Abstract
Plants grown in distinct soils typically harbor distinct microbial communities, but the degree of the soil microbiome influence on plant microbiome assembly remains largely undetermined. We also know that the microbes associated with seeds can contribute to the plant microbiome, but the magnitude of this contribution is likely variable. We quantified the influence of soil and seed microbiomes on the bacterial community composition of seedlings by independently inoculating seeds from a single cultivar of wheat (Triticum aestivum) with 219 unique soil slurries while holding other environmental factors constant, determining the composition of the seed, soil, and seedling bacterial communities via cultivation-independent methods. Soil bacterial communities exert a strong, but variable, influence on seedling bacterial community structure, with the extent of the soil bacterial contribution dependent on the soil in question. By testing a wide range of soils, we were able to show that the specific composition of the seedling microbiome is predictable from knowing which bacterial taxa are found in soil. Although the most ubiquitous taxa associated with the seedlings were seed derived, the contributions of the seed microbiome to the seedling microbiome were variable and dependent on soil bacterial community composition. Together this work improves our predictive understanding of how the plant microbiome assembles and how the seedling microbiome could be directly or indirectly manipulated to improve plant health.
Subject terms: Microbial ecology, Next-generation sequencing, Microbial ecology
Introduction
The structure and composition of microbial communities associated with plants (the plant microbiome) can influence plant health, with the plant microbiome capable of affecting plant growth, drought resistance, disease resistance, and flowering time among other phenotypes [1–5]. Despite the importance of microbes to plants, a thorough understanding of the drivers of plant microbiome assembly is still lacking, due in part to the strong and highly variable influence of a plant’s surrounding environment [4]. Specifically, we lack a predictive understanding of how variation in soil microbial communities drives differences in the assembly of a plant’s microbiome, and to what extent the soil and seed microbiome contribute to the emerging plant microbiome. While most previous work has focused on those microbiomes associated with adult plants, the impact of microbes can be particularly critical in the earliest life stages of plant development, as seed germination and seedling growth are vulnerable developmental stages that impact plant populations and agricultural productivity [6–9]. Understanding the determinants of microbiome composition in the early life stages of a plant (i.e., seedlings) is important for understanding the overall process of microbiome assembly in plants and for improving our ability to manipulate plant health outcomes.
The composition of the plant microbiome (here referring to microbes associated with internal and external plant tissues including the endosphere, phyllosphere, and rhizoplane) is determined by a combination of abiotic and biotic factors including environmental conditions (e.g., water and nutrient availability), host species or genotype, growth season, and plant growth stage [1, 10–15]. However, the soil or field site in which the plant is grown is often the most important determinant of plant microbiome composition [16–18]. One explanation for this is that soil represents the main source of microbes that colonize plants, with a large fraction of the plant microbiome represented by taxa derived from soil [19–21]. However, not all soil microbes are equivalent in their ability to colonize plants, and plant-associated microbial communities are quite distinct from soil communities [10, 17, 22] with plants actively or passively selecting for particular microbial taxa.
While the influence of soil on plant microbiomes has been documented across plant species and geography, we do not know how the magnitude of this soil microbiome effect varies across different soil types, and the degree to which the plant microbiome is predictable from the composition of the soil microbiome. This knowledge gap persists for two important reasons. First, our current understanding of plant microbiome assembly is based primarily on studies limited to one or a few soils, making it difficult to predict patterns of assembly across variable soil backgrounds and to identify the range of microbial taxa that can associate with plants. Second, soils with distinct edaphic characteristics typically also have distinct microbial communities. This makes it difficult to determine the extent to which the soil-specific plant microbiomes are a product of soil edaphic factors (e.g., soil pH, nutrient concentrations, organic C pools) or a product of the different soils having distinct microbial communities. A better understanding of how soil microbes influence plant microbial communities, independent of other soil variables, would improve our ability to determine the relative importance of biotic and abiotic soil variables to plant microbiome management and manipulation.
What also remains unclear is the extent to which microbial taxa on or inside seeds influence the germinating plant microbiome, and how the importance of the seed microbiome may vary depending on the soil in which the plant is grown. Seeds harbor living microbes that can include beneficial and pathogenic microbes that influence germination success [7, 23–27]. As potential early colonizers of plants, those microbes found in or on seeds may also directly or indirectly shape the composition of the microbial communities associated with plants as plants grow. However, the contribution of the seed microbiome to the plant microbiome is not often investigated in studies of plant host community assembly, as typically either the seed microbiome is not directly characterized, or seeds are sterilized prior to planting (but see refs [28–30]). Seed surface sterilization is a common practice to ensure a controlled (and pathogen reduced) background, but it may obfuscate the contribution of seed taxa to a plant’s microbiome. Exploring if, and how, seed-derived microbes persist in seedlings across a variety of soil microbiome backgrounds would advance our understanding of the relative importance of these two microbial sources (seed versus soil) in shaping the microbiomes of growing plants.
Investigating the importance of the soil microbiome to plant microbiome assembly requires growing plants exposed to a wide range of different soil community types while reducing variation in environmental and edaphic factors. We used a collection of over 200 distinct soils to make soil-free slurry microbial inocula and then investigated the community structure and distribution patterns of soil-derived and seed-derived bacterial taxa in inoculated seedlings. Here we focus on wheat (Triticum aestivum), an important agricultural crop that accounts for a large share of both global cropland area and global food trade [31].
The goal of this study was to understand the influences of soil and seed microbial communities in determining the emerging wheat plant microbiome. We measured the extent to which the seedling microbiome community composition varied with exposure to distinct soil microbial communities, hypothesizing that soil communities would have a non-random and predictable effect on seedling community composition independent of environmental factors. In other words, seedling communities should be more similar to the soil communities to which they are exposed than to other soil communities. We further sought to quantify the relative contributions of seed and soil taxa to the seedling microbiome and asked whether the importance of seed versus soil-derived communities varies depending on the soil microbiome in question. The relative strength of the soil influence likely depends on the composition of the soil microbiome, as we would not expect all soils to have the same numbers and types of taxa capable of associating with plants. Finally, we assessed the diversity of soil taxa that are capable of associating with seedlings and identified the soil and seed-derived taxa that were most commonly detected across seedlings.
Methods
Soil sample collection and characterization
To capture a range of distinct soil microbial communities, we collected 219 unique soil samples from across the continental United States. Soils were collected in the summer of 2018 following a standardized collection protocol. Sample locations were chosen to span a wide variety of cultivated and natural systems (including farms, forests, grasslands, and gardens). At each sampling location, approximately 30 volumetric ounces of soil were collected by excavating to 10 cm depth (following removal of the litter layer, if present). Information on site characteristics, including the dominant vegetation and soil amendments, was collected at the time of sampling (Supplementary Table S1). All soil samples were transported at ambient temperature to the University of Colorado Boulder where they were stored at 4 ˚C until further processing. Each soil was sieved to 2 mm, homogenized, and then eight 5.5 g (±0.25 g) soil sub-aliquots were stored at –20 ˚C. These aliquots were used for subsequent soil slurry preparation and DNA extraction. Remaining soil was stored at 4 ˚C for soil analyses with these analyses conducted by the Soil, Water and Plant Testing Laboratory at Colorado State University (Supplementary Table S1).
Seedling germination assay
To test the influence of distinct soil microbial communities on the wheat seedling microbiome while minimizing climate and edaphic effects, we inoculated wheat seeds with soil slurries generated from our soil collection, and germinated the seeds on autoclaved germination paper in the controlled conditions of a growth chamber. We prepared microbial inoculants by creating a slurry of each soil sample. Conical tubes containing 5.5 g (±0.25 g) frozen soil were thawed at room temperature. We then added 10 mL of sterile phosphate-buffered saline (PBS) to each tube, and then mixed tubes for 1 h via 360˚ rotation on a Hula mixer, with a 10 s shake step every 45 s. The tubes were centrifuged at low speed (600 × g) for 4 min to separate larger soil particles from the PBS-microbe suspension. The soil-free supernatant (“slurry,” ~ 9 mL recovered) was transferred to fresh sterile tubes. A 0.5 mL subsample of each slurry was immediately frozen for later DNA analysis. We confirmed that live microbes were present in the soil slurries by culturing 100 µL of slurry from four test slurries on LB agar for 24 h at 30 ˚C after which we observed complete “lawn” coverage, with no colonies detected in the corresponding soil-free PBS control samples. We compared the bacterial communities in a subset of the original soils and the corresponding soil slurries and found no notable differences in community composition, the slurry communities generally reflected the communities in bulk soil (Supplementary Fig. S1).
