, based on the presence of lipopolysaccharide biosynthesis genes across Ca. Eremiobacterota (Supplementary Table S5). All cells are shown as coccoid, based on observed cell morphology of Antarctic Ca. Eremiobacteria cells, even though the rod-cell shape determinant MreB was encoded across both classes (Supplementary Table S5). ABC ATP-binding cassette, BCAA branched-chain amino acids, BCKDC branched-chain 2-oxoacid dehydrogenase complex, BMC bacterial microcompartment, CBB cycle Calvin–Benson–Bassham cycle, CoA coenzyme A, CODH carbon monoxide dehydrogenase, cyt bd cytochrome bd, DMS dimethylsulfide, DMSO dimethylsulfoxide, DNRA dissimilatory nitrate reduction to ammonia, Dtp di-/tripeptide transporter, F420 coenzyme F420 (8-hydroxy-5-deazaflavin), GDH glutamate dehydrogenase, GS glutamine synthetase, GOGAT glutamate synthase, ox oxidized, PAPS 3′-phosphoadenosine-5′-phosphosulfate, PEP phosphoenolpyruvate, PHA polyhydroxyalkanoate (storage product), PitA inorganic phosphate transporter, POT proton-dependent oligopeptide transporter, PTS phosphotransferase, PYP photoactive yellow protein, red reduced, RuBisCO ribulose-bisphosphate carboxylase/oxygenase, TCA cycle tricarboxylic acid cycle, 6PGL 6-phosphogluconolactone.
Organic compounds
Ca. Eremiobacteria possess genes for a complete glycolysis (Embden–Meyerhof–Parnas [EMP] pathway), pentose phosphate pathway, and tricarboxylic acid (TCA) cycle (Supplementary Table S5). CAZy analysis revealed glycoside hydrolases (GH) for the degradation of complex polysaccharides, including cellulose, xyloglucan, xylan, and possibly chitin in certain Ca. Eremiobacteria (Table 1 and Supplementary Table S7). This could be especially relevant to Ca. Baltobacterales from Stordalen Mire, given that high molecular-weight, plant-derived polysaccharides (primarily cellulose and hemicellulose) comprise a large proportion of carbon in this system [10]. Aromatic compounds, present in soil as degradation products of plant material [61], could potentially be used as carbon and energy sources by Ca. Eremiobacteria (see “Supplementary Results and discussion: catabolism of plant metabolites and aromatic compounds”). Aromatic compound transporters and catabolic enzymes (including those capable of cleavage of aromatic rings) are encoded across Ca. Eremiobacteria genera (Supplementary Table S5; see “Supplementary Results and discussion: oxygen as an agent for substrate degradation”).
A number of Ca. Eremiobacteria encode pathways for glycogen synthesis, by either the classical pathway or the mycobacterial-type pathway sourced from trehalose (Supplementary Table S5) [62]. Certain Ca. Eremiobacteria encode the ability to synthesize trehalose, which can be accumulated for osmotic stress [63] or converted to 2-sulfotrehalose, a precursor for sulfolipid biosynthesis [64], as well as a compatible solute to protect against osmotic stress (Supplementary Table S5) [65]. Polyhydroxyalkanoate (PHA) synthase (PhaC) is encoded throughout the Ca. Eremiobacteria, indicating the capacity for carbon storage as PHA (Table 1, Fig. 4, and Supplementary Table S5).
The primary transporters in Ca. Eremiobacteria were dominated by those for oligopeptides and branched-chain amino acids (BCAAs) (Supplementary Table S5). Ca. Eremiobacteria genera encode numerous peptidases, both cytoplasmic and extracytoplasmic (Supplementary Table S5). Among these diverse peptidases, the majority of Ca. Eremiobacteria encode d-aminopeptidases (Peptidase M55) and β-peptidyl aminopeptidases (Peptidase S58), suggesting that they can utilize d-alanyl-d-alanine dipeptides from peptidoglycans [66] and natural peptides that contain β-amino acids (e.g., microcystins, nodularins, carnosine) [67], respectively. Combined with the prevalence of genes for peptide ATP-dependent cassette (ABC) transporters, this suggests that amino acids are important sources of carbon, nitrogen, and sulfur. Further, certain amino acids could be utilized for pH homeostasis (see “Acid tolerance and resistance,” below). Ca. Eremiobacteria are notable for the prevalence of BCAA aminotransferases and branched-chain 2-oxoacid dehydrogenase complexes; along with BCAA ABC transporters, this suggests that BCAAs are a major source of nitrogen and reductant (Fig. 4). Across Ca. Eremiobacteria, other potential organic nitrogen sources include urea, cyanate, sarcosine, creatine, and putrescine (Table 1 and Supplementary Table S5). In general, Ca. Eremiobacteria appear to rely predominantly on organic nitrogen and sulfur sources. Organosulfonates (including taurine and alkanesulfonates) appear to be an important source of sulfur for many Ca. Baltobacterales. There was no evidence of nitrogenase in the Ca. Eremiobacteria surveyed here, despite the high abundance of Ca. Eremiobacteria in nitrogen-limited Antarctic soils [5]. Only one Ca. Eremiobacteria genus (Ca. Velthaea) showed genomic evidence of reductive nitrate assimilation (Fig. 4d). Only a relatively small number of Ca. Eremiobacteria genera possess identifiable genes for the complete reductive assimilation of sulfate to sulfide (Supplementary Table S5). Phosphate ABC transporters, for the primary import of phosphate, and polyphosphate kinases, for high-energy phosphate storage, are encoded throughout the Ca. Eremiobacteria (Supplementary Table S5).
