Abstract
We report striking differences in the substrate specificities of two human SR proteins, SF2/ASF and SC35, in constitutive splicing. β-Globin pre-mRNA (exons 1 and 2) is spliced indiscriminately with either SR protein. Human immunodeficiency virus tat pre-mRNA (exons 2 and 3) and immunoglobulin μ-chain (IgM) pre-mRNA (exons C3 and C4) are preferentially spliced with SF2/ASF and SC35, respectively. Using in vitro splicing with mutated or chimeric derivatives of the tat and IgM pre-mRNAs, we defined specific combinations of segments in the downstream exons, which mediate either positive or negative effects to confer SR protein specificity. A series of recombinant chimeric proteins consisting of domains of SF2/ASF and SC35 in various combinations was used to localize trans-acting domains responsible for substrate specificity. The RS domains of SF2/ASF and SC35 can be exchanged without effect on substrate specificity. The RNA recognition motifs (RRMs) of SF2/ASF are active only in the context of a two-RRM structure, and RRM2 has a dominant role in substrate specificity. In contrast, the single RRM of SC35 can function alone, but its substrate specificity can be influenced by the presence of an additional RRM. The RRMs behave as modules that, when present in different combinations, can have positive, neutral, or negative effects on splicing, depending upon the specific substrate. We conclude that SR protein-specific recognition of specific positive and negative pre-mRNA exonic elements via one or more RRMs is a crucial determinant of the substrate specificity of SR proteins in constitutive splicing.
Pre-mRNA splicing is an essential step in the expression of eukaryotic genes (see review chapters in references 17 and 20). Introns are excised with a high degree of precision in two successive transesterification reactions, despite the enormous variability in intron number, size, and sequence in higher eukaryotic genes. The critical sequences involved in the transesterification reactions, at the 5′ splice site, the branch site, and the 3′ splice site, are relatively short and only weakly conserved. Many silent or cryptic splice site signals are present in both exons and introns, but they are normally ignored in the presence of the authentic signals. On the other hand, a number of pre-mRNAs show flexibility in the choice of alternative splice sites, often in response to tissue-specific, physiologically, or developmentally regulated states. Alternative splicing is a common strategy for the regulation of cellular and viral gene expression.
Pre-mRNA splicing takes place within a large complex, the spliceosome, which includes the small nuclear ribonucleoprotein particles (snRNPs) U1, U2, U4/U6, and U5 and a large number of non-snRNP splicing factors. Biochemical characterization of the spliceosome, together with genetic studies in budding yeast, predicts that over 50 proteins are essential for constitutive splicing. Considerable effort has been devoted to dissecting the cis elements and trans-acting factors involved in the complex splicing reaction, but little is known about the molecular mechanisms responsible for accuracy and specificity.
The members of the SR protein family are well-studied non-snRNP protein factors required for general pre-mRNA splicing (reviewed in references 4, 10, and 22). A prototype of the family, SF2/ASF, was originally purified by biochemical complementation of splicing-deficient cytosolic S100 extract from HeLa cells. This assay has been commonly used as a functional criterion to show the requirement of SF2/ASF or other SR proteins for constitutive pre-mRNA splicing. Another characteristic property of SR proteins is the concentration-dependent regulation of splice site selection in alternatively spliced pre-mRNAs. SR proteins also share a distinctive domain structure, which consists of one or two copies of an RNA recognition motif (RRM), followed by a characteristic C-terminal arginine/serine-rich (RS) domain. Nine authentic human SR proteins have been identified to date, and these belong to two subgroups, based on domain structure. The members of one subgroup, which comprises SF2/ASF, SRp30c, SRp40, SRp55, and SRp75, have two RRMs, of which the second one is somewhat atypical and includes a distinctive heptapeptide signature. The members of the other subgroup, which includes SC35, SRp20, 9G8, and p54, have only a single, N-terminal RRM. All of these SR proteins have been shown to complement the S100 extract for splicing in vitro. Therefore, SR proteins appear to have similar, and in some cases redundant, biochemical functions in general splicing, at least in vitro.
In contrast to the apparently interchangeable properties of different SR proteins in constitutive splicing in vitro, distinct effects of these proteins in alternative and enhancer-dependent splicing have been reported. For example, individual SR proteins promote the use of different alternative 5′ splice sites with certain pre-mRNAs, both in vitro and in vivo (5, 31, 45–47). Likewise, certain exonic splicing enhancers (ESEs) interact functionally and sequence specifically with distinct subsets of SR proteins (12, 28, 36, 40). High-affinity RNA-binding sites for several SR proteins were identified by iterative in vitro binding selection (SELEX) (13, 32, 37, 38). Recently, a selection procedure based on in vitro splicing was employed to derive consensus ESEs specific for SF2/ASF, SRp40, and SRp55 (21). SF2/ASF and SC35 are prototypical SR proteins representative of the two subgroups, and they have distinct functional properties in some assays. For example, SC35 can restore splicing in 9G8-depleted nuclear extract but SF2/ASF cannot (7). Moreover, SF2/ASF and SC35 have antagonistic effects on alternative splicing of chicken β-tropomyosin pre-mRNA (11). Genetic studies in Drosophila melanogaster demonstrated that SRp55/B52 is essential for development; however, it does not appear to be required for splicing of all pre-mRNAs, suggesting that it has specific pre-mRNA substrates in vivo (27, 29). Recently, SF2/ASF was shown to be essential for cell viability by targeted gene disruption in a chicken B-cell line; the lethality could be rescued by expression of cDNAs encoding human SF2/ASF, but not SC35 or SRp40, again showing that individual SR proteins have at least some unique functions (41).
Early studies showed that splicing of a human immunodeficiency virus type 1 (HIV-1) tat/rev minigene transcript is highly SF2/ASF dependent (18), and this dependence is manifested at an early step of spliceosome assembly, corresponding to the formation of an SR protein commitment complex (9). Subsequently, it was shown that RRM2 of SF2/ASF, but not RRM1 or the RS domain, plays a crucial role in formation of a specific commitment complex (8). In the present study, the different substrate specificities of two SR proteins, SF2/ASF and SC35, are demonstrated with three representative pre-mRNAs, β-globin, HIV-1 tat, and immunoglobulin μ chain (IgM). The roles of specific elements on the pre-mRNAs and of individual domains of SF2/ASF and SC35 in determining substrate specificity in constitutive splicing are reported.
MATERIALS AND METHODS
Plasmid constructions.