Immediately following soil slurry preparation, the slurries were used to inoculate wheat seeds for the germination assays. Eight Red Winter Monument wheat seeds were evenly spaced in ethanol-cleaned 245 mm2 plastic plate (Corning Untreated 245 mm Square BioAssay Dishes), lined with two layers of autoclaved germination paper (Anchor Heavy Weight Seed Germination Paper SD7615L, 10” × 15”), pre-wet with 75 mL deionized water. Each seed was then treated with 400 µL of inoculant (microbial soil slurry or PBS buffer) by pipetting liquid on seed and nearby surrounding paper. All eight seeds in an individual plate received the same inoculant.
Inoculated seeds were gently pressed between layers of germination paper to secure their position, and plates were stacked vertically in clear plastic bins holding 12 plates each. Plate position was randomized in bins, but an empty plate was placed on top of the assay plate stack to minimize differential light exposures. One buffer control-treated plate was included in every bin. All plates were incubated in a growth chamber (Percival Scientific AR36L) for 7 days on a diurnal cycle with the following conditions: day (lights on) 6 am–8 pm, 22 ˚C; night (lights off) 18 ˚C. One plate of eight seedlings was used for each of the 219 soil slurry inocula treatments. We also replicated a subset of the soil slurry inocula treatments (N = 15) an additional three times for a total of four replicates per soil slurry treatment. These 15 replicated soils were chosen to capture different types of microbial communities, and were thus selected to span a range of pH values, as pH is known to be strongly associated with differences in bacterial community composition [32]. Together the germination assays represented over 2500 treated wheat seedlings.
Following a week of growth, seedlings ranged in size with roots an average of 9.0 cm (range 0.5–14.3 cm) and shoots an average of 6.7 cm in length (range 0.5–10.4 cm). The rates of seed germination for this wheat cultivar averaged 97.5% across all treatments. To process seedling tissue for DNA extraction, first seeds and seed casings were removed from plant tissue with sterile forceps. The remaining plant tissue (root and shoot only) from all germinated seedlings was then transferred to one piece of parchment paper and placed in a drying bag. Seedling material was dried at 45 ˚C for 48 h in a drying oven. Dried seedling tissue was then transferred to –20 ˚C freezer for storage until DNA extraction. As our samples were dried prior to DNA extraction and the drying process may influence the plant microbiomes, we compared the bacterial communities between oven dried and fresh-to-frozen seedling plant tissue for the replicated seedlings to determine if the drying process introduced biases in our assessment of bacterial community composition (Supplementary Fig. S2). Fresh and dry-frozen seedlings retained similar bacterial communities and we found a minimal effect of sample processing compared to the effect of soil slurry treatment (Supplementary Fig. S2).
Bacterial community assessment: DNA library preparation and sequencing
To characterize the microbial communities in our 219 soil slurries, we extracted DNA from all slurries and a subset (n = 34) of soils and included procedural PBS blank and extraction kit negative controls to identify any potential contaminating taxa in experimental materials. DNA extraction was performed on soils and thawed slurry aliquots (400 µL of soil slurry per sample) using the Qiagen PowerSoil Kit in 96-well plates following the manufacturer’s instructions.
To characterize the microbial communities of the wheat seedlings, we extracted DNA from soil slurry-treated and control-treated seedlings. Seedling tissue (root and shoot combined) was first pulverized, then sterile swabs of ground plant tissue were transferred to Qiagen PowerSoil Kit 96-well plates. To characterize the microbial communities of the wheat seeds, we extracted genomic DNA (gDNA) from pools of ground seed tissue. We used one lot of wheat seeds for all experiments and sequencing. Six separate pools of approximately 3.5 g seeds each were ground with a sterile mortar and pestle using liquid N2. Subsamples were drawn from these six pools to yield a total of 18 seed samples. However, due to low biomass only nine seed samples passed through our data processing steps for inclusion in downstream analyses.
We amplified and sequenced the hypervariable V4–V5 region of the 16S rRNA gene using 515f/806r barcoded primers as performed previously [33, 34]. PCR amplification was performed in duplicate for 219 slurries with 6 PBS blank controls, 35 DNA extraction blank negative controls, and 6 no-template control (PCR) negative controls. In a separate duplicate PCR procedure, we used the same methods on 358 plant samples and 34 soil samples, with 10 procedural blank controls (PBS), 44 DNA extraction blank negative controls, and 5 no-template PCR negative controls. Amplicons were pooled, cleaned, and normalized using SequalPrep Normalization plates (Thermo Fisher Scientific, Waltham, MA). We sequenced all samples at the University of Colorado Next Generation Sequencing Facility on a MiSeq (Illumina) platform with 2 × 150 bp paired-end chemistry. Pooled amplicons were run on two separate MiSeq runs with the resulting data from both runs combined prior to downstream processing.
Sequence processing and data cleaning
Raw reads were processed using a DADA2-based bioinformatic pipeline (DADA2 version 1.10.1 [35], Fierer Lab pipeline v.0.1.0 [36]). Briefly, raw reads were demultiplexed using idemp and primers removed using cutadapt (version 1.8.1). Sequences were filtered and trimmed using the following settings: (truncLen=c(140,145) for plants, c(145,145) for slurries, maxEE=c[2, 2], truncQ=2, maxN=0, rm.phix=TRUE), and inferred using the DADA2 algorithm on pooled samples (pool=true). Error learning and sequence inference were performed independently for each sequencing run as recommended, and sequence tables were merged together using the mergeSequenceTables command. We then removed chimeras and used the DADA2 naïve Bayesian classifier method with the SILVA database v132 [37] for taxonomic identification of the resulting amplicon sequence variants (ASVs).
Prior to downstream analyses, we first removed all taxa not classified as bacteria. We then measured and subsequently removed chloroplast and mitochondrial reads from the dataset. We note that for the plant samples, plant host reads represented a majority of reads (75% of reads on average). We further filtered the data and removed samples with low- or poor-quality sequence data (samples with less than 1000 bacterial reads). We also removed instances of ASVs represented by less than ten reads in a given sample. After removing chloroplast and mitochondrial reads and imposing these quality filtering steps, our dataset of seed, seedling, and soil slurry inocula communities included ~5 million total bacterial reads, with an average of 10,606 reads per sample (range 929–35098 reads). We opted not to rarefy to avoid discarding additional information, and instead converted data to relative abundance values. After quality filtering, we were left with 208 slurry-treated seedling samples, 218 soil slurry samples, 25 control-treated seedling samples, and 9 seed samples. We included a number of negative controls to check for potential contaminants introduced during the experimental procedure, DNA extraction, and PCR amplification steps. There were no taxa consistently detected in the blanks, and the blanks typically had far fewer reads than in the actual samples (median of 1816 reads in the “blank” samples versus a median of 11241 reads in soil, seed, and plant tissue samples).
Tree construction
To generate a phylogenetic tree of the most abundant bacterial taxa in our dataset, we calculated phylogenetic relationships with maximum likelihood using RaxML [38]. To limit the size of the tree, we first restricted the number of ASVs in the tree by removing ASVs with a total relative abundance of less than 0.08 across the filtered dataset, which left 533 ASVs in total. This filtering threshold was applied only for the ASVs used in the tree visualization (and Supplementary Fig. S2 visualization), and was not used in any other analyses. The 16S rRNA gene sequences of these abundant ASVs were aligned with MUSCLE (version 3.8.31). Aligned reads were used to construct a tree with RaxML (version 7.3.9) using 100 Bootstrap searches followed by 20 ML searches to return the best tree. The resulting tree was annotated in iTol [39].
Quantitative PCR of soil slurry samples
We used 16S rRNA gene primers targeting the same 515f/806r region of the 16S rRNA gene for the quantitative PCR (qPCR) analyses as used for the sequencing described above, but the primers used for qPCR did not include Illumina adapters (515F: 5’-GTGCCAGCMGCCGCGGTAA-3’; 806R: 5’-GGACTACHVGGGTWTCTAAT-3’). For the DNA standard we used Escherichia coli gDNA purchased from the American Type Culture Collection (ATCC #700926D5). To generate a standard curve for each qPCR plate we created seven tenfold serial dilutions with the E. coli gDNA. Each qPCR reaction comprised of 1.25 µL forward and reverse primers, 12.5 µL master mix (ABsolute QPCR SYBR Green Mix, Thermo Fisher #AB-1159/A), 5 µL PCR-grade water, and 5 µL template DNA. Standards were run in duplicate on each plate, and all soil slurry samples were duplicated across separate plates to control for plate-to-plate variability. We used the CFX Connect Real-Time System (Bio-Rad) with the following cycling conditions: 95 °C 15 min [94 °C 45 sec, 50 °C 1 min, 72 °C 1:30 min] × 40 cycles, 72 °C 10 min.