Acid tolerance and resistance
The globally distributed Ca. Eremiobacteria is most often found in acidic and aerobic environments (Fig. 3) [12, 18]. The proton permeability of the cell membrane increases with temperature [68], so cooler environments would minimize proton permeation. Hopanoid lipids reduce membrane permeability to protons, and therefore are an important adaptation for life in acidic environments [69, 70]. All Ca. Baltobacterales genera encode squalene-hopene cyclase for hopanoid synthesis, with isoprenoids synthesized via the 1-deoxy-d-xylulose-5-phosphate pathway (Supplementary Table S5) [71]. Additional genomic features were identified that facilitate maintenance of pH homeostasis under acidic conditions. Proton influx under acidic conditions is offset by maintaining an inside-positive membrane potential as a charge barrier, achieved by uptake of potassium and other cations [69, 72]. Potassium uptake systems were present in most Ca. Eremiobacterota, including potassium channels and active potassium uptake systems (Supplementary Table S5) [73–75]. Ca. Eremiobacterota encode Na+/H+ antiporters (Nha), which may function to export excess protons and simultaneously import sodium ions [70].
Another acid resistance mechanism is to consume excess cytoplasmic protons using amino acid carboxylases, such as arginine carboxylase and glutamate carboxylase, which generate agmatine and γ-aminobutyrate (GABA), respectively; specific antiporters then release these products in exchange for import of exogenous precursors [76]. Arginine carboxylase is more widely distributed across Ca. Eremiobacteria, although the cognate arginine/agmatine antiporter could only be identified in three Ca. Baltobacterales genera (Supplementary Table S5). In addition, genes for glutamate carboxylase and/or glutamate/GABA antiporters were identified in five Ca. Baltobacterales genera (Supplementary Table S5).
Visualization of Ca. Eremiobacteria cells isolated from Antarctic soil
CARD-FISH was employed to visualize Ca. Eremiobacteria cells present in Antarctic desert soils [17]. Given the slow-growing nature of cold-adapted soil bacteria and presumably low ribosomal content [46] (Supplementary Fig. S9), combined with natural autofluorescence of soil particles (Supplementary Fig. S10), we elected to use CARD-FISH incorporating a horse radish peroxidase-labeled Erem-289 probe, tyramide signal amplification with Cy3 and counterstaining with DAPI to enhance fluorescence signal strength. Epifluorescence microscopy was used to visualize the morphology of members of this class for the first time, with the majority of Ca. Eremiobacteria cells being coccoid in shape, with 7.7% of cells analysed exhibiting a larger “spindle-shaped” morphology (Fig. 5; Supplementary Methods). The lower volume/surface area quotient of coccoid cells permits rapid and efficient nutrient uptake [77].
Fig. 5. Visualization of Candidatus Eremiobacteria cells isolated from an Antarctic soil.
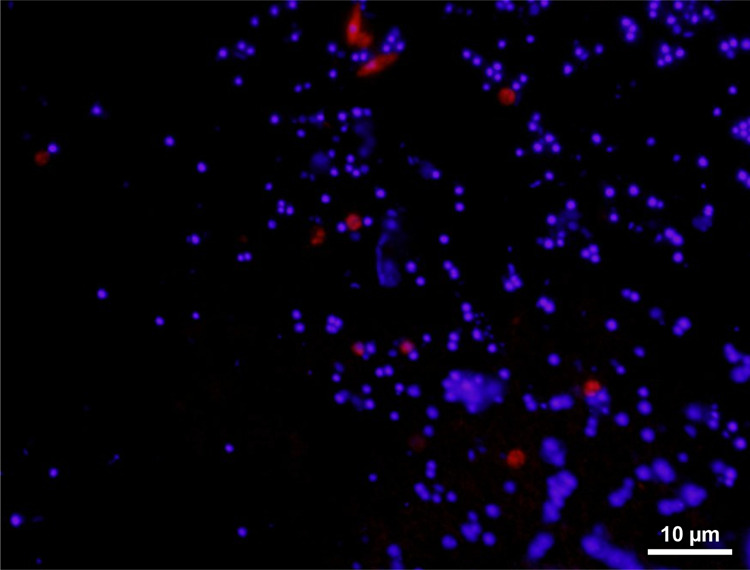
CARD-FISH was employed using the class-level Ca. Eremiobacteria-specific probe (Erem-289) labeled with Cy3 (red) and counterstained with DAPI (blue). The majority of Ca. Eremiobacteria cells are of coccoid morphology, although note the more “spindle-shaped” cells at the top of the field.
