Abstract
Microbial community circadian rhythms have a broad influence on host health and even though light-induced environmental fluctuations could regulate microbial communities, the contribution of light to the circadian rhythms of rhizosphere microbial communities has received little attention. To address this gap, we monitored diel changes in the microbial communities in rice (Oryza sativa L.) rhizosphere soil under light–dark and constant dark regimes, identifying microbes with circadian rhythms caused by light exposure and microbial circadian clocks, respectively. While rhizosphere microbial communities displayed circadian rhythms under light–dark and constant dark regimes, taxa possessing circadian rhythms under the two conditions were dissimilar. Light exposure concealed microbial circadian clocks as a regulatory driver, leading to fewer ecological niches in light versus dark communities. These findings disentangle regulation mechanisms for circadian rhythms in the rice rhizosphere microbial communities and highlight the role of light-induced regulation of rhizosphere microbial communities.
Subject terms: Microbial ecology, Community ecology
Introduction
The Earth’s rotation, and thus daily cycles of light and dark, has influenced living organisms throughout evolutionary time [1]. As a result, most organisms have an inherent ability to measure the passage of time at a circa 24-h scale by using daily biological oscillations called circadian clocks [2]. Circadian clocks synchronize an organism’s behavioral and physiological processes to periodic environmental factors and anticipate future rhythmic environmental changes [3]. These rhythms allow temporal mutualism and reduce competition among sympatric species, thereby promoting the survival of organisms in rhythmic environments [4].
The short reproductive time (<24 h) of most prokaryotes has been used as evidence that these organisms might not possess circadian clocks [4]. Even so, various prokaryotes have been observed with circadian growth patterns on agar plates and in complex natural ecosystems [5, 6], suggesting the existence of prokaryotic circadian clocks. Cyanobacteria were the first prokaryotes reported to have a core circadian oscillator consisting of KaiA, KaiB, and KaiC [7]. Evolutionarily, KaiC and its homologs are the oldest oscillators among the three [8, 9], which have been found in Proteobacteria, Thermotogae, Chloroflexi and Euryarchaeota [10–13]. Furthermore, oscillations in peroxiredoxin proteins observed in model organisms suggest that these proteins constitute a universal and conserved marker for circadian rhythms [14].
Light regulates numerous physiological processes in plants [15]. It influences enzyme expression in many plant pathways, including chlorophyll biosynthesis, electron transport photosystems, starch synthesis and degradation, and nitrogen/sulfur assimilation [16]. Plant carbon partitioning and root-soil nutrient exchange are also controlled by light [17, 18]. Plant responses to diurnal changes in light naturally lead to rhizosphere diurnal fluctuations in soil pH, oxygen content, nutrient contents, and antimicrobial compounds due to the diel consumption/production of resources and waste materials [19–21].
Light exposure is an upstream factor that governs the circadian rhythms of host microbiomes in vivo [22, 23]. Gut microbial communities exhibit circadian rhythms in response to the time of day under normal light conditions [24–27] and eventually lose their circadian rhythms when the host is exposed to constant light or dark conditions. These dynamics indicate complex relationships between host exposure to diurnal variation and microbial circadian rhythms [22, 28]. Similarly, rhizosphere microbial communities show circadian rhythms under cyclic light–dark (LD) conditions [29, 30]. Although several microbial taxa display diurnal fluctuations in abundance, it is not clear if this is a response to changes in the rhizosphere environment caused by root activities or microbial endogenous circadian clocks [30].
The molecular circadian clock is a conserved attribute that is protected from environmental changes within physiologically permissible limits [2]. In order to identify prokaryotic taxa with circadian clocks, we used the most common indicator: a circa 24-h oscillation of microbial abundance under otherwise constant environmental conditions [4]. Rhizosphere soil samples were collected at the stem elongation growth stage of rice grown under a 24-h LD cycle for 3 days or under constant dark (DD) conditions for 72 h to identify microorganisms with circadian rhythms in LD or DD regimes. Our results indicate that rhizosphere microbial communities differ between day and night in each treatment, and show that circadian microorganisms perform differently under altered light conditions. These findings expand our knowledge of the adaptive capacity of circadian rhythms and provide a foundation for controlling rhizosphere microbial community structure and function through light exposure.
Materials and methods
Experimental materials and rice cultivation
Experimental soil was collected from a rice field in Jiaxing, China (30° 50′ 8.74″ N, 120° 43′ 3.68″ E); basic physicochemical properties have been previously described [31]. For our experiment, soil was sieved with a 2-mm mesh and used as a media in plastic pots (diameter = 57 mm, height = 155 mm, 200 g dry soil per pot). Seeds of Oryza sativa L. cultivar Yongyou 12 were surface-sterilized, germinated and cultivated for 2 weeks as previously reported [31], after which uniform seedlings were selected and transplanted into the potted soil at a density of one plant per pot for a total of 84 pots (2 treatments × 6 sampling times × 7 replicates). Unplanted soil was used as a negative control for an additional 30 pots (6 sampling times × 5 replicates). All pots were kept flooded and under greenhouse conditions; greenhouse conditions were 12 h light (9 am to 9 pm) and 12 h dark (9 pm to 9 am) with constant temperature and relative humidity (28 °C; 70%).