Plate-wise comparisons to the standard curve yielded an estimate of genome copy number in each sample. To relate our standard to template genome copies we used 4.64 Mb as the E. coli genome size and assumed the average weight of a base pair to be 650 Daltons. Across the six qPCR plates all standard curves displayed a strong linear relationship between cycle threshold and log of gene copy number (R2 > 0.99 in all cases). We excluded samples whose coefficient of variance across the duplicate pair was greater than 15% (n = 187 remaining). We report the results in E. coli genome equivalents per µL soil slurry, which can be inferred as an index of total bacterial biomass.
Comparing bacterial community composition in seed, seedlings, and soil slurries
Statistical analyses and data visualizations were performed in the R environment (version 3.6.3 [40]), with all plots generated using ggplot2 (version 3.2.1 [41]). The sample map was created with the maps package (version 3.3.0 [42]).
We measured community dissimilarity between bacterial communities using Bray–Curtis distances in Vegan [43]. We visualized the overall differences in bacterial community composition by using nonmetric multidimensional scaling (NMDS) ordination plots. We tested for differences in microbial communities with permutational multivariate analyses of variance (adonis), and tested for differences in community dispersion using dispersion analysis (betadisper) and ANOVA on resulting dispersion values. We compared community distance and community richness values with Welch’s two-sample t-test or Wilcoxon rank-sum test as appropriate. Community richness was calculated by summing the number of distinct ASVs in each sample in the quality-filtered dataset.
ASV categorization
We defined seed-associated taxa as those ASVs found in more than three (out of nine) homogenized seed samples (29 ASVs from 55 total ASVs detected in seeds). These 29 “seed-associated” ASVs represented 96% of bacterial reads recovered from seed samples. Similar thresholding approaches have been used elsewhere to categorize ASV sources [44]. In addition, given the low biomass of the seed samples there was a higher possibility of well-to-well contamination during DNA extraction and PCR amplification [45], and thus we chose to exclude those ASVs detected infrequently in the homogenized seed samples.
For the predictive and correlative analyses, we used the full cleaned dataset of ASVs, with no further abundance thresholding. The “potential plant colonizers” used in the beta regression models and Random Forest model were defined as all ASVs in the soil slurry samples that were also detected in one or more plants, or put another way, all slurry-derived ASVs that had demonstrated potential to associate with plants. The “common” plant colonizers used in the rank correlation analysis were defined as ASVs detected in more than three plants, a threshold designed to restrict analysis to the more common and consistent plant-associating taxa and to make the correlation strategy more robust.
To limit number of ASVs used for the visualization of the phylogenetic tree and in Supplementary Fig. S3, we applied a relative abundance threshold of 0.08 that resulted in 533 ASVs. This limited dataset was used for visualizations only. To categorize the association and source in Supplementary Fig. S3, we used the sample number thresholds listed below to highlight those taxa we were most confident in categorizing, as our goal was to identify taxa that were more commonly associated with a given plant host microbial habitat. In Supplementary Fig. S3, seed-association is defined as ASVs detected in >3 seeds, plant-association is defined as ASVs detected in >3 plants, and slurry association is defined as ASVs detected in any of the soil slurries. The ASVs that did not meet these criteria were defined as being of undetermined origin. There were 14 of these “undetermined” ASVs in the visualization dataset, and they were detected in 11 seedling samples on average, representing 0.06% of total reads. These undetermined taxa likely either came from the soil but were in such low abundances as to be undetectable [44] or were derived from the experimental environment.
Predicting the proportion of seed-associated taxa in seedlings from soil microbiome features
To relate soil bacterial community characteristics to the proportion of seed taxa in seedling microbiomes, we used beta regressions (betareg version 3.1-3 [46]) with the proportion of seed taxa as the response variable. We elected to use beta regressions because the response variable was a continuous proportion [47]. To independently identify soil slurry taxa that influenced the seedling microbiome composition (proportion of seed-associated taxa), we developed a Random Forest model using Caret (version 6.0-86 [48]) with the Ranger package (version 0.12.1[49]) (ranger(), 1000 trees, mtry=96, split rule = “variance,” importance = “impurity,” five cross-fold validations). We used 80% of the data (n = 166) to train the model (prediction error (MSE) = 0.040, R2 = 0.31), and the remaining 20% of samples (n = 42) to test the model predictions.
We identified specific taxa that were commonly detected across inoculated seedlings (i.e., bacterial ASVs that were present in more than three inoculated plant samples, less than 5 out of 9 seed samples, less than 10 out of 25 untreated blank samples; n = 101), and individually compared the rank of these taxa in the soil slurry community with the rank in seedling community using Spearman rank correlations. These “common” plant colonizers represented 45% of the proportional abundance across all seedling samples. We chose to use rank correlations because relative abundance values were non-normally distributed and highly skewed, and also because relative abundance values of individual ASVs were so different between soils and seedlings due to the high diversity of the soil slurry communities. The rank abundance comparisons were performed at different levels of taxonomic resolution (order, family, genus, ASV). Multiple test corrections of the rank correlations were performed with the False Discovery Rate method (p.adjust, method = “fdr”).
Results
To characterize the influence of soil microbiome on the development of the seedling microbiome, we inoculated sets of wheat seeds with 219 distinct soil slurries harboring unique microbial communities. To isolate the effects of the soil inocula from edaphic and climatic variables, we grew seeds on sterile germination paper under controlled growth chamber conditions, varying only the soil slurry inocula applied (Fig. 1). To identify the relative importance of seed-derived microbes in the assembly of the seedling microbiome, we opted not to surface sterilize the wheat seeds and we characterized the bacterial taxa associated with the seeds used in this study. While the seedling microbiome can be considered to include bacteria, archaea, fungi, viruses, and protists, we focus here on bacteria only. We characterized the bacterial communities in the wheat seeds, seedlings, and soil slurry inocula using 16S rRNA gene amplicon sequencing. Although the PCR primers amplify the 16S rRNA gene from both bacteria and archaea, archaea were typically in relatively low abundance across the soils analyzed (average 1.6%, range 0–16%) and no archaea were detected in the wheat seed or seedling microbiomes.
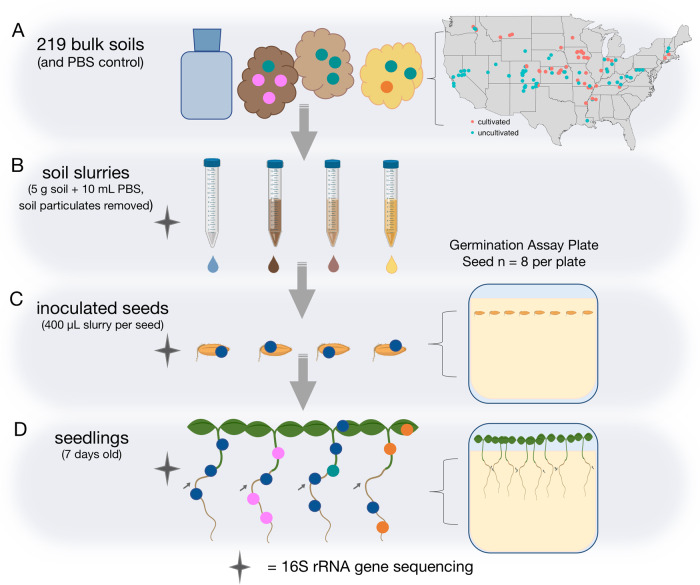
Fig. 1. Experimental design.
A Soil samples (n = 219) were collected from across the continental United States. B Soil slurries were created from each soil by mixing 5 g soil with 10 mL PBS, and slowly centrifuging to remove particulates. C Eight seeds were inoculated with each soil slurry, and grown on sterilized germination paper in a growth chamber. D After 1 week, all eight seedlings from one plate were combined and destructively sampled. In panels B–D, the stars indicate soil, seed, and seedling samples were analyzed with 16S rRNA gene sequencing to determine microbial community composition. Created with BioRender.com.