The wild-type minigene pre-mRNAs (Fig. 1) were transcribed from previously described linearized plasmids. β-Globin pre-mRNA (exon 1, 158 nucleotides [nt]; intron 1, 130 nt; exon 2, 209 nt) was transcribed from pSP64-HβΔ6 (19); tat pre-mRNA (tat/rev coding exon 2, 270 nt; intron with an internal deletion, 163 nt; coding exon 3, 101 nt) was transcribed from pSP64-HIV-1/tat23 (18); and IgM pre-mRNA (constant exon C3, 154 nt; intron, 107 nt; constant exon C4, 119 nt) was transcribed from pμC3-C4 (43). The exon swap (Fig. 2), exon segment deletion (Fig. 3 and 5), and exon segment replacement (Fig. 4 and 6) derivatives of these tat and IgM minigenes were constructed by one or more rounds of overlap-extension PCR with appropriate primers. The resulting amplification products were cleaved with HindIII and EcoRI and subcloned into the corresponding sites of the pCRII vector (Invitrogen). The RNA sequences of the Ta, Tb, and Tc segments from the tat pre-mRNA and of the Ca, Cb, and Cc segments from the IgM pre-mRNA are shown below (Fig. 3 and 5). To delete the previously reported exonic splicing silencer (ESS) region of the tat construct (Fig. 4) (1, 34), pSP64-HIV-1/tat23 DNA was amplified by PCR with an upstream SP6 promoter primer and a downstream primer containing the desired deletion. The amplified product was cleaved with HindIII and BamHI and subcloned into the corresponding sites of the pSP73 vector (Promega). The resulting plasmid, pSP73-tatΔESS, has a 20-bp deletion of the sequence, GATCCATTCGATTAGTGAAC, in the downstream exon 3. All new plasmid constructs were verified by sequencing. To generate templates for runoff in vitro transcription, pSP64-HβΔ6, pSP64-HIV-1/tat23, pSP73-tatΔESS, pCRII-CaTbTc, and pCRII-CaCbTc were linearized with BamHI. pμC3-C4 and all other pCRII-derivative plasmids were linearized with HindIII and EcoRI, respectively.
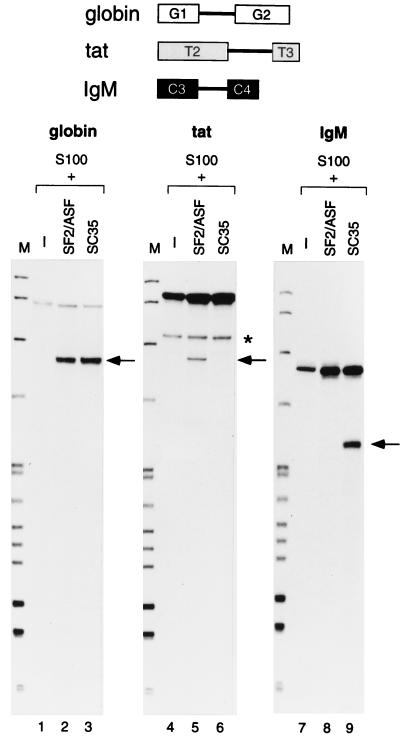
FIG. 1.
In vitro splicing of three representative pre-mRNAs in S100 extract complemented with SF2/ASF or SC35. The structures of the β-globin, tat, and IgM minigene pre-mRNAs (see Materials and Methods) are shown schematically at the top. The positions of the spliced mRNAs are indicated by arrows. The asterisk indicates a cleavage product unrelated to splicing (18). The splicing products were previously characterized in detail (18, 30, 43). pBR322/HpaII DNA size markers are shown (lanes M).
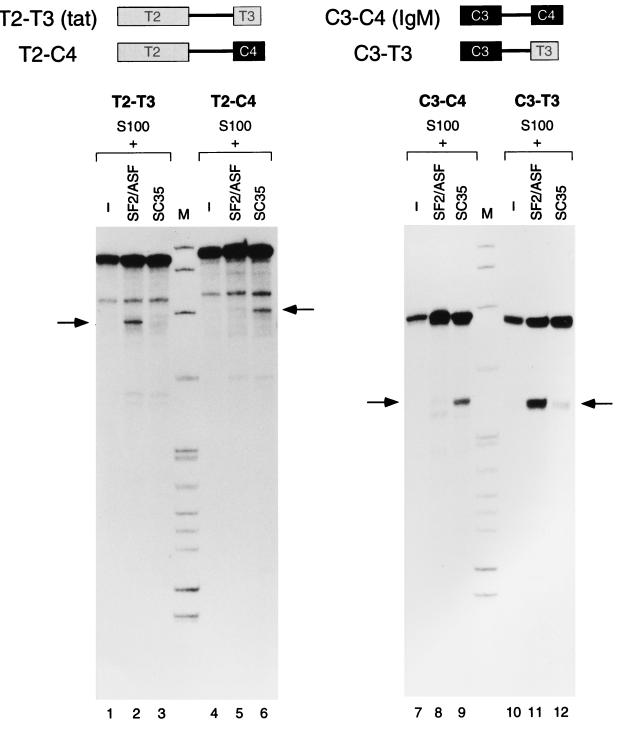
FIG. 2.
In vitro splicing of hybrid tat and IgM pre-mRNAs with swapped downstream exons. The structures of the control wild-type minigene pre-mRNAs (tat and IgM) and swap pre-mRNAs (T2-C4 and C3-T3) are shown schematically at the top. The positions of the spliced mRNAs are indicated by arrows. DNA size markers (lanes M) are as described for Fig. 1.
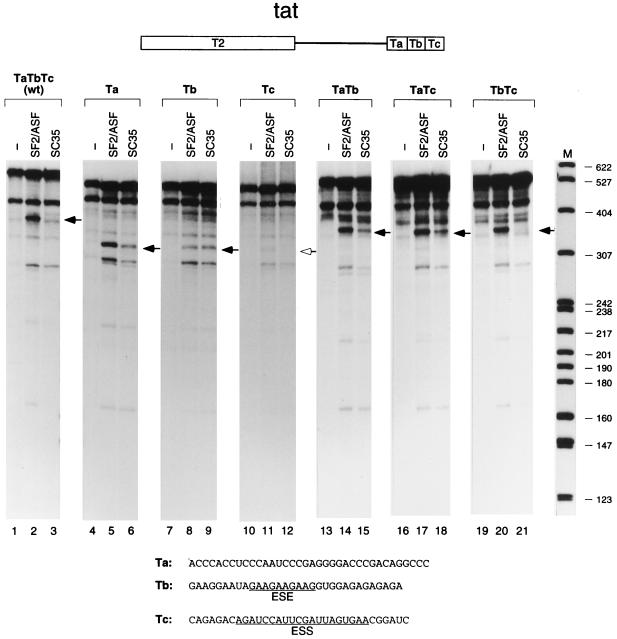
FIG. 3.