Experimental design and sampling
After 60 days of growth, plants were divided into two groups: one was given the regular LD cycle as described above and the other received a DD environment for 3 days. For both groups, seven replicates were harvested every 12 h at 8 am (AM) and 8 pm (PM); harvest occurred 1 h prior to the light condition change in the LD group. Rhizosphere soil was defined as soil tightly adhering to the roots (n = 7) and while bulk soil was collected from unplanted soil (n = 5) [32, 33]; plant matter was excluded from the rhizosphere soil. Soil was collected in an RNase-free tube with 3 mL LifeGuard Soil Preservation Solution (MoBio Laboratories, Carlsbad, CA, USA) per gram of sample. Samples were homogenized by hand-mixing, then immediately frozen with liquid nitrogen and stored at −80 °C until RNA extraction.
During the 72-h sampling period, in situ rhizosphere oxygen and pH profiles were determined using oxygen and pH microelectrode sensors (Unisense OXY25 and pH-N; Science Park, Aarhus, Denmark). The microsensor tip was inserted into the soil adhering to the roots and measured every 90 s. For each harvest time point, rhizosphere soil was sampled for dissolved organic carbon (DOC) concentration using a 1:5, soil:water extraction and quantification by TOC analyzer (Multi N/C 3100, Analytik Jena AG, Jena, Germany).
RNA extraction and 16S cDNA amplicon sequencing
Soil total RNA was extracted with E.Z.N.A. Soil RNA Mini Kits and purified with E.Z.N.A. RNase-Free DNase I Set according to the manufacturer’s protocols (Omega Bio-tek, Norcross, GA, USA). After extraction, RNA was used in reverse transcription to generate complimentary DNA (cDNA) by PrimeScript II 1st strand cDNA Synthesis Kits (Takara, Dalian, China). 16S cDNA was amplified by primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 907R (5′-CCGTCAATTCCTTTGAGTTT-3′) [34]. PCR conditions were as follows: 98 °C for 1 min; 30 cycles at 98 °C (10 s), 50 °C (30 s) and 72 °C (30 s); and 72 °C for 5 min. Amplified PCR products were sequenced on an Illumina HiSeq PE250 sequencing platform (Illumina, San Diego, CA, USA). Sequences were clustered into operational taxonomic units (OTUs) with VSEARCH-2.11.1 [35] with a sequence similarity threshold of 0.97. Ribosomal database project training set v16 [36] was used for taxonomy annotations at a threshold of 0.8 using the SINTAX algorithm [37]. All sequence data were deposited into Genome Sequence Archive under PRJCA003001 and PRJCA003009.
Determination of 16S cDNA gene copies by real-time quantitative PCR
To calculate the absolute abundance of prokaryotes, qPCR was performed on the cDNA samples to determine the copy number of 16S cDNA [38] by LightCycler 480II (Roche, Mannheim, Germany). Concentration of cDNA was determined by a Nanodrop ND-2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). PCR primers were chosen as above and PCR conditions were as follows: 95 °C for 2 min; 40 cycles at 95 °C (1 min), 49 °C (30 s) and 72 °C (50 s); and 72 °C for 5 min. The qPCR reaction mixture profiles and data analysis methods were conducted as before [39]. Amplification efficiencies of standard curves were 91–97% with R2 values of 0.992–0.999.
Microbial community analysis
Microbial community analysis used absolute abundance and was conducted using R (version 3.6.1; R Core Team, Vienna, Austria). The community matrix was normalized using R package DESeq [40]. Principal coordinates analysis of Weighted UniFrac distance [41] was chosen to show microbial community variations between samples. Genera with statistically significant differences in normalized abundance between day and night, that also displayed the same change trends between day and night were regarded as taxa with circadian rhythms. That is, if one genus is significantly lower in the evening, it must be lower than its adjacent time points in order to be considered a ‘circadian taxon’. A random forests approach [42] identified the top 30 (rhizosphere) and top 40 (bulk soil) important genera responsible for day–night differences in the two experimental groups using R package randomForest [43, 44]. To show the interactions among species in different communities, microbial co-occurrence networks were constructed has been previously described [45]. Network Enhancement [46] was used to denoise undirected weighted biological networks using the R package neten. The cutoff of correlation coefficients was determined through random matrix theory-based methods [47] using the R package RMThreshold. We generated sub-networks from four meta-community networks by preserving genera presented at each time point by the subgraph function in the R package igraph [48]. To compare our networks against random effects on network generation, we also generated random networks with same node number for each time point using the erdos.renyi.game function in igraph [48]. Network plots were visualized using Gephi [49] and topological features for each network were calculated using igraph [48].
Statistical analysis
We assessed significant differences for 16S cDNA copy number at the community level and genus level and α-diversity across different groups using one-way analysis of variance (ANOVA) and Tukey’s Honest Significant Difference (HSD). Permutational multivariate analysis of variance (PERMANOVA) was employed to evaluate significant differences between groups by the R package Vegan [50].