Diversity and community composition of soil inocula, seeds, and seedlings
The bacterial communities in the soil slurries were highly variable in composition, reflecting the wide range of environments from which they were collected (Fig. 1). These soils came from natural and cultivated ecosystems that included desert soils, temperate forest soils, and cropland soils, and varied accordingly in their edaphic characteristics and bacterial community compositions. For example, soil pH values ranged from 4.5 to 10.5 and organic matter concentrations ranged from 0.6 to 7.9%. Likewise, the relative abundances of the dominant bacterial phyla such as Proteobacteria and Actinobacteria ranged from 0.07 to 89% and 0.0 to 62%, respectively (Supplementary Fig. S4). There were no ASVs detected in all soil slurries, and few ASVs were detected in most soil inocula with individual ASVs found in only three distinct soils, on average.
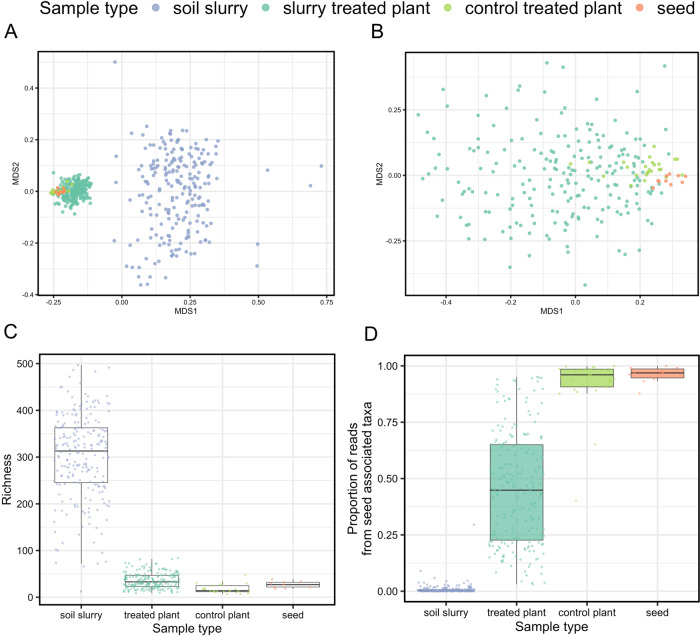
The inoculation of wheat seeds with different soil slurries had a strong influence on the bacterial communities associated with the week-old seedlings. Seedlings inoculated with soil slurries had distinct, more diverse, and more highly dispersed bacterial communities compared to seedling inoculated with PBS buffer only (control-treated), and compared to seeds (Fig. 2A, B, PERMANOVA R2 = 0.09, p < 0.001). A total of 546 ASVs were detected across the soil slurry-inoculated seedlings, with an average of 36 bacterial ASVs detected per slurry-inoculated seedling sample (range 9–86 ASVs, Fig. 2C). Control-treated seedlings had lower diversity than inoculated seedlings with an average of 18 ASVs per sample (range 7–51 ASVs per seedling sample, Wilcoxon rank-sum test p < 0.001, Fig. 2C). Interestingly, the control-treated seedlings had bacterial communities more similar to seeds than to slurry-treated seedlings, as observed in an NMDS ordination (Fig. 2B) and via statistical comparison of community distances (Bray–Curtis mean community distance: control plant versus treated plant = 0.78, control plant versus seed = 0.70; Welch t-test p < 0.001). Together, these results demonstrate that inoculation with soil bacteria has a strong influence on the composition of seedling microbiomes.
Fig. 2. Community composition and richness of soil slurries, wheat seedlings, and wheat seeds.
A,B NMDS ordination plots (first two of three dimensions) of bacterial community structure in soil slurries, soil slurry-treated seedlings (“treated plant”), PBS buffer control-treated seedlings (“control plant”), and seeds. Panel B includes the same samples as panel A with the soil slurry samples excluded. Panel A stress: 0.11; Panel B stress: 0.18. PERMANOVA R2 = 0.09, p < 0.001; homogeneity of dispersions test, ANOVA p = 171, p < 0.001. C Bacterial richness per sample type calculated as number of distinct ASVs out of 1000 bacterial reads per sample. D The proportion of reads from “seed-associated” taxa (ASVs detected in >3 seed samples) across different sample types.
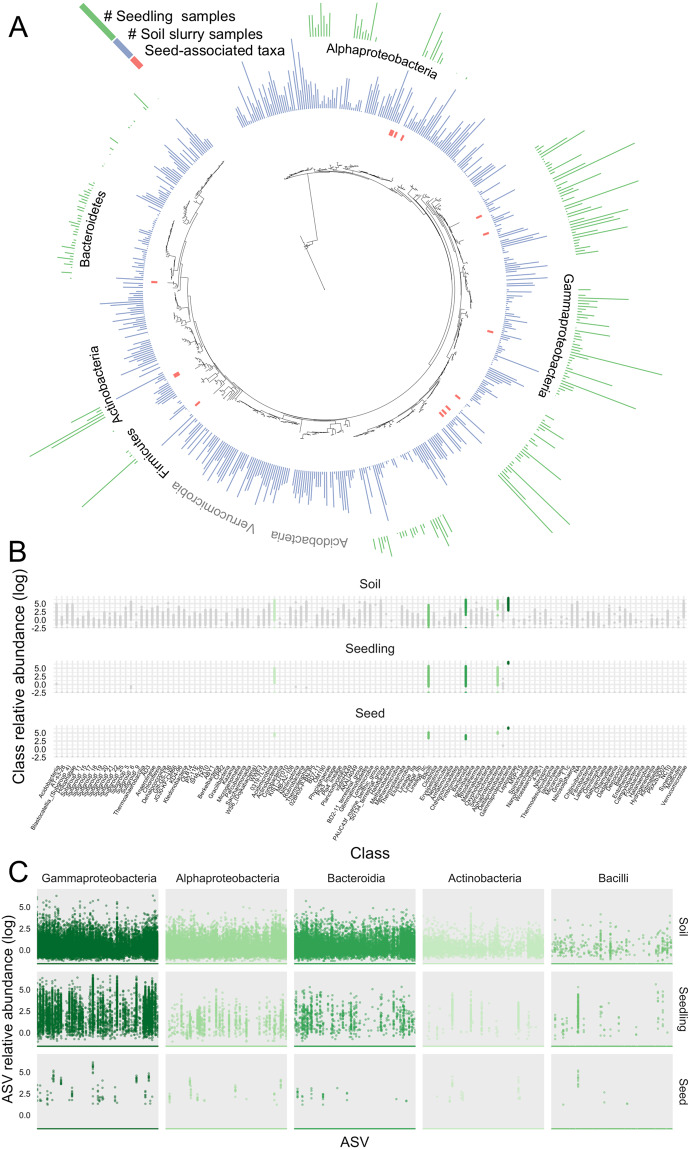
The composition of the soil slurry-treated seedling bacterial communities was variable, but broadly corresponded with previous studies of plant-associated bacterial communities in that the seedlings were dominated by the following bacterial phyla: Gammaproteobacteria (61–100% of bacterial reads), Bacteroidetes (0–28%), Alphaproteobacteria (0–20%), and Actinobacteria (0–15%) [50–52]. As expected based on previous plant and wheat microbiome studies [9, 19], the bacterial taxa associated with the inoculated seedlings represented a small fraction of the bacterial taxa and lineages found in soil, with five classes of bacteria accounting for >99% of 16S rRNA gene reads in seedlings (Fig. 3A–C). Bacterial taxa that were relatively abundant in the soil inocula, including Verrucomicrobia, WPS-2, and Acidobacteria phyla, were not detected in any seedlings (Fig. 3B). We note that many bacterial taxa that were relatively abundant in inoculated seedlings had relatively low abundances in the corresponding soil inocula (e.g., Burkholderiaceae Massilia ASV 5, and Pseudomonadaceae spp. ASV 6), while few taxa were relatively abundant both in the soil inocula and the corresponding inoculated seedlings (e.g., Pseudomonadaceae spp. ASV 10, Enterobacteriaceae Klebsiella ASV 4) (Fig. 3A and Supplementary Table S2). We did, however, find a small number of soil-dwelling ASVs (n = 7) that were detected in seedlings across a majority of soil slurry treatments. Members of the Pseudomonas genus were the most common taxa found to colonize plants from soil, accounting for 10 of the top 20 most common soil-derived taxa recovered in seedlings (Supplementary Table S2).
Fig. 3. Bacterial diversity in soil slurries, seedlings, and seeds.