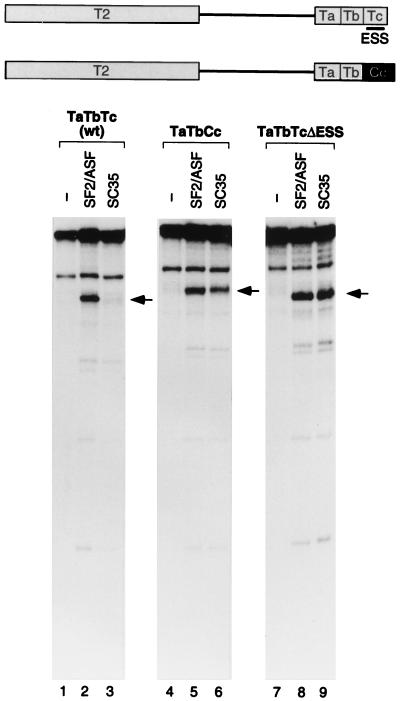
In vitro splicing of tat pre-mRNA derivatives with downstream exon deletions. The structure of the wild-type (wt) tat minigene pre-mRNA, with the T3 exon divided into three segments, Ta, Tb, and Tc, is shown schematically at the top. The RNA sequence of each segment is shown at the bottom. Previously identified ESE and ESS elements are underlined (1, 33, 34). The positions of the spliced mRNAs are indicated by arrows; the open arrowhead indicates the expected position in the case of an mRNA that is at the limit of detection. DNA size markers (lane M) are as described for Fig. 1 (sizes in nucleotides).
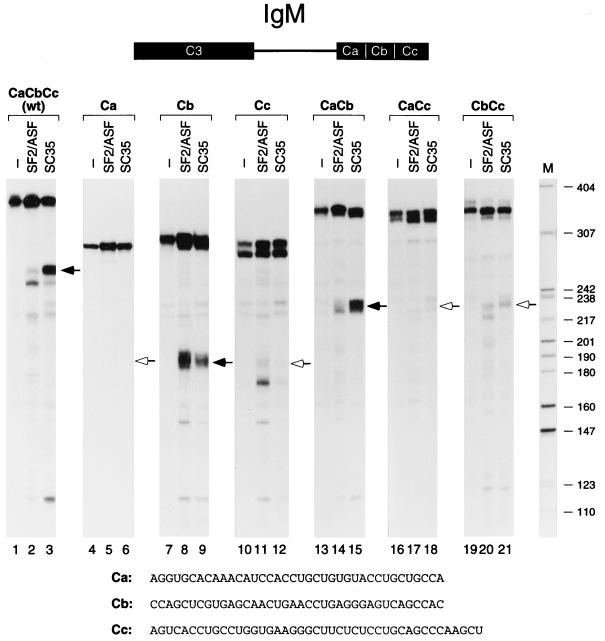
FIG. 5.
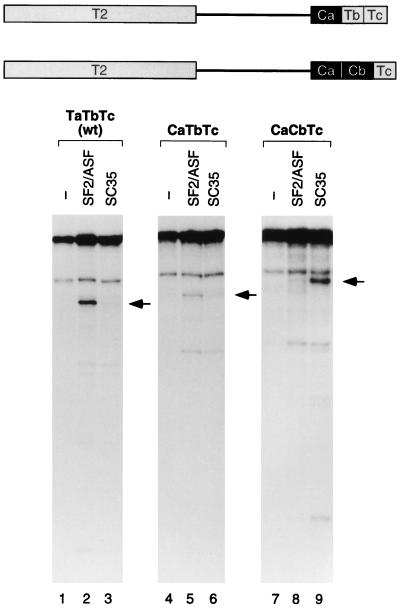
In vitro splicing of IgM pre-mRNA derivatives with downstream exon deletions. The structure of the wild-type (wt) IgM minigene pre-mRNA, with the C4 exon divided into three segments, Ca, Cb, and Cc, is shown schematically at the top. The RNA sequence of each segment is shown at the bottom. The positions of the spliced mRNAs are indicated by arrows; the open arrowheads indicate the expected positions in the cases of mRNAs that are at or below the limit of detection. The identity of the aberrant processing product in lane 11 has not been determined. DNA size markers (lane M) are as described for Fig. 1 (sizes in nucleotides).
FIG. 4.
In vitro splicing of tat pre-mRNA derivatives with replacement or internal deletion of exon segment Tc. The structures of the pre-mRNA derivatives are shown schematically at the top. The deleted ESS element (Fig. 3) is indicated by a horizontal bar, and the black box shows the Cc segment of the IgM C4 exon (Fig. 5) replacing the Tc segment. The positions of the spliced mRNAs are indicated by arrows. wt, wild type.
FIG. 6.
In vitro splicing of tat pre-mRNA derivatives with replacements of downstream exon segments. The structures of the pre-mRNAs are shown schematically at the top. The positions of the spliced mRNAs are indicated by arrows. wt, wild type.
In vitro splicing assays.
32P-labeled pre-mRNA substrates were prepared by runoff in vitro transcription with SP6 or T7 (only for the pSP73-tatΔESS template) RNA polymerase as described elsewhere (23). HeLa cell S100 extract and chimeric SF2/ASF and SC35 glutathione S-transferase-tagged recombinant proteins expressed in baculovirus were prepared as described elsewhere (8, 24). Purified recombinant, nontagged SF2/ASF and SC35 proteins expressed in baculovirus were a generous gift from K. Lynch and T. Maniatis. In vitro splicing complementation reactions were carried out in 25 μl with 9 μl of S100 extract, 10 pmol (0.4 μM final concentration) of a wild-type or chimeric recombinant protein, and 20 fmol of 32P-labeled pre-mRNA substrate, followed by incubation at 30°C for 1.5 to 4 h as described elsewhere (23). RNA products were analyzed by electrophoresis on a 5.5% polyacrylamide–7 M urea gel followed by autoradiography with an intensifying screen at −70°C.
RESULTS
SF2/ASF and SC35 have distinct pre-mRNA substrate specificities.