Ca. Xenobia is metabolically divergent from Ca. Eremiobacteria
Ca. Xenobium metabolism
Ca. Xenobium MAGS were recovered from separate and geographically diverse industrial waste samples (Table 1 and Supplementary Table S1) [16]. Both Ca. Xenobia genera, Ca. Xenobium (Fig. 4c) and Ca. Bruticola (Fig. 4d), encode the same acidophilic adaptations as Ca. Eremiobacteria (hopanoid biosynthesis, arginine decarboxylase, Ktr potassium uptake system, Nha sodium/proton antiporters) (Supplementary Table S5), suggesting that an acidophilic or acidotolerant lifestyle is common to this terrestrial phylum. Ca. Xenobium has a relatively large genome (>5 Mbp), comparable in size to the largest genomes identified in Ca. Eremiobacteria (e.g., Ca. Velthaea), and far larger than that of Ca. Bruticola (Supplementary Table S1).
Ca. Xenobium is inferred to be capable of both microaerobic and anaerobic respiration (Fig. 4c and Supplementary Table S5). Both Ca. Xenobium species encode a complete glycolytic (EMP) pathway, TCA cycle, pentose phosphate pathway, Complex I, and cytochrome bd oxygen reductase, the latter suggesting they are adapted to microaerobic conditions [57]. A potential for anaerobic respiration via dissimilatory nitrate reduction to ammonium (DNRA) is suggested by a putative respiratory complex for nitrate reduction to nitrite (NarGH), and a nitrite reductase (NrfAH) for the direct reduction of nitrite to ammonium (Fig. 4c and Supplementary Table S5) [78, 79]. DNRA generates nitric oxide and hydroxylamine as toxic intermediates, and Ca. Xenobium encodes enzymes for protection against oxidative and nitrosative stress, including F420-dependent glucose-6-dehydrogenase, nitrite reductase, and hydroxylamine reductase (Supplementary Table S5) [78]. Certain polysaccharides and oligosaccharides appear to be utilized as substrates, based on genes for secreted GH (see “Supplementary Results and discussion: additional metabolic properties of Ca. Xenobia”). Other potential sources of reductant include the oxidation of amino acids (including derived from peptide degradation), alcohols, and aldehydes (Supplementary Table S5) [80]. Ca. Xenobium also has the capacity for both PHA and glycogen synthesis (Supplementary Table S5). Ca. Xenobium encodes proteins required for the construction of bacterial microcompartments (also encoded in Ca. Tityobacter from Ca. Baltobacterales), likely for sequestering toxic aldehydes [81, 82] (see “Supplementary Results and discussion: additional metabolic properties of Ca. Xenobia”).
Ca. Bruticola metabolism
All five MAGs (81.41–85.65% completeness) that were derived from the baboon fecal metagenome belong to a single genus and species (Table 1 and Supplementary Table S1). Ca. Bruticola has a smaller genome (~2.8 Mbp) compared to Ca. Xenobium (Supplementary Table S1). There is no evidence of genomic streamlining [83], and the genome encodes biosynthetic abilities for lipids and certain amino acids, but not nucleosides (presumably imported) (Fig. 4d). Overall, Ca. Bruticola is inferred to have an extremely simplified, heterotrophic metabolism, with substrates that include oligopeptides, amino acids, and sugars (Supplementary Information; Supplementary Tables S5 and S7).
Ca. Bruticola lacks any evidence for genes associated with respiration, and this genus is therefore predicted to be obligately fermentative. A glycolytic (EMP) pathway is evident, possibly initiated by a phosphoenolpyruvate-phosphotransferase system for concomitant uptake and phosphorylation of glucose. Ca. Bruticola is predicted to lack a TCA cycle, with the only two identifiable genes being those for aconitase and isocitrate dehydrogenase (Fig. 4d and Supplementary Table S5). The truncated central metabolism is sufficient to synthesize the five universal anabolic precursors: acetyl-CoA, pyruvate, PEP, oxaloacetate, and 2-oxoglutarate [84, 85]. Unique among Ca. Eremiobacterota, Ca. Bruticola encodes all the components of citrate lyase, which catalyzes the acyl-carrier-protein (ACP)-dependent cleavage of citrate into oxaloacetate and acetate (Supplementary Table S5) [86]. The genes for citrate lyase (citGCXFED) form a cluster with genes for aconitase, isocitrate lyase, and 3-oxoacyl-ACP-reductase (fabG), the last of which catalyzes the first step in the chain elongation cycle of fatty acid biosynthesis. Thus, we posit that citrate has two possible fates: conversion to 2-oxoglutarate via isocitrate; or generation of oxaloacetate and acetate. Acetyl-CoA can be converted to acetate, to generate ATP by substrate-level phosphorylation (phosphate acetyltransferase, acetate kinase), as in Ca. Xenobium. A Na+-translocating oxaloacetate decarboxylase [87] is encoded by Ca. Bruticola; as well as generating pyruvate, this enzyme extrudes Na+ across the cytoplasmic membrane to create a Na+-motive force. One possibility is that Ca. Bruticola has a Na+-dependent ATP synthase [88], meaning oxaloacetate decarboxylation would be a source of ATP generation for the cell (Fig. 4d).