A A maximum-likelihood phylogenetic tree of the top 533 most abundant bacterial ASVs detected in this study. Colored bar height indicates prevalence (number of samples) in which each ASV was detected in seedlings (green), and soil slurries (blue); red bars indicate seed-associated taxa (ASVs detected in >3 seed samples). B Relative abundances of bacterial classes shown for seed, seedling, and soil samples. Bacterial classes that were detected in seedlings are highlighted in green shades, all other classes are colored gray. C Relative abundance of ASVs in the five plant-associating classes shown for seed, seedling, and soil samples.
Overall, few individual bacterial ASVs were shared across seedlings, likely due to the distinct nature of the soil inocula. Only 16 ASVs were detected in more than half of the seedling samples, while 235 ASVs were found in fewer than 10 of the 208 seedling samples (Supplementary Fig. S3). This low rate of occupancy is likely the result of the soil inocula harboring such distinct bacterial communities (Supplementary Fig. S4) and highlights the importance of surveying large numbers of distinct soil communities to capture the breadth of potential plant-associated taxa. Put another way, if we had only focused on three distinct soils, we would have identified only ~30–80 seedling-associated bacterial ASVs, a number far below the total number detected upon examining all 208 distinct soil inocula (546 ASVs). The number of distinct wheat-associated bacterial taxa identified increased with each additional slurry inocula tested (Supplementary Fig. S5).
The most commonly detected ASVs across the wheat seedling samples were found to be seed associated as opposed to ASVs originating from the soil slurries (Supplementary Fig. S3). Nine of the 29 ASVs identified as being seed associated (see Methods) were among the 11 most commonly detected ASVs across the seedlings, including the only ASV detected in all seedling samples (Enterobacteriaceae Pantoea, ASV 3) [53]. The seed-associated ASVs were members of the Gammaproteobacteria, Alphaproteobacteria, Actinobacteria, Bacilli, and Bacteroidia bacterial classes (Fig. 3A). The seed-associated ASVs were either absent or found in low abundances in the soil inocula (Fig. 3A and Supplementary Fig. S3). We note that these taxa identified as “seed associated” could have been vertically inherited from the parent plant (i.e., seed endophytes) or introduced to the seed from external sources.
Inoculation with soil bacteria decreases the contributions of seed-associated bacteria to the seedling microbiome
Given that the soil slurry communities demonstrably influenced community structure in seedlings, but that the most ubiquitous taxa detected across seedlings appeared to come from seeds, we next sought to determine the relative contributions of soil slurry taxa versus seed-associated taxa to the seedling bacterial community. The proportion of seed taxa detected in seedlings was calculated by summing the relative abundances of the seed-associated ASVs in each seedling sample. The seed-associated ASVs accounted for a median of 45% of bacterial 16S rRNA gene reads obtained from the soil slurry-inoculated seedling samples (range 3–95%), and represented approximately a third of inoculated seedling ASV diversity (mean 33% ASVs, range 7–73% seed-associated ASVs in seedlings) (Fig. 2D). This variance in the proportion of seed taxa detected across seedlings was notable, as we highlight in more detail below. As expected, the control-treated seedlings were more strongly dominated by seed taxa (median of 92% of 16S rRNA gene reads, Wilcoxon rank-sum test p < 0.001) (Fig. 2D), a pattern corroborated by their community similarity mentioned previously (Fig. 2B). The ASVs detected in the control-treated seedlings that accounted for the remaining <8% of 16S rRNA gene reads were presumably introduced from the air inside the lab or growth chamber, or from experimental materials like the assay plates or the deionized water used to wet the germination paper, and were not consistently found across the control-treated seedlings.
Together these results show that seed-associated taxa represent an important, but highly variable, component of the week-old seedling microbiome. However, it is important to note that a few seed-derived ASVs were the most commonly detected ASVs across inoculated seedlings (Supplementary Fig. S3), suggesting a restricted but consistent role of microbe transmission from seed to seedling regardless of soil community exposure.
Soil bacterial contributions to the seedling microbiome are determined by soil microbial community characteristics
We next assessed whether the variation in the proportion of seed-associated taxa detected across inoculated seedlings (3–95%; Supplementary Fig. S6) could be explained by differences in the soil slurry inocula. We hypothesized that the proportion of seed taxa remaining on seedlings would be higher in seedlings inoculated with soil slurries that contained lower proportions of taxa that could potentially associate with plants. Here, we defined “potential plant colonizers” as soil slurry taxa (ASVs) that were detected in one or more seedling samples (see Methods). We used beta regressions to detect relationships between soil bacterial community composition and the proportion of seed taxa remaining in seedlings, and also used Random Forest modeling to independently verify these results by predicting the proportion of seed taxa in seedlings from soil slurry bacterial community composition. In addition, we assessed whether total bacterial biomass in the soil slurries (estimated from qPCR) was related to the observed variation in seed-associated taxa found across seedlings.
We found that the proportion of seed-associated taxa in the seedling microbiome was partially explained by the proportion of potential plant colonizers found in the soil slurries. If the soil slurry communities harbored more taxa with the potential to colonize seedlings, the proportion of seed taxa remaining on week-old seedlings was lower (ASV model phi = 3.9, pseudo R2 = 0.17, p < 0.001; Pearson r = –0.41) (Supplementary Fig. S6). While this relationship was strongest when considering bacterial taxa at the ASV level (i.e., the sum relative abundance of all ASVs found to associate with one or more plants), the pattern held across levels of taxonomic resolution up to the class level (the sum relative abundance of all five classes found to associate with one or more plants) (class model phi = 3.7, pseudo R2 = 0.12, p < 0.001) (Supplementary Fig. S6). Particularly, the relative abundance of Gammaproteobacteria in the soil inocula was negatively correlated with the proportion of seed taxa in seedlings (Pearson r = –0.33, p < 0.001), meaning that the higher the gammaproteobacterial relative abundances in soil slurries, the lower the abundances of seed-associated taxa on the seedlings (Supplementary Fig. S6). These findings suggest that the likelihood of seed taxa persisting in a growing seedling depends on the relative abundances of specific taxa in the surrounding soil.
To complement the beta regressions and to test the predictive power of soil microbiome information for determining the relative contributions of seed taxa to the seedling microbiomes, we built a Random Forest model to predict the proportion of seed-associated taxa in seedlings using the relative abundances of bacterial families in the soil slurry inocula as predictors. We used a Random Forest model to independently identify soil taxa important for determining the magnitude of seed community influence without any a priori knowledge of whether bacterial taxa were plant or seed associated. The Random Forest model described over 30% of the variation in proportion of seed taxa in seedlings (R2 = 0.32, p < 0.001), and the model predictions correlated strongly with the true values of seed taxa abundances in the test set (Pearson r = 0.64, p < 0.001) (Supplementary Fig. S6). The most important bacterial families for generating these predictions were families within the Gammaproteobacteria and Actinobacteria phyla (Supplementary Table S3). Together these results show that the relative contribution of seed taxa to seedling communities is strongly influenced by the soil community, with the magnitude of colonization from seed taxa dependent on the relative abundance of specific lineages (especially Gammaproteobacteria) in the soil. The success of seedling colonization by soil or seed microbes is strongly related to the composition of the soil bacterial communities.
To test whether the bacterial biomass in soil slurries influenced the proportion of seed or soil slurry-associated taxa proliferating in seedlings (a “mass effect”), we related the qPCR-based estimates of bacterial genome copies detected in each soil slurry to the proportion of seed-associated taxa detected in seedlings (Supplementary Fig. S7). We found a weak negative relationship between the qPCR-based estimates of bacterial biomass in the soil slurries and the proportion of seed taxa found to be associated with the seedlings (beta regression phi = 3.4, pseudo R2 = 0.07, p < 0.001, Pearson r = –0.28, p < 0.001). Thus, the biomass of soil slurry communities also influenced the relative success of soil and seed-derived bacteria in seedlings, but to a lesser extent than the taxonomy of those taxa.
The soil microbiome is predictive of the seedling microbiome
We next determined the degree to which the soil slurry microbiome had a predictable influence on the structure or membership of seedling microbiomes. First, to determine the reproducibility of bacterial community assembly in our seedling system, we inoculated seeds with four replicate slurries derived from each of 15 soils (see Methods). We found that, across the replicate seedling samples (those inoculated with the same soil slurry), soil slurry inoculum explained 70% of the variation among seedling communities (PERMANOVA R2 = 0.70, p < 0.001; Supplementary Fig. S8). In addition, pairwise community distances across replicates that received the same soil slurry inocula were significantly lower than distances between replicates of different soil slurry treatments (Welch’s t-test p < 0.0001; Supplementary Fig. S8). The consistency and similarity of community composition within replicates highlights the reproducibility of community assembly in this system.