Results
Rhythms in rice rhizosphere physicochemical properties
Rhizosphere physicochemical properties displayed regular rhythms under LD cycles. Oxygen concentration increased to ~200 μmol L−1 after 3 h under light conditions and decreased to ~5 μmol L−1 in 3 h under dark conditions (Fig. 1a). Soil pH decreased from 6.4 to 6.0 during the day and recovered to 6.4 during the night; there was a circa 1-h delay in pH response to the LD cycle. DOC concentration was significantly higher (Tukey HSD, n = 21, P < 0.001) during the day (231–270 mg kg−1 soil) than at night (161–201 mg kg−1 soil, Fig. 1c). The oxygen, pH and DOC rhythms were not present in the DD treatment; DD soils maintained a constant oxygen concentration of ~5 μmol L−1, a slow increase of pH to 6.8, and a continuous decrease in DOC (to ~140 mg kg−1 soil) over the 72-h experimental period.
Fig. 1. Diurnal changes in rice rhizosphere environment for the light-dark cycle and constant dark treatments.
a Oxygen content. b Rhizosphere pH. c Dissolved organic carbon concentration. d Experimental design.
Circadian rhythms in the microbial communities in rice rhizosphere and bulk soils
Microbial activity in rice rhizosphere soil, as quantified by the copy number of 16S rRNA genes using metatranscriptomics, fluctuated with the LD cycle (Fig. 2a and Fig. S1). Microbial activity was significantly higher by day than night in the LD regime (Tukey HSD, n = 21, P < 0.001), a response not observed in the DD regime. Rhizosphere microbial activity was significantly lower in the DD samples than the LD samples (Tukey HSD, n = 42, P < 0.001).
Fig. 2. Circadian rhythms in the rhizosphere microbial communities.
a 16S cDNA copy number in each sample. b Alpha diversity based on Shannon index in all four rhizosphere groups (n = 21 for each group). Boxes are vertically bounded by the 1st and 3rd quartiles, center line is median, and whiskers extend to ≤1.5x inter-quantile-range. c Alpha diversity based on Chao1 index. d Principal coordinate analysis (PCoA) of weighted unifrac dissimilarities (n = 21 for each group). Curvilinear polygons show estimations of frequency densities. PCoA ellipses indicate 95% confident interval.
Light exposure significantly decreased α-diversity of the rice rhizosphere microbial communities (Fig. 2b, c). The α-diversity was significantly lower in PM (end of light period) versus AM measurements of LD samples (Tukey HSD, n = 21, P = 0.005 for Shannon index and P = 0.017 for Chao1 index), but did not fluctuate in the DD regime (Tukey HSD, n = 21, P > 0.05 for both Shannon and Chao1 indices). Rhizosphere α-diversity during dark measurements was similar for both LD and DD samples (Tukey HSD, n = 21, pairwise P > 0.05 for both Shannon and Chao1 indices).
Rhizosphere microbial communities were significantly different between day and night under both LD and DD treatments (Fig. 2d, Fig. S2). The turnover rate of rhizosphere microbial communities was significantly higher under LD than DD (Supplementary Table S1). Night measurements were not statistically different between LD than DD treatments.
To differentiate plant-induced influences from internal microbiota changes, bulk soil was also collected. Compared to rhizosphere communities, bulk soil communities had a higher α-diversity but lower activity (Fig. S3a–c). However, similar to rhizosphere communities in the DD regime, bulk soil communities also had distinct community structures between day and night in both LD and DD treatments (Fig. S3d, Supplementary Table S2) even though there were no significant differences in community composition or activity between the four sampling conditions (i.e., DD, LD, AM, and PM).
Circadian rhythms in rhizosphere and bulk soil microbial genera
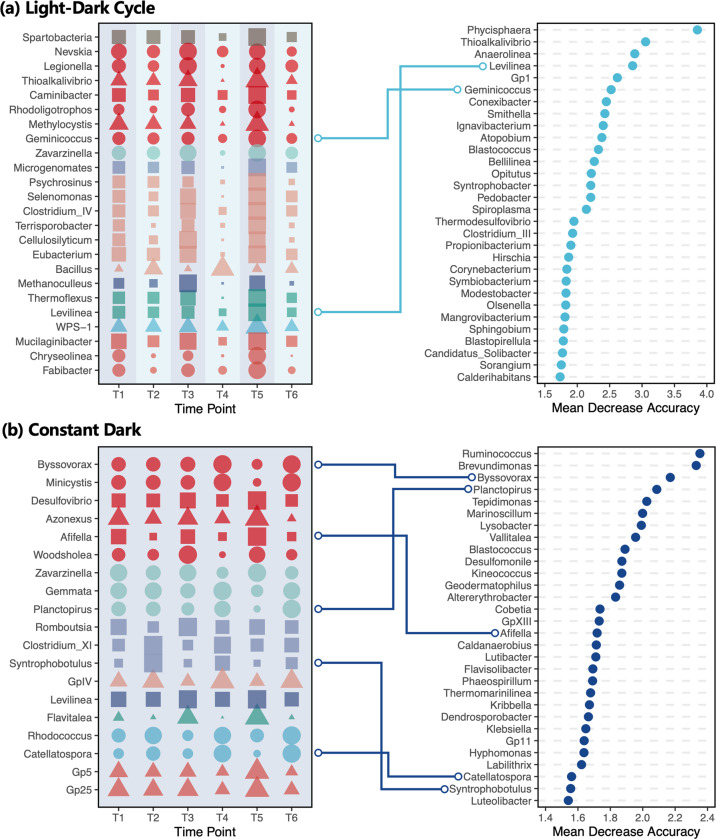
Within the rhizosphere communities, the proportion and classification of taxa with circadian rhythms differed between the LD and DD treatments: ~12.2% of the microbial communities (24 genera) showed daily variation in the LD group, while only ~7.4% (19 genera) responded similarly in the DD group (Fig. 3). Of the genera with identified circadian rhythms, only Levilinea was shared by both groups. Within the bulk soil communities, however, the taxa with circadian rhythms were much more similar between the LD and DD regimes: ~10.8% of the microbial communities (39 genera) showed daily variation in the LD group and ~9.7% (37 genera) responded similarly in the DD group (Fig. S4). There were genera with identified circadian rhythms shared between the two soil types, with rhizosphere DD samples sharing genera with bulk soil LD (15 genera) and DD (14 genera) samples. There was a smaller number of genera shared by the rhizosphere LD samples, with only two genera shared with bulk soil LD and three genera shared with bulk soil DD. The DD rhizosphere microbial communities were therefore more similar than the LD rhizosphere communities to the bulk soil communities.