Ca. Eremiobacterota ecology
Members of Ca. Eremiobacterota were found to be united by multiple adaptations for living in acidic environments, in terms of cell envelope composition, mechanisms for maintenance of a charge barrier, and efflux and consumption of cytoplasmic protons. However, Ca. Eremiobacterota are metabolically diverse, with disparate metabolic strategies exhibited across genera reflecting the localities from which they were recovered (Fig. 4, Table 1, and Supplementary Table S5). Members of Ca. Eremiobacterota have adaptations that allow cells to survive in a diverse range of ecosystems, including acidic, contaminated, and severely nutrient-poor environments. Class Ca. Eremiobacteria has genomic capacities for trace gas oxidation and carbon fixation, and is highly adapted for growth and survival in low-pH, aerobic environments, including polar soils, desert soils, and peatlands [1, 10, 11]. The majority of the Ca. Eremiobacteria MAGs assessed in the current study were derived from peatlands, which is replete with plant-derived organic matter, much of it recalcitrant [10], with Ca. Eremiobacteria from this system typically encoding enzymes for the degradation of a narrow range of polysaccharides (Supplementary Tables S5 and S7). However, the presence of some polysaccharide-degrading enzymes (especially for glucan and xyloglucan) in Antarctic Ca. Eremiobacteria suggests that they also have access to plant degradation products in their cold and arid habitats. Vegetation (lichens and mosses) are present in Antarctica, including in the Windmill Islands region [89, 90] from where Ca. Eremiobacteria MAGs were recovered.
Ca. Eremiobacteria encode abundant mechanisms for obtaining reduced nitrogen from amino acids and proteins, with abilities to obtain nitrogen from diverse peptides, including those that contain β- or d-amino acids (Fig. 4). The latter abilities are not unique among bacteria, but may give Ca. Eremiobacteria a competitive advantage in environments depleted in organic nitrogen sources [5]. Multiple genera within Ca. Eremiobacteria encode the capacity to scavenge H2, CO2, and CO from the atmosphere for use as carbon and/or energy sources (Fig. 4 and Supplementary Table S5) [1]. In addition, several members of the order Ca. Baltobacterales encode the capacity for bacteriochlorophyll-based anoxygenic photosynthesis and anaerobic respiration [12]. Thus, certain Ca. Baltobacterales encode impressive metabolic capacities, with Ca. Velthaea as an exemplar; in addition to the aforementioned capabilities, this genus has genes for dual glycolytic pathways, reductive nitrate assimilation, photoreception, and chemotaxis using flagella (Fig. 4b and Supplementary Table S5).
Given the presence of genes associated with the degradation of aromatic acids, organohalogens, alkanes, and alcohols (Supplementary Table S5), Ca. Eremiobacteria have the capacity to contribute toward the degradation of environmental contaminants. Class Ca. Xenobia (formerly UBP9, SHA-109) [12, 16], comprising two genera, Ca. Xenobium and Ca. Bruticola (Fig. 2), appears to be highly divergent compared to Ca. Eremiobacteria. Yet, the ability to survive in polluted habitats is shared by Ca. Xenobium, with MAGs being recovered from industrial waste sources [16]. Ca. Xenobium encodes mechanisms to deal with heavy metals, arsenic, oxidative stress, and nitrosative stress, with all but the last shared with Ca. Eremiobacteria. Members of genus Ca. Xenobium are inferred to be facultative anaerobes adapted to microaerobic conditions (based on the nature of encoded terminal reductases), with a preference for peptides and amino acids, and capable of fermentation (Fig. 4c). The related genus Ca. Bruticola has the capacity to metabolize a narrow repertoire of organic substrates, has an incomplete TCA cycle, and is incapable of respiration (Fig. 4d). Ca. Bruticola may require an animal host, given that MAGs were recovered from baboon fecal samples. However, the natural environments of Ca. Xenobium and Ca. Bruticola are currently not known, and the exact ecologies of both genera await future discoveries.
Supplementary information
Supplementary Table S1. MAGs information
Supplementary Table S3. Taxonomy 16S source for Figure S2
Supplementary Table S5. Pathways and proteins
Acknowledgements
This research was funded through an Australian Research Council Future Fellowship (FT170100341) and an Australian Antarctic Science project Grant (4406), both awarded to BCF. Thank you to the Australian Antarctic Program expedition teams between 2005 and 2019 for sampling of the Antarctic soils used in this study. We also thank B. Creisler for assistance with etymologies of genus and species names.
Data availability
Nine curated genome bins of Ca. Eremiobacterota are available in the NCBI sequence read archive under BioProject (PRJNA587560). Currently awaiting public identifiers for each MAG from NCBI. The metadata analysed in this study are available through the Australian Antarctic Data (AAD) Centre (10.4225/15/526F42ADA05B1) and the AusMicrobiome repository (https://data.bioplatforms.com/organization/about/australian-microbiome).
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Mukan Ji, Timothy J. Williams
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-00944-8.