We also compared the pairwise distances in community composition between soil slurry inocula and inoculated seedlings. We expected that seedlings inoculated with more similar soil communities would have more similar microbiomes. We found that a seedling sample inoculated with a given soil slurry community consistently harbored bacterial communities that were more similar in composition to that soil slurry community than to other (random) soil slurry communities to which it was not exposed (Supplementary Fig. S8). Even though seedlings and slurries had distinct bacterial communities with plants selecting for particular taxa (Fig. 2A), the influence of soil slurries on seedling microbiome composition was clearly evident.
Correspondence between soil bacterial abundances and abundances on inoculated seedlings
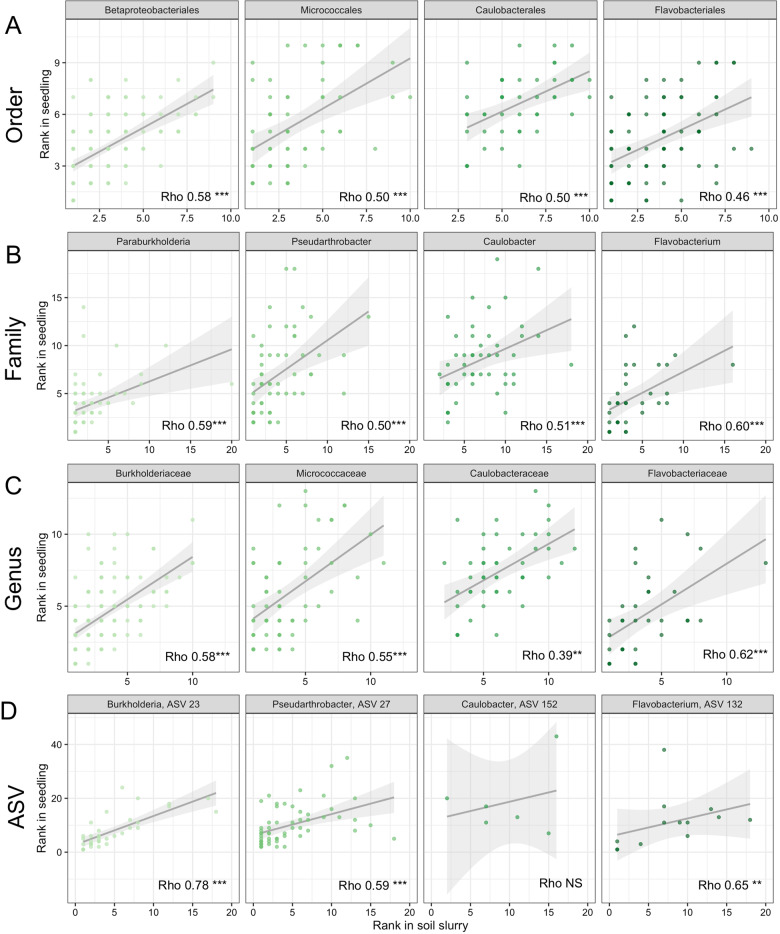
We next investigated whether there were specific taxa responsible for the community-level patterns of similarity between corresponding soil slurries and slurry-treated seedlings. We asked whether the abundance of the most common plant-associated ASVs in the soil slurries explained the presence or abundance of those ASVs in seedlings. Even though many plant-associating taxa were detected at relatively low relative abundances in the soil slurries, within a restricted group of around 100 soil taxa that were most commonly found across seedlings, their rank abundances in soil slurries corresponded with their rank abundances in seedlings. The correlations between taxon abundances in soil slurries and the abundances in inoculated seedlings were evident at varying levels of taxonomic resolution, and were particularly strong for several individual ASVs (Fig. 4 and Supplementary Table S4). For example, the rank abundance of family Burkholderiaceae in slurries and on seedlings correlated at a Rho value of 0.59 (df = 169, p < 0.001, Fig. 4), while Burkholderia ASV 23 correlated at a Rho value 0.78 (df = 60, p < 0.001, Fig. 4). These results suggest that, for those taxa capable of associating with wheat seedlings, their initial abundance in the soil slurries can be an important determinant of their relative abundance on the corresponding plant.
Fig. 4. The relationship between the rank abundance of plant-associating bacterial taxa in soil slurry inocula and in seedlings across different levels of taxonomic resolution.
A Common plant-associating orders, B families within the orders from panel A directly above, C genera within families from panel B directly above, D ASVs within genera from panel C directly above. Asterisks indicate Spearman correlation values significant at ***p < 0.001, **p < 0.01, *p < 0.05.
Discussion
Understanding the influence of the soil microbiome on plant microbiomes is challenging due to the difficulties of disentangling the effects of climactic and soil edaphic properties from the effects of changes in soil microbiome composition. Our experimental setup allowed us to test the predictability of seedling microbiome assembly as a function of soil community composition, although we acknowledge that the controlled nature of our system minimizes additional variation that could be important in more field-relevant conditions. By screening over 200 individual soils with a single wheat cultivar, we were able to show that the soil bacterial community exerts an important but variable influence on the wheat seedling bacterial community, with the seed microbiome contributing the most ubiquitous taxa. The contribution of the soil and seed bacteria to the seedling microbiome is predictable from characteristics of the soil microbiome.
The observed differences between microbiomes of seedlings treated with different soil slurries (Fig. 2) are in agreement with previous work showing that soil microbes exert an important influence on plant microbiomes [8, 10, 14, 54]. We found that soil-derived taxa to typically represent a majority of the week-old seedling microbiome, with seed-associated taxa contributing to approximately a third of bacterial reads in the slurry-inoculated wheat seedlings (Fig. 2D). Seedlings that were not inoculated with soil slurries bacteria harbored a larger fraction of seed-associated taxa. This result suggests that, while seed-associated microbes are capable of colonizing seedlings, the majority of microbes colonizing a plant originate from the soil as has been suggested previously [55, 56].
Few published studies that have investigated the assembly of plant microbiomes have included a characterization of the starting seed community (but see [7, 42]), and we quantified a variable, but consistently present, contribution of the seed microbiome to the seedling microbiome (Fig. 2 and Supplementary Fig. S3). However, since we used a single lot of seeds for all of our experiments, we acknowledge that the starting seed microbiome likely had limited variability. The most commonly detected taxa across seedlings were seed associated (Supplementary Fig. S3), a pattern that could be due to these seed taxa being highly evolved with the plant, potentially due to vertical inheritance, or due to the advantage of being the initial taxa present as plants start to grow [7, 26]. We acknowledge that the young age of the plants (7 days), and controlled conditions of the experimental setup may have made it easier for seed taxa to persist on seedlings, and it is likely this community would continue to shift as the plants matured [12, 57]. However, even if the seed-derived taxa were outcompeted as plants continue to grow, the presence of seed taxa in the earliest stages of a plant’s life could drive later patterns in community assembly, or be critical for the early stages of plant growth [58, 59]. Previous drop-out and arrival order experiments show that historical contingency and priority effects can influence plant microbiome community assembly [60]. Future work should investigate whether those seed-associated taxa that are highly ubiquitous across plants are vertically transmitted across generations, or are seed-mediated environmentally acquired taxa. Identifying the identities and traits of these “successful” seed taxa that effectively colonized emerging seedlings could have important implications for improving seed coat treatment technologies.
Despite the restricted phylogenetic diversity of the bacteria associated with wheat seedlings (Fig. 3 and Supplementary Fig. S4), we generally found few ASVs shared across inoculated seedlings outside of those that were seed associated. The community dispersion across treated seedlings (Fig. 2A, B) and ASV-level heterogeneity highlights the benefits of surveying multiple different soil types when designing experiments to characterize plant-microbe associations. Depending on the specific research questions posed, the number of soil types included in an experiment is an important consideration given that distinct soils can yield such distinct seedling microbiomes.
Despite the variation in community composition across seedling microbiomes, there were a few bacterial taxa that commonly occurred in soils and were consistently detected in seedlings (Fig. 3A and Supplementary Table S2). Those soil-derived taxa that were particularly effective at colonizing wheat seedlings (including members of the Pseudomonas, Pseudoarhthrobacter, and Novosphingobium genera) are likely to be good candidates for probiotic applications in wheat agriculture, as has been discussed previously [61–63]. More generally, the broad collection of paired soil and plant samples captured could inform the selection of other potential taxa that might successfully be used as plant probiotics due to the combination of their abundance across soil types and plant colonization efficacy.