Fig. 3. Rhizosphere microbiota with identified circadian rhythms (left) and the top 30 indicator taxa associated with AM and PM measurements (right).
a Light-dark cycle treatment. b Constant dark treatment. Within the left panel, bubble size indicates normalized abundance of a genus along six time points wherein the population of each genus at each time point was compared to the maximum population of each genera within all time points. Key to bubble shape: square, obligate anaerobic taxa; triangle, anaerobic, or anaerobic taxa; circle, obligate aerobic taxa. Bubble colors represent phyla. Polylines connect genus that are identified as both circadian taxa and indicator taxa.
Random forest analysis was further used to identify indicator taxa with AM versus PM differences within the LD and DD groups, and was ranked in order of Mean Decrease Accuracy (MDA) index to indicate the contribution of genera to community diel changes. Only two genera with circadian rhythms in the LD group were identified as indicator taxa. In contrast, five genera were identified as indicator taxa in DD; these genera also had a higher MDA value sum (Fig. 3, LD vs DD = 5.38 vs 9.09). Within the bulk soil communities, eight indicator genera with circadian rhythms were identified in the LD regime while nine genera were identified in the DD regime; the contribution of these indicator genera were similar for both treatments (Fig. S4, MDA value, LD vs DD = 14.89 vs 16.01). The number of circadian taxa identified as indicator taxa for each treatment over the two soil types suggests that while the plants influence microbial communities in the dark, microbial circadian clocks might still play a more important role than light-induced diurnal fluctuations in regulating the circadian rhythms of rice rhizosphere microbial communities under a DD regime.
Meta-community co-occurrence networks
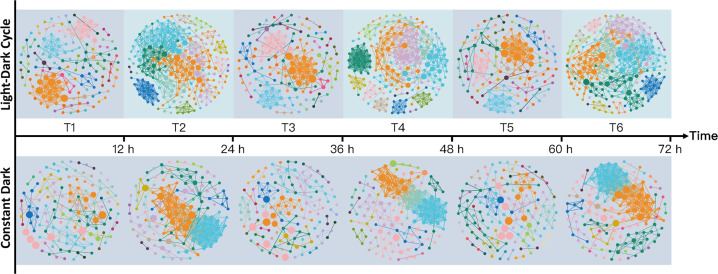
Light exposure significantly changed the topological features of the co-occurrence networks at the genus level (Fig. 4 and Supplementary Table S3). LD daytime networks had more nodes and edges than any dark sample group, and also included a larger proportion of OTUs, higher average degree, normalized closeness centrality and normalized betweenness centrality. Furthermore, LD networks had fewer modules, indicating that light samples had higher niche overlap than dark samples regardless of treatment. When compared to randomly generated networks containing the same node and edge numbers, differences in network topology indicate that the experiment networks were not random (Fig. S5 and Supplementary Table S4).
Fig. 4. Rhizosphere co-occurrence networks at genus taxonomy level.
Nodes represent unique genera; node size is proportional to abundance and node colors indicate modules.
Discussion
Rice rhizosphere microbial communities had observable diurnal cycles under both LD and DD treatments, a result that is in contrast to gut microbial communities which have been observed to lose their circadian rhythms when exposed to DD conditions [22]. In addition from the distinctly different environments that soil microbes and gut microbes live in, difference between these two systems may result from RNA-based microbial community analysis being a more sensitive measure for detecting exogenous and endogenous changes than a DNA-based community [51, 52]. Due to the inherent relationships between community composition and environmental factors such as oxygen, pH, and DOC, microbiota co-occurrence patterns varied less over the 3-day observation window in the DD compared to the LD group. The response of rhizosphere microbial communities suggests that there may be entrained rhythms in the LD group [53], whereas the response of bulk soil microbial communities suggests intrinsic rhythms in the DD group. Notably, the patterns of diel divergence between the LD and DD groups were completely different: microbes driven by circadian clocks (as observed in DD results) were masked in the LD treatment by the overall community responses to diel environmental drivers. As a result of this masking, only one taxon was observed with both entrained and endogenous rhythms.