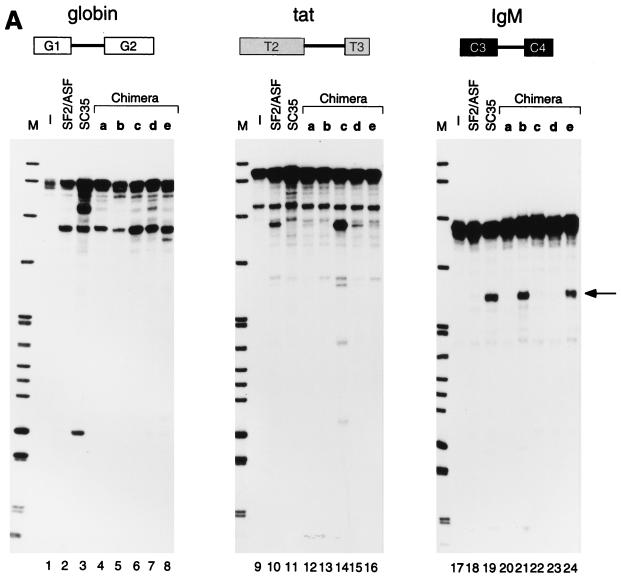
To investigate the substrate specificity of SR proteins, transcripts from various minigene constructs including δ-crystallin, IgM, HIV-1 tat, and Drosophila ftz, were tested for splicing in vitro. Using HeLa cell S100 extract, in which SR proteins are limiting for splicing, we assayed the activity of individual recombinant SR proteins, including SF2/ASF, SC35, SRp30c, SRp40, and SRp55 (data not shown). In each of these minigene transcripts, only one pair of authentic splice sites is present, and hence the assays measure constitutive splicing. We focused on the substrates and SR proteins that exhibited the most striking differences in specificity, namely, the tat and IgM pre-mRNAs spliced in the presence of SF2/ASF and SC35 (Fig. 1). The control β-globin pre-mRNA (G1-G2) gave comparable levels of splicing with either SR protein (lanes 1 to 3), as expected from previous studies. In contrast, the HIV tat pre-mRNA (T2-T3) spliced when the S100 extract was complemented with SF2/ASF but not when it was complemented with SC35 (lanes 4 to 6). Conversely, the IgM pre-mRNA (C3-C4) spliced in the presence of SC35 but not that of SF2/ASF (lanes 7 to 9).
The downstream exons are responsible for substrate specificity.
As an initial approach to mapping segments of each pre-mRNA that are responsible for substrate specificity, we first constructed two hybrid pre-mRNAs, in which the upstream or downstream half of β-globin pre-mRNA (G1-G2) was replaced by the corresponding segment from the IgM pre-mRNA (C3-C4), with the fusion point located in the middle of each intron. The hybrid G1-C4 pre-mRNA showed SC35 specificity, as did the parent IgM pre-mRNA, whereas the C3-G2 hybrid pre-mRNA was spliced with either SF2/ASF or SC35, similar to the parent β-globin pre-mRNA (data not shown). To determine whether an SC35-responsive element is localized exclusively within the IgM C4 exon or within the 3′ portion of the preceding intron, downstream exon swapping constructs were made (Fig. 2). When just the downstream T3 exon of the tat pre-mRNA was replaced with the IgM C4 exon, the SR protein specificity switched completely from SF2/ASF to SC35 (lanes 1 to 6). With the reciprocal construct, in which the downstream C4 exon of the IgM pre-mRNA was replaced with the tat T3 exon, a full specificity switch from SC35 to SF2/ASF was observed (lanes 7 to 12). We conclude that specific SF2/ASF- and SC35-responsive elements are present in the T3 and C4 downstream exons, respectively.
Mapping of tat pre-mRNA downstream exon elements responsible for SF2/ASF specificity.
To further map the SF2/ASF-responsive element in the T3 exon of the tat pre-mRNA, a series of mutants was constructed. Since there is a highly purine-rich sequence in the middle of the T3 exon and purine-rich elements frequently function as ESEs (reviewed in reference 4), we divided the T3 exon into three similarly sized segments, designated Ta, Tb (purine rich), and Tc. Pre-mRNAs in which the T3 exon was replaced by each of these segments alone or in combination were generated and spliced in vitro (Fig. 3). The three segments mediated very different SR protein specificities in splicing. The Ta and Tb segments individually promoted splicing activity with either SF2/ASF or SC35, whereas the Tc fragment alone was essentially inactive (lanes 4 to 12). Splicing was still observed when the purine-rich Tb segment was deleted (TaTc), indicating that the purine-rich segment is not an essential enhancer element in the T3 exon (lanes 13 to 15). Interestingly, only one combination of two segments, TbTc, resulted in SF2/ASF-specific splicing, as seen in the wild-type construct (lanes 1 to 3 and 19 to 21), whereas the TaTb and TaTc combinations behaved similarly to Ta alone (lanes 4 to 6 and 13 to 18). With the TbTc substrate, splicing activity in the presence of SF2/ASF was stronger than that with the Tb substrate, and splicing activity with SC35 was completely repressed. Therefore, the Tc fragment, which is inactive by itself, plays an SC35-specific negative role that results in SF2/ASF-specific splicing.
To confirm that the specific negative effect of the Tc segment is due to specific sequences and not, for example, to exon length, we replaced this segment with a fragment from the 3′ end of the IgM C4 exon (Fig. 4). The resulting pre-mRNA, TaTbCc, regained splicing activity with SC35 and retained activity with SF2/ASF (lanes 1 to 6). An ESS was previously identified in this exon of the tat pre-mRNA, as assayed in vivo and by in vitro splicing in nuclear extract (1, 34). Within the Tc segment (Fig. 3), the mapped ESS sequence, AGAUCCAUUCGAUUAGUGAA, has been shown to comprise two functional core sequences, AGAUCC and UUAG (33). We therefore deleted most of the above 20-nt sequence from the T3 exon and found that splicing activity with SC35 was restored (Fig. 4, lanes 3 and 9). We conclude that the previously identified ESS acts by repressing the activity of SC35 but has no effect on the activity of SF2/ASF (lanes 2 and 8).
Mapping of IgM pre-mRNA downstream exon elements responsible for SC35 specificity.
A similar analysis was performed with the IgM C4 exon, which was arbitrarily divided into three segments of similar lengths, Ca, Cb, and Cc. Six constructs with individual segments or combinations thereof in place of the intact C4 exon were generated and spliced in the presence of SC35 or SF2/ASF (Fig. 5). In contrast to the above results with the tat T3 exon, only one of the single-segment substrates, Cb, was spliced, and this was the case with either SF2/ASF or SC35 (lanes 4 to 12). Splicing with the Ca substrate was reproducibly below detection. The correct selectivity for SC35 required two adjacent segments, Ca and Cb, such that the CaCb substrate spliced much more efficiently in the presence of SC35 than in the presence of SF2/ASF, similar to the wild type (lanes 13 to 15). Comparison of the CaCb substrate with the Ca and Cb substrates suggests that the Ca segment has repressive activity toward SF2/ASF, although it is also possible that the distance between the Cb segment and the 3′ splice site differentially affects recognition by SF2/ASF and by SC35 (lanes 4 to 9 and 13 to 15). The other two-segment substrates, CaCc and CbCc, gave little to no splicing (lanes 16 to 21). Thus, the Cc segment, which by itself was inactive, inhibited the activity of the Cb segment (lanes 7 to 12 and 19 to 21). However, this effect was context dependent, since the Cc element was neutral in a heterologous context (Fig. 4, lanes 4 to 6). Two short purine-rich tracts are present within the Cb (GAGGGAG) and Cc (GUGAAGGG) segments, but disruption of either purine-rich sequence by mutagenesis had no effect on splicing efficiency or on SC35 specificity, compared to the wild-type pre-mRNA (data not shown).