Finally, several lines of evidence support the hypothesis that soil bacterial communities exert a predictable influence on seedling microbiome structure and membership. First, replicate exposure of seeds to soil slurries derived from the same parent soil assembled highly similar and reproducible communities (Supplementary Fig. S8). This reproducibility for a given soil slurry inoculum treatment suggests a dominance of deterministic factors over stochasticity in bacterial community assembly on plants [60, 64], though we acknowledge that the absence of environmental variability and the fact that we focused exclusively on wheat seedlings (not adult plants) may enhance the observed similarity among replicates. Second, bacterial communities in the soil slurries and the corresponding recipient inoculated seedlings were more similar to each other than to “non-paired” (or mismatched) plant and soil communities (Supplementary Fig. S8). The higher similarity between soil slurries and recipient plants shows that plant communities reflect their surrounding soil communities despite the stringent filtering imposed by the plant. Third, of the soil taxa identified as having the ability to associate with plants, their abundances in soil often corresponded to their abundance on seedlings (Fig. 4). The taxa detected as abundant in the soil slurries were not enriched by the wheat seedlings since the bacterial communities in the soil slurries were characterized prior to seed inoculation. We emphasize that, while some dominant soil taxa never colonized seedlings, of those taxa with a demonstrated ability to associate with seedlings, their abundance in soil slurries was often related to their abundance in the seedlings. Fourth, the composition of the soil slurry community influenced the degree to which seed-associated taxa proliferated in seedlings (Supplementary Fig. S6). Variation in the proportion of seed-associated taxa in seedling microbiomes was predictable from the presence and relative abundance of particular bacterial groups in the soil slurries (Supplementary Fig. S6). However, we note that the concentration of bacteria in the soil slurries was also correlated with the proportion of seed-associated taxa in seedlings (but this relationship was relatively weak), with higher bacterial biomass in soil slurries corresponding to seedlings that tended to be colonized by fewer seed taxa (Supplementary Fig. S7). This suggests that the “mass effect” of sheer bacterial cell numbers in soil could add additional explanatory value in considering seedling assembly patterns [65]. Together, these results suggest a hierarchy of assembly forces for seedling microbiome assembly, with more deterministic processes restricting the types of taxa that colonize seedlings and the abundances of those taxa in soil determining their abundances on seedlings. In general, which taxa ultimately associate with plants is largely determined by the abundance and composition of potential plant colonizing taxa found in soil. The role of soil community composition in determining the extent of the soil or seed influence has important implications for attempts to manage plant microbiomes: seed inoculants may not be as effective in soils that have higher abundances of taxa able to colonize plants.
Conclusions
By characterizing the bacterial communities of wheat seeds, over 200 soil slurries, and the seedlings inoculated with those soil slurries, we were able to quantify how the seed and soil microbiomes influence the wheat seedling microbiome. We found soils to have a strong, but variable, influence on the nascent wheat seedling microbiome, and identified a restricted, but important, contribution of the seed microbiome to the seedling microbiome. Since the composition of a plant’s microbiome likely influences plant health [61, 66, 67], determining how the plant microbiome varies across different soil types can ultimately improve our predictive understanding of how the plant microbiome could be directly or indirectly manipulated to improve plant health, a topic of intense interest in basic and applied agricultural research [2, 61, 66, 68].
Supplementary information
Acknowledgements
We thank Jessica Henley and Matthew Gebert for assistance with DNA extraction and DNA sequencing preparation. We thank Spencer Debenport and Charlie Hubbard for input on plant growth protocols and for help coordinating soil collection from agricultural sources. We also thank Lior Gross, Mike Strickland, Gary King, and Maya Montoya-Pimolwatana for their substantial contributions to the soil collection. Funding for this work was provided by a National Science Foundation Graduate Research Fellowship to CMW (DGE 1650115), a US National Science Foundation grant to NF from the Dimensions of Biodiversity program (DEB 1542609), and by Indigo Agriculture.
Data availability
The dataset generated and analyzed in this study along with representative sequences and sample metadata are available on FigShare (10.6084/m9.figshare.c.5323961) and further sample metadata is available in the Supplementary tables.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-00967-1.
References
- 1.Wagner MR, Lundberg DS, Coleman-Derr D, Tringe SG, Dangl JL, Mitchell-Olds T. Natural soil microbes alter flowering phenology and the intensity of selection on flowering time in a wild Arabidopsis relative. Ecol Lett. 2014;17:717–26. doi: 10.1111/ele.12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Vries FT, Griffiths RI, Knight CG, Nicolitch O, Williams A. Harnessing rhizosphere microbiomes for drought-resilient crop production. Science. 2020;368:270–4. doi: 10.1126/science.aaz5192. [DOI] [PubMed] [Google Scholar]
- 3.Compant S, Clément C, Sessitsch A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem. 2010;42:669–78. doi: 10.1016/j.soilbio.2009.11.024. [DOI] [Google Scholar]
- 4.Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK. Plant–microbiome interactions: from community assembly to plant health. Nat Rev Microbiol. 2020;18:607–21. doi: 10.1038/s41579-020-0412-1. [DOI] [PubMed] [Google Scholar]
- 5.Berendsen RL, Vismans G, Yu K, Song Y, de Jonge R, Burgman WP, et al. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018;12:1496–507. doi: 10.1038/s41396-018-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson EB. Microbial dynamics and interactions in the spermosphere. Annu Rev Phytopathol. 2004;42:271–309. doi: 10.1146/annurev.phyto.42.121603.131041. [DOI] [PubMed] [Google Scholar]
- 7.Nelson EB. The seed microbiome: origins, interactions, and impacts. Plant Soil Springe Int Publ. 2018;422:7–34. doi: 10.1007/s11104-017-3289-7. [DOI] [Google Scholar]
- 8.Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci USA. 2015;112:E911–20. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzpatrick CR, Salas-González I, Conway JM, Finkel OM, Gilbert S, Russ D, et al. The plant microbiome: from ecology to reductionism and beyond. Annu Rev Microbiol. 2020;74:81–100. doi: 10.1146/annurev-micro-022620-014327. [DOI] [PubMed] [Google Scholar]
- 10.Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 11.Naylor D, DeGraaf S, Purdom E, Coleman-Derr D. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 2017;11:2691–704. doi: 10.1038/ismej.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaparro JM, Badri DV, Vivanco JM. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014;8:790–803. doi: 10.1038/ismej.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzpatrick CR, Copeland J, Wang PW, Guttman DS, Kotanen PM, Johnson MTJ. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc Natl Acad Sci USA. 2018;115:E1157–65. doi: 10.1073/pnas.1717617115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grady KL, Sorensen JW, Stopnisek N, Guittar J, Shade A. Assembly and seasonality of core phyllosphere microbiota on perennial biofuel crops. Nat Commun. 2019;10:4135. doi: 10.1038/s41467-019-11974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlaeppi K, Dombrowski N, Oter RG, Ver Loren Van Themaat E, Schulze-Lefert P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci USA. 2014;111:585–92. doi: 10.1073/pnas.1321597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bulgarelli D, Garrido-Oter R, Münch PC, Weiman A, Dröge J, Pan Y, et al. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe. 2015;17:392–403. doi: 10.1016/j.chom.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, et al. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci USA. 2013;110:6548–53. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–38. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 20.Coleman-Derr D, Desgarennes D, Fonseca-Garcia C, Gross S, Clingenpeel S, Woyke T, et al. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. N Phytol. 2016;209:798–811. doi: 10.1111/nph.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Souza RSC, Okura VK, Armanhi JSL, Jorrín B, Lozano N, da Silva MJ, et al. Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome. Sci Rep. 2016;6:28774. doi: 10.1038/srep28774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy A, Salas Gonzalez I, Mittelviefhaus M, Clingenpeel S, Herrera Paredes S, Miao J, et al. Genomic features of bacterial adaptation to plants. Nat Genet. 2017;50:138–50. doi: 10.1038/s41588-017-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vannier N, Mony C, Bittebiere A-K, Michon-Coudouel S, Biget M, Vandenkoornhuyse P. A microorganisms’ journey between plant generations. Microbiome. 2018;6:79. doi: 10.1186/s40168-018-0459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tobias TB, Farrer EC, Rosales A, Sinsabaugh RL, Suding KN, Porras-Alfaro A. Seed-associated fungi in the alpine tundra: both mutualists and pathogens could impact plant recruitment. Fungal Ecol. 2017;30:10–18. doi: 10.1016/j.funeco.2017.08.001. [DOI] [Google Scholar]
- 25.Truyens S, Weyens N, Cuypers A, Vangronsveld J. Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environ Microbiol Rep. 2015;7:40–50. doi: 10.1111/1758-2229.12181. [DOI] [Google Scholar]
- 26.Shade A, Jacques M-A, Barret M. Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr Opin Microbiol. 2017;37:15–22. doi: 10.1016/j.mib.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Hardoim PR, van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, et al. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev. 2015;79:293–320. doi: 10.1128/MMBR.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Normander BO, Prosser JI. Bacterial origin and community composition in the barley phytosphere as a function of habitat and presowing conditions. Appl Environ Microbiol. 2000;66:4372–7. doi: 10.1128/AEM.66.10.4372-4377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green SJ, Inbar E, Michel FC, Hadar Y, Minz D. Succession of bacterial communities during early plant development: transition from seed to root and effect of compost amendment. Appl Environ Microbiol. 2006;72:3975–83. doi: 10.1128/AEM.02771-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ofek M, Hadar Y, Minz D. Colonization of cucumber seeds by bacteria during germination. Environ Microbiol. 2011;13:2794–807. doi: 10.1111/j.1462-2920.2011.02551.x. [DOI] [PubMed] [Google Scholar]
- 31.OECD-FAO. Agricultural Outlook 2020–2029.