Rice photosynthesis induced an increase in oxygen and DOC during the light period and led to unique microbial assemblies in light versus dark groups, possibly decreasing the contribution of microbial circadian clocks. Oxygen secreted by rice roots during photosynthesis alters the redox and electron supply status of an otherwise anaerobic environment [31, 54], leading to clear patterns in rhizosphere microbial communities [55]. Carbon resources are a well-known driver of microbial assembly, with taxa existing along a spectrum of copiotrophy to oligotrophy based on their adaption to resource availability [56]. Copiotrophs exhibit high growth rates when resource conditions are abundant, while oligotrophs have higher substrate affinities under conditions of low nutrient availability [57]. Our results indicate that the light group had fewer ecological niches and higher niche overlap, and therefore that microorganisms may have temporal complementarity due to changes in niche preferences [58]. This relationship implies that time is a key niche axis that minimizes interspecific competition across the diel cycle [59, 60].
Microbial communities were less stable under light conditions than dark, which is likely a direct response to environmental fluctuations. Light group co-occurrence networks were characterized by properties that indicate low stability, such as higher connectivity and normalized betweenness centrality, and lower modularity [61]. Of these properties, normalized betweenness centrality values were significantly higher in nodes from the light versus dark networks, indicating that the network structures of the light networks depends more on individual taxa and thus potentially have decreased community resilience to environmental stressors [62].
Circadian rhythms are expected to enhance organism fitness by improving their ability to adapt to external influences [63–65]. It is therefore reasonable to infer that entrained rhythms would be less advantageous to organisms living under constant environments [1]. Although it was assumed that organisms might lose their rhythms in the absence of periodicity, circadian rhythms are observed even in organisms that have spent many generations in apedriodic environments [2]. Nevertheless, circadian rhythms are not necessarily beneficial to the organism if the external force is removed or otherwise altered [66–68]. In our study, a majority of genera appeared to lose their rhythms under constant darkness, a pattern also observed in Cyanobacteria [69]. Although such as response suggests an adaptive property of a circadian clock in Cyanobacteria [69], it actually indicates that this adaption is activated by rhythmic environments, an argument that is supported by mathematical modeling of Cyanobacterial fitness under arbitrary light/dark schedules [70]. Furthermore, there is evidence of endogenous redox rhythms driven by oxidative stress and controlled by circadian oscillators which then disappear when measured under DD conditions [71–73]. However, the loss of a particular circadian rhythm does not necessarily mean damage to the circadian system; it is likely that circadian oscillators are functioning normally when involved in other processes, but that they become partially uncoupled from their circadian-related activities [53].
In contrast to studies showing a loss of rhythms under dark conditions, our results show that microbiota without observable circadian rhythms under LD cycles appeared to acquire them when exposed to the DD treatment. The apparent development of circadian rhythms in otherwise non-circadian organisms after entrainement by light cycles is thought to be controlled by circadian clocks [74]. Cave dwelling millipedes, for example, demonstrated a circadian rhythm in locomotive activity after entrainment with a LD cycle [75]. The capacity for circadian entrainment of organisms in otherwise aperiodic environments indicates an intrinsic adaptation value of circadian clocks, possibly due to a requirement to coordinate internal metabolic processes or regulate seasonal breeding [53]. It is also possible that biological clocks exist in steady states ranging from arrhythmic to rhythmic. Therefore, a small fluctuation or stimulus could change an existing arrhythmic state into a rhythmic state or vice versa [1, 76]. Further research is required to identify consistently reliable markers for circadian rhythms in prokaryotes, such as peroxiredoxins, that are more conserved and widespread than KaiA, KaiB, and KaiC [14, 77]. Moreover, our study only provides a short time scale perspective on microbial and plant–microbe interactions; further research examining the effects of contrasting time scales and other rhythmic factors are required to minimize the impacts of hidden deterministic and stochastic factors.
In summary, our study confirmed that light exposure mediates changes in the circadian rhythms of rice rhizosphere microbial communities, and suggests that light-induced diurnal fluctuations in the rhizosphere are a stronger determinant than microbial circadian clocks in regulating circadian rhythms. Our results reveal a distinct diurnal niche differentiation under different light conditions; microbial communities in light conditions had fewer ecological niches and higher niche overlap than those in dark conditions, but were also less stable. Importantly, as differences in rhizosphere community structure could lead to functional variation [29, 78], our results raise the possibility that rhizosphere microbial community functions could be regulated by above ground light exposure.
Supplementary information
Supplemental tables and legends of supplemental figures
Acknowledgements
The authors wish to thank Prof. Randy A. Dahlgren from University of California, Davis for his advice on the manuscript. The authors also thank the editors and two anonymous reviewers for their helpful and constructive comments on this work. This work was supported by the National Natural Science Foundation of China (41721001, 41991334), Zhejiang Natural Science Foundation (LD19D060001), and the 111 Project (B17039).
Author contributions
BM and JX designed the study. KZ and YX performed the experiments. KZ and BM performed the data visualization. KZ, BM, ES and JX contributed to scientific discussion and wrote the manuscript. All authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kankan Zhao, Bin Ma.
Contributor Information
Bin Ma, Email: bma@zju.edu.cn.
Jianming Xu, Email: jmxu@zju.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-00957-3.