References
- 1.Ji M, Greening C, Vanwonterghem I, Carere CR, Bay SK, Steen JA. Atmospheric trace gases support primary production in Antarctic desert surface soil. Nature. 2017;552:400–3. doi: 10.1038/nature25014. [DOI] [PubMed] [Google Scholar]
- 2.Grostern A, Alvarez-Cohen L. RubisCO-based CO2 fixation and C1 metabolism in the actinobacterium Pseudonocardia dioxanivorans CB1190. Environ Microbiol. 2013;15:3040–53. doi: 10.1111/1462-2920.12144. [DOI] [PubMed] [Google Scholar]
- 3.Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB, et al. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 2016;10:761–77. doi: 10.1038/ismej.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bay S, Ferrari BC, Greening C. Life without water: how do bacteria generate biomass in desert ecosystems? Microbiol Austral. 2018;39:28–32. doi: 10.1071/MA18008. [DOI] [Google Scholar]
- 5.Ray A, Zhang E, Terauds A, Ji M, Kong W, Ferrari BC. Soil microbiomes with the genetic capacity for atmospheric chemosynthesis are widespread across the poles and are associated with moisture, carbon and nitrogen limitation. Front Microbiol. 2020;11:1–13. doi: 10.3389/fmicb.2020.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greening C, Berney M, Hards K, Cook GM, Conrad R. A soil actinobacterium scavenges atmospheric H2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases. Proc Natl Acad Sci. 2014;111:4257–61. doi: 10.1073/pnas.1320586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nogales B, Moore ERB, Llobet-Brossa E, Rossello-Mora R, Amann R, Timmis KN. Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl Environ Microbiol. 2001;67:1874–84. doi: 10.1128/AEM.67.4.1874-1884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nessner Kavamura V, Taketani RG, Lançoni MD, Andreote FD, Mendes R, Soares de Melo I. Water regime influences bulk soil and rhizosphere of Cereus jamacaru bacterial communities in the Brazilian Caatinga biome. PLoS ONE. 2013;8:e73606. doi: 10.1371/journal.pone.0073606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serkebaeva YM, Kim Y, Liesack W, Dedysh SN. Pyrosequencing-based assessment of the bacteria diversity in surface and subsurface peat layers of a northern wetland, with focus on poorly studied phyla and candidate divisions. PLoS ONE. 2013;8:e63994. doi: 10.1371/journal.pone.0063994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodcroft BJ, Singleton CM, Boyd JA, Evans PN, Emerson JB, Zayed AAF, et al. Genome-centric view of carbon processing in thawing permafrost. Nature. 2018;560:49–54. doi: 10.1038/s41586-018-0338-1. [DOI] [PubMed] [Google Scholar]
- 11.Holland-Moritz H, Stuart J, Lewis LR, Miller S, Mack MC, McDaniel SF, et al. Novel bacterial lineages associated with boreal moss species. Environ Microbiol. 2018;20:2625–38. doi: 10.1111/1462-2920.14288. [DOI] [PubMed] [Google Scholar]
- 12.Ward LM, Cardona T, Holland-Moritz H. Evolutionary implications of anoxygenic phototrophy in the bacterial phylum Candidatus Eremiobacterota (WPS-2) Front Microbiol. 2019;10:1658. doi: 10.3389/fmicb.2019.01658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheremet A, Jones GM, Jarett J, Bowers RM, Bedard I, Culham C, et al. Ecological and genomic analyses of candidate phylum WPS-2 bacteria in an unvegetated soil. Environ Microbiol. 2020;22:3143–57. doi: 10.1111/1462-2920.15054. [DOI] [PubMed] [Google Scholar]
- 14.Dewhirst FE, Klein EA, Thompson EC, Blanton JM, Chen T, Milella L, et al. The canine oral microbiome. PLoS ONE. 2012;7:e36067. doi: 10.1371/journal.pone.0036067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camanocha A, Dewhirst FE. Host-associated bacterial taxa from Chlorobi, Chloroflexi, GN02, Synergistetes, SR1, TM7, and WPS-2 Phyla/candidate divisions. J Oral Microbiol. 2014;6. 10.3402/jom.v6.25468. [DOI] [PMC free article] [PubMed]
- 16.Parks DH, Rinke C, Chuvochina M, Chaumeil PA, Woodcroft BJ, Evans PN, et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–42. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 17.Ji M, van Dorst J, Bissett A, Brown MV, Palmer AS, Snape I, et al. Microbial diversity at Mitchell Peninsula, Eastern Antarctica: a potential biodiversity “hotspot”. Pol Biol. 2015;39:237–49. doi: 10.1007/s00300-015-1776-y. [DOI] [Google Scholar]
- 18.Ferrari BC, Bissett A, Snape I, van Dorst J, Palmer AS, Ji M, et al. Geological connectivity drives microbial community structure and connectivity in polar, terrestrial ecosystems. Environ Microbiol. 2016;18:1834–49. doi: 10.1111/1462-2920.13034. [DOI] [PubMed] [Google Scholar]
- 19.Bissett A, Fitzgerald A, Meintjes T, Mele PM, Reith F, Dennis PG, et al. Introducing BASE: the biomes of Australian soil environments soil microbial diversity database. Gigascience. 2016;5:21. doi: 10.1186/s13742-016-0126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siciliano SD, Palmer AS, Winsley T, Lamb E, Bissett A, Brown MV, et al. Polar soil bacterial and fungal biodiversity survey, Ver. 1. Australian Antarctic Data Centre; 2014. 10.4225/15/526F42ADA05B1. Accessed 11 Feb 2021.