- 32.Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol. 2017;15:579–90. doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- 33.Rath KM, Fierer N, Daniel, Murphy V, Rousk J. Linking bacterial community composition to soil salinity along environmental gradients. ISME J. 2018;13:836–46. doi: 10.1038/s41396-018-0313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108:4516–22. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliverio A, Holland-Moritz H. dada2 tutorial with MiSeq dataset for Fierer Lab. 2019.
- 37.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–63. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 39.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–8. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 40.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2020.
- 41.Wickham H. ggplot2: elegant graphics for data analysis. Springer-Verlag: New York; 2016.
- 42.Becker RA, Wilks AR, Minka TP, Deckmyn A. maps: draw geographical maps. 2018.
- 43.Jari Oksanen F, Blanchet G, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: community ecology package. R. 2019. p. R package version 2.5-6.
- 44.Rochefort A, Simonin M, Marais C, Guillerm-Erckelboudt A-Y, Barret M, Sarniguet A. Asymmetric outcome of community coalescence of seed and soil microbiota during early seedling growth. bioRxiv. 2020.11.19.390344.
- 45.Minich JJ, Sanders JG, Amir A, Humphrey G, Gilbert JA, Knight R. Quantifying and understanding well-to-well contamination in microbiome research. mSystems. 2019;4:e00186–19. doi: 10.1128/mSystems.00186-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cribari-Neto F, Zeileis A. Beta regression in R. J Stat Software; Vol 1, Issue 2. 2010.
- 47.Douma JC, Weedon JT. Analysing continuous proportions in ecology and evolution: a practical introduction to beta and Dirichlet regression. Methods. Ecol Evol. 2019;10:1412–30. [Google Scholar]
- 48.Kuhn M. caret: classification and regression training. 2020.
- 49.Wright MN, Ziegler A. Ranger: a fast implementation of random forests for high dimensional data in C++ and R. J Stat Softw. 2017;77:1–17. doi: 10.18637/jss.v077.i01. [DOI] [Google Scholar]
- 50.Thiergart T, Durán P, Ellis T, Vannier N, Garrido-Oter R, Kemen E, et al. Root microbiota assembly and adaptive differentiation among European Arabidopsis populations. Nat Ecol Evol. 2019;4:122–31. doi: 10.1038/s41559-019-1063-3. [DOI] [PubMed] [Google Scholar]
- 51.Simonin M, Dasilva C, Terzi V, Ngonkeu ELM, Diouf D, Aboubacry K, et al. Influence of plant genotype and soil on the wheat rhizosphere microbiome: evidences for a core microbiome across eight African and European Soils. FEMS Microbiol Ecol. 2016;96:fiaa067. doi: 10.1093/femsec/fiaa067. [DOI] [PubMed] [Google Scholar]
- 52.Robinson RJ, Fraaije BA, Clark IM, Jackson RW, Hirsch PR, Mauchline TH. Wheat seed embryo excision enables the creation of axenic seedlings and Koch’s postulates testing of putative bacterial endophytes. Sci Rep. 2016;6:25581. doi: 10.1038/srep25581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walterson AM, Stavrinides J. Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol Rev. 2015;39:968–84. doi: 10.1093/femsre/fuv027. [DOI] [PubMed] [Google Scholar]
- 54.Hardoim PR, Hardoim CCP, van Overbeek LS, van Elsas JD. Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS One. 2012;7:e30438. doi: 10.1371/journal.pone.0030438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu F, Hewezi T, Lebeis SL, Pantalone V, Grewal PS, Staton ME. Soil indigenous microbiome and plant genotypes cooperatively modify soybean rhizosphere microbiome assembly. BMC Microbiol. 2019;19:201. doi: 10.1186/s12866-019-1572-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nelson EB, Simoneau P, Barret M. Editorial special issue: the soil, the seed, the microbes and the plant. Plant Soil. 2018;422:1–5. doi: 10.1007/s11104-018-3576-y. [DOI] [Google Scholar]
- 57.Shmida A, Wilson MV. Biological determinants of species diversity. J Biogeogr. 1985;12:1–20. doi: 10.2307/2845026. [DOI] [Google Scholar]
- 58.Hannula SE, Kielak AM, Steinauer K, Huberty M, Jongen R, De Long JR, et al. Time after time: temporal variation in the effects of grass and forb species on soil bacterial and fungal communities. MBio. 2019;10:e02635–19. doi: 10.1128/mBio.02635-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jack ALH, Nelson EB. A seed-recruited microbiome protects developing seedlings from disease by altering homing responses of Pythium aphanidermatum zoospores. Plant Soil. 2018;422:209–22. doi: 10.1007/s11104-017-3257-2. [DOI] [Google Scholar]
- 60.Verma SK, Kharwar RN, White JF. The role of seed-vectored endophytes in seedling development and establishment. Symbiosis. 2019;78:107–13. doi: 10.1007/s13199-019-00619-1. [DOI] [Google Scholar]
- 61.Carlström CI, Field CM, Bortfeld-Miller M, Müller B, Sunagawa S, Vorholt JA. Synthetic microbiota reveal priority effects and keystone strains in the Arabidopsis phyllosphere. Nat Ecol Evol. 2019;3:1445–54. doi: 10.1038/s41559-019-0994-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Busby PE, Soman C, Wagner MR, Friesen ML, Kremer J, Bennett A, et al. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 2017;15:e2001793. doi: 10.1371/journal.pbio.2001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Finkel OM, Castrillo G, Herrera Paredes S, Salas González I, Dangl JL. Understanding and exploiting plant beneficial microbes. Curr Opin Plant Biol. 2017;38:155–63. doi: 10.1016/j.pbi.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Toju H, Peay KG, Yamamichi M, Narisawa K, Hiruma K, Naito K, et al. Core microbiomes for sustainable agroecosystems. Nat Plants. 2018;4:247–57. doi: 10.1038/s41477-018-0139-4. [DOI] [PubMed] [Google Scholar]
- 65.Maignien L, DeForce EA, Chafee ME, Murat Eren A, Simmons SL. Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. MBio. 2014;5:e00682–13. doi: 10.1128/mBio.00682-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wei Z, Gu Y, Friman V-P, Kowalchuk GA, Xu Y, Shen Q, et al. Initial soil microbiome composition and functioning predetermine future plant health. Sci Adv. 2019;5:eaaw0759. doi: 10.1126/sciadv.aaw0759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Durán P, Thiergart T, Garrido-Oter R, Agler M, Kemen E, Schulze-Lefert P, et al. Microbial interkingdom interactions in roots promote arabidopsis survival. Cell. 2018;175:973–83. doi: 10.1016/j.cell.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hamonts K, Trivedi P, Garg A, Janitz C, Grinyer J, Holford P, et al. Field study reveals core plant microbiota and relative importance of their drivers. Environ Microbiol. 2018;20:124–40. doi: 10.1111/1462-2920.14031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset generated and analyzed in this study along with representative sequences and sample metadata are available on FigShare (10.6084/m9.figshare.c.5323961) and further sample metadata is available in the Supplementary tables.