References
- 1.Sharma VK. Adaptive significance of circadian clocks. Chronobiol Int. 2003;20:901–19. doi: 10.1081/CBI-120026099. [DOI] [PubMed] [Google Scholar]
- 2.Paranjpe DA, Kumar Sharma V. Evolution of temporal order in living organisms. J Circadian Rhythms. 2005;3:1–13. doi: 10.1186/1740-3391-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nobs SP, Tuganbaev T, Elinav E. Microbiome diurnal rhythmicity and its impact on host physiology and disease risk. EMBO Rep. 2019;20:e47129. doi: 10.15252/embr.201847129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sartor F, Eelderink-Chen Z, Aronson B, Bosman J, Hibbert LE, Dodd AN, et al. Are there circadian clocks in non-photosynthetic bacteria? Biology. 2019;8:41. doi: 10.3390/biology8020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soriano M, Roibas B, Garcia A, Espinosa-Urgel M. Evidence of circadian rhythms in non-photosynthetic bacteria? J Circadian Rhythms. 2010;8:1–4. doi: 10.1186/1740-3391-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–29. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 7.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–23. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 8.Aylward FO, Boeuf D, Mende DR, Wood-Charlson EM, Vislova A, Eppley JM, et al. Diel cycling and long-term persistence of viruses in the ocean’s euphotic zone. Proc Natl Acad Sci USA. 2017;114:11446–51. doi: 10.1073/pnas.1714821114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dvornyk V, Vinogradova O, Nevo E. Origin and evolution of circadian clock genes in prokaryotes. Proc Natl Acad Sci USA. 2003;100:2495–500. doi: 10.1073/pnas.0130099100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maniscalco M, Nannen J, Sodi V, Silver G, Lowrey PL, Bidle KA. Light-dependent expression of four cryptic archaeal circadian gene homologs. Front Microbiol. 2014;5:79. doi: 10.3389/fmicb.2014.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernal P, Allsopp LP, Filloux A, Llamas MA. The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 2017;11:972–87. doi: 10.1038/ismej.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmelling NM, Lehmann R, Chaudhury P, Beck C, Albers S-V, Axmann IM, et al. Minimal tool set for a prokaryotic circadian clock. BMC Evol Biol. 2017;17:1–20. doi: 10.1186/s12862-017-0999-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong L, Vani BP, Thiede EH, Rust MJ, Dinner AR. Molecular dynamics simulations of nucleotide release from the circadian clock protein KaiC reveal atomic-resolution functional insights. Proc Natl Acad Sci USA. 2018;115:11475–84. doi: 10.1073/pnas.1812555115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edgar RS, Green EW, Zhao Y, Ooijen Gvan, Olmedo M, Qin X, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–64. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–3. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 16.Farré EM, Weise SE. The interactions between the circadian clock and primary metabolism. Curr Opin Plant Biol. 2012;15:293–300. doi: 10.1016/j.pbi.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 17.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–77. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 18.Haydon MJ, Mielczarek O, Robertson FC, Hubbard KE, Webb AAR. Photosynthetic entrainment of the Arabidopsis circadian clock. Nature. 2013;502:689–92. doi: 10.1038/nature12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeAngelis KM, Brodie EL, DeSantis TZ, Andersen GL, Lindow SE, Firestone MK. Selective progressive response of soil microbial community to wild oat roots. ISME J. 2009;3:168–78. doi: 10.1038/ismej.2008.103. [DOI] [PubMed] [Google Scholar]
- 20.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhalnina K, Louie KB, Hao Z, Mansoori N, da Rocha UN, Shi S, et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol. 2018;3:470–80. doi: 10.1038/s41564-018-0129-3. [DOI] [PubMed] [Google Scholar]
- 22.Wu G, Tang W, He Y, Hu J, Gong S, He Z, et al. Light exposure influences the diurnal oscillation of gut microbiota in mice. Biochem Biophys Res Commun. 2018;501:16–23. doi: 10.1016/j.bbrc.2018.04.095. [DOI] [PubMed] [Google Scholar]
- 23.Teichman EM, O’Riordan KJ, Gahan CGM, Dinan TG, Cryan JF. When rhythms meet the blues: circadian interactions with the microbiota-gut-brain axis. Cell Metab. 2020;31:448–71. doi: 10.1016/j.cmet.2020.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014;20:1006–17. doi: 10.1016/j.cmet.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015;17:681–9. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang X, Bushman FD, FitzGerald GA. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc Natl Acad Sci USA. 2015;112:10479–84. doi: 10.1073/pnas.1501305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaczmarek JL, Musaad SM, Holscher HD. Time of day and eating behaviors are associated with the composition and function of the human gastrointestinal microbiota. Am J Clin Nutr. 2017;106:1220–31. doi: 10.3945/ajcn.117.156380. [DOI] [PubMed] [Google Scholar]
- 28.Deaver JA, Eum SY, Toborek M. Circadian disruption changes gut microbiome taxa and functional gene composition. Front Microbiol. 2018;9:737. doi: 10.3389/fmicb.2018.00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hubbard CJ, Brock MT, van Diepen LT, Maignien L, Ewers BE, Weinig C. The plant circadian clock influences rhizosphere community structure and function. ISME J. 2018;12:400–10. doi: 10.1038/ismej.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Staley C, Ferrieri AP, Tfaily MM, Cui Y, Chu RK, Wang P, et al. Diurnal cycling of rhizosphere bacterial communities is associated with shifts in carbon metabolism. Microbiome. 2017;5:1–13. doi: 10.1186/s40168-016-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng J, Xu Y, Ma B, Tang C, Brookes PC, He Y, et al. Assembly of root-associated microbiomes of typical rice cultivars in response to lindane pollution. Environ Int. 2019;131:104975. doi: 10.1016/j.envint.2019.104975. [DOI] [PubMed] [Google Scholar]
- 32.Gremion F, Chatzinotas A, Harms H. Comparative 16S rDNA and 16S rRNA sequence analysis indicates that Actinobacteria might be a dominant part of the metabolically active bacteria in heavy metal-contaminated bulk and rhizosphere soil. Environ Microbiol. 2003;5:896–907. doi: 10.1046/j.1462-2920.2003.00484.x. [DOI] [PubMed] [Google Scholar]
- 33.Lavecchia A, Curci M, Jangid K, Whitman WB, Ricciuti P, Pascazio S, et al. Microbial 16S gene-based composition of a sorghum cropped rhizosphere soil under different fertilization managements. Biol Fertil Soils. 2015;51:661–72. doi: 10.1007/s00374-015-1017-0. [DOI] [Google Scholar]
- 34.Wang B, Zhao J, Guo Z, Ma J, Xu H, Jia Z. Differential contributions of ammonia oxidizers and nitrite oxidizers to nitrification in four paddy soils. ISME J. 2015;9:1062–75. doi: 10.1038/ismej.2014.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:633–42. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar RC. SINTAX: a simple non-Bayesian taxonomy classifier for 16S and ITS sequences. 2016. https://www.biorxiv.org/content/10.1101/074161v1.