- 21.Lane D. Nucleic acid techniques in bacterial systematics. In: Stackebrandt E, Goodfellow M, editors. Chichester NY: John Wiley and Sons; 1991. p. 115–75.
- 22.Siciliano SD, Palmer A, Winsley T, Lamb E, Bissett A, Brown M, et al. Soil fertility is associated with fungal and bacterial richness, whereas pH is associated with community composition in polar soil microbial communities. Soil Biol Biochem. 2014;78:10–20. doi: 10.1016/j.soilbio.2014.07.005. [DOI] [Google Scholar]
- 23.Archer E. R package. 2016.
- 24.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed]
- 25.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27:824–34. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bushnell B. BBMap: a fast, accurate, splice-aware aligner. Berkeley, CA, United States: Lawrence Berkeley National Laboratory; 2014.
- 27.Imelfort M, Parks D, Woodcroft BJ, Dennis P, Hugenholtz P, Tyson GW. GroopM: an automated tool for the recovery of population genomes from related metagenomes. PeerJ. 2014;2:e603. doi: 10.7717/peerj.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu YW, Simmons BA, Singer SW. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2016;32:605–7. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 29.Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ. Binning metagenomic contigs by coverage and composition. Nat Methods. 2014;11:1144–6. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 31.Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019;7:e7359. doi: 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 33.Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2020;36:1925–7. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray AE, Freudenstein J, Gribaldo S, Hatzenpichler R, Hugenholtz P, Kämpfer P, et al. Roadmap for naming uncultivated Archaea and Bacteria. Nat Microbiol. 2020;5:987–94. doi: 10.1038/s41564-020-0733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 37.Tabita FR, Hanson TE, Li H, Satagopan S, Singh J, Chan S. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev. 2007;71:576–99. doi: 10.1128/MMBR.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tabita FR, Hanson TE, Satagopan S, Witte BH, Kreel NE. Phylogenetic and evolutionary relationships of RubisCO and the RubisCO-like proteins and the functional lessons provided by diverse molecular forms. Philos Trans R Soc Lond B Biol Sci. 2008;363:2629–40. doi: 10.1098/rstb.2008.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–9. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sondergaard D, Pedersen CN, Greening C. HydDB: a web tool for hydrogenase classification and analysis. Sci Rep. 2016;6:34212. doi: 10.1038/srep34212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–71. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuchs BM, Wallner G, Beisker W, Schwippl I, Ludwig W, Amann R. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 1998;64:4973–82. doi: 10.1128/AEM.64.12.4973-4982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schramm A, Fuchs BM, Nielsen JL, Tonolla M, Stahl DA. Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ Microbiol. 2002;4:713–20. doi: 10.1046/j.1462-2920.2002.00364.x. [DOI] [PubMed] [Google Scholar]
- 45.Lindahl V. Improved soil dispersion procedures for total bacterial counts, extraction of sandy clayey silt soil bacteria and cell survival. J Microbiol Meth. 1996;25:279–86. doi: 10.1016/0167-7012(95)00102-6. [DOI] [Google Scholar]
- 46.Ferrari BC, Tujula N, Stoner K, Kjelleberg S. Catalysed reporter deposition-FISH allows for enrichment independent detection of microcolony forming soil bacteria. Appl Environ Microbiol. 2006;72:918–22. doi: 10.1128/AEM.72.1.918-922.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim M, Lim HS, Hyun CU, Cho A, Noh HJ, Hong SG, et al. Local-scale variation of soil bacterial communities in ice-free regions of maritime Antarctica. Soil Biol Biochem. 2019;133:165–73. doi: 10.1016/j.soilbio.2019.03.011. [DOI] [Google Scholar]
- 48.Islam ZF, Welsh C, Bayly K, Grinter R, Southam G, Gagen EJ, et al. A widely distributed hydrogenase oxidises atmospheric H2 during bacterial growth. ISME J. 2020;14:2649–58. doi: 10.1038/s41396-020-0713-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Myers MR, King GM. Isolation and characterization of Acidobacterium ailaaui sp. nov., a novel member of Acidobacteria subdivision 1, from a geothermally heated Hawaiian microbial mat. Int J Syst Evol Microbiol. 2016;66:5328–35. doi: 10.1099/ijsem.0.001516. [DOI] [PubMed] [Google Scholar]