The properties of the Ca and CaCb segments were further tested in a heterologous context, i.e., the tat pre-mRNA. Introduction of the Ca segment in place of the corresponding Ta segment in exon T3 resulted in a reduction in splicing efficiency, although SR protein specificity was retained (Fig. 6, lanes 2 and 5). This result confirms that the Ca segment has a repressive effect with SF2/ASF. When the tat TaTb portion of the exon was replaced by the corresponding IgM CaCb fragment, a complete switch in SR protein specificity was observed (lanes 1 to 3 and 7 to 9). Thus, the CaCb region of exon C4 is a bipartite element that mediates SC35-specific splicing in either a homologous or a heterologous pre-mRNA context. We conclude that the Cb segment includes a cis-acting element(s) for general splicing activity, whereas the Ca segment is necessary, although not sufficient, for SC35 specificity.
Domains of SF2/ASF and SC35 responsible for substrate specificity.
To determine which of the modular domains of SF2/ASF and SC35 mediates substrate specificity by recognition of the above elements, we used a collection of recombinant chimeric proteins that consist of domains from these two SR proteins in different combinations (Fig. 7). We previously used these chimeric proteins to show synergy between the two RRMs of SF2/ASF, but not its RS domain, in mediating formation of a specific SR protein commitment complex with the tat pre-mRNA (8). In the present study, we have systematically analyzed the role of the individual domains of SF2/ASF and SC35 in substrate specificity during constitutive splicing, by using complementation of SR protein-deficient S100 extract and three representative substrates, β-globin, tat, and IgM pre-mRNAs (Fig. 7A).
FIG. 7.
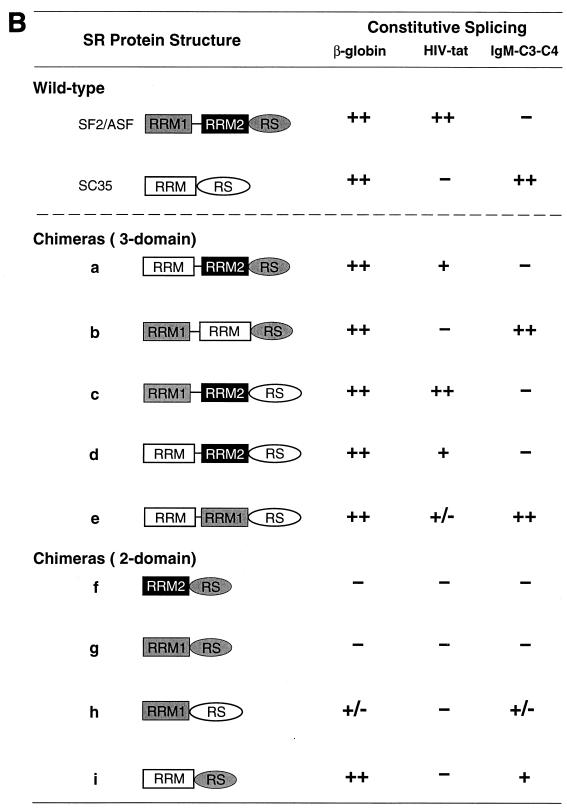
(A) In vitro splicing of β-globin, tat, and IgM pre-mRNAs with wild-type or chimeric glutathione S-transferase-tagged SR proteins assembled from different combinations of SF2/ASF and SC35 domains. See panel B for the designation of each protein. The structures of the pre-mRNAs are shown schematically at the top. The positions of the spliced mRNAs are indicated by arrows. DNA size markers (lanes M) are as described for Fig. 1. (B) Summary of the splicing activities of the chimeric SR proteins with each pre-mRNA. The domain structure of each protein is shown schematically, with the designation used in panel A. The relative splicing complementation activities are indicated (−, no detectable activity; +/−, trace activity; +, weak activity; ++, strong activity) and are based on two independent experiments, one of which is shown in panel A.
Efficient splicing of β-globin pre-mRNA was obtained by complementation with any of the chimeric proteins with a three-domain structure (Fig. 7A, lanes 2 to 8). This observation serves as a useful control for the proper folding and functional integrity of these chimeric proteins. In the case of the tat pre-mRNA, splicing was observed only with the chimeric proteins possessing the atypical RRM2 of SF2/ASF (lanes 12 and 15), but efficient activity comparable to that of wild-type SF2/ASF was seen only with the chimera that includes both RRM1 and RRM2 (lanes 10 and 14). Thus, RRM2 of SF2/ASF plays a key role in tat splicing specificity and it synergizes with the otherwise neutral RRM1, resulting in much higher splicing efficiency. The source of RS domain, either from SF2/ASF or SC35, had no significant influence on SF2/ASF splicing specificity. However, an RS domain is necessary for splicing activity, since deletion of the RS domain abolished the activity of the proteins in the S100 complementation assay (data not shown).
The domain requirements for IgM pre-mRNA splicing were very different from those observed for tat pre-mRNA splicing. We found that the chimeric proteins that include the RRM of SC35 in conjunction with RRM1 of SF2/ASF, but not with RRM2, were as active as wild-type SC35 (lanes 19, 21, and 24). However, inclusion of SF2/ASF RRM2 completely abolished IgM splicing and activated tat splicing (lanes 20, 22, and 23). Thus, in the three-domain chimeras, the SC35 RRM confers substrate specificity only when combined with RRM1 of SF2/ASF. As with the tat pre-mRNA, the source of RS domain had no effect on splicing specificity. These data are summarized in Fig. 7B. Taken together, these results demonstrate that the RRMs, rather than the RS domain, define the substrate specificity of SF2/ASF and SC35.
To define the role of individual RRMs, we also analyzed chimeric proteins with a two-domain structure (Fig. 7B). Chimeric proteins with a single RRM derived from SF2/ASF, RRM1 or RRM2, were essentially inactive, whereas the single RRM of SC35 joined to the RS domain of SF2/ASF (chimeric protein i) was active and had the same specificity as wild-type SC35 (Fig. 7B and data not shown), again confirming the crucial role of the SC35 RRM for the substrate specificity of SC35. These observations indicate that RRM1 or RRM2 from SF2/ASF cannot function as a single RRM in conjunction with an RS domain, in contrast to the RRM from SC35, which can, as it does in wild-type SC35.