- 38.Deng Y, Ruan Y, Ma B, Timmons MB, Lu H, Xu X, et al. Multi-omics analysis reveals niche and fitness differences in typical denitrification microbial aggregations. Environ Int. 2019;132:105085. doi: 10.1016/j.envint.2019.105085. [DOI] [PubMed] [Google Scholar]
- 39.Yu M, Meng J, Yu L, Su W, Afzal M, Li Y, et al. Changes in nitrogen related functional genes along soil pH, C and nutrient gradients in the charosphere. Sci Total Environ. 2019;650:626–32. doi: 10.1016/j.scitotenv.2018.08.372. [DOI] [PubMed] [Google Scholar]
- 40.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–12. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Zhang N, Liu Y-X, Zhang X, Hu B, Qin Y, et al. Root microbiota shift in rice correlates with resident time in the field and developmental stage. Sci China Life Sci. 2018;61:613–21. doi: 10.1007/s11427-018-9284-4. [DOI] [PubMed] [Google Scholar]
- 43.Liaw A, Wiener M. Classification and regression by random Forest. R News. 2002;2:18–22. [Google Scholar]
- 44.Breiman L. Random forests. Mach Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 45.Ma B, Wang H, Dsouza M, Lou J, He Y, Dai Z, et al. Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J. 2016;10:1891–901. doi: 10.1038/ismej.2015.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang B, Pourshafeie A, Zitnik M, Zhu J, Bustamante CD, Batzoglou S, et al. Network enhancement as a general method to denoise weighted biological networks. Nat Commun. 2018;9:1–8. doi: 10.1038/s41467-017-02088-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo F, Zhong J, Yang Y, Scheuermann RH, Zhou J. Application of random matrix theory to biological networks. Phys Lett A. 2006;357:420–3. doi: 10.1016/j.physleta.2006.04.076. [DOI] [Google Scholar]
- 48.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal. Complex Syst. 2006;1695:1–9. [Google Scholar]
- 49.Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. Icwsm. 2009;8:361–2. [Google Scholar]
- 50.Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14:927–30. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 51.Li H, Su J-Q, Yang X-R, Zhu Y-G. Distinct rhizosphere effect on active and total bacterial communities in paddy soils. Sci Total Environ. 2019;649:422–30. doi: 10.1016/j.scitotenv.2018.08.373. [DOI] [PubMed] [Google Scholar]
- 52.Vieira S, Sikorski J, Dietz S, Herz K, Schrumpf M, Bruelheide H, et al. Drivers of the composition of active rhizosphere bacterial communities in temperate grasslands. ISME J. 2019; 1–13. [DOI] [PMC free article] [PubMed]
- 53.Yerushalmi S, Green RM. Evidence for the adaptive significance of circadian rhythms. Ecol Lett. 2009;12:970–81. doi: 10.1111/j.1461-0248.2009.01343.x. [DOI] [PubMed] [Google Scholar]
- 54.Pedersen O, Sand‐Jensen K, Revsbech NP. Diel pulses of O2 and CO2 in sandy lake sediments inhabited by Lobelia dortmanna. Ecology. 1995;76:1536–45. doi: 10.2307/1938155. [DOI] [Google Scholar]
- 55.Hernandez ME, Beck DAC, Lidstrom ME, Chistoserdova L. Oxygen availability is a major factor in determining the composition of microbial communities involved in methane oxidation. PeerJ. 2015;3:e801. doi: 10.7717/peerj.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saifuddin M, Bhatnagar JM, Segrè D, Finzi AC. Microbial carbon use efficiency predicted from genome-scale metabolic models. Nat Commun. 2019;10:1–10. doi: 10.1038/s41467-019-11488-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354–64. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- 58.Saleem M, Hu J, Jousset A. More than the sum of its parts: microbiome biodiversity as a driver of plant growth and soil health. Annu Rev Ecol Evol Syst. 2019;50:145–68. doi: 10.1146/annurev-ecolsys-110617-062605. [DOI] [Google Scholar]
- 59.Cozzi G, Broekhuis F, McNutt JW, Turnbull LA, Macdonald DW, Schmid B. Fear of the dark or dinner by moonlight? Reduced temporal partitioning among Africa’s large carnivores. Ecology. 2012;93:2590–9. doi: 10.1890/12-0017.1. [DOI] [PubMed] [Google Scholar]
- 60.Kohl MT, Ruth TK, Metz MC, Stahler DR, Smith DW, White PJ, et al. Do prey select for vacant hunting domains to minimize a multi-predator threat? Ecol Lett. 2019;22:1724–33. doi: 10.1111/ele.13319. [DOI] [PubMed] [Google Scholar]