- 50.Cordero PRF, Bayly K, Leung PM, Huang C, Islam ZF, Schittenhelm RB, et al. Atmospheric carbon monoxide oxidation is a widespread mechanism supporting microbial survival. ISME J. 2019;13:2868–81. doi: 10.1038/s41396-019-0479-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tremblay PL, Lovley DR. Role of the NiFe hydrogenase Hya in oxidative stress defense in Geobacter sulfurreducens. J Bacteriol. 2012;194:2248–53. doi: 10.1128/JB.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greening C, Cook GM. Integration of hydrogenase expression and hydrogen sensing in bacterial cell physiology. Curr Opin Microbiol. 2014;18:30–8. doi: 10.1016/j.mib.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 53.English RS, Lorbach SC, Qin X, Shively JM. Isolation and characterization of a carboxysome shell gene from Thiobacillus neapolitanus. Mol Microbiol. 1994;12:647–54. doi: 10.1111/j.1365-2958.1994.tb01052.x. [DOI] [PubMed] [Google Scholar]
- 54.Bonomi HR, Toum L, Sycz G, Sieira R, Toscani AM, Gudesblat GE, et al. Xanthomonas campestris attenuates virulence by sensing light through a bacteriophytochrome photoreceptor. EMBO Rep. 2016;17:1565–77. doi: 10.15252/embr.201541691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gamiz-Hernandez AP, Kaila VRI. Conversion of light-energy into molecular strain in the photocycle of the photoactive yellow protein. Phys Chem Chem Phys. 2016;18:2802–9. doi: 10.1039/C5CP05244K. [DOI] [PubMed] [Google Scholar]
- 56.Zhang E, Thibaut LM, Terauds A, Raven M, Tanaka MM, van Dorst J, et al. Lifting the veil on arid-to-hyperarid Antarctic soil microbiomes: a tale of two oases. Microbiome. 2020;8:37. doi: 10.1186/s40168-020-00809-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta. 2011;1807:1398–413. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCrindle SL, Kappler U, McEwan AG. Microbial dimethylsulfoxide and trimethylamine-N-oxide respiration. Adv Micro Physiol. 2005;50:147–98. doi: 10.1016/S0065-2911(05)50004-3. [DOI] [PubMed] [Google Scholar]
- 59.Bogachev AV, Bertsova YV, Bloch DA, Verkhovsky MI. Urocanate reductase: identification of a novel anaerobic respiratory pathway in Shewanella oneidensis MR-1. Mol Microbiol. 2012;86:1452–63. doi: 10.1111/mmi.12067. [DOI] [PubMed] [Google Scholar]
- 60.Hopper AC, Li Y, Cole JA. A critical role for the cccA gene product, cytochrome c2, in diverting electrons from aerobic respiration to denitrification in Neisseria gonorrhoeae. J Bacteriol. 2013;195:2518–29. doi: 10.1128/JB.02300-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nichols NN, Harwood CS. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J Bacteriol. 1997;179:5056–61. doi: 10.1128/jb.179.16.5056-5061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fraga J, Maranha A, Mendes V, Pereira PJB, Empadinhas N, Macedo-Ribeiro S, et al. Structure of mycobacterial maltokinase, the missing link in the essential GlgE-pathway. Sci Rep. 2015;5:8026. doi: 10.1038/srep08026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reina-Bueno M, Argandoña M, Nieto JJ, Hidalgo-García A, Iglesias-Guerra F, Delgado MJ, et al. Role of trehalose in heat and desiccation tolerance in the soil bacterium Rhizobium etli. BMC Microbiol. 2012;12:207. doi: 10.1186/1471-2180-12-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mougous JD, Petzold CJ, Senaratne RH, Lee DH, Akey DL, Lin FL, et al. Identification, function and structure of the mycobacterial sulfotransferase that initiates sulfolipid-1 biosynthesis. Nat Struct Mol Biol. 2004;11:721–9. doi: 10.1038/nsmb802. [DOI] [PubMed] [Google Scholar]
- 65.Oren A. Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. J Ind Microbiol Biotechnol. 2002;28:56–63. doi: 10.1038/sj/jim/7000176. [DOI] [PubMed] [Google Scholar]
- 66.Cheggour A, Fanuel L, Duez C, Joris B, Bouillenne F, Devreese B, et al. The dppA gene of Bacillus subtilis encodes a new D-aminopeptidase. Mol Microbiol. 2000;38:504–13. doi: 10.1046/j.1365-2958.2000.02117.x. [DOI] [PubMed] [Google Scholar]
- 67.Geueke B, Heck T, Limbach M, Nesatyy V, Seebach D, Kohler HPE. Bacterial β-peptidyl aminopeptidases with unique substrate specificities for β-oligopeptides and mixed β,α-oligopeptides. FEBS J. 2006;273:5261–72. doi: 10.1111/j.1742-4658.2006.05519.x. [DOI] [PubMed] [Google Scholar]
- 68.Driessen AJM, van de Vossenberg JLM, Konings WN. Membrane composition and ion-permeability in extremophiles. FEMS Microbiol Rev. 1996;18:139–48. doi: 10.1111/j.1574-6976.1996.tb00232.x. [DOI] [Google Scholar]
- 69.Jones DS, Albrecht HL, Dawson KS, Schaperdoth I, Freeman KH, Pi Y, et al. Community genomic analysis of an extremely acidophilic sulfur-oxidizing biofilm. ISME J. 2012;6:158–70. doi: 10.1038/ismej.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nguyen NL, Yu WJ, Gwak JH, Kim SJ, Park SJ, Herbold CW, et al. Genomic insights into the acid adaptation of novel methanotrophs enriched from acidic forest soils. Front Microbiol. 2018;9:1982. doi: 10.3389/fmicb.2018.01982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xue J, Ahring BK. Enhancing isoprene production by genetic modification of the 1-deoxy-d-xylulose-5-phosphate pathway in Bacillus subtilis. Appl Environ Microbiol. 2011;77:2399–405. doi: 10.1128/AEM.02341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baker-Austin C, Dopson M. Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 2007;15:165–71. doi: 10.1016/j.tim.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 73.Siebers A, Altendorf K. The K+-translocating Kdp-ATPase from Escherichia coli. Purification, enzymatic properties and production of complex- and subunit-specific antisera. Eur J Biochem. 1988;178:131–40. doi: 10.1111/j.1432-1033.1988.tb14438.x. [DOI] [PubMed] [Google Scholar]