DISCUSSION
Coordinate effects of downstream exon segments mediate SR protein-specific splicing.
Using in vitro splicing with deletion and chimeric derivatives of tat and IgM minigene transcripts, we defined specific segments in the downstream exons that act in a coordinated manner to determine SR protein-specific splicing. SF2/ASF-specific splicing, as seen with wild-type tat pre-mRNA, was observed exclusively with the TbTc substrate. The Tb and Tc segments act in concert to confer SR protein specificity, since splicing activity in the presence of SF2/ASF is stronger than that with the Tb segment alone, whereas splicing in the presence of SC35 is repressed. In the case of the IgM pre-mRNA, only two kinds of deleted substrates were spliced at significant levels. The Cb substrate was spliced indiscriminately with either SF2/ASF or SC35, whereas the CaCb pre-mRNA displayed SC35-specific splicing, similar to the wild-type IgM pre-mRNA. Therefore, the Ca and Cb segments also act in concert to achieve SR protein specificity.
The Tb segment of tat pre-mRNA is highly purine rich, except for two uridines (Fig. 3), and indeed, in vivo and in vitro splicing studies previously revealed that a longer region including this segment functions as an ESE (1, 34). However, we found that a substrate containing the Ta segment alone as the downstream exon spliced more efficiently with SF2/ASF than did a substrate containing the purine-rich Tb segment alone. Therefore, in this case the purine-rich segment is not the ESE, at least with SF2/ASF. Indeed, not all purine-rich sequences function as ESEs, and conversely, not all ESEs are purine rich (21, 35, 38, 39). An SF2/ASF-specific splicing enhancer element consensus, (C/G)(A/G)(C/G)A(C/G)GA, was recently identified by functional SELEX and comprises sequences that are not exclusively purine rich (21). Analysis of the tat T3 exon with an SF2/ASF motif scoring matrix (21) showed a higher density of high-score matches to the 7-nt consensus in the Ta segment than in the Tb segment (data not shown; see Fig. 3 for the sequences). In addition, replacement of the original tat ESE with an ESE from Rous sarcoma virus (RSV) causes a dramatic increase of splicing efficiency in vivo (34); the RSV ESE sequence has a better match to the above 7-nt consensus than the exclusively purine-rich tat ESE (data not shown). In the IgM pre-mRNA, only one high-score motif match to the SF2/ASF consensus was found in the Cb segment (data not shown; see Fig. 5 for the sequences), which is consistent with splicing activity being found exclusively with the Cb and CaCb substrates. The CaCb segment is necessary and sufficient for SC35-specific splicing, since the heterologous tat pre-mRNA behaved like the IgM pre-mRNA upon insertion of this segment.
Interestingly, both the Tc and the Ca segments have negative effects, i.e., they are silent by themselves but they can prevent SC35- and SF2/ASF-specific splicing, respectively, in combination with other segments. The Ca segment includes sequences that prevent splicing in the presence of SF2/ASF, and this effect was verified in a heterologous pre-mRNA. The Tc segment in the tat T3 exon includes a previously identified ESS (1, 33, 34). The role of this ESS element was tested by deletion, and we found that splicing with SF2/ASF was not affected, whereas splicing with SC35 was largely restored by deletion of the 20-nt ESE fragment. Therefore, the repressive effect of this ESS may be SC35 specific, and this is consistent with the fact that only partial inhibition of splicing by the ESS was observed in vivo and in vitro (33, 34). Thus, other SR proteins, e.g., SF2/ASF in this case, may be involved in recognition of this exon. However, the function of the ESS is sensitive to the sequence context. We did not observe a strong SC35-repressive effect of the Tc segment when it was joined to the Ta segment, i.e., the Ta and TaTc substrates showed similar SR protein specificities. Furthermore, we failed to observe SC35-specific splicing repression with the CaCbTc pre-mRNA. In this case, the positive effect of the CaCb segment for SC35-specific splicing is dominant over the negative effect of the Tc segment, which includes the ESS. The context dependence of the negative effect by the Tc segment does not appear to be a function of the distance to the upstream 3′ splice site, since this distance is very similar in the TaTbTc and CaCbTc substrates, but repression is seen only with TaTbTc (wild type). Our observation is consistent with, and extends, previous in vivo splicing experiments, which showed that this ESS does not repress splicing in the context of pre-mRNAs containing an ESE from the fibronectin gene or a β-globin intron (34). Likewise, it was recently shown that this ESS is less active in the context of an optimal or strong 3′ splice site (33).
The existence of a cellular inhibitory factor(s) that binds to the ESS elements of the tat pre-mRNA exon 2 and exon 3, which have similar sequences, was suggested by splicing assays performed in the presence of competitor RNA (1, 33). We observed that complementation reactions with SF2/ASF and SC35 together resulted in decreased splicing efficiency with the tat pre-mRNA; this negative effect was not observed with other pre-mRNAs (data not shown). In fact, with the β-globin and TaTbTcΔESS pre-mRNAs, additive effects of SF2/ASF and SC35 were observed, suggesting the presence of two or more ESEs and in agreement with the previous observation that splicing efficiency increases linearly with the number of ESEs present in an exon (14). SELEX experiments identified sequences that are bound with high affinity by SC35 lacking the RS domain but which do not function as ESEs (38). Interestingly, the sequence AUUCGAUUA, which has a 7- of 9-nt match to one of the SELEX winners identified in that study, GUUCGAGUA, is present in the tat exon 3 ESS (Fig. 3).
Together, the above observations suggest that SC35 itself may bind directly to the tat ESS and behave in this context as an inhibitor, rather than an activator. This suggestion is consistent with recent data showing that the ESS inhibits splicing by blocking the formation of a functional spliceosome at an early step (33). In the published experiments with competitor RNAs, SC35 may be titrated out, so that it can no longer bind to the functional SC35 binding site on the pre-mRNA. This is reminiscent of the sequestration of SR proteins by binding to viral RNAs possessing high-affinity binding sites for SR proteins (15). When the ESS is present in cis, there are several potential reasons why SC35 may be inhibitory. First, binding may be too tight to be compatible with function; second, the geometry of binding may be inappropriate; third, SC35 binding may prevent binding of other required coactivators. Inhibitory effects of SR proteins, including SF2/ASF and SC35, were previously reported in adenovirus and RSV src pre-mRNAs, although in these cases the negative elements are present in the introns (16, 26). In the chicken β-tropomyosin pre-mRNA, SC35 also acts as an inhibitor, antagonizing inclusion of an alternative exon mediated by SF2/ASF, which recognizes an intronic splicing enhancer (11).