- 61.de Vries FT, Griffiths RI, Bailey M, Craig H, Girlanda M, Gweon HS, et al. Soil bacterial networks are less stable under drought than fungal networks. Nat Commun. 2018;9:1–12. doi: 10.1038/s41467-018-05516-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidt JE, Kent AD, Brisson VL, Gaudin ACM. Agricultural management and plant selection interactively affect rhizosphere microbial community structure and nitrogen cycling. Microbiome. 2019;7:1–18. doi: 10.1186/s40168-019-0756-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DeCoursey PJ, Walker JK, Smith SA. A circadian pacemaker in free-living chipmunks: essential for survival? J Comp Physiol A. 2000;186:169–80. doi: 10.1007/s003590050017. [DOI] [PubMed] [Google Scholar]
- 64.Worden BD, Skemp AK, Papaj DR. Learning in two contexts: the effects of interference and body size in bumblebees. J Exp Biol. 2005;208:2045–53. doi: 10.1242/jeb.01582. [DOI] [PubMed] [Google Scholar]
- 65.Yerushalmi S, Bodenhaimer S, Bloch G. Developmentally determined attenuation in circadian rhythms links chronobiology to social organization in bees. J Exp Biol. 2006;209:1044–51. doi: 10.1242/jeb.02125. [DOI] [PubMed] [Google Scholar]
- 66.Lone SR, Sharma VK. Exposure to light enhances pre-adult fitness in two dark-dwelling sympatric species of ants. BMC Dev Biol. 2008;8:1–11. doi: 10.1186/1471-213X-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yadav P, Choudhury D, Sadanandappa MK, Sharma VK. Extent of mismatch between the period of circadian clocks and light/dark cycles determines time-to-emergence in fruit flies. Insect Sci. 2015;22:569–77. doi: 10.1111/1744-7917.12126. [DOI] [PubMed] [Google Scholar]
- 68.Yadav P, Thandapani M, Sharma VK. Interaction of light regimes and circadian clocks modulate timing of pre-adult developmental events in Drosophila. BMC Dev Biol. 2014;14:1–12. doi: 10.1186/1471-213X-14-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr Biol. 2004;14:1481–6. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 70.Lambert G, Chew J, Rust MJ. Costs of clock-environment misalignment in individual cyanobacterial cells. Biophys J. 2016;111:883–91. doi: 10.1016/j.bpj.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lai AG, Doherty CJ, Mueller-Roeber B, Kay SA, Schippers JHM, Dijkwel PP. CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc Natl Acad Sci USA. 2012;109:17129–34. doi: 10.1073/pnas.1209148109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tanaka K, Ishikawa M, Kaneko M, Kamiya K, Kato S, Nakanishi S. The endogenous redox rhythm is controlled by a central circadian oscillator in cyanobacterium Synechococcus elongatus PCC7942. Photosynth Res. 2019;142:203–10. doi: 10.1007/s11120-019-00667-0. [DOI] [PubMed] [Google Scholar]
- 73.Tanaka K, Nakanishi S Time-of-day dependent responses of cyanobacterial cellular viability against oxidative stress. 2019. https://www.biorxiv.org/content/10.1101/851774v2. [DOI] [PMC free article] [PubMed]
- 74.Krittika S, Yadav P. Circadian clocks: an overview on its adaptive significance. Biol Rhythm Res. 2019;0:1–24. [Google Scholar]
- 75.Koilraj AJ, Sharma VK, Marimuthu G, Chandrashekaran MK. Presence of circadian rhythms in the locomotor activity of a cave-dwelling millipede Glyphiulus cavernicolus sulu (Cambalidae, Spirostreptida) Chronobiol Int. 2000;17:757–65. doi: 10.1081/CBI-100102111. [DOI] [PubMed] [Google Scholar]
- 76.Roenneberg T, Merrow M. Life before the clock: modeling circadian evolution. J Biol Rhythms. 2002;17:495–505. doi: 10.1177/0748730402238231. [DOI] [PubMed] [Google Scholar]
- 77.Espinasa L, Jeffery WR. Conservation of retinal circadian rhythms during cavefish eye degeneration. Evol Dev. 2006;8:16–22. doi: 10.1111/j.1525-142X.2006.05071.x. [DOI] [PubMed] [Google Scholar]
- 78.Hubbard CJ, McMinn RL, Weinig C. Rhizosphere microbes influence host circadian clock function. 2018. https://www.biorxiv.org/content/10.1101/444539v1.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental tables and legends of supplemental figures