- 74.Milkman R. An Escherichia coli homologue of eukaryotic potassium channel proteins. Proc Natl Acad Sci. 1994;91:3510–4. doi: 10.1073/pnas.91.9.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Holtmann G, Bakker EP, Uozumi N, Bremer E. KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J Bacteriol. 2003;185:1289–98. doi: 10.1128/JB.185.4.1289-1298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Castanie-Cornet MP, Penfound TA, Smith D, Elliott JF, Foster JW. Control of acid resistance in Escherichia coli. J Bacteriol. 1999;181:3525–35. doi: 10.1128/JB.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Geisseler D, Horwath WR. Regulation of extracellular protease activity in soil in response to different sources and concentrations of nitrogen and carbon. Soil Biol Biochem. 2008;40:3040–8. doi: 10.1016/j.soilbio.2008.09.001. [DOI] [Google Scholar]
- 78.Einsle O, Messerschmidt A, Stach P, Bourenkov GP, Bartunik HD, Huber R, et al. Structure of cytochrome c nitrite reductase. Nature. 1999;400:476–80. doi: 10.1038/22802. [DOI] [PubMed] [Google Scholar]
- 79.Simon J, Pisa R, Stein T, Eichler R, Klimmek O, Gross R. The tetraheme cytochrome c NrfH is required to anchor the cytochrome c nitrite reductase (NrfA) in the membrane of Wolinella succinogenes. Eur J Biochem. 2001;268:5776–82. doi: 10.1046/j.0014-2956.2001.02520.x. [DOI] [PubMed] [Google Scholar]
- 80.Nair RV, Bennett GN, Papoutsakis ET. Molecular characterization of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824. J Bacteriol. 1994;176:871–85. doi: 10.1128/jb.176.3.871-885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Axen SD, Erbilgin O, Kerfeld CA. A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comput Biol. 2014;10:e1003898. doi: 10.1371/journal.pcbi.1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Erbilgin O, McDonald KL, Kerfeld CA. Characterization of a planctomycetal organelle: a novel bacterial microcompartment for the aerobic degradation of plant saccharides. Appl Environ Microbiol. 2014;80:2193–205. doi: 10.1128/AEM.03887-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Giovannoni SJ, Cameron Thrash J, Temperton B. Implications of streamlining theory for microbial ecology. ISME J. 2014;8:1553–65. doi: 10.1038/ismej.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Srinivasan V, Morowitz HJ. The canonical network of autotrophic intermediary metabolism: minimal metabolome of a reductive chemoautotroph. Biol Bull. 2009;216:126–30. doi: 10.1086/BBLv216n2p126. [DOI] [PubMed] [Google Scholar]
- 85.Nunoura T, Chikaraishi Y, Izaki R, Suwa T, Sato T, Harada T, et al. A primordial and reversible TCA cycle in a facultatively chemolithoautotrophic thermophile. Science. 2018;359:559–63. doi: 10.1126/science.aao3407. [DOI] [PubMed] [Google Scholar]
- 86.Bekal S, Van Beeumen J, Samyn B, Garmyn D, Henini S, Diviès C, et al. Purification of Leuconostoc mesenteroides citrate lyase and cloning and characterization of the citCDEFG gene cluster. J Bacteriol. 1998;180:647–54. doi: 10.1128/JB.180.3.647-654.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dimroth P, Jockel P, Schmid M. Coupling mechanism of the oxaloacetate decarboxylase Na(+) pump. Biochim Biophys Acta. 2001;1505:1–14. doi: 10.1016/S0005-2728(00)00272-3. [DOI] [PubMed] [Google Scholar]
- 88.Mulkidjanian AY, Dibrov P, Galperin MY. The past and present of sodium energetics: may the sodium-motive force be with you. Biochim Biophys Acta. 2008;1777:985–92. doi: 10.1016/j.bbabio.2008.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lewis Smith RI. Plant community dynamics in Wilkes Land, Antarctica, vol. 3. Proceedings of the NIPR Symposium on Polar Biology. 1990. p. 229–44.
- 90.Seppelt RD. Plant communities at Wilkes Land. In: Geoecology of Antarctic ice-free coastal landscapes. Ecological studies (Analysis and synthesis), vol. 154. Springer; 2002. p. 233–48.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1. MAGs information
Supplementary Table S3. Taxonomy 16S source for Figure S2
Supplementary Table S5. Pathways and proteins
Data Availability Statement
Nine curated genome bins of Ca. Eremiobacterota are available in the NCBI sequence read archive under BioProject (PRJNA587560). Currently awaiting public identifiers for each MAG from NCBI. The metadata analysed in this study are available through the Australian Antarctic Data (AAD) Centre (10.4225/15/526F42ADA05B1) and the AusMicrobiome repository (https://data.bioplatforms.com/organization/about/australian-microbiome).