In contrast to our current, as well as previous, in vitro splicing studies (references 9, 18, and 44 and this study), in vivo depletion of SF2/ASF was recently shown to increase splicing of tat pre-mRNA from a stably transfected minigene in a chicken B-lymphocyte cell line (42). There are many possible explanations for this apparent discrepancy, including the use of cell-free versus in vivo systems, cell-type-specific differences, differences in the abundance and ratios of various SR proteins and other splicing factors, and potential compensatory mechanisms that may operate in vivo when the abundance of an SR protein is manipulated.
Unique properties of the RRMs mediate substrate specificity.
The substrate specificity of several chimeric recombinant SR proteins with various combinations of RRM or RS domains derived from SF2/ASF or SC35 was determined with three reference substrates. We demonstrated that the substrate specificity of these SR proteins is defined by their RRMs, of which two are present in SF2/ASF and one is present in SC35, whereas the C-terminal RS domains of these proteins do not affect the substrate specificity and are interchangeable.
The function of each RRM in constitutive splicing is unique, and when two RRMs are present, they can markedly influence each other. Neither RRM1 nor RRM2 from SF2/ASF can function in constitutive splicing as a single RRM when joined to an RS domain. In contrast, the single RRM from SC35 has evolved to function in this context, and when joined to a heterologous RS domain, it is sufficient to confer SC35 specificity. RRM1 and RRM2 of SF2/ASF have naturally evolved as part of a double-RRM structure, and they synergize to give maximal splicing activity with the substrate specificity of SF2/ASF. The distinctive RRM2 of SF2/ASF is required for activity with the tat pre-mRNA, and it abrogates the activity of the SC35 RRM with the IgM pre-mRNA. Therefore, RRM2 can affect other RRMs positively and negatively. Of the three RRMs analyzed here, RRM1 of SF2/ASF has the most neutral character and does not affect the specificity conferred by the SC35 RRM. These data support the idea that each RRM in double-RRM SR proteins has been fine-tuned during evolution to function as part of a bipartite RNA-binding domain while retaining significant modular character. Similar conclusions have been reached in the case of the SR protein antagonist in alternative splicing, hnRNP A1 (25).
The RS domains of SF2/ASF and SC35 are interchangeable in the constitutive in vitro splicing assays with β-globin, IgM, and tat pre-mRNAs. This observation is consistent with the recent finding that the RS domain of SF2/ASF can be replaced by those of three other SR proteins tested, including SC35, to maintain the viability of a chicken B-lymphocyte cell line (42). On the other hand, the specific sequences of the RS domains of each SR protein are highly conserved phylogenetically, which is strongly indicative of specific roles for each RS domain (2). Individual RS domains have been shown to mediate different subnuclear targeting and nuclear-cytoplasmic shuttling properties (5, 6), and these unique functions may be important at the organismal level or in certain cell types.
Substrate specificity appears to be determined in the early stages of spliceosome assembly, since the three-domain chimeric proteins that include RRM2 from SF2/ASF can form specific commitment complexes with the tat pre-mRNA, despite the presence of other SR proteins in the nuclear extract (8). However, in the case of two-domain chimeric proteins including RRM1 from SF2/ASF, but not those including RRM2, stable commitment complexes can also form, allowing splicing of β-globin and IgM pre-mRNAs. In the S100 complementation assay, however, single-RRM proteins with either of the SF2/ASF RRMs were inactive. The SR protein commitment assay may be more permissive because of the presence of endogenous SR proteins, which may interact with the mutant or chimeric proteins in the preassembled commitment complex.
It is striking that of the six chimeric proteins that had splicing activity in the complementation assay (of nine tested), all of them displayed substrate specificity patterns that qualitatively resembled either that of wild-type SF2/ASF or that of SC35. Thus, although all these proteins were very active with at least one substrate, none were active with tat or IgM but not β-globin, active with β-globin but neither tat nor IgM, or active with all three transcripts. The exclusion of these patterns suggests that each of the transcripts possesses a distinctive set of recognition sequences that can be productively recognized in only a limited number of ways by the RRMs. There is already considerable evidence that individual SR proteins interact specifically with ESE elements (12, 21, 28, 36–38, 40). Although how individual RRMs interact with specific segments of pre-mRNA, resulting in splicing specificity, is not yet known, the present results suggest that each RRM interacts directly or indirectly with specific exon segments, including splicing enhancers or silencers.
Modular nature of SR proteins in the context of different functions.
The roles of the structural domains of several SR proteins were previously studied in other contexts. Unique roles of the RRMs in alternative splicing regulation were previously documented in vivo with adenovirus E1A pre-mRNA, which has three alternative 5′ splice sites. In contrast to the present and to previous results with constitutive splicing activity (3, 48), deletion of either RRM from SF2/ASF yields proteins that retain activity in alternative 5′ splice-site switching, although different 5′ splice sites are selected depending upon which RRM is deleted (5). The fact that single-RRM proteins possessing either RRM1 or RRM2 from SF2/ASF function in alternative splicing but not in constitutive splicing assays suggests that the mechanisms of SR protein involvement in these two contexts are distinct. It is possible that the more permissive domain requirements for alternative splicing reflect interactions between the mutant protein and endogenous SR proteins, since the alternative splicing assays are carried out in vivo or in nuclear extract, i.e., in the presence of multiple, wild-type SR proteins (3, 5, 31, 48). This would also be consistent with the more permissive domain requirements for SR protein function in the commitment assay, which is carried out in nuclear extract as well (8).
The RS domain of SR proteins is essential for an exclusive nuclear localization although it is not sufficient for subnuclear targeting in double-RRM SR proteins (5). Some but not all SR proteins shuttle continuously between the nucleus and the cytoplasm, a process for which the RS domain is a critical determinant (6). On the other hand, RS domains are interchangeable between SR proteins, at least in terms of the viability requirements of a B cell grown in culture (42). In these previous studies, as well as in the present study, chimeric proteins with specific combinations of SR protein domains were shown to be functional in a variety of assays, underscoring the modular character of SR proteins and the contribution of individual domains to different functions.
ACKNOWLEDGMENTS
We thank K. Lynch and T. Maniatis for baculovirus recombinant SC35 and SF2/ASF and A. Watakabe and Y. Shimura for the pμC3-C4 plasmid. We thank H.-X. Liu and M. Zhang for computer analysis of enhancer motif scores and S. H. Munroe for valuable comments on the manuscript.
A.M. and A.R.K. were supported by grant GM42699 from the NIH; G.R.S. was supported by the Wellcome Trust and Arthritis and Rheumatism Council; S.D.C. was supported by an NIH predoctoral fellowship; X.-D.F. is a Leukemia Society of America Scholar and was supported by grant GM49369 from the NIH.
REFERENCES
- 1.Amendt B A, Si Z H, Stoltzfus C M. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol Cell Biol. 1995;15:4606–4615. doi: 10.1128/mcb.15.8.4606. . (Erratum, 15:5480.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birney E, Kumar S, Krainer A R. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cáceres J F, Krainer A R. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cáceres J F, Krainer A R. Mammalian pre-mRNA splicing factors. In: Krainer A R, editor. Eukaryotic mRNA processing. Oxford, United Kingdom: Oxford University Press; 1997. pp. 174–212. [Google Scholar]
- 5.Cáceres J F, Misteli T, Screaton G R, Spector D L, Krainer A R. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cáceres J F, Screaton G R, Krainer A R. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavaloc Y, Popielarz M, Fuchs J P, Gattoni R, Stévenin J. Characterization and cloning of the human splicing factor 9G8: a novel 35 kDa factor of the serine/arginine protein family. EMBO J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandler S D, Mayeda A, Yeakley J M, Krainer A R, Fu X-D. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc Natl Acad Sci USA. 1997;94:3596–3601. doi: 10.1073/pnas.94.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu X-D. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- 10.Fu X-D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 11.Gallego M E, Gattoni R, Stévenin J, Marie J, Expert-Bezançon A. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the β-tropomyosin alternative exon 6A. EMBO J. 1997;16:1772–1784. doi: 10.1093/emboj/16.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gontarek R R, Derse D. Interactions among SR proteins, an exonic splicing enhancer, and a lentivirus Rev protein regulate alternative splicing. Mol Cell Biol. 1996;16:2325–2331. doi: 10.1128/mcb.16.5.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinrichs V, Baker B S. The Drosophila SR protein RBP1 contributes to the regulation of doublesex alternative splicing by recognizing RBP1 RNA target sequences. EMBO J. 1995;14:3987–4000. doi: 10.1002/j.1460-2075.1995.tb00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hertel K J, Maniatis T. The function of multisite splicing enhancers. Mol Cell. 1998;1:449–455. doi: 10.1016/s1097-2765(00)80045-3. [DOI] [PubMed] [Google Scholar]
- 15.Himmelspach M, Cavaloc Y, Chebli K, Stévenin J, Gattoni R. Titration of serine/arginine (SR) splicing factors during adenoviral infection modulates E1A pre-mRNA alternative splicing. RNA. 1995;1:794–806. [PMC free article] [PubMed] [Google Scholar]
- 16.Kanopka A, Mühlemann O, Akusjärvi G. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature. 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- 17.Krainer A R, editor. Eukaryotic mRNA processing. Oxford, United Kingdom: Oxford University Press; 1997. [Google Scholar]
- 18.Krainer A R, Conway G C, Kozak D. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 1990;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- 19.Krainer A R, Maniatis T, Ruskin B, Green M R. Normal and mutant human β-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- 20.Lamond A I, editor. Pre-mRNA processing. R. G. Austin, Tex: Landes Company; 1995. [Google Scholar]
- 21.Liu H-X, Zhang M, Krainer A R. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 23.Mayeda, A., and A. R. Krainer. Mammalian in vitro splicing assays. Methods Mol. Biol., in press. [DOI] [PubMed]
- 24.Mayeda, A., and A. R. Krainer. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol., in press. [DOI] [PubMed]
- 25.Mayeda A, Munroe S H, Xu R-M, Krainer A R. Distinct functions of the closely related tandem RNA-recognition motifs of hnRNP A1. RNA. 1998;4:1111–1123. doi: 10.1017/s135583829898089x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNally L M, McNally M T. SR protein splicing factors interact with the Rous sarcoma virus negative regulator of splicing element. J Virol. 1996;70:1163–1172. doi: 10.1128/jvi.70.2.1163-1172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng X, Mount S M. Genetic enhancement of RNA-processing defects by a dominant mutation in B52, the Drosophila gene for an SR protein splicing factor. Mol Cell Biol. 1995;15:6273–6282. doi: 10.1128/mcb.15.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramchatesingh J, Zahler A M, Neugebauer K M, Roth M B, Cooper T A. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ring H Z, Lis J T. The SR protein B52/SRp55 is essential for Drosophila development. Mol Cell Biol. 1994;14:7499–7506. doi: 10.1128/mcb.14.11.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruskin B, Krainer A R, Maniatis T, Green M R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984;38:317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- 31.Screaton G R, Cáceres J F, Mayeda A, Bell M V, Plebanski M, Jackson D G, Bell J I, Krainer A R. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi H, Hoffman B E, Lis J T. A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol Cell Biol. 1997;17:2649–2657. doi: 10.1128/mcb.17.5.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Si Z-H, Rauch D, Stoltzfus C M. The exon splicing silencer in human immunodeficiency virus type 1 tat exon 3 is bipartite and acts early in spliceosome assembly. Mol Cell Biol. 1998;18:5404–5413. doi: 10.1128/mcb.18.9.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staffa A, Cochrane A. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol Cell Biol. 1995;15:4597–4606. doi: 10.1128/mcb.15.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Q, Mayeda A, Hampson R K, Krainer A R, Rottman F M. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 37.Tacke R, Chen Y, Manley J L. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc Natl Acad Sci USA. 1997;94:1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tacke R, Manley J L. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka K, Watakabe A, Shimura Y. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol Cell Biol. 1994;14:1347–1354. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian M, Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Takagaki Y, Manley J L. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev. 1996;10:2588–2599. doi: 10.1101/gad.10.20.2588. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Xiao S-H, Manley J L. Genetic analysis of the SR protein ASF/SF2: interchangeability of RS domains and negative control of splicing. Genes Dev. 1998;12:2222–2233. doi: 10.1101/gad.12.14.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watakabe A, Inoue K, Sakamoto H, Shimura Y. A secondary structure at the 3′ splice site affects the in vitro splicing reaction of mouse immunoglobulin μ chain pre-mRNAs. Nucleic Acids Res. 1989;17:8159–8169. doi: 10.1093/nar/17.20.8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao S H, Manley J L. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 45.Zahler A M, Neugebauer K M, Lane W S, Roth M B. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- 46.Zahler A M, Roth M B. Distinct functions of SR proteins in recruitment of U1 small nuclear ribonucleoprotein to alternative 5′ splice sites. Proc Natl Acad Sci USA. 1995;92:2642–2646. doi: 10.1073/pnas.92.7.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang W J, Wu J Y. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol Cell Biol. 1996;16:5400–5408. doi: 10.1128/mcb.16.10.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuo P, Manley J L. Functional domains of the human splicing factor ASF/SF2. EMBO J. 1993;12:4727–4737. doi: 10.1002/j.1460-2075.1993.tb06161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]